Abstract
Clinical and preclinical studies suggest that some of the behavioral alterations observed in schizophrenia (SZ) may be mechanistically linked to synaptic dysfunction of glutamatergic signaling. Recent genetic and proteomic studies suggest alterations of cortical glutamate receptors of the AMPA-type (AMPARs), which are the predominant ligand-gated ionic channels of fast transmission at excitatory synapses. The impact of gene and protein alterations on the electrophysiological activity of AMPARs is not known in SZ. In this proof of principle work, using human postmortem brain synaptic membranes isolated from the dorsolateral prefrontal cortex (DLPFC), we combined electrophysiological analysis from microtransplanted synaptic membranes (MSM) with transcriptomic (RNA-Seq) and label-free proteomics data in 10 control and 10 subjects diagnosed with SZ. We observed in SZ a reduction in the amplitude of AMPARs currents elicited by kainate, an agonist of AMPARs that blocks the desensitization of the receptor. This reduction was not associated with protein abundance but with a reduction in kainate’s potency to activate AMPARs. Electrophysiologically-anchored dataset analysis (EDA) was used to identify synaptosomal proteins that linearly correlate with the amplitude of the AMPARs responses, gene ontology functional annotations were then used to determine protein-protein interactions. Protein modules associated with positive AMPARs current increases were down-regulated in SZ, while protein modules that were up-regulated in SZ were associated with decreased AMPARs currents. Our results indicate that transcriptomic and proteomic alterations, frequently observed in the DLPFC in SZ, converge at the synaptic level producing a functional electrophysiological impairment of AMPARs.
Keywords: AMPA receptors, microtransplantation of synaptic membranes, schizophrenia, synaptic dysfunction
1. Introduction
Alterations of excitatory synaptic function have been found in transcriptomic, genetic, and proteomic studies of schizophrenia (SZ) (Akbarian et al., 1996; Banerjee et al., 2015; Catts et al., 2015; Catts et al., 2016; Gluck et al., 2002; Hall et al., 2020; Harrison, 1999; Harrison and Weinberger, 2005; Hu et al., 2015; Mirnics et al., 2000; Pinner et al., 2016; Sequeira et al., 2012; Vawter et al., 2002). Clinical and preclinical studies suggest that abnormal synaptic transmission and several behavioral abnormalities in SZ may be mechanistically linked to abnormal glutamatergic neurotransmission, probably through hypofunction of N-methyl-D-aspartate (NMDA) receptors (NMDARs), in brain regions affected in SZ (Coyle et al., 2003; Gilmour et al., 2012; Hardingham and Do, 2016; Jadi et al., 2016; Krystal et al., 2002; Krystal et al., 2003; Krystal et al., 1994). However, the limited clinical efficacy of pharmacological modulation of NMDARs in clinical and preclinical trials indicate that synaptic dysfunction in SZ is a far more complex process (Fleischhacker and Miyamoto, 2016; Miyamoto et al., 2012). For instance, the function of NMDARs at the synapse is interdependent with the activity of α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors (AMPARs) (Cull-Candy and Leszkiewicz, 2004). At rest, NMDARs are blocked by extracellular Mg2+; activation of postsynaptic AMPARs removes the Mg2+ blockade allowing NMDARs to be permeable and electrically active (Mayer and Westbrook, 1987; Mayer et al., 1984; Nowak et al., 1984). Thus, abnormal responses of AMPARs may lead to hypofunction of NMDARs. Indeed, a recent genetic study found that mutations in GRIA2, the gene for the most expressed cortical AMPARs subunit GluA2, is strongly associated with SZ (Gulsuner et al., 2020). Similarly, synaptic GluA2 and GluA3 expression were mostly, but not uniformly, reduced in the auditory cortex of people with SZ (MacDonald et al., 2019). Taken together these recent studies strongly suggests a primary role of AMPARs in SZ.
Although AMPARs are constructed as heterotetramers from a pool of only four subunits (GluA1-GluA4), a large functional heterogeneity arises from diverse heteromeric assemblies, alternative splicing (Sommer et al., 1990), RNA editing (Wright and Vissel, 2012), post-translational modifications (Lussier et al., 2015), and transmembrane accessory proteins (Carbone and Plested, 2016; Diaz, 2010). Notably, transcriptional and proteomic changes of AMPARs have been demonstrated in the dorsolateral prefrontal cortex (DLPFC) of brains from SZ patients; although some studies have also found no changes (Corti et al., 2011; Hammond et al., 2010; Pinner et al., 2016; Rubio et al., 2012). Importantly, the direct measurement of ion currents through synaptic receptors from fresh-frozen brains from subjects with SZ has never been undertaken, and the impact of reported transcriptional and proteomic alterations that can affect the physiology of synaptic receptors in SZ is not known. In this proof of principle work using human postmortem brain synaptic membranes, we show that the function of synaptic AMPARs is significantly altered in SZ. Our results indicate that transcriptomic and proteomic alterations observed in the dorsolateral prefrontal cortex (DLPFC) from SZ brains, converge at the synaptic level producing functional impairment of AMPARs currents.
2. Materials and Methods
This preliminary research was performed using gray matter from the DLPFC in a cohort of 10 SZ cases and 10 controls from which we first analysed expression using RNA-Seq; second, label-free proteomics was performed using liquid chromatography-mass spectrometry (LC-MS/MS) in enriched-synaptosome preparations, and finally, we studied two-electrode voltage clamp electrophysiological (TEVC) profiles using microtransplantation of synaptic membranes (MSM) (Fig. 1A). Postmortem brain specimens of the DLPFC (BA9 plus 46, without selection for the left or right hemisphere) were obtained from the University of California Irvine Brain Bank (UCIBB) following the university’s Institutional Review Board and after obtaining verbal and written consent from next of kin. A psychological autopsy was completed based on family informant interview, medical and psychiatric records, coroner’s toxicology reports, and the subject’s medication history. The UCIBB autopsy protocol is based largely on procedures validated by Kelly and Mann (Kelly and Mann, 1996) and includes questions concerning the decedents’ demographics, medical history, psychiatric symptoms, medication use, hospitalizations, substance use, and physical health. The human brain dissection and freezing protocol are described in detail elsewhere (Jones et al., 1992). Briefly, after collection, frontal cortex samples were frozen in isopentane at −40° C and then stored at −80° C for downstream studies. The subjects’ demographic comparison for age, gender, postmortem interval (PMI), and RNA integrity number (RIN) showed that pH and RIN were significantly different in subjects with SZ compared to controls (Table 1 and supplementary data 1 for demographics, use of drugs, and prescribed medications use). Correction for confounding variables is described in the transcription profile section. This cohort was free of known disorders that could potentially cause synaptic alterations due to neurodegeneration, such as Alzheimer’s disease, epilepsy, AIDS, or Parkinson’s disease. The DLPFC was selected because the altered activation of this region in functional imaging studies have shown that ‘hypofrontality’ is one of the best-replicated findings in patients with SZ (Barch et al., 2001; Bolkan et al., 2016; Perlstein et al., 2001; Weinberger and Berman, 1996; Weinberger et al., 1988).
Figure 1. Functional enrichment and network interactions of differentially regulated genes in RNA-Seq datasets.
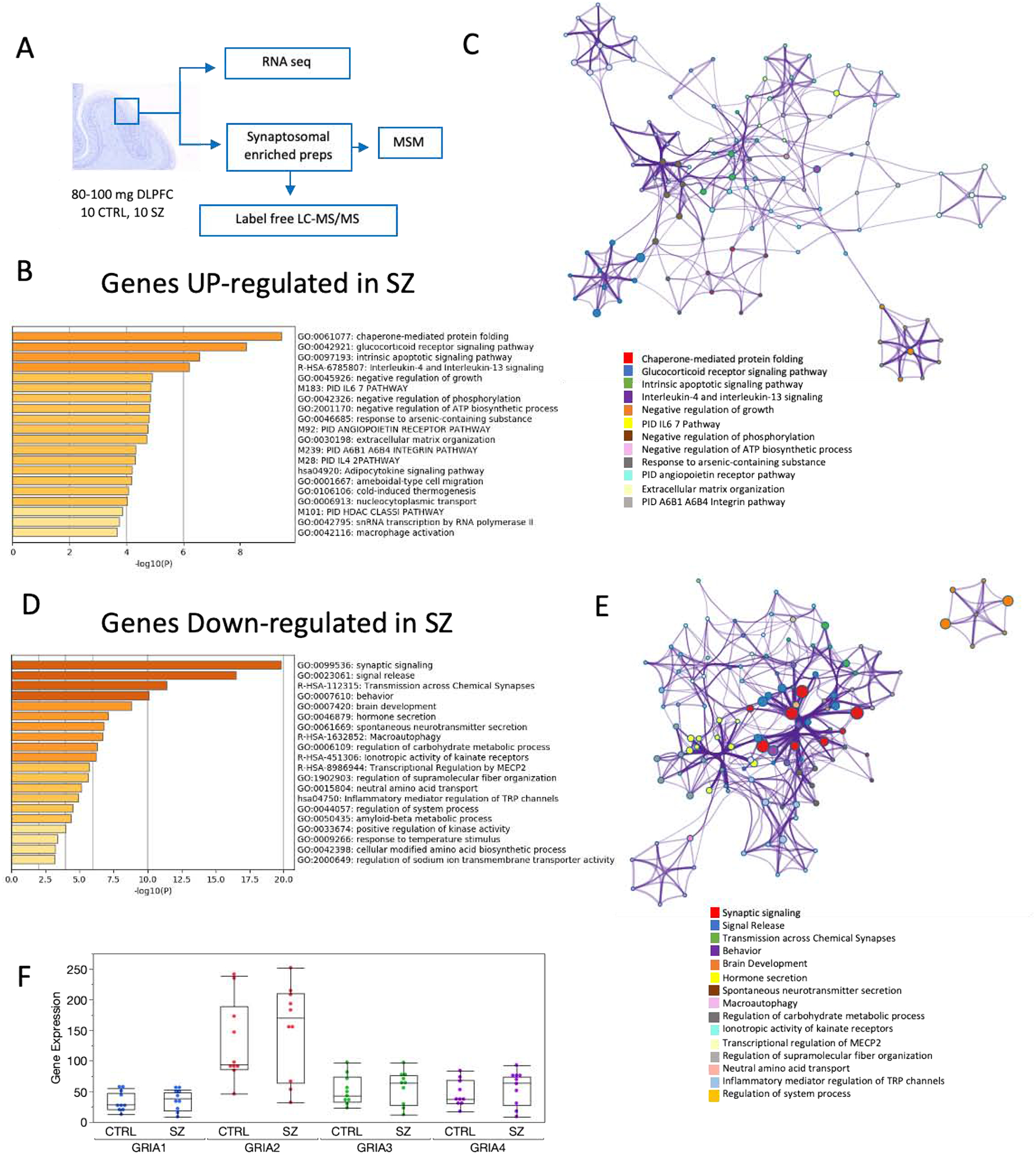
A. Diagram illustrating the methodological approach for processing fresh-frozen DLPFC for the different levels of analyses presented in this study. B. D. Enriched ontology clusters for genes found to be differentially expressed in SZ at the p < 0.01 level of significance using Metascape (Zhou et al., 2019). C, E. Network layout of gene interactions wherein each gene is a circle node. The node size is proportional to the number of input genes that fall in that gene and color is the enrichment identity, as per the color insert at the bottom of the network. F. Gene expression levels transcripts per million (TPM) for AMPARs subunits in control and schizophrenia (mean ± SEM).
Table 1.
Cohort demographics showing mean ± standard deviation.
| Diagnosis | Gender (F/M) | Age (years) | PMI (hr) | pH | RIN |
|---|---|---|---|---|---|
| SZ | 3/7 | 44 ± 10 | 22.8 ± 10 | 6.6 ±0.4 | 5.4 ± 1.2 |
| CTRL | 3/7 | 48 ±13 | 20.5 ± 8 | 6.2 ± 0.2* | 6.6 ±1.3* |
P<0.05 (Student t test), N = 10 each group. PMI, postmortem interval.
2.1. Transcriptomic Profile
We characterized the transcriptome from the gray matter of 20 DLPFC samples by RNA-Seq. RNA was prepared from 80 to 100 mg of frozen tissue samples, with Omni Prep Multi-Sample Homogenizer (Omni International, Kennesaw, GA). Total RNA isolation was performed with TRIZOL™ reagent (Invitrogen, Carlsbad, CA), following the manufacturer’s instructions. RNA was quantified by OD260/280 with a UV spectrophotometer and treated with RNase-free DNase using the RNeasy MinElute columns (Qiagen, Valencia, CA). The quality of the total RNA was finally evaluated using the Agilent 2100 Bioanalyzer RNA Chip (Santa Clara, CA). Libraries were prepared using Illumina TruSeq RNA Sample Prep kit v2., following manufacturer’s instructions. Pooled libraries (8–12 samples per lane) were then sequenced on a HiSeq 2000 platform, generating 100 bp paired-end reads, yielding an average of 299,831,258 reads per lane. For data analysis, first, TruSeq adapter sequences were trimmed using cutadapt (v2.5; with settings -n 3 --trim-n -m 40). Reads mapping to the human transcriptome (Gencode version 32) were then quantified using Salmon (Patro et al., 2017) (v0.14.1; indexed with -k 31 and quantified with settings --validateMappings --mimicBT2 --gcBias --seqBias). Mapping rates were 83.63% ± 1.05% (mean ± SD). Read counts were then summarized at the gene level, and genes with counts per million (CPM) >5 in at least two samples were considered expressed, yielding a total of 13,849 expressed genes used for all downstream analyses. Latent variables were estimated using surrogate variable analysis (SVA; svaseq approach (Leek, 2014; Leek et al., 2012). Differential gene expression was analyzed using the edgeR Bioconductor package (Robinson et al., 2010); gene-level counts were fit to a negative binomial model containing diagnosis and four surrogate variables representing latent variables. Two of the four surrogate variables (SVs) were highly correlated with RIN and pH respectively (supplementary figure 1). Differential gene expression between controls and SZ was assessed using Likelihood Ratio Tests. To adjust for multiple testing false-discovery rates were obtained using the Benjamini-Hochberg method.
2.2. Synaptosomes preparation
Human synaptoneurosomes were isolated from ≈50 mg frozen gray matter samples of the DLPFC using the Syn-PER method (Thermo Fisher Scientific) using the manufacturer instructions and as previously reported (Limon et al., 2019). The resultant pellet, enriched in synaptoneurosomes (P2 fraction), was resuspended in sterile distilled to break up the synaptosomes and to create small proteoliposomes containing synaptic membranes and their synaptic proteins. The protein concentration of the P2 fraction was determined by using Qubit protein assay reagent kit (Thermo Fisher Scientific). No differences in the total amount of protein in synaptic preparations was found between groups (2.756 mg/mL in controls vs 2.76 mg/mL in SZ (n= 10 ctrl, 10 SZ; t-test, double tailed, p-value = 0.920). These P2 membrane preparations were stored at −80°C until needed for downstream studies.
2.3. Proteomic Profile
One aliquote containing 50 μg of protein from each P2 membrane preparation of each subject was analyzed by LC-MS/MS to measure relative protein abundance in the synapto-proteome (English et al., 2015; Focking et al., 2015). The analysis was done on a Thermo Scientific Q Exactive mass spectrometer connected to a Dionex Ultimate 3000 (RSLCnano) chromatography system as has been previously described (English et al., 2015; Focking et al., 2015). All data were acquired with the mass spectrometer operating in positive ion and automatic data-dependent switching mode. A high resolution (70,000) MS scan (300–1600 m/z) was performed using the Q Exactive to select the 12 most intense ions prior to MS/MS analysis using high energy collision-induced dissociation (HCD). We collected two types of mass spectrometry data in the experiments. It is formally a ‘shotgun’ experiment and the collected tandem mass spectra were used to identify the proteins. The parent ion signal (MS1 scan) on high-resolution instruments such as the Q Exactive is widely used to calculate protein abundance in so-called ‘label-free’ proteomics experiments (Cox and Mann, 2011). The signal for each peptide derived from a given protein is integrated over time (the time it takes to enter the instrument via HPLC), and the summed signals are used to estimate the relative abundance of each protein. To analyze the data, we used the MaxQuant program specifically for label-free experiments using high-resolution instruments supported by Andromeda as a database search engine for peptide identification (Cox et al., 2011). This program has several statistical control steps to ensure that only high-quality reliable ion signals are accepted. Label-free quantitation was performed as previously described (Luber et al., 2010). Carbamidomethylation was defined as a fixed modification, while oxidation and acetylation of the protein N-terminus were defined as variable modifications. Only peptides with seven or more amino acid residues were allowed for identification. Additionally, at least one unique peptide was required to identify a protein. The cut off for false discovery rate for peptide and protein identification was set to 0.01 (1%). To avoid bias associated with protein under-representation between groups, proteins were excluded in cases where there was less than 50% availability of the LFQ intensities in each biological group. The label-free algorithm takes the maximum number of identified peptides between any two samples and compares the intensity of these peptides to determine peptide ratios. Label-free quantitation (LFQ) intensity values were used for protein quantification across the groups. Raw LFQ intensities were extracted from the MaxQuant software and log base 2 transformed prior to analysis to eliminate distributional skew and to give approximate normality. Regression normalization was performed to remove technical variation between samples (Callister et al., 2006). Significance testing was performed at the 5% level between SZ and control samples. Fold changes were obtained from an exponentiation (power of 2) of the difference in group means. Fold changes <1 were inverted (−1/fold change) to present a consistent scale of measurement. Two control samples and one SZ sample that had poor chromatographic profiles as compared to the rest of the samples LC-MS/MS were excluded from the proteomic analyses. All 20 subjects were included in electrophysiological experiments using microtransplantation of synaptic membranes (MSM) procedures.
2.4. Two Electrode Voltage Clamp (TEVC) electrophysiology Profile
Aliquotes from the original P2 preparation were used in MSM experiments to record ion currents elicited by native AMPARs embedded in synaptic membranes (Limon, 2019; Mazzo et al., 2016). Briefly, synaptosome-enriched preparations were sonicated, 3 times for 5 seconds at 1 min intervals in ice, to break the synaptosomes and create small proteoliposomes. These sonicated preparations were injected into stage V-VI Xenopus laevis oocytes using protocols previously published for cellular membranes (Conti et al., 2013; Limon et al., 2008, 2011, 2012). Each oocyte was injected with 50 nL of synaptic proteoliposomes (2 mg/mL protein concentration) and recorded after18–36 hrs of the injection. This is the period of time when responses are most stable. For electrophysiological recordings, oocytes were placed in a recording chamber (volume ≈0.1 ml) and perfused continuously (5–10 ml/min) with Ringer’s solution [115 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 5 mM HEPES (pH 7.4)] at room temperature (19–21°C). Data acquisition and analyses were performed by using WinEDR v2.3.8 Strathclyde Electrophysiology software (John Dempster, Glasgow, United Kingdom). To determine the amplitude of AMPA receptors the maximum amplitude of ion currents activated by 100 μM kainate, which elicits non-desensitizing currents through AMPARs (Limon et al., 2010; Stern-Bach et al., 1998) was used. The use of kainate allows consistent recordings of ion currents via AMPARs and produces less intra-subject variation compared to currents elicited by the combination of AMPA and cyclothiazide, which are coapplied to remove the strong desensitization of AMPARs (Limon et al., 2007). Other chemicals were from Sigma-Aldrich. Electrophysiological results are expressed as mean ± SEM. Statistical comparisons to identify the effect of diagnosis and the contribution of demographic variables (age, RIN, PMI, and pH) used the mean of kainate current for each subject (n = 10–14 injected oocytes were technical replicates per subject) in an analysis of covariance (ANCOVA) model. Fourteen oocytes per subject were recorded to measure current amplitude, but only oocytes that showed a stable baseline and signal to noise ratio of at least 2 were included in the analysis. To determine differences in current amplitude an ANCOVA (between-subject factor: diagnosis; covariates: pH and RIN; interaction: pH*Diagnosis; (JMP, version 14) was used. The EC50 for kainate for microtransplanted oocytes was determined as previously reported (Limon, 2019). In all cases, p<0.05 was considered significant. Pearson product-moment was used for linear correlations using JMP version 14, Adjusted p-values are shown when multivariate analyses were implemented. Previously we have shown stability of the electrical recording with long PMIs (Limon et al., 2008).
2.5. Functional enrichment and protein-protein interaction analyses.
Functional enrichment analysis was done with Metascape web interface which incorporates access to 40 knowledgebases (Zhou et al., 2019). Transcriptomic analysis of differentially expressed genes was done against the whole genome. For pathway and protein enrichment a minimum overlap of 3, p-value cutoff of 0.01, minimum enrichment of 1.5, and gene prioritization by evidence counting (GPEC) was used (Zhou et al., 2019). For protein-protein interaction enrichment, the minimum network size was 3 using the databases BioGrid, InWeb_IM (human) and OmniPath (human). Proteomics data was analyzed by obtaining dysregulated proteins using GPEC and the total list of identified proteins by LC-MS/MS as background. For the integrated analysis of transcriptomic, proteomic and subset of proteins that correlated with ion currents from AMPARs, we used the multi-list feature of Metascape using pathway and protein enrichment with a minimum overlap of 3, p-value cutoff of 0.00001, minimum enrichment of 3 and GPEC; the proteomics dataset was used as background.
3. Results
3.1. Whole transcriptomic and synaptosome-proteomic profiles
Gene expression levels measured by RNA-Seq were significantly upregulated in SZ for 151 genes at the p<0.01 level (supplementary data 2). Functional and protein-protein interaction analyses of these genes showed significant enrichment for chaperone-mediated folding, positive regulation of inflammation, stress response, negative regulation of phosphorylation and ATP biosynthesis (Fig. 1, supplementary data 3). In contrast, 186 genes were downregulated in SZ. These downregulated genes were found to be significantly involved in synaptic signaling, signal release and transmission across chemical synapses (supplementary data 4). These results are in agreement with a reduction of synaptic communication and loss of dendritic spines that have been observed in the DLPFC of SZ subjects (Glantz and Lewis, 2000). However, the expression of AMPARs subunits (GRIA1, GRIA2, GRIA3 and GRIA4), the principal gates for synaptic input at synaptic spines, was not different between control and SZ subjects (Fig. 1F).
To explore the possible effect of medications we reanalyzed the non-human primate CommonMind Consortium (CMC) Knowledge Portal macaque collection consisting of clozapine (n=9), and high haloperidol dosage (n=7) and low dosage (n=10) compared with placebo (n=10). The collection was obtained from Synapse (MP Vawter, PI) and analyzed by using 3 surrogate variables in an SVA analysis, and drug group as a factor. The details of the CMC Macaque collection can be found on the CMC website (https://www.synapse.org//#!Synapse:syn2759792/wiki/69613). The analysis of the macaque frontal cortex showed that none of the GRIA1,−2,−3,−4 genes were differentially expressed in the low Haldol or Clozapine drug group compared to the control group (supplementary data). GRIA2 and GRIA3 were nominally decreased (p = 0.04, not significant following FDR correction) in expression in the high Haldol dose group compared to controls by 7%.
Comparative studies between transcriptome and proteome in animal models have shown that levels of transcripts and proteins do not necessarily correlate (Ghazalpour et al., 2011); therefore, we determined the relative abundance of AMPARs subunits within the context of the synapto-proteome. In agreement with transcriptomic analysis, the abundance of AMPARs subunits in the proteomics analysis was not different between controls and SZ subjects (Fig. 2). However, GluA4 was not found in our synaptic preparations. This was probably due to its very low abundance in the cortex (Schwenk et al., 2014; Schwenk et al., 2012) and the detection limits of label-free proteomics (Sandberg et al., 2014). Interestingly, only mRNA and protein levels for GRIA3 and GluA3 correlated (r=0.59 p=0.013; supplementary figure 2).
Figure 2. Functional enrichment of differentially regulated proteins in synaptosomal-enriched preparations.
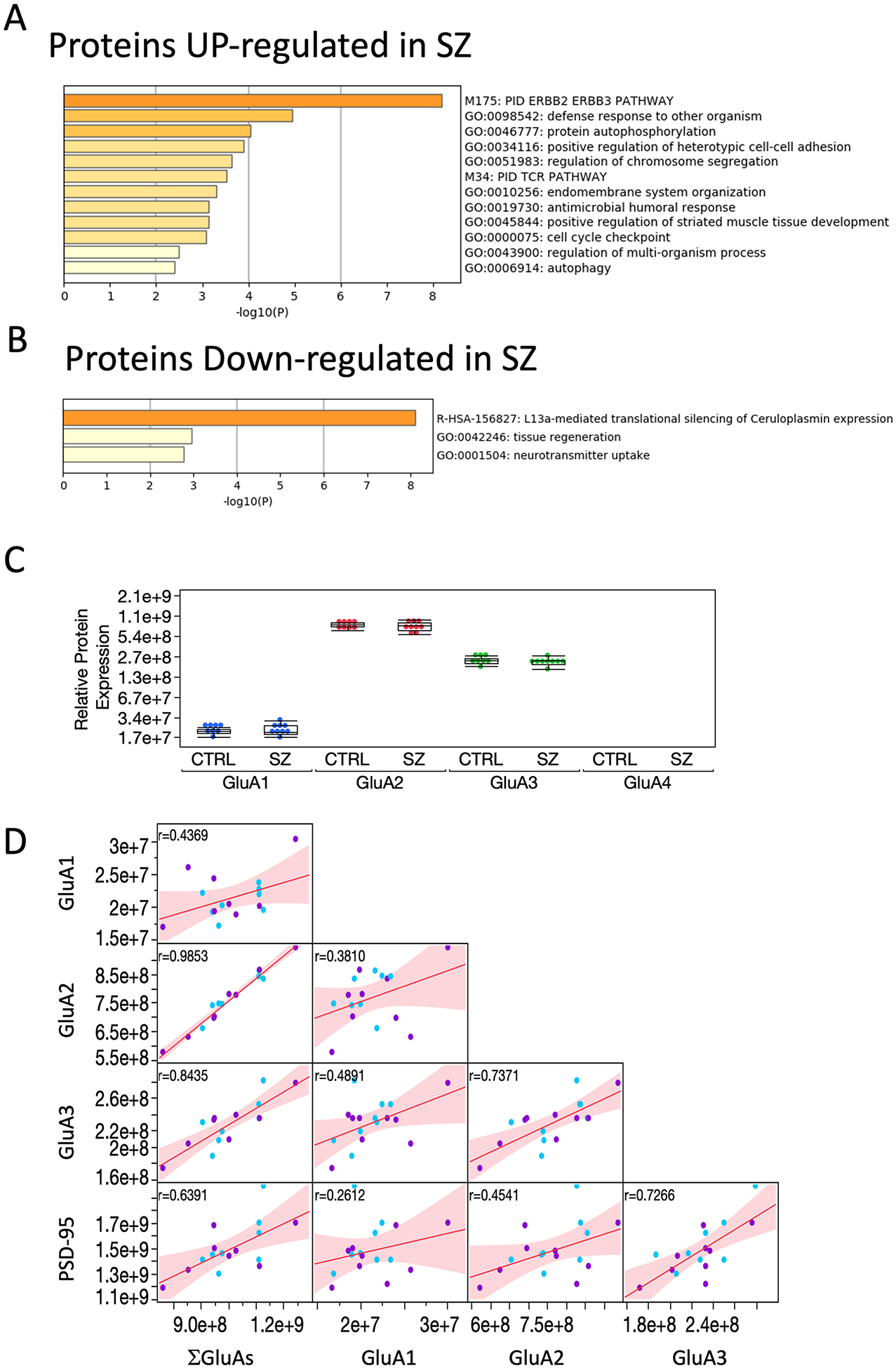
A. B. Enriched ontology clusters for proteins differentially expressed in SZ at the P < 0.05 level of statistical significance. C. Label-free quantitation (LFQ) intensity of expression levels for GluA subunits in control and schizophrenia (mean ± SEM). D. Multivariate correlation between GluA, their sum that represents the combination of all available AMPARs heteromers, and the postsynaptic density protein PSD-95 that anchors AMPARs to the synaptic cell membrane.
In both groups the protein levels for GluA2 was the most abundant subunit, accounting for 75.3 ± 1.7 % of the total protein available to assemble in AMPARs (mean ± SD; n=17 subjects), followed by GluA3 (22.6 ± 1.6 %) and GluA1 (2.1 ± 0.3 %). The relative proportion of GluA2 > GluA3 > GluA1 > GluA4 has the same order of that reported in the cortex of rodents, although the proportion in rodent for those subunits are approximately 45%, 27%, 21%, and 6%, respectively (Schwenk et al., 2014).
No differences in the relative proportions of AMPARs subunits were found between controls and SZ subjects. The proportions and high correlation between GluA2 and GluA3 (r (17) = 0.737; p=0.0007; Pearson’s correlation) compared to that of GluA2 and GluA1 (r (17) = 0.381; p =0.13) suggest the presence of heteromeric GluA2/3 and homomeric GluA2 receptors as the major drivers of synaptic inputs in the DLPFC and a significantly lower proportion of GluA1/2 receptors (Fig. 2D).
Further analysis of the 2487 identified proteins in the P2 fractions identified 128 proteins that were differentially expressed between SZ and control subjects (P<0.05; supplementary data 5), 54 and 74 proteins were upregulated and downregulated in SZ, respectively. Protein to protein interaction (PPI) and functional annotation analyses found that upregulated proteins participate in ERBB2/ERBB3 signaling, stress response, and autophosphorylation pathways (supplementary data 6). Downregulated proteins were found to be involved in translational silencing of gene expression, particularly LI3a-mediated (e.g. EIF4B, RPL28), tissue regeneration (GAP43, GJA1), and neurotransmitter uptake and release (SLC6A1, SNAP25) (supplementary data 7). These results also indicate convergence between RNA-Seq and synaptic-proteomic data on the downregulation of synaptic function, although the total abundance of AMPARs subunits is maintained in the DLPFC of SZ subjects.
3.2. Electrophysiology of native AMPA receptors profile.
AMPA receptors are a complex of proteins which in addition to the four principal subunits (GluA1-GluA4), they are co-assembled with a large number of auxiliary proteins (Schwenk et al., 2012; Schwenk et al., 2009) that modulate the gating, permeability and pharmacology of the channel (Cho et al., 2007; Gill et al., 2011; Herring et al., 2013; Kato et al., 2010a; Kato et al., 2010b; Milstein et al., 2007). Similarly, AMPA receptors activity is modulated by many intracellular pathways (Wang et al., 2005). To determine whether proteomic changes in SZ impact the electrophysiological activity of AMPARs, aliquots of the original P2 samples used for proteomics were microtransplanted into Xenopus oocytes. All samples from the cohort exhibited functional agonist-induced responses when tested by TEVC in MSM, confirming the viability of AMPARs. Figure 3A shows ion currents elicited by 100 μM kainate from DLPFC synaptosome membranes injected into oocytes. Kainate is a low-affinity agonist of AMPARs that keeps the channel open in a non-desensitized state, allowing the measurement of kainate-induced AMPARs responses in the steady-state (Limon et al., 2010). Because Xenopus oocytes do not express endogenous AMPARs (Limon et al., 2008; Limon, 2019), electrophysiological responses elicited by AMPARs agonists in microtransplanted oocytes are from functional and successfully incorporated, receptor protein complexes docked into the surface of the oocyte. This is consistent with previous work indicating that synaptic function and intracellular signaling is preserved in fresh-frozen and thawed human postmortem brain slices (Hahn et al., 2006).
Figure. 3. Altered AMPARs responses in SZ.
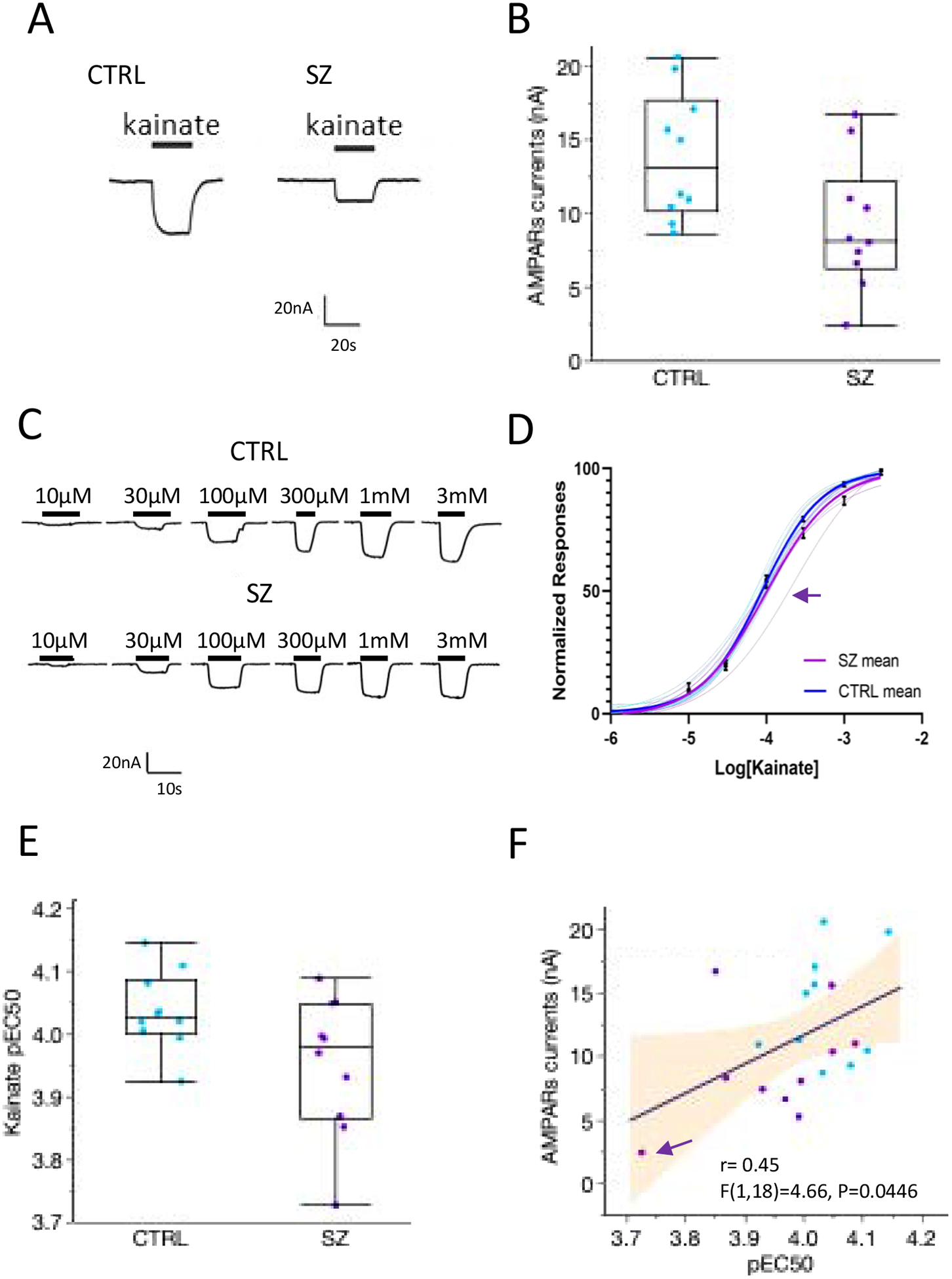
A. Functional responses of AMPARs elicited by 100 μM kainate of oocytes microtransplanted with DLPFC synaptic membranes from a control and an SZ brain. B. AMPARs responses in SZ cases were reduced by 34% (n = 10 CTRL, 10 SZ; F (4,15) = 5.85, p = 0.0048; ANCOVA correcting for pH, RIN and the interaction between diagnosis and pH). Each point in the graph represents the mean for each subject (n = 10–14 oocytes measured per each subject). C. Representative concentration-dependent responses to kainate in oocytes microtransplanted with SZ and control synaptic membranes. D. Hill equation fits to normalized concentration-response data for each subject (thin lines) and for the group average (thick lines). Each curve for a single subject was done with a minimum of 3 oocytes per subject. Symbols indicate the mean ± SEM of responses of each group, the arrow indicates a subject with reduced affinity respect to the whole cohort. E. F. The affinity AMPARs was reduced in SZ controls (P = 0.03, one sided tail), and the differences in affinity were positively correlated with the amplitude of AMPARs. pEC50 indicates the negative logarithm of the EC50 for each subject. Arrow shows a subject with no detectable CACGN8 protein levels.
The mean of kainate-induced AMPARs responses (AMPARs currents) for each subject was not significantly correlated with age or PMI (r (20) = 0.0245, p = 0.918 for age, and r (20) = 0.3, p = 0.19 for PMI), but found significant correlations with pH and RIN in the controls, but not in SZ. A screening effect for sources of variation confirmed the presence of an interaction between diagnosis and pH for the amplitude of AMPARs currents. Therefore, an ANCOVA model (between-subject factor: diagnosis; covariates: pH and RIN; interaction: pH*Diagnosis; N=20, F (4,15) = 5.85, p = 0.0048) was used to test for diagnosis effects. This analysis found a significant effect of diagnosis for the reduction of AMPARs currents in SZ (Diagnosis LogWorth = 3.1; p = 0.0008). Corrected AMPARs current was found to correlate with the protein levels of GluA2 and GluA3 in the cohort (r (17) = 0.61, p = 0.038 for GluA2 and (r (17) = 0.61, p = 0.01 for GluA3) and the sum of all AMPARs subunits (ΣGluA; (r(17) = 0.54, p = 0.03). Notably, the correlation between AMPARs currents and protein levels was higher in SZ than in the control group e.g. AMPA responses vs GluA3 in control (r(8) = 0.583, p = 0.129, and AMPA responses vs GluA3 in SZ (r (9) = 0.708, p = 0.033). Similarly, GluA1 was correlated with AMPARs currents in SZ (r (9) = 0.77, p = 0.0149) but not in the control (r (8) = 0.126, p = 0.75), suggesting a higher participation of GluR1 in AMPARs responses in SZ.
Because gene and proteomic analysis did not find changes in the abundance of AMPARs subunits, we hypothesized that lower amplitude of AMPARs currents in SZ was caused by a reduction in the EC50 for kainate. In agreement with this hypothesis, AMPARs receptors in SZ had lower EC50 for kainate than controls (p= 0.03, one-sided tail testing a priori for lower sensitivity, Fig 3E). The lower amplitude of AMPARs currents correlated with lower kainate potency (Fig. 3F). These results are consistent with a reported hypofunction of AMPARs (Coyle et al., 2003; Hammond et al., 2010; Pinner et al., 2016) and a loss of excitatory connectivity in the DLPFC of SZ patients (Glantz and Lewis, 2000; Harrison, 1999; Penzes et al., 2011).
3.3. Electrophysiologically-anchored dataset analysis (EDA) in SZ
Transcriptomic and proteomic datasets provide converging evidence indicating synaptic dysfunction in SZ; however, loss of synaptic communication in schizophrenia and compensatory rearrangements are cellular processes that involve a large number of structural and accessory proteins at the synapse. As a first examination of the utility in using MSM data to unravel this complexity, and to better understand the relationship between protein expression and physiological activity of AMPARs, we implemented what we called an EDA approach, to generate lists of proteins that correlate with the physiological activity of AMPARs (supplementary data 8 for all correlations). We found that in addition to the positive correlation with GluA subunit protein counts and AMPARs current, the AMPARs currents were also positively correlated with 118 proteins representing the excitatory postsynaptic density, and mitochondrial and ribosomal complexes (Fig. 4; supplementary data 9 shows all the modules assessed). A molecular complex detection algorithm MCODE, that identifies densely connected regions in large PPI networks (Bader and Hogue, 2003), identified 5 molecular modules: 1) Nonsense mediated decay (NMD) complex, which is an mRNA surveillance system that maintains synaptic architecture and synaptic vesicle efficacy (Long et al., 2010); 2) Ca2+ signaling pathway, which is fundamental for buffering of synaptic Ca2+, neurotransmitter release and activity-dependent regulation of synaptic function (Greer and Greenberg, 2008); 3) the mitochondria electron transport respiratory chain complex I, 4) the mitochondria F1F0-ATP synthase of Complex V which is fundamental to supply the high metabolic demand of active synapses, and 5) activation of AMPARs and synaptic plasticity, which are the proteins responsible for generating the AMPARs currents. In contrast, AMPARs currents were negatively correlated with 107 proteins involved in intracellular signaling, phosphorylation, and DNA biosynthetic processes (Fig. 5, supplementary data 10 shows all the modules assessed). Three MCODE modules that correlated negatively with AMPARs were representative of complexes involved in 1) formation of tubulin folding intermediates by CCT/TRiC, which are chaperonin mechanisms important for synaptic proteostasis (Gorenberg and Chandra, 2017), 2) glucagon signaling pathway, and 3) AURKA activation by TPX2 pathway, which is important for neurite elongation and dendrites differentiation (Kahn et al., 2015; Mori et al., 2009).
Figure 4. Functional enrichment and MCODE modules of proteins positively correlated with AMPARs responses.
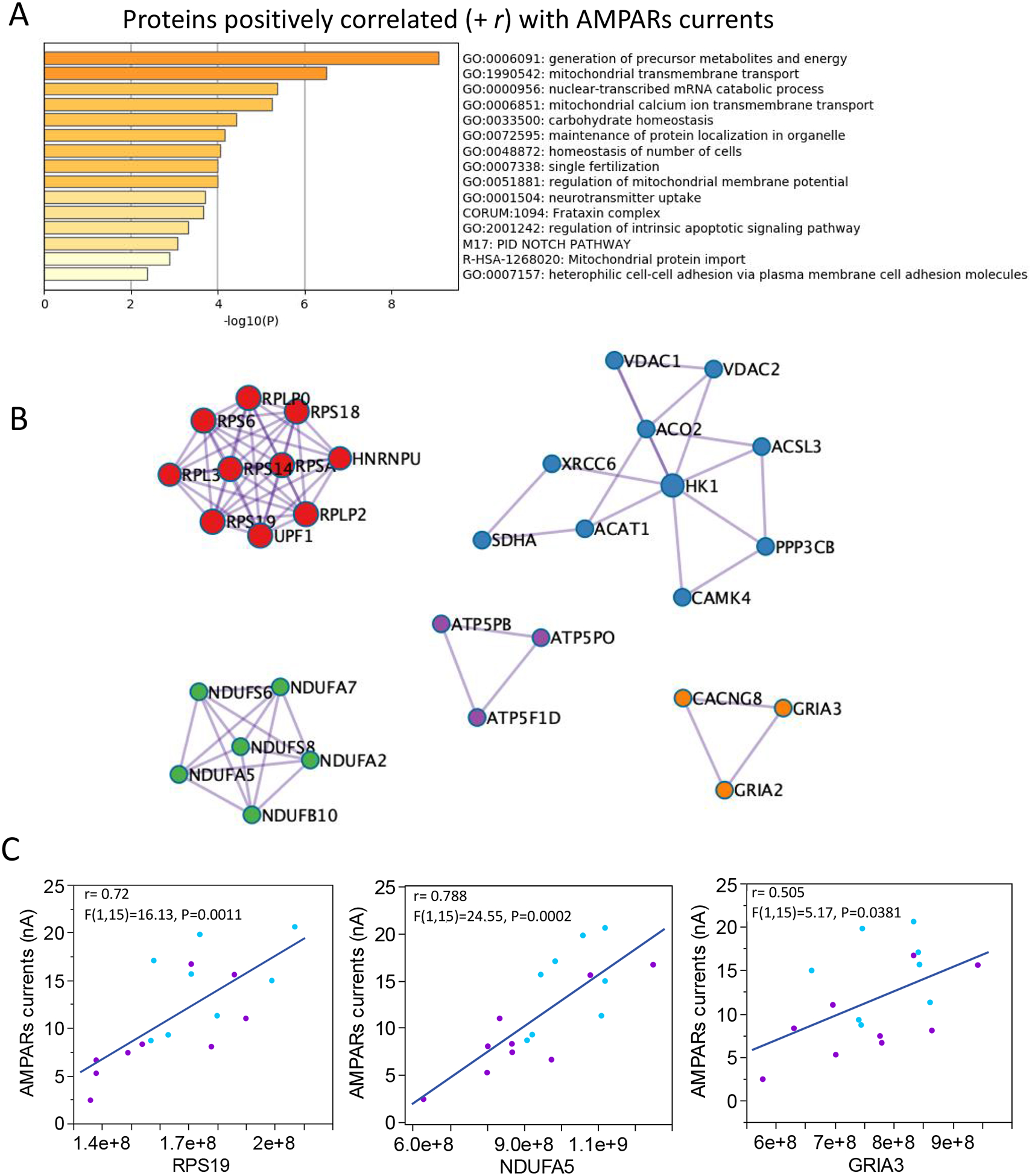
A. Enriched ontology clusters for proteins differentially expressed in SZ at the P < 0.05 level. B. Proteins densely connected identified by molecular complex detection algorithm (MCODE). Protein complexes involved with postsynaptic densities and AMPARs subunits, energy production in mitochondria, and protein translation in ribosomes were positively correlated with AMPARs currents. C. Linear correlations between AMPARs currents and proteins representative of MCODE modules shown in B.
Figure 5. Functional enrichment and MCODE modules of proteins negatively correlated with AMPARs responses.
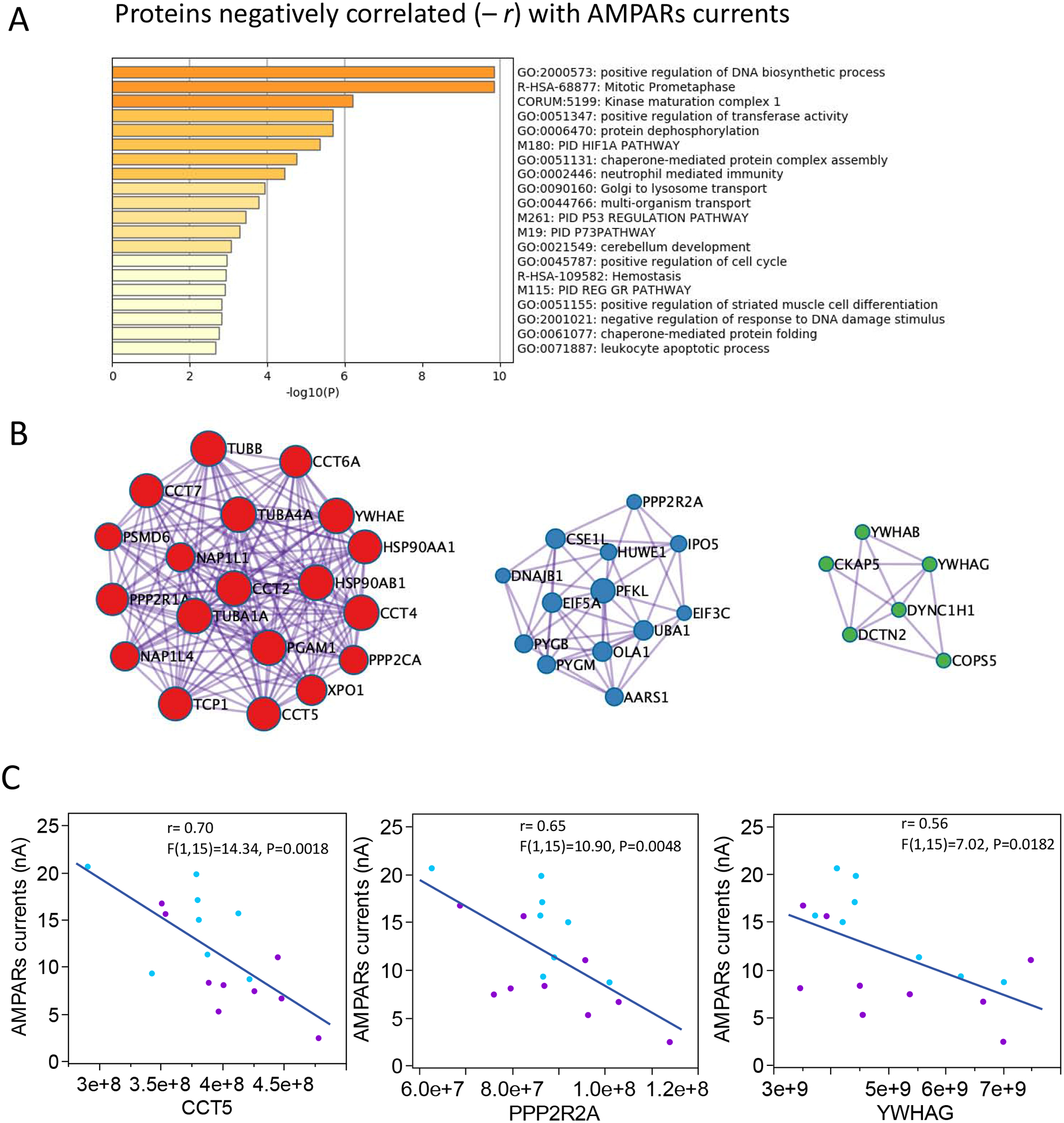
A. Enriched ontology clusters for proteins differentially expressed in SZ at the p < 0.05 level. B. Proteins densely connected identified by molecular complex detection algorithm (MCODE). Proteins complexes typically involved in intracellular signaling and DNA biosynthesis were negatively correlated with AMPARs currents. C. Linear correlations between AMPARs currents and proteins representative of MCODE modules shown in B.
A metanalysis including transcriptomic, proteomic, and EDA-generated lists of proteins shows that 15.3% of proteins that positively correlate with the amplitude of AMPARs currents are downregulated in SZ (Fig. 6); in contrast, 10% of proteins with negative correlation with AMPARs currents are upregulated in SZ (supplementary data 11). Interestingly, functional convergence of upregulated transcriptome and synapto-proteome indicated that the MAPK pathway (Table 2), which has been consistently implicated in SZ studies, had a negative association with AMPARs currents (Figure 6).
Fig. 6. Transcriptomic and synaptic-proteomic convergence on the amplitude of AMPARs currents in SZ.
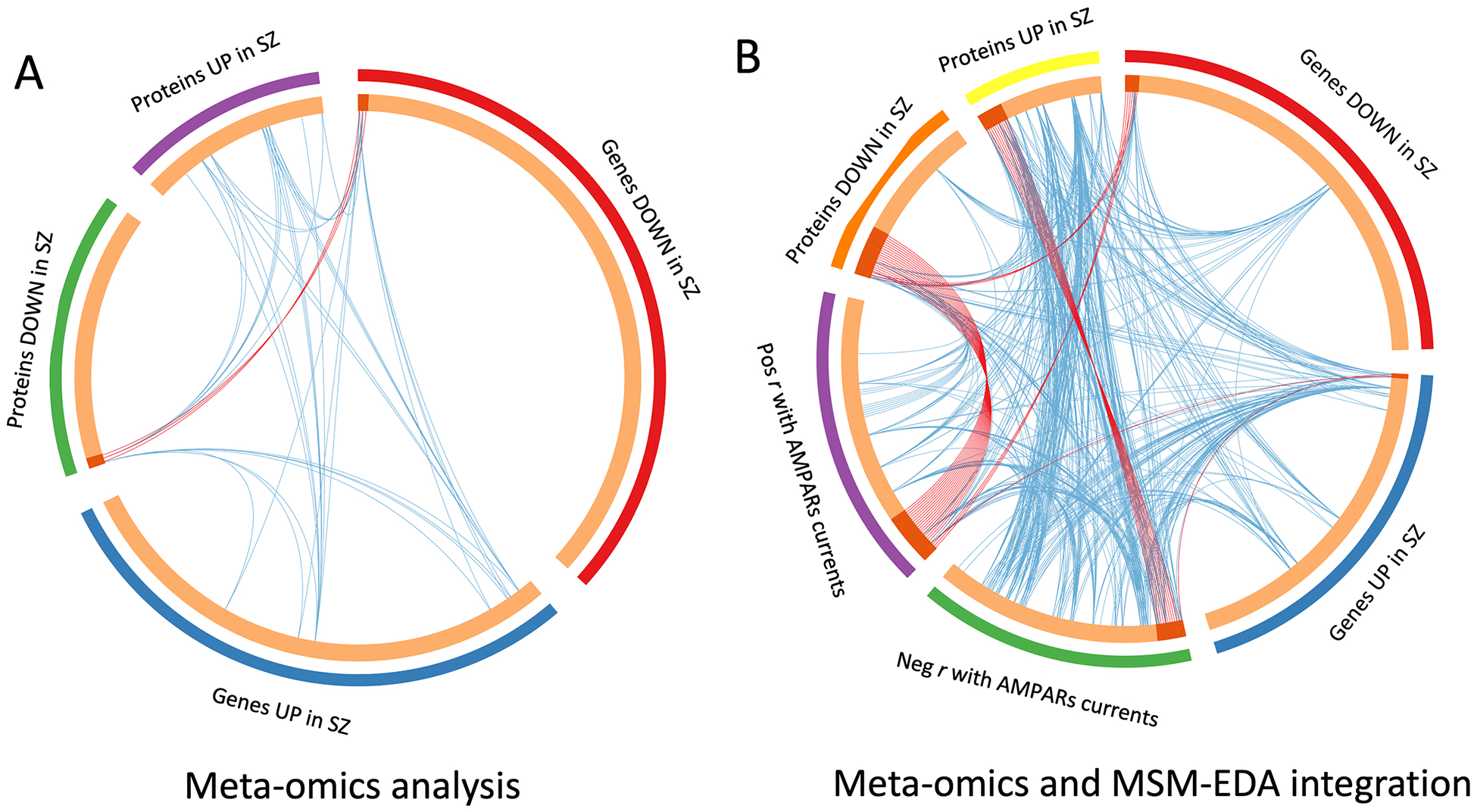
A. Circos plot showing gene/protein overlap across different levels of analysis. Outside arches represent the different gene/protein lists generated during this study. Inside arches contain genes/proteins. Dark orange arches indicate the genes/proteins that appear in multiple lists. Purple lines connect the same gene that is shared by multiple lists. Blue links indicate the amount of functional overlap among the input gene lists. B. Similar analysis as in A, including proteins that correlate with the amplitude of AMPARs currents. Notice that genes and proteins that are downregulated in SZ overlap with proteins that are involved in the generation of the amplitude of AMPARs currents measured in microtransplanted membranes. In contrast, genes and proteins upregulated in SZ overlap with proteins that negatively impact the amplitude of the responses measured. Functional overlap is also stronger between lists of genes that negatively impact the amplitude of the responses in SZ, indicating a convergence of mechanisms of synaptic dysfunction.
Table 2.
Functional convergence between upregulation of MAPK-involved signaling pathways and lower AMPARs responses in SZ
| SOURCE | GENE/PROTEIN | NAME |
|---|---|---|
| RNA-Seq | CDKN1A | Cyclin Dependent Kinase Inhibitor 1A |
| RNA-Seq | GADD45B | Growth Arrest and DNA Damage Inducible Beta |
| RNA-Seq | MAPK9 | Mitogen-Activated Protein Kinase 9 |
| MSM-EDA | PLCB1 | Phospholipase C Beta 1 |
| LC-MS/MC | PLCD1 | Phospholipase C Delta 1 |
| MSM-EDA | PPP3CB | Protein Phosphatase 3 Catalytic Subunit Beta |
| LC-MS/MC;RNA-Seq | PRKCB | Protein Kinase C Beta |
| RNA-Seq | PRKCE | Protein Kinase C Epsilon |
| RNA-Seq | SHC1 | SHC Adaptor Protein 1 |
| LC-MS/MC | CAPN1 | Calpain 1 |
| LC-MS/MC | CDC42 | Cell Division Cycle 42 |
| LC-MS/MC | COL4A2 | collagen type IV alpha 2 chain |
| LC-MS/MC | EIF2S1 | Eukaryotic Translation Initiation Factor 2 Subunit Alpha |
| LC-MS/MC | HRAS | HRas proto-oncogene, GTPase |
| LC-MS/MC | KRAS | KRAS proto-oncogene, GTPase |
| LC-MS/MC | MAPK1 | mitogen-activated protein kinase 1 |
| LC-MS/MC | MAPK3 | mitogen-activated protein kinase 3 |
| RNA-Seq; MSM-EDA | HSPB1 | Heat Shock Protein Family B (Small) Member 1 |
| MSM-EDA | MAPK10 | mitogen-activated protein kinase 10 |
| MSM-EDA | PPP2CA | Protein Phosphatase 2 Catalytic Subunit Alpha |
| MSM-EDA | PPP2R1A | Protein Phosphatase 2 Scaffold Subunit Aalpha |
| MSM-EDA | PTPN11 | Protein Tyrosine Phosphatase Non-Receptor Type 11 |
| MSM-EDA | TUBA1A | Tubulin Alpha 1a |
| MSM-EDA | TUBA4A | Tubulin Alpha 4a |
| MSM-EDA | YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta |
| LC-MS/MC; MSM-EDA | FGA | Fibrinogen Alpha Chain |
| LC-MS/MC; MSM-EDA | MAP2K2 | mitogen-activated protein kinase kinase 2 |
4. Discussion
In this preliminary study, we used a variant of the method of microtransplantation of plasma membranes from postmortem tissue (Miledi et al., 2004) to record, for the first time, the activity of synaptic AMPARs from the DLPFC of people diagnosed with SZ. It was previously shown that microtransplanting synaptic membranes from autopsy tissue allows the pharmacological and electrophysiological characterization of synaptic receptor complexes in human brain regions; a prerequisite for drug discovery (Limon, 2019; Mazzo et al., 2016; Zwart et al., 2019). MSM opens the door to use biophysical parameters of ion currents as functional endpoints for more sophisticated analysis in neurological and neuropsychiatric disorders. In this study, by integrating multi-dimensional data with physiological responses of postsynaptic receptors our results suggest a reduced function of AMPARs in SZ. This reduction may be at least partially explained by changes in potency and by alterations in protein modules that impact the trafficking and function of AMPARs. However, we cannot rule out reverse causation. It may be possible that altered AMPAR responses cause changes in protein subunit levels since previous studies have shown reductions (Beneyto and Meador-Woodruff, 2006; Vawter et al., 2002), no changes (Healy et al., 1998), or elevations (Chinopoulos et al., 2007; Dracheva et al., 2005) in the expression levels of AMPARs in postmortem studies of SZ.
Our transcriptomic and proteomic analysis of this cohort found no changes in the DLPFC levels of transcripts or proteins for any of the AMPARs subunits in SZ DLPF; however, we observed functional deficits in the receptors’ electrophysiological activity (EC50) that is supportive of AMPARs deficits in SZ. Normal abundance of mRNA and protein of AMPARs subunits in SZ may be a compensatory attempt to normalize synaptic function by homeostatic mechanisms (Turrigiano, 2017). Taken together, the data shows that alterations in cellular pathways and altered AMPARs function in the DLPFC are part of a molecular alteration of synaptic signaling which may lead to a decreased excitatory input and functional dysconnectivity in the DLPFC in SZ.
The convergence of different lines of evidence suggests that synaptic functional alterations of AMPARs may underlie the hypofunction and decreased activation that has been consistently observed in the DLPFC of SZ patients (Berman, 2002). Reduced excitation and the subsequent potential shunt of NMDARs activation may underlie the generation of psychoses in individuals with SZ due to the complementary causes for NMDA receptor hypofunction that our data suggest. While this hypothesis is far from new, it does represent a functional measure that augments previous investigations that relied solely on singular methods that counted abundance of genes or proteins.
The analysis of the electrophysiological activity and relative abundances of native receptors from human brain tissue provides a glimpse of functional information that warrants further studies. For example, we observed that postmortem pH was negatively correlated with the amplitude of AMPARs currents in the control but not the SZ group. While it has been reported that patients with SZ may exhibit extracellular acidification in magnetic resonance spectroscopy (MRS) studies and postmortem brains associated with impaired oxidative phosphorylation and mitochondria dysfunction (Dogan et al., 2018; Hagihara et al., 2018; Sullivan et al., 2019b), and also that AMPARs are negatively modulated by extracellular H+ (Ihle and Patneau, 2000; Traynelis and Cull-Candy, 1991), it is not known whether AMPARs in a chronically acidified milieu have compensatory modifications that make them less sensitive to agonal changes of pH. However, the experimental setting in which we studied native receptors was constant in terms of buffer and pH. This does not rule out the possibility that there are post-translational modifications or alternative splicing changes to the AMPARs that we have not accounted for in our proteomic analysis.
Further analyses studying the role of auxiliary subunits, which are known to modify the kinetics, pharmacology and polyamine block of AMPA receptors depending on the cellular needs (Kato et al., 2010b), in this process are needed. Auxiliary subunits may also be involved in the pharmacological differences that we observed in MSM experiments. Previous work by Meador-Woodruff’s lab had shown a reduction in the transmembrane AMPA regulator protein TARP-γ8 in the anterior cingulate in SZ (Drummond et al., 2013); although we did not observe group differences in TARP-γ8 in the DLPFC in our cohort, in physiological conditions TARP-γ8 is known to increase glutamate and kainate affinity (Kato et al., 2010b; Tomita et al., 2007), and a reduction in this protein may explain the reduced potency of AMPARs that we observed in the DLPFC of some SZ subjects. In support of this hypothesis, the subject marked with an arrow in MSM experiments (Figure 3F) had the lowest gene expression for TARP-γ8 (CACGN8) in the SZ group and had no detectable protein quantities in LC-MS/MS experiments. Future studies in larger cohorts should aid in the categorization of SZ subtypes based on the specific alterations of genes or proteins that correlate with the excitatory electrical activity of the synapses.
Our analysis indicates that additional mechanisms converge into producing deficits of AMPARs. For instance, both, RNA-Seq and LC-MS/MS data implicate MAPK3 signaling in SZ, which is in agreement with de novo mutations and alterations in copy number studies of SZ (Costain et al., 2013; Kirov et al., 2012), transcriptomic-wide association studies (Gusev et al., 2018), proteomic alterations of isolated postsynaptic densities from the anterior cingulate (Focking et al., 2015) and altered levels of MAPK signaling in postmortem tissue (Funk et al., 2012). Abnormally increased signaling may underlie some of the previously described alterations in posttranslational modifications (Tucholski et al., 2013), and impaired intracellular transport of AMPARs (Hammond et al., 2010). We found correlations in AMPARs with mitochondria Complex I and Complex V protein abundance, supporting prior evidence that dysregulation of these genes in SZ would have consequences in neurotransmission (Rollins et al., 2018; Schulmann et al., 2019). However, it is clear that medications are playing a role in modulation of the mitochondria functions (Chan et al., 2019), thus an analysis of medication-free patients is essential in answering this question. We did not find alterations in GRIA1, −2,−3,−4 by RNA-Seq cortex by a low chronic dosing with Haldol or clozapine, while there was a 7% increase in GRIA3, −4 expression with high dosage of Haldol in pharmacological analysis of the macaque. The small number of subjects with schizophrenia in this preliminary study did not allow us to subgroup by concurrent prescriptions for antipsychotics (3), antidepressants (4), antianxiety (5), anticonvulsant (2), and no medications (1). Thus further replication of this study with subjects that have a complete toxicological profile is underway and to compare with drug-free patients.
Another limitation of our study is that we cannot determine cell-specific changes associated to subpopulations of neurons whether these synaptosomes come from excitatory or inhibitory cells; however, because excitatory pyramidal cells account for 75–90% of cells in the cortex (DeFelipe et al., 2002) a large proportion of synapses will likely come from excitatory neurons. Importantly, there are layer-specific differences of excitatory neurons (Sullivan et al., 2019a) that our study cannot detect.
This work represents a novel method to measure a new functional electrophysiological endophenotype in synaptoneurosomal preparations directly obtained from the brains of patients with schizophrenia. While we cannot yet discern cause and consequence in this postmortem study, we are confident that the AMPARs current alterations are most likely present as a continuum in healthy controls and those with psychiatric disorders, and reference ranges will be the subject of further study in larger cohorts. These findings represent the synaptic compartment, as opposed to extra-synaptic locations, and this specificity can be altered by applying different fractionation methods for membrane isolation and proteomic and electrophysiological analysis. Since mitochondria are part of synaptosomes, this study showed support for correlation with the protein amount of mitochondria in the synaptosome and excitatory current.
The correlation between protein abundance of synaptic components involved in local synthesis protein, energy production, and synaptic proteins with the actual amplitude of AMPARs currents provides a novel method to integrate RNA and protein levels with functional data from large datasets. The EDA approach can be expanded and be used to find SNPs, genes, proteins, isoforms, metabolites and other elements in large pharmacological and clinical datasets that correlate positively or negatively with functional data in diverse schizophrenia relevant brain regions at a subcellular specific level using enriched fractions.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbarian S, Sucher NJ, Bradley D, Tafazzoli A, Trinh D, Hetrick WP, Potkin SG, Sandman CA, Bunney WE Jr., Jones EG, 1996. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci 16(1), 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW, 2003. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Wang HY, Borgmann-Winter KE, MacDonald ML, Kaprielian H, Stucky A, Kvasic J, Egbujo C, Ray R, Talbot K, Hemby SE, Siegel SJ, Arnold SE, Sleiman P, Chang X, Hakonarson H, Gur RE, Hahn CG, 2015. Src kinase as a mediator of convergent molecular abnormalities leading to NMDAR hypoactivity in schizophrenia. Mol Psychiatry 20(9), 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barch DM, Carter CS, Braver TS, Sabb FW, MacDonald A 3rd, Noll DC, Cohen JD, 2001. Selective deficits in prefrontal cortex function in medication-naive patients with schizophrenia. Arch Gen Psychiatry 58(3), 280–288. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Meador-Woodruff JH, 2006. Lamina-specific abnormalities of AMPA receptor trafficking and signaling molecule transcripts in the prefrontal cortex in schizophrenia. Synapse 60(8), 585–598. [DOI] [PubMed] [Google Scholar]
- Berman KF, 2002. Functional Neuroimaging in Schizophrenia, in: Davis KL, Charney, Coyle JT, Nemeroff C (Eds.), Neuropsychopharmacology- %th Generation of Progress. Lippincott, Williams, & Williams, Pennsylvania [Google Scholar]
- Bolkan SS, Carvalho Poyraz F, Kellendonk C, 2016. Using human brain imaging studies as a guide toward animal models of schizophrenia. Neuroscience 321, 77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister SJ, Barry RC, Adkins JN, Johnson ET, Qian WJ, Webb-Robertson BJ, Smith RD, Lipton MS, 2006. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res 5(2), 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone AL, Plested AJ, 2016. Superactivation of AMPA receptors by auxiliary proteins. Nat Commun 7, 10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts VS, Derminio DS, Hahn CG, Weickert CS, 2015. Postsynaptic density levels of the NMDA receptor NR1 subunit and PSD-95 protein in prefrontal cortex from people with schizophrenia. NPJ Schizophr 1, 15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts VS, Lai YL, Weickert CS, Weickert TW, Catts SV, 2016. A quantitative review of the postmortem evidence for decreased cortical N-methyl-d-aspartate receptor expression levels in schizophrenia: How can we link molecular abnormalities to mismatch negativity deficits? Biol Psychol 116, 57–67. [DOI] [PubMed] [Google Scholar]
- Chan ST, McCarthy MJ, Vawter MP, 2019. Psychiatric drugs impact mitochondrial function in brain and other tissues. Schizophr Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinopoulos C, Connor JA, Shuttleworth CW, 2007. Emergence of a spermine-sensitive, non-inactivating conductance in mature hippocampal CA1 pyramidal neurons upon reduction of extracellular Ca2+: dependence on intracellular Mg2+ and ATP. Neurochem Int 50(1), 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CH, St-Gelais F, Zhang W, Tomita S, Howe JR, 2007. Two families of TARP isoforms that have distinct effects on the kinetic properties of AMPA receptors and synaptic currents. Neuron 55(6), 890–904. [DOI] [PubMed] [Google Scholar]
- Conti L, Limon A, Palma E, Miledi R, 2013. Microtransplantation of cellular membranes from squid stellate ganglion reveals ionotropic GABA receptors. Biol Bull 224(1), 47–52. [DOI] [PubMed] [Google Scholar]
- Corti C, Xuereb JH, Crepaldi L, Corsi M, Michielin F, Ferraguti F, 2011. Altered levels of glutamatergic receptors and Na+/K+ ATPase-alpha1 in the prefrontal cortex of subjects with schizophrenia. Schizophr Res 128(1–3), 7–14. [DOI] [PubMed] [Google Scholar]
- Costain G, Lionel AC, Merico D, Forsythe P, Russell K, Lowther C, Yuen T, Husted J, Stavropoulos DJ, Speevak M, Chow EW, Marshall CR, Scherer SW, Bassett AS, 2013. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet 22(22), 4485–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M, 2011. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem 80, 273–299. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M, 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10(4), 1794–1805. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Tsai G, Goff D, 2003. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci 1003, 318–327. [DOI] [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN, 2004. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE 2004(255), re16. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Alonso-Nanclares L, Arellano JI, 2002. Microstructure of the neocortex: comparative aspects. J Neurocytol 31(3–5), 299–316. [DOI] [PubMed] [Google Scholar]
- Diaz E, 2010. Regulation of AMPA receptors by transmembrane accessory proteins. Eur J Neurosci 32(2), 261–268. [DOI] [PubMed] [Google Scholar]
- Dogan AE, Yuksel C, Du F, Chouinard VA, Ongur D, 2018. Brain lactate and pH in schizophrenia and bipolar disorder: a systematic review of findings from magnetic resonance studies. Neuropsychopharmacology 43(8), 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, McGurk SR, Haroutunian V, 2005. mRNA expression of AMPA receptors and AMPA receptor binding proteins in the cerebral cortex of elderly schizophrenics. J Neurosci Res 79(6), 868–878. [DOI] [PubMed] [Google Scholar]
- Drummond JB, Tucholski J, Haroutunian V, Meador-Woodruff JH, 2013. Transmembrane AMPA receptor regulatory protein (TARP) dysregulation in anterior cingulate cortex in schizophrenia. Schizophr Res 147(1), 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JA, Fan Y, Focking M, Lopez LM, Hryniewiecka M, Wynne K, Dicker P, Matigian N, Cagney G, Mackay-Sim A, Cotter DR, 2015. Reduced protein synthesis in schizophrenia patient-derived olfactory cells. Transl Psychiatry 5, e663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker WW, Miyamoto S, 2016. Pharmacological Treatment of Schizophrenia: Current Issues and Future Perspectives. Clinical Neuropsychopharmacology and Therapeutics 7, 1–8. [Google Scholar]
- Focking M, Lopez LM, English JA, Dicker P, Wolff A, Brindley E, Wynne K, Cagney G, Cotter DR, 2015. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry 20(4), 424–432. [DOI] [PubMed] [Google Scholar]
- Funk AJ, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH, 2012. Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology 37(4), 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG 2nd, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ, 2011. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 7(6), e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Roberts MF, Yu H, Wang H, Tomita S, Bredt DS, 2011. Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J Neurosci 31(18), 6928–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour G, Dix S, Fellini L, Gastambide F, Plath N, Steckler T, Talpos J, Tricklebank M, 2012. NMDA receptors, cognition and schizophrenia--testing the validity of the NMDA receptor hypofunction hypothesis. Neuropharmacology 62(3), 1401–1412. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA, 2000. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry 57(1), 65–73. [DOI] [PubMed] [Google Scholar]
- Gluck MR, Thomas RG, Davis KL, Haroutunian V, 2002. Implications for altered glutamate and GABA metabolism in the dorsolateral prefrontal cortex of aged schizophrenic patients. Am J Psychiatry 159(7), 1165–1173. [DOI] [PubMed] [Google Scholar]
- Gorenberg EL, Chandra SS, 2017. The Role of Co-chaperones in Synaptic Proteostasis and Neurodegenerative Disease. Front Neurosci 11, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME, 2008. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59(6), 846–860. [DOI] [PubMed] [Google Scholar]
- Gulsuner S, Stein DJ, Susser ES, Sibeko G, Pretorius A, Walsh T, Majara L, Mndini MM, Mqulwana SG, Ntola OA, Casadei S, Ngqengelele LL, Korchina V, van der Merwe C, Malan M, Fader KM, Feng M, Willoughby E, Muzny D, Baldinger A, Andrews HF, Gur RC, Gibbs RA, Zingela Z, Nagdee M, Ramesar RS, King MC, McClellan JM, 2020. Genetics of schizophrenia in the South African Xhosa. Science 367(6477), 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A, Schizophrenia Working Group of the Psychiatric Genomics, C., McCarroll S, Neale BM, Ophoff RA, O’Donovan MC, Crawford GE, Geschwind DH, Katsanis N, Sullivan PF, Pasaniuc B, Price AL, 2018. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 50(4), 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H, Catts VS, Katayama Y, Shoji H, Takagi T, Huang FL, Nakao A, Mori Y, Huang KP, Ishii S, Graef IA, Nakayama KI, Shannon Weickert C, Miyakawa T, 2018. Decreased Brain pH as a Shared Endophenotype of Psychiatric Disorders. Neuropsychopharmacology 43(3), 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, Bakshi K, Kamins J, Borgmann-Winter KE, Siegel SJ, Gallop RJ, Arnold SE, 2006. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med 12(7), 824–828. [DOI] [PubMed] [Google Scholar]
- Hall LS, Medway CW, Pain O, Pardinas AF, Rees EG, Escott-Price V, Pocklington A, Bray NJ, Holmans PA, Walters JTR, Owen MJ, O’Donovan MC, 2020. A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum Mol Genet 29(1), 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JC, McCullumsmith RE, Funk AJ, Haroutunian V, Meador-Woodruff JH, 2010. Evidence for abnormal forward trafficking of AMPA receptors in frontal cortex of elderly patients with schizophrenia. Neuropsychopharmacology 35(10), 2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Do KQ, 2016. Linking early-life NMDAR hypofunction and oxidative stress in schizophrenia pathogenesis. Nat Rev Neurosci 17(2), 125–134. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, 1999. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain 122 (Pt 4), 593–624. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR, 2005. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Molecular psychiatry 10(1), 40–68; image 45. [DOI] [PubMed] [Google Scholar]
- Healy DJ, Haroutunian V, Powchik P, Davidson M, Davis KL, Watson SJ, Meador-Woodruff JH, 1998. AMPA receptor binding and subunit mRNA expression in prefrontal cortex and striatum of elderly schizophrenics. Neuropsychopharmacology 19(4), 278–286. [DOI] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW, Nicoll RA, 2013. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77(6), 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, MacDonald ML, Elswick DE, Sweet RA, 2015. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Ann N Y Acad Sci 1338, 38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle EC, Patneau DK, 2000. Modulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor desensitization by extracellular protons. Mol Pharmacol 58(6), 1204–1212. [DOI] [PubMed] [Google Scholar]
- Jadi MP, Behrens MM, Sejnowski TJ, 2016. Abnormal Gamma Oscillations in N-Methyl-D-Aspartate Receptor Hypofunction Models of Schizophrenia. Biol Psychiatry 79(9), 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG, Hendry SH, Liu XB, Hodgins S, Potkin SG, Tourtellotte WW, 1992. A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J Neurosci Methods 44(2–3), 133–144. [DOI] [PubMed] [Google Scholar]
- Kahn OI, Ha N, Baird MA, Davidson MW, Baas PW, 2015. TPX2 regulates neuronal morphology through kinesin-5 interaction. Cytoskeleton (Hoboken) 72(7), 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, Bredt DS, 2010a. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68(6), 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Yu H, Nisenbaum ES, Bredt DS, 2010b. TARPs differentially decorate AMPA receptors to specify neuropharmacology. Trends Neurosci 33(5), 241–248. [DOI] [PubMed] [Google Scholar]
- Kelly TM, Mann JJ, 1996. Validity of DSM-III-R diagnosis by psychological autopsy: a comparison with clinician antemortem diagnosis. Acta Psychiatr Scand 94(5), 337–343. [DOI] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O’Donovan MC, Owen MJ, 2012. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 17(2), 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Anand A, Moghaddam B, 2002. Effects of NMDA receptor antagonists: implications for the pathophysiology of schizophrenia. Archives of general psychiatry 59(7), 663–664. [DOI] [PubMed] [Google Scholar]
- Krystal JH, D’Souza DC, Mathalon D, Perry E, Belger A, Hoffman R, 2003. NMDA receptor antagonist effects, cortical glutamatergic function, and schizophrenia: toward a paradigm shift in medication development. Psychopharmacology 169(3–4), 215–233. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Karper LP, Seibyl JP, Freeman GK, Delaney R, Bremner JD, Heninger GR, Bowers MB Jr., Charney DS, 1994. Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Archives of general psychiatry 51(3), 199–214. [DOI] [PubMed] [Google Scholar]
- Leek JT, 2014. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 42(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD, 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6), 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, Delbruck E, Yassine A, Pandya D, Myers RM, Barchas JD, Lee F, Schatzberg, Watson SJ, Akil H, Bunney WE, Vawter MP, Sequeira A, 2019. Electrophysiological evaluation of extracellular spermine and alkaline pH on synaptic human GABAA receptors. Transl Psychiatry 9(1), 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, Reyes-Ruiz JM, Eusebi F, Miledi R, 2007. Properties of GluR3 receptors tagged with GFP at the amino or carboxyl terminus. Proc Natl Acad Sci U S A 104(39), 15526–15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, Reyes-Ruiz JM, Miledi R, 2008. Microtransplantation of neurotransmitter receptors from postmortem autistic brains to Xenopus oocytes. Proc Natl Acad Sci U S A 105(31), 10973–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, Reyes-Ruiz JM, Miledi R, 2011. GABAergic drugs and Alzheimer’s disease. Future Med Chem 3(2), 149–153. [DOI] [PubMed] [Google Scholar]
- Limon A, Reyes-Ruiz JM, Miledi R, 2012. Loss of functional GABA(A) receptors in the Alzheimer diseased brain. Proc Natl Acad Sci U S A 109(25), 10071–10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, Reyes-Ruiz JM, Vaswani RG, Chamberlin AR, Miledi R, 2010. Kaitocephalin antagonism of glutamate receptors expressed in Xenopus oocytes. ACS Chem Neurosci 1(3), 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, D. E Yassine A Pandya D Myers RM Barchas JD, Lee F Schatzberg AF, Watson SJ Akil H Bunney WE Vawter MP, Sequeira A, 2019. Electrophysiological evaluation of extracellular spermine and alkaline pH on synaptic human GABAA receptors. Translational Psychiatry, Accepted, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AA, Mahapatra CT, Woodruff EA 3rd, Rohrbough J, Leung HT, Shino S, An L, Doerge RW, Metzstein MM, Pak WL, Broadie K, 2010. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci 123(Pt 19), 3303–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O’Keeffe M, Mann M, 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32(2), 279–289. [DOI] [PubMed] [Google Scholar]
- Lussier MP, Sanz-Clemente A, Roche KW, 2015. Dynamic Regulation of N-Methyl-d-aspartate (NMDA) and alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors by Posttranslational Modifications. J Biol Chem 290(48), 28596–28603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Garver M, Newman J, Sun Z, Kannarkat J, Salisbury R, Glausier J, Ding Y, Lewis DA, Yates N, Sweet RA, 2019. Synaptic Proteome Alterations in the Primary Auditory Cortex of Individuals With Schizophrenia. JAMA Psychiatry, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, 1987. The physiology of excitatory amino acids in the vertebrate central nervous system. Prog Neurobiol 28(3), 197–276. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, Guthrie PB, 1984. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309(5965), 261–263. [DOI] [PubMed] [Google Scholar]
- Mazzo F, Zwart R, Serratto GM, Gardinier KM, Porter W, Reel J, Maraula G, Sher E, 2016. Reconstitution of synaptic Ion channels from rodent and human brain in Xenopus oocytes: a biochemical and electrophysiological characterization. J Neurochem 138(3), 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miledi R, Duenas Z, Martinez-Torres A, Kawas CH, Eusebi F, 2004. Microtransplantation of functional receptors and channels from the Alzheimer’s brain to frog oocytes. Proc Natl Acad Sci U S A 101(6), 1760–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AD, Zhou W, Karimzadegan S, Bredt DS, Nicoll RA, 2007. TARP subtypes differentially and dose-dependently control synaptic AMPA receptor gating. Neuron 55(6), 905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P, 2000. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron 28(1), 53–67. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Miyake N, Jarskog LF, Fleischhacker WW, Lieberman JA, 2012. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol Psychiatry 17(12), 1206–1227. [DOI] [PubMed] [Google Scholar]
- Mori D, Yamada M, Mimori-Kiyosue Y, Shirai Y, Suzuki A, Ohno S, Saya H, Wynshaw-Boris A, Hirotsune S, 2009. An essential role of the aPKC-Aurora A-NDEL1 pathway in neurite elongation by modulation of microtubule dynamics. Nat Cell Biol 11(9), 1057–1068. [DOI] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A, 1984. Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307(5950), 462–465. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14(4), 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM, 2011. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 14(3), 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein WM, Carter CS, Noll DC, Cohen JD, 2001. Relation of prefrontal cortex dysfunction to working memory and symptoms in schizophrenia. Am J Psychiatry 158(7), 1105–1113. [DOI] [PubMed] [Google Scholar]
- Pinner AL, Tucholski J, Haroutunian V, McCullumsmith RE, Meador-Woodruff JH, 2016. Decreased protein S-palmitoylation in dorsolateral prefrontal cortex in schizophrenia. Schizophr Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK, 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1), 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins BL, Morgan L, Hjelm BE, Sequeira A, Schatzberg AF, Barchas JD, Lee FS, Myers RM, Watson SJ, Akil H, Potkin SG, Bunney WE, Vawter MP, 2018. Mitochondrial Complex I Deficiency in Schizophrenia and Bipolar Disorder and Medication Influence. Mol Neuropsychiatry 3(3), 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MD, Drummond JB, Meador-Woodruff JH, 2012. Glutamate receptor abnormalities in schizophrenia: implications for innovative treatments. Biomol Ther (Seoul) 20(1), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A, Branca RM, Lehtio J, Forshed J, 2014. Quantitative accuracy in mass spectrometry based proteomics of complex samples: the impact of labeling and precursor interference. J Proteomics 96, 133–144. [DOI] [PubMed] [Google Scholar]
- Schulmann A, Ryu E, Goncalves V, Rollins B, Christiansen M, Frye MA, Biernacka J, Vawter MP, 2019. Novel Complex Interactions between Mitochondrial and Nuclear DNA in Schizophrenia and Bipolar Disorder. Mol Neuropsychiatry 5(1), 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, Fakler B, Schulte U, 2014. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84(1), 41–54. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, Klocker N, Schulte U, Fakler B, 2012. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74(4), 621–633. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, Klocker N, 2009. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323(5919), 1313–1319. [DOI] [PubMed] [Google Scholar]
- Sequeira PA, Martin MV, Vawter MP, 2012. The first decade and beyond of transcriptional profiling in schizophrenia. Neurobiology of Disease 45(1), 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH, 1990. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249(4976), 1580–1585. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, Rosenmund C, 1998. A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron 21(4), 907–918. [DOI] [PubMed] [Google Scholar]
- Sullivan CR, Koene RH, Hasselfeld K, O’Donovan SM, Ramsey A, McCullumsmith RE, 2019a. Neuron-specific deficits of bioenergetic processes in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry 24(9), 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CR, Mielnik CA, Funk A, O’Donovan SM, Bentea E, Pletnikov M, Ramsey AJ, Wen Z, Rowland LM, McCullumsmith RE, 2019b. Measurement of lactate levels in postmortem brain, iPSCs, and animal models of schizophrenia. Sci Rep 9(1), 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, Overman KM, Atz ME, Myers RM, Jones EG, Watson SJ, Akil H, Bunney WE Jr., 2004. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry 55(4), 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Byrd RK, Rouach N, Bellone C, Venegas A, O’Brien JL, Kim KS, Olsen O, Nicoll RA, Bredt DS, 2007. AMPA receptors and stargazin-like transmembrane AMPA receptor-regulatory proteins mediate hippocampal kainate neurotoxicity. Proc Natl Acad Sci U S A 104(47), 18784–18788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Cull-Candy SG, 1991. Pharmacological properties and H+ sensitivity of excitatory amino acid receptor channels in rat cerebellar granule neurones. J Physiol 433, 727–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucholski J, Simmons MS, Pinner AL, Haroutunian V, McCullumsmith RE, Meador-Woodruff JH, 2013. Abnormal N-linked glycosylation of cortical AMPA receptor subunits in schizophrenia. Schizophr Res 146(1–3), 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, 2017. The dialectic of Hebb and homeostasis. Philos Trans R Soc Lond B Biol Sci 372(1715). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, Freed WJ, 2002. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophrenia Research 58(1), 11–20. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Arora A, Yang L, Parelkar NK, Zhang G, Liu X, Choe ES, Mao L, 2005. Phosphorylation of AMPA receptors: mechanisms and synaptic plasticity. Mol Neurobiol 32(3), 237–249. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Berman KF, 1996. Prefrontal function in schizophrenia: confounds and controversies. Philos Trans R Soc Lond B Biol Sci 351(1346), 1495–1503. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Berman KF, Illowsky BP, 1988. Physiological dysfunction of dorsolateral prefrontal cortex in schizophrenia. III. A new cohort and evidence for a monoaminergic mechanism. Arch Gen Psychiatry 45(7), 609–615. [DOI] [PubMed] [Google Scholar]
- Wright A, Vissel B, 2012. The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1), 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart R, Mazzo F, Sher E, 2019. Microtransplantation of human brain receptors into oocytes to tackle key questions in drug discovery. Drug Discov Today 24(2), 533–543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


