Abstract
Extracellular vesicles (EVs) represent promising circulating biomarkers for cancers, but their high-throughput analyses in clinical settings prove challenging due to lack of simple, fast, and robust EV assays. Here, we describe a bead-based EV assay detected by flow cytometry, which integrates EV capture using microbeads with EV protein analyses by flow cytometry. The assay is fast (< 4 hrs for 48 samples), robust and compatible with conventional flow cytometry instruments for high-throughput EV analysis. With the method, we successfully analyzed a panel of pancreatic cancer biomarkers in EVs from plasma samples of pancreatic cancer patients. The assay is readily translatable to other biomarkers or cancer types and can be run with standard materials on conventional flow cytometers, making it highly flexible and adaptable to diverse research and clinical needs.
Keywords: extracellular vesicles, pancreatic cancer, flow cytometry, biomarkers, diagnosis
Graphical Abstract
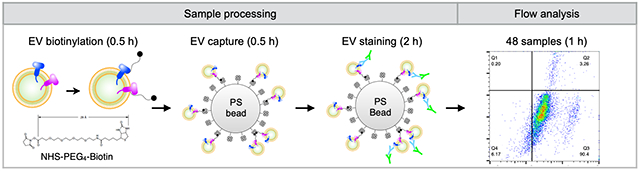
A bead-based EV assay detected by flow cytometry enables fast, robust, compatible with conventional flow cytometry instruments for high-throughput EV analysis. The method combines EV capture using polystyrene (PS) beads with EV protein analyses by flow cytometry. The assay can be run with standard materials on conventional flow cytometers, making it highly flexible and adaptable to diverse research and clinical needs.
Given the advanced and incurable disease often found with pancreatic ductal adenocarcinoma (PDAC) at initial presentation, timely and sensitive diagnostic methods are needed[1]. Current diagnostic methods are typically invasive and expensive[2], relying heavily on the use of imaging modalities such as CT, MRI, and PET, which often miss early disease even in high risk patients. Serum CA19-9 levels are clinically used as diagnostic and predictive biomarkers of PDAC[3], although their poor sensitivity and specificity (< 80%) often lead to false negative or positive results[4]. Research methods for noninvasive and early PDAC diagnoses have focused on circulating tumor DNA and cells or pancreatic fluid to identify rare epigenetic changes[5]. Recent work has examined extracellular vesicles (EVs) as potentially invaluable biomarker sources within bodily fluids[5–7].
EVs reflect a heterogeneous population of particles, such as exosomes or microvesicles, continuously shed into circulation by all cell types whether benign or malignant[8]. EVs contain proteins and RNA cargo highly reflective of their cell of origin, making them invaluable sources of potential biomarkers for liquid biopsy diagnostics[9–11]. Despite the ready accessibility of EVs within bodily fluids, currently no standard EV protein analysis methods exist that are fast and sensitive yet straightforward enough to implement across diverse laboratory settings. Conventional methods such as ELISA and Western blot have poor sensitivity for EV analysis due to their large sample amount requirements for measurement[10]. Extremely sensitive and novel methodologies are under development[7,10–13], but they often require specialized setups that are not yet commercialized for widespread use.
Flow cytometry is an alternative powerful tool widely used across laboratories and clinics for cellular analyses. Yet, EVs’ sub-micron sizes, polydispersity, limited surface areas, and low scattering intensities obviate use of conventional flow cytometry for reliable EV analyses[14,15]. Recently, several groups have reported single EV flow cytometry using custom built/dedicated flow cytometers [16–22]. Commercial systems capable of reliable single EV flow cytometry are also beginning to emerge[23–26]. While providing invaluable information about single EV heterogeneity, both custom built and commercial nanoscale flow cytometers are not yet readily available to clinical research settings. Additionally, direct labeling of EVs with antibodies involves extensive and time-consuming staining and washing processes, which could result in additional sample loss. Such challenges run counter to clinical workflow and biospecimen repository needs, respectively.
In response, we sought to develop an accessible and standardized method to rapidly analyze EV proteins from human samples in high-throughput. Notably, we developed a bead-based EV assay detected by flow cytometry that combined EV capture using polystyrene (PS) beads with EV protein analyses by flow cytometry. The bead-based flow assay offers several advantages over existing methods: i) improved EV capture efficiency using a high affinity biotin-streptavidin interaction; ii) an inexpensive, simplified assay easy to implement in laboratories equipped with standard flow cytometers; and iii) relatively quick turnaround time (~4 hours for 48 samples). Using the bead-based flow cytometry, we demonstrated sensitive detection of tumor-derived EVs directly from plasma samples of PDAC patients.
Figure 1A shows a schematic workflow of the bead-based flow cytometry assay for EV protein analysis. Isolated EVs were first biotinylated using NHS-PEG4-Biotin (30 min), followed by capture on 5 μm streptavidin-coated polystyrene (PS) beads in another 30 min reaction. Bead-bound EVs (shown in Figure S1 by scanning electron microscopy) were then stained with primary antibodies (60 min) followed by Alexa Fluor 488 secondary antibodies (30 min) for flow cytometry measurements. Analysis was done using a portable benchtop flow cytometer (CytoFLEX, Beckman Coulter Inc.) equipped with automatic handling of samples in a 96-well plate. We found that increasing the EV binding or primary antibody incubation time up to 24 hrs does not significantly improve the detection signal (Figure S2). For EV isolation, we used size-exclusion chromatography for human plasma samples and ultracentrifugation for cell-culture supernatants to process large volumes (see Experimental Section for details). Once EVs were isolated, the process from EV preparation to analyses took about 4 hrs for up to 48 samples.
Figure 1. Bead-based EV assay detected by flow cytometry.
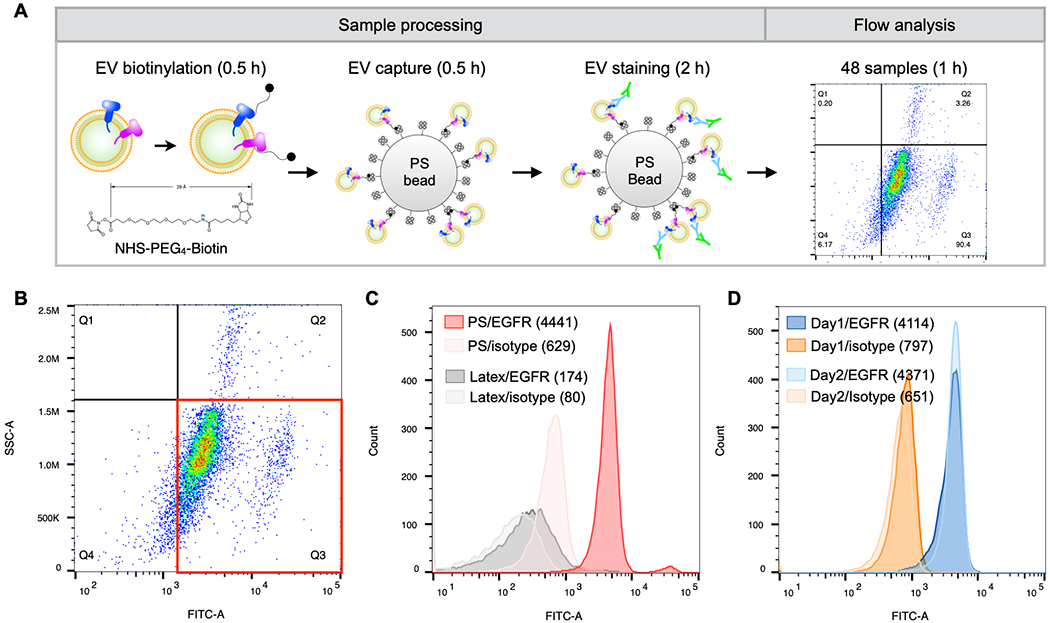
A. Following isolation from plasma, EVs were biotinylated and captured on 5 μm streptavidin-coated polystyrene (PS) beads. The captured EVs were then stained with primary and AlexaFluor 488 conjugated secondary antibodies. Samples were analyzed on a CytoFlex (Beckman Coulter) 96-well plate flow cyotmeter. The entire workflow is complete within 4 hrs for 48 samples. B. Dot plot of EVs from a patient-derived xenograft cell line (1617 PDAC) stained with a FITC-EGFR antibody. C. EVs from 1617 PDAC cells were adsorbed onto 3.9 μm aldehyde/sulfate latex beads (gray, 1.3 x 107 particles) or biotinylated and captured on 5 μm streptavidin PS beads (red, 1.8 x 106 particles). EV-bead conjugates were then stained using an EGFR antibody and AlexaFluor 488 secondary antibody for flow cytometry analysis. D. EVs from 1617 PDAC cells were isolated on different days and were stained with an isotype control (orange) or EGFR (blue) antibody for bead-based flow analysis on different days to assess assay reproducibility. All dot plots are gated on isotype control stained samples.
While new nanoscale flow cytometers have become available for single EV analysis[25,26], direct labeling and measurement of individual EVs in conventional flow cytometers is not trivial. We used the CytoFLEX violet side scatter for small particle detection and found only ~10% of measured EVs from a patient-derived xenograft cell line (1617 PDAC), exhibiting high EGFR expression[7], were positive for EGFR by flow cytometry (Figure S3). Moreover, single EV flow cytometry cab be difficult to interpret, making it cumbersome to incorporate into a clinical workflow. In contrast, attaching EVs to large PS beads with biotin-streptavidin, results in a signal over background readout that is simple to interpret. For example, 1617 EVs captured on 5 μm PS beads and stained with EGFR antibodies, resulted in a 7-fold increase in median fluorescence intensity (MFI) of EV-bead conjugates over isotype control (Figure 1B).
This highlights how the difficulty of direct EV analyses using conventional flow cytometry can be circumvented with the standardized and robust readouts of our bead-based flow cytometry assay.
Previous studies demonstrated bead-based EV capture using either affinity ligands (e.g. antibodies) for marker-specific EV capture or passive adsorption of EVs on aldehyde/sulfate latex beads with a hydrophobic surface[15,27]. The immunobead capture method requires identification of a set of antibodies for efficient capture. The passive EV adsorption on latex beads often requires a minimum 2.5 hr incubation. We reasoned that capturing biotinylated EVs could obviate the need to identify a set of capture antibodies and improve capture efficiency compared to the passive adsorption method. To test the capture efficiencies, we incubated EVs from the 1617 PDAC cell line with latex or streptavidin-coated PS beads for 30 min and labeled them with EGFR antibodies. EVs captured on latex beads by passive adsorption showed only ~2-fold increase in the MFI for EGFR whereas streptavidin-coated PS beads showed ~7-fold increase for EGFR (Figure 1C). The nonspecific binding of unlabeled EVs to streptavidin-PS beads or of biotinylated EVs to unmodified PS beads is negligible (Figure S4). The bead-based flow cytometry assay also showed good reproducibility with a variation of 6% in independently repeated experiments (Figures 1D and S5).
We expanded the comparison tests for other PDAC markers (e.g. EpCAM, MUC1, WNT2) and a PDACEV signature, comprising a cocktail of antibodies (EpCAM, EGFR, MUC1, WNT-2, GPC1, Figure 2A), that previously showed high PDAC detection accuracy[7]. In comparison with 3.9 and 5 μm aldehyde/sulfate latex beads, the 5 μm streptavidin-coated PS beads showed better signal for the EV markers tested, likely due to better EV capture on beads through the high affinity biotin-streptavidin interaction. Using 5 μm streptavidin-coated PS beads, we have performed titration experiments with increasing amounts of 1617 EVs and examined the flow cytometry signals in terms of MFI fold change over isotype control background (Figure 2B). With 500 ng of biotinylated 1617 EVs (~1.5 × 108 particles) incubated with 10 μl streptavidin-PS beads (1.8 × 106 beads), MFI fold change is saturated. For lower concentrations, the MFI fold change increases linearly. We tested limit of detection (LOD) values in the bead-based flow cytometry assay for individual EV markers and PDACEV signature (i.e. marker combination); the LODs for EV protein and particle counts were estimated to 41.3 ng and 1.3 × 107 particles, respectively (Figures 2C and S6).
Figure 2. Validation of the bead-based flow method.
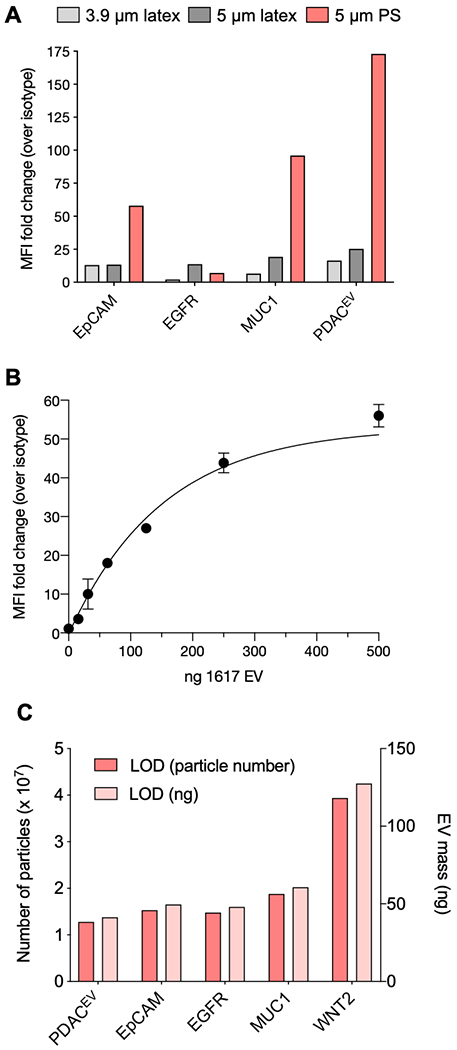
A. 1617 PDAC EVs were adsorbed onto 3.9 μm (light gray, 1.3 x 107 particles), 5 μm (dark gray, 4.9 x 106 particles) aldehyde/sulfate latex beads, or biotinylated and captured on 5 μm streptavidin polystyrene beads (red, 1.8 x 106 particles). EV-bead conjugates were stained with antibodies against EpCAM, EGFR, MUC1, or a five antibody cocktail (PDACEV: EpCAM, EGFR, MUC1, WNT-2, GPC1). Median fluorescence intensity (MFI) from each antibody was compared to an identical EV-bead conjugate stained with an isotype control antibody and the resulting fold change in MFI is shown. B. 1617 EV dilution in the BEAD flow assay. Beads were incubated with increasing ng amount of 1617 EV and then stained with either mouse IgG1k isotype control or EpCAM antibody. Increasing EV amount (in ng) was compared to MFI of isotype control antibody staining. C. The limit of detection (LOD) for the PDACEV antibodies as single markers and an antibody cocktail is shown in both the minimum number of particles and ng amount of EVs needed for bead-based flow.
We used Bead-based flow cytometry to test our PDACEV biomarker signature, a combination of EpCAM, EGFR, MUC1, GPC1, WNT-2[7], on EVs collected from various PDAC cell lines (Figure 3). We measured protein signal on EVs from five different cell lines for both individual markers and the marker combination in comparison with corresponding IgG controls. For example, EVs from 1617 PDAC cell line showed high expression for all individual markers, except GPC1 (Figures 3A). The low background signal of negative controls in the absence of EVs indicated low non-specific binding (Figure S7). The highest signal was obtained when we use the PDACEV signature, supporting that the marker combination improves the detection sensitivity for PDAC-derived EVs.
Figure 3. PDACEV biomarkers in cell line-derived EVs.
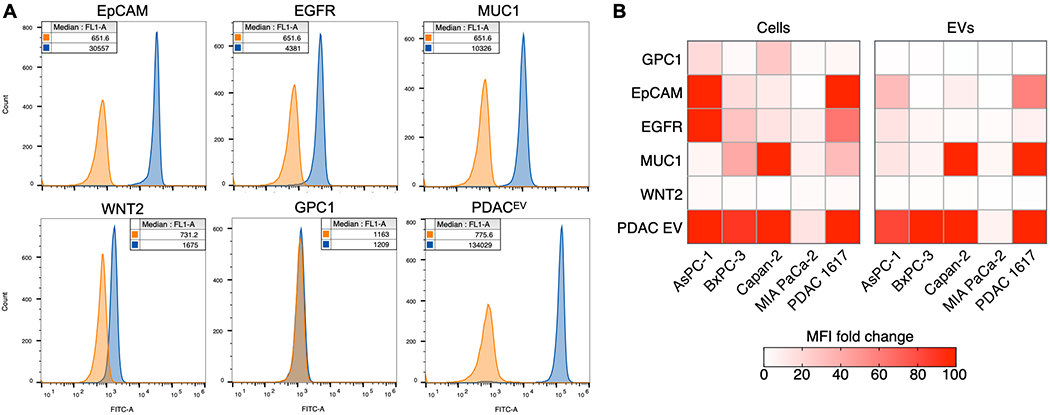
A. Biotinylated EVs were captured on 5 μm streptavidin polystyrene beads and stained with isotype control (orange) or primary antibodies (blue) for EpCAM, EGFR, MUC1, WNT-2, GPC1, and the PDACEV signature (mixture of EpCAM, EGFR, MUC1, WNT-2, GPC1), followed by AlexaFluor 488 secondary antibodies. Median fluorescence intensity values are shown in the legend. B. Cells and EVs isolated from the same cell lines were stained for surface biomarkers using the same antibodies as in A. Data are shown as the fold change in median fluorescence intensity of antibody over isotype control.
We next compared protein expression between EVs and their parental cells. The bead-based flow cytometry method was used for EV analyses. Cells were directly labeled with the same sets of antibodies (see Methods for details) and analyzed by the same flow cytometer as the bead-based flow cytometry assay. Comparing protein expression levels between EVs and their parental cells, we observed a moderate correlation (Spearman correlation coefficient r = 0.74, p < 0.0001; Figure 3B) for five PDAC cell lines, including one patient-derived xenograft cell line (PDAC 1617). For outliers, such as EGFR on 1617 EVs, single marker expression levels could have fallen below our method’s limit of detection. In the case of GPC1, low to moderate GPC1 expression was also observed in cell lines, with little to no expression evident on EV surfaces. To ensure the functionality of the GPC1 antibodies, we tested antibodies from two different vendors with purified GPC1 protein as well as EVs from the Capan-2 cell line (Figure S8). While we observed high signal (> 1000 fold increase over isotype) with purified GPC1 protein, little or no signal (< 1 fold) was observed in EVs, possibly because expression was below the detection limit of this assay[28].
To validate the clinical utility, we applied the bead-based flow cytometry assay to detect and analyze EVs from clinical samples. In this pilot clinical test, EVs were isolated from plasma in a cohort of 10 PDAC patients and 3 age-matched controls. On average, we used 0.5 mL of plasma samples. Due to these small volumes, we used size-exclusion columns (qEVoriginal, iZON Science) to isolate EVs (see methods for details). In addition to the five markers comprising the PDACEV signature, five additional markers for pancreatic cancer based on recent literature reports (CD73[29], TIMP1[30], EphA2[13], LRG1[30], and Mesothelin[31]) were also analyzed while consuming <1 mL plasma (Figure 4). Using bead-based flow cytometry, the PDACEV signature was able to differentiate PDAC patients from the control group, while none of the new markers outperform the PDACEV signature. To test the robustness of the bead-based flow cytometry assay, we compared MUC1 expression on aliquots of the same patient sample (P2), using two different flow cytometers for analysis. In this test, both measurements show comparable fold change in median fluorescence intensity over isotype control staining (Figure S9). The strategy of utilizing a marker combination for bead-based flow would allow us to analyze EVs from very small volumes of plasma (~100 μL, isolated with small volume size exclusion columns) with high sensitivity and specificity for cancer versus non-cancerous states.
Figure 4. Bead-based flow distinguishes PDAC from non-cancer patients.
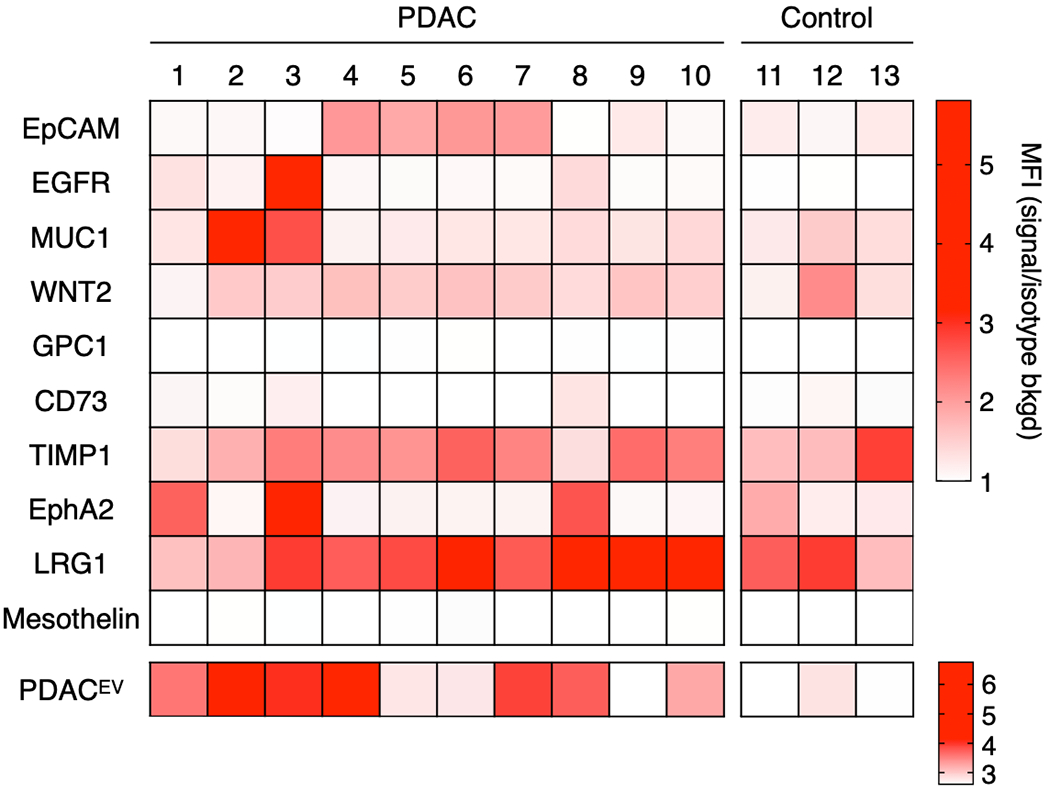
EVs were isolated from patient plasma using qEV columns (iZON), biotinylated, and captured on 5 μm streptavidin polystyrene beads. Beads were stained with the indicated primary antibodies (top heatmap) or with a PDACEV antibody cocktail (mixture of EpCAM, EGFR, MUC1, WNT-2, and GPC1; bottom heatmap). The fold change in median fluorescence over isotype control is depicted in the heatmaps with P1-10 representing pancreatic cancer EVs and C1-3 representing age-matched controls.
Discussion
EVs have emerged as potential circulating biomarkers for cancers, but high-throughput analysis in routine clinical settings has been challenging. Here, we describe a new EV protein analysis assay utilizing conventional flow cytometry. There are several challenges to directly apply standard operating procedures of flow cytometry used in cell analysis for EVs, mainly due to their small sizes. EVs range in size from 50 - 1000 nm, but the vast majority are smaller than 200 nm. The small sizes result in weak light scatter and fluorescence signals, due to limited amounts of antigen available per EVs, often below detection limits of conventional flow cytometers. Recent studies have suggested signal amplification strategies[32] and specialized nanoscale cytometers optimized for nanosized particles[17–24,33][25,26] to overcome the challenges. Other novel approaches that integrate EV isolation and detection in a single device have been suggested for direct EV analysis from plasma samples[34]. Although promising, these nascent approaches are not commercially available or widely adopted in clinical environments. To promote wider uptake, we implemented EV capture on microbeads. This offers an assay compatible with standard flow cytometers that is robust, standardized, and enables high-throughput analyses. Compared to immunobead-capture or adsorption methods, the bead-based flow cytometry assay: a) obviates the need to identify sets of capture and detection antibodies; b) performs well across different sample volumes, and c) completes the EV capture in under an hour.
Using BEAD flow, we analyzed EVs from 13 patient samples for a PDACEV biomarker signature (EGFR, EPCAM, MUC1, GPC1, and WNT2) with excellent sensitivity and reproducibility. We favored the multi-marker approach given tumoral heterogeneity and its demonstrated superior diagnostic accuracy over relying on single EV protein markers (e.g. GPC1). To further validate this proof-of-concept work, larger cohorts are needed to better assess statistical significance.
The current study focused on a developing a novel and versatile assay for conventional flow cytometers to eventually position it for rigorous clinical studies. There are opportunities to improve the bead-based flow cytometry technology: (i) currently limited to bulk measurements which cannot capture intrinsic heterogeneity of EV samples; (ii) the need for EV biotinylation; (iii) accurate quantification for tumor-derived EV counts and marker expression per EVs; (iv) modifying marker panels to further improve accuracy (e.g. KRAS mutants); (v) validation with larger cohorts across different institutions and varying pre-analytical handling. With new chemistry and signal amplification strategies, further optimization and validation, we envision that these points could be addressed and further improved.
While we focused on PDAC to develop this method, it can be expanded to include additional biomarkers and cancer types. Combining bead-based EV analysis with minimal sample consumption aligns with scientific workflow needs for both clinical evaluation and research (e.g. biorepositories where precious samples are sought to be leveraged to their fullest potential).
Experimental Section
EV Biotinylation:
We isolated EVs using size-exclusion columns for human plasma samples and ultracentrifugation for in vitro cell culture (Please see supporting information for detailed EV isolation protocols). Total EV protein was measured using 5-10 μl EVs and the Qubit protein assay kit (ThermoFisher, Q33212). Samples were biotinylated for 30 min in PBS (Mediatech, 21-040-CV) with a 20-fold molar excess of EZ-Link NHS-PEG4-Biotin (ThermoFisher, 21330) in 100-200 μl total volume. The following equation was used to calculate mmol biotin per reaction: mL EV*(mg EV/mL EV)*(mmol EV/150,000mg EV)*(20mmol biotin/mmol EV). Excess biotin was removed using MW 3000 Exosome Spin Columns (ThermoFisher, 4484449). Total biotinylated EV protein was again measured with the Qubit protein assay.
EV Capture on Beads:
10 μl (1.8 x 106 particles) of 5.0-5.9 μm streptavidin coated polystyrene particles (Spherotech, SVP-50-5, 0.5% w/v, 1.8 x 108 particles/mL) were diluted in 1 mL PBS/1% BSA (Mediatech 21-040-CV; Fisher, BP16051-00), centrifuged at 3000 × g for 2 min and supernatant was removed. Beads were mixed in a total of 10 μl with 500 ng EVs and PBS. For more than one sample of the same EV population, EVs were captured on beads in batch (ex: 5 μg EVs in 100 μl PBS/beads) and later split into multiple tubes for antibody staining. EVs were incubated for 30 min with beads on a HulaMixer (ThermoFisher, 15920D) at room temperature. Excess reacted EVs were removed by centrifuging at 3000 x g for 2 min and washing twice with PBS/1% BSA. EV-beads were diluted in 100 μl PBS/1% BSA and transferred to a 96-well u-bottom plate for antibody staining.
Flow Cytometry Staining and Analysis:
EVs and cells were stained using the same antibodies and procedure. Cells were prepared for flow staining by fixing 500,000 cells per antibody condition in 4% formaldehyde (ThermoFisher, 28908) in PBS for 15 min at room temp on a nutating mixer. Cells were washed in PBS/1% BSA and aliquoted to a 96-well u-bottom plate for staining. Cells or EVs were pelleted by centrifugation at 400 x g for 3 min (cells) or 1000 x g for 1 min (EVs on beads) in the 96-well plate and excess buffer was removed. Samples were resuspended in 100 μl PBS/1% BSA or in 10 μg/ml primary antibody diluted in the same buffer/volume and incubated for 30 min on a plate shaker set to medium speed (see Table S1 for a complete list of antibodies used in this study). For the PDAC EV antibody cocktail, all antibodies were mixed at a final concentration of 10 μg/ml. Cells or EVs were pelleted and washed twice with 150 μl PBS/1% BSA. Cells or EVs were then resuspended in 100 μl of the appropriate AlexaFluor 488 secondary antibody diluted 1:1000 in PBS/1% BSA and incubated for 30 min (protected from light) on a plate shaker. Samples were again pelleted, washed twice with 150 μl PBS/1% BSA, and resuspended in 200 μl PBS/1% BSA for flow analysis. Samples were measured using a Beckman Coulter CytoFlex flow cytometer (C09752, B2-R0-V2 configuration) with 96-well plate handling using the 525/40 nm bandpass filter (Beckman, A01-1-0051) and the following settings: cells FSC 49V, SSC 104V, FITC 20V; EVs FSC 201V, SSC 90V, FITC 159V. Sample backwash and mixing were turned off and the threshold was set automatically on FSC. Pre-gating was done to ensure single cells or beads were analyzed and a total of 10,000 events were collected within the gated area. FlowJo (v10) was used to analyze samples by measuring the median fluorescence intensity for proteins of interest and corresponding isotype controls. For comparisons, fold change in median fluorescence intensity was calculated by dividing the signal for a protein of interest over the isotype control. Prism 8 was used for statistical analyses.
Supplementary Material
Acknowledgements
The authors acknowledge funding from the U.S. National Institutes of Health (NIH) R01CA204019 (R.W.), P01CA069246 (R.W.), R00CA201248 (H.I.), R21CA217662 (H.I.), R01HL113156 (H.L.), R21CA205322 (H.L.); Lustgarten Foundation (R.W.); Dana Farber / Harvard Cancer Center GI SPORE (C.M.C.); and the Andrew L. Warshaw, M.D. Institute for Pancreatic Cancer Research at MGH (K.S.Y. and H.I.). Hsing-Ying Lin is supported by the Massachusetts General Hospital Fund for Medical Discovery (FMD) Postdoctoral Fellowship Award.
References
- [1].Lennon AM, Wolfgang CL, Canto MI, Klein AP, Herman JM, Goggins M, Fishman EK, Kamel I, Weiss MJ, Diaz LA, Papadopoulos N, Kinzler KW, Vogelstein B, Hruban RH, Cancer Res 2014, 74, 3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kaur S, Baine MJ, Jain M, Sasson AR, Batra SK, Biomark Med 2012, 6, 597.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Fogel EL, Shahda S, Sandrasegaran K, DeWitt J, Easler JJ, Agarwal DM, Eagleson M, Zyromski NJ, House MG, Ellsworth S, El Hajj I, O’Neil BH, Nakeeb A, Sherman S, Am J Gastroenterol 2017, 112, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maithel SK, Maloney S, Winston C, Gönen M, D’Angelica MI, Dematteo RP, Jarnagin WR, Brennan MF, Allen PJ, Ann Surg Oncol 2008, 15, 3512. [DOI] [PubMed] [Google Scholar]
- [4].Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W, Int J Clin Exp Med 2015, 8, 11683.; [PMC free article] [PubMed] [Google Scholar]; Kawai S, Suzuki K, Nishio K, Ishida Y, Okada R, Goto Y, Naito M, Wakai K, Ito Y, Hamajima N, Int J Cancer 2008, 123, 2880. [DOI] [PubMed] [Google Scholar]
- [5].Herreros-Villanueva M, Bujanda L, Ann Transl Med 2016, 4, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R, Nature 2015, 523, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang KS, Im H, Hong S, Pergolini I, Del Castillo AF, Wang R, Clardy S, Huang CH, Pille C, Ferrone S, Yang R, Castro CM, Lee H, Del Castillo CF, Weissleder R, Sci Transl Med 2017, 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Raposo G, Stoorvogel W, J Cell Biol 2013, 200, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rak J, Guha A, Bioessays 2012, 34, 489.; [DOI] [PubMed] [Google Scholar]; Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R, Nat Commun 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H, Nat Biotechnol 2014, 32, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H, Nat Med 2012, 18, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM, J Extracell Vesicles 2015, 4, 26659.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, Zöller M, Int J Cancer 2015, 136, 2616.; [DOI] [PubMed] [Google Scholar]; Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, Chiocca EA, Breakefield XO, Lee H, Weissleder R, ACS Nano 2018, 12, 494.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H, ACS Nano 2016, 10, 1802.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, Nonaka R, Yamamoto H, Ishii H, Mori M, Furuta K, Nakajima T, Hayashi H, Sugisaki H, Higashimoto H, Kato T, Takeshita F, Ochiya T, Nat Commun 2014, 5, 3591.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhao Z, Yang Y, Zeng Y, He M, Lab on a Chip 2016, 16, 489.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Ko J, Hemphill MA, Gabrieli D, Wu L, Yelleswarapu V, Lawrence G, Pennycooke W, Singh A, Meaney DF, Issadore D, Sci Rep 2016, 6, 31215.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Sevenler D, Daaboul GG, Ekiz Kanik F, Ünlü NL, Ünlü MS, ACS Nano 2018, [DOI] [PubMed] [Google Scholar]
- [13].Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z, Hu Y, Nat Biomed Eng 2017, 1, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Szatanek R, Baj-Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J, Int J Mol Sci 2017, 18, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Morales-Kastresana A, Jones JC, Methods Mol Biol 2017, 1545, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Headland SE, Jones HR, D’Sa AS, Perretti M, Norling LV, Sci Rep 2014, 4, 5237.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Kibria G, Ramos EK, Lee KE, Bedoyan S, Huang S, Samaeekia R, Athman JJ, Harding CV, Lötvall J, Harris L, Thompson CL, Liu H, Sci Rep 2016, 6, 36502.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, Wang S, Li Z, Chen C, Li L, Wu L, Yan X, ACS Nano 2018, 12, 671. [DOI] [PubMed] [Google Scholar]
- [17].Yang L, Zhu S, Hang W, Wu L, Yan X, Anal Chem 2009, 81, 2555. [DOI] [PubMed] [Google Scholar]
- [18].Steen HB, Cytometry A 2004, 57, 94. [DOI] [PubMed] [Google Scholar]
- [19].Nolan JP, Duggan E, Methods Mol Biol 2018, 1678, 79. [DOI] [PubMed] [Google Scholar]
- [20].Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, Bryja V, J Extracell Vesicles 2015, 4, 25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stoner SA, Duggan E, Condello D, Guerrero A, Turk JR, Narayanan PK, Nolan JP, Cytometry A 2016, 89, 196. [DOI] [PubMed] [Google Scholar]
- [22].Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, Rosner A, Demberg T, Watson DC, Karpova TS, Freeman GJ, DeKruyff RH, Pavlakis GN, Terabe M, Robert-Guroff M, Berzofsky JA, Jones JC, Sci Rep 2017, 7, 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lacroix R, Robert S, Poncelet P, Dignat-George F, Semin Thromb Hemost 2010, 36, 807. [DOI] [PubMed] [Google Scholar]
- [24].van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH, Nat Protoc 2012, 7, 1311. [DOI] [PubMed] [Google Scholar]
- [25].Görgens A, Bremer M, Ferrer-Tur R, Murke F, Tertel T, Horn PA, Thalmann S, Welsh JA, Probst C, Guerin C, Boulanger CM, Jones JC, Hanenberg H, Erdbrügger U, Lannigan J, Ricklefs FL, El-Andaloussi S, Giebel B, J Extracell Vesicles 2019, 8, 1587567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brittain GC, Chen YQ, Martinez E, Tang VA, Renner TM, Langlois MA, Gulnik S, Sci Rep 2019, 9, 16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Théry C, Amigorena S, Raposo G, Clayton A, Curr Protoc Cell Biol 2006, Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- [28].Lucien F, Lac V, Billadeau DD, Borgida A, Gallinger S, Leong HS, Oncotarget 2019, 10, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang B, Cancer Res 2010, 70, 6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL, Maitra A, Tempero MA, Brand R, Firpo MA, Mulvihill SJ, Katz MH, Brennan P, Feng Z, Taguchi A, Hanash SM, J Natl Cancer Inst 2017, 109, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Clin Cancer Res 2001, 7, 3862.; [PubMed] [Google Scholar]; Li M, Bharadwaj U, Zhang R, Zhang S, Mu H, Fisher WE, Brunicardi FC, Chen C, Yao Q, Mol Cancer Ther 2008, 7, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shen W, Guo K, Adkins GB, Jiang Q, Liu Y, Sedano S, Duan Y, Yan W, Wang SE, Bergersen K, Worth D, Wilson EH, Zhong W, Angew Chem Int Ed Engl 2018, 57, 15675.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Löf L, Ebai T, Dubois L, Wik L, Ronquist KG, Nolander O, Lundin E, Söderberg O, Landegren U, Kamali-Moghaddam M, Sci Rep 2016, 6, 34358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hercher M, Mueller W, Shapiro HM, J Histochem Cytochem 1979, 27, 350. [DOI] [PubMed] [Google Scholar]
- [34].Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T, Aiello NM, McKenzie L, O’Hara M, Redlinger C, Romeo J, Carpenter EL, Stanger BZ, Issadore D, ACS Nano 2017, 11, 11182.; [DOI] [PubMed] [Google Scholar]; Reátegui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, Floyd FP, Khankhel AH, Thapar V, Hochberg FH, Sequist LV, Nahed BV, Carter BS, Toner M, Balaj L, Ting DT, Breakefield XO, Stott SL, Nat Commun 2018, 9, 175.; [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y, Huang TJ, Proc Natl Acad Sci U S A 2017, 114, 10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


