Abstract
Human milk contains a dynamic and complex site-specific microbiome, which is not assembled in an aleatory way, formed by organized microbial consortia and networks. Presence of some genera, such as Staphylococcus, Streptococcus, Corynebacterium, Cutibacterium (formerly known as Propionibacterium), Lactobacillus, Lactococcus and Bifidobacterium, has been detected by both culture-dependent and culture-independent approaches. DNA from some gut-associated strict anaerobes has also been repeatedly found and some studies have revealed the presence of cells and/or nucleic acids from viruses, archaea, fungi and protozoa in human milk. Colostrum and milk microbes are transmitted to the infant and, therefore, they are among the first colonizers of the human gut. Still, the significance of human milk microbes in infant gut colonization remains an open question. Clinical studies trying to elucidate the question are confounded by the profound impact of non-microbial human milk components to intestinal microecology. Modifications in the microbiota of human milk may have biological consequences for infant colonization, metabolism, immune and neuroendocrine development, and for mammary health. However, the factors driving differences in the composition of the human milk microbiome remain poorly known. In addition to colostrum and milk, breast tissue in lactating and non-lactating women may also contain a microbiota, with implications in the pathogenesis of breast cancer and in some of the adverse outcomes associated with breast implants. This and other open issues, such as the origin of the human milk microbiome, and the current limitations and future prospects are addressed in this review.
Keywords: human milk, microbiota, microbiome, vertical transfer, biological functions, mastitis, breast tissue, breast cancer
Introduction
Historically human milk was considered sterile under physiological conditions and, therefore, the presence of microbes was considered either as an infection or as a contamination. The mammary glands are made up of a moist intra-mammary mucosal ecosystem which, during late pregnancy and throughout the lactation period, becomes an ideal environment for bacterial growth due to the availability of a wide range of nutrients, and the optimum temperature for many microbes. In addition, the extremely complex duct system may favor the growth and spreading of biofilm-forming bacteria, a property that is common among Staphylococcus strains isolated from human milk (Delgado et al., 2009; Delgado et al., 2011; Begović et al., 2013). In addition, the mammary ecosystem is exposed to the external environment (via the ducts/nipple) and, also, to the internal environment since tight junctions remain open for a few days after birth. Since the first culture-dependent and -independent reports of the human milk microbiome (Heikkilä and Saris, 2003; Martin et al., 2003), all published studies have provided evidence of the presence of bacteria or bacterial DNA in milk collected from healthy women under hygienic conditions. In contrast, no modern study has found evidence of their absence.
The Microbiota of Human Milk: Historical Perspective
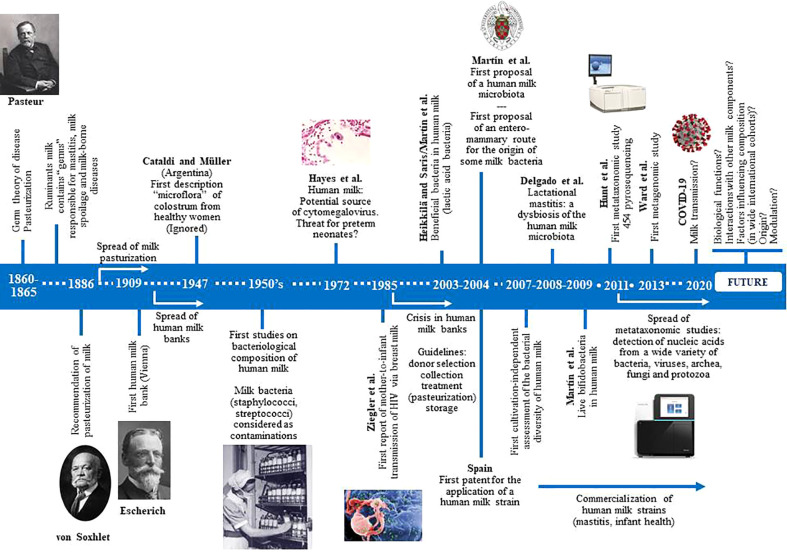
Knowledge of the bacterial content of fresh mammalian milk is as old as Pasteur’s “germ” theory ( Figure 1 ). Numerous studies, dating back more than a century ago, demonstrated that bacteria are common in milk of healthy ruminants. However, milk microbes were traditionally viewed from three perspectives: (a) as a potential cause of mastitis, leading to economic losses in farms (Macfadyen, 1891; French, 1903; Jones, 1918); (b) as a potential cause of milk spoilage (Hall and Trout, 1968); or (c) as a potential threat to human health because of the potential transfer of pathogenic microbes, including those with a zoonotic origin and those arising from a non-hygienic handling (Anonymous, 1889; Park, 1901). Many milk-borne diseases, including tuberculosis, brucellosis, typhoid fever, diphtheria or scarlet fever, had been recognized before 1900 (Holsinger et al., 1997), and that it is estimated that approximately 65,000 people died of milk-borne tuberculosis in England alone between 1912 and 1937 (Wilson, 1943). More than half a century later (mid-1950s), the first studies dealing with the bacteriological composition of human milk were published (Lindemann and Rupp, 1954; Novel and Pongratz, 1954; Lindemann, 1955; Mocquot, 1955; Meyer and Potel, 1956; Sager, 1956; Mossel and Weijers, 1957; Lindemann, 1958). Most of them reported an unexpected fact: the abundant and widespread bacterial “contamination” of milk donated to human milk banks. Since these institutions were rapidly spreading in Western countries, the number of studies proposing either bacterial criteria for acceptance of donor milk, or collection, storage and processing procedures to reduce the milk bacterial load, also increased rapidly from the 1950s to the early 1980s (Bachmann and Pulverer, 1962; Krieg, 1966; Liebhaber et al., 1978; Carroll et al., 1979; Davidson et al., 1979; Eidelman and Szilagyi, 1979; Jones et al., 1979; Lucas and Roberts, 1979; West et al., 1979; Björkstén et al., 1980; Carroll et al., 1980).
Figure 1.
Historical perspectives of the main milestones in research focused in human milk microbes.
In parallel, a number of studies warned about the presence of some potential pathogenic bacteria among such “contaminants”, which posed a risk for infant health. They included mainly Staphylococcus aureus but, also, Streptococcus pyogenes, Streptococcus agalactiae and some enterobacteria (Salmonella spp., Klebsiella pneumoniae) (Dubois, 1954; Cayla et al., 1955; Rantasalo and Kauppinen, 1959; Foster and Harris, 1960; Ottenheimer et al., 1961; Dluzniewska, 1966; Burbianka and Dluzniewska, 1971; Fleischrocker et al., 1972; Kenny, 1977; Ryder et al., 1977; Schreiner et al., 1977; Lucas and Roberts, 1978; Williamson et al., 1978; Donowitz et al., 1981; Cooke et al., 1987; Lemoine, 1987). Finally, a few studies were focused on the relationship between milk bacteria and mastitis (Dorr and Sittel, 1953; Cherkasskaia et al., 1980; Thomsen, 1982).
In 1985, the first report of a case of presumed mother-to-infant transmission of human immunodeficiency virus (HIV) via milk was published (Ziegler et al., 1985). In the following years, fear of HIV transmission led human milk banks to a strong crisis and to the closing of many of them because of the financial burden of serological testing of donors (Haiden and Ziegler, 2016). In addition, a few years earlier, human milk had also been recognized as a source of cytomegalovirus (CMV) (Hayes et al., 1972). This worsened the situation since most banked milk was intended for preterm neonates, a population where CMV may be particularly harmful, and no routine procedure for CMV screening was available at that time. As a result, most microbial studies of human milk published in the second half of the 80’s and throughout the 90’s were related to its role as a vehicle of HIV and/or CMV, and to the search for the best methods for donors screening and for viral inactivation (Stagno et al., 1980; Friis and Andersen, 1982; Cheeseman and McGraw, 1983; Dworsky et al., 1983; Boyes, 1987; Anonymous, 1988). At this stage, presence of any kind of microbe in milk was generally seen as a potential threat for infant health.
This disease-centric view of milk changed in 2003 following the publication of two articles that described the presence of lactic acid bacteria in human milk (Heikkilä and Saris, 2003; Martin et al., 2003). Such bacteria were generally recognized as safe and beneficial for infant health. One year later, it was proposed that human milk contains its own site-specific microbiota (Martín et al., 2004). Soon, it was found that some strains isolated from human milk displayed a wide array of probiotic traits (Beasley and Saris, 2004; Martín et al., 2005; Martín et al., 2006; Olivares et al., 2006a). In fact, the first description of the presence of lactobacilli (Lactobacillus acidophilus) and bifidobacteria (then the so-called Lactobacillus bifidus) in colostrum from healthy women was made in 1947 (Cataldi and Müller, 1947); however, such work was (and still is) ignored by the medical and scientific community, likely due to the fact that it was published in an Argentine journal with no diffusion outside of Spanish-talking countries.
Presence of microorganisms in human milk is no longer valid as a predictor of the risk of infant infection (Boer et al., 1981; Schanler et al., 2011; Zimmermann et al., 2017), despite the fact that additional cases of human milk-related infant infections have been reported (Qutaishat et al., 2003; Kayıran et al., 2014; Weems et al., 2015).
The Composition of the Human Milk Microbiota
From 2003, the study of the human milk microbiota has attracted the interest of many research groups worldwide (Fernandez et al., 2013; Jost et al., 2015; Ojo-Okunola et al., 2018), enabling the detection of approximately 200 different bacterial, archeal and fungal species from more than 50 different genera (Fernandez et al., 2013), including new species (Martín et al., 2011), and new genera, including Lactomassilus, Lactimicrobium, Anaerolactibacter, Galactobacillus, and Acidipropionibacterium (Togo et al., 2017; Togo et al., 2019a).
Culture-based methods have revealed that some species of the genera Staphylococcus (Staphylococcus epidermidis and other coagulase-negative species [CNS]), Streptococcus (S. salivarius, S. mitis and other species of the mitis group), Corynebacterium, Cutibacterium and other taxonomically-related Gram-positive bacteria are usually the dominant cultivable bacteria in samples of milk from healthy women (Jiménez et al., 2008a; Jiménez et al., 2008b; Solís et al., 2010; Schanler et al., 2011; Ding et al., 2019). Less frequently, lactic acid bacteria (Lactococcus, Enterococcus, Lactobacillus, Leuconostoc, and Weissella) and bifidobacteria are isolated from this biological fluid (Martin et al., 2003; Martín et al., 2006; Abrahamsson et al., 2009; Martín et al., 2009; Solís et al., 2010; Arboleya et al., 2011; Makino et al., 2015; Murphy et al., 2017). Some Lactobacillus (L. salivarius, L. reuteri, L. gasseri, L. fermentum) and Bifidobacterium (B. breve and B. longum) species have received particular interest because of the potential of the strains belonging to such species to be employed as probiotics. It must be highlighted that the different species of the genera Lactobacillus and Leuconostoc have been recently reclassified into 25 different genera (Zheng et al., 2020).
Under physiological conditions, milk bacterial concentrations may range from <1 to 4 log10 colony-forming units (cfu)/mL if the samples are obtained by either manual expression or through sterile pumps following hygienic practices (Espinosa-Martos et al., 2016). In contrast, bacterial concentrations can rise up to 6 log10 cfu/mL, or even higher, in mastitis cases (Fernández et al., 2014) or if milk is collected through the use of non-sterile pumps (Boo et al., 2001; Brown et al., 2005; Marín et al., 2009; Jiménez et al., 2017). At present, cell count methods, from classic counting chambers to flow cytometry, are the best methods for an accurate quantification of (live) bacterial cells in milk while quantitative PCR methods usually lead to an overestimation due to the presence of dead bacterial cells, exosomes and/or free bacterial DNA but, also, to mispriming with human DNA (Walker et al., 2020), and copy number bias (Větrovský and Baldrian, 2013; Louca et al., 2018).
Introduction of new media, supplements and incubation conditions have allowed the isolation of bacteria that were previously unnoticed. However, recent developments in culturomics have revealed that many bacteria previously regarded as non-cultivable can now be isolated from complex ecosystems when a proper combination of culture conditions is provided (Lagier et al., 2012; Lagier et al., 2015; Lau et al., 2016; Lagier et al., 2018; Schwab et al., 2019). These new methodologies have served as critical tests to validate data derived from metagenomic studies regarding the gut ecosystem (Lau et al., 2016). Culture-based techniques enable the isolation, preservation and characterization of strains (Bäckhed et al., 2005; Lara-Villoslada et al., 2007; Jiménez et al., 2008b; Delgado et al., 2009; Jiménez et al., 2010a; Jiménez et al., 2010b; Arboleya et al., 2011; Delgado et al., 2011; Gueimonde et al., 2012; Jiménez et al., 2012; Langa et al., 2012; Martín et al., 2012a; Martín et al., 2013; Cárdenas et al., 2014; Cárdenas et al., 2015).
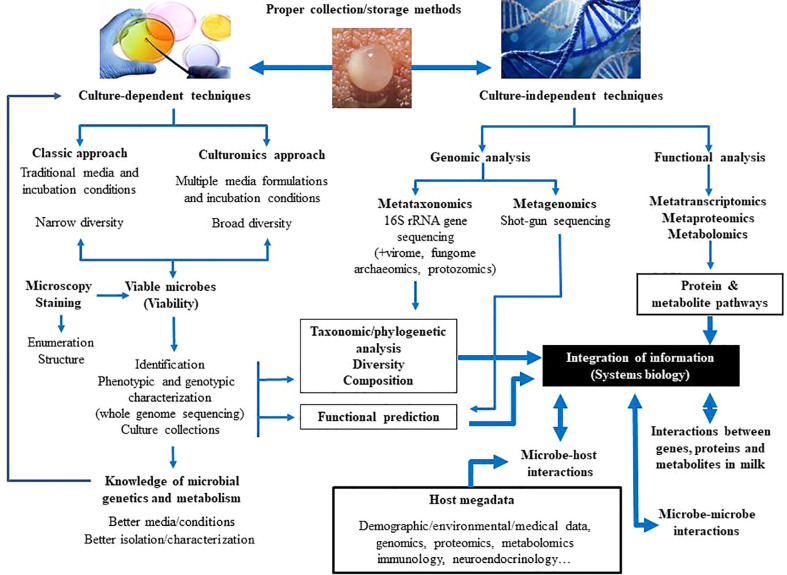
In relation to the milk microbiome, the use of first generation of culture-independent techniques, including PCR, combined or not with creation of bacterial gene libraries, denaturing gradient gel electrophoresis (DGGE), and temperature gradient gel electrophoresis (TGGE), allowed a better knowledge of the milk bacterial populations (Gueimonde et al., 2007; Martín et al., 2007a; Martín et al., 2007b; Delgado et al., 2008; Collado et al., 2009). Nowadays, they have been replaced by high-throughput Next Generation Sequencing (NGS) techniques, including metataxonomics (16S rRNA amplicon analysis) and metagenomics (total DNA sequencing). Sequencing of bacterial 16S rRNA genes has revealed that milk contains DNA of diverse microbial groups that were previously undetected with conventional culture-based techniques (Hunt et al., 2011; Cabrera-Rubio et al., 2012; Jost et al., 2013; Jost et al., 2014; Cabrera-Rubio et al., 2016; Boix-Amorós et al., 2017; Pannaraj et al., 2017). Although results from such studies do not provide evidence for viability, they have been useful to highlight the complexity of the milk microbiota and its role in modulating mammary homeostasis and gut colonization during early life. Shotgun sequencing of milk microbial DNA has been rarely performed to date (Ward et al., 2013; Jiménez et al., 2015), despite the fact that it can reveal the presence and potential functions of neglected members of this microbiota (archaea, viruses, fungi, protozoa). In addition, although DNA-based studies cannot demonstrate function, they can imply functional capacity based on the genes present in the samples. Comprehensive metagenomic, metatranscriptomic and metabolomic investigations are required for a holistic understanding of genetic diversity and functionality within the milk ecosystem ( Figure 2 ).
Figure 2.
Current and future approaches to study the composition and functions of the human milk microbiota.
Human milk microbiota is not aleatory assembled and contains organized bacterial consortia and networks (Ma et al., 2015; Drago et al., 2017). The milk microbiota of a woman is very stable throughout the lactation period, particularly in relation to the most abundant genera (Hunt et al., 2011; Williams et al., 2017a). Culture-independent studies have shown a wide diversity of bacterial signatures belonging to more than 800 different bacterial species, mainly from four major phyla (Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria) (Togo et al., 2019b), including all the cultivable species (Gueimonde et al., 2007; Martín et al., 2007a; Martín et al., 2007b; Delgado et al., 2008; Collado et al., 2009; Hunt et al., 2011; Cabrera-Rubio et al., 2012; Jost et al., 2013; Ward et al., 2013; Jost et al., 2014; Jiménez et al., 2015; Boix-Amorós et al., 2016; Cabrera-Rubio et al., 2016; Sakwinska et al., 2016; Chen et al., 2018; Simpson et al., 2018; Tuominen et al., 2018; Ding et al., 2019; Lackey et al., 2019). Additionally, DNA from some gut-associated strict anaerobes (Faecalibacterium, Bacteroides, Clostridium, Blautia, Coprococcus, Ruminococcus, Roseburia, Eubacterium, Veillonella, and others) has also been repeatedly detected (Cabrera-Rubio et al., 2012; Jost et al., 2013; Jost et al., 2014; Jiménez et al., 2015; Gómez-Gallego et al., 2016).
Different studies have also reported the frequent detection of DNA sequences from soil- and water-related bacteria, including Bradyrhizobium, Novosphingobium, Methylobacterium, Pseudomonas, Sphingobium, Sphingopyxis, Stenotrophomonas, Sphingomonas or Xanthomonas, in human milk (Hunt et al., 2011; Cabrera-Rubio et al., 2012; Urbaniak et al., 2014a; Urbaniak et al., 2014b; Li et al., 2017). It is possible that a high proportion of such sequences are the result of technical artifacts. DNA from the bacterial genera cited above is generally present in molecular biology reagents, solutions and kits (Grahn et al., 2003; Mühl et al., 2010; Salter et al., 2014; Lauder et al., 2016). This represents a major challenge assessing the composition of microbiotas characterized by a low microbial biomass, as is the case of the human milk microbiota in healthy women. Upon amplification, contaminating DNA may overcome the low amount of starting material in the biological sample and lead to incorrect results (Laurence et al., 2014). Sequences belonging to such genera (e.g., Pseudomonas) may be so abundant that they can be wrongly included into the milk core microbiome. There are several measures that can be implemented to minimize this problem, such as sequencing of negative (blank) controls and contaminant removal procedures (Salter et al., 2014; Hornung et al., 2019; Karstens et al., 2019; Stinson et al., 2019), while providing microbial DNA-free sampling containers and molecular reagents is still a pending challenge for companies working in this field.
Differences in the techniques and procedures employed in different studies may account, at least partly, for conflicting results regarding the frequency and abundance of sequences belonging to Lactobacillus, Bifidobacterium and strict anaerobes in human milk (Lagier et al., 2012). These kind of controversies have also happened in relation to the microbiota of the infant gut (Palmer et al., 2007; Turroni et al., 2012). Other limitations and biases of molecular techniques include the lack of discrimination between live or dead organisms when techniques compatible with viability assessments are not selected (Emerson et al., 2017), and the over- or underestimation of some microbial groups because of the composition of their plasmatic membranes, outer membranes or cell walls, methods used for extraction of nucleic acids, copy number of the target gene, the specific 16S rRNA region(s) targeted by the selected primers, and the pipelines used for the bioinformatic analysis (McGuire and McGuire, 2015; Gómez-Gallego et al., 2016; McGuire and McGuire, 2017).
Some studies have revealed the presence of cells, DNA and/or RNA from viruses, archea, fungi and protozoa in human milk (Jiménez et al., 2015; Pannaraj et al., 2018; Boix-Amorós et al., 2019). Although some pathogenic viruses, such as HIV, CMV, Ebola and Zika viruses may be found in human milk (Kourtis et al., 2003; Van de Perre et al., 2012; Bardanzellu et al., 2019; Sampieri and Montero, 2019; Ververs and Arya, 2019), other viruses, including bacteriophages, are also present (Mohandas and Pannaraj, 2020). The human milk virome is distinct from other body anatomical sites and includes eukaryotic viruses, bacteriophages, and viral elements integrated in the host chromosomes (Duranti et al., 2017; Pannaraj et al., 2018). The most abundant eukaryotic viruses belong to the families Herpesviridae, Poxviridae, Mimiviridae and Iridoviridae. Bacteriophages comprise 95% of the human milk viruses and have the ability of modulating the bacterial ecology by killing specific bacteria or by supplying them with additional gene functions (Pannaraj et al., 2018). Human endogenous retroviruses, accounting for 0.06 to 3.63% of all reads, have also been identified in milk samples (Jiménez et al., 2015). Both pathogenic and non-pathogenic viruses can be vertically transferred from mother to infant (Kuehn et al., 2013; Lugli et al., 2016; Duranti et al., 2017; Blohm et al., 2018; Pannaraj et al., 2018).
Some studies have identified archaeal sequences in human milk (Ward et al., 2013; Jiménez et al., 2015; Togo et al., 2019c). In addition, Togo et al. (2019b) were able to isolate Methanobrevibacter smithii from 3 colostrum and 5 milk samples out of a total of 20 samples while Methanobrevibacter oralis was cultured from one milk sample. Methanogenic archaea have remained largely underestimated in human microbiome studies due to technical difificulties in their assessment (Bang and Schmitz, 2015). However, they are particularly adapted to the human gut, participating actively in metabolism and health through methanogenesis (Samuel et al., 2007; Dridi et al., 2009). Presence of M. smithii in the human gut is a feature of healthy lean adults while there is a depletion of this species in obese adults (Le Chatelier et al., 2013; Goodrich et al., 2014). In this context, M. smithii was less frequently detected by either culture or PCR in the milk samples obtained from obese mothers in the study of Togo et al. (2019c). The human gastrointestinal tract is colonized by M. smithii early in life (Grine et al., 2017), and colostrum and milk may represent relevant sources of methanogenic archaea.
In relation to fungi, a metagenomic analysis detected fungal-related sequences in 17 out of the 20 milk samples included in the study (Jiménez et al., 2015). More specifically, the reads belonged to the phyla Basidiomycota and Ascomycota, and to the species Calocera cornea, Candida dubliniensis, Guepiniopsis buccina, Malassezia globosa, Malassezia restricta, Podospora anserina, Sordaria macrospora, Talaromyces stipitatus, and Yarrowia lipolytica, with M. globosa being the most widespread species (Jiménez et al., 2015). Boix-Amorós et al. (2017) could visualize and isolate yeasts from 17 out of 41 milk samples from healthy women, and most of the isolates belonged to the species Candida parapsilosis and Rhodotorula mucilaginosa. Later, the same group analyzed 80 milk samples from women of 4 different countries by sequencing of the ITS1 region of the fungal rDNA gene, and found that Malassezia and Davidiella were the most prevalent genera independenty of the country, while delivery mode and geographic location were associated with shifts in the milk mycobiome composition (Boix-Amorós et al., 2017). Sequencing of the ITS2 region allowed the identification of Candida and Saccharomyces sequences in 6 milk samples from women whose infant stayed in a neonatal intensive care unit (NICU) althouh sequences of the same genera were also identified in samples from the NICU environment (Heisel et al., 2019). Overall, these studied suggest that milk may be a source of fungi for the infant gut, thus contributing to the acquisition and development of the gut mycobiota. However, more studies are required to confirm this role, and to elucidate the potential interactions with other microbes. The significance of the protozoa (Toxoplasma gondii, Giardia intestinalis) detected in milk from some mothers remains unclear (Jiménez et al., 2015).
Factors Affecting the Composition of the Human Milk Microbiota
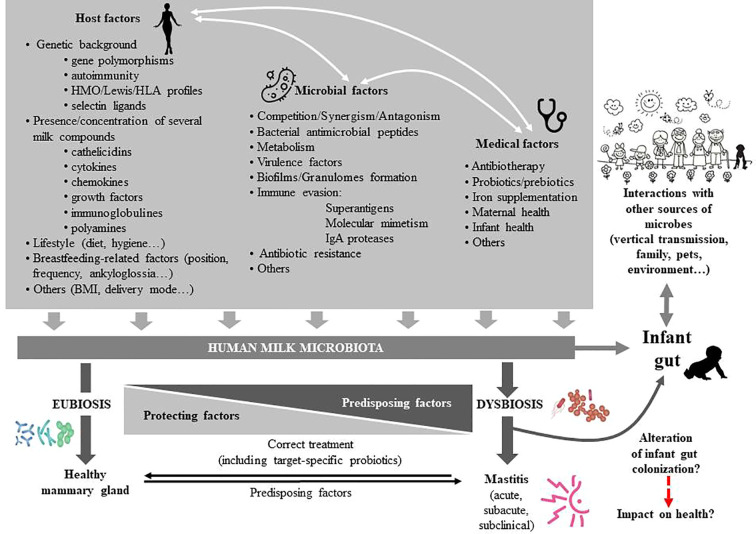
The milk microbiome is characterized by a certain degree of interindividual variability (Martín et al., 2007a; Martín et al., 2007b; Hunt et al., 2011; Boix-Amorós et al., 2016; Cabrera-Rubio et al., 2016; Avershina et al., 2018; Chen et al., 2018). Modifications in its composition may have biological implications for infant colonization, metabolism, immune and neuroendocrine development and for mammary health. However, the current knowledge about the impact of a wide variety of factors (genetic background, ethnicity, milk sampling, geographical location, circadian rhythm, maternal age, diet and body mass index [BMI], delivery mode, gestational age, therapies and food supplements, infant and maternal health status, and others) on human milk microbial communities is very limited (Fernández et al., 2014; Gómez-Gallego et al., 2016) ( Figure 3 ).
Figure 3.
Factors affecting the composition of the human milk microbiota. HMO, human milk oligosaccharides; HLA, human leukocyte antigen; BMI, body mass index; IgA, immunoglobulin A.
Although some studies have tried to elucidate the influence of some of these factors, most of them involved a low number of samples/women and/or have relied on short amplicon sequencing, a technology which poor resolution at the lower taxonomical levels may mask differences or overinflate them; these limitations together with the fact that many factors with a potential impact on the composition of the milk microbiome may interact, makes it difficult to evaluate their true impact on the milk microbiome (LeMay-Nedjelski et al., 2018).
Sample Collection
When milk is collected through the use of domestic (non-single use) milk pumps, a high concentration of yeasts and Gram-negative bacteria (Enterobacteriaceae, Stenotrophomonas, Pseudomonas) may arise from the water used to rinse the devices or as a consequence of unhygienic practices (Knoop et al., 1985; Moloney et al., 1987; Thompson et al., 1997; Boo et al., 2001; Brown et al., 2005; Marín et al., 2009; Jiménez et al., 2017); in fact, some cases of neonatal and infant sepsis and infections initially associated with human milk were the result of contamination of breast pumps (Gransden et al., 1986). A recent study comparing milk samples from mothers who regularly used pumps versus those who never used pumps showed that pumped milk was consistently related to an enrichment of potential pathogens compared to hand expressed milk (Moossavi et al., 2019a). This observation suggests that regular pump use may alter the milk microbiome over time. In fact, it has been shown that when sterile single-use pumps are used, no difference is detected between hand-expressed and pump-expressed samples, while the use of the mother’s own multi-use pump does result in a significant change in the apparent milk microbiome (Rodríguez-Cruz et al., 2020).
Human Milk Oligosaccharides
Human milk oligosaccharides (HMOs) exert prebiotic effects on some bacteria that are frequently detected in human milk, including Bifidobacterium spp. and Staphylococcus epidermidis (Hunt et al., 2012; Thongaram et al., 2017). Milk oligosaccharides and microbes confer a highly personalized “symbiotic” complex which may be crucial for the development of the infant gut microbiota (Jost et al., 2015; Lewis et al., 2015; Wang et al., 2015; Williams et al., 2017a; Moossavi et al., 2019b). Positive correlations between HMO concentration and the abundance of Staphylococcus (Williams et al., 2017b), between B. breve and sialylated HMOs, between B. longum group and non-fucosylated/non-sialylated HMOs, between fucosylated HMOs and Akkermansia muciniphila, and between fucosylated/sialylated HMOs and S. aureus have been described (Aakko et al., 2017). A study found that the maternal secretor status was associated with the composition of the milk microbiota for the first 4 weeks after parturition (Cabrera-Rubio et al., 2019). Although no differences on diversity and richness were detected, Lactobacillus spp., Enterococcus spp., Streptococcus spp., and Bifidobacterium spp. were lower or less prevalent in non-secretor samples than in secretor samples (Cabrera-Rubio et al., 2019).
There are several mechanisms by which HMOs may modulate the human milk microbiota, including prebiotic, antimicrobial, or immunomodulatory properties (Ackerman et al., 2017; Ackerman et al., 2018; Triantis et al., 2018; Bode, 2020). The ability to assimilate, metabolize or use HMOs is conserved among the Bifidobacterium species that are most frequently detected in the infant gut and in human milk (Sela and Mills, 2010; Underwood et al., 2015), although the pathways vary among species and strains (Sakanaka et al., 2019). However, a recent study has shown that there is not a strict correlation between bifidobacterial populations in human milk and their ability to metabolize HMOs (Lugli et al., 2020). The exact growth-promoting effect of HMOs on Staphylococcus isolates from human milk remains unknown, since these bacteria do not metabolize these compounds (Hunt et al., 2012). Recently, novel pathways enabling the growth of Roseburia and Eubacterium (two butyrate-producing genera that have been repeteadly detected in human milk) on HMOs have been published (Pichler et al., 2020). More studies are required to elucidate how HMOs can contribute to shape the microbiota of milk.
Other Human Milk Metabolites
The complexity and dynamic chemical composition of human milk represents a challenge for researchers (Ojo-Okunola et al., 2020). In addition to HMOs, human milk contains a plethora of metabolites produced by either human or bacterial cells that are relevant for mammary and infant health (Gay et al., 2018). Some of them have the potential to shape the composition of the human milk and/or the infant gut microbiotas because of their roles in changing environmental conditions that are relevant for bacterial growth, such as the redox potential or by other mechanisms that promote or inhibit the growth of certain species or strains. Up to 68 metabolites were identified in one study assessing the metabolic and microbiota profiles of milk from 79 women from Finland, Spain, South Africa, and China. Bacilli, Actinobacteria. and Proteobacteria were the bacterial groups displaying more positive or negative associations with milk metabolites (Gómez-Gallego et al., 2018).
Short chain fatty acids (SCFA), including butyrate, acetate, and propionate, are the result of bacterial metabolism in the maternal gut and may reach the mammary gland through the bloodstream. These metabolites are key players in human homeostasis because of their interactions with the microbiota, the immune system, and the neuroendocrine system, contributing to the immunological, metabolic, and neurological programming of the host (Stinson et al., 2020). In this context, the concentrations of acetate and butyrate in milk samples from atopic mothers are significantly lower than in samples from non-atopic mothers (Stinson et al., 2020). Levels of butyrate in milk have also been negatively associated with infant BMI and this fact may program healthy adiposity outcomes later in life (Prentice et al., 2019). Interestingly, administration of some Lactobacillus strains isolated from human milk (Lactobacillus salivarius CECT 5713, Lactobacillus fermentum CECT 5716) to infants has shown ability for increasing the fecal concentration of butyrate (Maldonado et al., 2010; Gil-Campos et al., 2012). Variations in the concentrations of some hormones present in human milk (leptin, insulin) have also been associated with changes in the composition of the infant gut microbiota, SCFA concentrations, and gut permeability (Lemas et al., 2016). Current knowledge on the interactions between the metabolome and the microbiota of human milk is scarce despite the fact that they may have a paramount relevance for infant and mammary outcomes. Integrative studies are required to address these complex interactions.
Maternal Diet and Body Mass Index
Maternal diet is associated with the abundance of some bacterial genera (Williams et al., 2017a; Williams et al., 2017b). Protein intake is positively correlated with the abundance of Gemella while consumption of monounsaturated and saturated fatty acids is negatively related with the abundance of Corynebacterium; similarly, a negative correlation was found between Firmicutes and total carbohydrates, disaccharides, and lactose ingestion (Williams et al., 2017a). A previous study had revealed that monounsaturated fatty acids were positively associated with Lactobacillus genus, while the contrary was observed for Proteobacteria (Kumar et al., 2016).
An association between vitamin C intake during pregnancy and the bacterial diversity of milk has also been reported (Padilha et al., 2019a). The same study found a positive relationship between ingestion of some fatty acids (linoleic and polyunsaturated fatty acids) during lactation and the level of Bifidobacterium in milk.
In relation to BMI, prepregnancy BMI has been associated with a higher milk microbial diversity and a lower abundance of Streptococcus (Davé et al., 2016). Another study found that the abundance of Granulicatella in milk from overweight and obese mothers was higher than in milk from normoweight women (Williams et al., 2017a). A recent study showed that the milk of mothers with a high postpartum BMI contained more Staphylococcus and less Lactobacillus and Streptococcus sequences than milk from normoweight mothers (Ding et al., 2019).
Immune Cells
The lactating mammary gland is a component of the mucosal-associated immune system that displays unique features when compared with other mucosal sites. For example, it contains a low biomass microbiota in contrast to the high bacterial concentration that characterizes the intestinal or the upper respiratory tracts (Mestecky, 2020). In addition, the mammary gland is considered a relevant component of the infant immune system since this link enables maternal-infant immune dialogue (Brandtzaeg, 2010; Hassiotou and Geddes, 2015). It is long known that the gut microbiota is essential for programming the infant immune system and vice versa (Milani et al., 2017a). However, while the development of the immune system and the development of the microbiota are coordinated in the gut, they remain independently regulated in the breast (Niimi et al., 2018). Interactions between milk microbiota and the mammary immune system are poorly known despite the fact that maternal mononuclear cells can transport gut-derived bacteria and bacterial components to the breast during pregnancy and lactation (Perez et al., 2007). It has been speculated that this process facilitates the discrimination between pathogens and commensal microbes by the neonatal immune system (Perez et al., 2007).
Studies dealing with potential associations between the milk concentrations of bacteria and immune cells are scarce and have provided conflicting results from no correlation (Boix-Amorós et al., 2016) to a negative association between the relative abundance of Serratia and both the somatic cell count and the neutrophil concentration (Williams et al., 2017b). As in the case of HMOs and other milk metabolites, there is a need for integrative approaches to clarify the interactions between the immune system and the microbiota in the mammary ecosystem. Information on the potential roles of specific bacterial strains isolated from human milk on the infant immune system is provided in Moving From Composition to Function.
Gestational Age, Mode of Delivery, and Postpartum Period
Controversial results have been obtained from studies comparing the human milk microbiome composition among women delivering preterm or term neonates. So, while Urbaniak et al. (2016a) did not detect differences in bacterial profiles between preterm and term births, Soeorg et al. (2017a) reported that milk of mothers of preterm infants had higher staphylococcal counts but lower species diversity compared with term mothers. The same authors found that the S. epidermidis strains that colonized the gastrointestinal tract and skin of preterm neonates were different to those found in milk while the opposite was observed in term neonates (Soeorg et al., 2017b). However, the gut of breast-fed preterm infants was gradually enriched with strains present in the milk of their mothers.
Concerning the delivery mode, Urbaniak et al. (2016a) found no significant differences in bacterial profiles between Cesarean section (either elective or non-elective) and vaginal deliveries, which is in contrast with the data provided by other authors (Khodayar-Pardo et al., 2014; Cabrera-Rubio et al., 2016; Kumar et al., 2016; Ding et al., 2019). Toscano et al. (2017) described many differences in the microbiome of colostrum depending on the mode of delivery. Compared to Cesarean section, vaginal delivery was associated with a lower abundance of Staphylococcus, Pseudomonas, and Prevotella. In addition, colostrum from women delivering by Cesarean section was associated with a higher number of bacterial hubs and was richer in environmental bacteria. In practice, it is difficult to separate the influence of the mode of delivery with other factors since, as an example, antibiotherapy administered per protocol to women delivering by Cesarean section may be responsable for some of the differences in the microbial population. A recent work reported that the effect of intrapartum antibiotic exposure was less decisive than that of delivery mode on the milk microbiome assessed one month after delivery (Hermansson et al., 2019).
In relation to the postpartum period, Hoashi et al. (2016) found changes in bacterial abundance and glycosylation patterns associated with the amount of time passed since delivery but only in milk from women who delivered vaginally. Drago et al. (2017) detected differences between the microbiomes of colostrum and milk since abundance of anaerobic intestinal bacteria was lower in colostrum than in mature milk. The permeability of the tight junctions in the mammary epithelium is greater in the first days after parturition and this fact determines major differences in the biochemical and immunological composition of colostrum with respect to mature milk. Such differences may be responsible for differences in the microbial composition of colostrum and mature milk. In addition, increasing exposures of the breast to the infant microbiota may also determine microbial changes over time.
Another study showed that the relative abundance of Staphylococcus in samples collected at 3 months postpartum was lower in comparison to samples collected at 10 days postpartum which, in turn, had a lower diversity of operational taxonomic units (OTUs) from the genera Rothia, Veillonella and Granulicatella (Simpson et al., 2018). Biagi et al. (2018) reported that the composition of the milk microbiota changes after the first infant latching, becoming more diverse and being dominated by oral microbes (Rothia and Streptococcus). In contrast, Li et al. (2017) found that neither the lactation stage nor the maternal BMI influenced the milk microbiota of Taiwanese and Chinese women.
Geographical Location
Geographical location of the mother seems to exert a strong impact on the microbiota composition of human milk. A study including samples from mothers of Spain, Finland, South Africa and China found location-related differences in the relative abundance of Bacteroidetes, Actinobacteria and Proteobacteria (Kumar et al., 2016). Geographical differences in the microbiota of samples obtained from 133 mothers in seven regions of China and Taiwan have also been described (Li et al., 2017). Similarly, differences in the alpha diversity and Lactobacillus occurrence in milk were also observed depending on the region of China where the mothers were recruited (Ding et al., 2019). A study involving a higher number of samples from mothers living in Ethiopia, Gambia, US, Ghana, Kenya, Peru, Spain, and Sweden provided evidence of substantial variability within and across cohorts (Lackey et al., 2019).
There are several reasons that may explain why geographical location, even within the same country, might influence the bacterial composition of human milk. Ethnicity, genetic background, diet and climate vary across regions, but people with different ethnicity, genetic background, age, diet, housing, or contact with other people and animals usually coexist in the same village or town. The environmental microbiome (air, surfaces, water, plants, animals, food, waste, etc.) reflects the influences existing in a given place and might drive differences in the composition of the microbiome in any body site. Experiments to elucidate the impact of the geographical location on the milk microbiome are very challenging because of the high number of factors that may bias the interpretation of the data. They should have to take into account the factors cited above but also use identical protocols from sample collection, storage and shipping to data analysis, which is very difficult in practice.
Maternal Treatments (Antibiotics, Probiotics, Prebiotics, and Chemotherapy) and Infections
Antibiotic administration is one of the main drivers of dysbiosis in mucosal surfaces and the lactating mammary gland does not seem to be an exception. Lactobacilli and bifidobacteria are more abundant in milk from women who are not treated with antibiotics during pregnancy, delivery or lactation (Soto et al., 2014). Similarly, the Bifidobacterium load in milk obtained in the first week after delivery was lower among women receiving antibiotic prophylaxis in comparison to the control group (Padilha et al., 2019b). It is interesting to note that significantly lower amounts of bifidobacteria have been found in milk of allergic mothers compared with non-allergic ones (Grönlund et al., 2007).
Soeorg et al. (2017a) found that the probability of finding mecA-positive CNS in milk increases if the mother is hospitalized during the first month after delivery or if the neonate received antibiotherapy or needed an arterial catheter. These authors suggested that the presence in milk of pathogenic Staphylococcus could be reduced by limiting the exposition of women to the hospital environment. Intrapartum antibiotic prophylaxis is associated with a higher presence of transmissible genes conferring resistance to antibiotics in milk, which are subsequently shared with their infants (Pärnänen et al., 2018)
In relation to probiotics, oral administration of the probiotic VSL#3 (a mix of 8 strains belonging to the species B. breve, B. longum, L. acidophillus, Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus paracasei, Lactobacillus plantarum, and Streptococcus thermophilus) to pregnant and lactating women led to higher concentrations of Lactobacillus and Bifidobacterium in colostrum and milk of those women who ingested the product when compared to the placebo group (Mastromarino et al., 2015). However, this effect was only seen among women with vaginal deliveries. In contrast, ingestion of milk containing Bifidobacterium animalis ssp. lactis Bb-12, L. acidophilus La-5 and Lactobacillus rhamnosus GG during late pregnancy and early lactation, did not modified significantly the composition of the milk microbiota (Simpson et al., 2018). Recently, Padilha et al. (2020) reported that the intake of prebiotics (fructooligosaccharides) affects the composition of the milk microbiota and found that this effect was influenced by the age of the mother.
Two studies studied the impact of chemotherapy employed during the treatment of Hodgkin’s lymphoma in the milk microbiome. The first study included milk samples obtained regularly during four months from a single treated woman and from 8 healthy lactating women (Urbaniak et al., 2014b). Chemotherapy led to significant changes in the milk bacterial composition, including an increase of Xanthomonadaceae, Acinetobacter and Stenotrophomonas, and a drastic reduction in the presence of Eubacterium, Bifidobacterium, Cloacibacterium and Staphylococcus. Changes in the metabolic profile of milk, characterized by a decrease of inositol and docosahexaenoic, were also observed (Urbaniak et al., 2014b). The second study provided contradictory results since no negative effect of chemotherapy on community diversity was found (Ma et al., 2016).
Maternal and infant infections exert a strong influence on the immunological composition of milk (Bryan et al., 2007; Riskin et al., 2012; Hassiotou et al., 2013) and, therefore, they likely impact on its microbiological composition. However, there is an almost complete absence of studies addresing this question. Human papilloma virus (HPV) infection was not correlated with a modification of the bacterial composition of milk (Tuominen et al., 2018) although the authors stated that this result might be due to the low number of HPV positive milk samples (3 out of 35) in the analyzed population.
Transfer of Milk Bacteria to the Infant Gut
The bacteria present in colostrum and milk are among the first microbes to enter the neonatal gastrointestinal tract and, as a consequence, they may have a paramount relevance as drivers in the acquisition and development of a healthy microbiota. The role of milk as a source of bacteria, including strict anaerobes, to the infant gut has been repeteadly reported at the species and/or the strain level, by classical culture techniques (Martin et al., 2003; Martín et al., 2006; Martín et al., 2009; Solís et al., 2010; Albesharat et al., 2011; Makino et al., 2011; Martín et al., 2012b; Kozak et al., 2015; Makino et al., 2015; Murphy et al., 2017), and by culture-independent approaches (Milani et al., 2015; Asnicar et al., 2017; Duranti et al., 2017; Milani et al., 2017b). In addition, mother-to-infant transfer of Bifidobacterium phages through human milk has also been described (Duranti et al., 2017). However, the significance and contribution to infant gut colonization of milk bacteria remains an open question. Clinical studies trying to elucidate the question are confounded by the profound impact of non-microbial human milk components to intestinal microecology.
The microbiota of healthy breastfed infants is related to that present in the milk of their respective mothers. Pannaraj et al. (2017) suggested that approximately a quarter of the bacteria detected in infant feces during early life may derive from milk while Murphy et al. (2017) reported that the genera responsible for most of the bacterial abundance (>70%) in infant feces are shared with human milk. However, these two studies achieved genus level resolution, and only strain level analysis is valid for identifying shared taxa. Asnicar et al. (2017) described the vertical milk transmission of microbial strains, including some belonging to strict anaerobic species, and characterized their transcriptional activity once in the infant gut. Bacterial-host networks are different when breast-fed infants are compared to formula-fed infants (Martín et al., 2016), a fact that is reflected in the host transcriptome (Praveen et al., 2015). Some studies have also found long lasting differences in the fecal microbiota when comparing exclusively breastfed and non-exclusively breastfed infants (Pannaraj et al., 2017; Ho et al., 2018; Moossavi et al., 2019a). Overall, bacterial diversity and microbial pathways implied in the metabolism of carbohydrates are higher in non-exclusively breastfed infants while the contrary is observed for those pathways involved in the metabolism of vitamins, lipids and xenobiotic compounds (Ho et al., 2018).
Although the introduction of solid food during weaning was once thought to be associated with a sharp increase in the diversity of the gut microbiota in breastfed infants (Favier et al., 2002), the fecal microbiota of infants are dominated by milk bacteria, independently of the introduction or not of other foods, as long as they are breastfed (Bäckhed et al., 2015).
Moving From Composition to Function
Human milk is a source of a wide spectrum of beneficial microorganisms that might play a role in priming the development and function of many infant systems (Ojo-Okunola et al., 2018). However, studies dealing with the functions of such microbiota/microbiome are very scarce and, as stated by Theobald Smith, (1904), “it is what bacteria do rather than what they are that commands attention, since our interest centers in the host rather than in the parasite”. Human colostrum and milk contain a complex array of bioactive molecules and cells, including the microbiota, which may act synergistically to preserve infant and maternal health through a wide variety of mechanisms (Morrow and Rangel, 2004).
The human milk microbiota may contribute to, at least, some of the functional properties and health benefits that epidemiological studies have associated with breastfeeding (Renfrew et al., 2012), including protection against infections, metabolic programming, immunomodulation and neuromodulation. Human milk bacteria may provide a certain degree of protection against infections caused by viruses, bacteria or fungi through a variety of mechanisms: (a) biosynthesis of compounds with antimicrobial activity, including organic acids (lactic acid, acetic acid, ethanol), bacteriocins, reuterin or hydrogen peroxide (Heikkilä and Saris, 2003; Beasley and Saris, 2004; Martín et al., 2005; Martín et al., 2006; Cárdenas et al., 2016; Cárdenas et al., 2019; Angelopoulou et al., 2020; García-Gutierrez et al., 2020); (b) coaggregation with pathobionts, impeding their access to the gut epithelial cells (Cárdenas et al., 2019); (c) competitive exclusion with pathobionts for nutrients or host receptors (Olivares et al., 2006a; Martín et al., 2010; Langa et al., 2012); (d) reinforcement of the infant gut barrier by preserving and decreasing intestinal permeability and increasing mucin biosynthesis (Olivares et al., 2006a; Vanhaecke et al., 2017; Liu et al., 2020); and (e) inmmunomodulation (Liu et al., 2020).
The analysis of the genome of some strains isolated from human milk provides some clues that may explain their anti-infectious properties. As an example, L. salivarius CECT 5713 is able to inhibit HIV-1 infectivity in vitro and its genome contains a gene encoding a protein containing a recognition motif of the high mannose N-linked oligosaccharides displayed by many pathogen antigens, such as gp120, which is essential for HIV pathogenesis (Langa et al., 2012). In developing countries, the World Health Organization (WHO) recommends exclusive breastfeeding among HIV-infected women during the first six months after birth “unless replacement feeding is acceptable, feasible, affordable, sustainable, and safe for them and their infants”, and to continue breastfeeding thereafter, with gradual introduction of solid foods (WHO, 2016). In these settings, the advantages of breastfeeding for mother and infant health compensate the potential risk of viral transmission (Barthel et al., 2013). Unfortunately, the contribution of the human milk microbiota in protecting from infant infections caused by this or other life-threatening viruses which are relatively frequent in developing (dengue, Ebola, and zika) or developed countries (CMV) remains largely unexplored. Interestingly, the outcome of neonatal rotavirus infections is influenced by the complex interplay between HMOs, the milk microbiome and the infant gut microbiome (Ramani et al., 2018). In the context of the ongoing pandemic caused by the SARS-CoV-2 virus, a recent study did not detect the virus in human milk (Chambers et al., 2020), and COVID-19-positive women are recommended to continue breastfeeding by the WHO and the United Nations Children’s Fund (UNICEF) (United Nations Children’s Fund, 2020).
Although most of the information about potential antimicrobial properties of human milk bacteria has arisen from in vitro studies and animal models, the beneficial effects of some strains have been confirmed in human clinical trials. Daily intake of a formula containing L. fermentum CECT5716 by 6-month-old children significantly decreased the rates of upper respiratory tract infections, gastrointestinal infections and total infections in the following 6 months (Maldonado et al., 2012). L. salivarius PS7, a human milk strain with a strong antimicrobial activity against several otopathogens, has been shown to be efficient in preventing recurrent acute otitis media in children (Cárdenas et al., 2019).
Studies addressing the potential role of the human milk microbiota on infant metabolism are scarce but promising. Bacterial diversity in milk is positively correlated with metabolites that are known to exert beneficial effects, including docosahexaenoic acid (DHA), as assessed by the construction of diversity-metabolites networks (Ma et al., 2016). Additionally, some species seem to be critical to regulate the concentrations of relevant milk metabolites, including inositol, DHA or butanal (Ma et al., 2016). Previous studies revealed that some strains originally isolated from human milk display metabolic activivity once in the human gut and are able to participate in the biosynthesis of functional metabolites, such as butyrate, leading to better bowel habits (Olivares et al., 2006c). More recently, administration of L. fermentum CECT5716 to pregnanat and lactating rats induced beneficial changes in the fatty acid composition of milk by increasing total polyunsaturated fatty acids including linoleic and α-linolenic acids and decreasing the proportion of palmitic acid (Azagra-Boronat et al., 2020).
Human milk bacteria may also be involved in immune programming of infants through different mechanisms, impacting both innate and acquired immunity (Díaz-Ropero et al., 2006; Olivares et al., 2006b; Pérez-Cano et al., 2010; Azagra-Boronat et al., 2020). Such mechanisms seem to be complementary and to exhibit a high degree of flexibility depending on factors related to the gut environment, such as the exposition to lipopolysaccharide (Díaz-Ropero et al., 2006). L. fermentum CECT5716 and L. salivarius CECT5713 behave as activators of NK, CD4+, CD8+, and regulatory T cells, and their immunomodulatory effects are different to those displayed by other strains of the same species but isolated from other sources (Pérez-Cano et al., 2010). In fact, L. fermentum CECT5716 is able to significantly ameliorate the inflammatory response and the intestinal damage in an animal model of intestinal inflammation (Peran et al., 2005; Peran et al., 2006; Peran et al., 2007).
As cited above, immunomodulation is involved in the anti-infectious protection conferred by some human milk bacteria or in the restoration of the damage caused by infections. In vivo assays have shown that L. rhamnosus SHA113 inhibits the expression of TNF-α and IL-6 caused by a multi-drug resistant S. aureus strain, restoring the concentration of leukocytes in blood (Liu et al., 2020).
Bacterial strains originally isolated from milk of healthy women are appealing as probiotic candidates because of their origin, which implies a history of safe intake by infants (Lara-Villoslada et al., 2009; Maldonado et al., 2010; Gil-Campos et al., 2012; Maldonado-Lobón et al., 2015a), and complex symbiotic interactions from the time we are born (Lara-Villoslada et al., 2007; Fernández et al., 2013; Jeurink et al., 2013). Among them, those species belonging to the genera Bifidobacterium and Lactobacillus (B. longum, B. breve, L. salivarius, L. fermentum, L. reuteri, L. gasseri, L. plantarum, and L. rhamnosus), have received special attention since many of them enjoy the GRAS (Generally Recognised As Safe) status (Food and Drug Administration, USA) and the QPS (Qualified Presumption of Safety) of the European Food Safety Authority (EFSA). In contrast, S. mitis and other mitis streptococci, S. salivarius and CNS have received marginal attention because of theoretical safety concerns despite of the fact that they are among the dominant bacteria both in this biological fluid (Jiménez et al., 2008a; Hunt et al., 2012; Martín et al., 2012b; Cacho et al., 2017) and in the feces of breast-fed infants (Jiménez et al., 2008a). However, they may provide relevant probiotic functions in practice (Kirjavainen et al., 2001; Uehara et al., 2001; Otto, 2009; Iwase et al., 2010; Park et al., 2011), and therefore, their potential beneficial roles in infants deserve future studies.
Desciphering the genomes of representative collections of human milk isolates and analysis of the human milk metagenome will allow the expansion of our knowledge on the functions that a “healthy” human milk microbiome should provide to the mother-infant dyad. The design of human milk bacterial consortia specifically tailored to meet early life requeriments is an attractive strategy for those infants that are devoid of the benefits of breastfeeding (Fernández et al., 2018).
The Origins of the Human Milk Microbiota
While suckling, some oral bacteria from the mouth or nasopharynx of the infant may seed milk (Ramsey et al., 2004; Moossavi et al., 2019a); however, pre-colostrum expressed during late pregnancy contain some of the bacteria usually detected in human milk (Martín et al., 2004). In fact, oral-associated bacteria have also been isolated in precolostrum collected at the end of the first pregnancy and, therefore, before any contact with the newborn (Ruiz et al., 2014).
The origin of the microbes that constitute the oral microbiome remains widely unknown (Zaura et al., 2014) but streptococci are already present in edentulous infants (Li et al., 1997; Caufield et al., 2000; Bearfield et al., 2002; Cephas et al., 2011). The fact that these bacteria are also frequently detected in human milk (Jiménez et al., 2008a; Jiménez et al., 2008b; Hunt et al., 2011; Martín et al., 2015), might suggest a role in the acquisition or modulation of the oral bacterial communities. A recent study investigating potential relationships among bacterial communities in samples of milk, maternal and infant feces, and maternal and infant oral swabs from some mother-infant pairs found strong associations among these three complex microbial communities and Streptococcus was the most abundant genus not only in infant and maternal oral samples but also in milk (Williams et al., 2019). Similar results had been previously obtained by Davé et al. (2016). Biagi et al. (2017) showed that only a limited number of OTUs were shared between milk and the mouth of the infants, but they included specific Streptococcus and Staphylococcus OTUs.
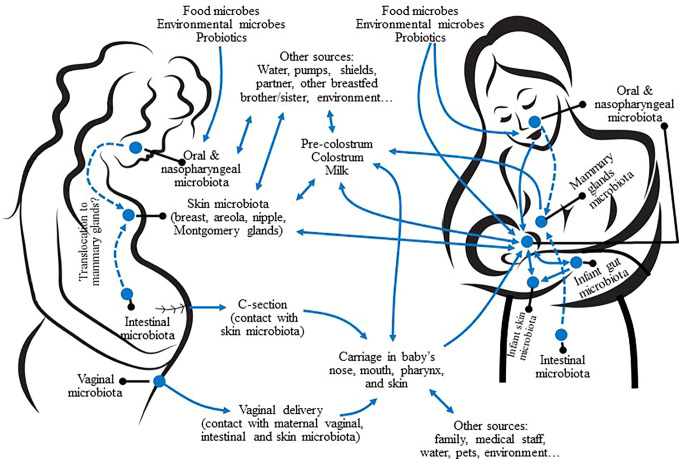
Some species and genera commonly detected from human milk, such as Corynebacterium C. acnes and, especially, S. epidermidis, are also inhabitants of the human skin (Oh et al., 2014). Therefore, breast skin, nipples, and mammary areolas may be a source of such bacteria for human breast and milk ( Figure 4 ). However, no studies on the specific transfer of skin bacterial strains to human milk or breast tissue have been performed yet. CNS, Cutibacterium and Corynebacterium are also present in all the human mucosal surfaces, including that of the digestive tract, which is, very likely, a relevant source of these bacteria for the mammary environment ( Figure 4 ).
Figure 4.
Potential sources of the microbes present in human milk and interactions with other mother-infant microbiotas. Dashed arrows represent potential translocation through an endogenous pathway.
Although milk, breast skin and the infant oral cavity may share some phylotypes, there are major differences between their respective microbial communities (Hunt et al., 2011; Cabrera-Rubio et al., 2012; Jost et al., 2013; Jiménez et al., 2015). A study that compared the bacteriome of areolar skin, milk, and feces of 107 mother-infant dyads found differences in diversity and composition among the bacterial communities of the three ecosystems (Pannaraj et al., 2017).
The mucosal surfaces of the maternal digestive tract (oral cavity, gut) may be a source of bacteria for the mammary ecosystem from late pregnancy to the end of lactation (Martín et al., 2004; Rodríguez, 2014; Mira and Rodríguez, 2017; Fernández and Rodríguez, 2020). These oral- and entero-mammary routes imply a highly regulated cross-talk between bacterial cells, immune cells (dendritic cells [DCs] and macrophages) and epithelial cells. These complex cross-talks would drive the physiological translocation of certain bacteria without compromising the integrity of the gut epithelium (Vazquez-Torres et al., 1999; Rescigno et al., 2001; Macpherson and Uhr, 2004). Upon translocation, the mammary gland would exert a homing effect on the immune cells that act as bacterial carriers (Perez et al., 2007).
Low level bacterial translocation from the digestive tract to extra-digestive locations is a relatively common process (Ouwehand et al., 2001; Vankerckhoven et al., 2004; Begier et al., 2005; Dasanayake et al., 2005), while an increase in the rates of bacterial translocation from the gastrointestinal tract to the mammary environment has been observed in pregnant and lactating rodents without compromising host health (Perez et al., 2007; Treven et al., 2015; de Andrés et al., 2017; Azagra-Boronat et al., 2020). In addition, the existence of an entero-mammary traffic of immune cells during late pregnancy and lactation has long been known (Bertotto et al., 1991; Roitt, 2001; Newburg, 2005). Such efflux is responsible for the integration of the lactating breast into the mucosal immune system and its transformation in a formidable organ from the immunological point of view (Pabst, 1987; Brandtzaeg, 2010). It has also been reported that some bacterial strains isolated from human milk can cross Caco-2 monolayer by using a mechanism that involves interactions with DCs while this ability is lost in the absence of DC-like cells (Langa, 2006; Langa et al., 2012). Carrying of bacterial cells, including streptococci, lactobacilli, and bifidobacteria, by blood and milk maternal mononuclear cells has already been reported (Perez et al., 2007).
In fact, presence in milk of specific lactic acid bacteria after their per os administration has been reported in lactating rodents (Treven et al., 2015; de Andrés et al., 2017; Azagra-Boronat et al., 2020), lactating women (Jiménez et al., 2008c; Abrahamsson et al., 2009; Arroyo et al., 2010), or pregnant women (Fernández et al., 2016). Similarly, mothers who had ingested L. rhamnosus GG, L. acidophilus La-5, and B. animalis ssp. lactis Bb-12 can transfer such strains to their infants through breastfeeding (Avershina et al., 2018; Simpson et al., 2018).
Previously, it has been shown a Lactobacillus strain ingested during pregnancy could be detected in the feces of the breastfed infants, including those who were born by Caesarean section (Schultz et al., 2004). However, the authors did not investigate if the strain was present in the milk of the mothers. More recently, a B. breve strain detected in a rectal sample and in the milk of a woman could be also detected in the feces of her infant who was delivered via Caesarean section, suggesting a direct mother-to-infant transmission and supporting the possibility of a microbial translocation through an entero-mammary pathway (Kordy et al., 2020). In addition, the possibility of a shared environmental source, including the indoor microbiome, must not be ruled out.
Many transient anatomical and physiological changes may increase the translocation rate from the digestive system to the mammary glands in pregnant and lactating women (Rodríguez, 2014; Mira and Rodríguez, 2017; Fernández and Rodríguez, 2020). Such a homing effect may involve a physiological immunodepression to tolerate the fetus, a formidable angiogenesis process in the breast and the filling of the mammary duct system with pre-colostrum during late pregnancy, which would be an excellent source of nutrients for bacteria.
Although more studies are necessary to elucidate the existence of oral- and entero-mammary pathways, if this was confirmed, it would provide new approaches to manipulate the microbiota of mothers and infants inorder to improve infant health and development (Rautava et al., 2002; Martín et al., 2004).
Mammary Dysbiosis and Lactational Mastitis
Mastitis constitutes a common feeding problem for most mammalian species and humans are not an exception (Contreras and Rodríguez, 2011). This condition is the major cause of undesired weaning from the medical point of view and represents a relevant public health issue since such premature weaning prevents the health benefits that breastfeeding provides to infants and mothers (U.S. Department of Health and Human Services, 2011; American Academy of Pediatrics, 2012; Renfrew et al., 2012).
A wide variety of bacteria may inhabit the mammary gland ecosystem during a healthy lactation period, including potential mastitis-causing species; however, if there is a disturbance of this balanced state, milk dysbiosis may occur, eventually leading to mastitis (Delgado et al., 2008; Fernández et al., 2014; Mediano et al., 2017) ( Figure 3 ). Patel et al. (2017) analyzed milk samples from women suffering either subacute or acute mastitis and, also, from healthy controls. In comparison to controls, the microbiome of samples from acute and subacute cases were distinct: their diversity was drastically reduced, and they were significantly enriched in some aerotolerant bacteria, including Staphylococcus, while depleted in obligate anaerobes, such as Ruminococcus, Faecalibacterium, or Eubacterium. Similar alterations in the milk microbiome have also been found in cases of mastitis involving other mammalian species (Kuehn et al., 2013; Oikonomou et al., 2014; Derakhshani et al., 2018).
The etiology and pathogenesis of acute and subacute lactational mastitis have been reviewed by Fernández et al. (2014) and Rodríguez and Fernández (2017). Empiric use of antibiotics has been, and still is, the most common approach to treat mastitis. However, many cases do not respond to such therapy since mastitis agents are becoming increasingly resistant to antimicrobials through different mechanisms, including intrinsic resistances, presence of transmissible antibiotic resistance genes and/or the formation of biofilms (Marín et al., 2017). The high rates of antimicrobial resistance among mastitis-causing bacteria have clinical relevance in relation to treatment options. In addition, wide-spectrum antibiotics may alter the bacterial composition of milk, impairing vertical transmission of microbes through breastfeeding (Soto et al., 2014). Therefore, new strategies for the management of mastitis are needed, and in this context, those based in the modulation of the mammary bacterial communities through the selection and application of probiotic strains that were originally isolated from human milk seem particularly suited for this target (Fernández et al., 2014).
Several human trials have shown that oral administration of some human milk strains (L. salivarius CECT5713, L. salivarius PS2, Lactobacillus gasseri CECT5714, L. fermentum CECT5716) provoke relevant changes in a variety of milk microbiological, biochemical and immunological parameters, including a significant decrease in the concentration of mastitis-causing agents (Jiménez et al., 2008c; Maldonado-Lobón et al., 2015b; Espinosa-Martos et al., 2016). In fact, such an approach has been found to be more efficient than empiric antibiotherapy for the treatment of this condition (Arroyo et al., 2010). Metabolomic studies have revealed that the impact of the probiotic treatment can be also observed in the urine of the treated women (Vazquez-Fresno et al., 2014). As an example, lactose was present in urine samples before the treatment but it was no longer detected after the probiotic treatment, indicating a restoration of the integrity of the mammary epithelium. Assessment of transcriptomic changes in milk somatic cells associated with the intake of L. salivarius PS2 by women with mastitis has also provided valuable information about the potential mechanisms responsible for the efficacy of specific probiotics in treating mastitis (de Andrés et al., 2018). Finally, a few strains (L. salivarius PS2 and L. fermentum CECT5716) have been successfully applied as a preventive strategy compared to a placebo, when administered either during late pregnancy (Fernández et al., 2016) or during lactation (Hurtado et al., 2017) to women with a history of mastitis after previous pregnancies.
Significant increases in the milk concentrations of TGF-β2 and IgA have been observed after intake of other probiotics during pregnancy or lactation (Rautava et al., 2002; Prescott et al., 2008; Nikniaz et al., 2013), suggesting that the probiotic approach may control the growth of mastitis-causing bacteria in the mammary gland preventing potential damage to the mammary epithelium.
Microbiota of the Breast Tissue of Non-Lactating Women
Most microbial studies addressing the human mammary ecosystem have been focused on the lactation period and limited to the analysis of colostrum and milk samples. However, the number of studies dealing with the microbiota of breast tissue in non-lactating women is rapidly increasing, particularly in the frame of breast cancer research. Breast cancer ranks as the most common malignancy in women. Some studies have observed a correlation between dysbiosis of the gut microbiota and breast cancer (Goedert et al., 2015; Goedert et al., 2018; Laborda-Illanes et al., 2020). In addition, changes in the microbiota of breast tissue have also been linked to breast cancer.
In a pioneering study, samples of breast tissue from 81 women with and without cancer were analyzed by sequencing of the V6 region of the 16S rRNA gene (Urbaniak et al., 2014a). Sequences corresponding to Staphylococcus, Cutibacterium, Acinetobacter, Enterobacteriaceae, Bacillus, Pseudomonas, or Prevotella were detected in a high percentage of the samples, including some obtained from women without a previous history of lactation. Cultures confirmed the presence of viable bacteria in some of the samples. In parallel, Xuan et al. (2014) employed 454 pyrosequencing of the V4 region of 16S rRNA gene to analyze breast tissue samples from 20 patients with breast cancer, each one providing tumor tissue and normal adjacent tissue. These authors found that Methylobacterium radiotolerans was enriched in tumor tissue, while Sphingomonas yanoikuyae was enriched in normal tissue. However, some doubts arise from their results since this technique does not discriminate at the species level, the DNA of the two genera that discriminated between both groups are typically found in DNA extraction reagents and kits, and no blank controls were included in the study. Later, Urbaniak et al. (2016b) reported that the bacteriome of breast tissue adjacent to breast cancer differs from that found in breast tissue from healthy controls undergoing cosmetic surgery. Compared to healthy controls, the relative abundances of Bacillus, Enterobacteriaceae and Staphylococcus were higher in samples from women with breast cancer while the abundance of lactic acid bacteria was lower (Urbaniak et al., 2016b).
Hieken et al. (2016) found different bacterial communities in breast tissue from women with breast cancer in comparison to women suffering from a benign breast disease. Malignancy was associated with an increase in some taxa that were present at a low abundance level, including the genera Atopobium, Fusobacterium, Gluconacetobacter and Hydrogenophaga. Chan et al. (2016) investigated the presence of bacteria in the nipple aspirate fluid obtained from women with a previous history of breast cancer and from healthy women and found a higher incidence of the genus Alistipes in the first group. In addition, the bacteria associated with breast cancer shared β-glucuronidase activity and, therefore, the authors suggested that this enzymatic activity may promote breast cancer. Recently, two studies described racial differences in the microbiome of breast tumors (Smith et al., 2019; Thyagarajan et al., 2020) while another study identified differences in diversity and composition not only between tumor and normal tissue but also among women and between the breasts of the same woman (Klann et al., 2020).
The microbiota may promote or inhibit tumorigenesis by several mechanisms, including the alteration of immune responses or the effect of bacterial-derived enzymes (e.g., β-glucuronidase and β-glucosidase activities) and metabolites, such as short-chain fatty acids, lipopolysaccharides, secondary bile acids, estrogens, or genotoxins (Kovács et al., 2020). In addition, the microbiota and its metabolites may exert a strong influence on the efficacy of chemotherapy and radiation therapies (Kovács et al., 2020). Overall, the results of previous studies warrant further research to elucidate the relationships between breast microbiota and breast cancer.
The breast microbiome may also play a key role in the outcomes of breast plastic or cosmetic surgery, including breast reconstruction, breast reduction or breast augmentation. Breast augmentation is one of the most frequent cosmetic surgical procedures practiced worldwide and can lead to several complications, capsular contracture being the most common one (Rieger et al., 2013; Cook et al., 2020). Bacterial growth, and subsequent biofilm formation, is one of the main risk factors for capsular contracture (Dancey et al., 2012; Ajdic et al., 2016; Walker et al., 2019). It has been suggested that chronic biofilm infection of breast implants may be implicated in the development of breast implant-associated lymphoma (Hu et al., 2015; Hu et al., 2016). Bacteria that are often associated with human milk, breast tissue and breast skin (S. epidermidis and other CNS species; C. acnes) have been repeatedly isolated within or surrounding breast implants from patients with capsular contracture by using classic culture-based approaches (Virden et al., 1992; Dobke et al., 1995; Ahn et al., 1996; Pajkos et al., 2003; Galdiero et al., 2018) and culture-independent techniques (Cook et al., 2020). At present, the origin of the bacteria detected in the explants (breast tissue and skin contamination) remains unclear (Bachour et al., 2019), and more studies are required to elucidate the role of mammary-related bacteria in the tolerance toward these devices or in the adverse outcomes that are relatively frequently associated with their implantation.
Future Trends and Conclusions
Conflicting results when trying to analyze the factors that may play a role in shaping the composition of the human milk microbiota can be explained, at least partially, by the low number of samples/women analyzed in most of the studies carried out so far and, also, by many host, environmental, perinatal, and technical factors (LeMay-Nedjelski et al., 2018). International and collaborative studies, involving a high number of participants and performed under identical conditions, are required in order to elucidate the impact of these factors and their interactions.
The origin of the microbes found in the mammary ecosystem remains a largely open question. Some of the bacteria detected in milk most likely originate from the maternal skin and areola. During lactation, shared features between the microecology in the infant mouth and milk suggest interactions but the precise directions and significance of which are yet to be determined. Perhaps most intriguingly, experimental and human data indicates that some bacteria in milk may originate in the maternal digestive tract and that an enteromammary pathway for microbes may exist. Further assessment of this hypothesis demands sophisticated experimental and clinical studies and state-of-the-art methods to ensure accurate strain-level identification of specific bacteria in not only the intestine and milk but also in the bloodstream and within the immune cells thought to be responsible for the transfer.
The emerging data indicating that various maternal characteristics and exposures, including BMI, antibiotic exposure, gestational age or delivery mode, are associated with the composition of the milk microbiota suggest that the microbes in milk are linked to health and disease in the mother and perhaps, also, in the infant. As of present, however, the biological role and clinical significance of the bacteria in human milk remain poorly characterized and several fundamental questions require elucidation. Detailed metagenomic, metatranscriptomic and metabolomic studies are paramount for understanding the functionality of the milk microbiota. It is also important to appreciate the fact that most published studies describing the bacterial communities of human milk are based on culture-independent molecular methods, such as qPCR and sequencing of either of the 16S rRNA gene or the whole bacterial genome. While these tools are highly sensitive, detection of bacterial DNA does not entail the presence of viable or even intact bacteria. Furthermore, there are published data indicating that at least some bacteria in milk may actually be found on and inside immune cells (Perez et al., 2007). Distinguishing between intact and viable free or intracellular bacteria and the mere presence of bacterial fragments is a crucial step in understanding their biological function. The presence of bacterial components such as DNA in milk may be sufficient to induce immune responses in the mammary epithelium, immune cells, or the neonatal gut. Milk immune cells interacting with bacteria, on the other hand, may mediate immune responses specific to these bacteria. Given the role of human milk in establishing immune tolerance towards antigens in the maternal diet (Verhasselt et al., 2008), it is conceivable that milk may serve as a vehicle for inducing tolerance in the newborn to colonizing microbes from the mother. Currently, no direct evidence to corroborate or refute this hypothesis exists.
Live bacteria in human milk serve biological purposes in the mother. The relationships between the bacterial composition of milk and the risk of mastitis suggest that the indigenous bacteria may be necessary for mammary gland health. Mammary bacteria may also play some roles in the non-lactating mammary gland and the development of breast carcinoma, which is a subject of substantial clinical significance and an area of active research.
In addition to implications to maternal health, the human milk microbiota may be transferred to the infant, potentially acting as a driver in early oral and gut colonization. As reviewed above, shared bacterial taxa detected in maternal stool, human milk and the infant gut suggest that milk may be a vehicle for early colonization. This has to be interpreted with caution since there is the possibility of other routes of bacterial transfer and taxonomic discrimination should be performed at the strain level to confirm identity. As in the case of the origin of the milk bacteria, experimental studies in animal models offer a more reductionist means of dissecting the role of milk microbes for gut colonization. A translational approach complementing clinical studies with basic science is therefore needed.
So far, most studies dealing with the human milk microbiome have been focused on the taxonomical composition and only a few of them have dealt with its potential functions for the infant-mother dyad. Breastfeeding has been associated with reduced risk of chronic conditions such as obesity (Victora et al., 2016), which has also been linked to aberrant early gut colonization. The extent to which the beneficial health effects of breastfeeding are mediated by modulation of the developing gut microbiota remains an open question. Even less clear is the impact of human milk bacteria on child health. Indeed, studying the roles of the human milk microbiota in health and disease is a difficult task since there are synergistic activities among different milk molecules and cells, and several non-microbial components in human milk have the potential to modify the infant gut microbiota. However, we have a rapidly increasing variety of powerful in vitro and in vivo tools, techniques, and procedures to address such a question from cell biology to well-designed clinical trials, from human breast organoids to human milk microbiota-associated mouse models (Wang et al., 2017) and from true metagenome (integrating data from microbiome, microbial genomes and human genome projects) to systems biology approaches.
Author Contributions
All the authors contributed equally to the review of the literature and to the writing and revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
PSP receives grant support from R01 HD100542-01. JMR and LF are recipients of grant PID2019-105606RB-I00 (Ministerio de Ciencia e Innovación, Spain).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aakko J., Kumar H., Rautava S., Wise A., Autran C., Bode L., et al. (2017). Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef. Microbes 8, 563–567. 10.3920/BM2016.0185 [DOI] [PubMed] [Google Scholar]
- Abrahamsson T. R., Sinkiewicz G., Jakobsson T., Fredrikson M., Björkstén B. (2009). Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J. Pediatr. Gastroenterol. Nutr. 49, 349–354. 10.1097/MPG.0b013e31818f091b [DOI] [PubMed] [Google Scholar]
- Ackerman D. L., Doster R. S., Weitkamp J. H., Aronoff D. M., Gaddy J. A., Townsend S. D. (2017). Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against group B Streptococcus . ACS Infect. Dis. 3, 595–605. 10.1021/acsinfecdis.7b00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman D. L., Craft K. M., Doster R. S., Weitkamp J. H., Aronoff D. M., Gaddy J. A., et al. (2018). Antimicrobial and antibiofilm activity of human milk oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii . ACS Infect. Dis. 4, 315–324. 10.1021/acsinfecdis.7b00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn C. Y., Ko C. Y., Wagar E. A., Wong R. S., Shaw W. W. (1996). Microbial evaluation: 139 implants removed from symptomatic patients. Plast. Reconstr. Surg. 98, 1225–1229. 10.1097/00006534-199612000-00016 [DOI] [PubMed] [Google Scholar]
- Ajdic D., Zoghbi Y., Gerth D., Panthaki Z. J., Thaller S. (2016). The relationship of bacterial biofilms and capsular contracture in breast implants. Aesthet. Surg. J. 36 (3), 297–309. 10.1093/asj/sjv177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albesharat R., Ehrmann M. A., Korakli M., Yazaji S., Vogel R. F. (2011). Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 34, 148–155. 10.1016/j.syapm.2010.12.001 [DOI] [PubMed] [Google Scholar]
- American Academy of Pediatrics (2012). Breastfeeding and the use of human milk. Pediatrics 129, e827–e841. 10.1542/peds.2011-3552 [DOI] [PubMed] [Google Scholar]
- Angelopoulou A., Warda A. K., O’Connor P. M., Stockdale S. R., Shkoporov A. N., Field D., et al. (2020). Diverse bacteriocins produced by strains from the human milk microbiota. Front. Microbiol. 11, 788. 10.3389/fmicb.2020.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous (1889). Bacteria in milk and its products. Science 14, 116–118. 10.1126/science.ns-14.341.116 [DOI] [PubMed] [Google Scholar]
- Anonymous (1988). Breast-feeding/breast milk and human immunodeficiency virus (HIV). Bull. Pan. Am. Health Organ. 22, 87–88. [PubMed] [Google Scholar]
- Arboleya S., Ruas-Madiedo P., Margolles A., Solís G., Salminen S., de Los Reyes-Gavilán C. G., et al. (2011). Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 149, 28–36. 10.1016/j.ijfoodmicro.2010.10.036 [DOI] [PubMed] [Google Scholar]
- Arroyo R., Martin V., Maldonado A., Jiménez E., Fernández L., Rodríguez J. M. (2010). Treatment of infectious mastitis during lactation: antibiotics versus oral administration of lactobacilli isolated from breast milk. Clin. Infect. Dis. 50, 1551–1558. 10.1086/652763 [DOI] [PubMed] [Google Scholar]
- Asnicar F., Manara S., Zolfo M., Truong D. T., Scholz M., Armaninni F., et al. (2017). Studying vertical microbiome transmission from mothers to infants by strain-level metagenomics profiling. mSystems 2, e00164. 10.1128/mSystems.00164-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avershina E., Angell I. L., Simpson M., Storrø O., Øien T., Johnsen R., et al. (2018). Low maternal microbiota sharing across gut, breast milk and vagina, as revealed by 16S rRNA gene and reduced metagenomic sequencing. Genes 9, E231. 10.3390/genes9050231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azagra-Boronat I., Tres A., Massot-Cladera M., Franch À., Castell M., Guardiola F., et al. (2020). Lactobacillus fermentum CECT5716 supplementation in rats during pregnancy and lactation affects mammary milk composition. J. Dairy Sci. 103, 2982–2992. 10.3168/jds.2019-17384 [DOI] [PubMed] [Google Scholar]
- Bachmann K. D., Pulverer G. (1962). [Studies on the microorganism content of human milk. With a contribution to the problem of preservation by freeze drying]. Z. Kinderheilkd. 87, 52–59. 10.1007/BF00456354 [DOI] [PubMed] [Google Scholar]
- Bachour Y., Poort L., Verweij S. P., van Selms G., Winters H., Ritt M., et al. (2019). PCR characterization of microbiota on contracted and non-contracted breast capsules. Aesthetic Plast. Surg. 43, 918–926. 10.1007/s00266-019-01383-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Bang C., Schmitz R. A. (2015). Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol. Rev. 39, 631–648. 10.1093/femsre/fuv010 [DOI] [PubMed] [Google Scholar]
- Bardanzellu F., Fanos V., Reali A. (2019). Human breast milk-acquired cytomegalovirus infection: Certainties, Doubts and Perspectives. Curr. Pediatr. Rev. 15, 30–41. 10.2174/1573396315666181126105812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel A., Gourinat A. C., Cazorla C., Joubert C., Dupont-Rouzeyrol M., Descloux E. (2013). Breast milk as a possible route of vertical transmission of dengue virus? Clin. Infect. Dis. 57, 415–417. 10.1093/cid/cit227 [DOI] [PubMed] [Google Scholar]
- Bearfield C., Davenport E. S., Sivapathasundaram V., Allaker R. P. (2002). Possible association between amniotic fluid micro-organism infection and microflora in the mouth. Br. J. Obstet. Gynaecol. 109, 527–533. 10.1111/j.1471-0528.2002.01349.x [DOI] [PubMed] [Google Scholar]
- Beasley S. S., Saris P. E. J. (2004). Nisin-producing Lactococcus lactis strains isolated from human milk. Appl. Environ. Microbiol. 70, 5051–5053. 10.1128/AEM.70.8.5051-5053.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begier E. M., Barrett N. L., Mshar P. A., Johnson D. G., Hadler J. L., Connecticut Bioterrorism Field Epidemiology Response Team (2005). Gram-positive rod surveillance for early anthrax detection. Emerg. Infect. Dis. 11, 1483–1486. 10.3201/eid1109.041013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begović J., Jovčić B., Papić-Obradović M., Veljović K., Lukić J., Kojić M., et al. (2013). Genotypic diversity and virulent factors of Staphylococcus epidermidis isolated from human breast milk. Microbiol. Res. 168, 77–83. 10.1016/j.micres.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Bertotto A., Gerli R., Castellucci G., Scalise F., Vaccaro R. (1991). Human milk lymphocytes bearing the gamma/delta T-cell receptor are mostly delta TCS1-positive cells. Immunology 74, 360–361. [PMC free article] [PubMed] [Google Scholar]
- Biagi E., Quercia S., Aceti A., Beghetti I., Rampelli S., Turroni S., et al. (2017). The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front. Microbiol. 8, 1214. 10.3389/fmicb.2017.01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E., Aceti A., Quercia S., Beghetti I., Rampelli S., Turroni S., et al. (2018). Microbial community dynamics in mother’s milk and infant’s mouth and gut in moderately preterm infants. Front. Microbiol. 9, 2512. 10.3389/fmicb.2018.02512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkstén B., Burman L. G., De Château P., Fredrikzon B., Gothefors L., Hernell O., et al. (1980). Collecting and banking human milk: to heat or not to heat? Br. Med. J. 281, 765–769. 10.1136/bmj.281.6243.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blohm G. M., Lednicky J. A., Márquez M., White S. K., Loeb J. C., Pacheco C. A., et al. (2018). Evidence for mother-to-child transmission of Zika virus through breast milk. Clin. Infect. Dis. 66, 1120–1121. 10.1093/cid/cix968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L. (2020). Human milk oligosaccharides: structure and functions. Nestle Nutr. Inst. Workshop Ser. 94, 115–123. 10.1159/000505339 [DOI] [PubMed] [Google Scholar]
- Boer H. R., Anido G., Macdonald N. (1981). Bacterial colonization of human milk. South Med. J. 74, 716–718. 10.1097/00007611-198106000-00022 [DOI] [PubMed] [Google Scholar]
- Boix-Amorós A., Collado M. C., Mira A. (2016). Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 7, 492. 10.3389/fmicb.2016.00492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boix-Amorós A., Martinez-Costa C., Querol A., Collado M. C., Mira A. (2017). Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci. Rep. 7, 13016. 10.1038/s41598-017-13270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boix-Amorós A., Puente-Sánchez F., du Toit E., Linderborg K. M., Zhang Y., Yang B., et al. (2019). Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl. Environ. Microbiol. 85, e02994–e02918. 10.1128/AEM.02994-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boo N. Y., Nordiah A. J., Alfizah H., Nor–Rohaini A. H., Lim V. K. (2001). Contamination of breast milk obtained by manual expression and breast pumps in mothers of very low birth weight infants. J. Hosp. Infect. 49, 274–281. 10.1053/jhin.2001.1117 [DOI] [PubMed] [Google Scholar]
- Boyes S. M. (1987). AIDS virus in breast milk: a new threat to neonates and donor breast milk banks. Neonatal Netw. 5, 37–39. [PubMed] [Google Scholar]
- Brandtzaeg P. (2010). The mucosal immune system and its integration with the mammary glands. J. Pediatr. 156 (2 Suppl), S8–S15. 10.1016/j.jpeds.2009.11.014 [DOI] [PubMed] [Google Scholar]
- Brown S. L., Bright R. A., Dwyer D. E., Foxman B. (2005). Breast pump adverse events: reports to the Food and Drug Administration. J. Hum. Lact. 21, 169–174. 10.1177/0890334405275445 [DOI] [PubMed] [Google Scholar]
- Bryan D. L., Hart P. H., Forsyth K. D., Gibson R. A. (2007). Immunomodulatory constituents of human milk change in response to infant bronchiolitis. Pediatr. Allergy Immunol. 18, 495–502. 10.1111/j.1399-3038.2007.00565.x [DOI] [PubMed] [Google Scholar]
- Burbianka M., Dluzniewska I. (1971). [Enterotoxic staphylococci and enterotoxins in human milk]. Rocz. Panstw. Zakl. Hig. 22, 537–544. [PubMed] [Google Scholar]
- Cabrera-Rubio R., Collado M. C., Laitinen K., Salminen S., Isolauri E., Mira A. (2012). The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 96, 544–551. 10.3945/ajcn.112.037382 [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R., Mira-Pascual L., Mira A., Collado M. C. (2016). Impact of mode of delivery on the milk microbiota composition of healthy women. J. Dev. Orig. Health Dis. 7, 54–60. 10.1017/S2040174415001397 [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R., Kunz C., Rudloff S., García-Mantrana I., Crehuá-Gaudiza E., Martínez-Costa C., et al. (2019). Association of maternal secretor status and human milk oligosaccharides with milk microbiota: an observational pilot study. J. Pediatr. Gastroenterol. Nutr. 68, 256–263. 10.1097/MPG.0000000000002216 [DOI] [PubMed] [Google Scholar]
- Cacho N. T., Harrison N. A., Parker L. A., Padgett K. A., Lemas D. J., Marcial G. E., et al. (2017). Personalization of the microbiota of donor human milk with mother’s own milk. Front. Microbiol. 8, 1470. 10.3389/fmicb.2017.01470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas N., Martín V., Delgado S., Rodríguez J. M., Fernández L. (2014). Characterization of Lactobacillus gastricus strains isolated from human milk. Int. Dairy J. 39, 167–177. 10.1016/j.idairyj.2014.06.008 [DOI] [Google Scholar]
- Cárdenas N., Laiño J. E., Delgado S., Jiménez E., Juárez del Valle M., Savoy de Giori G., et al. (2015). Relationships between the genome and some phenotypical properties of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. Appl. Microbiol. Biotechnol. 99, 4343–4353. 10.1007/s00253-015-6429-0 [DOI] [PubMed] [Google Scholar]
- Cárdenas N., Arroyo R., Calzada J., Peirotén Á., Medina M., Rodríguez J. M., et al. (2016). Evaluation of technological properties of Enterococcus faecium CECT 8849, a strain isolated from human milk, for the dairy industry. Appl. Microbiol. Biotechnol. 100, 7665–7677. 10.1007/s00253-016-7616-3 [DOI] [PubMed] [Google Scholar]
- Cárdenas N., Martín V., Arroyo R., López M., Carrera M., Badiola C., et al. (2019). Prevention of recurrent acute otitis media in children through the use of Lactobacillus salivarius PS7, a target-specific probiotic strain. Nutrients 11, 376. 10.3390/nu11020376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L., Osman M., Davies D. P., McNeish A. S. (1979). Bacteriology of raw breast milk. Lancet 2, 1186. 10.1016/s0140-6736(79)92409-7 [DOI] [PubMed] [Google Scholar]
- Carroll L., Osman M., Davies D. P. (1980). Does discarding the first few millilitres of breast milk improve the bacteriological quality of bank breast milk? Arch. Dis. Child. 55, 898–899. 10.1136/adc.55.11.898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi M. S., Müller A. (1947). Microflora del calostro y de la superficie cutánea del seno materno en relación con la del tubo digestivo del recién nacido. El Día Médico 7 abril 1945, 473–475. [Google Scholar]
- Caufield P. W., Dasanayake A. P., Li Y., Pan Y., Hsu J., Hardin J. M. (2000). Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68, 4018–4023. 10.1128/IAI.68.7.4018-4023.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayla A., Perrot R., Bamberger J. (1955). [Etiological role of Staphylococcus aureus in erythroderma desquamativum in infants (Leiner-Moussous disease); the presence of Staphylococcus aureus in the breast milk]. Arch. Fr. Pediatr. 12, 410–413. [PubMed] [Google Scholar]
- Cephas K. D., Kim J., Mathai R. A., Barry K. A., Dowd S. E., Meline B. S., et al. (2011). Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PloS One 6, e23503. 10.1371/journal.pone.0023503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers C., Krogstad P., Bertrand K., Contreras D., Tobin N. H., Bode L., et al. (2020). Evaluation for SARS-CoV-2 in breast milk from 18 infected women. JAMA, 324, 1347–1348. 10.1001/jama.2020.15580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. A., Bashir M., Rivas M. N., Duvall K., Sieling P. A., Pieber T. R., et al. (2016). Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 6, 28061. 10.1038/srep28061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman S., McGraw B. R. (1983). Studies on cytomegalovirus in human milk. J. Infect. Dis. 148, 615–616. 10.1093/infdis/148.3.615a [DOI] [PubMed] [Google Scholar]
- Chen P.-W., Lin Y.-L., Huang M.-S. (2018). Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J. Food Drug Anal. 26, 1235–1244. 10.1016/j.jfda.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasskaia R. S., Dodonov V. N., Darbeeva O. S., Severov A. M., Khodoc V. G. (1980). [Microbiologic characteristics of postpartum lactation mastitis]. Zh. Mikrobiol. Epidemiol. Immunobiol. 3, 100–105. [PubMed] [Google Scholar]
- Collado M. C., Delgado S., Maldonado A., Rodríguez J. M. (2009). Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 48, 523–528. 10.1111/j.1472-765X.2009.02567.x [DOI] [PubMed] [Google Scholar]
- Contreras G. A., Rodríguez J. M. (2011). Mastitis: comparative etiology and epidemiology. J. Mammary Gland Biol. Neoplasia 16, 339–356. 10.1007/s10911-011-9234-0 [DOI] [PubMed] [Google Scholar]
- Cook J., Holmes C. J., Wixtrom R., Newman M. I., Pozner J. (2020). Characterizing the microbiome of the contracted breast capsule using next generation sequencing. Aesthet. Surg. J. sjaa097. Epub ahead of print. 10.1093/asj/sjaa097 [DOI] [PubMed] [Google Scholar]
- Cooke R. P., Devlin J., Robinson M. J. (1987). Breast milk and methicillin-resistant Staphylococcus aureus . J. Hosp. Infect. 10, 312. 10.1016/0195-6701(87)90016-8 [DOI] [PubMed] [Google Scholar]
- Dancey A., Nassimizadeh A., Levick P. (2012). Capsular contracture - what are the risk factors? A 14 year series of 1400 consecutive augmentations. J. Plast. Reconstr. Aesthetic Surg. 65, 213–218. 10.1016/j.bjps.2011.09.011 [DOI] [PubMed] [Google Scholar]
- Dasanayake A. P., Li Y., Wiener H., Ruby J. D., Lee M. J. (2005). Salivary Actinomyces naeslundii genospecies 2 and Lactobacillus casei levels predict pregnancy outcomes. J. Periodontol. 76, 171–177. 10.1902/jop.2005.76.2.171 [DOI] [PubMed] [Google Scholar]
- Davé V., Street K., Francis S., Bradman A., Riley L., Eskenazi B., et al. (2016). Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr. Res. 79, 846–854. 10.1038/pr.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D. C., Poll R. A., Roberts C. (1979). Bacteriological monitoring of unheated human milk. Arch. Dis. Child. 54, 760–764. 10.1136/adc.54.10.760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrés J., Jiménez E., Chico-Calero I., Fresno M., Fernández L., Rodríguez J. M. (2017). Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients 10, E14. 10.3390/nu10010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrés J., Jiménez E., Espinosa-Martos I., Rodríguez J. M., García-Conesa M. T. (2018). An exploratory search for potential molecular targets responsive to the probiotic Lactobacillus salivarius PS2 in women with mastitis: gene expression profiling vs. interindividual variability. Front. Microbiol. 9, 2166. 10.3389/fmicb.2018.02166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S., Arroyo R., Martín R., Rodríguez J. M. (2008). PCR-DGGE assessment of the bacterial diversity of breast milk in women with lactational infectious mastitis. BMC Infect. Dis. 8, 51. 10.1186/1471-2334-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S., Arroyo R., Jiménez E., Marín M. L., del Campo R., Fernández L., et al. (2009). Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol. 9, 82. 10.1186/1471-2180-9-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S., García P., Fernández L., Jiménez E., Rodríguez-Baños M., del Campo R., et al. (2011). Characterizationn of Staphylococcus aureus strains involved in human and bovine mastitis. FEMS Immunol. Med. Microbiol. 62, 225–235. 10.1111/j.1574-695X.2011.00806.x [DOI] [PubMed] [Google Scholar]
- Derakhshani H., Fehr K. B., Sepehri S., Francoz D., De Buck J., Barkema H. W., et al. (2018). Invited review: Microbiota of the bovine udder: Contributing factors and potential implications for udder health and mastitis susceptibility. J. Dairy Sci. 101, 10605–10625. 10.3168/jds.2018-14860 [DOI] [PubMed] [Google Scholar]
- Díaz-Ropero M. P., Martín R., Sierra S., Lara-Villoslada F., Rodríguez J. M., Xaus J., et al. (2006). Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J. Appl. Microbiol. 102, 337–343. 10.1111/j.1365-2672.2006.03102.x [DOI] [PubMed] [Google Scholar]
- Ding M., Qi C., Yang Z., Jiang S., Bi Y., Lai J., et al. (2019). Geographical location specific composition of cultured microbiota and Lactobacillus occurrence in human breast milk in China. Food Funct. 10 (2), 554–564. 10.1039/C8FO02182A [DOI] [PubMed] [Google Scholar]
- Dluzniewska I. (1966). [Pathogenic staphylococci in milk]. Ginekol. Pol. 37, 745–751. [PubMed] [Google Scholar]
- Dobke M. K., Svahn J. K., Vastine V. L., Landon B. N., Stein P. C., Parsons C. L. (1995). Characterization of microbial presence at the surface of silicone mammary implants. Ann. Plast. Surg. 34, 563–569. 10.1097/00000637-199506000-00001 [DOI] [PubMed] [Google Scholar]
- Donowitz L. G., Marsik F. J., Fisher K. A., Wenzel R. P. (1981). Contaminated breast milk: A source of Klebsiella bacteremia in a newborn intensive care unit. Rev. Infect. Dis. 3, 716–720. 10.1093/clinids/3.4.716 [DOI] [PubMed] [Google Scholar]
- Dorr H., Sittel I. (1953). [Bacteriological examination of human milk and its relation to mastitis]. Zentralbl. Gynakol. 75, 1833–1835. [PubMed] [Google Scholar]
- Drago L., Toscano M., De Grandi R., Grossi E., Padovani E. M., Peroni D. G. (2017). Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 11, 875–884. 10.1038/ismej.2016.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B., Henry M., El Khéchine A., Raoult D., Drancourt M. (2009). High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PloS One 4, e7063. 10.1371/journal.pone.0007063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M. (1954). Role of contamination by breast milk in staphylococcus infection of newborn]. Rev. Prat. 4, 493–495. [PubMed] [Google Scholar]
- Duranti S., Lugli G. A., Mancabelli L., Armanini F., Turroni F., James K., et al. (2017). Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5, 66. 10.1186/s40168-017-0282-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworsky M., Yow M., Stagno S., Pass R. F., Alford C. (1983). Cytomegalovirus infection of breast milk and transmission in infancy. Pediatrics 72, 295–299. [PubMed] [Google Scholar]
- Eidelman A., II, Szilagyi G. (1979). Patterns of bacterial colonization of human milk. Obstet. Gynecol. 53, 550–552. [PubMed] [Google Scholar]
- Emerson J. B., Adams R., II, Román C., Brooks B., Coil D. A., Dahlhausen K., et al. (2017). Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5, 86. 10.1186/s40168-017-0285-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Martos I., Jiménez E., de Andrés J., Rodríguez-Alcala L. M., Tavarez S., Manzano S., et al. (2016). Milk and blood biomarkers associated to the clinical efficacy of a probiotic for the treatment of infectious mastitis. Benef. Microbes 7, 305–318. 10.3920/BM2015.0134 [DOI] [PubMed] [Google Scholar]
- Favier C. F., Vaughan E. E., de Vos W. M., Akkermans A. D. L. (2002). Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68, 219–226. 10.1128/AEM.68.1.219-226.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L., Rodríguez J. M. (2020). Human milk microbiota: origin and potential uses. Nestle Nutr. Inst. Workshop Ser. 94, 75–85. 10.1159/000505031 [DOI] [PubMed] [Google Scholar]
- Fernández L., Langa S., Martin V., Maldonado A., Jiménez E., Martin R., et al. (2013). The Human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 69, 1–10. 10.1016/j.phrs.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Fernández L., Arroyo R., Espinosa I., Marín M., Jiménez E., Rodríguez J. M. (2014). Probiotics for human lactational mastitis. Benef. Microbes 5, 169–183. 10.3920/BM2013.0036 [DOI] [PubMed] [Google Scholar]
- Fernández L., Cardenas N., Arroyo R., Manzano S., Jiménez E., Martin V., et al. (2016). Prevention of infectious mastitis by oral administration of Lactobacillus salivarius PS2 during late pregnancy. Clin. Infect. Dis. 62, 568–573. 10.1093/cid/civ974 [DOI] [PubMed] [Google Scholar]
- Fernández L., Ruiz L., Jara J., Orgaz B., Rodríguez J. M. (2018). Strategies for the preservation, restoration and modulation of the human milk microbiota. Implications for human milk banks and neonatal intensive care units. Front. Microbiol. 9, 2676. 10.3389/fmicb.2018.02676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischrocker G., Vutuc C., Werner H. P. (1972). [Infection of a newborn infant by breast milk containing Salmonella typhimurium]. Wien Klin. Wochenschr. 84, 394–395. [PubMed] [Google Scholar]
- Foster W. D., Harris R. E. (1960). The incidence of Staphylococcus pyogenes in normal human breast milk. J. Obstet. Gynaecol. Br. Emp. 67, 463–464. 10.1111/j.1471-0528.1960.tb07027.x [DOI] [PubMed] [Google Scholar]
- French R. (1903). Epidemic sore throat and suppurative mammitis in cows. Br. Med. J. 2, 1492–1493. 10.1136/bmj.2.2240.1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis H., Andersen H. K. (1982). Rate of inactivation of cytomegalovirus in raw banked milk during storage at -20 degrees C and pasteurisation. Br. Med. J. (Clin. Res. Ed.) 285, 1604–1605. 10.1136/bmj.285.6355.1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M., Larocca F., Iovene M. R., Martora F., Pieretti G., D’Oriano V., et al. (2018). Microbial evaluation in capsular contracture of breast implants. Plast. Reconstr. Surg. 141 (1), 23–30. 10.1097/PRS.0000000000003915 [DOI] [PubMed] [Google Scholar]
- García-Gutierrez E., O’Connor P. M., Colquhoun I. J., Vior N. M., Rodríguez J. M., Mayer M. J., et al. (2020). Production of multiple bacteriocins, including the novel bacteriocin gassericin M, by Lactobacillus gasseri LM19, a strain isolated from human milk. Appl. Microbiol. Biotechnol. 104, 3869–3884. 10.1007/s00253-020-10493-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay M., Koleva P. T., Slupsky C. M., Toit E. D., Eggesbo M., Johnson C. C., et al. (2018). Worldwide variation in human milk metabolome: indicators of breast physiology and maternal lifestyle? Nutrients 10, 1151. 10.3390/nu10091151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Campos M., López M. A., Rodríguez-Benítez M. V., Romero J., Roncero I., Linares M. D., et al. (2012). Lactobacillus fermentum CECT 5716 is safe and well tolerated in infants of 1-6 months of age: a randomized controlled trial. Pharmacol. Res. 65, 231–238. 10.1016/j.phrs.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Goedert J. J., Jones G., Hua X., Xu X., Yu G., Flores R., et al. (2015). Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J. Natl. Cancer Inst. 107, djv147. 10.1093/jnci/djv147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert J. J., Hua X., Bielecka A., Okayasu I., Milne G. L., Jones G. S., et al. (2018). Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br. J. Cancer. 118, 471–479. 10.1038/bjc.2017.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gallego C., Garcia-Mantrana I., Salminen S., Collado M. C. (2016). The human milk microbiome and factors influencing its composition and activity. Semin. Fetal Neonatal Med. 21, 400–405. 10.1016/j.siny.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Gómez-Gallego C., Morales J., Monleón D., du Toit E., Kumar H., Linderborg K., et al. (2018). Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients 10, 1355. 10.3390/nu10101355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. K., Waters J. L., Poole A. C., Sutter J. L., Koren O., Blekhman R., et al. (2014). Human genetics shape the gut microbiome. Cell 159, 789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn N., Olofsson M., Ellnebo-Svedlund K., Monstein H. J., Jonasson J. (2003). Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol. Lett. 219, 87–91. 10.1016/S0378-1097(02)01190-4 [DOI] [PubMed] [Google Scholar]
- Gransden W. R., Webster M., French G. L., Phillips I. (1986). An outbreak of Serratia marcescens transmitted by contaminated breast pumps in a special care baby unit. J. Hosp. Infect. 7, 149–154. 10.1016/0195-6701(86)90057-5 [DOI] [PubMed] [Google Scholar]
- Grine G., Boualam M. A., Drancourt M. (2017). Methanobrevibacter smithii, a methanogen consistently colonising the newborn stomach. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2449–2455. 10.1007/s10096-017-3084-7 [DOI] [PubMed] [Google Scholar]
- Grönlund M. M., Gueimonde M., Laitinen K., Kociubinski G., Grönroos T., Salminen S., et al. (2007). Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin. Exp. Allergy 37, 1764–1772. 10.1111/j.1365-2222.2007.02849.x [DOI] [PubMed] [Google Scholar]
- Gueimonde M., Laitinen K., Salminen S., Isolauri E. (2007). Breast milk: A source of bifidobacteria for infant gut development and maturation? Neonatology 92, 64–66. 10.1159/000100088 [DOI] [PubMed] [Google Scholar]
- Gueimonde M., Bottacini F., van Sinderen D., Ventura M., Margolles A., Sánchez B. (2012). Genome sequence of Parascardovia denticolens IPLA 20019, isolated from human breast milk. J. Bacteriol. 194, 4776–4777. 10.1128/JB.01035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiden N., Ziegler E. E. (2016). Human Milk Banking. Ann. Nutr. Metab. 69 (2), 8–15. 10.1159/000452821 [DOI] [PubMed] [Google Scholar]
- Hall C. W., Trout G. M. (1968). Milk pasteurization (Westport, Connecticut: AVI Publishing Co.). [Google Scholar]
- Hassiotou F., Geddes D. T. (2015). Immune cell-mediated protection of the mammary gland and the infant during breastfeeding. Adv. Nutr. 6, 267–275. 10.3945/an.114.007377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassiotou F., Hepworth A. R., Metzger P., Lai C.-T., Trengove N., Hartmann P. E., et al. (2013). Maternal and infant infections stimulate a rapid leukocyte response in breastmilk. Clin. Transl. Immunol. 2, e3. 10.1038/cti.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes K., Danks D. M., Gibas H., Jack I. (1972). Cytomegalovirus in human milk. N. Engl. J. Med. 287, 177–178. 10.1056/NEJM197207272870407 [DOI] [PubMed] [Google Scholar]
- Heikkilä M. P., Saris P. E. J. (2003). Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 95, 471–478. 10.1046/j.1365-2672.2003.02002.x [DOI] [PubMed] [Google Scholar]
- Heisel T., Nyaribo L., Sadowsky M. J., Gale C. A. (2019). Breastmilk and NICU surfaces are potential sources of fungi for infant mycobiomes. Fungal Gen. Biol. 128, 29–35. 10.1016/j.fgb.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson H., Kumar H., Collado M. C., Salminen S., Isolauri E., Rautava S. (2019). Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front. Nutr. 6, 4. 10.3389/fnut.2019.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieken T. J., Chen J., Hoskin T. L., Walther-Antonio M., Johnson S., Ramaker S., et al. (2016). The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci. Rep. 6, 30751. 10.1038/srep30751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N. T., Li F., Lee-Sarwar K. A., Tun H. M., Brown B. P., Pannaraj P. S., et al. (2018). Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Comm 9, 4169. 10.1038/s41467-018-06473-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoashi M., Meche L., Mahal L. K., Bakacs E., Nardella D., Naftolin F., et al. (2016). Human milk bacterial and glycosylation patterns differ by delivery mode. Reprod. Sci. 23, 902–907. 10.1177/1933719115623645 [DOI] [PubMed] [Google Scholar]
- Holsinger V. H., Rajkowski K. T., Stabel J. R. (1997). Milk pasteurisation and safety: a brief history and update. Rev. Sci. Tech. 16, 441–451. 10.20506/rst.16.2.1037 [DOI] [PubMed] [Google Scholar]
- Hornung B. V. H., Zwittink R. D., Kuijper E. J. (2019). Issues and current standards of controls in microbiome research. FEMS Microbiol. Ecol. 95, pii: fiz045. 10.1093/femsec/fiz045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jacombs A., Vickery K., Merten S. L., Pennington D. G., Deva A. K. (2015). Chronic biofilm infection in breast implants is associated with an increased T-cell lymphocytic infiltrate: implications for breast implant-associated lymphoma. Plast. Reconstr. Surg. 135, 319–329. 10.1097/PRS.0000000000000886 [DOI] [PubMed] [Google Scholar]
- Hu H., Johani K., Almatroudi A., Vickery K., Van Natta B., Kadin M. E., et al. (2016). Bacterial biofilm infection detected in breast implant-associated anaplastic large-cell lymphoma. Plast. Reconstr. Surg. 137, 1659–1669. 10.1097/PRS.0000000000002010 [DOI] [PubMed] [Google Scholar]
- Hunt K. M., Foster J. A., Forney L. J., Schütte U. M. E., Beck D. L., Abdo Z., et al. (2011). Characterization of the diversity and temporal stability of bacterial communities in human milk. PloS One 6, e21313. 10.1371/journal.pone.0021313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt K. M., Preuss J., Nissan C., Davlin C. A., Williams J. E., Shafii B., et al. (2012). Human milk oligosaccharides promote the growth of staphylococci. Appl. Environ. Microbiol. 78, 4763–4770. 10.1128/AEM.00477-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado J. A., Maldonado-Lobón J. A., Díaz-Ropero M. P., Flores-Rojas K., Uberos J., Leante J. L., et al. (2017). Oral administration to nursing women of Lactobacillus fermentum CECT5716 Prevents lactational mastitis development: a randomized controlled trial. Breastfeed. Med. 12, 202–209. 10.1089/bfm.2016.0173 [DOI] [PubMed] [Google Scholar]
- Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., et al. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- Jeurink P. V., van Bergenhenegouwen J., Jimenez E., Knippels L. M., Fernandez L., Garssen J., et al. (2013). Human milk: a source of more life than we imagine. Benef. Microbes 4, 17–30. 10.3920/BM2012.0040 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Delgado S., Maldonado A., Arroyo R., Albujar M., García N., et al. (2008. a). Staphylococcus epidermidis: A differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. 8, 143. 10.1186/1471-2180-8-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Delgado S., Fernández L., García N., Albújar M., Gómez A., et al. (2008. b). Assessment of the bacterial diversity of human colostrum and screening of staphylococcal and enterococcal populations for potential virulence factors. Res. Microbiol. 159, 595–601. 10.1016/j.resmic.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Fernández L., Maldonado A., Martin R., Olivares M., Xaus J., et al. (2008. c). Oral administration of lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl. Environ. Microbiol. 74, 4650–4655. 10.1128/AEM.02599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Langa S., Martín V., Arroyo R., Martín R., Fernández L., et al. (2010. a). Complete genome sequence of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. J. Bacteriol. 192, 4800. 10.1128/JB.00702-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Martín R., Maldonado A., Martín V., Gomez de Segura A., Fernández L., et al. (2010. b). Complete genome sequence of Lactobacillus salivarius CECT5713, a probiotic strain isolated from human milk and infant feces. J. Bacteriol. 192, 5266–5267. 10.1128/JB.00703-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Villar-Tajadura M. A., Marín M., Fontecha J., Requena T., Arroyo R., et al. (2012). Complete genome sequence of Bifidobacterium breve CECT 7263, a strain isolated from human milk. J. Bacteriol. 194, 3762–3763. 10.1128/JB.00691-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., de Andrés J., Manrique M., Pareja-Tobes P., Tobes R., Martínez-Blanch J. F., et al. (2015). Metagenomic Analysis of milk of healthy and mastitis-suffering women. J. Hum. Lact. 31, 406–415. 10.1177/0890334415585078 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Arroyo R., Cárdenas N., Marín M., Serrano P., Fernández L., et al. (2017). Mammary candidiasis, a medical condition without scientific evidence? PloS One 12, e0181071. 10.1371/journal.pone.0181071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. L., Jennison R. F., D’Souza S. W. (1979). Bacterial contamination of expressed breast milk. Br. Med. J. 2, 1320–1322. 10.1136/bmj.2.6201.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. S. (1918). Studies in bovine mastitis: II. The relation of hemolytic streptococci to udder infections. J. Exp. Med. 28, 253–267. 10.1084/jem.28.3.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C., Chassard C. (2013). Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 110, 1253–1262. 10.1017/S0007114513000597 [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C. P., Rochat F., Chassard C. (2014). Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16, 2891–2904. 10.1111/1462-2920.12238 [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C., Chassard C. (2015). Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr. Rev. 73, 426–437. 10.1093/nutrit/nuu016 [DOI] [PubMed] [Google Scholar]
- Karstens L., Asquith M., Davin S., Fair D., Gregory W. T., Wolfe A. J., et al. (2019). Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems 4, e00290–e00219. 10.1128/mSystems.00290-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayıran P. G., Can F., Kayıran S. M., Ergonul O., Gürakan B. (2014). Transmission of methicillin-sensitive Staphylococcus aureus to a preterm infant through breast milk. J. Matern. Fetal Neonatal Med. 27, 527–529. 10.3109/14767058.2013.819332 [DOI] [PubMed] [Google Scholar]
- Kenny J. F. (1977). Recurrent group B streptococcal disease in an infant associated with the ingestion of infected mother’s milk. J. Pediatr. 91, 158–159. 10.1016/S0022-3476(77)80473-3 [DOI] [PubMed] [Google Scholar]
- Khodayar-Pardo P., Mira-Pascual L., Collado M. C., Martínez-Costa C. (2014). Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 34, 599–605. 10.1038/jp.2014.47 [DOI] [PubMed] [Google Scholar]
- Kirjavainen P. V., Apostolou E., Arvola T., Salminen S. J., Gibson G. R., Isolauri E. (2001). Characterizing the composition of intestinal microflora as a prospective treatment target in infant allergic disease. FEMS Immunol. Med. Microbiol. 32, 1–7. 10.1111/j.1574-695X.2001.tb00526.x [DOI] [PubMed] [Google Scholar]
- Klann E., Williamson J. M., Tagliamonte M. S., Ukhanova M., Asirvatham J. R., Chim H., et al. (2020). Microbiota composition in bilateral healthy breast tissue and breast tumors. Cancer Causes Control. 31, 1027–1038. 10.1007/s10552-020-01338-5 [DOI] [PubMed] [Google Scholar]
- Knoop U., Matheis G., Schütt-Gerowitt H. (1985). [Bacterial contamination of pump-collected breast milk]. Monatsschr Kinderheilkd. 133, 537–541. [PubMed] [Google Scholar]
- Kordy K., Gaufin T., Mwangi M., Li F., Cerini C., Lee D. J., et al. (2020). Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve . PloS One 15, e0219633. 10.1371/journal.pone.0219633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis A. P., Butera S., Ibegbu C., Belec L., Duerr A. (2003). Breast milk and HIV-1: vector of transmission or vehicle of protection? Lancet Infect. Dis. 3, 786–793. 10.1016/s1473-3099(03)00832-6 [DOI] [PubMed] [Google Scholar]
- Kovács T., Mikó E., Ujlaki G., Sári Z., Bai P. (2020). The microbiome as a component of the tumor microenvironment. Adv. Exp. Med. Biol. 1225, 137–153. 10.1007/978-3-030-35727-6_10 [DOI] [PubMed] [Google Scholar]
- Kozak K., Charbonneau D., Sanozky-Dawes R., Klaenhammer T. (2015). Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 6, 341. 10.1080/19490976.2015.1103425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg H. (1966). The bacterial content of raw and freeze-dried women’s milk. Med. Klin. 61, 953–956. [PubMed] [Google Scholar]
- Kuehn J., Kuhn L., Kim H. Y., Walter J., Thea D. M., Sinkala M., et al. (2013). HIV-1 concentrations in human breast milk before and after weaning. Sci. Trans. Med. 5, 181ra51. 10.1126/scitranslmed.3005113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., du Toit E., Kulkarni A., Aakko J., Linderborg K. M., Zhang Y., et al. (2016). Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 7, 1619. 10.3389/fmicb.2016.01619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborda-Illanes A., Sanchez-Alcoholado L., Dominguez-Recio M. E., Jimenez-Rodriguez B., Lavado R., Comino-Méndez I., et al. (2020). Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers 12, E2465. 10.3390/cancers12092465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey K. A., Williams J. E., Meehan C. L., Zachek J. A., Benda E. D., Price W. J., et al. (2019). What’s Normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: The INSPIRE study. Front. Nutr. 6, 45. 10.3389/fnut.2019.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J. C., Million M., Hugon P., Armougom F., Raoult D. (2012). Human gut microbiota: repertoire and variations. Front. Cell Infect. Microbiol. 2, 136. 10.3389/fcimb.2012.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J. C., Hugon P., Khelaifia S., Fournier P. E., La Scola B., Raoult D. (2015). The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 28, 237–264. 10.1128/CMR.00014-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J. C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 1, 540–550. 10.1038/s41579-018-0041-0 [DOI] [PubMed] [Google Scholar]
- Langa S., Maldonado-Barragán A., Delgado S., Martín R., Martín V., Jiménez E., et al. (2012). Characterization of Lactobacillus salivarius CECT 5713, a strain isolated from human milk: from genotype to phenotype. Appl. Microbiol. Biotechnol. 94, 1279–1287. 10.1007/s00253-012-4032-1 [DOI] [PubMed] [Google Scholar]
- Langa S. (2006). Interactions between lactic acid bacteria, intestinal epithelial cells and immune cells. Development of in vitro models. PhD Thesis (Madrid: Complutense University of Madrid; ). [Google Scholar]
- Lara-Villoslada F., Sierra S., Díaz-Ropero M. P., Olivares M., Xaus J. (2007). Safety assessment of the human milk-isolated probiotic Lactobacillus salivarius CECT5713. J. Dairy Sci. 90, 3583–3589. 10.3168/jds.2006-685 [DOI] [PubMed] [Google Scholar]
- Lara-Villoslada F., Sierra S., Díaz-Ropero M. P., Rodríguez J. M., Xaus J., Olivares M. (2009). Safety assessment of Lactobacillus fermentum CECT5716, a probiotic strain isolated from human milk. J. Dairy Res. 76, 216–221. 10.1017/S0022029909004014 [DOI] [PubMed] [Google Scholar]
- Lau J. T., Whelan F. J., Herath I., Lee C. H., Collins S. M., Bercik P., et al. (2016). Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 8, 72. 10.1186/s13073-016-0327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder A. P., Roche A. M., Sherrill-Mix S., Bailey A., Laughlin A. L., Bittinger K., et al. (2016). Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4, 29. 10.1186/s40168-016-0172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence M., Hatzis C., Brash D. E. (2014). Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PloS One 9, e97876. 10.1371/journal.pone.0097876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Lemas D. J., Young B. E., Baker P. R., Tomczik A. C., Soderborg T. K., Hernandez T. L., et al. (2016). Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am. J. Clin. Nutr. 103, 1291–1300. 10.3945/ajcn.115.126375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMay-Nedjelski L., Copeland J., Wang P. W., Butcher J., Unger S., Stintzi A., et al. (2018). Methods and strategies to examine the human breastmilk microbiome. Methods Mol. Biol. 1849, 63–86. 10.1007/978-1-4939-8728-3_5 [DOI] [PubMed] [Google Scholar]
- Lemoine L. (1987). Possible transmission of methicillin-resistant Staphylococcus aureus by expressed human breast milk. J. Hosp. Infect. 9, 93–94. 10.1016/0195-6701(87)90105-8 [DOI] [PubMed] [Google Scholar]
- Lewis Z. T., Totten S. M., Smilowitz J. T., Popovic M., Parker E., Lemay D. G., et al. (2015). Maternal fucosyl transferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3, 13. 10.1186/s40168-015-0071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Teague E., Zhuang Z., Caufield P. W. (1997). Screening for the spaP gene of Streptococcus mutans in predentate infants. J. Dental Res. 76, 102. [Google Scholar]
- Li S.-W., Watanabe K., Hsu C.-C., Chao S.-H., Yang Z.-H., Lin Y.-J., et al. (2017). Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and mainland China. Front. Microbiol. 8, 965. 10.3389/fmicb.2017.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhaber M., Lewiston N. J., Asquith M. T., Sunshine P. (1978). Comparison of bacterial contamination with two methods of human milk collection. J. Pediatr. 92, 236–237. 10.1016/S0022-3476(78)80014-6 [DOI] [PubMed] [Google Scholar]
- Lindemann G., Rupp H. (1954). [Bacteriological control test in human milk collection center]. Zentralbl. Gynakol. 76, 1167–1176. [PubMed] [Google Scholar]
- Lindemann G. (1955). [Possibilities of improving the bacteriologic quality of human milk]. Zentralbl. Gynakol. 77, 1383–1389. [PubMed] [Google Scholar]
- Lindemann G. (1958). [Studies on samples from human milk banks]. Monatsschr. Kinderheilkd. 106, 52–55. [PubMed] [Google Scholar]
- Liu G., Pang B., Li N., Jin H., Li J., Wu W., et al. (2020). Therapeutic effect of Lactobacillus rhamnosus SHA113 on intestinal infection by multi-drug-resistant Staphylococcus aureus and its underlying mechanisms. Food Funct. 11, 6226–6239. 10.1039/d0fo00969e [DOI] [PubMed] [Google Scholar]
- Louca S., Doebeli M., Parfrey L. W. (2018). Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6, 41. 10.1186/s40168-018-0420-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A., Roberts C. D. (1978). Group B streptococci in pooled human milk. Br. Med. J. 1 (6117), 919–920. 10.1136/bmj.1.6117.919-b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A., Roberts C. D. (1979). Bacteriological quality control in human milk-banking. Br. Med. J. 1 (6156), 80–82. 10.1136/bmj.1.6156.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G. A., Milani C., Turroni F., Tremblay D., Ferrario C., Mancabelli L., et al. (2016). Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ. Microbial. 18, 2196–2213. 10.1111/1462-2920.13154 [DOI] [PubMed] [Google Scholar]
- Lugli G. A., Duranti S., Milani C., Mancabelli L., Turroni F., Alessandri G., et al. (2020). Investigating bifidobacteria and human milk oligosaccharide composition of lactating mothers. FEMS Microbiol. Ecol. 96, fiaa049. 10.1093/femsec/fiaa049 [DOI] [PubMed] [Google Scholar]
- Ma Z. S., Guan Q., Ye C., Zhang C., Foster J. A., Forney L. J. (2015). Network analysis suggests a potentially ‘evil’ alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci. Rep. 5, 8275. 10.1038/srep08275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z. S., Li L., Li W., Li J., Chen H. (2016). Integrated network-diversity analyses suggest suppressive effect of Hodgkin’s lymphoma and slightly relieving effect of chemotherapy on human milk microbiome. Sci. Rep. 6, 28048. 10.1038/srep28048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfadyen A. (1891). Observations on a Mastitis Bacillus . J. Anat. Physiol. 25, 571–577. [PMC free article] [PubMed] [Google Scholar]
- Macpherson A. J., Uhr T. (2004). Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665. 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- Makino H., Kushiro A., Ishikawa E., Muylaert D., Kubota H., Sakai T., et al. (2011). Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 77, 6788–6793. 10.1128/AEM.05346-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H., Martin R., Ishikawa E., Gawad A., Kubota H., Sakai T., et al. (2015). Multilocus sequence typing of bifidobacterial strains from infant’s faeces and human milk: are bifidobacteria being sustainably shared during breastfeeding? Benef. Microbes 6, 563–572. 10.3920/BM2014.0082 [DOI] [PubMed] [Google Scholar]
- Maldonado J., Lara-Villoslada F., Sierra S., Sempere L., Gómez M., Rodríguez J. M., et al. (2010). Safety and tolerance of the human milk probiotic strain Lactobacillus salivarius CECT5713 in 6-month-old children. Nutrition 26, 1082–1087. 10.1016/j.nut.2009.08.023 [DOI] [PubMed] [Google Scholar]
- Maldonado J., Cañabate F., Sempere L., Vela F., Sánchez A. R., Narbona E., et al. (2012). Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J. Pediatr. Gastroenterol. Nutr. 54, 55–61. 10.1097/MPG.0b013e3182333f18 [DOI] [PubMed] [Google Scholar]
- Maldonado-Lobón J. A., Gil-Campos M., Maldonado J., López-Huertas E., Flores-Rojas K., Valero A. D., et al. (2015. a). Long-term safety of early consumption of Lactobacillus fermentum CECT5716: A 3-year follow-up of a randomized controlled trial. Pharmacol. Res., 95–96, 12-9. 10.1016/j.phrs.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Maldonado-Lobón J. A., Díaz-López M. A., Carputo R., Duarte P., Díaz-Ropero M. P., Valero A. D., et al. (2015. b). Lactobacillus fermentum CECT 5716 reduces Staphylococcus load in the breastmilk of lactating mothers suffering breast pain: a randomized controlled trial. Breastfeed. Med. 10, 425–432. 10.1089/bfm.2015.0070 [DOI] [PubMed] [Google Scholar]
- Marín M. L., Arroyo R., Jiménez E., Gómez A., Fernández L., Rodríguez J. M. (2009). Cold storage of human milk, effect on its bacterial composition. J. Ped. Gastroenterol. Nutr. 49, 343–348. 10.1097/MPG.0b013e31818cf53d [DOI] [PubMed] [Google Scholar]
- Marín M., Arroyo R., Espinosa-Martos I., Fernández L., Rodríguez J. M. (2017). Identification of emerging human mastitis pathogens by MALDI-TOF and assessment of their antibiotic resistance patterns. Front. Microbiol. 8, 1258. 10.3389/fmicb.2017.01258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Langa S., Reviriego C., Jiminez E., Marin M. L., Xaus J., et al. (2003). Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 143, 754–758. 10.1016/j.jpeds.2003.09.028 [DOI] [PubMed] [Google Scholar]
- Martín R., Langa S., Reviriego C., Jiménez E., Marín M., Olivares M., et al. (2004). The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. 15, 121–127. 10.1016/j.tifs.2003.09.010 [DOI] [Google Scholar]
- Martín R., Olivares M., Marín M. L., Fernández L., Xaus J., Rodríguez J. M. (2005). Probiotic potential of 3 lactobacilli strains isolated from breast milk. J. Hum. Lact. 21, 8–17. 10.1177/0890334404272393 [DOI] [PubMed] [Google Scholar]
- Martín R., Jiménez E., Olivares M., Marín M. L., Fernández L., Xaus J., et al. (2006). Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 112, 35–43. 10.1016/j.ijfoodmicro.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Martín R., Heilig G. H. J., Zoetendal E. G., Jiménez E., Fernández L., Smidt H., et al. (2007. a). Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol; 158, 31–37. 10.1016/j.resmic.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Martín R., Heilig G. H. J., Zoetendal E. G., Smidt H., Rodríguez J. M. (2007. b). Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J. Appl. Microbiol. 103, 2638–2644. 10.1111/j.1365-2672.2007.03497.x [DOI] [PubMed] [Google Scholar]
- Martín R., Jiménez E., Heilig H., Fernandez L., Marin M. L., Zoetendal E. G., et al. (2009). Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-DGGE and qRTi-PCR. Appl. Environ. Microbiol. 75, 965–969. 10.1128/AEM.02063-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Maldonado A., Fernández L., Rodríguez J. M., Connor R. I. (2010). Inhibition of human immunodeficiency virus type 1 by lactic acid bacteria from human breast milk. Breastfeed. Med. 5, 153–158. 10.1089/bfm.2010.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Mañés-Lázaro R., Rodríguez J. M., Maldonado A. (2011). Streptococcus lactarius sp. nov., isolated from breast milk of healthy women. Int. J. Syst. Evol. Microbiol. 61, 1048–1052. 10.1099/ijs.0.021642-0 [DOI] [PubMed] [Google Scholar]
- Martín V., Maldonado A., Jiménez E., Ruas-Madiedo P., Fernández L., Rodríguez J. M. (2012. a). Complete genome sequence of Streptococcus salivarius PS4, a strain isolated from human milk. J. Bacteriol. 194, 4466–4467. 10.1128/JB.00896-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Maldonado A., Moles L., Rodríguez-Baños M., Del Campo R., Fernández L., et al. (2012. b). Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 28, 36–44. 10.1177/0890334411424729 [DOI] [PubMed] [Google Scholar]
- Martín V., Cárdenas N., Jiménez E., Maldonado A., Rodríguez J. M., Fernández L. (2013). Genome sequence of Lactobacillus gastricus PS3, a strain isolated from human milk. Genome Announc. 1, e00489–e00413. 10.1128/genomeA.00489-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Mediano P., Del Campo R., Rodríguez J. M., Marín M. (2015). Streptococcal diversity of human milk and comparison of different methods for the taxonomic identification of streptococci. J. Hum. Lact. 32, NP84–NP94. 10.1177/0890334415597901 [DOI] [PubMed] [Google Scholar]
- Martín R., Makino H., Cetinyurek Yavuz A., Ben-Amor K., Roelofs M., Ishikawa E., et al. (2016). Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PloS One 11, e0158498. 10.1371/journal.pone.0158498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastromarino P., Capobianco D., Miccheli A., Praticò G., Campagna G., Laforgia N., et al. (2015). Administration of a multistrain probiotic product (VSL#3) to women in the perinatal period differentially affects breast milk beneficial microbiota in relation to mode of delivery. Pharmacol. Res. 95-96, 63–70. 10.1016/j.phrs.2015.03.013 [DOI] [PubMed] [Google Scholar]
- McGuire M. K., McGuire M. A. (2015). Human milk: mother nature’s prototypical probiotic food? Adv. Nutr. 6, 112–123. 10.3945/an.114.007435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire M. K., McGuire M. A. (2017). Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr. Opin. Biotechnol. 44, 63–68. 10.1016/j.copbio.2016.11.013 [DOI] [PubMed] [Google Scholar]
- Mediano P., Fernández L., Jiménez E., Arroyo R., Espinosa-Martos I., Rodríguez J. M., et al. (2017). Microbial diversity in milk of women with mastitis: potential role of coagulase-negative staphylococci, viridans group streptococci, and corynebacteria. J. Hum. Lact. 33, 309–318. 10.1177/0890334417692968 [DOI] [PubMed] [Google Scholar]
- Mestecky J. (2020). The mammary gland as an integral component of the common mucosal immune system. Nestle Nutr. Inst. Workshop Ser. 94, 27–37. 10.1159/000505336 [DOI] [PubMed] [Google Scholar]
- Meyer J., Potel J. (1956). [Qualitative improvement of human milk by bacteriological studies]. Dtsch. Gesundheitsw. 11, 1254–1256. [PubMed] [Google Scholar]
- Milani C., Mancabelli L., Lugli G. A., Duranti S., Turroni F., Ferrario C., et al. (2015). Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 81, 7078–7087. 10.1128/AEM.02037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., et al. (2017. a). The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 81, e00036–e00017. 10.1128/MMBR.00036-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Mangifesta M., Mancabelli L., Lugli G. A., James K., Duranti S., et al. (2017. b). Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 11, 2834–2847. 10.1038/ismej.2017.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A., Rodriguez J. (2017). “The origin of human milk bacteria,” in Prebiotics and Probiotics in Human Milk: Origins and Functions of Milk-Borne Oligosaccharides and Bacteria. Eds. McGuire M., McGuire M., Bode L. (London: Academic Press; ), 349–364. [Google Scholar]
- Mocquot G. (1955). Technical and hygienic aspects of the problem of milk destined for human consumption]. Ann. Nutr. Aliment. 9, 251–270. [PubMed] [Google Scholar]
- Mohandas S., Pannaraj P. S. (2020). Beyond the bacterial microbiome: virome of human milk and effects on the developing Infant. Nestle Nutr. Inst. Workshop Ser. 94, 86–93. 10.1159/000504997 [DOI] [PubMed] [Google Scholar]
- Moloney A. C., Quoraishi A. H., Parry P., Hall V. (1987). A bacteriological examination of breast pumps. J. Hosp. Infect. 9, 169–174. 10.1016/0195-6701(87)90056-9 [DOI] [PubMed] [Google Scholar]
- Moossavi S., Sepehri S., Robertson B., Bode L., Goruk S., Field C. J., et al. (2019. a). Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 25, 324–335.e4. 10.1016/j.chom.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Moossavi S., Atakora F., Miliku K., Sepehri S., Robertson B., Duan Q. L., et al. (2019. b). Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the CHILD Cohort. Front. Nutr. 6, 58. 10.3389/fnut.2019.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow A. L., Rangel J. M. (2004). Human milk protection against infectious diarrhea: implications for prevention and clinical care. Semin. Pediatr. Infect. Dis. 15, 221–228. 10.1053/j.spid.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Mossel D. A., Weijers H. A. (1957). [Results of bacteriological analysis of human milk & its significance for pediatric practice]. Maandschr. Kindergeneeskd. 25, 37–63. [PubMed] [Google Scholar]
- Mühl H., Kochem A.-J., Disqué C. S. S. (2010). Activity and DNA contamination of commercial polymerase chain reaction reagents for the universal 16SrDNA real-time polymerase chain reaction detection of bacterial pathogens in blood. Diagn. Microbiol. Infect. Dis. 66, 41–49. 10.1016/j.diagmicrobio.2008.07.011 [DOI] [PubMed] [Google Scholar]
- Murphy K., Curley D., O’Callaghan T. F., O’Shea C. A., Dempsey E. M., O’Toole P. W., et al. (2017). The composition of human milk and infant fecal microbiota over the first three months of life: A pilot study. Sci. Rep. 7, 40597. 10.1038/srep40597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg D. S. (2005). Innate immunity and human milk. J. Nutr. 135, 1308–1312. 10.1093/jn/135.5.1308 [DOI] [PubMed] [Google Scholar]
- Niimi K., Usami K., Fujita Y., Abe M., Furukawa M., Suyama Y., et al. (2018). Development of immune and microbial environments is independently regulated in the mammary gland. Mucosal Immunol. 11, 643–653. 10.1038/mi.2017.90 [DOI] [PubMed] [Google Scholar]
- Nikniaz L., Ostadrahimi A., Mahdavi R., Hejazi M. A., Hosseini Salekdeh G. (2013). Effects of synbiotic supplementation on breast milk levels of IgA, TGF-β1, and TGF-β2. J. Hum. Lact. 29, 591–596. 10.1177/0890334413490833 [DOI] [PubMed] [Google Scholar]
- Novel E., Pongratz E. (1954). [Three years of bacteriological analysis of the human milk]. Mitt. Geb. Lebensmittelunters Hyg. 45, 33–37. [PubMed] [Google Scholar]
- Oh J., Byrd A. L., Deming C., Conlan S., NISC Comparative Sequencing Program. Kong H. H., et al. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64. 10.1038/nature13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G., Bicalho M. L., Meira E., Rossi R. E., Foditsch C., Machado V. S., et al. (2014). Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PloS One 9, e85904. 10.1371/journal.pone.0085904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojo-Okunola A., Nicol M., du Toit E. (2018). Human breast milk bacteriome in health and disease. Nutrients 10, 1643. 10.3390/nu10111643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojo-Okunola A., Cacciatore S., Nicol M. P., du Toit E. (2020). The determinants of the human milk metabolome and its role in infant health. Metabolites 10, 77. 10.3390/metabo10020077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M., Díaz-Ropero M. P., Martín R., Rodríguez J. M., Xaus J. (2006. a). Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J. Appl. Microbiol. 101, 72–79. 10.1111/j.1365-2672.2006.02981.x [DOI] [PubMed] [Google Scholar]
- Olivares M., Díaz-Ropero M. P., Gómez N., Lara-Villoslada F., Sierra S., Maldonado J. A., et al. (2006. b). The consumption of two new probiotic strains, Lactobacillus gasseri CECT5714 and Lactobacillus coryniformis CECT5711, boost the immune system of healthy adults. Int. Microbiol. 9, 47–52. 10.1016/j.ijfoodmicro.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Olivares M., Díaz-Ropero M. P., Gómez N., Lara-Villoslada F., Sierra S., Maldonado J. A., et al. (2006. c). Oral administration of two probiotic strains, Lactobacillus gasseri CECT5714 and Lactobacillus coryniformis CECT5711, enhances the intestinal function of healthy adults. Int. J. Food Microbiol. 107, 104–111. 10.1016/j.ijfoodmicro.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Ottenheimer E. J., Jr, Minchew B. H., Cohen L. S., Cluff L. E. (1961). Studies of the epidemiology of staphylococcal infection. II. Isolation of staphylococci from the milkof nursing mothers under non-epidemic conditions. Bull. Johns Hopkins Hosp. 109, 114–123. [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis, the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwehand A. C., Isolauri E., He F., Hashimoto H., Benno Y., Salminen S. (2001). Differences in Bifidobacterium flora composition in allergic and healthy infants. J. Allergy Clin. Immunol. 108, 144–145. 10.1067/mai.2001.115754 [DOI] [PubMed] [Google Scholar]
- Pabst R. (1987). The anatomical basis for the immune function of the gut. Anat. Embryol. (Berl) 176, 135–144. 10.1007/BF00310046 [DOI] [PubMed] [Google Scholar]
- Padilha M., Danneskiold-Samsøe N. B., Brejnrod A., Hoffmann C., Cabral V. P., Iaucci J. M., et al. (2019. a). The human milk microbiota is modulated by maternal diet. Microorganisms 7, 502. 10.3390/microorganisms7110502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilha M., Iaucci J. M., Cabral V. P., Diniz E. M. A., Taddei C. R., Saad S. M. I. (2019. b). Maternal antibiotic prophylaxis affects Bifidobacterium spp. counts in the human milk, during the first week after delivery. Benef. Microbes 10, 155–163. 10.3920/BM2018.0046 [DOI] [PubMed] [Google Scholar]
- Padilha M., Brejnrod A., Danneskiold-Samsøe N. B., Hoffmann C., de Melo Iaucci J., Cabral V. P., et al. (2020). Response of the human milk microbiota to a maternal prebiotic intervention is individual and influenced by maternal age. Nutrients 12, 1081. 10.3390/nu12041081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajkos A., Deva A. K., Vickery K., Cope C., Chang L., Cossart Y. E. (2003). Detection of subclinical infection in significant breast implant capsules. Plast. Reconstr. Surg. 111 (5), 1605–1611. 10.1097/01.PRS.0000054768.14922.44 [DOI] [PubMed] [Google Scholar]
- Palmer C., Bik E. M., Digiulio D. B., Relman D. A., Brown P. O. (2007). Development of the human infant intestinal microbiota. PloS Biol. 5, e177. 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannaraj P. S., Li F., Cerini C., Bender J. M., Yang S., Rollie A., et al. (2017). Association between breast milk bacterial communities and establishment and development of the infnat gut microbiome. JAMA Pediatr. 171, 647–654. 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannaraj P. S., Ly M., Cerini C., Saavedra M., Aldrovandi G. M., Saboory A. A., et al. (2018). Shared and distinct features of human milk and infant stool viromes. Front. Microbiol. 9, 1162. 10.3389/fmicb.2018.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B., Iwase T., Liu G. Y. (2011). Intranasal application of S. epidermidis prevents colonization by methicillin-resistant Staphylococcus aureus in mice. PloS One 6, e25880. 10.1371/journal.pone.0025880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W. H. (1901). The bacterial condition of city milk and the need of health authorities to prevent the sale of milk containing excessive numbers of bacteria. J. Boston Soc Med. Sci. 5, 370–371. [PMC free article] [PubMed] [Google Scholar]
- Pärnänen K., Karkman A., Hultman J., Lyra C., Bengtsson-Palme J., Larsson D., et al. (2018). Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 9, 3891. 10.1038/s41467-018-06393-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. H., Vaidya Y. H., Patel R. J., Pandit R. J., Joshi C. G., Kunjadiya A. P. (2017). Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 7, 7804. 10.1038/s41598-017-08451-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peran L., Camuesco D., Comalada M., Nieto A., Concha A., Diaz-Ropero M. P., et al. (2005). Preventative effects of a probiotic, Lactobacillus salivarius ssp. salivarius, in the TNBS model of rat colitis. World J. Gastroenterol. 11, 5185–5192. 10.3748/wjg.v11.i33.5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peran L., Camuesco D., Comalada M., Nieto A., Concha A., Adrio J. L., et al. (2006). Lactobacillus fermentum, a probiotic capable to release glutathione, prevents colonic inflammation in the TNBS model of rat colitis. Int. J. Colorectal Dis. 21, 737–746. 10.1007/s00384-005-0773-y [DOI] [PubMed] [Google Scholar]
- Peran L., Sierra S., Comalada M., Lara-Villoslada F., Bailón E., Nieto A., et al. (2007). A comparative study of the preventative effects exerted by two probiotics, Lactobacillus reuteri and Lactobacillus fermentum, in the trinitrobenzenesulfonic acid model of rat colitis. Br. J. Nutr. 97, 96–103. 10.1017/S0007114507257770 [DOI] [PubMed] [Google Scholar]
- Perez P. F., Doré J., Leclerc M., Levenez F., Benyacoub J., Serrant P., et al. (2007). Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119, e724–e732. 10.1542/peds.2006-1649 [DOI] [PubMed] [Google Scholar]
- Pérez-Cano F. J., Dong H., Yaqoob P. (2010). In vitro immunomodulatory activity of Lactobacillus fermentum CECT5716 and Lactobacillus salivarius CECT5713: two probiotic strains isolated from human breast milk. Inmunobiology 12, 996–1004. 10.1016/j.imbio.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Pichler M. J., Yamada C., Shuoker B., Alvarez-Silva C., Gotoh A., Leth M. L., et al. (2020). Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat. Commun. 11, 3285. 10.1038/s41467-020-17075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen P., Jordan F., Priami C., Morine M. J. (2015). The role of breast-feeding in infant immune sytem: a systems perspective on the intestinal microbiome. Microbiome 3, 41. 10.1186/s40168-015-0104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice P. M., Schoemaker M. H., Vervoort J., Hettinga K., Lambers T. T., van Tol E., et al. (2019). Human milk short-chain fatty acid composition is associated with adiposity outcomes in infants. J. Nutr. 149, 716–722. 10.1093/jn/nxy320 [DOI] [PubMed] [Google Scholar]
- Prescott S. L., Wickens K., Westcott L., Jung W., Currie H., Black P. N., et al. (2008). Supplementation with Lactobacillus rhamnosus or Bifidobacterium lactis probiotics in pregnancy increases cord blood interferon-gamma and breast milk transforming growth factor-beta and immunoglobin A detection. Clin. Exp. Aller. 38, 1606–1614. 10.1111/j.1365-2222.2008.03061.x [DOI] [PubMed] [Google Scholar]
- Qutaishat S. S., Stemper M. E., Spencer S. K., Borchardt M. A., Opitz J. C., Monson T. A., et al. (2003). Transmission of Salmonella enterica serotype typhimurium DT104 to infants through mother’s breast milk. Pediatrics 111, 1442–1446. 10.1542/peds.111.6.1442 [DOI] [PubMed] [Google Scholar]
- Ramani S., Stewart C. J., Laucirica D. R., Ajami N. J., Robertson B., Autran C. A., et al. (2018). Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 9, 5010. 10.1038/s41467-018-07476-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey D. T., Kent J. C., Owens R. A., Hartmann P. E. (2004). Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 113, 361–367. 10.1542/peds.113.2.361 [DOI] [PubMed] [Google Scholar]
- Rantasalo I., Kauppinen M. A. (1959). The occurrence of Staphylococcus aureus in mother’s milk. Ann. Chir. Gynaecol. Fenn. 48, 246–258. [PubMed] [Google Scholar]
- Rautava S., Kalliomäki M., Isolauri E. (2002). Probiotics during pregnancy and breast-feeding might confer immunomodulatory protection against atopic disease in the infant. J. Allergy Clin. Immunol. 109, 119–121. 10.1067/mai.2002.120273 [DOI] [PubMed] [Google Scholar]
- Renfrew M. J., Pokhrel S., Quigley M., McCormick F., Fox-Rushby J., Dodds R., et al. (2012). Preventing disease and saving resources: the potential contribution of increasing breastfeeding rates in the UK (London: UNICEF UK; ). [Google Scholar]
- Rescigno M., Urbano M., Valzasina B., Francolín M., Rotta G., Bonasio R., et al. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367. 10.1038/86373 [DOI] [PubMed] [Google Scholar]
- Rieger U. M., Mesina J., Kalbermatten D. F., Haug M., Frey H. P., Pico R., et al. (2013). Bacterial biofilms and capsular contracture in patients with breast implants. Br. J. Surg. 100, 768–774. 10.1002/bjs.9084 [DOI] [PubMed] [Google Scholar]
- Riskin A., Almog M., Peri R., Halasz K., Srugo I., Kessel A. (2012). Changes in immunomodulatory constituents of human milk in response to active infection in the nursing infant. Pediatr. Res. 71, 220–225. 10.1038/pr.2011.34 [DOI] [PubMed] [Google Scholar]
- Rodríguez J. M., Fernández L. (2017). “Infectious mastitis during lactation: a mammary dysbiosis model,” in Prebiotics and Probiotics in Human Milk. Eds. McGuire M., McGuire M., Bode L. (London: Academic Press; ), 401–428. [Google Scholar]
- Rodríguez J. M. (2014). The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 5, 779–784. 10.3945/an.114.007229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cruz M., Alba C., Aparicio M., Checa M. Á., Fernández L., Rodríguez J. M. (2020). Effect of sample collection (manual expression vs. pumping) and skimming on the microbial profile of human milk using culture techniques and metataxonomic analysis. Microorganisms 8, E1278. 10.3390/microorganisms8091278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitt I. (2001). Essential Immunology (Oxford: Blackwell Scientific Publications; ). [Google Scholar]
- Ruiz L., Bacigalupe R., García-Carral C., Boix-Amoros A., Argüello H., Silva C. B., et al. (2019). Microbiota of human precolostrum and its potential role as a source of bacteria to the infant mouth. Sci. Rep. 9, 8435. 10.1038/s41598-019-42514-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder R. W., Crosby-Ritchie A., McDonough B., Hall W. J. 3. (1977). Human milk contaminated with Salmonella kottbus. A cause of nosocomial illness in infants. JAMA 238, 1533–1534. 10.1001/jama.1977.03280150103039 [DOI] [PubMed] [Google Scholar]
- Sager C. A. (1956). Biology of human milk. II. Decrease of germ content in human milk after freeze-drying. Monatsschr. Kinderheilkd. 104, 223–228. [PubMed] [Google Scholar]
- Sakanaka M., Gotoh A., Yoshida K., Odamaki T., Koguchi H., Xiao J. Z., et al. (2019). Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients 12, 71. 10.3390/nu12010071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakwinska O., Moine D., Delley M., Combremont S., Rezzonico E., Descombes P., et al. (2016). Microbiota in breast milk of chinese lactating mothers. PloS One 11, e0160856. 10.1371/journal.pone.0160856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampieri C. L., Montero H. (2019). Breastfeeding in the time of Zika: a systematic literature review. Peer J. 7, e6452. 10.7717/peerj.6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel B. S., Hansen E. E., Manchester J. K., Coutinho P. M., Henrissat B., Fulton R., et al. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. U. S. A. 104, 10643–10648. 10.1073/pnas.0704189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanler R. J., Fraley J. K., Lau C., Hurst N. M., Hovarth L., Rossmann S. N. (2011). Breastmilk cultures and infection in extremely premature infants. J. Perinatol. 31, 335–338. 10.1038/jp.2011.13 [DOI] [PubMed] [Google Scholar]
- Schreiner R. L., Coates T., Shackelford P. G. (1977). Possible breast milk transmission of group B streptococcal infection. J. Pediatr. 91, 159. 10.1016/S0022-3476(77)80474-5 [DOI] [PubMed] [Google Scholar]
- Schultz M., Göttl C., Young R. J., Iwen P., Vanderhoof J. A. (2004). Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J. Ped. Gastroenterol. Nutr. 38, 293–297. 10.1097/00005176-200403000-00012 [DOI] [PubMed] [Google Scholar]
- Schwab C., Voney E., Ramirez Garcia A., Vischer M., Lacroix C. (2019). Characterization of the cultivable microbiota in fresh and stored mature human breast milk. Front. Microbiol. 10, 2666. 10.3389/fmicb.2019.02666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D. A., Mills D. A. (2010). Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307. 10.1016/j.tim.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M. R., Avershina E., Storrθ O., Johnsen R., Rudi K., θjen T. (2018). Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J. Dairy Sci. 101, 889–899. 10.3168/jds.2017-13411 [DOI] [PubMed] [Google Scholar]
- Smith A., Pierre J. F., Makowski L., Tolley E., Lyn-Cook B., Lu L., et al. (2019). Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 9, 11940. 10.1038/s41598-019-48348-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. (1904). Some problems in the life-history of pathogenic microörganisms. Amer. Med. viii, 711–718. [DOI] [PubMed] [Google Scholar]
- Soeorg H., Metsvaht T., Eelmäe I., Metsvaht H. K., Treumuth S., Merila M., et al. (2017. a). Coagulase-negative staphylococci in human milk from mothers of preterm compared with term neonates. J. Hum. Lact. 33, 329–340. 10.1177/0890334417691505 [DOI] [PubMed] [Google Scholar]
- Soeorg H., Metsvaht T., Eelmäe I., Merila M., Treumuth S., Huik K., et al. (2017. b). The role of breast milk in the colonization of neonatal gut and skin with coagulase-negative staphylococci. Pediatr. Res. 82, 759–767. 10.1038/pr.2017.150 [DOI] [PubMed] [Google Scholar]
- Solís G., de Los Reyes-Gavilan C. G., Fernández N., Margolles A., Gueimonde M. (2010). Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16, 307–310. 10.1016/j.anaerobe.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Soto A., Martín V., Jiménez E., Mader I., Rodríguez J. M., Fernández L. (2014). Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 59, 78–88. 10.1097/MPG.0000000000000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno S., Reynolds D. W., Pass R. F., Alford C. A. (1980). Breast milk and the risk of cytomegalovirus infection. N. Engl. J. Med. 302, 1073–1076. 10.1056/NEJM198005083021908 [DOI] [PubMed] [Google Scholar]
- Stinson L. F., Keelan J. A., Payne M. S. (2019). Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass microbiome analyses. Lette. Appl. Microbiol. 68, 2–8. 10.1111/lam.13091 [DOI] [PubMed] [Google Scholar]
- Stinson L. F., Gay M., Koleva P. T., Eggesbø M., Johnson C. C., Wegienka G., et al. (2020). Human milk from atopic mothers has lower levels of short chain fatty acids. Front. Immunol. 11, 1427. 10.3389/fimmu.2020.01427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N., Pickler R. H., Munro C., Shotwell J. (1997). Contamination in expressed breast milk following breast cleansing. J. Hum. Lact. 13, 127–130. 10.1177/089033449701300213 [DOI] [PubMed] [Google Scholar]
- Thomsen A. C. (1982). Infectious mastitis and occurrence of antibody-coated bacteria in milk. Am. J. Obstet. Gynecol. 144, 350–351. 10.1016/0002-9378(82)90590-7 [DOI] [PubMed] [Google Scholar]
- Thongaram T., Hoeflinger J. L., Chow J., Miller M. J. (2017). Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. J. Dairy Sci. 100, 7825–7833. 10.3168/jds.2017-12753 [DOI] [PubMed] [Google Scholar]
- Thyagarajan S., Zhang Y., Thapa S., Allen M. S., Phillips N., Chaudhary P., et al. (2020). Comparative analysis of racial differences in breast tumor microbiome. Sci. Rep. 10, 14116. 10.1038/s41598-020-71102-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo A. H., Camara A., Konaté S., Doumbo O. K., Raoult D., Million M. (2017). ‘Lactomassilus timonensis,’ a new anaerobic bacterial species isolated from the milk of a healthy African mother. New Microbes New Infect. 21, 122–124. 10.1016/j.nmni.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo A. H., Diop A., Camara A., Kuete E., Konate S., Brevaut V., et al. (2019. a). Lactimicrobium massiliense gen. nov., sp. nov.; Anaerolactibacter massiliensis gen. nov., sp. nov.; Galactobacillus timonensis gen. nov., sp. nov. and Acidipropionibacterium timonense sp. nov. isolated from breast milk from healthy breastfeeding African women. New Microbes New Infect. 29, 100537. 10.1016/j.nmni.2019.100537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo A., Dufour J. C., Lagier J. C., Dubourg G., Raoult D., Million M. (2019. b). Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol. 14, 623–641. 10.2217/fmb-2018-0317 [DOI] [PubMed] [Google Scholar]
- Togo A. H., Grine G., Khelaifia S., des Robert C., Brevaut V., Caputo A., et al. (2019. c). Culture of methanogenic archaea from human colostrum and milk. Sci. Rep. 9, 18653. 10.1038/s41598-019-54759-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano M., De Grandi R., Peroni D. G., Grossi E., Facchin V., Comberiati P., et al. (2017). Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. 17, 205. . 10.1186/s12866-017-1109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treven P., Mrak V., Bogovič Matijašić B., Horvat S., Rogelj I. (2015). Administration of probiotics Lactobacillus rhamnosus GG and Lactobacillus gasseri K7 during pregnancy and lactation changes mouse mesenteric lymph nodes and mammary gland microbiota. J. Dairy Sci. 98, 2114–2128. 10.3168/jds.2014-8519 [DOI] [PubMed] [Google Scholar]
- Triantis V., Bode L., van Neerven R. (2018). Immunological effects of human milk oligosaccharides. Front. Pediatr. 6, 190. 10.3389/fped.2018.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H., Rautava S., Collado M. C., Syrjänen S., Rautava J. (2018). HPV infection and bacterial microbiota in breast milk and infant oral mucosa. PloS One 13, e0207016. 10.1371/journal.pone.0207016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., Peano C., Pass D. A., Foroni E., Severgnini M., Claesson M. J., et al. (2012). Diversity of bifidobacteria within the infant gut microbiota. PloS One 7, e36957. 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services (2011). The Surgeon General’s Call to Action to Support Breastfeeding (Washington, DC: U.S. Department of Health and Human Services, Office of the Surgeon General; ). [Google Scholar]
- Uehara Y., Kikuchi K., Nakamura T., Nakama H., Agematsu K., Kawakami Y., et al. (2001). H2O2 produced by viridans group streptococci may contribute to inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin. Infect. Dis. 32, 1408–1413. 10.1086/320179 [DOI] [PubMed] [Google Scholar]
- Underwood M. A., German J. B., Lebrilla C. B., Mills D. A. (2015). Bifidobacterium longum subspecies. infantis: champion colonizer infant gut. Pediatr. Res. 77, 229–235. 10.1038/pr.2014.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations Children’s Fund (UNICEF) (2020). Coronavirus disease (COVID-19): What parents should 177 know. (New York: United Nations Children’s Fund, UNICEF; ). Available at: https://www.unicef.org/stories/novel-coronavirus-outbreak178what-parents-should-know. [Google Scholar]
- Urbaniak C., Cummins J., Brackstone M., Macklaim J. M., Gloor G. B., Baban C. K., et al. (2014. a). Microbiota of human breast tissue. Appl. Environ. Microbiol. 80, 3007–3014. 10.1128/AEM.00242-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., McMillan A., Angelini M., Gloor G. B., Sumarah M., Burton J. P., et al. (2014. b). Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome 2, 24. 10.1186/2049-2618-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Angelini M., Gloor G. B., Reid G. (2016. a). Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 4, 1. 10.1186/s40168-015-0145-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Gloor G. B., Brackstone M., Scott L., Tangney M., Reid G. (2016. b). The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 82, 5039–5048. 10.1128/AEM.01235-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Perre P., Rubbo P. A., Viljoen J., Nagot N., Tylleskär T., Lepage P., et al. (2012). HIV-1 reservoirs in breast milk and challenges to elimination of breast-feeding transmission of HIV-1. Sci. Transl. Med. 4, 143sr3. 10.1126/scitranslmed.3003327 [DOI] [PubMed] [Google Scholar]
- Vanhaecke T., Aubert P., Grohard P. A., Durand T., Hulin P., Paul-Gilloteaux P., et al. (2017). L. fermentum CECT5716 prevents stress-induced intestinal barrier dysfunction in newborn rats. Neurogastroenterol. Motil. 29, e13069. 10.1111/nmo.13069 [DOI] [PubMed] [Google Scholar]
- Vankerckhoven V. V., Autgaerden T. A., Huys G., Vancanneyt M., Swings J., Goossens H. (2004). Establishment of the PROSAFE collection of probiotic and human lactic acid bacteria. Microb. Ecol. Health Dis. 16, 131–136. 10.1080/08910600410032349 [DOI] [Google Scholar]
- Vazquez-Fresno R., Llorach R., Marinic J., Tulipani S., Garcia-Aloy M., Espinosa-Martos I., et al. (2014). Urinary metabolomic fingerprinting after consumption of a probiotic strain in women with mastitis. Pharmacol. Res. 87, 160–165. 10.1016/j.phrs.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A., Jones-Carson J., Baumler A. J., Falkow S., Valdivia R., Brown W., et al. (1999). Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401, 804–808. 10.1038/44593 [DOI] [PubMed] [Google Scholar]
- Verhasselt V., Milcent V., Cazareth J., Kanda A., Fleury S., Dombrowicz D., et al. (2008). Breast milk-mediated transfer of an antigen induces tolerance and protection from allergic asthma. . Nat. Med. 14, 170–175. 10.1038/nm1718 [DOI] [PubMed] [Google Scholar]
- Ververs M., Arya A. (2019). Ebola virus disease and breastfeeding: time for attention. Lancet 394, 825. 10.1016/S0140-6736(19)32005-7 [DOI] [PubMed] [Google Scholar]
- Větrovský T., Baldrian P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS One 8, e57923. 10.1371/journal.pone.0057923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora C. G., Bahl R., Barros A. J., França G. V., Horton S., Krasevec J., et al. (2016). Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet 387, 475–490. 10.1016/S0140-6736(15)01024-7 [DOI] [PubMed] [Google Scholar]
- Virden C. P., Dobke M. K., Stein P., Parsons C. L., Frank D. H. (1992). Subclinical infection of the silicone breast implant surface as a possible cause of capsular contracture. Aesthetic Plast. Surg. 16, 173–179. 10.1007/BF00450610 [DOI] [PubMed] [Google Scholar]
- Walker J. N., Pinkner C. L., Pinkner J. S., Hultgren S. J., Myckatyn T. M. (2019). The detection of bacteria and matrix proteins on clinically benign and pathologic implants. Plast. Reconstr. Surg. Glob. Open 7, e2037. 10.1097/GOX.0000000000002037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. P., Barrett M., Hogan G., Flores Bueso Y., Claesson M. J., Tangney M. (2020). Non-specific amplification of human DNA is a major challenge for 16S rRNA gene sequence analysis. Sci. Rep. 10, 16356. 10.1038/s41598-020-73403-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Li M., Wu S., Lebrilla C. B., Chapkin R. S., Ivanov I., et al. (2015). Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J. Pediatr. Gastroenterol. Nutr. 60, 825–833. 10.1097/MPG.0000000000000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu H., Feng Z., Cao J., Fang C., Xu X., et al. (2017). Development of human breast milk microbiota-associated mice as a method to identify breast milk bacteria capable of colonizing gut. Front. Microbiol. 8, 1242. 10.3389/fmicb.2017.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. L., Hosid S., Ioshikhes I., Altosaar I. (2013). Human milk metagenome: a functional capacity analysis. BMC Microbiol. 13, 116. 10.1186/1471-2180-13-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems M. F., Dereddy N. R., Arnold S. R. (2015). Mother’s milk as a source of Enterobacter cloacae sepsis in a preterm infant. Breastfeed. Med. 10, 503–504. 10.1089/bfm.2015.0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West P. A., Hewitt J. H., Murphy O. M. (1979). Influence of methods of collection and storage on the bacteriology of human milk. J. Appl. Bacteriol. 46, 269–277. 10.1111/j.1365-2672.1979.tb00820.x [DOI] [PubMed] [Google Scholar]
- WHO (2016). Guideline: updates on HIV and infant feeding: the duration of breastfeeding, and support from health services to improve feeding practices among mothers living with HIV (Geneva: World Health Organization; ). [PubMed] [Google Scholar]
- Williams J. E., Carrothers J. M., Lackey K. A., Beatty N. F., York M. A., Brooker S. L., et al. (2017. a). Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J. Nutr. 147, 1739–1748. . 10.3945/jn.117.248864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. E., Price W. J., Shafii B., Yahvah K. M., Bode L., McGuire M. A., et al. (2017. b). Relationships among microbial communities, maternal cells, oligosaccharides, and macronutrients in human milk. J. Hum. Lact. 33, 540–551. 10.1177/0890334417709433 [DOI] [PubMed] [Google Scholar]
- Williams J. E., Carrothers J. M., Lackey K. A., Beatty N. F., Brooker S. L., Peterson H. K., et al. (2019). Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J. Nutr. 149, 902–914. 10.1093/jn/nxy299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S., Finucare E., Gamsu H. R., Hewitt J. H. (1978). Staphylococcus aureus in raw milk for neonates. Br. Med. J. 1 (6120), 1146. 10.1136/bmj.1.6120.1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. S. (1943). The Pasteurization of Milk. Br. Med. J. 1 (4286), 261–262. 10.1136/bmj.1.4286.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan C., Shamonki J. M., Chung A., Dinome M. L., Chung M., Sieling P. A., et al. (2014). Microbial dysbiosis is associated with human breast cancer. PloS One 9, e83744. 10.1371/journal.pone.0083744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaura E., Nicu E. A., Krom B. P., Keijser B. J. (2014). Acquiring and maintaining a normal oral microbiome: current perspective. Front. Cell Infect. Microbiol. 24 85. 10.3389/fcimb.2014.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wittouck S., Salvetti E., Franz C. M. A. P., Harris H. M. B., Mattarelli P., et al. (2020). A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae . Int. J. Syst. Evol. Microbiol. 70, 2782–2858. 10.1099/ijsem.0.004107 [DOI] [PubMed] [Google Scholar]
- Ziegler J. B., Cooper D. A., Johnson R. O., Gold J. (1985). Postnatal transmission of AIDS-associated retrovirus from mother to infant. Lancet 1 (8434), 896–898. 10.1016/S0140-6736(85)91673-3 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Gwee A., Curtis N. (2017). The controversial role of breast milk in GBS late-onset disease. J. Infect. 74 (1), S34–S40. 10.1016/S0163-4453(17)30189-5 [DOI] [PubMed] [Google Scholar]