Abstract
Infectious diseases caused by pathogens, such as SARS-COV, H7N9, severe fever with thrombocytopenia syndrome virus, and human immunodeficiency virus, have fatal outcomes with common features of severe fever and subsequent bacterial invasion progressing to multiorgan failure. Gene biomarkers are promising to distinguish specific infections from others with similar presenting symptoms for the prescription of correct therapeutics, preventing pandemics. While routine laboratory methods based on polymerase chain reaction (PCR) to measure gene biomarkers have provided highly sensitive and specific viral detection techniques over the years, they are still hampered by their precision and resource intensity precluding their point-of-care use. Recently, there has been growing interest in employing microfluidic technologies to advance current methods for infectious disease determination via gene biomarker measurements. Here, based on the requirement of infection detection, we will review three microfluidic approaches to compartmentalize gene biomarkers: (1) microwell-based PCR platforms; (2) droplet-based PCR; and (3) point-of-care devices including centrifugal chip, SlipChip, and self-powered integrated microfluidic point-of-care low-cost enabling chip. By capturing target genes in microwells with a small sample volume (∼μl), sensitivity can be enhanced. Additionally, with the advance of significant sample volume minimization (∼pl) using droplet technology, gene quantification is possible. These improvements in cost, automation, usability, and portability have thereby allowed point-of-care applications to decentralize testing platforms from laboratory-based settings to field use against infections.
INTRODUCTION
In the several months that have passed since the first reported infection cluster, coronavirus disease 2019 (COVID-19) has become a vicious worldwide pandemic. Within three months there were more than 3.6 × 106 people infected.1 This pandemic has resulted in severe healthcare and economic setbacks, which could have long lasting implications for many years. Infectious diseases such as COVID-19 are a major threat to the continued existence of humankind. For example, more than one million deaths per year were caused by human immunodeficiency virus (HIV)/acquired immune deficiency syndrome (AIDS), tuberculosis, and malaria.2 AIDS due to HIV is responsible for nearly ∼1 × 106 deaths per year despite implemented target prevention programs.3,4 The death toll of tuberculosis, caused by the bacterium Mycobacterium tuberculosis (MTB), is the highest among all infectious diseases, a problem exacerbated by the increasing number of antimicrobial-resistant variants of the bacterium.5 Malaria, a parasitic infection, has afflicted humans for thousands of years and continues to do so today. In light of the severity of the above-mentioned diseases and other infectious diseases, further efforts in device development for diagnosis must be directed toward the continued management of the global disease burden.
The COVID-19 pandemic has highlighted many questions that are relevant to effectively managing infectious diseases overall. How can infectious spreading be minimized in the early stages by technology development? How can patients with mild symptoms be recognized to prevent the spread of infection? How can infectious patients be rapidly identified to maintain economic activities, minimizing the effects of pandemics? To address these issues in public healthcare, available new technology for infection detection plays a central role. Laboratory-based instruments have been developed for infection analysis applications, and various pathogen analysis methods have been previously demonstrated. Infection detection is dependent on measuring specific responding antibodies in blood or viral materials, such as proteins, DNAs, and RNAs. Although protein biomarkers can be rapidly measured by using conventional binding assays, the detection sensitivity is limited due to the lack of biomarker sample amplification via polymerase chain reaction (PCR).
To diagnose infections confidently, most of the current instruments are based on gene biomarker identification via PCR.6 Quantitative PCR (qPCR) allows us to quantify PCR products in real time during the amplification process to determine biomarker concentrations.7 This technology can be further developed to measure RNAs via reverse transcription in real time (RT-PCR).8 In contrast to the advantages, time-consuming sample preparation (such as nucleic acid preparation, multiple-analyte separation, and preamplification) requires professional skills. Moreover, the measurements need to be processed by using costly laboratory-based instruments, which limit their applications in personal medicine at home. Indeed, although PCR is widely used to measure biomarkers of pathogens in serum, bodily fluids, and clinical samples, there is a necessity to improve their sensitivity, replicability, and scalability.9
In this study, we introduce the recent development of microfluidic devices to improve current instruments for gene biomarker detection, which can prove potent in rapidly determining infections and thus preventing pandemics. In the first part, the microvalve system for high-sensitivity gene detection via PCR is introduced. By encapsulating samples into microwells (volume ∼ 3 nl), the gene biomarker concentration can be increased due to small-volume compartmentalization. The pressure-driven actuation of a soft polydimethylsiloxane (PDMS) layer can regulate sample flow into the microwells. This microvalve system can be integrated with functionalized modules, such as sample mixers, sensors, and multimixing components, to approach a range of bioassays. Accordingly, high-sensitivity PCR is appropriate for the potential early diagnosis of infections. This device can be further modified for precise pathogen measurement and multiple gene biomarker determination. In the second part, droplet-based microfluidic technology for PCR, so-called digital PCR, will be discussed. This microfluidic manipulation method offers the possibility of generating streams of droplets with volumes ranging from nanoliters to picoliters and with a throughput of up to tens of thousands of droplets per second, creating isolated microreactors. By producing monodispersed water-in-oil droplets (volume ∼5 pl), the ultrahigh sensitivity of PCR can be approached. With the advance in low abundance gene biomarker detection, this technology can be conducted to evaluate patient disease progress and the immune capability against infections. In the third part, the recent microfluidic device design for point-of-care (PoC) diagnosis via portable PCR is introduced. By integrating the user-friendly interface for sample loading and microcomponents for thermal cycles, all-in-one PoC microfluidic PCR can be applied for infection detection at home. In addition to its advantages, the contradiction of sensitivity measurement is discussed. Notably, microfluidic device development is a rapidly developing research topic. This summary of recent progress is a foundation for future advanced microfluidic protocol design to identify gene biomarkers against infections, such as COVID-19.
MICROWELL PLATFORMS
In 2000, the microvalve platform was first developed to upload samples into a microwell array for bioassay with programable controls.10,11 By the pressure-driven deformation of a soft PDMS layer, the valves can be actuated to regulate the fluidic flows. With logical controls of multiple microvalves, the samples and reagents were able to be mixed with each other in the microwells for the multiplexed reactions. Notably, the volume of a microwell (∼36 nl)10 is at least ∼1000-fold smaller than the volume of conventional microtubes (20–100 μl). As the concentration is defined by the number of molecules per volume, when randomly capturing single molecules in the microwells, the concentration increases for high-sensitivity measurement. In 2003, this microvalve platform was further improved by minimizing the volumes of microwells (3 nl) to simultaneously process 400 reactions by loading a drop of DNA polymerase (2 μl).12 The uses of these platforms have also been extended to multigene (formyl-tetrahydrofolate synthetases) analysis and amplification of genes in single bacterial species.13 In 2006, this platform was used to process RT-PCR. Compared with conventional RT-PCR, 24-fold enhancement of throughput was approached.14
In 2018, a microwell-based diagnostic system (sample-in-digital-answer-out, SIDAO) was developed by integrating reagents prestored in sterilized tubes with automated pipetting modules.15 As a show case, it was used to determine the target genes of Mycobacterium tuberculosis from both spiked saliva and serum samples, demonstrating analytical accuracy and sensitivity with a measured copy number concentration as low as 6.63E + 4 copies/μl. In 2019, influenza viruses were determined by identifying target genes by sequencing in microwells.16 Currently, commercially available platforms, such as Fluidigm C1, can process up to 800 cells in parallel.17 By using the precise control of the integrated fluidic circuits (IFCs), automatic sample loading, reverse transcription, and cDNA PCR amplification is now possible in nanoliter reaction volumes. The Fluidigm IFC system was further developed to simultaneously analyze large-scale multiple reactions (192 samples, 24 assay conditions for each, 4608 reactions).18 This detection limitation of this system approaches one copy of RNA per ml. It is now employed for RT-PCR to determine SARS-COV2-synthetic RNA.19,20
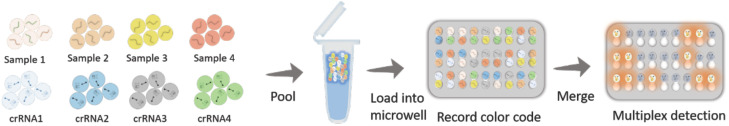
Recently, there has been growing interest in using clustered regularly interspaced short palindromic repeat (CRISPR)-based amplifications for viral diagnosis against infectious diseases. In 2020, combinatorial arrayed reactions for multiplexed evaluation nucleic acids (CARMEN) were developed by uploading water-in-oil droplets into microwells to simultaneously measure multiple pathogens in a scalable manner21 (Fig. 1). The samples containing Cas-13 reagents, PCR or recombinase polymerase amplification (RPA), and cleavage reporter 7 with a combination of distinct, fluorescent optical identifiers were uploaded to simultaneously measure 4500 crRNA-target pairs to determine infections. The CARMEN multiplexed assay was processed to differentiate 169 human-associated viruses with at least ten known genome sequences and with an additional crRNA to detect the causative agent of the 2020 COVID-19 pandemic.21
FIG. 1.
Microwell-based platform, CARMEN, for viral detection. Sample uploading of CARMEN-Cas13 consists of RPA-amplified DNA or RNA mixed with Cas13 within 1 nl droplets for pathogen detection. The droplets with samples and detection mixes are then pooled together and loaded into a microwell array, where each microwell accommodates two droplets at random, which generates pairwise combinations of physically isolated inputs to determine the reactions via fluorescence changes.
Droplet technology
Droplet digital PCR (DDPCR) uses a combination of microfluidics and proprietary surfactant chemistries to partition PCR samples into water-in-oil droplets for assaying.22 By uploading two immiscible fluids, oil and aqueous solutions, into a microfluidic junction with hydrophobic walls, monodispersed water-in-oil droplets are generated. To stabilize the droplets, a nonionic biocompatible surfactant (such as Pico-Surf 1, Dolomite Ltd.), is dispersed in the oil phase with an optimized concentration of 2%–5%. The diameter of the fabricated droplets can be controlled precisely in the range of 30 –250 μm.23,24 As the volume of a droplet (∼5 pl)25 is much smaller than the volume of the bulk solution (∼1 ml), when encapsulating single target molecules within the droplets, the concentration increases significantly for high-sensitivity PCR assays.
Notably, to quantify the target sequence in a solution by using conventional qPCR, it is necessary to evaluate amplified fluorescence signals at certain time points and to compare the signals to the standard curves. With DDPCR, as the single target molecules are compartmentalized within the droplets, by counting the number of PCR-positive droplets (Fig. 2), the number of target sequence copies can be directly quantified. DDPCR was previously developed as a commercial device. For example, the RainDrop system (Rain Dance) was demonstrated to quantify 1 in 200 000 mutants.26 For example, in 2012, the Bio-Rad QX100 Droplet Digital PCR system was employed for the detection of rare DNA from among 1 000 000-fold excess wild-type (WT) DNA.22
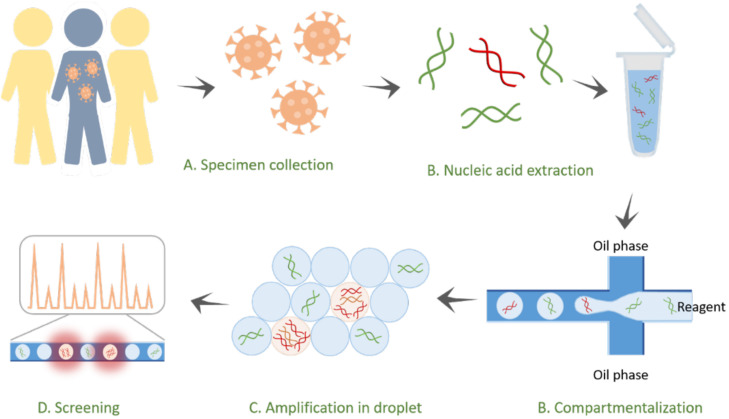
FIG. 2.
Droplet digital PCR to determine infections. The pathogens from an infected subject are extracted and encapsulated with primers, probes, or supermixes in droplets for the diagnosis of infectious diseases. As the target nucleic acids are compartmentalized within small droplets (∼5 pl), the concentrations are significantly enhanced for high-sensitivity measurements. After the reaction, the fluorescence droplets are screened via a fluorescence sensor to quantify the infectious pathogens in the samples.
DDPCR has been conducted to measure biomarkers in a range of human diseases, such as herpes, influenza, cytomegalovirus infection, and SARS-COV-2.27–30 In the case of hepatitis B virus (HBV), a DDPCR-based system detected template DNA input at the single-copy level (∼10−5 pg of plasmid HBV DNA). This allowed the quantification of covalently closed circular HBV DNA (cccDNA), which is an important parameter to monitor during antiviral therapy of liver-infected patients. For HIV RNA detection, compared with conventional qPCR, DDPCR demonstrated a significant increase in precision with an average fivefold decrease in the coefficient of variation of pol copy numbers (total HIV DNA) and a >20-fold accuracy improvement for 2-LTR circles (long terminal repeats). The genes of HIV-related ligands (CCL4) were determined. By comparing the number of copies from two genes (CCL41 and CCL42), it was found that the results obtained by DDPCR showed a stronger correlation (r = 0.99) than the results obtained by conventional qPCR (r = 0.44).31,32
Notably, compared with microwell-based platforms, the advantages of droplet platforms are in two main parts: (a) smaller volume of chambers (better sensitivity)33 and (b) higher throughput (∼1000-fold enrichment).34,35 The volume of a microwell is ∼1–10 nl, while the volume of a water-in-oil droplet generated is ∼5 pl. Since volume of droplets is ∼200 times smaller than the volume of microwells, the concentration of molecules captured is higher and the sensitivity is better. Moreover, the droplets can be generated in a throughput ∼1000 droplets per second, enabling to screen ∼107 reactions within hours.36 However, in a microwell-based platform, the throughput is restricted to ∼10 000–60 00036,37 chambers embedded in a device. By using droplet technology, it is easy to achieve higher order multiplexing through: amplitude multiplexing and probe mixing multiplexing, which is not easily achieved by using a microwell-based platform.38 This allows for improved sensitivity and precision as well as increased low-level detection in limited sample types like biopsies.39 On another hand, in a microwell-based platform, the ordering of the microchambers is remained. Therefore, it is easier to observe the reaction dynamics. Moreover, the washing process for immunobinding assays can be processed in the microwell-based platforms, while it cannot be well-processed within the droplets.
Point-of-care microfluidic diagnosis
For point-of-care diagnosis (PoC), the cost effective portable microfluidic devices are fabricated with which laboratory analysis can be performed at home. There are, however, considerable challenges to be faced before PoC devices become routine in clinical practice. For instance, although most microfluidic chips are small, the whole systems containing the optical components are bulky and are not suitable for PoC applications. Another challenge is the integration with pumps and fluid regulatory elements, where the flow of reagent can be precisely controlled. To address these challenges, it is necessary to develop the devices that are considerably simpler to allow the PoC testing.
At present, a popular approach to fabricate low-cost and portable PoC device is based on lateral flow assay (LFA) (Fig. 3). LFAs, which are paper-based devices, have these traits and have been used for PoC testing with proven utility for the detection and quantification of analytes in complex mixtures, where the sample is placed on the device, and the results are displayed within 30 min. LFAs have been used widely for qualitative and quantitative detection of gene biomarkers in a variety of physiological samples, including urine, saliva, sweat, serum, plasma, whole blood, and other fluids.40–44 Recently, LFAs have been adapted for nucleic acid testing, nucleic acid analysis, and gene identification.45,46 By using tagged primers, the amplicons of interest could be sandwiched between a membrane and gold nanoparticle (AuNP) probes for capture and signal readout. Notably, LFAs may employ colorimetric sensors that are optical sensors that measure a change in color occurring in the presence of certain analytes.47 Colorimetric sensors are widespread in PoC applications due to their high accessibility, ease of use, and low costs. Additionally, in CRISPR sensing, the use of colorimetric sensors is popular due to direct visualization of target sequences of interest and easy interpretation of results.48 One example of an LFA colorimetric assay includes paper-based sensors that combine RNA amplification with CRISPR/Cas9 modules to discriminate Zika virus strains at single-base resolution in 35 min.49 Above to its advantages, LFAs lack accuracy and efficiency, which makes these conventional paper-based platforms unsuitable for more quantitative analysis, which is desired in clinical applications.50
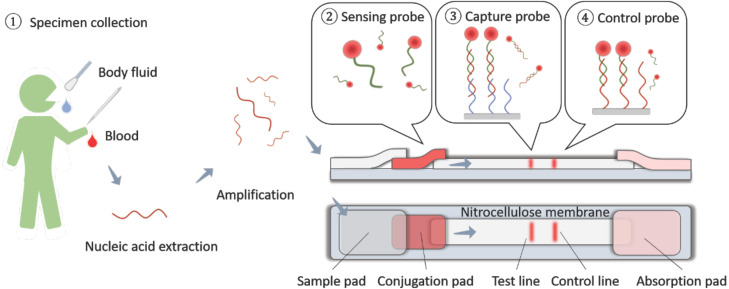
FIG. 3.
Lateral flow assay chips. Specimens in the form of bodily fluids, such as saliva, blood, and urine, are extracted for sample loading. The sample wicks through a conjugated pad with nanoparticle-antibody conjugation (or variants) to capture the biomarkers. After binding, the optical signal can be read for point-of-care diagnosis.
With the current COVID-19 pandemic, there has been an increasing focus on microfluidics PoC devices which have tremendous potential to determine pathogens in resource-limited condition. For example, closed-tube penn-ramp that employs loop-mediated isothermal amplification (LAMP) was developed to effectively measure target genes by mixing the reagents in microchambers without applying the thermocycles. This technology was conducted to determine COVID-19 with sensitivity as low as ∼7 target copies in 1 ml, which is ten times higher than the sensitivity of standard RT-PCR.51 Another noteworthy microfluidic PoC setup is Epidax that does not require using nucleic acid extraction step to amplify viral nucleic acid for the detections.52 Its detection sensitivity is ∼10 target copies in 1 ml. The testing result can be obtained in 1 h. In this section, we attempt to cover three PoC microfluidic compartmentalization designs, SlipChips, centrifuge chips, and self-powered integrated microfluidic PoC low-cost enabling (SIMPLE) chips. Our overarching goal is to comprehensively elaborate how they can be used as effective strategies and possible frameworks for effective for infectious gene biomarker detection.
Disk-based centrifugal microfluidics
Centrifugal microfluidics exploit centrifugal force and capillary force to control the liquid flow.53,54 This group of devices is designed in the format of compact disks (CDs) that house the reaction chambers and other components for PCR55 (Fig. 4). Centrifugal microfluidics has several advantages over conventional stationary microfluidics. Not only is the use of external pumps not required but also varying the rotation of speed of the disk offers a combination of microfluidic sample preparation steps, such as liquid mixing, metering, aliquoting, switching, valving, and storage.56 The first centrifugal chips were used for selective detection of Salmonella enterica from a mixture of S. enterica and Escherichia coli, where using centrifugal force, the sample under analysis could be distributed to microchambers in which on-chip PCR was performed.57 More recently, centrifugal microfluidic chips described by have enabled fast and parallel detection of Ebola virus.58 The system is based on reverse transcription loop-mediated isothermal amplification (RT-LAMP) and consists of four specific LAMP primers, a disk microfluidic chip, and a portable real-time fluorescence detector. The LOD is as small as ten copies per reaction, while the total consumption of sample and reagent is 0.94 μl per reaction. The final results could be obtained in 50 min after one addition of sample and reagent mixture. Other similar systems have enabled the simultaneous multiplexed detection of HBV, hepatitis C virus (HCV), and cytomegalovirus.59,60
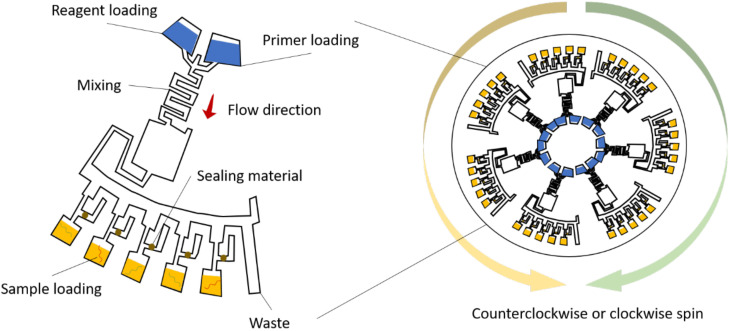
FIG. 4.
Centrifugal microfluidics. To perform centrifugal microfluidics, PCR mastermix, reagents, and primers are preloaded and distributed through the mixing area. The samples with nucleic acids (DNAs/RNAs) are then uploaded to the subchambers. By controlling the rotation of the disk, the samples and reagents can be mixed to initiate PCR for infection determination. To prevent contamination and evaporation, sealing materials are often used, including uncured PDMS, mineral oil, or pressure-sensitive tape.
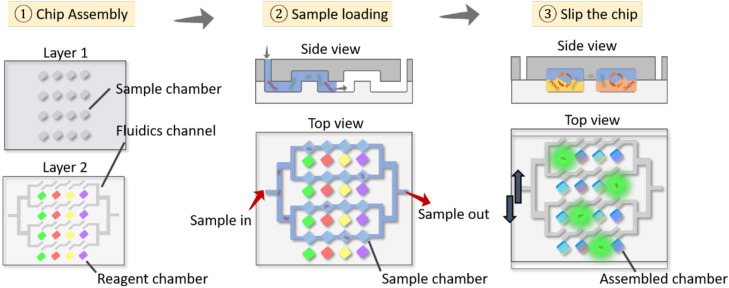
Slipchip microfluidics
While various microfluidic systems can perform multiplexed experiments at the nanoliter scale, many of them, such as disk-based centrifugal microfluidics, still require instrumentation for hydraulic control and gas-permeable materials together with sophisticated valve-based manipulations. Microplate-based systems and plug-based systems have partially addressed these instrumentation challenges because of their simplification in controlling surface chemistry, reducing the need for high-precision mechanics. However, these methods lack scalability and mass production and require further simplification for fabrication and operation. The SlipChip is a microfluidic device that compartmentalizes the reagents/samples into multiple microchambers to process multiple reactions by manually operating the chip without using pumps or valves in a resource-limited environment61 (Fig. 5). Consisting of two plates with arrays and channels, one plate is moved relative to the other, allowing reagents and sample to mix for reaction. Several improvements have been made since then, notably, a SlipChip PCR that contained 1280 droplets of 2.6 nl capacity capable of detecting the template DNA at the single-copy level.62 Among others is the nanoliter multiplex SlipChip PCR, which has robust performance and reduced reagent contamination and is able to screen up 384 primers.63 Although the SlipChip has been primarily used for the detection of the resistance gene mecA in methicillin-resistant Staphylococcus aureus (MRSA) and to identify different bacterial or fungal species, it is believed that the SlipChip can lead the way for new strategies for diagnostic PoC assays, especially for virus detection.
FIG. 5.
SlipChip microfluidics. Reagents are preloaded in one of the layers (layer 2 in the figure). After that, the sample is loaded into the device, and the target nucleic acids are compartmentalized into the microwells in layer 1. The top plate (layer 1) is slipped to align the microwells in layer 1 and layer 2 for sample mixing. Accordingly, the target nucleic acids react with the reagents to initiate PCR, producing fluorescence signals in the microwells for diagnosis.
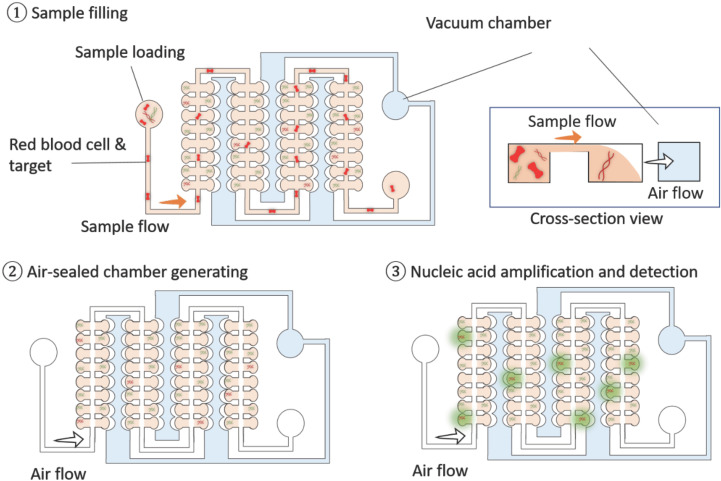
Self-powered integrated microfluidic point-of-care low-cost enabling device
Recently, a portable low-cost microfluidic device, self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE), for point-of-care diagnostics via quantitative nucleic acid detection was developed64 (Fig. 6). To lower the device complexity, instead of processing thermocycles to perform PCR, loop-medicated isothermal amplification (LAMP) technology was used to expand the number of gene copies for PCR assaying in the SIMPLE device. To remove the RBCs and other particles in unprocessed whole blood for direct blood measurements, a microfluidic reservoir component is fabricated in the inlet of the SIMPLE device. This reservoir was reported to be 5 mm in diameter. Most RBCs (∼95%) were removed from the blood through this component. By simply applying negative pressure in a side PDMS chamber, the air flow can be generated to drive fluidic flows for sample uploading. It is, therefore, not necessary to integrate bulky pumps and control components into the SIMPLE device. Notably, to effectively remove RBCs in the blood samples, deterministic lattice displacement (DLD) microfluidic component can be potentially integrated with a SIMPLE device. By designing the ratio of lateral-gap (GL) to downstream-gap (GD), efficient separation of RBCs can be approached.65 However, the pumps are required to operate DLD components making the system bulky limiting their applications for PoC applications. After removing RBCs, by compartmentalizing the samples in the microwells in the SIMPLE device with minimal manual operation, nucleic acids from human whole blood samples can be obtained via isothermal recombinase polymerase amplification (RPA).
FIG. 6.
SIMPLE chip. A SIMPLE chip can be used for whole blood analysis. The blood sample is uploaded into the device. With the microbarrier design, red blood cells (RBCs) are removed from the blood for plasma extraction. After sample loading, air is flowed through the device for sample compartmentalization. The target nucleic acids are then isolated into the microwells (100 nl) for high-sensitivity measurement with concentration enhancement. The isothermal nucleic acid assay is carried out by using this microchip to effectively quantify the target nucleic acids for point-of-care diagnosis.
To fabricate a SIMPLE device, the amplification initiator [magnesium acetate (MgOAc)] is prepatterned on the chip, and then the plasma is coated into the microwells for sensing. The self-powered pumping component is integrated by conducting a vacuum battery on the chip. The SIMPLE chip has a small footprint similar to that of glass slides (25 × 75 × 6 mm3), making it possible to transport easily in airtight aluminum vacuum-sealed pouches. The SIMPLE chip has demonstrated its ability to detect copies of methicillin-resistant Staphylococcus aureus DNA with sensitivity (10–105 copies/μl) in ∼30 min. The SIMPLE chip can be conducted in the future to measure infectious genes to rapidly determine the infection of pathogens, such as MRSA, Ebola, malaria, and coronavirus COVID-19.
CONCLUSION
Among their many ways of spreading, Infectious diseases may spread through microdroplets of saliva with concentrated virus when an infected person coughs or sneezes. Because of their fast spread, infectious diseases can cause vicious worldwide pandemics. For example, due to its rapid spread, COVID-19 is significantly impacting society, causing a healthcare crisis. This pandemic reminds us that our current technology should be improved to support the healthcare system against emerging infectious diseases. In this study, we introduced a series of microfluidic devices to measure gene biomarkers for effective infection diagnosis. To compare different microfluidic compartmentalization device design, a table is provided (Table I). In this table, the working mechanism, detection sensitivity, the size of the system, assay cost, and the suitable applications are provided. The central concept of compartmentalization to design these microfluidic devices is highlighted. Based on the compartmentalization methods, three types of devices are introduced. In the first part, the microvalve devices are discussed, which capture the target genes in microwells (μl–nl volume) for PCR. This device contains a two-port component to regulate fluidic transport to encapsulate the samples within the chamber for detection with concentrated gene biomarkers. In the second part, droplet microfluidics are introduced. Droplet-based microfluidics is a technology that addresses the formation and manipulation of uniform and micron-sized droplets at extremely high rates. Microdroplets (∼5 pl) are produced as reactors to compartmentalize biomarkers with high concentrations for infection diagnosis with high sensitivity. Notably, both microwell platforms and droplet-based systems are bulky and are used for laboratory-based tests. For PoC diagnosis, in the third part, microfluidic devices that integrate sample loading components and microchambers are introduced, enabling rapid capture of the biomarkers for assaying. Blood is directly uploaded into these devices for biomarker extraction and compartmentalized processing for PCR. Although the detection sensitivity might be affected by minimizing the sensing components integrated in the all-in-one devices, the low-cost tests can be processed without professional technicians to provide effective remote healthcare at home against infectious diseases. In summary, with the strategy of microfluidic compartmentalization, gene biomarkers can be effectively determined with ultrahigh sensitivity for infection diagnosis to minimize the impact of infection outbreaks in a community.
TABLE I.
Comparison of microfluidic devices for genomic biomarker detections.
| Working mechanism | Sensitivity (copies/reactor volume) | Throughput | Size and portability | Operation procedure | Cost | Application | |
|---|---|---|---|---|---|---|---|
| Microwell-based PCR platforms | Microwell for sample loading | High (∼40 copies/nl)12 | Medium (10 000–60 000 cells per experiment) | Bulky (∼122 × 66 × 100 cm, 100 kg) not portable66 | Automatic operation (Fluidigm machine) | High ($0.37 per sample per reaction) | Lab based diagnosis, digital PCR |
| Droplet-based PCR | Droplet for sample loading | High (7 copies/nl)67 | Very high (∼100–1000 droplets per second) | Bulky (∼1 m3);68 not portable | Automatic operation (Bio-Rad machine, etc.) | High | Lab based diagnosis, digital PCR |
| Centrifugal microfluidics | Centrifugal microwell chamber | High (10 copies/ul)58 | Low (∼35 samples per experiment) | Medium size58 (32 × 30 × 26 cm3, the diameter is ∼165 mm); may be portable | Automatic operation | Medium | Point-of-care diagnosis |
| SlipChip microfluidics | Microwell for sample loading | High (∼75 copies/ul)69,70 | Medium (∼100 samples per experiment) | Small size (4 × 2 × 0.07 cm3);63 highly portable | Easy manual operation | Low | Point-of-care diagnosis (e.g., HPV, HIV, HCV, etc.) |
| SIMPLE device | Microwell for sample loading | High (∼10 copies/ul) | Medium (∼224 samples per experiment) | Small size (25 × 75 × 3 mm3); highly portable | Easy manual operation | Low | Point–of-care diagnosis |
ACKNOWLEDGMENTS
We gratefully acknowledge the funding provided by the City University of Hong Kong (Nos. 9610467 and 7005436); National Natural Science Foundation of China (NNSFC) (No. 22074129); National Research Foundation Singapore, Synthetic Biology Research Program and Development Project Award (No. NRF SBP-P5); National Research Foundation Singapore, Competitive Research Program (No. NRF-CRP17-2017-03); National Medical Research Council Singapore, Open Fund - Individual Research Grant (No. NMRC/OFIRG/0061/2017); and Ministry of Education (MOE) Singapore (No. MOE2016-T2-2-016). Support provided by the Institute for Health Innovation and Technology (iHealthtech) at the National University of Singapore is also gratefully acknowledged.
Note: This paper is part of the special issue on Microfluidic Detection of Viruses for Human Health
DATA AVAILABILITY
The data that support the findings of this study are available within the article and the references cited.
REFERENCES
- 1.Al-Sadeq D. W. and Nasrallah G. K., “The incidence of the novel coronavirus SARS-CoV-2 among asymptomatic patients: A systematic review,” Int. J. Infect. Dis. 98, 372–380 (2020). 10.1016/j.ijid.2020.06.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Academies of Sciences, Engineering and MH and MDB on GHC on GH and the F of the US, Global Health and the Future Role of the United States (The National Academy of Sciences, Engineering, and Medicine, 2017). [Google Scholar]
- 3.WHO, “Number of deaths due to HIV/AIDS,” see https://www.who.int/gho/hiv/epidemic_status/deaths_text/en/.
- 4.Kempton J., Hill A., Levi J. A., Heath K., and Pozniak A., “Most new HIV infections, vertical transmissions and AIDS-related deaths occur in lower-prevalence countries,” J. Virus Erad. 5(2), 92–101 (2019). 10.1016/S2055-6640(20)30058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes K. K., Bertozzi S., Bloom B. R. et al. , Major Infectious Diseases: Key Messages from Disease Control Priorities, 3rd ed. (The International Bank for Reconstruction and Development/The World Bank, 2017). [PubMed] [Google Scholar]
- 6.Udugama B., Kadhiresan P., Kozlowski H. N. et al. , “Diagnosing COVID-19: The disease and tools for detection,” ACS Nano 14(4), 3822–3835 (2020). 10.1021/acsnano.0c02624 [DOI] [PubMed] [Google Scholar]
- 7.Peirson S. N. and Butler J. N., “Quantitative polymerase chain reaction,” Methods Mol. Biol. 362, 349–362 (2007). 10.1007/978-1-59745-257-1_25 [DOI] [PubMed] [Google Scholar]
- 8.Mackay I. M., Arden K. E., and Nitsche A., “Real-time PCR in virology,” Nucleic Acids Res. 30(6), 1292–1305 (2002). 10.1093/nar/30.6.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S. and Rothman R. E., “PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings,” Lancet Infect. Dis. 4(6), 337–348 (2004). 10.1016/S1473-3099(04)01044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang Y.-H., Kwon C. H., Kim S. B. et al. , “Deep wells integrated with microfluidic valves for stable docking and storage of cells,” Biotechnol. J. 6(2), 156–164 (2011). 10.1002/biot.201000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., “Monolithic microfabricated valves and pumps by multilayer soft lithography,” Science 288(5463), 113–116 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Hansen C., and Quake S. R., “Solving the ‘world-to-chip’ interface problem with a microfluidic matrix,” Anal. Chem. 75(18), 4718–4723 (2003). 10.1021/ac0346407 [DOI] [PubMed] [Google Scholar]
- 13.Ottesen E. A., Hong J. W., Quake S. R., and Leadbetter J. R., “Microfluidic digital PCR enables multigene analysis of individual environmental bacteria,” Science 314(5804), 1464–1467 (2006). 10.1126/science.1131370 [DOI] [PubMed] [Google Scholar]
- 14.Morrison T., Hurley J., Garcia J. et al. , “Nanoliter high throughput quantitative PCR,” Nucleic Acids Res. 34(18), e123 (2006). 10.1093/nar/gkl639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H., Chen Z., Cao X. et al. , “A sample-in-digital-answer-out system for rapid detection and quantitation of infectious pathogens in bodily fluids,” Anal. Bioanal. Chem. 410(27), 7019–7030 (2018). 10.1007/s00216-018-1335-9 [DOI] [PubMed] [Google Scholar]
- 16.Ahrberg C. D., Lee J. M., and Chung B. G., “Microwell array-based digital PCR for influenza virus detection,” BioChip J. 13(3), 269–276 (2019). 10.1007/s13206-019-3302-8 [DOI] [Google Scholar]
- 17.Pollen A. A., Nowakowski T. J., Shuga J. et al. , “Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex,” Nat. Biotechnol. 32(10), 1053–1058 (2014). 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamas A. Franco C. M., Regal P. et al. , “High-throughput platforms in real-time PCR and applications,” in Polymerase Chain Reaction for Biomedical Applications (InTechOpen, 2016), see https://www.intechopen.com/books/polymerase-chain-reaction-for-biomedical-applications/high-throughput-platforms-in-real-time-pcr-and-applications. [Google Scholar]
- 19.See https://www.fluidigm.com/singlearticles/covid-19-diagnostics for Fluidigm website for a high-throughput, extraction-free diagnostic test for the detection of SARS-CoV-2 in saliva for use under FDA EUA; accessed 4 October 2020.
- 20.See https://www.fluidigm.com/binaries/content/documents/fluidigm/resources/biomark-fluidigm-covid-19-faq-flyer-fldm-00152-rev-01/biomark-fluidigm-covid-19-faq-flyer-fldm-00152-rev-01/fluidigm%3Afile for Fluidigm website for real-time PCR for viral RNA detection on biomark HD frequently asked questions (2020).
- 21.Ackerman C. M., Myhrvold C., Thakku S. G. et al. , “Massively multiplexed nucleic acid detection with Cas13,” Nature. 582(7811), 277–282 (2020). 10.1038/s41586-020-2279-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindson B. J., Ness K. D., Masquelier D. A. et al. , “High-throughput droplet digital PCR system for absolute quantitation of DNA copy number,” Anal. Chem. 83(22), 8604–8610 (2011). 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi B., Wu D., Jiang Y., An J., and Wu W., “Off-chip vertical step emulsification droplets preparation device applied for droplet digital PCR,” Adv. Mater. Interfaces 7, 2001074 (2020). 10.1002/admi.202001074 [DOI] [Google Scholar]
- 24.Ye W., Tang X., Liu C., Wen C., Li W., and Lyu J., “Accurate quantitation of circulating cell-free mitochondrial DNA in plasma by droplet digital PCR,” Anal. Bioanal. Chem. 409(10), 2727–2735 (2017). 10.1007/s00216-017-0217-x [DOI] [PubMed] [Google Scholar]
- 25.Zonta E., Garlan F., Pécuchet N. et al. , “Multiplex detection of rare mutations by Picoliter droplet based digital PCR: Sensitivity and specificity considerations,” PLoS One 11(7), e0159094 (2016). 10.1371/journal.pone.0159094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pekin D., Skhiri Y., Baret J.-C. et al. , “Quantitative and sensitive detection of rare mutations using droplet-based microfluidics,” Lab Chip. 11(13), 2156–2166 (2011). 10.1039/c1lc20128j [DOI] [PubMed] [Google Scholar]
- 27.Suo T., Liu X., Feng J. et al. , “DdPCR: A more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens,” Emergin Microbes Infect. 9, 1259–1268 (2020). 10.1101/2020.02.29.20029439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sedlak R. H., Cook L., Cheng A., Magaret A., and Jerome K. R., “Clinical utility of droplet digital PCR for human cytomegalovirus,” J. Clin. Microbiol. 52(8), 2844–2848 (2014). 10.1128/JCM.00803-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts C. H., Last A., Molina-Gonzalez S. et al. , “Development and evaluation of a next-generation digital PCR diagnostic assay for ocular Chlamydia trachomatis infections,” J. Clin. Microbiol. 51(7), 2195–2203 (2013). 10.1128/JCM.00622-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalinina O., Lebedeva I., Brown J., and Silver J., “Nanoliter scale PCR with TaqMan detection,” Nucleic Acids Res. 25(10), 1999–2004 (1997). 10.1093/nar/25.10.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiselinova M., Pasternak A. O., De Spiegelaere W., Vogelaers D., Berkhout B., and Vandekerckhove L., “Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA,” PLoS One 9(1), e85999 (2014). 10.1371/journal.pone.0085999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruelle J., Yfantis V., Duquenne A., and Goubau P., “Validation of an ultrasensitive digital droplet PCR assay for HIV-2 plasma RNA quantification,” J. Int. AIDS Soc. 17(4 Suppl 3), 19675 (2014). 10.7448/IAS.17.4.19675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaushik A. M., Hsieh K., and Wang T.-H., “Droplet microfluidics for high-sensitivity and high-throughput detection and screening of disease biomarkers,” Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 10(6), e1522–e1522 (2018). 10.1002/wnan.1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia H., Strachan B. C., Gifford S. C., and Shevkoplyas S. S., “A high-throughput microfluidic approach for 1000-fold leukocyte reduction of platelet-rich plasma,” Sci. Rep. 6(1), 35943 (2016). 10.1038/srep35943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X., Shao C., Luo C. et al. , “Microfluidics-based enrichment and whole-genome amplification enable strain-level resolution for airway metagenomics,” Am. Soc. Microbiol. 4(4), e00198-19 (2019). 10.1128/mSystems.00198-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teh S.-Y., Lin R., Hung L.-H., and Lee A. P., “Droplet microfluidics,” Lab Chip 8(2), 198–220 (2008). 10.1039/b715524g [DOI] [PubMed] [Google Scholar]
- 37.Dimov I. K., Lu R., Lee E. P. et al. , “Discriminating cellular heterogeneity using microwell-based RNA cytometry,” Nat. Commun. 5, 3451 (2014). 10.1038/ncomms4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whale A. S., Huggett J. F., and Tzonev S., “Fundamentals of multiplexing with digital PCR,” Biomol. Detect Quantif. 10, 15–23 (2016). 10.1016/j.bdq.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belmonte F. R., Martin J. L., Frescura K. et al. , “Digital PCR methods improve detection sensitivity and measurement precision of low abundance mtDNA deletions,” Sci. Rep. 6(1), 25186 (2016). 10.1038/srep25186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno M de L., Cebolla Á, Muñoz-Suano A. et al. , “Detection of gluten immunogenic peptides in the urine of patients with coeliac disease reveals transgressions in the gluten-free diet and incomplete mucosal healing,” Gut 66(2), 250–257 (2017). 10.1136/gutjnl-2015-310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrio A., Sampedro C., Sanchez-Lopez J. L., Pimienta M., and Campoy P., “Automated low-cost smartphone-based lateral flow saliva test reader for drugs-of-abuse detection,” Sensors 15(11), 29569–29593 (2015). 10.3390/s151129569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pacifici R., Farré M., Pichini S. et al. , “Sweat testing of MDMA with the Drugwipe analytical device: A controlled study with two volunteers,” J. Anal. Toxicol. 25(2), 144–146 (2001). 10.1093/jat/25.2.144 [DOI] [PubMed] [Google Scholar]
- 43.De Giovanni N. and Fucci N., “The current status of sweat testing for drugs of abuse: A review,” Curr. Med. Chem. 20(4), 545–561 (2013). 10.2174/0929867311320040006 [DOI] [PubMed] [Google Scholar]
- 44.Magambo K. A., Kalluvya S. E., Kapoor S. W. et al. , “Utility of urine and serum lateral flow assays to determine the prevalence and predictors of cryptococcal antigenemia in HIV-positive outpatients beginning antiretroviral therapy in Mwanza, Tanzania,” J. Int. AIDS Soc. 17(1), 19040 (2014). 10.7448/IAS.17.1.19040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardee K., Green A. A., Ferrante T. et al. , “Paper-based synthetic gene networks,” Cell 159(4), 940–954 (2014). 10.1016/j.cell.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green A. A., Silver P. A., Collins J. J., and Yin P., “Toehold switches: De-novo-designed regulators of gene expression,” Cell 159(4), 925–939 (2014). 10.1016/j.cell.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye H. and Xia X., “Enhancing the sensitivity of colorimetric lateral flow assay (CLFA) through signal amplification techniques,” J. Mater. Chem. B. 6(44), 7102–7111 (2018). 10.1039/C8TB01603H [DOI] [PubMed] [Google Scholar]
- 48.van Dongen J. E., Berendsen J. T. W., Steenbergen R. D. M., Wolthuis R. M. F., Eijkel J. C. T., and Segerink L. I., “Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities,” Biosens. Bioelectron. 166, 112445 (2020). 10.1016/j.bios.2020.112445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardee K., Green A. A., Takahashi M. K. et al. , “Rapid, low-cost detection of Zika virus using programmable biomolecular components,” Cell 165(5), 1255–1266 (2016). 10.1016/j.cell.2016.04.059 [DOI] [PubMed] [Google Scholar]
- 50.Kasetsirikul S., Shiddiky M. J. A., and Nguyen N.-T., “Challenges and perspectives in the development of paper-based lateral flow assays,” Microfluid. Nanofluid. 24(2), 17 (2020). 10.1007/s10404-020-2321-z [DOI] [Google Scholar]
- 51.El-Tholoth M., Bau H. H., and Song J., “A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry,” chemRxiv:10.26434/chemrxic.11860137.v1 (2020).
- 52.Simmons C., “Singapore scientists construct portable micro-PCR coronavirus test device that gives off results in one hour,” The Science Times, see https://www.sciencetimes.com/articles/26265/20200630/singapore-scientists-construct-portable-micro-pcr-coronavirus-test-device-gives.htm; accessed 2 November 2020.
- 53.Cho H., Kim H.-Y., Kang J. Y., and Kim T. S., “How the capillary burst microvalve works,” J. Colloid Interface Sci. 306(2), 379–385 (2007). 10.1016/j.jcis.2006.10.077 [DOI] [PubMed] [Google Scholar]
- 54.Chen J. M., Huang P.-C., and Lin M.-G., “Analysis and experiment of capillary valves for microfluidics on a rotating disk,” Microfluid. Nanofluid. 4(5), 427–437 (2008). 10.1007/s10404-007-0196-x [DOI] [Google Scholar]
- 55.Sayad A., Ibrahim F., Mukim Uddin S., Cho J., Madou M., and Thong K. L., “A microdevice for rapid, monoplex and colorimetric detection of foodborne pathogens using a centrifugal microfluidic platform,” Biosens. Bioelectron. 100, 96–104 (2018). 10.1016/j.bios.2017.08.060 [DOI] [PubMed] [Google Scholar]
- 56.Clime L., Daoud J., Brassard D., Malic L., Geissler M., and Veres T., “Active pumping and control of flows in centrifugal microfluidics,” Microfluid. Nanofluid. 23(3), 29 (2019). 10.1007/s10404-019-2198-x [DOI] [Google Scholar]
- 57.Furutani S., Nagai H., Takamura Y., and Kubo I., “Compact disk (CD)-shaped device for single cell isolation and PCR of a specific gene in the isolated cell,” Anal. Bioanal. Chem. 398(7–8), 2997–3004 (2010). 10.1007/s00216-010-4205-7 [DOI] [PubMed] [Google Scholar]
- 58.Lin X., Jin X., Xu B. et al. , “Fast and parallel detection of four ebola virus species on a microfluidic-chip-based portable reverse transcription loop-mediated isothermal amplification system,” Micromachines 10(11), 777 (2019). 10.3390/mi10110777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang H.-C., Chao Y.-T., Yen J.-Y. et al. , “A turbidity test based centrifugal microfluidics diagnostic system for simultaneous detection of HBV, HCV, and CMV,” Adv. Mater. Sci. Eng. 2015, 306708 10.1155/2015/306708 [DOI] [Google Scholar]
- 60.Li L., Miao B., Li Z., Sun Z., and Peng N., “Sample-to-answer hepatitis B virus DNA detection from whole blood on a centrifugal microfluidic platform with double rotation axes,” ACS Sens. 4(10), 2738–2745 (2019). 10.1021/acssensors.9b01270 [DOI] [PubMed] [Google Scholar]
- 61.Du W., Li L., Nichols K. P., and Ismagilov R. F., “Slipchip,” Lab Chip 9(16), 2286–2292 (2009). 10.1039/b908978k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen F., Du W., Kreutz J. E., Fok A., and Ismagilov R. F., “Digital PCR on a SlipChip,” Lab Chip 10(20), 2666–2672 (2010). 10.1039/c004521g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen F., Du W., Davydova E. K., Karymov M. A., Pandey J., and Ismagilov R. F., “Nanoliter multiplex PCR arrays on a SlipChip,” Anal. Chem. 82(11), 4606–4612 (2010). 10.1021/ac1007249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeh E.-C., Fu C.-C., Hu L., Thakur R., Feng J., and Lee L. P., “Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip,” Sci. Adv. 3(3), e1501645 (2017). 10.1126/sciadv.1501645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ranjan S. et al. , “DLD pillar shape design for efficient separation of spherical and non-spherical bioparticles,” Lab Chip 14(21), 4250–4262 (2014). 10.1039/C4LC00578C [DOI] [PubMed] [Google Scholar]
- 66.See https://www.fluidigm.com/binaries/content/documents/fluidigm/resources/c1-srg-100-5201/c1-srg-100-5201/fluidigm%3Afile for “Fluidigm C1 System.”
- 67.Falzone L., Musso N., Gattuso G. et al. , “Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection,” Int. J. Mol. Med. 46(3), 957–964 (2020). 10.3892/ijmm.2020.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qx-One, QX ONE Droplet Digital PCR (ddPCR) System (Biorad, 2020), p. 2.
- 69.Shen F., Sun B., Kreutz J. E. et al. , “Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load,” J. Am. Chem. Soc. 133(44), 17705–17712 (2011). 10.1021/ja2060116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farka Z., Mickert M. J., Pastucha M., Mikušová Z., Skládal P., and Gorris H. H., “Advances in optical single-molecule detection: En route to supersensitive bioaffinity assays,” Angew. Chem. Int. Ed. Engl. 59(27), 10746–10773 (2020). 10.1002/anie.201913924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available within the article and the references cited.