Abstract
Extracellular vesicles (EVs)—nanoscale phospholipid vesicles secreted by cells—present new opportunities for molecular diagnosis from non-invasive liquid biopsies. Single EV protein analysis could be extremely valuable in studying EVs as circulating cancer biomarkers, but it is technically challenging due to weak detection signals associated with limited amounts of epitopes and small surface areas for antibody labeling. Here, we report a new, simple method that enables multiplexed analyses of EV markers with improved sensitivities. Specifically, we implement plasmon-enhanced fluorescence detection that amplifies fluorescence signals using surface plasmon resonances excited by periodic gold nanohole structures. We showed fluorescence signals in multiple channels are amplified by one order of magnitude, and both transmembrane and intravesicular markers can be detected at the single EV level. This approach could offer additional insight into understanding subtypes, heterogeneity, and production dynamics of EVs during disease development and progression.
Keywords: Plasmon-enhanced fluorescence, Nanoholes, Extracellular vesicles, Biosensing, Cancer
Graphical Abstract
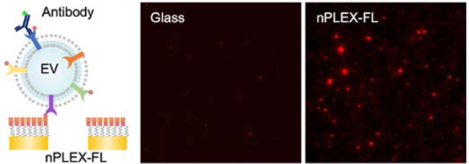
Plasmon-enhanced fluorescence detection on plasmonic Au nanohole arrays (nPLEX-FL) enables sensitive, multiplexed analysis of extracellular vesicles. Cell-derived EVs are captured on Au nanohole arrays and labeled by fluorophore-conjugated antibodies. The fluorescence signals are then amplified by surface plasmon resonances excited by the underlying Au nanoholes, which lead to sensitive single EV detection.
Extracellular vesicles (EVs) present new opportunities as circulating biomarkers for cancers[1], cardiovascular[2], neurodegenerative[3], and infectious diseases[4] among others. These cell-derived phospholipid vesicles are abundantly present in various bodily fluids (e.g., blood, cerebrospinal fluid, urine, saliva)[5]. More importantly, they carry a variety of biomolecules (lipids, proteins, and genetic materials) originating from their parental cells, which can be harnessed as a minimally invasive means to probe the molecular status of their cellular origins[6–8].
In further exploiting EVs’ potential and accelerating their clinical adaptation, a critical unmet need is to develop sensitive, robust, and standardized assays that can determine the composition and molecular profiles of EVs in clinical samples. However, their unique sizes (50–1000 nm) impose technical challenges in conventional analytical methods, which often lead to variable findings. Conventional methods for protein analyses (e.g., Western blotting, enzyme-linked immunosorbent assay/ELISA) require large amounts of samples and involve time-consuming and extensive processing steps, making them impractical in the clinical settings. Developing new EV molecular profiling platforms is thus a pivotal mandate to ultimately translate EVs into clinically relevant biomarkers[9]. To address these challenges, we previously developed a nanoplasmonic sensing platform, termed nPLEX (nano-plasmonic exosome), based on transmission surface plasmon resonance through periodic nanohole gratings[7,8,10]. In the previous studies, we showed that the nPLEX sensors could rapidly and sensitively detect tumor-derived EVs directly from clinical samples. Although promising, fundamental limitations remain in the current nPLEX system and other state-of-the-art EV sensing technologies: i) sensitivity limited to bulk analyses; ii) necessities of EV lysis for detecting markers inside of EVs; iii) lack of multiplexed analysis in single EVs.
Analyzing single EVs could reveal unique molecular profiles of cell-specific EVs, which will further promote clinical use of these vesicles and allow us to construct a comprehensive EV atlas per different biological parameters (e.g., cellular origin, cell state). Fluorescence detection and interferometric imaging now enable visualization of individual EVs captured on a solid substrate[11–14]. However, multiplexed molecular profiling of single EVs, especially for low abundant protein and intravesicular markers, often requires sophisticated multi-step signal amplification strategies, such as branched DNA probes[15] or enzymatic reactions[16], otherwise undetected due to weak signals associated with limited amounts of epitopes and small surface areas for antibody labeling.
Here, we report a next-generation nPLEX assay that enables multiplexed single EV analyses of target membrane and intravesicular markers with improved sensitivities. Specifically, we implement plasmon-enhanced fluorescence detection, which can amplify fluorescence signals using plasmonic metallic nanostructures[17], for sensitive, multi-channel EV biomarker profiling. The enhancement is achieved by simply using a substrate with periodic gold (Au) nanoholes instead of conventional glass or other plain substrates. Periodic Au nanohole arrays offer several advantages: 1) periodic nanohole gratings support surface plasmon resonances extended in a long range (~100 nm), more suitable for EVs, while many novel nanostructures (e.g., nanoparticles, nanostructures) support localized surface plasmon resonance (LSPR) tightly confined on the surface (<20 nm)[18]; 2) the resonance wavelength can be readily tuned by adjusting the nanohole periodicity[19]; 3) wafer-scale, high-throughput chip fabrication methods have been established [7,10]. Named nPLEX-FL (nano-plasmonic extracellular vesicle analysis with enhanced fluorescence detection), the new system provides a simple, robust signal amplification strategy that is crucial to improve the detection sensitivity and achieve multiplexed single EV analysis.
nPLEX-FL technology
Figure 1A shows the nPLEX-FL strategy for multiplexed single EV analysis. We first capture EVs on the Au nanohole surface via affinity ligands (e.g., capturing biotinylated EVs on avidin-coated Au nanohole surface). The captured EVs are then immunostained by fluorescently-labeled antibodies in different color channels (typically 3 – 4 colors). Depending on the absorption and emission spectra of fluorophores, the fluorescence signals are amplified by surface plasmon resonances (SPR) excited by the underlying Au nanohole structures. We chose previously optimized nanohole structures[7] as a SPR substrate. The hole diameter is 200 nm and the periodicity is 500 nm in a 100-nm thick Au film (Figure 1B). The periodic nanohole granting on the chip surface concentrates electromagnetic fields with the maximum field intensity up to 300-fold (Figure 1C). The resonance fields extend to 110 nm in the z-direction, which mostly covers small EVs (e.g., exosomes with an average diameter of 100 nm). In addition to the localized near field, the fluorescence radiation can be further enhanced by the interaction of the Au nanostructure with proximal fluorophores in the resonance range[20].
Figure 1.
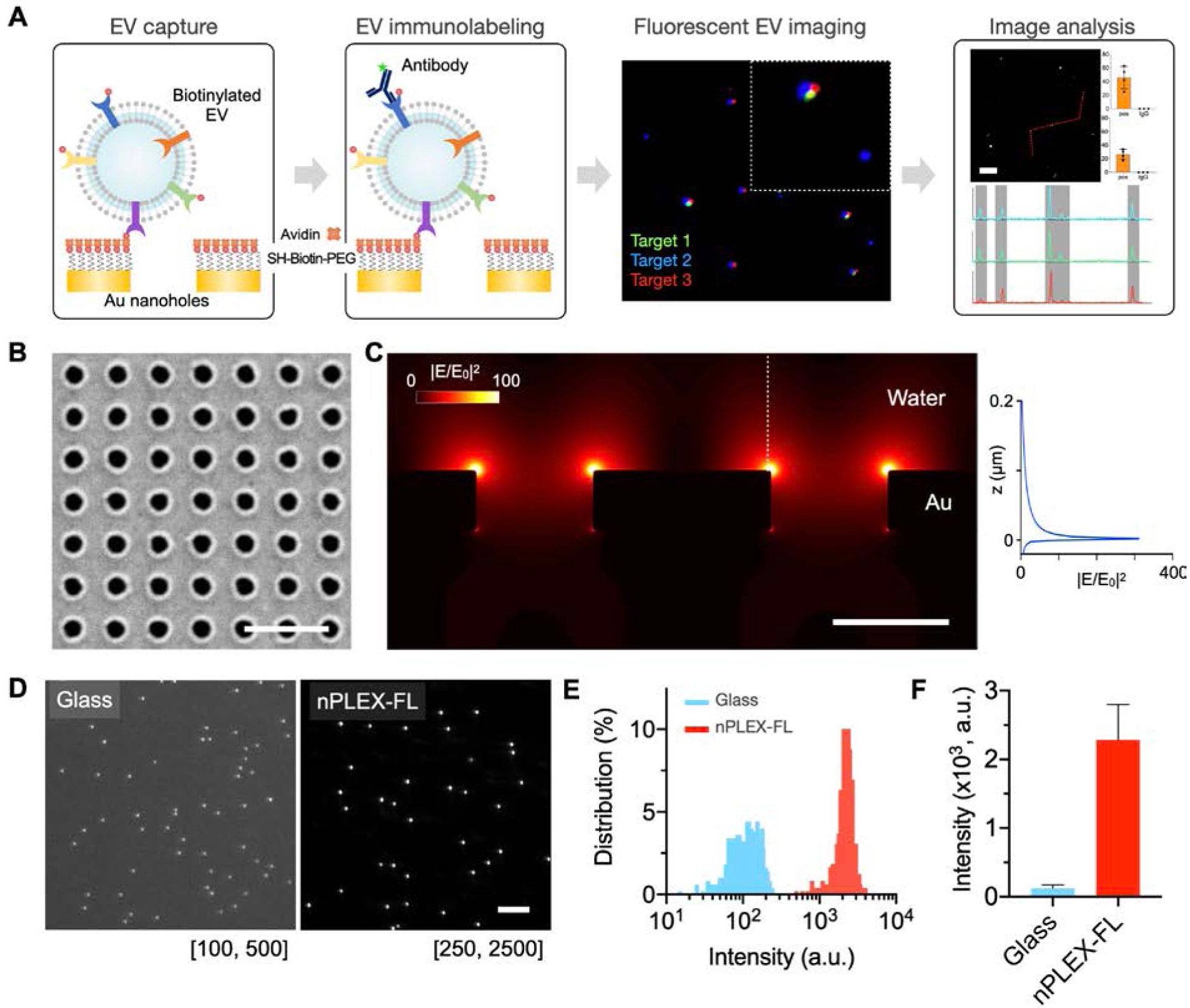
nPLEX-FL technology for single EV analysis. A) Overview of nPLEX-FL assay procedures. EVs are captured on the nanohole surface and immunostained by fluorescent detection probes. The labeled EVs are imaged in different fluorescence channels, and their intensities are analyzed. B) A scanning electron micrograph of periodic nanoholes in the nPLEX-FL sensor chip. The hole diameter is 200 nm and the periodicity is 500 nm. Scale bar, 1 μm. C) Finite-difference time-domain simulation shows the enhanced electromagnetic fields confined on the nanohole surface. The strong fields are responsible for plasmon-enhanced fluorescence signals. D) Representative images of fluorescent nanospheres (Cy5, 200 nm) on glass and nPLEX-FL substrates. Scale bar, 10 μm. E) Histograms of pixel intensities. F) Mean fluorescence intensity of fluorescent nanospheres on glass and nPLEX-FL substrates.
We first examined the plasmon-enhanced fluorescence by Au nanohole structures using fluorescent nanospheres (Cy5, 200 nm) in comparison with a glass substrate (Figure 1D). We showed that the fluorescence intensities of individual nanospheres were significantly higher on the nPLEX-FL substrate than those on the glass substrate (two-tailed t-test, p < 0.0001, Figure 1E); the mean fluorescence intensity of nanospheres was increased by a factor of 18 (Figure 1F), and the signal-to-noise ratio (given by a signal divided by 3-times standard deviation of blank) was increased by a factor of 20, from 17.7 (glass) to 358 (nPLEX-FL). There is no significant difference in the coefficient of variation (the ratio of the standard deviation to the mean) for fluorescence intensities between the glass (36.2%) and nPLEX-FL substrates (33.6%), indicating the signal amplification does not increase the intensity variation.
Characterization of nPLEX-FL chips
We next investigated the plasmon enhancement in different fluorescence channels using a molecular monolayer. We functionalized the Au nanohole surface using thiolated biotin polyethylene glycol derivatives (thiol-PEG-biotin) and then incubated fluorophore-conjugated streptavidin molecules on the biotinylated Au surface. To prevent fluorescence quenching by underlying Au substrates, we functionalized Au surface with thiol-PEG-biotin (1kDa, 6–8 nm) and avidin (60kDa, 4–5 nm), which resulted in an adhesion layer of 10–13 nm in thickness[21]. Figure 2A shows the fluorescence images of nPLEX-FL chips coated with four different colors of fluorophore-conjugated streptavidin (AF488, Cy3, Cy5, Cy5.5). Strong signal enhancements were observed in the 100 × 100 μm2 sized square area of nanohole gratings (highlighted by a white dashed box) compared to the flat Au area (outside of the square, Figure 2B). The signal enhancement was most dominated in the Cy5 channel; the Cy5 fluorescence signals on the nanoholes were 23-fold higher than signals on the flat Au area (Figure 2C). The Cy5.5 and Cy3 intensities were also increased by 17 and 9-fold, respectively. On the contrary, the AF488 signal was only increased by 3-fold. The observed various enhancement factors in the different channels could be explained by spectral overlaps[22] between the plasmon-supported light transmission through nanoholes and the absorption/emission spectra of fluorophores (Figure 2D). The light transmission peak (667 nm) of the nanohole array coincided with the Cy5 spectral peaks (absorption/emission peaks at 649/666 nm) the most, followed by Cy5.5 and Cy3.
Figure 2.
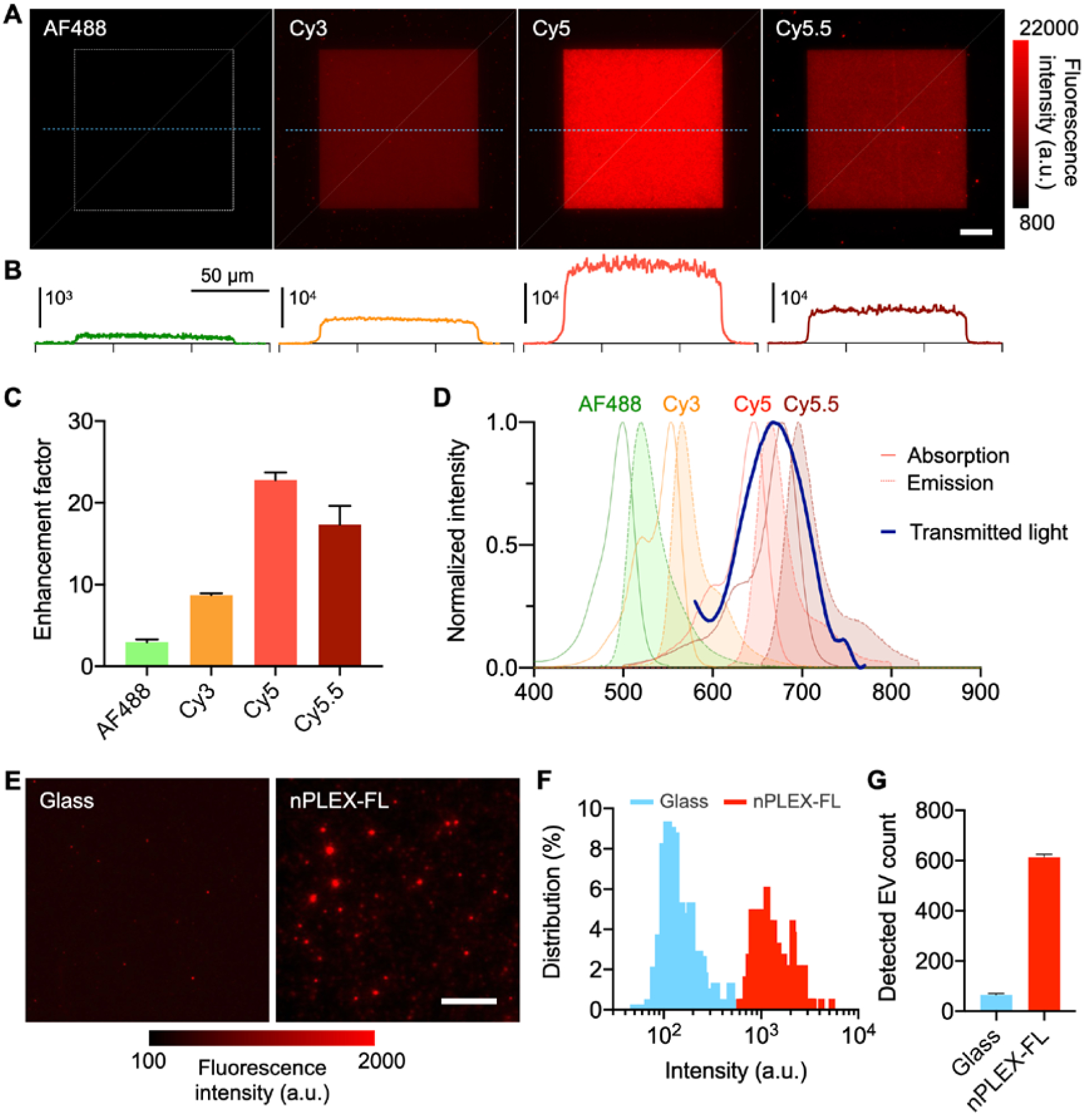
System characterization. A) Fluorescence images of nPLEX-FL chips coated with four different colors of fluorophore-conjugated streptavidin (streptavidin with AF488, Cy3, Cy5, or Cy5.5). Scale bar, 20 μm. Au nanoholes are made in the 100 × 100 μm2 sized square area highlighted by a white dashed box. B) Cross-sectional intensity profiles along the blue dashed lines. C) Enhancement factors of fluorescence intensity in different fluorescence channels (the nanohole area vs. flat Au areas). D) Plasmon-supported light transmission spectrum through periodic nanoholes overlaid with absorption/emission spectra of fluorophores E) Biotinylated EVs were captured on glass and nPLEX-FL substrates coated with the DOPA-based bioadhesive. The captured EVs were labeled with Cy5-conjugated streptavidin and imaged. Scale bar, 10 μm. F-G) Histograms of pixel intensities (F) and the number of detected EVs. G) between glass and nPLEX-FL substrates.
We further investigated plasmonic enhancements in EVs. We captured biotinylated EVs on glass and nPLEX-FL substrates, and subsequently labeled the captured EVs with streptavidin-conjugated dyes (Cy5, Figure 2E and AF488, Figure S1). We used a polyphenolic proteins-based bioadhesive layer to capture the same amounts of EVs on different substrates (glass and Au) and investigated fluorescence intensities and detectable EV counts. The averaged signal enhancement factors in terms of fluorescence intensity after background correction were measured to be 1.54 for AF488 and 8.60 for Cy5 (Figures 2F). The overall signal enhancement in the captured EVs was less prominent than the streptavidin monolayer coating (c.f. Figure 2c and f), likely because of the thickness difference between EVs and streptavidin monolayer; the electromagnetic fields are stronger near the surface (Figure 1c). Nevertheless, we could detect an order-of-magnitude larger number of Cy5 labeled EVs on the nPLEX-FL chip compared to a glass substrate, indicating higher sensitivity attained by the plasmon-enhanced signal amplification (Figure 2G). We observed comparable mean pixel intensities and EV counts for the AF488-labeled EVs on both nanohole chip and glass (Figure S1). This indicates that the plasmon enhancement on Cy5 dyes unveils EVs with weak fluorescence signals otherwise undetected without signal enhancement (glass substrates) or with weak enhancement (AF488). Hence, we assign low abundant or key EV markers in the Cy5 channel in the subsequent validation study for the maximum signal enhancement.
Multiplexed single EV analyses
Next, we applied the nPLEX-FL technology to demonstrate its feasibility on the multiplexed single EV analysis. We used glioblastoma cell lines for testing: Gli36-WT and Gli36-EGFRvIII (overexpressing human EGFRvIII[23]). EGFR and EGFRvIII are biomarkers of interest for glioblastoma as amplification of EGFR and its variant (EGFRvIII) occur frequently in glioblastoma. The presence of protein markers including i) ubiquitous EV tetraspanin combination named CD-pan (CD9, CD63, and CD81), ii) GAPDH, iii) EGFR and iv) EGFRvIII was examined by nPLEX-FL and benchmarked against Western blotting analysis as a standard method (Figure S2).
EVs were isolated from conditioned cell culture media. Nanoparticle tracking analysis showed that the isolated EVs used in this study have a size distribution ranging 50–200 nm with an average diameter of 100 nm, also confirmed by transmission electron micrographs (Figure S3). The isolated EVs were biotinylated, diluted in pure buffer (1–10 × 108 EVs per mL PBS), and captured on the neutravidin-coated gold nanohole surface (Figure S4). The captured EVs were immunolabeled against membrane (i.e., CD63, EGFR) and/or intravesicular markers (i.e., GAPDH) and imaged under a fluorescence microscope. Because most EVs are smaller than the diffraction limit, the average blob size of the detected vesicles in fluorescence images was about 500 nm (8 pixels with a pixel size of 63 nm, Figure S5). Single EVs generated detectable fluorescence signals, confirmed by scanning electron micrograph (Figure S6); some doublet EV showed a higher intensity in the streptavidin channel. Particles imaged larger than 1 μm (or 16 pixels) were considered large aggregates and excluded in our analysis.
We chose well established EV markers for a proof-of-principle demonstration of EV profiling and subpopulation sorting based on marker signals. In consideration of fluorescence signal enhancement, we assigned (i) green dye (AF488) to high abundance/easy-to-detect markers and (ii) far-red dye (Cy5) to low abundance/hard-to-detect markers. Figure 3A shows representative nPLEX-FL images of biotinylated EVs labeled against CD-pan (AF488), streptavidin (Cy3), and GAPDH (Cy5). We chose GAPDH as a representative intravesicular marker, which is commonly used as a control for many other quantitative methods (e.g., Western blotting, qPCR). We varied EV concentrations and counted the number of captured EV (Figure S7). Line scan (Figure 3B) shows high signal-to-noise ratios and signal heterogeneity for the chosen markers on individual vesicles. We then analyzed the raw intensity data for marker profiling of EVs. For a given marker, we identified two subpopulations— marker-positive and marker-negative—which can be separated by the intensity cutoff (mean + 2 × standard deviation of negative controls). Roughly 40% the captured streptavidin-positive vesicles was CD-pan positive, and of the CD-pan-positive EVs, a fraction expressed GAPDH (25%) (Figure 3C). The false-positive rate in a control sample (no EV) was negligible for both streptavidin staining (<1%) and antibody staining (<0.2%). Based on the negative control data, we set a threshold of 1% for positivity.
Figure 3.
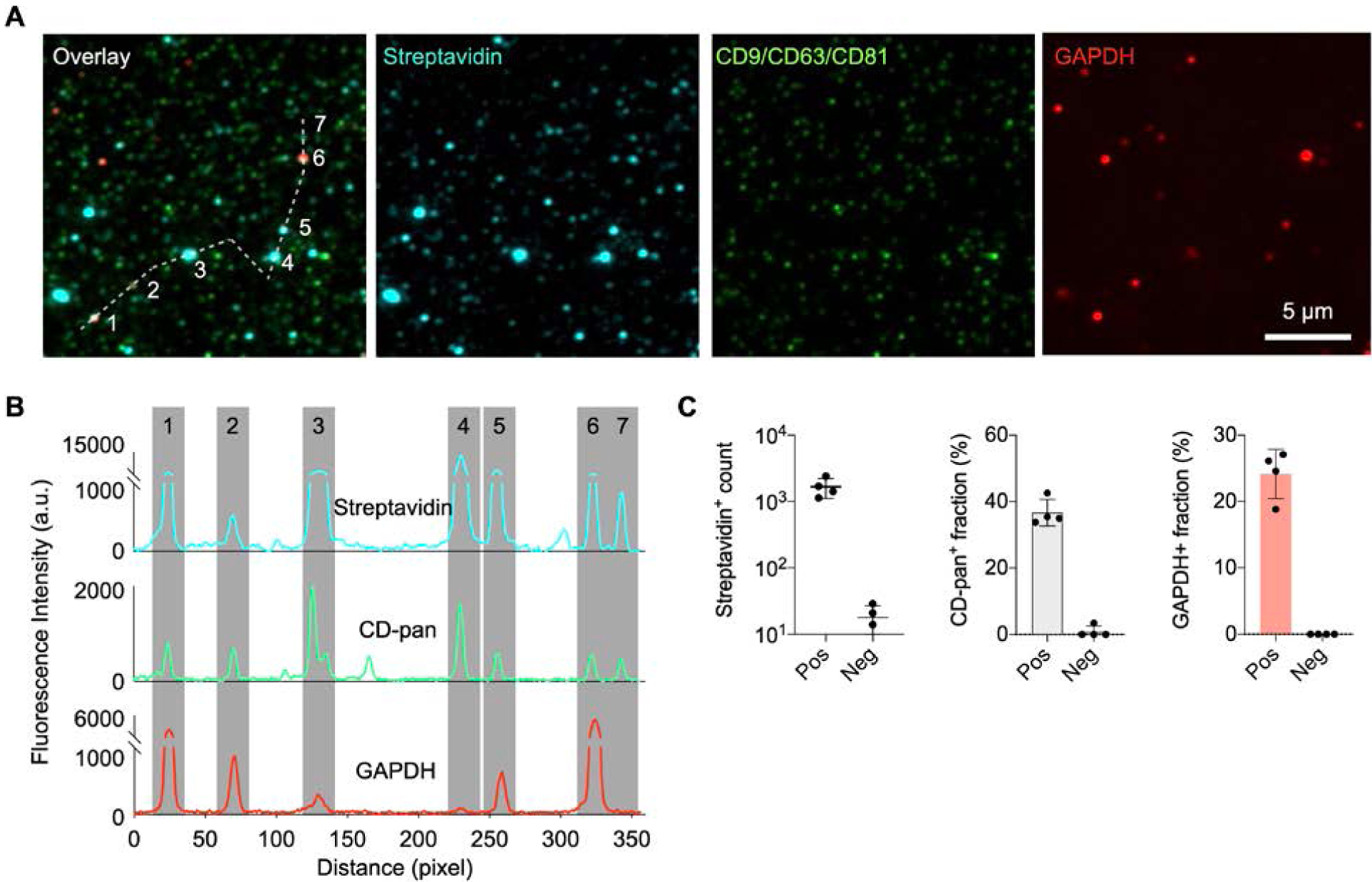
Single EV measurements using the nPLEX-FL assay. A) EVs from the Gli36-WT cell line were biotinylated and captured on the nPLEX-FL substrate. Individual EVs were detected through staining with Cy3-streptavidin. For molecular profiling, EVs were labeled with fluorescent antibodies against CD9/CD63/CD81 EV markers (CD-pan, AF488) and intravesicular markers (GAPDH, Cy5). Three-channel fluorescence images were overlaid on the top left. To help visualize, EVs were artificially color-coded. B) Line profiles of fluorescence intensity through a shite dashed line in (A) showing high signal-to-noise for the selected markers. Gray shading highlights EV positions. C) EV detection and marker profiling. A negative control was prepared with the same procedure with no EV incubation.
Proof-of-principle testing
To test the diagnostic potential for clinical applications, we spiked ~1010 EVs from Gli36-WT and Gli36-EGFRvIII cell lines into 1 mL human plasma samples. EVs were isolated from the spiked plasma samples using a size exclusion column (IZON column), biotinylated, and then loaded onto the chip (1–5 μL). The captured EVs were labeled against CD-pan (AF488), streptavidin (Cy3), and EGFR or EGFRvIII (Cy5). We implemented our decision tree algorithm with a nested gating strategy to classify EV populations based on EGFR and EGFRvIII signals (Figure 4A). Briefly, particles labeled with Cy3-conjugated streptavidin were first detected and prescreened by size exclusion (< 1 μm) to exclude large aggregates from the analysis. Among particles positive for streptavidin, we defined EVs positive for CD-pan markers (CD9, CD63, CD81). Then, the prescreened EVs were sub-gated with target glioblastoma markers of EGFR or EGFRvIII. We conducted the power analysis for a Mann Whitney test using two independent groups (EV positive and negative) to calculate the necessary EV sample size (n > 100) given the statistical power of 0.9 and the effect size of 0.43. Given the EV surface coverage of 0.1–0.5 EV per μm2, the minimum area required is roughly 200–1000 μm2. Yet, we used fluorescence images (n = 4) in a full FOV (120 μm × 100 μm) and sampled thousands of vesicles per measurement to ensure statistical significance and robust analysis.
Figure 4.
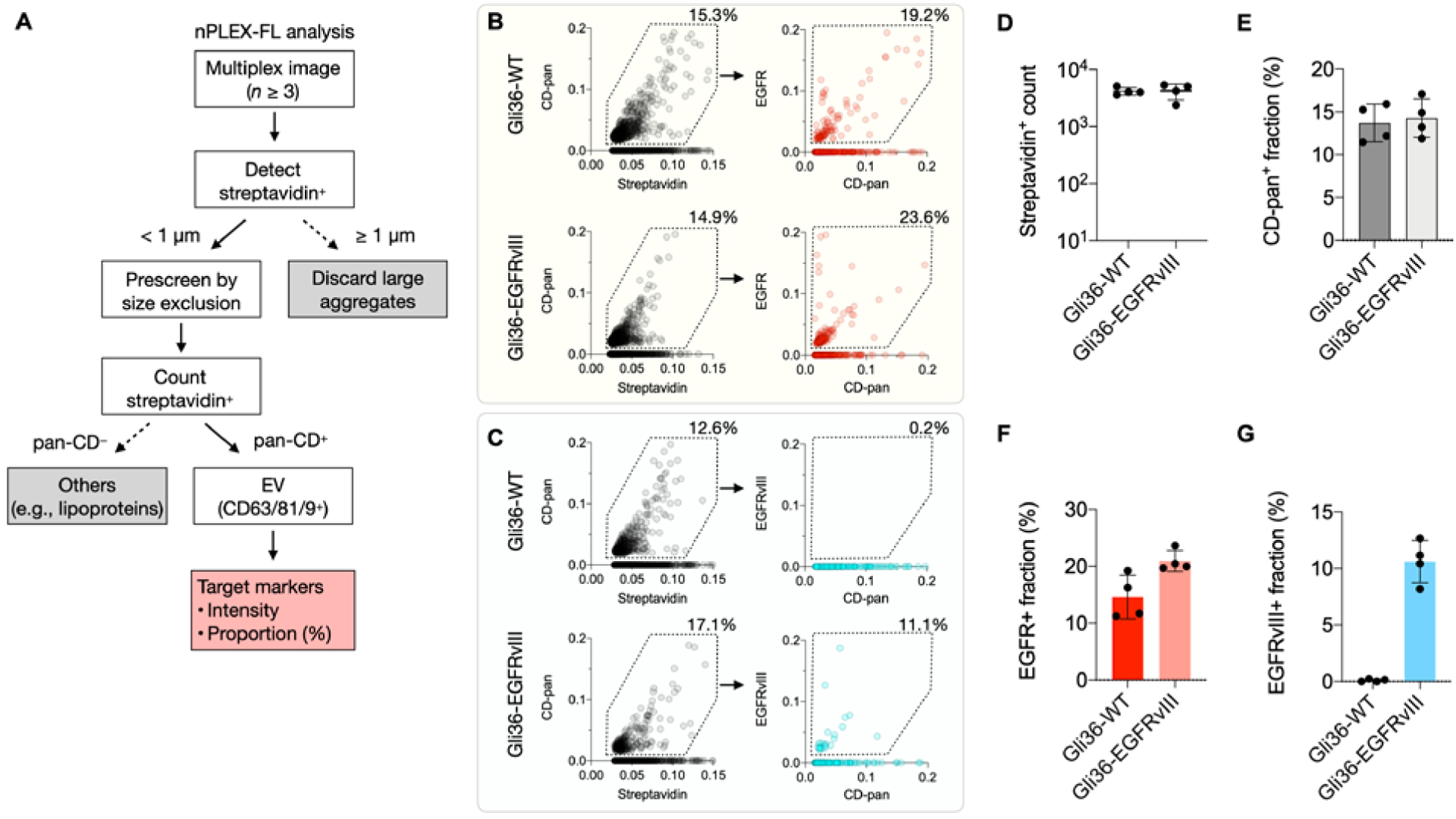
Measurements of tumor markers in EV-spiked plasma samples. A) Decision tree algorithm to classify EV populations. Briefly, captured biotinylated EVs positive for streptavidin were first detected and prescreened by size exclusion (< 1 μm). The prescreened EVs positive for tetraspanin CD-pan (CD9/CD63/CD81) were defined as EVs and their target markers were analyzed. B-C) Nested gating strategy illustrating EV population positive for streptavidin, CD-pan and target marker of EGFR (B) or EGFRvIII (C). EVs from Gli36-WT and Gli36-EGFRvIII cell lines were spiked in human plasma samples and used for testing. D-G) EV detection and marker profiling. Vesicles positive for streptavidin (D) and EV fraction positive for CD-pan (E), EGFR (F), EGFRvIII (G) were analyzed.
Figures 4B and C show biomarker distribution analyses on a single-EV level. We plotted bivariate histograms from 3-channel fluorescence images with a field-of-view (FOV) of 120 μm × 100 μm (representative images in Figure S8). On average, we detect about 4,200 particles positive for streptavidin in single images (minimum = 3,604, maximum = 5,057 EVs, Figure 4D). We observed 10–15% positivity of streptavidin-positive particles for CD-pan markers (Figure 4E). The lower fraction of CD-pan+ streptavidin+ particles in the plasma samples compared to that in the pure-buffer (PBS, Figure S9) could be attributed to the presence of lipoproteins and plasma protein aggregates in human plasma. For marker profiling, the detected EVs positive for CD-pan were screened for target markers of EGFR and EGFRvIII. For plasma samples spiked with EVs from Gli36-WT and Gli36-EGFRvIII cell lines, about 10–20% of detected EVs were positive for EGFR in both samples (Figure 4F). However, roughly 10% of EVs were positive for EGFRvIII only in the plasma samples with Gli36-EGFRvIII EVs, while the other sample with Gli36-WT EVs showed less than 1% positive EV fraction, which is below the threshold (Figure 4G). Comparable biomarker positivity for EGFR and EGFRvIII was observed between the plasma samples and the pure-buffer samples (c.f., Figure 4 and S9).
Lastly, we benchmarked the nPLEX-FL results against the standard quantification methods, including Western blotting analysis (Figure S3) and droplet digital PCR measurements for EV RNAs (Figure S10); these methods measure the bulk signals of EGFR or EGFR-vIII from the lysed EVs. Our results showed good agreement with the standard methods for biomarker expression (EGFR, EGFRvIII), indicating the assay’s potential capability in detecting and molecularly profiling cancer-derived EVs in human plasma samples.
Conclusion
In this proof-of-concept study, we applied plasmon-enhanced fluorescence detection for multiplexed single EV analysis. Given the EVs’ small sizes and a limited amount of biomarkers available in individual EVs, a simple and robust signal amplification strategy is crucial to improve the detection sensitivity and achieve multiplexed analysis. Here, we showed signal amplification in multiple fluorescence channels can be achieved by simply using Au nanohole substrates rather than conventional glass substrates. The plasmon enhancement enables more accurate and sensitive single EV analysis, especially for low-abundant markers. This strategy can be readily adapted as a plug-in to previously developed single EV imaging platforms for signal enhancement without adding the complexity of assay procedures[11,12,14].
The current study had a few limitations that we intend to further improve in future studies. First, the EV-spiked plasma samples may mimic clinical samples, yet they are limited to be representative of patient populations. Second, the current multiplexing level is limited by a fluorescence microscope system, typically enabling 3–4 colors. In the future, it may be possible to extend the multiplexing capabilities beyond the current capacity by harnessing image cycling approaches[24]. Third, we used biotin-SA chemistry to capture EVs and label them with fluorescently-conjugated streptavidin that showed specific capture on a substrate with a low false-positive rate (<1%). With the development of a simple, general method for EV labeling and tracking, we could further simplify the assay procedures and improve the assay accuracy. Finally, there is room for further system optimization (e.g., the distance between the nanohole surface and EVs, selection of nanohole periodicity, and fluorophores) that could lead to even higher signal enhancement. Combined with the previously developed spectral measurements[7,8], the new method could offer additional insight into understanding subtypes, heterogeneity, and production dynamics of EVs during disease development and progression.
Experimental Section
Cell culture
Gli36-WT (ATCC), Gli36-EGFRvIII (generated from Gli36-WT through lentivirus transduction by Leonora Balaj in the Breakefield lab), and MCF-7 cells (ATCC) were grown in DMEM (Cellgro). OVCA429 cells (ATCC) were cultured in RPMI-1640 medium (Cellgro). All media were supplemented with 10% fetal bovine serum (FBS, Thermo Fisher), 100 U/mL penicillin, and 100 μg/mL streptomycin (Cellgro) at 37 °C in 5% CO2. All cell lines were tested and were free of mycoplasma contamination (MycoAlert mycoplasma detection kit, Lonza).
EV isolation and biotinylation
Before EV collection, cells were incubated in DMEM with 1% exosome-depleted FBS (Thermo Fisher) for 48 h. The conditioned medium was collected and centrifuged at 300 × g for 5 min and then supernatant was filtered through a 0.2 μm membrane filter (Millipore Sigma). EV isolation was conducted using both standard ultracentrifugation (UC) and size-exclusion chromatography (SEC) methods: (i) For UC, the filtrates were concentrated by 100,000 × g for 1 h. After the supernatant was removed, the EV pellet was washed with PBS and centrifuged at 100,000 × g for 1 h. The EV pellet was resuspended in PBS. (ii) For SEC, the filtrates were loaded onto Centricon Plus-70 Centrifugal Filter (MWCO = 10 kDa, Millipore Sigma) and centrifuged at 3,500 × g for 30 min at 4°C. After concentration, the volume was adjusted to 1 mL with PBS. SEC was performed as previously described with modifications[25]. Briefly, 10 mL syringe (BD Biosciences) with a nylon net with 20 μm pore size (Millipore Sigma) at the bottom was prepared and packed with 10 mL of Sepharose CL-4B (GE healthcare). The concentrates were loaded on top and 6 fractions of 1 mL were collected under constant gravitational flow by adding PBS on top of the column. The fraction 4 and 5 were used for EV isolation. These were loaded onto Amicon Ultra-2 Centrifugal Filter (MWCO = 10 kDa, Millipore Sigma) and centrifuged at 3,500 × g for 30 min at 4°C. The isolated EVs were stored at −80°C until nPLEX-FL measurement.
For EV biotinylation, the isolated EVs were resuspended in 300 μL of PBS and incubated with 333 μM EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher) for 30 min at room temperature. We used a 20-fold molar excess of sulfo-NHS-biotin to EV protein in 0.5 mL volume. Approximately 4–6 biotins were expected to be incorporated per molecule. Excess biotin was then removed utilizing the Exosome Spin Columns, MW3000 (Thermo Fisher) per the kit instructions. The prepared EVs were filtered using a 0.22 μm centrifugal filter (Ultrafree, Millipore).
EV-spiked plasma testing
Roughly 1010 EVs from Gli36-WT and Gli36-EGFRvIII cell lines were spiked into 1mL single-donor human plasma. The EV-spiked plasma was first purified with size exclusion chromatography (SEC) columns (Izon qEV), biotinylated, and then loaded onto the chip (1–5 μL) for further nPLEX-FL processing.
nPLEX-FL chip characterization
nPLEX-FL chips were prepared using the lithography methods as previously described[7,8]. The chip was incubated overnight at room temperature with thiolated biotin polyethylene glycol (PEG) (10 mM in PBS, PG2-BNTH-1k, Nanocs). After washing with PBS, an equimolar mixture of streptavidin molecules conjugated with either Alexa Fluor 488, Cy3, Cy5, or Cy5.5 (Biolegend) was incubated for 10 min. The concentration of each fluorescence dye was diluted to be 2.5 μg/ml, except Alexa Fluor 488-conjugated streptavidin (25 μg/ml in PBS) due to the weak fluorescence signal compared to other channels.
nPLEX-FL protocol
After biotinylation with 10 mM thiolated biotin polyethylene glycol (PEG), the Au substrates were incubated in neutravidin (0.05 mg/mL, in PBS with 0.2% BSA), a linker to capture biotinylated EVs, for 1 hr. Biotinylated EVs were captured on the neutravidin-coated surface, followed by EV fixation and permeabilization in a fix/perm solution (BD Science) for 10 min. The surface passivation was achieved by placing the Au surface (with or without EVs) in a blocking solution (Superblock PBS, Thermo Fisher) for 20 min. This step is important to minimize undesired non-specific binding. The captured EVs were stained via two-step indirect labeling: firstly with a cocktail of primary antibodies (20 min) then with compatible secondary antibodies (10 min, Table S1). Thorough washing was done between steps. The EVs were labeled with fluorescently labeled streptavidin. Assay buffer was a BD perm/wash buffer solution (BD Biosciences). All antibodies used in these studies are listed in table S1. Finally, the labeled EVs were mounted with a mounting solution (Prolong Au Antifade mountant, Thermo Fisher) and covered with a glass coverslip. Fluorescence images were acquired on an Olympus BX-63 upright automated epifluorescence microscope with 40× (NA = 0.95) and 100× (NA = 1.40) objectives.
Image Processing
Image analyses were performed using ImageJ and CellProfiler. We used the streptavidin imaging channel to create masks at EV locations. For each molecular target, the corresponding fluorescent micrograph was aligned using ImageJ plugins (Align slices in the stack). At each mask position, we obtained average pixel intensities. The signal was corrected by subtracting background signal surrounding the mask.
Numerical Simulation
Electrodynamic computation was performed using the finite-difference time-domain (FDTD) method. For electric field distribution, x-polarized plane wave was illuminated along -z direction. 2-nm mesh size was used for the volume of 0.3 × 0.3 × 0.2 μm3 locating at the center of nanohole. Periodic boundary condition was imposed along x and y direction and perfect match layers were used for z direction. A z-polarized dipole source was used for radiative decay rate simulation. The position of the dipole is set to x = 100 nm, y = 0 nm and z = 6 nm to locate it at the edge of nanohole and 6 nm above the Au surface.
Statistics
Statistical analyses and data plotting were performed in GraphPad Prism 7. Group differences were tested using the nonparametric Mann-Whitney test for two groups and ANOVA with post-hoc analysis for more than two groups. The power analysis for Mann Whitney test was performed using G*Power to compute the necessary EV sample size given the statistical power (1- β) of 0.9 and the effect size of 0.43. All tests were two-sided, and a P-value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
J.M and T.S. contributed equally to this work. The authors thank Seung-Min Bang and Moon-Jae Chung (Severance Hospital) for assisting in electron microscopy and Koushik Muralidharan, Anudeep Yekula, and Leonora Balaj for droplet digital PCR measurements and helpful discussion. This work was supported in part by U.S. NIH Grants R00CA201248 (H.I.), R21CA217662 (H.I.), P30AG062421 (H.I), R01CA204019 (R.W.), P01CA069246 (R.W.), R01CA229777 (H.L.), U01CA233360 (H.L.), DoD-W81XWH1910199 (H.L.), DOD-W81XWH1910194 (H.L.); MGH Scholar Fund (H.L.); the Institute for Basic Science IBS-R026-D1 (H.L.), South Korea.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library.
Contributor Information
Pike See Cheah, Department of Neurology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Human Anatomy, Faculty of Medicine & Health Sciences, Universiti Putra Malaysia, Serdang, Selangor 43400, Malaysia..
Andreas Wegemann, Center for Systems Biology, Massachusetts General Hospital, Boston, MA 02114, USA; Institute of Medical Engineering (IMETUM), Technical University of Munich, Munich 80333, Germany.
Ralph Weissleder, Center for Systems Biology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Radiology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Systems Biology, Harvard Medical School, Boston, MA 02115, USA.
References
- [1].a) Verma M, Lam TK, Hebert E, Divi RL, BMC Clin Pathol 2015, 15, 6.; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W, J Hematol Oncol 2015, 8, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Ailawadi S, Wang X, Gu H, Fan GC, Biochim Biophys Acta 2015, 1852, 1.; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ibrahim A, Marbán E, Annu Rev Physiol 2016, 78, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Properzi F, Logozzi M, Fais S, Biomark Med 2013, 7, 769.; [DOI] [PubMed] [Google Scholar]; b) Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, Jackson B, McKee AC, Alvarez VE, Lee NC, Hall GF, J Biol Chem 2012, 287, 3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fuhrmann G, Neuer AL, Herrmann IK, Eur J Pharm Biopharm 2017, 118, 56. [DOI] [PubMed] [Google Scholar]
- [5].a) Alderton GK, Nat Rev Cancer 2015, 15, 453.; [DOI] [PubMed] [Google Scholar]; b) Raposo G, Stoorvogel W, J Cell Biol 2013, 200, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J, Nat Commun 2011, 2, 180.; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R, Nat Commun 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang KS, Im H, Hong S, Pergolini I, Del Castillo AF, Wang R, Clardy S, Huang CH, Pille C, Ferrone S, Yang R, Castro CM, Lee H, Del Castillo CF, Weissleder R, Sci Transl Med 2017, 9, eaal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H, Nat Biotechnol 2014, 32, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Jia S, Zocco D, Samuels ML, Chou MF, Chammas R, Skog J, Zarovni N, Momen-Heravi F, Kuo WP, Expert Rev Mol Diagn 2014, 14, 307; [DOI] [PubMed] [Google Scholar]; b) Shao, Im H, Castro CM, Breakefield X, Weissleder R, Lee H, Chem Rev 2018, 118, 1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Park J, Im H, Hong S, Castro CM, Weissleder R, Lee H, ACS Photonics 2018, 5, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, Chiocca EA, Breakefield XO, Lee H, Weissleder R, ACS Nano 2018, 12, 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fraser K, Jo A, Giedt J, Vinegoni C, Yang KS, Peruzzi P, Chiocca EA, Breakefield XO, Lee H, Weissleder R, Neuro Oncol 2019, 21, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Daaboul GG, Gagni P, Benussi L, Bettotti P, Ciani M, Cretich M, Freedman DS, Ghidoni R, Ozkumur AY, Piotto C, Prosperi D, Santini B, Ünlü MS, Chiari M, Sci Rep 2016, 6, 37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang Y, Shen G, Wang H, Li H, Zhang T, Tao N, Ding X, Yu H, Proc Natl Acad Sci U S A 2018, 115, 10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai Y, Wik L, Shen Q, Oelrich J, Qian X, Dubois KL, Ronquist KG, Nilsson M, Landegren U, Kamali-Moghaddam M, Nat Commun 2019, 10, 3854.; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shen W, Guo K, Adkins GB, Jiang Q, Liu Y, Sedano S, Duan Y, Yan W, Wang SE, Bergersen K, Worth D, Wilson EH, Zhong W, Angew Chem Int Ed Engl 2018, 57, 15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, An T, Yao S, Zheng L, Nano Lett 2018, 18, 4226. [DOI] [PubMed] [Google Scholar]
- [17].a) Zhang Q, Wu L, Wong TI, Zhang J, Liu X, Zhou X, Bai P, Liedberg B, Wang Y, Int J Nanomedicine 2017, 12, 2307.; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bauch M, Toma K, Toma M, Zhang Q, Dostalek J, Plasmonics 2014, 9, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mayer KM, Hafner JH, Chem Rev 2011, 111, 3828. [DOI] [PubMed] [Google Scholar]
- [19].Monteiro JP, Carneiro LB, Rahman MM, Brolo AG, Santos MJL, Ferreira J, Girotto EM, Sensors and Actuators B: Chemical 2013, 178, 366. [Google Scholar]
- [20].a) Jeong Y, Kook YM, Lee K, Koh WG, Biosens Bioelectron 2018, 111, 102.; [DOI] [PubMed] [Google Scholar]; b) Aslan K, Gryczynski I, Malicka J, Matveeva E, Lakowicz JR, Geddes CD, Curr Opin Biotechnol 2005, 16, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Anger P, Bharadwaj P, Novotny L, Phys Rev Lett 2006, 96, 113002.; [DOI] [PubMed] [Google Scholar]; b) Fu Y, Lakowicz JR, Laser Photon Rev 2009, 3, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Munechika K, Ginger DS, Nano Lett 2007, 7, 690. [DOI] [PubMed] [Google Scholar]
- [23].Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H, Nat Med 2012, 18, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].a) Ko J, Oh J, Ahmed MS, Carlson JCT, Weissleder R, Angew Chem Int Ed Engl 2020, 59, 6839.; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Giedt RJ, Pathania D, Carlson JCT, McFarland PJ, Del Castillo AF, Juric D, Weissleder R, Nat Commun 2018, 9, 4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Benedikter BJ, Bouwman FG, Vajen T, Heinzmann ACA, Grauls G, Mariman EC, Wouters EFM, Savelkoul PH, Lopez-Iglesias C, Koenen RR, Rohde GGU, Stassen FRM, Sci Rep 2017, 7, 15297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


