Abstract
Medulloblastoma (MB) is among the most common malignant brain tumors in children. Recent studies have identified at least four subgroups of the disease that differ in terms of molecular characteristics and patient outcomes. Despite this heterogeneity, most MB patients receive similar therapies, including surgery, radiation, and intensive chemotherapy. Although these treatments prolong survival, many patients still die from the disease and survivors suffer severe long-term side effects from therapy. We hypothesize that each MB patient is sensitive to different therapies and that tailoring therapy based on the molecular and cellular characteristics of patient tumors will improve outcomes. To test this, we assembled a panel of orthotopic patient-derived xenografts (PDX) and subjected them to DNA sequencing, gene expression profiling, and high-throughput drug screening. Analysis of DNA sequencing revealed that most MB do not have actionable mutations that point to effective therapies. In contrast, gene expression and drug response data provided valuable information about potential therapies for every tumor. For example, drug screening demonstrated that actinomycin D, which is used for treatment of sarcoma but rarely for MB, was active against PDX representing Group 3 MB, the most aggressive form of the disease. Functional analysis of tumor cells was succesfully used in a clinical setting to identify more treatment options than sequencing alone. These studies suggest that it should be possible to move away from a one-size-fits-all approach and begin to treat each patient with therapies that are effective against their specific tumor.
INTRODUCTION
Medulloblastoma (MB) is a highly malignant brain tumor that occurs predominantly in children. Genomic and epigenomic studies have revealed four major subgroups of the disease – WNT, Sonic hedgehog (SHH), Group 3 and Group 4 – that differ in terms of molecular characteristics, demographics and patient outcomes (1–4). Recently, heterogeneity within these subgroups has been recognized, and it has been suggested that MB may consist of up to 14 molecular subtypes (5–7). But so far the impact of this heterogeneity on therapy has been limited, with trials testing Smoothened antagonists for patients with SHH-associated MB (8), and efforts to reduce therapy for patients with WNT driven tumors (9), who have a relatively favorable prognosis. Outside of these trials, most MB patients still receive similar therapies, including surgery, craniospinal radiation (except in young children, for whom radiotherapy has devastating neurocognitive side effects) and multi-agent chemotherapy. For patients who fail frontline treatment, there are few curative options, and recurrent MB is frequently lethal. Overall, ~1/3 of MB patients die from the disease, and survivors suffer severe long-term side effects from treatment. We hypothesize that tailoring therapy based on characteristics of individual MB patients will result in improved outcomes.
The notion of tailoring therapies based on genomic characteristics is not new (10). “Basket trials,” in which a targeted therapy is matched to patients with a specific genetic lesion across a variety of tumor types, have led to FDA approval of treatments such as pembrolizumab in mismatch repair-deficient cancers and BRAF/MEK inhibition in metastatic BRAF V600E-mutated non-small-cell lung cancer. However, heterogeneity in overall response rate has been observed based on tumor type or histology (10). “Umbrella trials” such as NCI’s Molecular Analysis for Therapy of Choice (MATCH) trial include multiple targeted therapies, and assign patients to a therapy based on the presence of specific genetic lesions in their tumors (11). Although umbrella trials have shown some promise (12), adequate enrollment of patients into each therapeutic arm, heterogeneity of patients within each arm, and efficacy of single agent therapy in highly-pretreated patients, have been major challenges (10). “N of 1” trials use genomic and molecular characteristics to guide therapy on an individualized basis without a predetermined set of therapies but rather a host of therapies that may be chosen based on molecular predictors (13). In one such trial, tumors were sequenced and up to five targeted agents were chosen for therapy; targeting a larger fraction of molecular alterations was associated with better outcomes (14). Although such trials have improved responses in select patients, in many cases gains have been modest.
Precision medicine has also been evaluated in pediatric oncology. For example, a basket trial using larotrectinib showed promising results in Trk-fusion-positive tumors of a variety of histologies (15). Pediatric MATCH, an umbrella trial with 10 arms into which patients are assigned based on genetic lesions, has demonstrated the feasibility of this approach, but the prevalence of target lesions in the pediatric population is low, and evidence for efficacy of recommended drugs has been limited (16). The value of looking at genetic lesions on an individualized basis was previously shown by Gröbner et al. (17). However, in the individualized Cancer Therapy (iCAT) trial, an N of 1 trial for advanced, extra-cranial, solid tumors, 31 of 100 patients received a recommendation based on genetic alterations, but only three received a matched therapy and none of these showed an objective response (18). This and other evidence has emphasized the paucity of targetable genetic lesions in the pediatric population. Thus, the use of DNA alone to identify targeted therapies has been disappointing.
Other types of data that could point to appropriate therapies include gene expression, protein expression, epigenetic analysis and empirical drug screening. Although these approaches have been evaluated extensively in the lab (17,19), their application in the clinic has been much more limited. Here, using a panel of MB PDXs, we show that considering gene expression and drug response along with DNA variants may better inform therapeutic decisions than sequencing alone. In addition, we demonstrate that these approaches are feasible in a clinical setting, raising the possibility that they could be incorporated into current precision medicine protocols.
MATERIALS AND METHODS
Animals
NOD-SCID IL2R-gamma null (NSG) mice used for intracranial tumor transplantation were purchased from Jackson Labs (Bar Harbor, ME). Mice were maintained in the animal facilities at the Sanford Consortium for Regenerative Medicine. All experiments were performed in accordance with national guidelines and regulations, and with the approval of the animal care and use committees at the Sanford Burnham-Prebys Medical Discovery Institute and University of California San Diego (UCSD).
Establishment and Maintenance of PDXs
PDX lines were generated by implanting 0.5–1×106 dissociated patient cells directly into the cerebellum of NSG mice, and propagated from mouse to mouse without in vitro passaging. The identity and subgroup of each line was validated by DNA methylation analysis. PDX lines used for this study include BT084 (SHH) from the Milde lab (20); MB002 and MB009 (G3) from the Cho lab (21,22); ICb-984 (SHH), ICb-1572 (G3), ICb-1487 (G3) and ICb-1299 (G3), from the Li lab (23); MED1712-FH (SHH), MED411-FH (G3), MED211-FH (G3), MED1911-FH(G3), and MED2312-FH (G4) from the Olson lab (24,25); and RCMB28 (G3), RCMB18 (SHH), RCMB32 (SHH), RCMB38 (G4), RCMB20 (G3), RCMB40 (G3), RCMB24 (SHH), and DMB006 (G4) from the Wechsler-Reya lab (26,27). No WNT subgroup PDXs were available for these studies. For all experiments, cells were isolated from tumor-bearing mice, resuspended in NeuroCult with proliferation supplement and penicillin/streptomycin (StemCell Technologies, CAT# 05702), and assayed as described below. All PDXs described in this study will be made available to investigators at other institutions upon reasonable request.
Data Availability
Short-read sequencing data are available at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/), hosted by the European Bioinformatics Institute, under accession number EGAS00001004698. Methylation and gene expression data have been deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE151344 and GSE151343, respectively. Gene expression analyses (Affymetrix processing, gene expression quantification, and gene expression analyses using DiSCoVER) can be reproduced following the documentation and using the code in the GenePattern GitHub Repository.
Identification of Actionable Mutations from DNA
To determine what drugs would be predicted based on mutational analysis, we merged two databases containing mutation-drug-outcome associations: CIViC (28) and OncoKB (29). This created a searchable list of drugs targeting specific genes. We then ran PHIAL (30) to cross-reference the mutations found in our PDXs with mutations reported in the TARGET database (https://software.broadinstitute.org/cancer/cga/target as of February 2015). PHIAL returned a list of actionable mutations present in our samples. For each mutated gene, drugs identified by OncoKB and CIViC were added to our list of drug candidates. This compiled list of drug-mutation associations was reduced to include only genes mutated in our PDX lines.
Identification of Drug Candidates from RNA analysis.
The DiSCoVER method (31) extracts a highly expressed gene expression signature from a tumor sample and leverages publicly available databases to identify cell lines with a similar signature, and the compounds to which they are sensitive. Briefly, the method proceeds as follows:
The signature of the tumor consists of the 150 most highly differentially expressed genes in that tumor compared to a control. Here the tumors are PDXs and human cerebellar stem cells are used as the control.
A single sample version of Gene Set Enrichment Analysis (GSEA) (32,33) is used to score the activation of signature genes in the expression profiles from two cell line collections: the Cancer Cell Line Encyclopedia (CCLE) (34) and the Sanger Cell Line Project (35). The expression activation scores are then compared to the cell lines’ corresponding viability profiles for compounds in the appropriate screening databases: the Cancer Therapeutics Response Portal (CTRP) (36) for the CCLE cell lines and the Genomics of Drug Sensitivity in Cancer (GDSC) database for the Sanger Cell lines.
The association of the viability and the signature activation of the cell lines yields a score from which the probability of a compound’s effectiveness on the PDX can be inferred. The compounds are ranked according to the quantification of these associations.
High-Throughput Drug Screening
The screen used 7,729 compounds from the following libraries: StemSelect, InhibitorSelect Pathway, Kinase Inhibitors (all from EMD,), Spectrum, US and International Drug Collections (both from MicroSource), LOPAC (Sigma), Prestwick Chemical Library (Prestwick), LeadGen Collection, Epigenetics library (both from Enzo Life Sciences), NIH Clinical Collection (NIH), NCI Oncology Drugs (NCI), Kinase Inhibitors (Cayman), Kinase Inhibitors (SelleckChem). Many drugs were represented in multiple libraries; thus only 4,683 unique compounds were tested.
Prior to screening, 2.5 nL of compound-containing solution was pin-transferred into 384 well plates (Greiner Bio-One, CAT# 781098). Tumor tissue was harvested from tumor bearing mice and dissociated using 10U/ml papain (Worthington, CAT# LS003126) to create a single cell suspension. Into each drug-containing well, we plated 104 tumor cells in 25 μl Neurocult with proliferation supplement, resulting in a final drug concentration of 1 μM. Each drug was tested in triplicate, with tumor cells from at least 3 separate mice. Since testing the entire library required approximately 80 million cells, and since the yield of tumor cells from a given mouse ranged from 10 to 150 million cells, testing the entire library in triplicate required 3 to 24 mice for each PDX line. Every set of plates included 12 wells of DMSO (negative control) and each tumor replicate included 12 wells of 1 μM YM155 (positive control, Cayman, CAT# 11490). Viable cell number in each well (as indicated by ATP content) was determined using the CellTiter-Glo assay (Promega, CAT# G7571) and read in an automated Envision plate reader (Perkin-Elmer) after 48 hr incubation. Percent inhibition was calculated using the formula [sample result/mean value of the entire plate containing cells and compound × 100].
Treatment of Tumor Bearing Mice
For treatment of intracranial tumor-bearing mice in pilot experiments, actinomycin D, bortezomib, gambogic acid, idarubicin (all from Cayman, CAT# 11421, 10008822, 14761, 14176 respectively), and oleandrin (MedChemExpress, CAT# HY-13719) were dissolved in 10% DMSO, 10% Tween80, and sterile water (vehicle, n=8). Actinomycin D was administered at 0.03 mg/kg daily or 0.06 mg/kg biweekly (intraperitoneal, i.p., n=8 for each). Bortezomib was administered at 1 mg/kg biweekly (i.p., n=8). Oleandrin was administered at 2 mg/kg daily or 3 mg/kg biweekly (i.p., n=8 for each). Idarubicin was administered at 1 mg/kg daily or 1.5 mg/kg biweekly (i.p., n=8 for each). Gambogic acid was administered at 2 mg/kg daily or 4 mg/kg biweekly (i.p., n=8 for each).
For in vivo comparison to standard of care drugs, actinomycin D (Cayman), vincristine, and cisplatin (both from Sigma, CAT# V8879, PHR1624) were dissolved in 5% DMSO in PBS and cyclophosphamide (Sigma, CAT# C7397) was dissolved in 10% DMSO in PBS. Pilot experiments demonstrated that the maximum tolerated weekly doses were 130 mg/kg cyclophosphamide (i.p.) for 13 weeks with one week off in the 7th week, 4.5 mg/kg cisplatin (i.p.) for 8 weeks with 2 weeks off the 6th and 7th week, 1 mg/kg vincristine (i.p.) for eight weeks, and 0.3 mg/kg actinomycin D (retro-orbital intravenous, i.v.) for 10 weeks. Two weeks after transplantation, mice were randomly separated into five groups of 8, and treated with vehicle (10% DMSO); cyclophosphamide, cisplatin, vincristine or actinomycin D. Mice that received actinomycin D were given 1 million allogenic bone marrow cells i.v. every week, two days post drug treatment. Animals were drug treated every seven days, with weeks off as described above, until they displayed signs of morbidity or toxicity (>30% weight loss), whereupon they were euthanized.
Primary Patient Samples
Studies were conducted in accordance with recognized ethical guidelines and were approved by an institutional review board Written informed consent was obtained from patients.
RESULTS
Molecular characterization of PDXs
To identify novel therapies for MB, we assembled a panel of 20 PDX lines that had been passaged only in vivo. To determine which subgroups of MB these lines most closely resemble, we performed DNA methylation profiling and analyzed the data using the Heidelberg brain tumor classifier (www.molecularneuropathology.org; (37), which calculates a score between 0 and 1 indicating the similarity of that sample to one of the 82 CNS tumor classes present in the reference cohort. Samples classified as Group 3 or Group 4 MB were further subtyped using the Group 3/4 classifier described in Sharma et al. (7), which distinguishes between eight subtypes, designated I-VIII. Scores for each PDX, and when available, the primary tumor from which it was generated, are shown in Supp. Table 1.
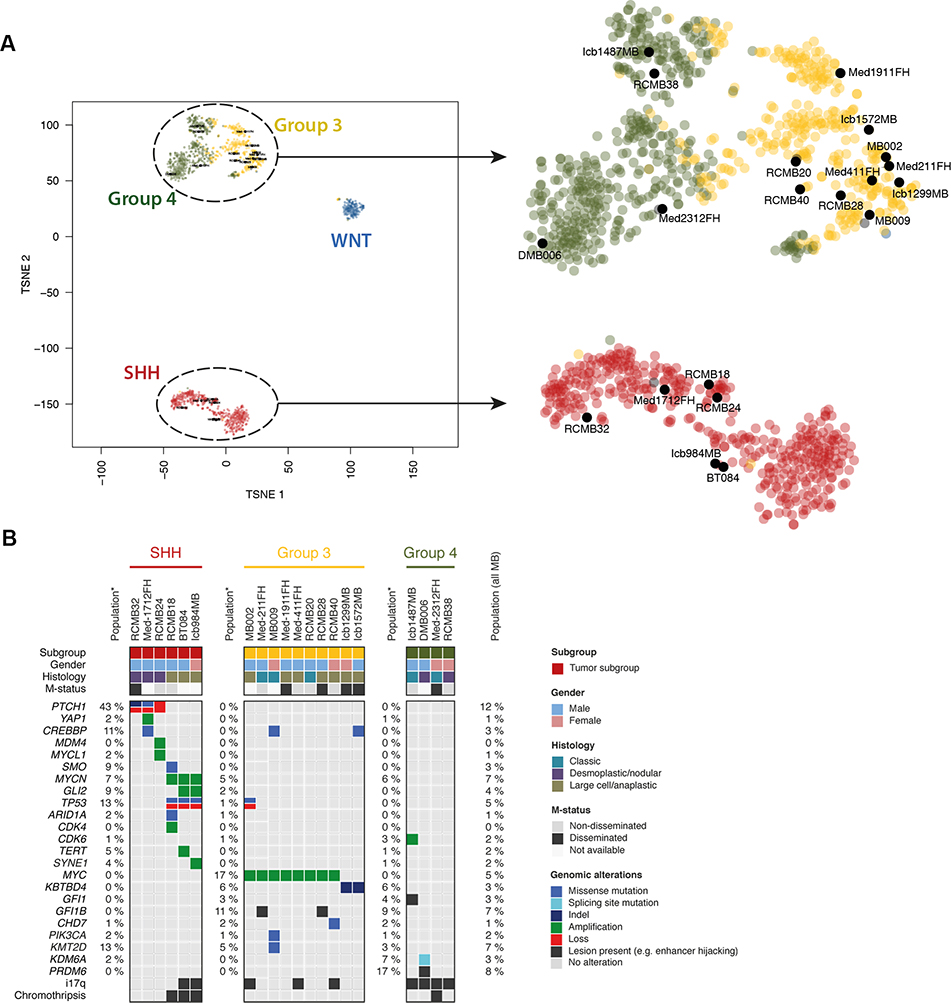
Methylation analysis identified six SHH, ten Group 3 and four Group 4 lines within our cohort. In most cases, the subgroup of primary tumors was consistent with that of the corresponding PDX models. One exception was MED-2312FH, for which the primary tumor was classified as Group 4 and the PDX was classified as Group 3. Subtype analysis indicated that both samples belonged to subtype V, which consists of tumors that are intermediate between Groups 3 and 4 (6). Given the high score supporting Group 4 classification of the primary tumor (0.936) and the relatively low score supporting the Group 3 classification of the PDX (0.627), for the analyses below we considered it a Group 4 tumor. t-distributed stochastic neighbor embedding (t-SNE) clustering of the DNA methylation profiles of the PDX models with a reference cohort of MBs representing all four subgroups (25) shows that PDX models cluster with the subgroups predicted by the classifier (Fig. 1A).
Figure 1: Molecular characterization of PDXs reveals subgroups and genetic lesions.
(A) tSNE clustering of DNA methylation profiles of PDX models with a reference cohort of MBs representing all four major subgroups shows that PDX models cluster with the subgroups as predicted by the Heidelberg brain tumor classifier (see Supp. Table 1). (B) Summary of subgroup, gender, histology and mutation status (m-status) of patients from whom PDXs were generated, along with the genomic alterations found in each PDX. Genomic alterations listed are those identified as previously observed in MB as a cancer gene and found in our PDXs (see Supp Table 2A,B). Numbers on the left side of each subgroup represent the percentage of patients from that subgroup with the corresponding genetic lesion; numbers on the far right represent the percentage of the total MB population with the lesion.
To identify the genetic lesions present in our PDX cohort we performed deep whole exome sequencing (WES) and low depth whole genome sequencing (WGS) (Fig. 1B and Supp. Table 2A–C). Among the six SHH lines, three (MED-1712FH, RCMB24, and RCMB32) had alterations in PTCH1, which occurs in about 43% of SHH-MB patients (27). The remaining three SHH lines, RCMB18, ICb-984 and BT084, exhibited loss or inactivation of both alleles of TP53, amplification of MYCN and/or GLI2, and chromothripsis, a genotype associated with extremely poor prognosis (38). Notably, alterations in TP53 account for only 13% of all SHH-MB cases, but were found in three out of six (50%) of our SHH PDXs, consistent with the notion that more aggressive tumors are more likely to take in mice (23,25). Likewise, among the ten Group 3 lines, eight (80%) exhibited MYC amplification, a biomarker of poor prognosis that is present in only 17% of Group 3 patients. Mutations in CREBBP, PIKC3A, KMT2D (MLL2), CHD7, and activation of GFI1B by enhancer hijacking were among the additional lesions observed in our Group 3 lines. Among the four Group 4 lines, one (ICb-1487) exhibited GFI1 enhancer hijacking and CDK6 amplification, and another (DMB006) had PRDM6 enhancer hijacking and a mutation in KDM6A. Gender, metastatic (M) status, histology, age, and prevalence of each genetic lesion within the MB population for all 20 PDX lines are shown in Fig. 1B.
Identification of candidate therapies based on genetic data
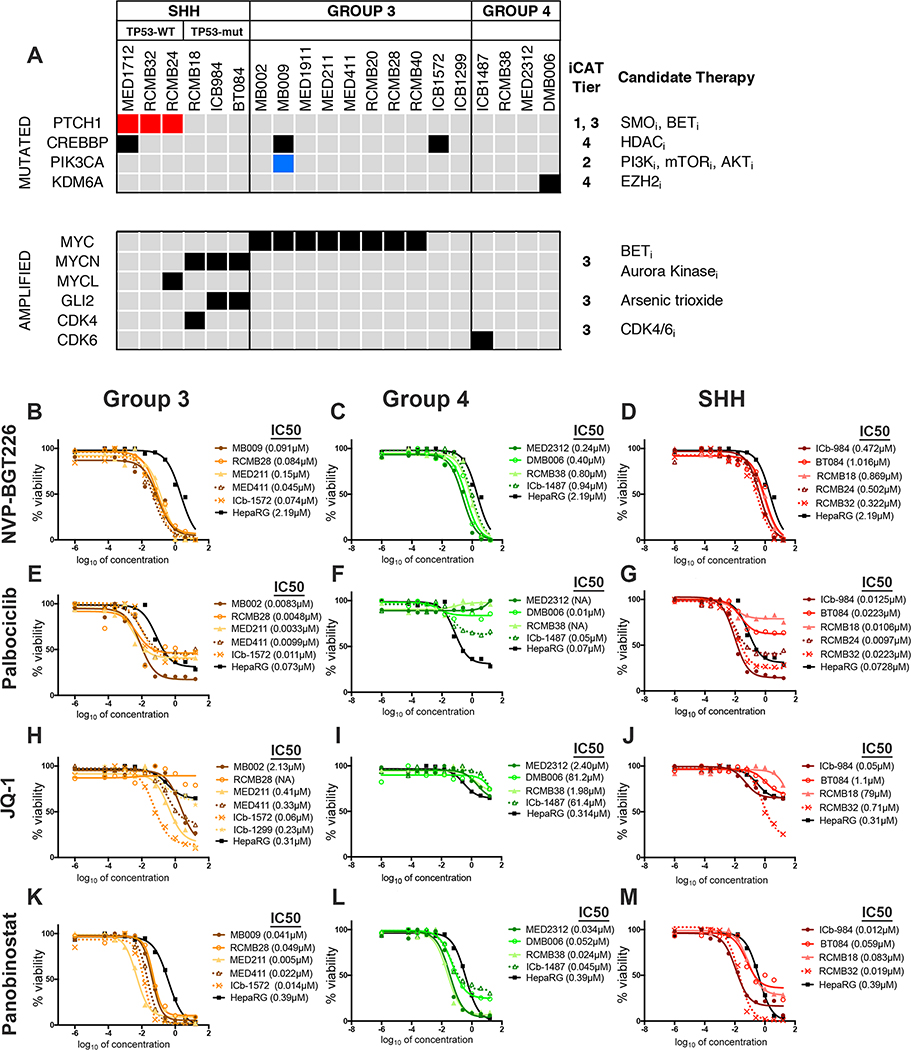
Most precision medicine trials use DNA sequencing as a basis for recommending therapies. To determine what therapies would be predicted to be effective using this approach, we analyzed DNA sequencing data from our PDXs (Supp. Table 2A–C) using mutation-drug-outcome associations from three databases: PHIAL2 (Precision Heuristics for Interpreting the Alteration Landscape) (30), CIViC (28), and OncoKB (29), and additional relevant publications (39–41). This allowed us to identify candidate therapies for several of our PDX lines (Fig. 2A). We categorized the strength of each of these mutation-drug predictions based on criteria used for the iCAT study: alterations associated with response to a therapeutic agent in clinical trials are considered Tier 1 (if the trial was in the same disease) or Tier 2 (for a different disease), and alterations associated with drug responses based on pre-clinical studies are considered Tier 3 (for studies in the same disease) or Tier 4 (for a different disease) (18).
Figure 2: Genomic data point to few actionable events in MB PDXs.
(A) Grid shows actionable genetic lesions (filled squares) for each PDX. Red and blue squares indicate lesions with iCAT Tier 1 and 2 lesions, respectively. Candidate therapies and iCAT tiers of evidence for those therapies are listed to the right of the grid. (B-M) Drugs suggested in (A) were tested on PDX lines for effects on cell viability in vitro. The PI3K-mTOR inhibitor NVP-BGT226 (B-D), the CDK4/6 inhibitor Palbociclib (E-G), the BET inhibitor JQ-1 (H-J) and the HDAC inhibitor Panobinostat (K-M) were tested on Group 3 (B, E, H, K), Group 4 (C, F, I, L), and SHH (D, G, J, M) PDX lines. The non-transformed hepatocyte line HepaRG was used as a control. Data are shown as percent viability, calculated as described in Methods. IC50 values are listed next to each PDX name. For each PDX, three biological replicates were performed, and one representative replicate is shown.
Using this framework, the only Tier 1 mutations were in PTCH1 (in the SHH lines RCMB24, RCMB32 and MED1712), which has been shown in clinical trials of MB patients to predict responsiveness to SMO antagonists. Notably, one PDX line (RCMB18) had a SMO mutation (R145C). However, it is unknown if the resulting protein remains sensitive to SMO antagonists. Moreover, this line also had amplification of MYCN, a downstream target of the SHH pathway that would be expected to render the tumor resistant to this therapy (27). Thus, we did not consider RCMB18 to have a Tier 1 lesion.
The only Tier 2 lesion we observed was a PIK3CA (E545K) mutation in MB009, which could be susceptible to PI3K, mTOR, or AKT inhibitors. Phase I-III clinical trials in various cancers have suggested that PI3K pathway inhibitors have greater clinical benefit in PIK3CA mutant than in PIK3CA WT tumors (42). However, there is evidence that E545K mutations have an equivalent response to wild type PI3K (43), raising questions about the predictive value of this lesion. Although pre-clinical studies have suggested that PI3K inhibitors are effective against Group 3 as well as SHH MB (44,45) no clinical data are available for MB, so this lesion was considered Tier 2.
The majority of lesions were Tier 3 or Tier 4. Among these were the CDK4/6 amplifications found in ICb-1487 and RCMB18. In principle, amplification of these genes should lead to overexpression of the corresponding proteins, and this could reflect an increased dependency of the tumor on the kinases. On the other hand, higher levels of the kinases could necessitate increased levels of inhibitors to block kinase activity, suggesting that amplified tumors would be more resistant to these inhibitors. In fact, both phenomena have been observed clinically (46,47). Although preclinical studies have suggested that palbociclib is effective in SHH and Group 3 MB (31,48) these studies were done using models lacking amplification of CDK4/6. Thus, there is limited evidence to suggest that CDK4/6 amplification represents a biomarker for sensitivity to CDK4/6 inhibitors in MB.
Additional mutation-drug associations assigned Tier 3 status were Aurora kinase inhibitors for MYC, MYCN, and MYCL amplification (49), BET inhibitors for PTCH1 and SMO mutation and GLI2 amplification (40) and MYC, MYCN, and MYCL amplification (21) and arsenic trioxide for GLI2 amplification (39). HDAC inhibitors for CREBBP mutations (50) and EZH2 inhibitors for KDM6A mutations (41) were each assigned Tier 4 status, as preclinical evidence has been shown in other cancer(s) with a similar lesions (Fig. 2A).
To examine the validity of the DNA-based predictions, we tested some of the suggested drugs on PDX lines in vitro. Although SMO inhibitors have been shown to be effective against PTCH1 mutant MB tumors in vivo (25,27), several studies have suggested that MB cells lose dependency on the SHH pathway when they are placed in culture (51). Consistent with this, the PTCH1-mutant PDXs in our study were not inhibited by the SMO antagonist NVP-LDE225. PI3K inhibitors such as NVP-BGT226, BKM120, and GSK2126458, which were predicted to work on the PIK3CA mutant line MB009, were found to inhibit survival of all Group 3 lines, regardless of PIK3CA mutational status (Fig. 2B–D, Supp. Fig. 1A–F). These drugs were active against some SHH and Group 4 PDXs, but notably, were also toxic to the non-transformed hepatocyte line, HepaRG. These results suggest that PI3K inhibitors are effective on Group 3 MB, but are not selectively active against PIK3CA-mutant MB cells, and may also be toxic to normal cells.
A similar trend was seen with other drugs predicted to work based on genetic lesions. For example, the CDK4/6 inhibitor palbociclib was predicted to work on the CDK4/6-amplified lines RCMB18 and ICb-1487, but showed minimal activity against these lines. Conversely, we observed inhibition of most Group 3 and SHH lines, including several lines previously shown to be sensitive to these inhibitors (48) (Med-211FH, Med-411FH and Med-1712FH) (Fig. 2E–G). BET inhibitors have been suggested to be effective for tumors with PTCH1, SMO, or MYC/MYCN/MYCL alterations (among our PDXs, this includes all SHH lines and all Group 3 lines except for ICb-1572 and ICb-1299). However, these drugs were not effective against several of the amplified or mutant lines as well as all Group 4 lines, but were effective against the non-MYC-amplified line ICb-1572 (Fig. 2H–J, Supp. Fig. 1G–I). The EZH2 inhibitor, GSK126, worked similarly in all Group 4 lines and two Group 3 lines, despite being predicted to work only on DMB006, the Group 4 line with a KDM6A mutation (Supp. Fig. 1J–L). Finally, HDAC inhibitors, which were predicted to be particularly effective against the CREBBP-mutated lines Med1712, MB009 and ICb1572, were effective against all lines tested, regardless of CREBBP mutation status (Fig. 2K–M, Supp. Fig. 1M–R). Thus, for our MB PDXs, mutations were not predictive of drug response.
Identification of candidate therapies using gene expression data
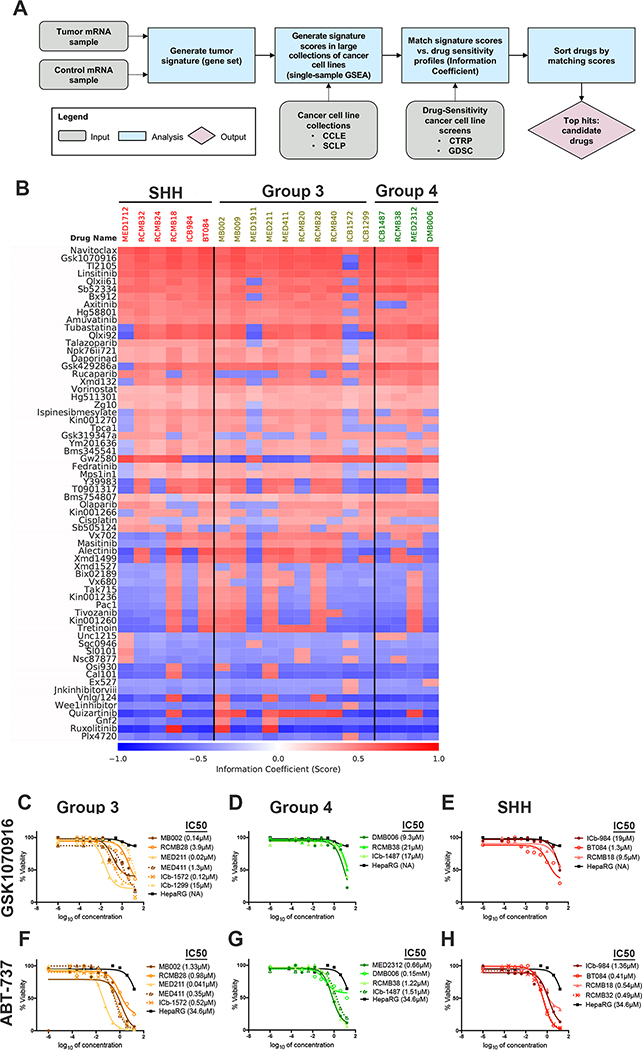
Changes in gene expression can also provide insight into pathways that drive tumor growth, and suggest therapies that might be used to disrupt these pathways (19,31). To determine whether this approach might be applicable to our PDXs, we performed gene expression profiling, and then used the DiSCoVER algorithm (31) to generate predictions of drug sensitivity (Fig. 3A). Briefly, the gene expression profile of each PDX (see Supp. Table 3A) was compared to that of normal cerebellar stem cells, and the most highly differentially expressed genes were used as a signature for that PDX (see Supp. Table 3B). DiSCoVER was then used to predict drug sensitivity, by comparing the signature of each PDX to the gene expression profiles of cell lines for which drug response data are publicly available (for details, see Methods). For each PDX, each drug was given a score that reflects the PDX’s predicted sensitivity to that drug: high scores suggest the PDX is likely to be sensitive and low or negative scores suggest that it is likely to be insensitive.
Figure 3: Expression data suggest additional drugs that may be useful against MB PDXs.
(A) The workflow for DiSCoVER analysis is depicted. By comparing gene expression between PDXs and control cells (human cerebellar stem cells), DiSCoVER created signatures for each PDX line and their activation in cells in cancer cell line collections were estimated. These scores were matched, via an information theoretic metric (IC), to drug sensitivity profiles of the cancer cell lines revealing a list of drugs to which the cell lines with activation of the signature were sensitive. (B) A heat map showing the top 25 drugs, suggested by DiSCoVER for each subgroup based on the information coefficient (IC) for each PDX. The IC quantifies how effective a given drug is expected to be on each PDX line. High values of the IC (maximum of 1, red) indicate predicted high effectiveness and low values of the IC (minimum of −1, blue) indicate predicted low effectiveness. Each drug’s target or mechanism of action is also listed. (C-K) Drugs suggested in (B) were tested in dose response on PDX lines for effects on viability in vitro compared to HepaRG control cells and plotted against percent viability. IC50 values are listed after each PDX name. For each PDX, three biological replicates were performed, and one representative replicate is shown. The Aurora kinase inhibitor GSK1070916 (C-E), the BCL2 inhibitor ABT-737 (F-H) were tested on Group 3 (C, F), Group 4 (D, G), and SHH (E, H) PDX lines.
Using the Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Therapeutics Response Portal (CTRP) databases, DiSCoVER analysis suggested drugs that would be effective for each PDX (Fig. 3B and Supp. Table 4A–B). Notably, the top 30 drugs predicted to be effective for each PDX were highly similar across most PDXs (Fig. 3B and Supp. Table 4). These included inhibitors of BCL2, Aurora kinases, receptor tyrosine kinases (IGF1R, EGFR, PDGFR), histone deacetylases, retinoic acid receptors (RARs) and poly (ADP-ribose) polymerase (PARP). Nonetheless, some drugs were predicted to be selectively active against a subset of PDXs. For example, GSK319347a and tpca1, both of which target IKK family members, were predicted to be more effective against SHH PDXs, whereas vx702, a p38 MAPK inhibitor, and xmd1499, an inhibitor of ALK, CDK7 and LTK, were predicted to work more selectively on Group 3 tumors. Rucaparib, a PARP inhibitor, was predicted to work on SHH and Group 4, and axitinib, an inhibitor of VEGFR and other RTKs, was predicted to work selectively on SHH and G3.
To determine if the drugs predicted based on gene expression were effective on the corresponding PDXs, we tested several of the top-ranked drugs in vitro. Based on this analysis, some of the drugs predicted to work on all subgroups were confirmed to be effective on most PDX lines tested. For example, the aurora kinase inhibitor GSK1070916 was among the top ranked predictions for all three subgroups (Supp. Table 4A), and in our in vitro studies we found it to be effective on all Group 3 and most SHH lines tested. It did not however, meet our efficacy criteria (IC50s were > 5μM) for Group 4 lines and for the SHH lines RCMB18 and ICb-984 (Fig. 3C–E). In contrast, the aurora kinase inhibitors alisertib and barasertib, suggested to work on all lines (Supp. Table 4B), showed efficacy on a subset of Group 3 lines, but were ineffective on PDXs derived from SHH and Group 4 tumors (Supp. Fig. 2A–F). BCL2 inhibitors, including ABT-737, navitoclax, and brd-m00053801, were predicted to work on all MB subgroups based on GDSC and CTRP (Supp. Table 4A,B), and efficacy was observed on all lines with ABT-737 and TW-37 (Fig. 3F–H, Supp. Fig. 2G–I). Notably, these drugs showed minimal activity on control HepaRG cells, suggesting that they did not exhibit non-specific toxicity at the doses tested.
In contrast, several drugs predicted to work on all PDXs or on specific subgroups showed no efficacy on most lines tested. Linsitinib, an IGFR1 inhibitor with high rank in all three subgroups (Supp. Table 4A,B), showed efficacy on all Group 3 lines but was less effective (maximal inhibition <50%) against SHH and Group 4 PDXs (Supp. Fig. 2J–L). SB52334, a TGF beta receptor I (ALK5) inhibitor, was a top-ranked prediction for all three subgroups (Supp Table 4A) but showed very little activity in all lines tested except the Group 3 line, ICb-1572, which was modestly inhibited (Supp. Fig. 2M–O). Likewise, the PARP inhibitors olaparib, rucaparib, and talazoparib were predicted to work on all subgroups (Supp. Table 4A,B), but in our assays, olaparib showed variable efficacy on Group 3 lines and minimal effect on lines from the other subgroups (Supp. Fig. 2P–R).
These results show that gene expression can be used to suggest drugs for most tumors. However, the majority of the top ranked drugs were predicted to be effective on most PDXs despite their divergent molecular profiles, and responses in vitro did not consistently match predictions.
Identification of candidate therapies based on high-throughput drug screening
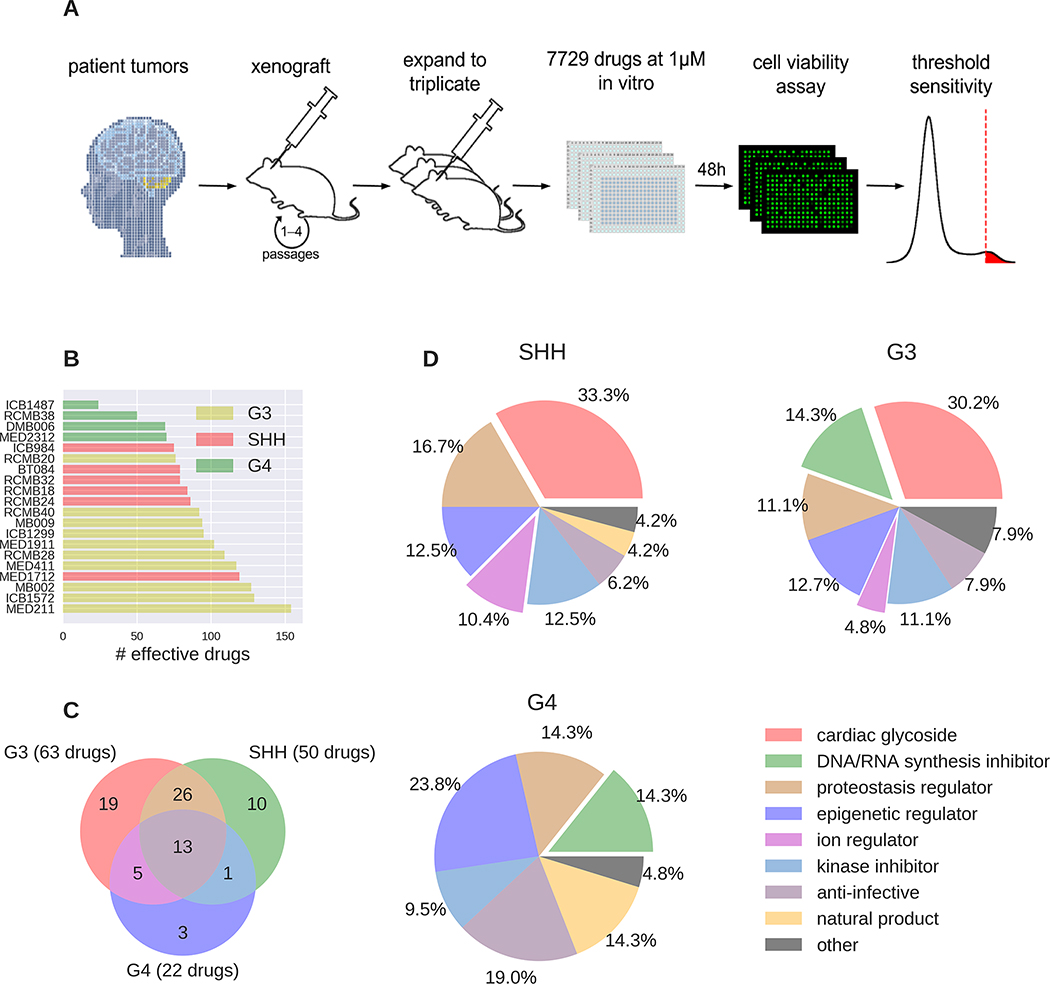
The strategies discussed above were used to infer sensitivity to drugs based on genomic or transcriptomic characteristics. To test for responsiveness empirically, we performed a drug screen on our cohort of PDXs using a library of 7729 compounds, including 4476 unique agents (Fig. 4A). Cells were cultured in the presence of compounds at a concentration of 1 μM for 48 hours, and cell viability was assessed using the CellTiter-Glo assay. Each drug was tested in triplicate, and average viability scores were calculated (Supp. Table 5). A drug was classified as effective against a particular PDX if cell viability scores for that drug were in the 0.1th percentile for that PDX (See Methods for details).
Figure 4: Drug screening reveals MB PDX vulnerabilities that were not predicted based on genome or transcriptome.
(A) Drug screening workflow is shown. PDXs were generated by orthotopic transplantation of patient tumor cells and expanded by “passaging” into new mice. At least 3 tumors from different mice of each PDX (biological triplicate) were dissociated and plated into 384 well plates containing the drugs for screening. Each well had a different drug, and the final concentration of each drug was 1μM. Forty-eight hrs after plating, a cell viability assay was performed. The threshold sensitivity was used to determine the viability cut-off and thus, which drugs were effective. (B) The bar graph represents the number of drugs found to be effective on each PDX line. The lines are color coded based on their subgroup (yellow = Group 3; red = SHH; green = Group 4). (C) The Venn diagram depicts the number of subgroup-effective drugs and shows the number of drugs that are effective in one or more subgroups. (D) All subgroup effective drugs were assigned a drug class. The percent of the effective drugs represented by each drug class is shown for drugs effective on SHH, Group 3, and Group 4 PDXs.
This analysis identified 375 drugs that were effective against at least one PDX line (Supp. Table 6A–C). Group 3 lines were generally sensitive to the highest number of drugs, whereas Group 4 lines tended to show sensitivity to fewer drugs (Fig. 4B,C). Efficacy in each subgroup was quantified using a binomial distribution (Supp. Table 7). We sorted the effective drugs for each subgroup into different drug classes (Fig. 4D). Among the most common classes of drugs effective against MB PDXs were cardiac glycosides (e.g. digoxin, digoxigenin, ouabain), inhibitors of DNA and RNA synthesis (e.g. idarubicin, daunorubicin, mitoxantrone), epigenetic regulators (e.g. apicidin) and regulators of protein homeostasis (e.g. bortezomib, MG132, gambogic acid). Interestingly, some classes of drugs showed specificity for certain subgroups. For example, SHH and Group 3 PDXs responded to cardiac glycosides and ion regulators whereas Group 4 PDXs did not. Likewise, Group 3 and Group 4 tumors responded to DNA/RNA synthesis inhibitors but SHH lines tended not to.
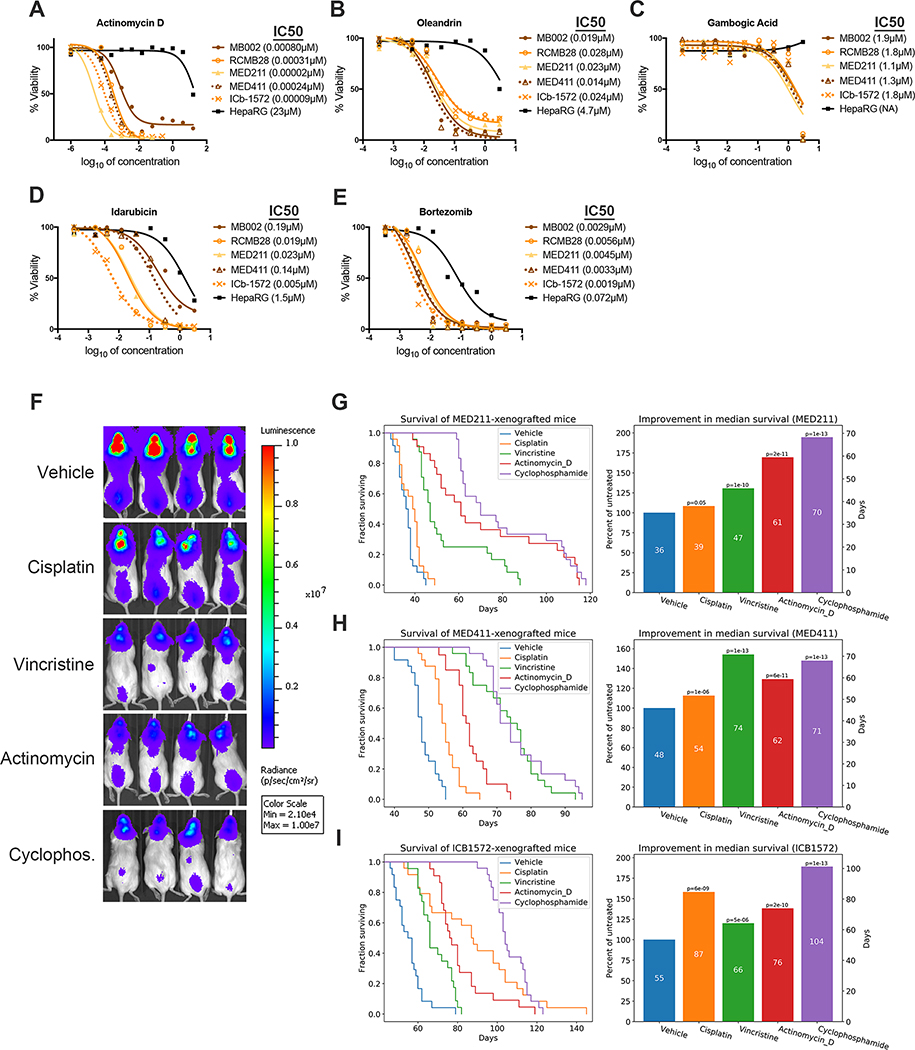
Since Group 3 MB is associated with particularly poor outcomes, we performed dose-response studies on several drugs predicted to be effective against Group 3 PDXs (Fig. 5A–E, Supp. Fig. 3A–O). Some of these drugs (e.g. Tyrphostin A9, PF-04691502, GSK2126458) were found to be toxic to HepaRG cells at doses similar to those that induced killing of Group 3 PDXs, and were therefore deprioritized. Among the remaining agents, we focused on those that are clinically available and have been used in patients, those for which there was available dosing information in mice, and those with novel mechanisms of action (for example, having already demonstrated efficacy of PI3K inhibitors and HDAC inhibitors in Group 3 MB (44), we did not focus on this class of drugs). Based on these criteria, we selected five agents – actinomycin D, oleandrin, gambogic acid, idarubicin and bortezomib – for further study.
Figure 5: Actinomycin D is at least as effective as standard-of-care drugs for Group 3 MB.
(A-E) Drugs were tested on Group 3 MB PDX lines to determine effects on viability in vitro. IC50 values are listed after each PDX name. For each PDX, three biological replicates with three technical replicates each were performed. One biological replicate is shown for each PDX. (A) Actinomycin D, (B) Oleandrin, (C) Gambogic Acid, (D) Idarubicin, (E) Bortezomib. (F-I) Mice with intracranial Group 3 PDX tumors were treated weekly with vehicle (10% DMSO i.p.), cisplatin (4.5 mg/kg i.p.), vincristine (1 mg/kg i.p.), actinomycin D (0.3 mg/kg i.v.), or cyclophosphamide (130 mg/kg i.p.) starting 2 weeks post-transplant, using the dosing schedule described in Methods. (F) Bioluminescent imaging of representative mice at 6 weeks post-transplant of MED411-FH. Survival curves and percent improvement in median survival above vehicle (100%) are shown for MED211-FH (G), MED411-FH (H), and ICb-1572 (I); data shown are averages of 3 experiments where each consisted of 40 mice with n=8 mice per group.
To test the efficacy of these agents in vivo, we performed pilot experiments (n = 8 mice per drug) on mice orthotopically transplanted with Med211-FH, which our drug screen had suggested was the most sensitive PDX line (see Fig. 4B). Notwithstanding their potency in vitro, idarubicin, gambogic acid, and oleandrin had no effect on survival of tumor-bearing mice, and bortezomib only marginally increased survival (Supp. Fig. 4A–D). In contrast, actinomycin D significantly prolonged survival (Supp. Fig. 4E), and was prioritized for further investigation. Notably, actinomycin D was much more effective on Group 3 PDXs (IC50s ranging from 0.02–0.31 nM, Fig. 5A) than on PDXs derived from Group 4 MB (10–57 nM) or SHH MB (IC50s from 9–33 nM) (Supp Fig. 4F–G). To optimize in vivo treatment with actinomycin D, we tested multiple dosing regimens, and found that weekly intravenous (i.v) administration was the most effective. We also tested combinations of actinomycin D with cyclophosphamide or radiation; while actinomycin D consistently prolonged survival as a single agent, toxicity was an issue when combined with other therapies.
We then compared the effects of actinomycin D to the standard chemotherapies used for MB – cisplatin, vincristine and cyclophosphamide – using three Group 3 PDX models. As shown in Fig. 5F–I, cyclophosphamide was generally the most effective drug in all PDX lines. Vincristine also prolonged survival in all PDX lines, but was particularly effective in MED411-FH, in which its activity was similar to cyclophosphamide. In contrast, cisplatin was highly effective on ICb-1572, but showed little activity on the other two lines. These studies demonstrated that drug response, even among standard of care therapies, is variable between PDX lines. Notably, actinomycin D was more effective than cisplatin and/or vincristine in all lines tested (Fig. 5F–I and Supp. Fig. 5A–C). These data suggest that actinomycin D could be an effective therapy for Group 3 MB.
Use of multimodal analysis for primary patient samples
The studies described above demonstrate the feasibility of using DNA sequencing, gene expression profiling, and empirical drug screening for predicting responses to therapeutic agents. To determine whether this approach could be used for patients in the clinic, we established a pipeline for acquiring DNA, RNA and drug response data on samples from brain tumor patients at Rady Children’s Hospital. WGS was done at 94x, and RNA sequencing was done at >30 million 150-base read pairs. Drug screening used a library of 120 drugs that are FDA-approved or in clinical trials for treating cancer (52).
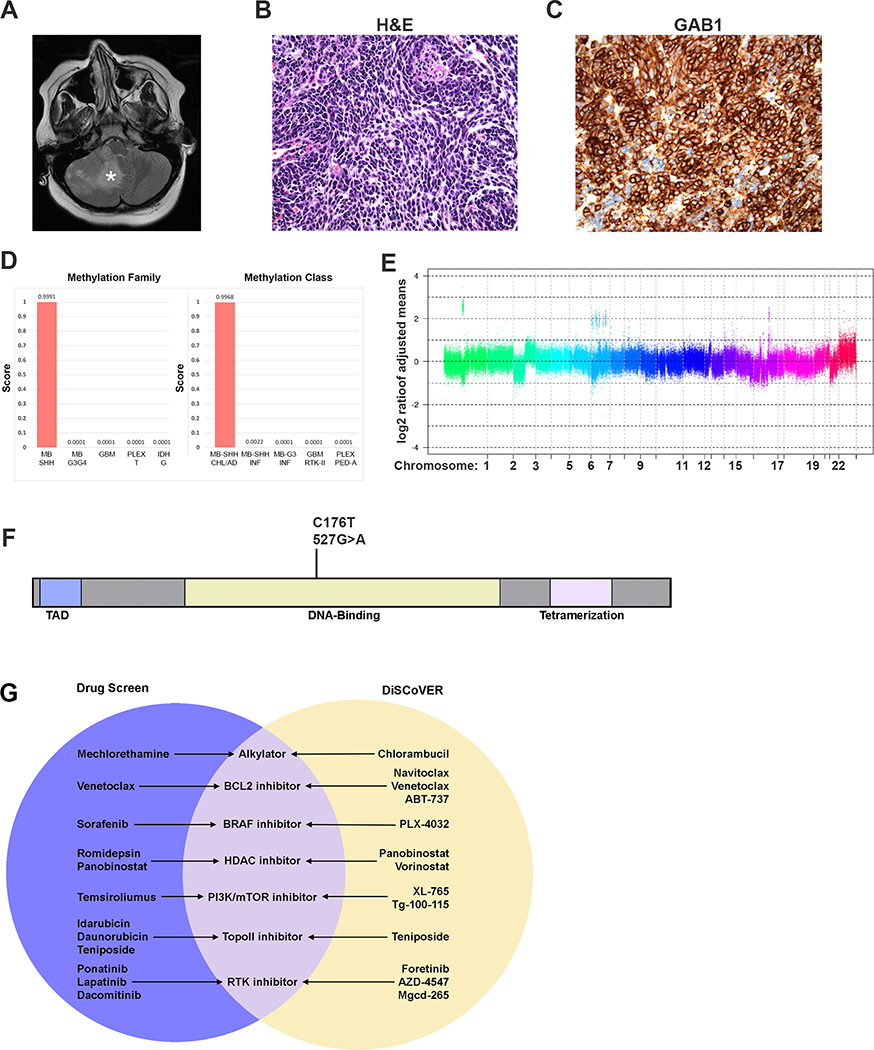
One case analyzed in this manner was an 8-year-old male with newly diagnosed, metastatic MB. The primary tumor was in the lateral cerebellum, as shown on T2-weighted MRI (Fig. 6A). Histopathology showed a densely cellular proliferation of small polygonal cells with high nuclear to cytoplasmic ratio, scant amphophilic cytoplasm, nuclear pleomorphism, molding, occasional cell-cell wrapping, and brisk mitotic activity, consistent with large cell/anaplastic MB (Fig. 6B). A majority of tumor cells were immunopositive for GAB1 (Fig. 6C), consistent with the immunophenotype of a SHH tumor (53). Moreover, subsequent DNA methylation analysis classified it as a SHH MB, subclass A (children and adults) (Fig. 6D).
Figure 6: A combination of DNA, RNA, and drug sensitivity analysis is feasible in patients.
(A) T2-weighted MRI reveals a right hemispheric cerebellar tumor with significant mass effect (B) Hematoxylin and Eosin staining of tumor sections reveals a small-round blue cell neoplasm with anaplastic features consistent with a diagnosis of large cell/anaplastic MB (C) GAB1 staining of tumor sections reveals diffuse positive staining. (D) DNA methylation family and class analysis scores. (E) SNP microarray analysis reveals chromosomal gains and losses. (F) The missense TP53 mutation at the nucleotide and amino acid level revealed by WGS. (G) The Venn diagram shows overlapping classes of drugs, predicted by both drug screening (purple) and DiSCoVER analysis (yellow). The middle (lavender) shows drug classes predicted by both approaches.
Genomic DNA sequencing of the tumor and SNP microarray analysis detected multiple complex copy number abnormalities and chromothripsis of chromosomes 1p, 7 and 17 (Fig. 6E and Supp. Table 8). Furthermore, a somatic p53 missense variant that translated to a p.Cys176Tyr substitution in the DNA binding domain was identified; this variant has been described in other cancer types (Fig. 6F) (54). The variant was not present in the patient’s germline DNA following targeted sequencing analysis. Copy number analysis by SNP microarray suggested loss of chromosome 17p (Supp. Table 8), which was confirmed by methylation profiling. Together, these data suggested that the patient had a TP53-mutated SHH tumor.
The patient’s tumor was also subjected to RNA sequencing and DiSCoVER analysis, and to high-throughput drug screening. The top 20 drugs predicted by DiSCoVER included BCL2 family inhibitors, and inhibitors of BRAF, PI3K/mTOR, HDACs and RTKs (Fig. 6G and Supp. Table 9–10). Drug screening identified distinct but overlapping classes of effective drugs, including RTK inhibitors, BCL2 inhibitors, HDAC inhibitors, a BRAF inhibitor, and mTOR inhibitors (Fig. 6G and Supp. Table 11). Our analysis suggested that it would be feasible to design a personalized treatment plan including drugs predicted by both gene expression and drug screening. However, consistent with standard practice, the patient was treated with high dose craniospinal proton therapy and adjuvant chemotherapy including cisplatin, cyclophosphamide and vincristine.
DISCUSSION
Genomic studies over the last decade have revealed that MB is a highly heterogeneous disease (4–6). However, most MB patients still receive the same therapy, comprised of surgical resection, craniospinal radiation and intensive chemotherapy. These treatments allow many patients to survive for five years or more, but ~1/3 of patients still die of their disease, and survivors suffer severe long-term side effects from therapy. Novel therapeutic approaches have been proposed for Group 3 tumors (24,31,44), but these have not yet shown efficacy in the clinic. Here, we test the hypothesis that DNA sequencing, RNA sequencing and empirical drug screening can help identify therapies that may be effective for a heterogeneous group of MB patients.
Most current efforts to find personalized therapies focus on genomic alterations. This offers the opportunity for targeted therapy and can sometimes suggest multiple therapies that can be used in combination. However, most pediatric cancers, including MB, have few actionable mutations. Instead, MB is often driven by copy number changes, structural variants or epigenetic changes (4), for which targeted therapies are often not available. Finally, many mutations that are considered actionable do not confer responsiveness to a therapy when tested empirically. For example, we observed that tumors with CDK4 or CDK6 amplifications do not necessarily show increased sensitivity to palbociclib, and tumors with CREBBP mutations are no more sensitive to HDAC inhibitors than those that lack such mutations. Thus, while sequencing may point to targeted therapies for some MB patients, most patients will not benefit from sequencing alone.
Expression-based strategies offer an alternative approach for identifying targeted therapies. Commonly used approaches include Connectivity map, DiSCoVER, and Ingenuity Pathway Analysis (19,31). One advantage of these approaches is that they almost always generate predictions for a given patient. However, as we have shown in our studies, drugs identified using expression-based approaches are not always validated when those drugs are tested empirically. One reason for this could be the discordance between the transcriptome (which is used to infer drug responsiveness) and the proteome (which is likely to mediate response, and resistance, to most anti-cancer drugs). In fact, recent studies have suggested that in Group 3 and 4 MB in particular, pathway activity predicted from RNA does not correlate well with that predicted from proteomic analysis (55,56). Another concern with expression-based prediction methods is that they are often based on the responses of cell lines that have been in culture for many years, and that are derived from other cancer types; for example, among the 1171 lines used for DiSCoVER analysis, only three (D283-Med, Daoy and ONS-76) are derived from MB patients. Since drug sensitivity may be context-specific, predictions based on a given tumor type may not be generalizable to other tumors, and this may explain why some RNA-based predictions were not validated by empirical drug testing on MB PDXs. Going forward, it may be helpful to generate MB-specific drug response datasets (from MB PDXs or patient samples) that can be used to predict responses in this disease.
Drug screening provides functional information about the susceptibility of tumor cells to therapeutic agents, and thus could be valuable for identifying effective therapies for patients in the clinic. But there are a number of caveats to this approach. For example, freshly isolated cells from a patient or PDX may not proliferate extensively in vitro, so the conditions used in our assay may underestimate the efficacy of drugs that inhibit proliferation without causing cell death. Furthermore, responses to drugs in vitro may not predict responses in vivo. This may be due to differences in drug metabolism or the ability of a drug to reach its target (for example due to the blood brain barrier), or to features of the tumor microenvironment (such as hypoxia, pH or the presence of other cell types) that modulate drug response. However, the fact that drug screening empirically measures responses rather than inferring them from other characteristics of tumor cells makes it a valuable source of information, particularly in cases where other approaches are not informative.
An example of the power of this approach is our identification of actinomycin D as a drug that is effective against the majority of Group 3 MBs. Actinomycin D was originally discovered as an antibiotic, but has been used for treatment of cancer – including childhood cancer – since the 1950s (57). Although it has been widely used for pediatric sarcomas (58,59), its use for MB has been very limited (60). One reason for this is the commonly held view that Actinomycin D does not cross the blood-brain barrier (BBB)(61). While this might be true, it is worth noting that many of the compounds we found to be active in vitro were not effective in orthotopic tumor-bearing mice (Figure 5B–E). Although there could be other reasons for this, it does that the tumors in our mice are not permeable to all compounds, and that actinomycin D might have some selective ability to accumulate in these tumors. It is also possible that actinomycin D does not cross the BBB efficiently, but that in our models the BBB is sufficiently disrupted to allow the drug to enter the tumor. To the extent that this is true, it is important to note that many patients with MB exhibit compromised blood-brain (or blood-tumor) barriers, so our models may accurately reflect how actinomycin D would behave in a patient.
A notable finding from our studies was that response to MB standard of care therapies varied among PDXs. Surprisingly, vincristine, whose efficacy has been called into question by some investigators (62), showed significant anti-tumor activity in all of the models we tested, and in one model appeared to be as effective as cyclophosphamide. Cisplatin, on the other hand, showed no activity in one line, modest activity in another, and was highly effective in only 1 out of 3 lines tested. Actinomycin D outperformed vincristine and/or cisplatin in all of our models. These findings call into question the utility of standard therapeutic regimens, and suggest that tailoring chemotherapies based on an individual patient’s response profile may be more effective. Our studies, along with recent work demonstrating the efficacy of actinomycin D in RelA-positive ependymoma, embryonal tumor with multilayered rosettes (ETMR) and glioblastoma (63–65), suggest the importance of reevaluating this agent in the context of pediatric brain tumors.
One of the most important conclusions from our study is that multimodal analysis, including DNA sequencing, RNA sequencing and drug screening, is feasible in a clinical setting. The patients studied at our center underwent surgical resection of their tumors, and excess tissue (beyond what was needed for diagnosis) was rapidly obtained and processed for sequencing and drug screening. Even for patients with recurrent tumors, there was usually sufficient tissue to perform all of these analyses. Moreover, while sequencing data were not available for a few weeks, drug screening was completed within a few days, making it one of the quickest sources of information regarding therapeutic responsiveness. Once all the data were collected, they were shared with a multidisciplinary molecular tumor board, and implications for diagnosis and therapy were discussed. As noted above, even for cases in which standard of care had been exhausted, multimodal analysis suggested possible therapies. Since most of the data were not obtained in a CLIA-certified setting, this information could not be used for clinical decision-making. But our results using this approach suggest that clinical grade versions of these tests could be extremely valuable and lead to significant improvements in therapy for both newly diagnosed and recurrent disease.
Supplementary Material
SIGNIFICANCE.
Findings show that high-throughput drug screening identifies therapies for medulloblastoma that cannot be predicted by genomic or transcriptomic analysis.
Acknowledgements
The authors gratefully acknowledge the Animal Facilities at UCSD and SBP for help with animal care and husbandry. We would also like to acknowledge the DKFZ Genomics and Proteomics Core Facility, DKFZ Heidelberg, Germany, and the AMC Department of Oncogenomics, Amsterdam, the Netherlands, for performing high-throughput sequencing and microarray analyses to a very high standard. We also thank the DKFZ data management group for their excellent support in processing the sequencing data. We are also indebted to the Conrad Prebys Center for Chemical Genomics, including Fu-Yue Zeng, Luis Orozco, and Michael Jackson, for their help with drug library selection and plating compounds and Sumeet Salaniwal for assistance with bioinformatics. We are most appreciative of Anthony Pinkerton and Robert Ardecky for their advice on in vivo drug testing. We gratefully acknowledge Brian James of the Genomics-DNA Analysis core at SBP for his help with PDX fingerprinting. This work was supported by funding from the National Cancer Institute (2R01 CA159859 (R.J.W.-R.), P30 CA30199 (R.J.W.-R.), U01CA184898 (J.P.M.), U24CA194107 (J.P.M.), U24CA220341 (J.P.M. and P.T.), U01CA217885 (J.P.M. and P.T.); the National Institute for Neurological Disorders and Stroke (R01 NS096368, R.J.W.-R.); the National Institute of General Medical Sciences (R01GM074024, J.P.M.), Deutsche Krebshilfe (111537) (M.K., S.M.P.), BMBF (01KT1605) (S.M.P.), and the Helmholtz International Graduate School for Cancer Research (S.Br.). SBP’s Shared Resources are supported by SBP’s NCI Cancer Center Support Grant, P30 CA030199. Methylation profiling at NYU was supported in part by a grant from the Friedeberg Charitable Foundation (M.S.). Work in R.J.W.-R.’s laboratory was also funded by the Pediatric Brain Tumor Foundation, Accelerate Brain Cancer Cure, Ian’s Friends Foundation, Alex’s Lemonade Stand Foundation, William’s Superhero Fund, the McDowell Charity Trust and the California Institute for Regenerative Medicine.
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 2011;29:1424–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 2011;29:1408–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 2008;3:e3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer 2012;12:818–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017;31:737–54 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma T, Schwalbe EC, Williamson D, Sill M, Hovestadt V, Mynarek M, et al. Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol 2019;138:309–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson GW, Orr BA, Wu G, Gururangan S, Lin T, Qaddoumi I, et al. Vismodegib Exerts Targeted Efficacy Against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J Clin Oncol 2015;33:2646–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goschzik T, Schwalbe EC, Hicks D, Smith A, Zur Muehlen A, Figarella-Branger D, et al. Prognostic effect of whole chromosomal aberration signatures in standard-risk, non-WNT/non-SHH medulloblastoma: a retrospective, molecular analysis of the HIT-SIOP PNET 4 trial. Lancet Oncol 2018;19:1602–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bui NQ, Kummar S. Evolution of early phase clinical trials in oncology. J Mol Med (Berl) 2018;96:31–8 [DOI] [PubMed] [Google Scholar]
- 11.Conley BA, Gray R, Chen A, O’Dwyer P, Arteaga C, Coffey B, et al. Abstract CT101: NCI-molecular analysis for therapy choice (NCI-MATCH) clinical trial: interim analysis. Cancer Research 2016;76:CT101–CT [Google Scholar]
- 12.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR Jr., Tsao A, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011;1:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Hoff DD, Stephenson JJ Jr., Rosen P, Loesch DM, Borad MJ, Anthony S, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010;28:4877–83 [DOI] [PubMed] [Google Scholar]
- 14.Sicklick JK, Kato S, Okamura R, Schwaederle M, Hahn ME, Williams CB, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med 2019;25:744–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol 2018;19:705–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen CE, Laetsch TW, Mody R, Irwin MS, Lim MS, Adamson PC, et al. Target and Agent Prioritization for the Children’s Oncology Group-National Cancer Institute Pediatric MATCH Trial. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7 [DOI] [PubMed] [Google Scholar]
- 18.Harris MH, DuBois SG, Glade Bender JL, Kim A, Crompton BD, Parker E, et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol 2016;2:608–15 [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Garrett Injac S, Cui K, Braun F, Lin Q, Du Y, et al. Systems biology-based drug repositioning identifies digoxin as a potential therapy for groups 3 and 4 medulloblastoma. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purzner T, Purzner J, Buckstaff T, Cozza G, Gholamin S, Rusert JM, et al. Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and a therapeutic target in medulloblastoma. Sci Signal 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res 2014;20:912–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gholamin S, Mitra SS, Feroze AH, Liu J, Kahn SA, Zhang M, et al. Disrupting the CD47-SIRPalpha anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci Transl Med 2017;9. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Liu Z, Yu L, Zhang Y, Baxter P, Voicu H, et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol 2012;14:574–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morfouace M, Shelat A, Jacus M, Freeman BB 3rd, Turner D, Robinson S, et al. Pemetrexed and gemcitabine as combination therapy for the treatment of Group3 medulloblastoma. Cancer Cell 2014;25:516–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brabetz S, Leary SES, Grobner SN, Nakamoto MW, Seker-Cin H, Girard EJ, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med 2018;24:1752–61 [DOI] [PubMed] [Google Scholar]
- 26.Brun SN, Markant SL, Esparza LA, Garcia G, Terry D, Huang JM, et al. Survivin as a therapeutic target in Sonic hedgehog-driven medulloblastoma. Oncogene 2015;34:3770–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014;25:393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet 2017;49:170–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 2014;20:682–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanaford AR, Archer TC, Price A, Kahlert UD, Maciaczyk J, Nikkhah G, et al. DiSCoVERing Innovative Therapies for Rare Tumors: Combining Genetically Accurate Disease Models with In Silico Analysis to Identify Novel Therapeutic Targets. Clin Cancer Res 2016;22:3903–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov 2015;5:1210–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhukova N, Ramaswamy V, Remke M, Pfaff E, Shih DJ, Martin DC, et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol 2013;31:2927–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Lee JJ, Kim J, Gardner D, Beachy PA. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc Natl Acad Sci U S A 2010;107:13432–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 2014;20:732–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezponda T, Dupere-Richer D, Will CM, Small EC, Varghese N, Patel T, et al. UTX/KDM6A Loss Enhances the Malignant Phenotype of Multiple Myeloma and Sensitizes Cells to EZH2 inhibition. Cell Rep 2017;21:628–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juric D, Krop I, Ramanathan RK, Wilson TR, Ware JA, Sanabria Bohorquez SM, et al. Phase I Dose-Escalation Study of Taselisib, an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov 2017;7:704–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res 2013;73:276–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, et al. HDAC and PI3K Antagonists Cooperate to Inhibit Growth of MYC-Driven Medulloblastoma. Cancer Cell 2016;29:311–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh AR, Joshi S, Zulcic M, Alcaraz M, Garlich JR, Morales GA, et al. PI-3K Inhibitors Preferentially Target CD15+ Cancer Stem Cell Population in SHH Driven Medulloblastoma. PLoS One 2016;11:e0150836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickson MA, Schwartz GK, Keohan ML, D’Angelo SP, Gounder MM, Chi P, et al. Progression-Free Survival Among Patients With Well-Differentiated or Dedifferentiated Liposarcoma Treated With CDK4 Inhibitor Palbociclib: A Phase 2 Clinical Trial. JAMA Oncol 2016;2:937–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 2017;36:2255–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cook Sangar ML, Genovesi LA, Nakamoto MW, Davis MJ, Knobluagh SE, Ji P, et al. Inhibition of CDK4/6 by Palbociclib Significantly Extends Survival in Medulloblastoma Patient-Derived Xenograft Mouse Models. Clin Cancer Res 2017;23:5802–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diaz RJ, Golbourn B, Faria C, Picard D, Shih D, Raynaud D, et al. Mechanism of action and therapeutic efficacy of Aurora kinase B inhibition in MYC overexpressing medulloblastoma. Oncotarget 2015;6:3359–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, et al. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov 2018;8:1422–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasai K, Romer JT, Lee Y, Finkelstein D, Fuller C, McKinnon PJ, et al. Shh pathway activity is down-regulated in cultured medulloblastoma cells: implications for preclinical studies. Cancer Res 2006;66:4215–22 [DOI] [PubMed] [Google Scholar]
- 52.Ding KF, Finlay D, Yin H, Hendricks WPD, Sereduk C, Kiefer J, et al. Analysis of variability in high throughput screening data: applications to melanoma cell lines and drug responses. Oncotarget 2017;8:27786–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol 2011;121:381–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oden-Gangloff A, Di Fiore F, Bibeau F, Lamy A, Bougeard G, Charbonnier F, et al. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br J Cancer 2009;100:1330–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Archer TC, Ehrenberger T, Mundt F, Gold MP, Krug K, Mah CK, et al. Proteomics, Post-translational Modifications, and Integrative Analyses Reveal Molecular Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2018;34:396–410 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forget A, Martignetti L, Puget S, Calzone L, Brabetz S, Picard D, et al. Aberrant ERBB4-SRC Signaling as a Hallmark of Group 4 Medulloblastoma Revealed by Integrative Phosphoproteomic Profiling. Cancer Cell 2018;34:379–95 e7 [DOI] [PubMed] [Google Scholar]
- 57.Tan CT, Dargeon HW, Burchenal JH. The effect of actinomycin D on cancer in childhood. Pediatrics 1959;24:544–61 [PubMed] [Google Scholar]
- 58.Arndt CAS, Bisogno G, Koscielniak E. Fifty years of rhabdomyosarcoma studies on both sides of the pond and lessons learned. Cancer Treat Rev 2018;68:94–101 [DOI] [PubMed] [Google Scholar]
- 59.Bernstein M, Kovar H, Paulussen M, Randall RL, Schuck A, Teot LA, et al. Ewing’s sarcoma family of tumors: current management. Oncologist 2006;11:503–19 [DOI] [PubMed] [Google Scholar]
- 60.Nathanson L, Kovacs SG. Chemotherapeutic response in metastatic medulloblastoma: report of two cases and a review of the literature. Med Pediatr Oncol 1978;4:105–10 [DOI] [PubMed] [Google Scholar]
- 61.Tattersall MH, Sodergren JE, Dengupta SK, Trites DH, Modest EJ, Frei E 3rd. Pharmacokinetics of actinoymcin D in patients with malignant melanoma. Clin Pharmacol Ther 1975;17:701–8 [DOI] [PubMed] [Google Scholar]
- 62.Boyle FM, Eller SL, Grossman SA. Penetration of intra-arterially administered vincristine in experimental brain tumor. Neuro Oncol 2004;6:300–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tzaridis T, Milde T, Pajtler KW, Bender S, Jones DT, Muller S, et al. Low-dose Actinomycin-D treatment re-establishes the tumoursuppressive function of P53 in RELA-positive ependymoma. Oncotarget 2016;7:61860–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt C, Schubert NA, Brabetz S, Mack N, Schwalm B, Chan JA, et al. Preclinical drug screen reveals topotecan, actinomycin D, and volasertib as potential new therapeutic candidates for ETMR brain tumor patients. Neuro Oncol 2017;19:1607–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor JT, Ellison S, Pandele A, Wood S, Nathan E, Forte G, et al. Actinomycin D Downregulates Sox2 and Improves Survival in Preclinical Models of Recurrent Glioblastoma. Neuro Oncol 2020; 22: 1289–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Short-read sequencing data are available at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/), hosted by the European Bioinformatics Institute, under accession number EGAS00001004698. Methylation and gene expression data have been deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE151344 and GSE151343, respectively. Gene expression analyses (Affymetrix processing, gene expression quantification, and gene expression analyses using DiSCoVER) can be reproduced following the documentation and using the code in the GenePattern GitHub Repository.