Abstract
Background
In acute respiratory distress syndrome (ARDS) unrelated to COVID-19, two phenotypes, based on the severity of systemic inflammation (hyperinflammatory and hypoinflammatory), have been described. The hyperinflammatory phenotype is known to be associated with increased multiorgan failure and mortality. In this study, we aimed to identify these phenotypes in COVID-19-related ARDS.
Methods
In this prospective observational study done at two UK intensive care units, we recruited patients with ARDS due to COVID-19. Demographic, clinical, and laboratory data were collected at baseline. Plasma samples were analysed for interleukin-6 (IL-6) and soluble tumour necrosis factor receptor superfamily member 1A (TNFR1) using a novel point-of-care assay. A parsimonious regression classifier model was used to calculate the probability for the hyperinflammatory phenotype in COVID-19 using IL-6, soluble TNFR1, and bicarbonate levels. Data from this cohort was compared with patients with ARDS due to causes other than COVID-19 recruited to a previous UK multicentre, randomised controlled trial of simvastatin (HARP-2).
Findings
Between March 17 and April 25, 2020, 39 patients were recruited to the study. Median ratio of partial pressure of arterial oxygen to fractional concentration of oxygen in inspired air (PaO2/FiO2) was 18 kpa (IQR 15–21) and acute physiology and chronic health evaluation II score was 12 (10–16). 17 (44%) of 39 patients had died by day 28 of the study. Compared with survivors, patients who died were older and had lower PaO2/FiO2. The median probability for the hyperinflammatory phenotype was 0·03 (IQR 0·01–0·2). Depending on the probability cutoff used to assign class, the prevalence of the hyperinflammatory phenotype was between four (10%) and eight (21%) of 39, which is lower than the proportion of patients with the hyperinflammatory phenotype in HARP-2 (186 [35%] of 539). Using the Youden index cutoff (0·274) to classify phenotype, five (63%) of eight patients with the hyperinflammatory phenotype and 12 (39%) of 31 with the hypoinflammatory phenotype died. Compared with matched patients recruited to HARP-2, levels of IL-6 were similar in our cohort, whereas soluble TNFR1 was significantly lower in patients with COVID-19-associated ARDS.
Interpretation
In this exploratory analysis of 39 patients, ARDS due to COVID-19 was not associated with higher systemic inflammation and was associated with a lower prevalence of the hyperinflammatory phenotype than that observed in historical ARDS data. This finding suggests that the excess mortality observed in COVID-19-related ARDS is unlikely to be due to the upregulation of inflammatory pathways described by the parsimonious model.
Funding
US National Institutes of Health, Innovate UK, and Randox.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel virus leading to COVID-19 that has resulted in a global pandemic and is associated with high mortality and morbidity.1, 2, 3 SARS-CoV-2 pneumonia in its most severe form can lead to profound hypoxia and acute respiratory distress syndrome (ARDS) requiring invasive mechanical ventilation.1, 3 Little is understood about the pathophysiology of COVID-19, though many have speculated that a central pathophysiological abnormality associated with severe COVID-19 is an exaggerated systemic inflammatory response or a so-called cytokine storm.4, 5, 6 However, no objective data-driven evidence supports this theory.7
Considerable evidence does exist for the presence of subgroups of ARDS with exaggerated inflammation. In secondary analyses of five ARDS randomised controlled trials, two phenotypes, termed hyperinflammatory and hypoinflammatory, have been consistently identified using latent class analysis (LCA).8, 9, 10, 11 The hyperinflammatory phenotype is associated with exaggerated inflammation evidenced by greatly increased levels of circulating proinflammatory cytokines and increased incidence of shock. Mortality rates in the phenotype with lower systemic inflammatory responses are about 20% and consistently 20% lower than in the hyperinflammatory phenotype. Further, in three of these randomised controlled trials, differential treatment responses to randomised interventions were observed in the two phenotypes.8, 9, 10 These findings suggest that the hyperinflammatory phenotype might be useful for prognostic and predictive enrichment in ARDS.
Research in context.
Evidence before this study
We searched PubMed and Google Scholar using the search terms “COVID-19”, “SARS-CoV2”, “inflammation”, “cytokines”, and “immune responses” for research published in 2020, with no language restrictions. Additionally, we considered work by co-authors and colleagues on the subject of ARDS phenotyping. Two phenotypes of acute respiratory distress syndrome (ARDS) have consistently been identified in randomised controlled trials with divergent characteristics, clinical outcomes, and treatment responses. The hyperinflammatory phenotypes had more severe plasma inflammatory responses and worse outcomes. It has been hypothesised that the cytokine storm is integral to the pathogenesis of severe COVID-19. The prevalence of this phenotype in COVID-19-related ARDS was unknown.
Added value of this study
Using a previously validated parsimonious model and a point-of-care biomarker analyser, in this preliminary report, we classified 39 patients with COVID-19 ARDS into hypoinflammatory and hyperinflammatory phenotypes. Compared with a matched cohort of patients from the HARP-2 study of patients with ARDS due to causes other than COVID-19, the prevalence of the hyperinflammatory phenotype in the COVID-19 cohort was lower, and mortality at day 28 was higher in both phenotypes.
Implications of all the available evidence
The findings of this exploratory study suggest that the hyperinflammatory phenotype of ARDS is less prevalent in COVID-19 than in previous ARDS cohorts, undermining the theory that the cytokine storm is disproportionately characteristic of COVID-19. Future studies are needed to confirm these findings and to better understand the pathophysiology driving poor outcomes in patients with COVID-19-associated ARDS.
LCA-derived phenotypes are usually identified using large datasets and the algorithms are dependent on research biomarkers. Parsimonious classifier models have been developed to identify ARDS phenotypes using a small number of variables.12 We used these models and novel point-of-care assays13 to identify ARDS phenotypes in patients with COVID-19 in real time. We aimed to describe the prevalence of ARDS phenotypes in COVID-19-associated ARDS; and to compare the clinical and biological characteristics of patients with COVID-19 and ARDS to a previously characterised population of patients with ARDS due to other causes—those enrolled in the Hydroxymethylglutaryl-CoA reductase inhibition with simvastatin in Acute lung injury to Reduce Pulmonary dysfunction (HARP-2) clinical trial.14
Methods
Study design and population
This was a prospective observational study done at two centres in Newport and London, UK. The study was a subset of an ongoing multicentre study, clinical evaluation of a point of care assay to identify PHenotypes IN the acute respiratory Distress syndrome (PHIND; NCT04009330). All patients were unable to provide consent themselves, so consent was gained using the appropriate emergency consent mechanisms in line with the ethical approval of the study by the Bromley Research Ethics Committee, UK (reference number 19/LO/0672). The study sites were the Royal Gwent Hospital, a district general hospital in Newport, Wales, and University College Hospital, a university hospital serving an inner-city population in London. Both intensive care units (ICUs) were operating at surge capacity for the duration of the study (appendix p 1).
Patients were eligible for recruitment if they were positive for SARS-CoV-2 and met the Berlin definition of ARDS.15 Patients were excluded from the study if they were younger than 18 years; if onset of ARDS was more than 48 h before screening; if they were receiving extracorporeal membrane oxygenation; or if they had a do not resuscitate order in place. Diagnosis of ARDS was established by the attending physicians caring for the patient.
The study protocol is available online.
Data collection
Comprehensive data were collected at baseline, including demographics, chronic health conditions, vital signs, and ventilatory and laboratory investigations. In addition to standard laboratory investigations, data were also available for acute markers of inflammation widely described for COVID-19. These were D-dimer, ferritin, C-reactive protein, procalcitonin, lactate dehydrogenase, fibrinogen, and troponin. Biospecimens were also collected at baseline to quantify additional protein biomarker levels. The study was censored at day 28 and vital status was adjudicated at this point.
Protein biomarker quantification and phenotype classification
Probabilities for belonging to the hyperinflammatory phenotype were generated using a novel rapid point-of-care platform. In a prespecified two-step process performed in real time, plasma samples were first used to quantify interleukin 6 (IL-6) and soluble tumour necrosis factor receptor superfamily member 1A (TNFR1) concentrations. Plasma levels of the two biomarkers were quantified at the time of study recruitment using a novel point-of-care assay measured using the Evidence Multistat Analyser (Randox Laboratories, Country Antrim, UK). Next, as per the PHIND study protocol,12 a three-variable parsimonious classifier model comprised of IL-6, serum bicarbonate, and soluble TNFR1 was used to generate the probabilities of phenotype assignment (appendix p 2).12 Values for serum bicarbonate were measured in clinical laboratories. Clinical staff at both sites were masked to the biomarker data and generated probabilities. The point-of-care platform-generated probabilities have been validated against probabilities generated using ELISA-based biomarker quantification and the same classifier model.13 The study showed good correlation between the probabilities generated by the two methods, and both methods classified ARDS phenotypes accurately.13 Details of assay-specific procedures are in the appendix (p 1).
As per the PHIND protocol, patients were classified into the hyperinflammatory phenotype using one of two prespecified probability cutoffs: (1) 0·5 or higher; and (2) the Youden index generated during model development (≥0·274). During previous model validation, classification based on a cutoff of 0·5 led to higher specificity, whereas the Youden index cutoff led to higher sensitivity.12 Once classified, differences in measured baseline variables and mortality at day 28 were compared between the phenotypes.
Previous findings from the secondary analysis using LCA of a phase 2b randomised trial of simvastatin for treatment of ARDS (the HARP-2 study)14 were used as a historical reference standard to compare proportions of phenotypes and clinical outcomes in the COVID-19 phenotypes. HARP-2 was specifically selected because data were available for IL-6 and soluble TNFR1 quantified by the Multistat analyser in a selection of patients and would allow direct comparison with the studied cohort. First, phenotype proportions, acute physiology and chronic health evaluation II (APACHE II) scores, ratio of partial pressure of arterial oxygen to fractional concentration of oxygen in inspired air (PaO2/FiO2), and clinical outcomes from this study were compared with the entire HARP-2 cohort (n=539). For HARP-2, phenotypes described are those derived using LCA. It was not possible to use the parsimonious model used in the COVID-19 cohort in HARP-2 because bicarbonate was not measured. Next, biomarker levels, phenotype proportions, APACHE II scores, and clinical outcomes in the COVID-19 cohort were compared with an equivalent number of matched patients from HARP-2 that had IL-6 and soluble TNFR1 levels measured using the Evidence Multistat Analyser (herein referred to as the HARP-2 matched cohort). This matched analysis permitted comparison of biomarker levels quantified using the same assay across two independent populations. Of the entire HARP-2 cohort, Multistat biomarker analysis was available in 98 patients. In an effort to compare aetiologically similar groups to COVID-19, only patients with pneumonia as the primary risk factor for ARDS were selected for matching from this subset. Matching of patients to the COVID-19 cohort was done on the basis of a logistic regression-derived score using age, gender, and PaO2/FiO2 as predictor variables (appendix p 2).
Statistical analysis
Clinical data from the time of study enrolment were used for analysis. Given the small sample size in the analysed subgroups, data are presented as median (IQR) for all continuous variables. Characteristics between groups were compared using Wilcoxon signed-rank test or Fisher's exact test depending on the nature of the variable. Spearman's rank correlation coefficient was used to assess association between biomarkers. All analyses were done on R Studio, version 1.1.453, using R, version 3.4.1.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
39 patients were recruited to the study between March 17 and April 25, 2020. Of these, 32 were from Royal Gwent Hospital and seven were from University College Hospital. All samples were collected within 2 h of enrolment into the study and within 24 h of diagnosis of ARDS and meeting study enrolment criteria. The median time from the onset of symptoms to study enrolment was 10 days (IQR 7–13). 35 (90%) of 39 patients were receiving invasive mechanical ventilation and four patients were non-invasively ventilated at the time of recruitment to the study (table 1 ). All four patients receiving non-invasive ventilation were subsequently intubated during their stay in the ICU. 24 (62%) of 39 patients were on vasopressors at baseline (median dose 0·08 μg/kg per min). The median APACHE II score was 12 (IQR 10–16) and median PaO2/FiO2 was 18 kpa (15–21). At day 28, 17 (44%) of 39 patients had died. Of the survivors, seven remained in the ICU on day 28 of the study and have subsequently been discharged alive. 12 (38%) of 32 died in the Royal Gwent Hospital cohort and five (71%) of seven died in the University College Hospital cohort (appendix p 4).
Table 1.
Baseline characteristics of the cohort
| Total population (n=39) | Survivors (n=22) | Non-survivors (n=17) | p value | ||
|---|---|---|---|---|---|
| Age, years | 57 (52–61) | 54 (45–57) | 60 (56–64) | 0·0036 | |
| Sex | 0·0490* | ||||
| Men | 25 (64%) | 11 (50%) | 14 (82%) | ||
| Women | 14 (36%) | 11 (50%) | 3 (18%) | ||
| Race | 0·40* | ||||
| White | 19 (49%) | 10 (45%) | 9 (53%) | ||
| Asian | 9 (23%) | 4 (18%) | 5 (29%) | ||
| Black | 4 (10%) | 2 (9%) | 2 (12%) | ||
| Other† | 7 (18%) | 6 (27%) | 1 (6%) | ||
| Diabetes | 9 (23%) | 6 (27%) | 3 (18%) | 0·70* | |
| Hypertension | 6 (15%) | 2 (9%) | 4 (24%) | 0·37* | |
| Heart rate, beats per min | 103 (81–142) | 106 (84–153) | 98 (79–130) | 0·34 | |
| Mean arterial pressure, mm Hg | 64 (61–72) | 64 (61–69) | 65 (61–72) | 0·60 | |
| PaO2/FiO2, kPa | 18 (15–21) | 20 (17–24) | 15 (11–18) | 0·0040 | |
| Minute ventilation, L/min | 10·5 (9·4–12·1) | 10·2 (9·3–12·2) | 10·8 (9·8–11·2) | 0·60 | |
| Plateau pressure, cm H2O | 31 (27–34) | 30 (27–34) | 31 (26–34) | 0·82 | |
| Positive end-expiratory pressure, cm H2O | 12 (6–20) | 13 (12–15) | 12 (10–15) | 0·37 | |
| Compliance, mL/cm H2O | 24 (20–28) | 24 (21–28) | 25 (20–29) | 0·79 | |
| White blood cells, × 109 per L | 10 (8–12) | 8·6 (7·8–12) | 10·4 (9·7–14·2) | 0·25 | |
| Lymphocytes, × 109 per L | 1 (0·6–1·1) | 0·90 (0·6–1·1) | 1 (0·6–1·4) | 0·56 | |
| Platelets, × 109 per L | 272 (213–330) | 285 (236–332) | 244 (177–319) | 0·16 | |
| Albumin, g/L | 23 (20–26) | 24 (20–27) | 23 (20–25) | 0·61 | |
| Bilirubin, μmol/L | 10 (6–23) | 8 (6–12) | 23 (9–40) | 0·0235 | |
| Bicarbonate, mmol/L | 26 (24–30) | 27 (24–31) | 25 (23–27) | 0·32 | |
| Creatinine, μmol/L | 84 (65–172) | 74 (63–165) | 94 (74–201) | 0·19 | |
| Troponin, ng/L | 18 (5–37) | 9 (5–21) | 23 (12–58) | 0·0549 | |
| Lactate dehydrogenase, units per L | 458 (336–591) | 439 (343–499) | 530 (307–732) | 0·24 | |
| Procalcitonin, ng/mL | 1·2 (0·4–2·9) | 1·2 (0·3–2·9) | 1·7 (0·9–7·1) | 0·28 | |
| Fibrinogen, g/L | 6·6 (5·8–6·8) | 6·4 (5·8–6·6) | 6·6 (6·2–7·1) | 0·0520 | |
| D-dimer, ng/mL | 1622 (888–3742) | 1089 (815–2262) | 3730 (1604–5640) | 0·0187 | |
| Ferritin, μg/L | 1196 (421–2825) | 806 (382–1613) | 2178 (471–2947) | 0·12 | |
| C-reactive protein, mg/L | 214 (154–320) | 199 (145–322) | 277 (205–293) | 0·19 | |
| Interleukin-6, pg/mL | 192 (112–556) | 149 (84–270) | 457 (192–1042) | 0·0048 | |
| Soluble TNFR1, pg/mL | 3150 (2455–4405) | 2735 (2323–3705) | 4200 (3030–4590) | 0·0197 | |
| Vasopressor use (baseline) | 24 (62%) | 14 (64%) | 10 (59%) | 0·99* | |
| Invasive ventilation (baseline) | 35 (90%) | 21 (95%) | 14 (82%) | 0·44* | |
| Sequential organ failure assessment score | 6 (5–8) | 6 (4–7) | 7 (6–9) | 0·09 | |
| APACHE II score | 12 (10–16) | 12 (10–15) | 14 (11–16) | 0·26 | |
Data are median (IQR) and n (%). The cohort (a COVID-19 subset of the PHIND cohort of patients with ARDS) is stratified into groups of survivors and non-survivors. p values show comparison of survivors versus non-survivors and were calculated by Wilcoxon signed-rank test unless noted otherwise. APACHE II=acute physiology and chronic health evaluation II. ARDS=acute respiratory distress syndrome. PaO2/FiO2=ratio of partial pressure of arterial oxygen to fractional concentration of oxygen in inspired air. PHIND=clinical evaluation of a point of care assay to identify PHenotypes IN the acute respiratory Distress syndrome. TNFR1=tumour necrosis factor receptor superfamily member 1A.
Fisher's exact test.
Includes Filipino and Romani.
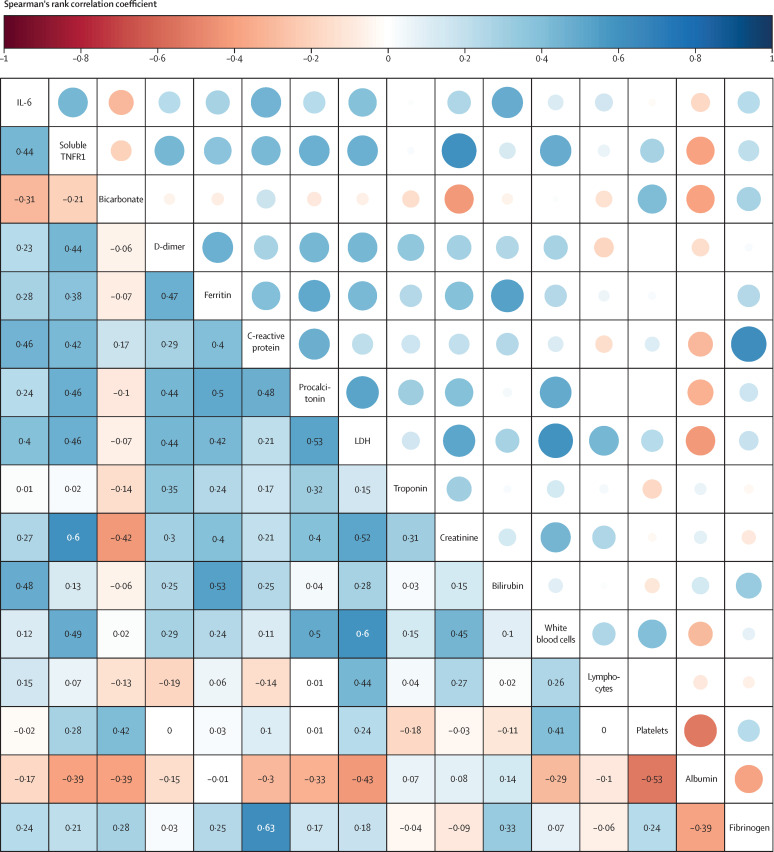
Median age of survivors at baseline (54 years [IQR 45–57]) was significantly lower than that of non-survivors (60 years [56–64], p=0·0036). Of the baseline respiratory variables, only the PaO2/FiO2 was significantly different, with lower levels in non-survivors (p=0·0040). Of the baseline biomarkers, IL-6 (p=0·0048), soluble TNFR1 (p=0·0197), D-dimer (p=0·0187), and bilirubin (p=0·0235) were all significantly higher in non-survivors than in survivors (table 1). Significant correlations were noted between many of the measured biomarkers (figure 1 ). D-dimer, ferritin, C-reactive protein, lactate dehydrogenase, and procalcitonin showed association with one another with some correlation coefficients between 0·4 and 0·5 for the more highly correlated variables. The highest correlations were observed between fibrinogen and C-reactive protein (r=0·63) and soluble TNFR1 and creatinine (r=0·60).
Figure 1.
Correlation matrix of the biomarkers measured at baseline in our cohort
Increased size of the circles shows stronger correlation. Coefficients are derived using the Spearman's rank correlation coefficient. IL-6=interleukin 6. LDH=lactate dehydrogenase. TNFR1=tumour necrosis factor receptor superfamily member 1A.
Applying the parsimonious classifier model to the COVID-19 cohort resulted in a median probability for the hyperinflammatory classification of 0·03 (IQR 0·01–0·2), suggesting low prevalence of the phenotype in this population. Using a probability cutoff of 0·5 to assign phenotype, four (10%) of 39 patients were in the hyperinflammatory phenotype. With this cutoff, mortality at day 28 in the hyperinflammatory phenotype was 75% (three of four patients) and 40% (14 of 35 patients) in the hypoinflammatory phenotype (appendix p 5). Using the Youden index cutoff (0·274) to assign class led to eight patients (21%) being classified as the hyperinflammatory phenotype (table 2 ). It is worth noting that without LCA-derived phenotypes, it is not possible to ascertain which of the two cutoffs is more accurate. Given that more patients were in the hyperinflammatory phenotype using the Youden index cutoff, to enhance interpretability of comparative statistics, for the remainder of the manuscript only classifications using this cutoff are presented.
Table 2.
Difference in baseline characteristics between hypoinflammatory and hyperinflammatory phenotypes using a probability cutoff of 0·274 (Youden index) to assign class
| Hypoinflammatory (n=31) | Hyperinflammatory (n=8) | p value | ||
|---|---|---|---|---|
| Age, years | 57 (53–61) | 57 (46–60) | 0·55 | |
| Sex | 0·69* | |||
| Men | 19 (61%) | 6 (75%) | .. | |
| Women | 12 (39%) | 2 (25%) | .. | |
| Race | 0·38* | |||
| White | 17 (55%) | 2 (25%) | ||
| Asian | 6 (19%) | 3 (38%) | .. | |
| Black | 3 (10%) | 1 (13%) | .. | |
| Other† | 5 (16%) | 2 (25%) | .. | |
| Diabetes | 7 (23%) | 2 (25%) | 0·99* | |
| Hypertension | 6 (19%) | 0 | 0·31* | |
| Heart rate, beats per min | 98 (77–141) | 104 (97–144) | 0·44 | |
| Mean arterial pressure, mm Hg | 64 (61–71) | 70 (60–75) | 0·64 | |
| PaO2/FiO2, kPa | 18 (16–22) | 17 (11–21) | 0·27 | |
| Minute ventilation, L/min | 10·2 (9·4–11·3) | 10·6 (9·3–13·0) | 0·75 | |
| Plateau pressure, cm H2O | 31 (26–34) | 31 (28–34) | 0·98 | |
| Positive end-expiratory pressure, cm H2O | 12 (12–15) | 12 (11–15) | 0·83 | |
| Compliance, mL/cm H2O | 24 (20–28) | 27 (21–29) | 0·68 | |
| White blood cells, × 109 per L | 9·9 (7·6–12·2) | 10·6 (9·1–12·7) | 0·30 | |
| Lymphocytes, × 109 per L | 0·8 (0·6–1·1) | 1·1 (1·0–1·4) | 0·06 | |
| Platelets, × 109 per L | 272 (216–314) | 259 (197–314) | 0·48 | |
| Albumin, g/L | 23 (20–27) | 24 (22–25) | 0·96 | |
| Bilirubin, μmol/L | 10 (6–21) | 12 (8–28) | 0·55 | |
| Creatinine, μmol/L | 78 (63–130) | 216 (104–275) | 0·0217 | |
| Troponin, ng/L | 18 (5–29) | 23 (8–220) | 0·34 | |
| Lactate dehydrogenase, units per L | 439 (315–534) | 597 (534–758) | 0·0392 | |
| Procalcitonin, ng/mL | 0·9 (0·4–2·9) | 2·6 (1·6–10·5) | 0·14 | |
| Fibrinogen, g/L | 6·6 (6·0–6·8) | 5·8 (5·4–6·8) | 0·39 | |
| D-dimer, ng/mL | 1601 (873–4081) | 1643 (1126–3226) | 0·91 | |
| Ferritin, μg/L | 807 (422–1855) | 2878 (1229–4225) | 0·21 | |
| C-reactive protein, mg/L | 206 (145–304) | 255 (145–348) | 0·78 | |
| Vasopressor use (baseline) | 19 (61%) | 5 (63%) | 0·99* | |
| Invasive ventilation (baseline) | 28 (90%) | 7 (88%) | 0·76 | |
| Sequential organ failure assessment score | 6 (5–8) | 8 (6–10) | 0·10 | |
| APACHE II score | 12 (10–15) | 17 (16–18) | 0·0223 | |
| Mortality at day 28 | 12 (39%) | 5 (63%) | 0·26* | |
p values calculated by Wilcoxon signed-rank test unless noted otherwise. APACHE II=acute physiology and chronic health evaluation II. PaO2/FiO2=ratio of partial pressure of arterial oxygen to fractional concentration of oxygen in inspired air.
Fisher's exact test.
Includes Filipino and Romani.
As with previous studies, baseline APACHE II score was higher in patients with the hyperinflammatory phenotype (17 [16–18]) than in those with the hypoinflammatory phenotype (12 [10–15]; p=0·0223). Five (63%) of eight individuals with the hyperinflammatory phenotype had died at day 28 compared with 12 (39%) of 31 individuals with the hypoinflammatory phenotype; the difference between the two groups was not statistically significant (p=0·26; table 2).
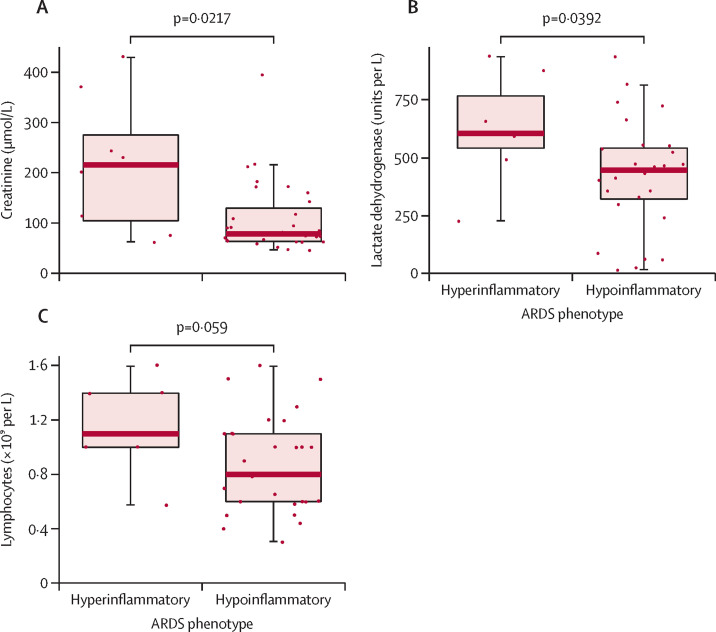
Baseline creatinine and lactate dehydrogenase were significantly higher in the hyperinflammatory than in the hypoinflammatory phenotype (figure 2A, B ). Lymphocyte counts were not significantly different between the groups, but were slightly lower in individuals with the hypoinflammatory phenotype (figure 2C). Values of D-dimer (1601 ng/mL [873–4081] in the hypoinflammatory subgroup vs 1643 ng/mL [1126–3226] in the hyperinflammatory subgroup; p=0·91) and C-reactive protein (206 mg/dL [145–304] vs 255 mg/dL [145–348]; p=0·78) were similar between the phenotypes. Vital signs and respiratory variables at baseline were also similar between the two phenotypes (table 2). In contrast to previous studies, in which vasopressor use was consistently greater on the hyperinflammatory phenotype,8, 9, 10, 11 use was similar between the two phenotypes: five (63%) of eight patients in the hyperinflammatory subgroup used vasopressors versus 19 (61%) of 31 in the hypoinflammatory subgroup (p=0·99).
Figure 2.
Comparison of measures of creatinine, lactate dehydrogenase, and lymphocytes in the hyperinflammatory and hypoinflammatory phenotypes of COVID-19-associated ARDS
Comparisons of creatinine (A), lactate dehydrogenase (B), and lymphocytes (C) between the hyperinflammatory and hypoinflammatory subgroups of the COVID-19 subset of the PHIND cohort. Phenotypes were assigned using the Youden index as the cutoff (≥0·274). Boxes show medians and IQRs; whiskers show the full range; and dots show individual observations. p values were calculated by Wilcoxon signed-rank test. ARDS=acute respiratory distress syndrome. PHIND=clinical evaluation of a point of care assay to identify PHenotypes IN the acute respiratory Distress syndrome.
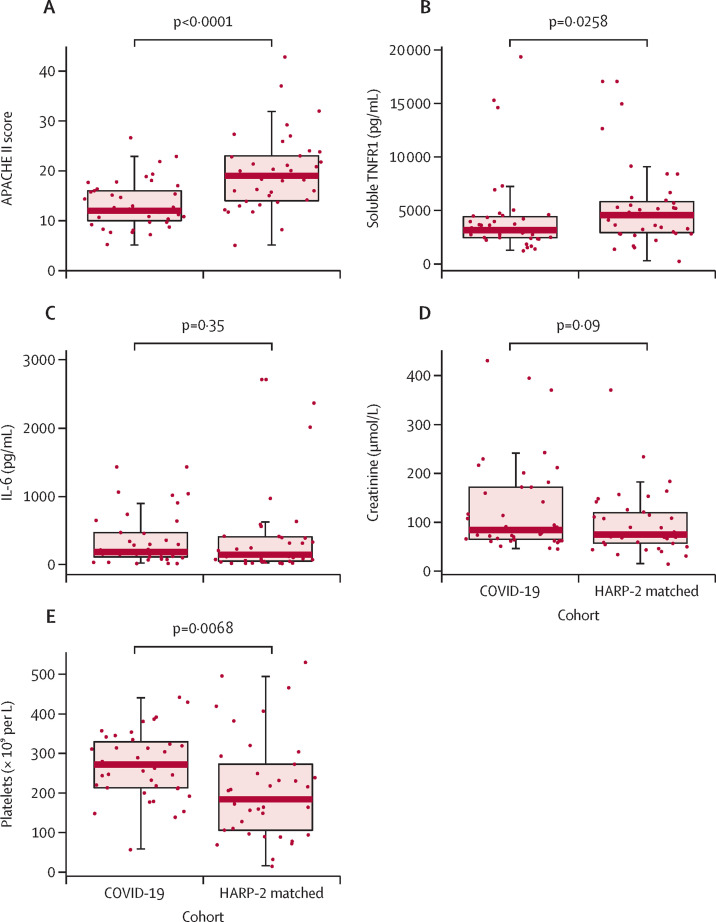
The entire HARP-2 cohort (n=539) had a similar age range (median 54 [IQR 42–66]) to the COVID-19 cohort (57 [52–61]). The median PaO2/FiO2 in HARP-2 was 15 kPa (11–21) compared with 18 kPa (15–21) in this study (p=0·07). Median APACHE II score in HARP-2 (18 [14–24]) was significantly higher than in this cohort (12 [10–16]; p<0·0001). Baseline PaO2/FiO2, sex, and age were used to match the COVID-19 cohort with patients in the HARP-2 cohort (n=39; appendix pp 6–7). Baseline characteristics of the entire HARP-2 cohort and the HARP-2 matched cohort are presented in the appendix (p 7). APACHE II score (p<0·0001; figure 3A ) and soluble TNFR1 (p=0·0258; figure 3B) were significantly higher in the HARP-2 matched cohort than in our COVID-19 cohort; IL-6 (p=0·35; figure 3C) and creatinine (p=0·09; figure 3D) were similar between the two cohorts; and platelets (p=0·0068; figure 3E) were significantly higher in our cohort than in the HARP-2 matched cohort.
Figure 3.
Comparison of patient characteristics in the COVID-19-related ARDS cohort and the HARP-214 matched cohort
Comparisons of APACHE II score (A) and measures of soluble TNFR1 (B), IL-6 (C), creatinine (D), and platelets (E) between the COVID-19 subset of the PHIND cohort and HARP-2 matched cohort. Boxes show medians and IQRs; whiskers show the full range; and dots show individual observations. p values were calculated by Wilcoxon signed-rank test. APACHE II=acute physiology and chronic health evaluation II. ARDS=acute respiratory distress syndrome. IL-6=interleukin 6. HARP-2=Hydroxymethylglutaryl-CoA reductase inhibition with simvastatin in Acute lung injury to Reduce Pulmonary dysfunction. PHIND=clinical evaluation of a point of care assay to identify PHenotypes IN the acute respiratory Distress syndrome. TNFR1=tumour necrosis factor receptor superfamily member 1A.
Despite the lower APACHE II score and similar PaO2/FiO2, mortality at day 28 in our COVID-19 cohort (17 [44%] of 39) was significantly higher than in the HARP-2 cohort (132 [24%] of 539; p=0·0128), and non-significantly higher than the HARP-2 matched cohort (11 [28%] of 39; p=0·16; table 3 ). Using the Youden index to assign phenotype, our COVID-19 cohort had a smaller proportion of patients classified in the hyperinflammatory phenotype (eight [21%] of 39) than both the entire HARP-2 cohort (186 [35%] of 539) and HARP-2 matched cohort (11 [28%] of 39). Mortality at day 28 in the hypoinflammatory phenotype in our COVID-19 cohort (12 [39%] of 31) was higher than in the two HARP-2 cohorts (59 [17%] of 353 in the whole cohort and six [21%] of 28 in the matched cohort; table 3). Notably, the mortality rate in the COVID-19 hypoinflammatory phenotype was similar to the rate in the hyperinflammatory phenotype in HARP-2 and HARP-2 matched (table 3). By contrast, the hyperinflammatory phenotype in the COVID-19 cohort had higher mortality rates than all other groups (five [63%] of eight).
Table 3.
Comparison of mortality at day 28 between the HARP-2 cohort,14 HARP-2 matched cohort, and COVID-19 PHIND cohort
|
Total cohort |
Hypoinflammatory |
Hyperinflammatory |
||||
|---|---|---|---|---|---|---|
| n | Mortality | n | Mortality | n | Mortality | |
| HARP-2 | 539 | 132/539 (24%) | 353/539 (65%) | 59/353 (17%) | 186/539 (35%) | 73/186 (39%) |
| HARP-2 matched | 39 | 11/39 (28%) | 28/39 (72%) | 6/28 (21%) | 11/39 (28%) | 5/11 (45%) |
| COVID-19 | 39 | 17/39 (44%) | 31/39 (79%) | 12/31 (39%) | 8/39 (21%) | 5/8 (63%) |
Data are n or n/N (%). In HARP-2 and HARP-2 matched cohorts, the phenotypes were derived from the original latent class analysis studies. In the COVID-19 subset of the PHIND cohort, the phenotypes were derived using the parsimonious model using a probability cutoff of 0·274 (Youden index). HARP-2=Hydroxymethylglutaryl-CoA reductase inhibition with simvastatin in Acute lung injury to Reduce Pulmonary dysfunction. PHIND=clinical evaluation of a point of care assay to identify PHenotypes IN the acute respiratory Distress syndrome.
A sensitivity analysis was done by excluding the patients from University College Hospital and the findings were similar to those presented (data not shown).
Discussion
To our knowledge, this study is the first that has sought to identify the prevalence of previously described ARDS phenotypes in patients with COVID-19-associated ARDS. The findings of this preliminary study of 39 patients with COVID-19-associated ARDS suggest that the prevalence of the hyperinflammatory phenotypes was low in our cohort (10–21%). Mortality rates were about 20% higher in patients with the hyperinflammatory phenotype than in those with the hypoinflammatory phenotype, which is similar to previous findings for patients with ARDS. However, although the magnitude of difference in mortality between the phenotypes was consistent, the mortality rate for both phenotypes was considerably higher in the COVID-19 cohort than in historical ARDS data.8, 9, 10, 11 A second novel feature of the study was the use of a rapid point-of-care assay to quantify both IL-6 and soluble TNFR1, the levels of which were similar or lower in our patients with COVID-19-associated ARDS than in patients with ARDS in HARP-2.
The hyperinflammatory phenotype of ARDS is associated with higher circulating levels of proinflammatory biomarkers such as IL-6, IL-8, and soluble TNFR1 and lower levels of vitamin K-dependent protein C.8, 9, 10, 11 Further, this phenotype is associated with increased evidence of multiorgan failure and shock.8, 9, 10, 11 The low prevalence of the hyperinflammatory phenotype in COVID-19 ARDS challenges the hypothesis of the cytokine storm in its pathogenesis and suggests that it might not be as ubiquitous as purported, and might be less frequently encountered than in ARDS secondary to other causes.
The high mortality rate in the hypoinflammatory phenotype in COVID-19 is a notable and novel finding of this study. In previous studies, mortality in patients with the hypoinflammatory phenotype was about 20%.8, 9, 10, 11, 16 However, the mortality of patients with COVID-19 and the hypoinflammatory phenotype in our study was nearly double that. Coupled with the lower burden of systemic inflammatory responses measured by IL-6 and TNFR1, the findings of higher mortality rates in COVID-19-associated ARDS suggests severity of pathogenesis not captured by these inflammatory biomarkers. The differences in mortality compared with patients with pneumonia in the HARP-2 matched cohort, in which the infective pathogen is more likely to be bacterial, might allude to the pathogenesis of SARS-CoV-2 and an absence of therapeutic options for source control in COVID-19 ARDS. A second factor to consider is whether attributable mortality in these patients differs. In ARDS unrelated to COVID-19, multiorgan failure is frequently encountered as the attributable factor for death,17 whereas in COVID-19, reports suggest that a greater proportion of patients die because of respiratory failure,13 a physiological abnormality that might be pathologically independent of systemic inflammation and subject to more localised injury to the lungs.
It is also worth noting that the APACHE II scores in our COVID-19 population were significantly lower than those in the HARP-2 cohort despite higher mortality in our cohort. All patients with COVID-19 in our study were managed in ICUs at surge capacity with a reduced nursing ratio, which might, in part, explain this finding. Overwhelmed ICU capacity might have an effect on outcomes in COVID-19 and lower mortality rates have been reported in ICUs that have operated under more conventional conditions and staffing ratios in patients with COVID-19 with similar APACHE II scores.18, 19 The low APACHE II scores are also in keeping with those reported by the Intensive Care National Audit and Research Centre in 9777 patients admitted to the ICU in the National Health Service hospitals in the UK,20 where the median APACHE II score in patients with COVID-19 was 14 (IQR 11–18) and the mortality rate was greater than 40%. These consistent findings suggest that the APACHE II score might not be valid for prognostication in COVID-19. Taken together, the findings of the low APACHE II score and high mortality suggest that alternative phenotyping approaches might be needed to identify biologically and clinically homogeneous clusters using novel biomarkers that might, in turn, enhance our understanding of pathogenesis and improve prognostication in COVID-19-related ARDS.
One advantage of specifically studying the COVID-19 population is that the heterogeneity of the cause, a common feature of ARDS unrelated to COVID-19, is largely negated. Notably, the prevalence of vasopressor use at baseline was similar between patients with the hyperinflammatory phenotype and those with the hypoinflammatory phenotype, whereas in previous studies of ARDS unrelated to COVID-19, vasopressor use was significantly higher in those with the hyperinflammatory phenotype.8, 9, 10, 11 This might in part be explained by the fact that in previous studies, the risk factor for ARDS differed between the phenotypes with sepsis predominantly featuring in the hyperinflammatory phenotype. In COVID-19, given the uniformity of cause, it might be that there are additional drivers of vasopressor use that are disease specific and extraneous to inflammatory phenotypes, such as cardiovascular complications.21
It is also known that cause is an important determinant of the signature of circulating biomarkers.22 For example, indirect causes of lung injury, such as sepsis, are associated with higher levels of endothelial injury, whereas direct lung injury is associated with higher levels of markers of epithelial injury.23 Biomarkers pertaining to severity of epithelial injury and cell death might be more informative in COVID-19-associated ARDS because the primary source of injury is presumed to be a viral pneumonitis. In two recent case series of autopsies of patients with severe COVID-19, the only common findings in all patients across both studies was diffuse alveolar damage.24, 25 However, this theory remains speculative, and it stands to reason that before phenotyping, comprehensive typing of COVID-19 and its biological signature using data is needed, preferably from large multinational collaboratives such as ISARIC 4C by the International Severe Acute Respiratory and Emerging Infection Consortium.
Another strength of this study has been to show the logistical feasibility of rapid point-of-care phenotyping of patients in a busy ICU using a novel bioanalyser. Precision-based care has been a promising yet elusive opportunity in critical care medicine.26 Although other specialties have more time, in the ICU, any phenotype-based decisions need to be made rapidly. The time taken to do ELISA-based assays is prohibitive in the clinical implementation of biomarker-driven phenotypes.22 Using this novel solid state-based analysing technology, we were able to classify patients into biomarker-driven phenotypes in less than 1 h from sample acquisition. Bicarbonate can be easily measured using standard clinical laboratory assays. The availability of such assays has important implications for future precision medicine studies in critical care.
Paradoxically, this strength is also a limitation of the study. The larger PHIND study, from which this COVID-19 subset was derived, was designed to further validate the point-of-care platform. The platform has only been validated using stored plasma samples, and its performance using real samples from patients in the ICU is yet to be formally validated. Given this uncertainty, the findings of this study should be interpreted with caution. The clinically measured biomarker component of the model, namely bicarbonate, can often be informative of the validity of the distribution of the phenotypes. In a previous ARDS cohort,11 in which the prevalence of the hyperinflammatory phenotype was 37%, the mean serum bicarbonate level was 22 mmol/L (SD 6) compared with the 27 mmol/L (6) in our COVID-19 cohort. On the basis of this comparison, the estimated prevalence of the hyperinflammatory phenotype between 10% and 20% in this cohort seems accurate.
The key limitation of this study is the small sample size. The even smaller number in the hyperinflammatory phenotype and the observed sample size imbalance when comparing phenotypes makes comparative statistics difficult to interpret, and differences between groups must be interpreted with caution. A further limitation of the study is that it is focused on baseline data only for phenotype classification. The natural progression of COVID-19 over time might lead to changing phenotypes and requires further study. Another important limitation is that only circulating levels of two biomarkers were studied, whereas in previous work we studied six to eight protein biomarkers. Inflammatory markers might differ more substantially in the lungs. In addition, if a larger number of plasma inflammatory biomarkers were studied in a larger population, more distinct patterns of differences in the inflammatory response might have been detected. Further, we were unable to validate the biomarkers quantified using the Multistat analyser against conventional ELISAs because of an absence of stored plasma samples from patients with COVID-19. Future studies of COVID-19 pneumonia, where feasible, should study the circulating plasma and lung compartments simultaneously and over the course of COVID-19 critical illness.
In summary, in this small exploratory analysis of 39 patients, the prevalence of the hyperinflammatory phenotype in patients with COVID-19-associated ARDS was lower than in patients with ARDS unrelated to COVID-19 in a previous study. This finding suggests that, compared with other causes of ARDS, the excessive mortality in COVID-19-related ARDS is unlikely to be due to upregulation of the inflammatory pathways described by the parsimonious model. Finally, with the caveat that the findings require validation with LCA-derived phenotypes, the point-of-care platform used to classify phenotypes at the bedside shows the feasibility of phenotype-informed trials in the ICU.
Data sharing
Requests for data will be reviewed on an individual basis by the PHIND chief investigator (DFM) and the Clinical Trials Unit (CTU). After the publication of primary and secondary outcomes from the overall PHIND trial, requests for data should be made in writing to the chief investigator via the CTU, who will discuss such requests with the study sponsor.
Acknowledgments
Acknowledgments
This publication presents independent research funded by the US NIH, grant numbers GM008440–21 (PS) and HL140026 (CSC), and Innovate UK (reference 104639). Randox funded the development of the point-of-care assay. MS-H is funded by an NIHR Clinician Scientist Award. ACG is funded by an NIHR Research Professorship (RP-2015-06-18) and supported by the NIHR Imperial Biomedical Research Centre. We thank the staff of the Northern Ireland Clinical Trials Unit for their support in conducting the study. We thank Jeremy Parker, John Lamont, and the staff at Randox for their role in the development of the point-of-care assay. We would also like to thank the patients and staff at the two hospitals from which these data originated: The Royal Gwent Hospital, Newport and University College Hospital, London. The views expressed are those of the author(s) and not necessarily those of the UK National Health Service, the NIHR, or the UK Department of Health and Social Care.
Contributors
All authors conceived and designed the study. SCh, DB, SCu, CKin, CKil, OR, YC, CB, and TS collected the data. All authors contributed to data analysis and interpretation. PS drafted the manuscript and all authors contributed to revisions and approved the final version.
Declaration of interests
CSC reports grants from the US National Institutes of Health (NIH), during the conduct of the study; grants from Roche/Genentech and Bayer, outside the submitted work; and personal fees for consultancy from Quark Pharmaceuticals, Vasomune, and Gen1e Life Sciences, outside the submitted work. ACG reports funding through a UK National Institute for Health Research (NIHR) Research Professorship, during the conduct of the study; and fees for consultancy, paid to his institution, from Bristol-Meyers Squibb and GlaxoSmithKline (GSK), outside the submitted work. CMO'K reports grants from Innovate UK, during the conduct of the study; and grants from NIHR, the Wellcome Trust, the UK Medical Research Council, Northern Ireland (NI) Health and Social care R&D Division, NI Chest Heart and Stroke, and Medical Research Council, outside the submitted work. DFM reports a grant from Innovate UK for the conduct of the phenotypes in the acute respiratory distress syndrome (PHIND) study; and personal fees for consultancy from GSK, Boehringer Ingelheim, and Bayer, outside the submitted work. DFM's institution has received grants from the NIHR, the Wellcome Trust, NI Health and Social care R&D Division, NI Chest Heart and Stroke, and Medical Research Council; DFM has a patent issued to his institution for a treatment for acute respiratory distress syndrome. DFM is a Director of Research for the Intensive Care Society and NIHR Efficacy and Mechanism Evaluation Programme Director. All other authors declare no competing interests.
Supplementary Material
References
- 1.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatraju PK, Ghassemieh BJ, Nichols M. Covid-19 in critically ill patients in the Seattle region—case series. N Engl J Med. 2020;382:2012–2022. doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G, Zangrillo A, Zanella A. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright DJM. Prevention of the cytokine storm in COVID-19. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30376-5. published online May 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Tan Y, Ling Y. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 6.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.3313. published online June 20. [DOI] [PubMed] [Google Scholar]
- 8.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calfee CS, Delucchi KL, Sinha P. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med. 2018;6:691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Famous KR, Delucchi K, Ware LB. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017;195:331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha P, Delucchi KL, Thompson BT, McAuley DF, Matthay MA, Calfee CS. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med. 2018;44:1859–1869. doi: 10.1007/s00134-018-5378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinha P, Delucchi KL, McAuley DF, O'Kane CM, Matthay MA, Calfee CS. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med. 2020;8:247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker JC, Higgins M, Torrisi M, et al. Development of a point-of-care (POC) theranostic assay to stratify patients with acute respiratory distress syndrome (ARDS). European Respiratory Society research seminar: personalised medicine in acute respiratory distress syndrome. Barcelona, Spain; Jan 30–31, 2020 (poster).
- 14.McAuley DF, Laffey JG, O'Kane CM. Simvastatin in the acute respiratory distress syndrome. N Engl J Med. 2014;371:1695–1703. doi: 10.1056/NEJMoa1403285. [DOI] [PubMed] [Google Scholar]
- 15.Ranieri VM, Rubenfeld GD, Thompson BT. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 16.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stapleton RD, Wang BM, Hudson LD, Rubenfeld GD, Caldwell ES, Steinberg KP. Causes and timing of death in patients with ARDS. Chest. 2005;128:525–532. doi: 10.1378/chest.128.2.525. [DOI] [PubMed] [Google Scholar]
- 18.Shime N, Bunya N, Endo T. Save the ICU and save lives during the COVID-19 pandemic. J Intensive Care. 2020;8:40. doi: 10.1186/s40560-020-00456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanderburg S, Alipanah N, Crowder R. Management and outcomes of critically-ill patients with COVID-19 pneumonia at a safety-net hospital in San Francisco, a region with early public health interventions: a case series. medRxiv. 2020 doi: 10.1101/2020.05.27.20114090. published online May 29. [DOI] [Google Scholar]
- 20.Intensive Care National Audit And Research Centre . Intensive Care National Audit and Research Centre; London: 2020. ICNARC report on COVID-19 in critical care 12 June 2020.https://www.icnarc.org/DataServices/Attachments/Download/7e1a720c-dcac-ea11-9126-00505601089b [Google Scholar]
- 21.Guo T, Fan Y, Chen M. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. Curr Opin Crit Care. 2019;25:12–20. doi: 10.1097/MCC.0000000000000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calfee CS, Janz DR, Bernard GR. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest. 2015;147:1539–1548. doi: 10.1378/chest.14-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann M, Verleden SE, Kuehnel M. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaller T, Hirschbühl K, Burkhardt K. Postmortem examination of patients with COVID-19. JAMA. 2020;323 doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabaudon M, Blondonnet R, Audard J. Recent directions in personalised acute respiratory distress syndrome medicine. Anaesth Crit Care Pain Med. 2018;37:251–258. doi: 10.1016/j.accpm.2017.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for data will be reviewed on an individual basis by the PHIND chief investigator (DFM) and the Clinical Trials Unit (CTU). After the publication of primary and secondary outcomes from the overall PHIND trial, requests for data should be made in writing to the chief investigator via the CTU, who will discuss such requests with the study sponsor.