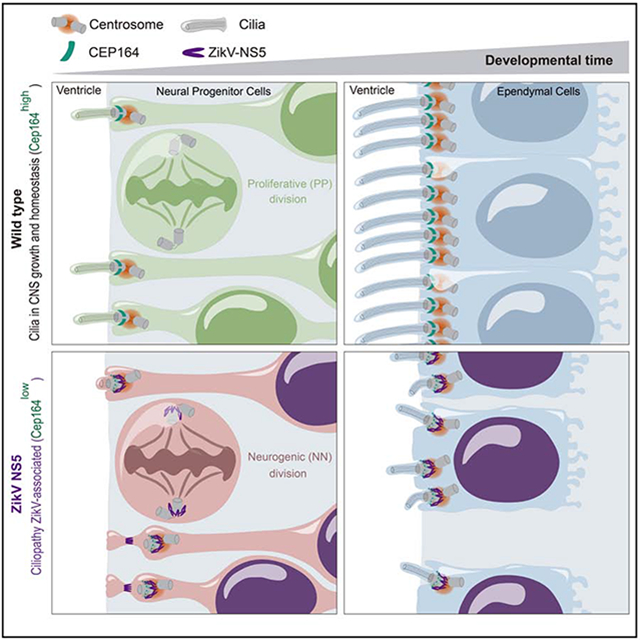
SUMMARY
Zika virus (ZikV) is a flavivirus that infects neural tissues, causing congenital microcephaly. ZikV has evolved multiple mechanisms to restrict proliferation and enhance cell death, although the underlying cellular events involved remain unclear. Here we show that the ZikV-NS5 protein interacts with host proteins at the base of the primary cilia in neural progenitor cells, causing an atypical non-genetic ciliopathy and premature neuron delamination. Furthermore, in human microcephalic fetal brain tissue, ZikV-NS5 persists at the base of the motile cilia in ependymal cells, which also exhibit severe ciliopathy. While the enzymatic activity of ZikV-NS5 appears to be dispensable, the amino acids Y25, K28 and K29 that are involved in NS5- oligomerization are essential for the localization and interaction with components of the cilium base, promoting ciliopathy and premature neurogenesis. These findings lay the foundation for therapies that target ZikV-NS5-multimerization and prevent the developmental malformations associated with congenital Zika syndrome.
Graphical Abstract
eTOC Blurb
Monociliated NPCs proliferation ensures normal brain growth. The NS5 protein encoded by Zika virus interacts with host proteins at the cilia base, such as CEP164, induces ciliopathy and promotes neurogenesis. Following primary neurogenesis, NPCs differentiate into multiciliated ependymal cells in which ZikV-NS5 depletes CEP164, provokes ciliopathy and ependymal layer disorganization.
INTRODUCTION
Zika virus (ZikV) is a flavivirus transmitted by the bite of the Aedes mosquito (Pierson and Kielian, 2013). The 11 kb positive-sense, single-stranded RNA genome of ZikV encodes ten mature viral proteins: three structural (S) proteins (C, prM and E) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Pierson and Kielian, 2013). A single amino acid substitution (S139N) in prM seems to be responsible for the change in virus tropism that provokes a dramatic increase in neonatal microcephaly (Yuan et al., 2017) that has led to this virus being declared a global threat to public health. ZikV can directly infect human neural progenitor cells (NPCs) in both 2D and 3D in vitro models of the developing cerebral cortex, resulting in defects resembling congenital microcephaly (Dang et al., 2016; Gabriel et al., 2017; Garcez et al., 2016; Li et al., 2016; Nowakowski et al., 2016; Qian et al., 2016; Retallack et al., 2016; Tang et al., 2016). However, the mechanisms by which ZikV disrupts neurogenesis have not yet been fully elucidated. Macaque monkey infection provokes regional disturbances in the brain that impair postnatal neurogenesis, provoking cognitive deficits and epilepsy (Adams Waldorf et al., 2018). This vulnerability of late neurogenic regions to ZikV also persists in adult mice (Li et al., 2016). As it is well known that human brain development extends for many years beyond birth and that adult neurogenesis influences cognitive functions, it is no longer safe to consider ZikV as a transient infection in adult humans without marked long-term effects.
The limited availability of human tissue to perform histological analyses at different developmental stages (Abbott, 2011) emphasizes the need to use in vivo animal models to understand congenital Zika syndrome. Here we used the chick embryo neural tube (NT) to screen ZikV protein components and assess their impact on neurogenesis, particularly given that this in vivo model has served to identify fundamental processes in mammalian neural development and human disease. Through this approach, ZikV-NS5 was seen to disturb the growth of the nervous system. ZikV-NS5 is the largest NS protein (~100 kDa) and it is essential to the viral life cycle. It contains two functional domains, a N-terminal methyltransferase (MTase) domain responsible for the catalysis of 5’-mRNA capping and methylation, and a RNA-dependent RNA polymerase (RdRP) domain at the C-terminus that drives genome replication (Duan et al., 2017; Ferrero et al., 2019; Wang et al., 2017; Zhou et al., 2007). ZikV-NS5 can form dimers and higher-order oligomers that regulate its enzymatic functions, as well as its association with other viral or host factors during infection, making it an extremely attractive anti-viral target.
To understand how ZikV-NS5 affects neurogenesis, we searched for the host ZikV-NS5 interactome in human NPCs in a yeast two-hybrid (Y2H) assay. This unbiased approach detected previously identified ZikV-NS host interactors (Coyaud et al., 2018; Scaturro et al., 2018) and it revealed that the ZikV-NS5 protein interacts with cellular components known to localize at the base of the cilium. We showed that primary cilia elongation is impaired in NPCs expressing ZikV-NS5 and that neural delamination is accelerated, two key events that promote premature neurogenesis and impair CNS growth. However, these effects were hindered when ZikV-NS5 multimerization was prevented by point mutations. In addition, we observed a severe ciliopathy in the ependymal cells (ECs) that line the ventricular cavities of a ZikV-infected human fetal microcephalic brain, a phenomenon that might impair the cerebrospinal fluid (CSF) flow necessary to maintain brain homoeostasis and for toxin washout. Together our data indicate that the ZikV-NS5 causes ciliopathies –both a motile ciliopathy and a primary ciliopathy- which contribute to the collection of developmental malformations provoked in congenital Zika syndrome, such as microcephaly and ventriculomegaly.
RESULTS
ZikV-NS5 interacts with basal ciliary proteins in NPCs
To investigate whether individual proteins encoded by the Suriname strain (human isolate Z1106033 sequence) ZikV can directly regulate neurogenesis (Figure 1A), we monitored the expression of the anti-proliferative gene Tis21 (PC3, BTG2) (Iacopetti et al., 1999) 24 hours after electroporation (hpe) of constructs encoding each of the ZikV structural (ZikV-S) and non-structural (ZikV-NS) proteins into Hamburger and Hamilton (HH) stage 12 chick embryo NTs (Hamburger, 1951) (Figures 1B, S1 I, J). The sub-cellular distribution of each ZikV protein was also studied in early NPCs (Figures S1A–H). ZikV-NS2 and ZikV-NS5 provoked an increase in luciferase activity (Figure 1B) and indeed, ZikV-NS2 has been reported to dampen NPC proliferation by directly interacting with components of the adherens junction (AJ) complex (Yoon et al., 2017). The expression of ZikV-NS5 also reduced the expression of the progenitor marker pSox2, enhancing the expression of additional pan-neural markers like pNeuroD and pTubb3 (Figure 1C, Figures S1K–L), without disrupting NT tissue integrity or affecting cell viability (Figures S1M–P). We further confirm the conserved capability of ZikV-NS5 to regulate neurogenesis from several strains such as the African strain MR766 (ZikV-NS5-AF) and the Asian strain H/PF/2013 (ZikV-NS5-AS) (Figure S2A–C).
Figure 1.-. ZikV-NS5 interacts with proteins at the cilia base.
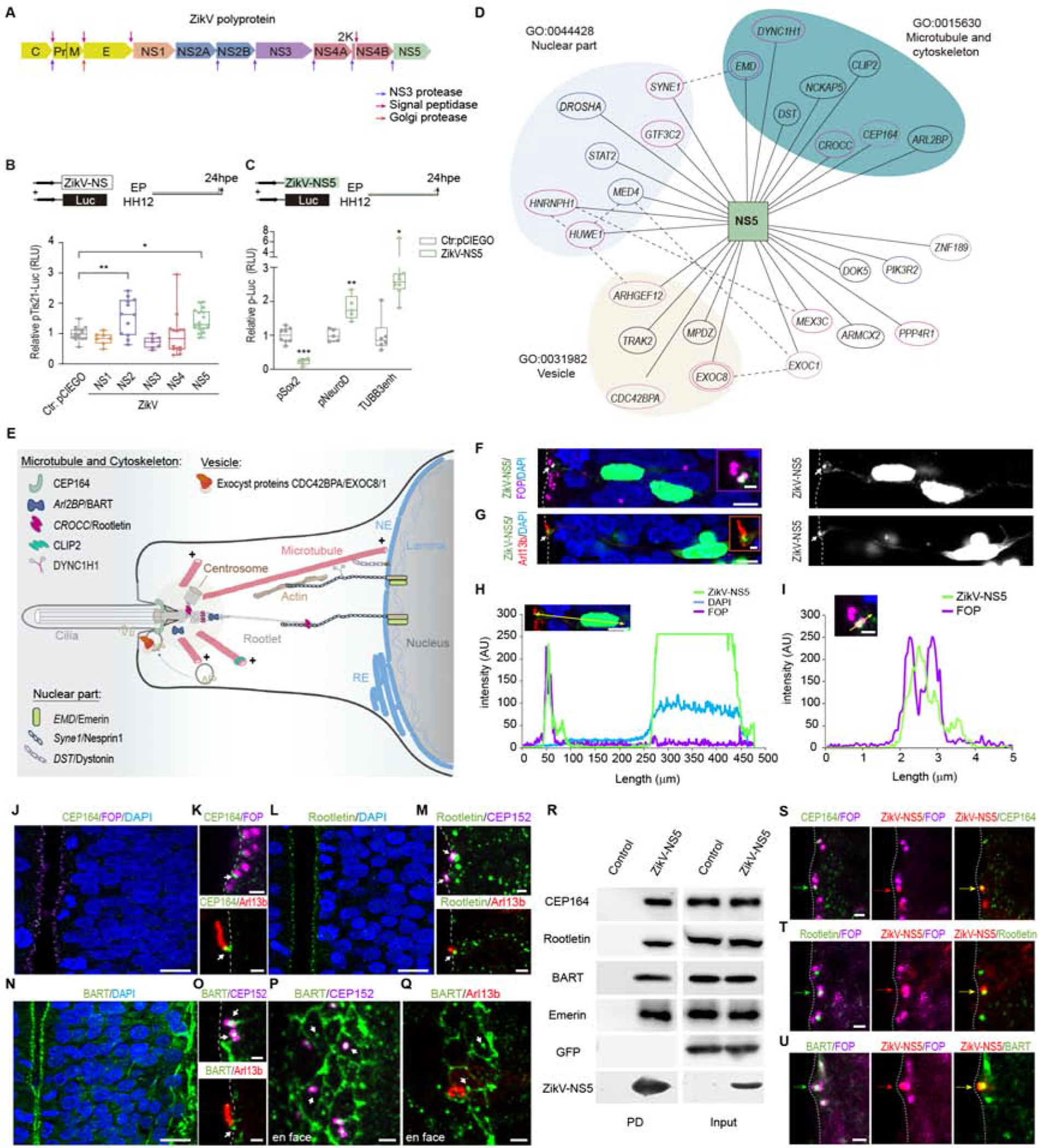
(A) The scheme depicts the structural and non-structural proteins of the ZikV polyprotein.Arrows indicate proteolytic cleavage sites.
(B) Luc/Renilla activity of the pTis21-Luc reporter after electroporation (elect.) of the empty vector pCIEGO or the ZikV-NS DNAs indicated (mean ± SD, n=6-8 embryos/condition).
(C) Luc/Renilla activity of the pSox2-Luc, the pNeuroD-Luc and the Tubb3enh-Luc reporters after elect. of the empty vector pCIEGO or ZikV-NS5 (mean ± SD, n=6-8 embryos/condition).
(D) Network representation of the ZikV-NS5-host interactome in human NPCs. NS5 is represented as a square and the 27 interacting host proteins as ellipses. The subcellular localization of the host proteins are taken from Gene Ontology. Proteins previously reported to interact with ZikV-NS5 in neural cells are in pink and those that interact in HEK291 cells are in blue. Published physical interactions of the host proteins are indicated by dotted lines.
(E) The scheme depicts the apical pole of a NPC where the primary cilium extends into the NT lumen. The centrosome at the cilium base organizes microtubules that link the nuclear envelope (NE) to the cilium base through the Rootlet. Key to proteins that interact with ZikV-NS5.
(F, G) Images show the subcellular distribution of ZikV-NS5 in elect. NPCs. (F) ZikV-NS5-FLAG visualised by anti-FLAG staining in the nucleus (DAPI) and in the centrosomes (arrow) lining the NT lumen labelled with anti-FOP. (G) ZikV-NS5-FLAG located at the base of the cilium (arrow) labelled with Arl13b-RFP.
(H, I) Plots of the fluorescence intensity F.I. (AU) at the distances from the NT lumen (zero): purple labelled centrosomes, blue labelled nuclei and green labelled ZikV-NS.
(J, K) sSections showing endogenous CEP164 lining the NT lumen and arrows pointing to the distal co-localization in the mother centriole with FOP at the base of the Arl13b-RFP labelled cilium.
(L, M) Sections showing endogenous Rootletin in which the arrows indicate its co-localization with the centrosome marker CEP152-GFP at the base of the Arl13b-RFP labelled cilia.
(N, O) Arrows point to endogenous BART co-localizing with CEP152-GFP at the base of the Arl13b-RFP labelled cilia.
(P, Q) En face imaging of the BART labelled apical belt of NPCs end-feet, with arrows pointing to the co-localization with CEP152 labelled centrosome and at the base of Arl13b-RFP labelled cilia.
(R) Pull-down (PD) assays show ZikV-NS5 binding to Cep164, Rootletin, BART and Emerin in transfected HEK-293 cells. Control is referred to a pull down with the resin in the absence of ZikV-NS5 recombinant protein and the empty vector pCIEGO-GFP was served as a negative control.
(S-U) Images of ZikV-NS5-FLAG co-localization with the FOP+ centrosome and with endogenous CEP164 (S), endogenous Rootletin (, T) and endogenous BART (U) lining the NT lumen (dotted line). The proteins are indicated with coloured arrows according to their fluorescence labelling.
***p<0.001, **p<0.01; exact P values 0.0091 (b), 0.0079 (c), *P<0.05 exact P values, 0.0416 (B), 0.0101 (C), ns: not significant. B, one-way ANOVA; c, two-sided unpaired t-test. Scale bars 5 μm (F, G inset 1 μm), 9 μm (J, L, N), 1 μm (inset F, inset G, K, M, O, S-U), 2 μm (P, Q).
See also Figures S1, S2 and Tables S1 and S2.
We searched for the cellular proteins that might interact with ZikV-NS5 and influence neurogenesis. As such, the ZikV-NS5 (1-904)-LexA fusion protein was used as bait in a Y2H screen of a human foetal brain library (HFBR_RP1_hgx4776v1_pB29). After analysing 99.6 million interactions and processing more than 141 colonies, a total of 27 ZikV-NS5-host interactors were defined (Table S1), ~70% of which were common to those interactors identified previously (Coyaud et al., 2018; Scaturro et al., 2018) (Table S2). Among the host proteins detected, enrichment of the microtubule/cytoskeleton, vesicle and nuclear compartments was revealed by gene ontology (GO) analysis (Figure 1D). Interestingly, several of these proteins are known to localize to the base of the cilium and/or, to link the nuclear envelope to the cytoskeleton (Figure 1E; Table S1).
A hallmark of NPCs is the single primary cilium at their apical surface, which is nucleated by the basal body where the mother centriole is docked to the plasma membrane through its appendage proteins (Kim and Dynlacht, 2013) (Figure 1E). This cilium extends into the NT lumen, where it is able to detect the growth factors in the CSF that control CNS growth (Lehtinen et al., 2011). Findings from the interactome prompted us to study the subcellular distribution of ZikV-NS5 in NPCs by introducing a plasmid encoding ZikV-NS5-FLAG tagged into the chick embryo NT. As expected given the nuclear localization signals encoded in this protein, the ZikV-NS5-FLAG protein accumulated in the nucleus (Ferrero et al., 2019) (Figures. 1F–H), a subcellular distribution unique among the ZikV proteins (Figures S1A–H). However, some ZikV-NS5 also accumulated at the base of the cilia, which was identified through the pan-cilia marker, the small GTPase ADP-ribosylation factor-like 13b fused to the red fluorescent protein (Arl13b-RFP) that specifically associates with the ciliary membrane (Paridaen et al., 2013; Saade et al., 2017). ZikV-NS5 localizes to the centrosome, identified as a pair of dots at the surface of the ventricular zone (VZ) where the centrosome proteins 152K (CEP152) (Dzhindzhev et al., 2010) and FGFR1 Oncogene Partner (FOP) (Yan et al., 2006) were detected as pan-centrosome markers (Figures 1F–I). As expected, ZikV-NS5-Flag form the African and Asian strains show a conserved subcellular localization associated to the nucleus and the primary cilia base (Figures S2F–J). Moreover, the docking of ZikV-NS5 to the centrosomes persisted throughout mitosis in these NPCs (Figures S1Q–U).
To confirm that ZikV-NS5 interacts directly with components of the cilium base in a cell context, endogenous centrosome protein 164K (CEP164) was detected in NPCs by immunofluorescence, lining the NT lumen where it co-localizes with the FOP-labelled mother centriole at the cilium base (Figures 1J,K). CEP164 is a mature centriole-specific protein that localizes to the basal body distal appendages and it is required for assembly of the primary cilium (Graser et al., 2007). ZikV-NS5 pulled down CEP164, in HEK293 cell extracts (Figure 1R, and Figure S1Y) and in NPCs, ZikV-NS5 co-localized with CEP164 and with centriole markers (Figure 1S, Figure S4D). In NPCs, the endogenous rootlet coiled-coil protein (CROCC/Rootletin) was detected by immunofluorescence in the rootlet fibres, as well as lining the NT lumen where it co-localized with centrosomal markers at the cilium base (Figures 1L, M). Rootletin localizes to the intercentriolar linker and it is required for the correct positioning of the cilium basal body relative to the cell nucleus (Yang et al., 2006; Yang et al., 2002). ZikV-NS5 also pulled down Rootletin in HEK293 cell extracts (Figure 1R, and Figure S1Y), and ZikV-NS5 co-localized with Rootletin and with centrosomal markers in NPCs (Figure 1T, Figure S4E). Moreover, immunofluorescence demonstrated the presence of the endogenous ADP ribosylation factor-like 2-binding protein (ARL2BP/BART) in NPCs that line the NT lumen, where it co-localized with the centrosomal marker CEP152 at the cilium base (Figures 1N–Q). BART localizes to the basal body and it is required for cilia elongation (Davidson et al., 2013). En face NT imaging revealed BART also localized to the apical belt (Figures 1P, Q) and the base of the primary cilium identified by Arl13b (Figure 1Q). ZikV-NS5 pulled down BART in transfected HEK293 cells (Figure 1R, and Figure S1Y) and in NPCs, ZikV-NS5 co-localized with BART and with centrosomal markers (Figure 1U, Figure S4H). In addition, the Y2H screen retrieved three ZikV-NS5 protein interactors that localize to the nuclear envelope (Emerin, Nesprin and Dystonin: Figure 1E). ZikV-NS5 pulled down Emerin from HEK293 cells (Figure 1R), a member of the nuclear lamina-associated protein family that controls anchorage to the cytoskeleton and that links centrosomes to the nuclear envelope through an association with microtubules (Virtanen and Vartiainen, 2017; Wilson, 2000). Together these data indicate the ZikV-NS5 interacts with protein components of the base of the primary cilium in NPCs.
ZikV-NS5 causes a ciliopathy and premature neuron delamination
In dividing NPCs, cilia length shortens prior to entry into mitosis and the ciliary membrane remnant is internalized along with the mother centrosome. The daughter cells that inherit the mother centrosome can reform long cilia faster and they remain as NPCs, while the differentiating daughter cells delaminate from the ventricular surface and migrate to the lateral NT (Figure 2A) (Paridaen et al., 2013; Saade et al., 2018). We assessed whether ZikV-NS5 affects cilia length, measuring the length of Arl13b-RFP labelled cilia protruding from the two FOP-labelled centrioles at the VZ surface in NPCs. In control cells the cilia length varied as a function of the cell cycle phase and nuclear position (median=2.08 ± 1.3 μm, n=100), as in electroporated NPCs where ZikV-NS5 is exclusively localized in the nucleus (Nu-ZikN-NS5, median=2 ± 1.1 μm, n=100: Figure 2D). However, the cilia were significantly shorter in electroporated NPCs showing ZikV-NS5 localization to the centrosome at the cilia base (Cs-ZikV-NS5, median=0.68 ± 0.3 μm, n=60: Figures 2B–D; Figures S3A–C). Co-expressing all ZikV associated NS proteins in NPCs did not affect the localization of NS5 to the cilia base or revert the ciliopathy (Figures S1V–X).
Figure 2.-. ZikV-NS5 localization to the cilium base impairs cilia elongation and promotes neural delamination.
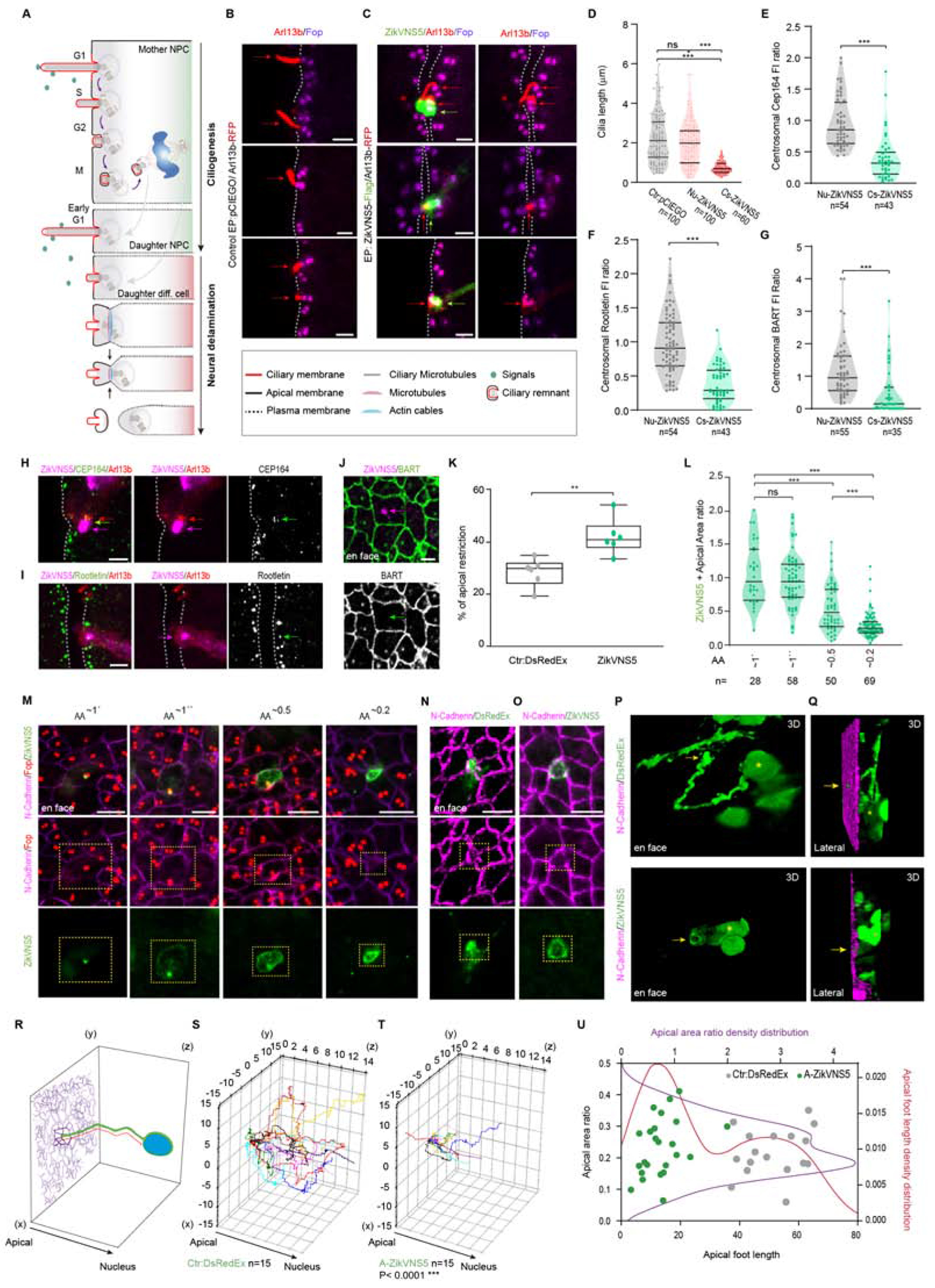
(A) The scheme depicts the variation in cilia length during the cell cycle in a dividing NPC. The daughter cell remaining as a NPC regrows a primary cilium, while the daughter cell entering differentiation delaminates from the epithelium.
(B, C) Images show the Arl13b-RFP labelled cilia (arrows) and FOP stained centrosomes lining the NT lumen (dotted line) in (B) control and (C) ZikV-NS5 elect. (green arrows) NPCs.
(D) Cilia length in NPCs elect.with empty vector (control), in NPCs elect. with ZikV-NS5, in which the protein remains localized within the nucleus (N-ZikV-NS5) and in NPCs elect. with ZikV-NS5, in which the protein is localized to the centrosome at the the cilia base (Cs-ZikV-NS5).
(E-G) Fluorescence average pixel intensity (FI) of the indicated endogenous proteins at the centrosome of NPCs, in which ZikV-NS5 remains localized within the nucleus (N-ZikV-NS5 ), compared to NPCs in which the ZikV-NS5 localized to the centrosome at the cilia base (Cs-ZikV-NS5). The ratio of FI was obtained dividing by the mean of FI of the indicated endogenous in four of non-elect. neighbour NPCs (spaced by one cell distance from the ZikV-NS5 electroporated NPCs).
(H, I) Images of ZikV-NS5-FLAG (purple arrow) co-localization with Arl13b (red arrows) and endogenous CEP164 (H) or endogenous Rootletin (I) (green arrows) (J) En face images of endogenous BART labelled apical ring and its absence from the centrosome where ZikV-NS5-FLAG is located (purple arrow).
(k) Plots of the proportion of cells with apically restricted areas in DsRedEx-control and ZikV-NS5 elect. NTs (n= 6 embryos; median ± S.D.).
(L) Plots of apical areas (AA) in relation to ZikV-NS5 positioning. The non-restricted areas (~1) correspond to ZikV-NS5 co-localization at the centrosome and the restricted areas (≤0.5) to ZikV-NS5 localization at the apical belt.
(M) En face images of N-cadherin labelled AJs, FOP labelled centrosomes and ZikV-NS5-FLAG subcellular localization according to the apical area (AA).
(N, O) En face images of N-cadherin showing restricted apical areas in DsRedEx-control (N) and ZikV-NS5-FLAG elect. NPCs.
(P, Q) 3D reconstructions of the apical foot in DsRedEx-control or ZikV-NS5-FLAG transfected NPCs. Arrows point to the apical end-foot area facing the NT lumen (N-cadherin, purple: asterisks correspond to nuclei).
(R) The scheme depicts the apical foot length (red line) as the distance between the restricted apical end-foot area facing the NT lumen (purple) and the basal nucleus (blue) in NPCs.
(S, T) 3D Python Plots of apical foot length in NPCs expressing DsRedEx (S, control) in comparison to NPCs presenting an apical belt distribution of ZikV-NS5 (T , A-ZikV-NS5) Both conditions show similar apical end-foot area restriction (0< AA >0.5) as shown in U.(U) Plots of the apical foot length relative to the apical end-foot area restriction in DsRedEx-control (grey dots) and apical belt ZikV-NS5 expressing NPCs (green dots). The density distribution of the apical end-foot area restriction presents a Gaussian distribution (in purple), while the density distribution of the apical foot length presents a bimodal distribution (Red).
In violin plots, the upper and lower lines indicate the interquartile range, and the middle line the median. ***p<0.001, **p<0.01; exact P value 0.0043 (K), *P<0.05, ns: not significant. D-K, Mann-Whitney U test; L, Kruskal-Wallis test. Scale bars 2 μm (B, C, H-J) 4 μm (M-O).
See also Figures S1–S3 and Videos 1–4.
We assessed whether ZikV-NS5 from the African and the Asian strains also affects cilia length according to the subcellular localization in electroporated NPCs. In electroporated NPCs where ZikV-NS5-AF and ZikV-NS5-AS are exclusively localized in the nucleus (Nu-ZikN-NS5-AF: median=2.2 ± 1.3 μm, n=65; Nu-ZikN-NS5-AS: median=2.1 ± 1 μm, n=45, Figures S2 D, E, F, I ), cilia length varied as in controls cells (Figures S2 D, E). However, the cilia were significantly shorter in electroporated NPCs showing ZikV-NS5-AF and ZikV-NS5-AS localization to the centrosome at the cilia base (Cs-ZikN-NS5-AF: median=0.7 ± 0.74 μm, n=36; Cs-ZikN-NS5-AS: median=0.7 ± 0.36 μm, n=32) (Figures S2D, E, G, H, I, J).
In order to show the conservation of defects caused by ZikV-NS5 protein, we cultured human induced pluripotent stem cells (iPSCs) following a standard neural induction protocol (Akizu et al., 2013). As the phenotype that we described in the chick embryo NT, hNPCs after transfection with ZikV-NS5 increased the expression of the anti-proliferative gene Tis21 (Figure 3A–D). Moreover, ZikV-NS5 pulled down endogenous CEP164, Rootletin and BART in hNPCs (Figure 3 E) and localizes at the base of Arl13B labelled primary cilia in Nestin positive hNPCs, and this subcellular localization cause cilia shorteneing (Figure 3F–M).
Figure 3.-. ZikV-NS5 localization to the cilium base impairs cilia elongation in human NPCs.
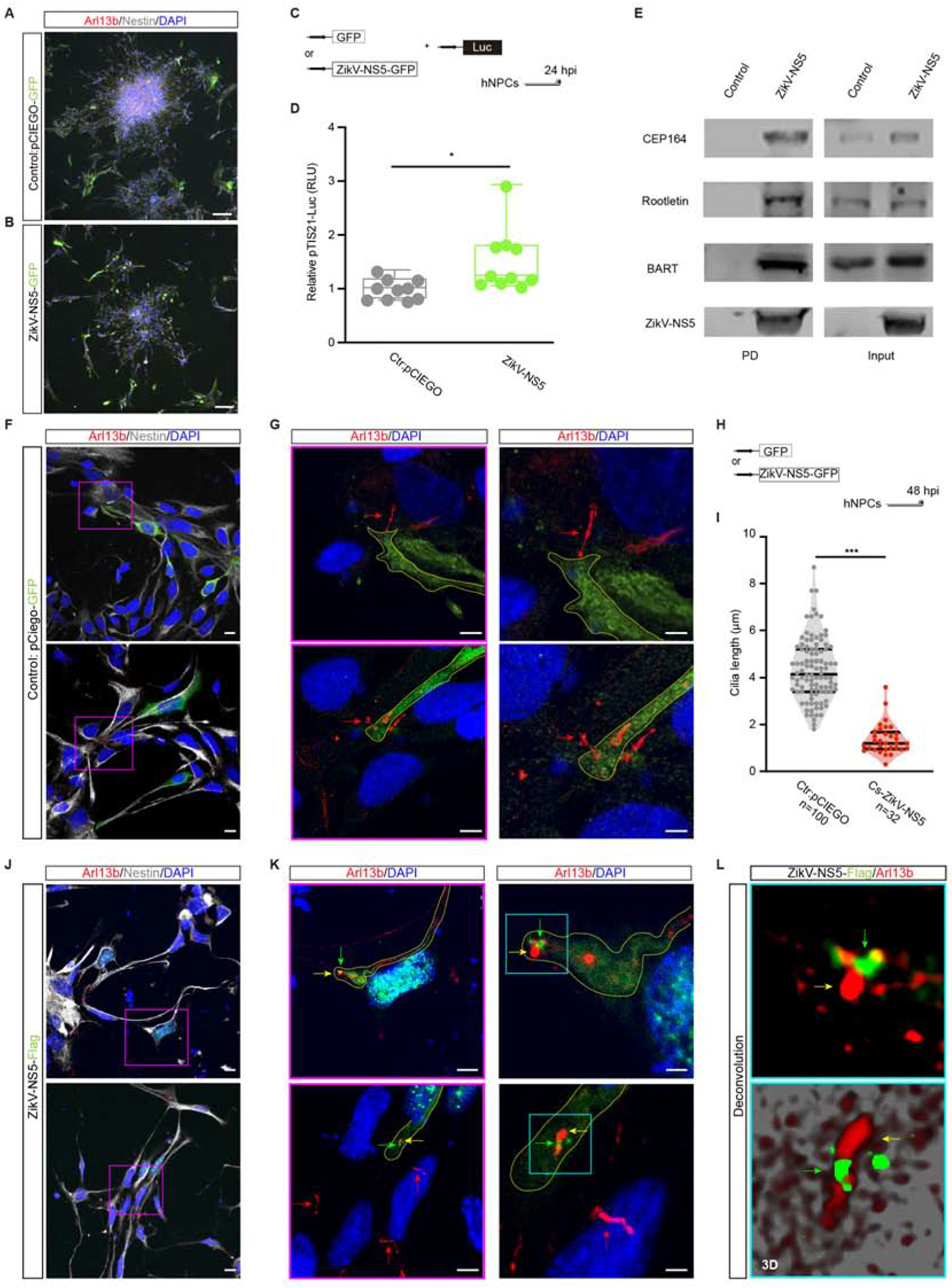
(A, B) Selected images show human NPCs positive for Nestin and transfected with the GFP control vector (A) or ZikV-NS5-GFP (B).
(C) DNAs transfection for luciferase assay.
(D) Luc/Renilla activity of the pTis21-Luc reporter 24 hours after transfection of the empty vector GFP or the ZikV-NS5 DNA (Box and Whiskers plot, n=10).
(E) hNPCs endogenous protein extracts were analysed by western blots. Pull-down (PD) assays show ZikV-NS5 binding to endogenous Cep164, Rootletin, BART. Control is referred to a pull down with the resin in the absence of ZikV-NS5 recombinant protein.
(F, G) Images show Arl13b labelled cilia in Nestin positive hNPCs transfected with the control vector encoding for GFP. Different inset magnifications of G (purple scare) show selected GFP positive hNPCs (dashed yellow line) with a normal Arl13b labelled cilia (red arrow; G).
(H) DNAs transfection for cilia length measurement using Arl13b as a pan-cilia marker.
(I) Cilia length in hNPCs transfected with the control vector encoding GFP, in hNPCs transfected with ZikV-NS5, in which the protein is localized to the centrosome at the cilia base (Cs-ZikV-NS5).
(J-L) Images show Arl13b labelled cilia in Nestin positive hNPCs transfected with ZikV-NS5 (J). Different inset magnification of J (purple scare) show ZikV-NS5 localization close to the cilia (green arrow) and Arl13b labelled defected cilia (yellow arrow; K). Normal cilia are shown with red arrows (K). Different inset magnifications of K (blue scare) show deconvolution images of ciliopathy (yellow arrow) associated to apical ZikV-NS5 (green arrow; L).
In violin plots, the upper and lower lines indicate the interquartile range, and the middle line the median. ***p<0.001, **p<0.01, *P<0.05, ns: not significant. Mann-Whitney U test D, G. Scale bars 75 μm (A, B), 10 μm (F, J), 5 μm (G, K), 2 μm (insets F, J, insets K).
To search for the mechanism by which ZikV-NS5 mediates cilia shortening, we quantified the endogenous target proteins in NPCs relative to the subcellular localization of ZikV-NS5. Compared to electroporated NPCs with ZikV-NS5 exclusive localization in the nucleus, there was a significant reduction in CEP164 protein when ZikV-NS5 localizes to the centrosome at the cilia base (Nu-Zikv-NS5 median=0.85 ± 0.05 AU, n=54; Cs-ZikV-NS5 median=0.32 ± 0.05, n=43: Figures 2E, H; Figures S3D, F). Interestingly, less CEP164 was reported previously in ZikV-infected NPCs forming brain organoids (Gabriel et al., 2017). In addition, there was less endogenous centrosomal Rootletin and BART in NPCs upon ZikV-NS5 localization to the centrosome at the cilia base (Rootletin Nu-Zikv-NS5 median=0.9 ± 0.05 AU, n=80; Cs-ZikV-NS5 median =0.3 ± 0.04 AU, n=52; BART Nu-Zikv-NS5 median=0.95 ± 0.1AU, n=55; Cs-ZikV-NS5 median=0.15 ± 0.1 AU, n=35: Figures 2F,G, I, J; Figures. S3E, G). Hence, the reduction in the endogenous CEP164, Rootletin and BART protein at the centrosome of cells expressing ZikV-NS5 correlated with the impaired cilia elongation in NPCs. In turn, cilia shortening might impair the reception of extracellular growth signals, such as that provided by Sonic Hedgehog (Shh), an important regulator of proliferation (Goetz and Anderson, 2010; Saade et al., 2018).
In NPCs, apical centrosomes organize a microtubule ring that, when aligned with the actin cables, plays an active role in reducing their apical surface and withdrawing their apical end-foot from the ventricular surface (Figure 2A) (Baek et al., 2018; Kasioulis et al., 2017). We evaluated whether ZikV-NS5 might influence neural delamination, since BART localizes to this apical belt (Figures. 1P–Q, 2J; Figures S3H, J–K). We measured the apical area (AA) of electroporated cells surrounded by non-electroporated neighbors in en face images of the NT, the latter used as a reference for AA restriction. The proportion of NPCs with a smaller AA is higher following ZikV-NS5 electroporation (control ratio=28.32 ± 5.13; ZikV-NS5 ratio=41.90 ± 6.7: Figure 2K; control median=0.7 ± 0.53 AU, n=163; ZikV-NS5 median =0.51 ± 0.46 AU, n=213: Figure S3L). The AA was delimitated by the AJ-associated protein N-cadherin and the centrosome was defined by FOP (Figure 2M). In the non-restricted AA, ZikV-NS5 localizes to either the central or lateral positioned centrosome, a feature that precedes neural delamination (AA~1′ ratio=0.92 ± 0.46; AA~1″ ratio=0.92 ± 0.39: Figures 2L–M, Figures S3I) (Wilsch-Brauninger et al., 2012). However, the localization of ZikV-NS5 to the apical belt was correlated with a reduction in the AA (AA~0.5 ratio=0.46 ± 0.35; AA~0.2 ratio=0.23 ± 0.19: Figures 2L–M, Figure S3L), coinciding with early apical to basal migration of the centrosome (Figure 2M, Figures S3I, M–N, Video 1). Moreover, a reduction in the AA at the ventricular surface is preceded by the basal migration of the new-born delaminating neuronal cell body (Kasioulis et al., 2017). Confocal images of DsRedEx or ZikV-NS5 electroporated cells were used to generate 3D reconstructions and to track the cell processes that extend from the apically end-foot restricted surface to the nucleus (Figures 2N–R). ZikV-NS5 electroporated cells have a short apical foot length (control length=51.5 ± 9.85μm, n=17; ZikV-NS5 length=13 ± 7μm, n=25: Figures 2O–Q, T, U, Videos 2, 3) compared to control (Figures 2N, P, Q, S, U, Video 4), indicating that ZikV-NS5 promotes apical end-foot restriction at the ventricular surface before the basal migration of the new-born delaminating neuronal cell body is completed.
Finally, when we assessed the expression of Tis21 to determine if these cellular events promote terminal neurogenesis, reporter activation was associated with the localization of ZikV-NS5 to the cilium base (Figure S3O). Quantification of Tis21p-RFP+ NPCs at 16 hpe (n=5 embryos) showed that 93 ± 7% of the NPCs expressed pTis21-RFP when ZikV-NS5 was localized to the cilium base, as opposed to 25 ± 10% of NPCs in which ZikV-NS5 localizes to the nucleus (n=5 embryos), or 25 ± 3% of the control NPCs (n=6 embryos: Figure S3P). Together, these data indicate that primary cilia elongation is impaired and apical end-foot restriction is accelerated in NPCs exposed to ZikV-NS5 protein, two key events that promote neural delamination. These results prompted us to study how the ZikV-NS5 protein affects primary neurogenesis and embryonic CNS growth.
The multimeric arrangement of ZikV-NS5 is required to promote terminal neurogenic divisions
Normal growth of the CNS requires a fine balance between the different modes of NPC division and a premature switch to neuron generating divisions may cause microcephaly (Saade et al., 2018). Symmetric proliferative divisions that leave two NPC daughter cells (PP) can be monitored in vivo with the pSox2-EGFP construct, whereas pSox2-EGFP and the neurogenic pTis21-RFP reporter are co-expressed in asymmetric divisions that produce one NPC and one differentiating neuron (PN). By contrast, symmetric neurogenic divisions generating two neurons (NN) that detach from the ventricular zone can be monitored by the expression of the pTis21-RFP reporter (Figure 4A) (Le Dreau et al., 2014; Saade et al., 2017; Saade et al., 2013). To determine whether ZikV-NS5 affects the mode of NPC division, stage HH10/12 embryos were co-electroporated with ZikV-NS5 together with the pSox2-EGFP/pTis21-RFP reporters (Figure 4B). When the embryos were analyzed at 24 hpe through fluorescent activated cells sorting (FACs), the rate of PP divisions (pSox2-EGFP+ cells) was seen to decrease significantly in ZikV-NS5 embryos (from 51 ± 2% in controls to 30 ± 2.8% in ZikV-NS5 embryos). This reduction in PP divisions took place at the expense of an increase in neurogenic divisions (from 18 ± 1.6% in controls to 46.4 ± 5.6% in ZikV-NS5 embryos: Figure 4C). Similarly, when embryos electroporated were immunostained at 24 hpe, there was again a clear decrease in the rate of PP divisions (in pH3+ cells) in ZikV-NS5 embryos (from 32± 3.4% in the controls to 16± 6.5% in ZikV-NS5-EP embryos), which was compensated by an increase in NN divisions (from 13± 3.4% in controls to 33 ± 6.4% in ZikV-NS5-EP embryos: Figures 4D–F). Together these data suggest that by preventing cilia elongation, the ZikV encoded NS5 protein alone is sufficient to switch the mode of NPC division and to promote neural delamination. Thus, ZikV-NS5 drives the exhaustion of the neural progenitor pool.
Figure 4. Multimeric arrangement of ZikV-NS5 is required for its localization to the cilia base and the promotion of terminal neurogenic divisions.
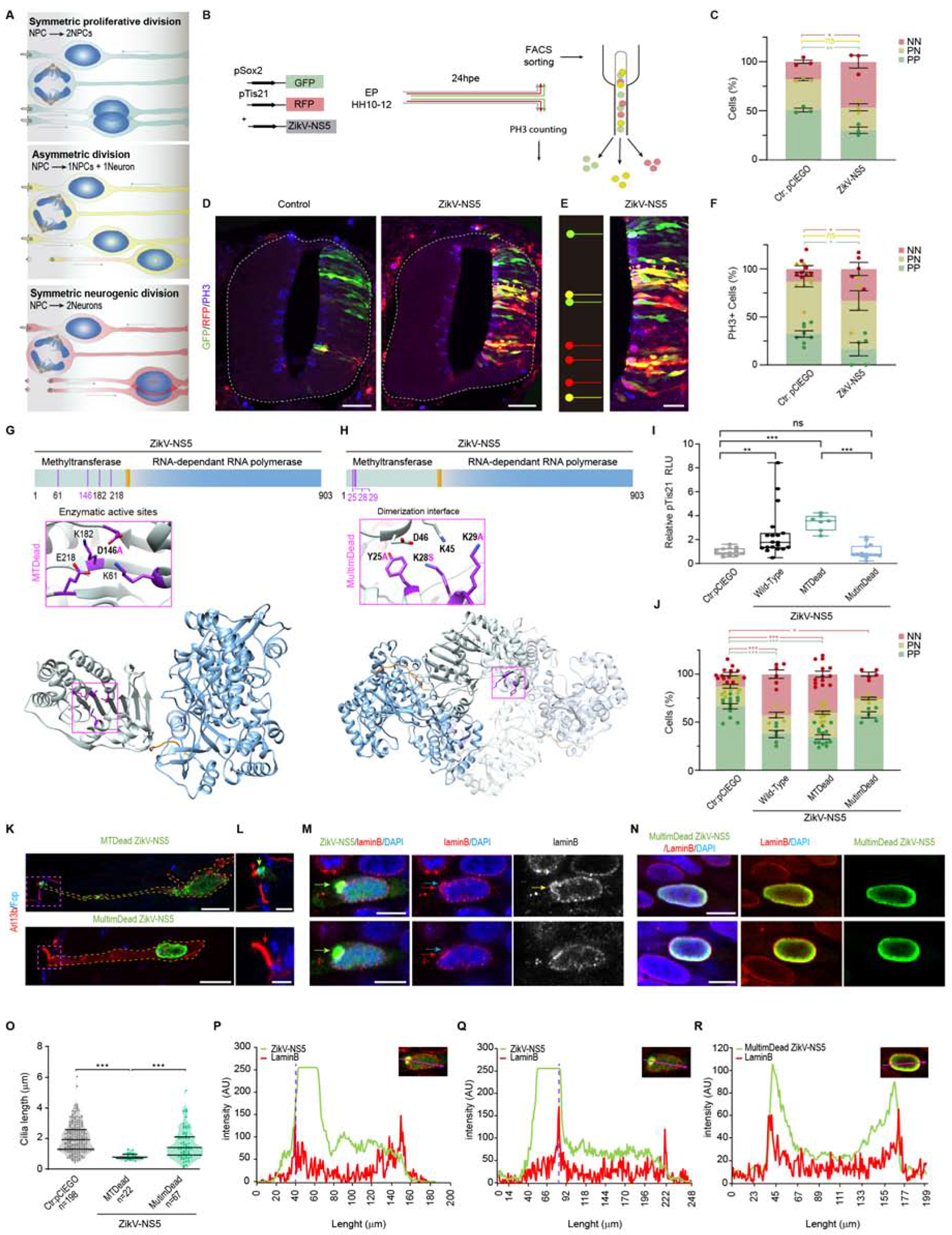
(A) Scheme showing the three modes of NPC division.
(B) Scheme representing the reporter co-electroporation experiments, harvested at 24 hpe, either for flow cytometry analysis (FACS) or for PH3 immunostaining.
(C) Quantification of the cells expressing the reporters corresponding to each division: PP (green), PN (yellow), and NN (red). ( mean ± s.e.m).
(D, E) Images show pSox2 (green) and pTis21 (red) NPCs, and pH3 stained mitoses (blue) in control or ZikV-NS5 (e) elect. NTs. Green, yellow and red lines in E indicate mitotic PP, PN and NN divisions, respectively.
(F) Quantification of reporter-expressing pH3+ dividing cells in each condition. The data represents mean ± s.e.m.: ***P<0.001 **P<0.0, *P<0.05.
(G) The scheme depicts the MTase and RdRP domains in the ZikV-NS5 protein. 3D representation of one NS5 monomer shown in an orientation from the top of RdRP with the MTase domain shown in grey and the RdRP finger, palm and thumb sub-domains in blue. Inset showing the amino acids involved in the enzymatic activity, with the amino acid substitution highlighted in purple.
(H) The scheme depicts the ZikV-NS5 protein and a 3D representation of one ZikV-NS5 dimer. Inset shows the amino acids involved in dimerization, highlighting the amino acid substitution in purple.
(I) Luc/Renilla activity of the pTis21-Luc reporter after elect. of the DNAs indicated (mean ± SD, n=6-8 embryos/condition).
(J) Quantification of reporter-expressing FACS sorted cells in each condition: PP (pSox2+/pTis21−, green), PN (pSox2+/pTis21+, yellow) and NN (pSox2−/pTis21+, red). ( means ± s.e.m).
(K, L) Images of Arl13b at cilia, the FOP labelled centrosomes at the cilia base and the localization of the ZikV-NS5 variants: ZikV-NS5-MTDead at the cilium base (green arrow in L), ZikV-NS5-MultimDead excluded from the cilium base (red arrows point cilia in L).
(M, N) Images of nuclear ZikV-NS5-FLAG in two separate z stacks. Upper image shows nuclear aggregates (green arrow) causing disruption of the nuclear envelope. Lower image shows ZikV-NS5-FLAG aggregates (green arrow) exit from the nuclear envelop. Nuclear Envelop disruption is highlighted by Lamin B (red arrow) and DAPI (blue arrow). ZikV-NS5-MultimDead shown in two separate cells in N, localizes at the vicinity of the nuclear envelop.
(O) Cilia length in control NPCs and those elect. with the mutant ZikV-NS5 constructs (upper and lower lines indicate the interquartile range, and the middle line the median).
(P, Q) Plots of the F. I. (AU) of green labelled ZikV-NS5 relative to the red lamin B labelling at the two separate Z-stacks shown in M. The purple dotted line depicts nuclear envelope disruption.
(R) Plots of the F. I. (AU) of green labelled ZikV-NS5-MultimDead with the same intensity profile as the red labelled lamin B.
***p<0.001, **p<0.01; exact P values 0.0054 (C) 0.0034 (I) , *P<0.05; exact P values 0.0129 (C) 0.0151 (NN, F) 0.0398 (PP, F), 0.0225 (J).ns: not significant. C, F two-sided unpaired t-test; I, J, one-way ANOVA; O, Kruskal-Wallis test. Scale bars 30 μm (D), 15 μm (E). 7 μm (K), 2 μm (L), 5 μm (M, N).
See also Figures S3 and S4.
To better define the neurogenic effects of ZikV-NS5, we monitored the expression of Tis21 after co-electroporation of the reporter and the ZikV-NS5 N-terminal (NTerm, residues 1–262; MTase) or ZikV-NS5 C-terminal (CTerm, residues 267–903; RdRP) domains into the chick embryo NT (Figures S4A, B). In contrast to the ZikV-NS5-CTerm domain, the ZikV-NS5-NTerm alone was sufficient to enhance pTis21-Luciferase reporter activity (Figure S4B). Moreover, the ZikV-NS5-NTerm drove an increase in the rate of NN divisions in the embryos, as witnessed in immunostained embryos (pH3+ cells; Figures S4C–E) and by FACs analysis (Figures S4C, F) Together these data suggest that the neurogenic capacity of the ZikV-NS5 protein resides in its N-terminal domain, which prompted us to assess whether enzymatic activity was required to promote neurogenesis.
The MTase domain of the NS5 flavivirus protein mediates the addition of a guanine cap to the 5’ end of the RNA genome, and the sequential methylation of guanine N-7 and ribose 2′-O to form a type 1 cap structure (m7NGpppA2′Om). This activity is essential to evade the host’s immune response and for replication, increasing the viral polyprotein translation efficiency. The ZikV MTase active site contains residues K61, D146, K182 and E218, which are in close contact with the S-adenosylmethionine (SAM) co-factor, the methyl group donor for the reaction (Figure 4G). However, a single amino acid substitution (D146A) was sufficient to abolish MTase activity (Zhou et al., 2007) (MTDead). To assess whether this enzymatic activity is required for the neurogenic capacity of ZikV-NS5, we co-electroporated the pTis21-Luciferase reporter together with the ZikV-NS5-MTDead construct. Consequently, the luciferase activity of the reporter increased in response to mutant ZikV-NS5 MTase in a similar manner to that produced by the ZikV-NS5 protein (Figure 4I). Indeed, the ZikV-NS5-MTDead construct also provoked an increase in the rate of NN divisions in embryos when analyzed by FACs (13 ± 3% in controls, 42.5 ± 4.5% in ZikV-NS5, and 40.5 ± 3% in ZikV-NS5-D146A embryos: Figure 4J).
We recently characterized the supramolecular arrangement of the full-length ZikV-NS5, highlighting the assembly of NS5 monomers into dimeric structures, as well as the interactions of dimers to form a higher order fibrillar structures (Ferrero et al., 2019). The main interface for dimer formation involves the MTase-MTase contacts, connecting amino acids Y25, K28 and K29 in one ZikV-NS5 molecule with residues K45 and D46 of the second ZikV-NS5 subunit (Ferrero et al., 2019) (Figure 4H). To test whether the multimeric arrangement of ZikV-NS5 is required to promote premature neurogenesis, we used the ZikV-NS5-Y25A/K28S/K29A mutant that fails to form dimers (MultimDead) (Ferrero et al., 2019). This MultimDead mutant protein did not induce premature differentiation of NPCs (Figure 4I) and in embryos analyzed by FACs at 24 hpe, dimerization was required for ZikV-NS5 to promote the neurogenic mode of NPC division (25 ± 2% NN divisions in ZikV-NS5-MultimDead embryos: Figure 4J). These data indicate that while the enzymatic activity of NS5 is dispensable, ZIKV-NS5 oligomerization is required for this protein to promote neurogenesis.
To define the cellular mechanisms altered by the ZikV-NS5 protein variants, we followed their subcellular distribution in chick embryo NPCs. The ZikV-NS5 MTDead protein was detected in the nucleus and at the cilium base, like the wild type protein, while the MultimDead protein variant was retained within the nucleus, concentrating in the vicinity of the nuclear envelope (Figures 4K–N). Moreover, the multimerization of ZikV-NS5 contributes to the formation of ring-like nuclear aggregates that exit the nucleus toward the apical foot and localize at the centrosome forming the cilia base (Figure S4G). Furthermore, the nuclear exit of multimerized ZikV-NS5 is associated to the transient disruption of the nuclear envelope evident as Lamin B blebs and protruding chromatin (Figures 4M, N, P–R). Indeed, this transient loss of nuclear integrity has already been reported in ZikV infection (Onorati et al., 2016), as well as in other viral infections (Hatch and Hetzer, 2014). Nuclear retention of the ZikV-NS5 MultimDead protein, might rely on its reduced capacity to bind the nuclear envelope components such as Emerin (Figures S4J, K). Hence, the trafficking of ZikV-NS5 to the cilia base appears to depend on its capacity to form multimers.
The retention of ZikV-NS5 within the nucleus was sufficient to prevent the ciliopathy, as evident by the length of the Arl13b-RPF labelled cilia in NPCs (control median=1.98 ± 0.97 μm, n=198; ZikV-NS5-MTDead median= 0.82 ± 0.2 μm, n=22; ZikV-NS5 MultimDead median= 1.44 ± 1 μm, n=67: Figure 4O). Even though the ZikV-NS5 MultimDead protein variant has a reduced capacity to bind to the nuclear envelop protein Emerin and to the centrosome protein CEP164 (Figures S4J, K), a high concentration of this protein can pass through the nuclear envelop (Figures S4L, M, Q, R), translocate to the cilia base and induce a ciliopathy (Figures S4N–P), ultimately promoting the neurogenic responses of the Tis21-Luc reporter as the non-mutated ZikV-NS5 (Figure S4S, T).
Microcephaly in a human post-mortem ZikV infected foetal brain is accompanied by a ciliopathy in ependymal cells
To support our hypothesis that the ciliopathy caused by ZikV-NS5 results in a microcephaly phenotype, we examined post-mortem tissue from a reported ZikV-infected microcephalic foetus from a ZikV infected mother. The pregnant woman reported possible exposure to the Aedes mosquito until the 12th gestational week (GW) and ZikV RNA was detected in the GW22. After ultrasound and MRI imaging, the foetus was diagnosed with severe microcephaly and the pregnancy was terminated in the GW22+6. A severe developmental delay was evident in forebrain coronal sections, with almost complete cortical plate agyria when compared to the control sections of a non-pathological GW21 foetal brain. A thinner cortical plate (CP), and thinner intermediate (IZ) and subventricular zones (SVZ) were also detected (Figures 5A, B). We examined the layered organization of neurons in two CP areas, which, not only showed a dramatic reduction in neuron numbers in the ZikV infected foetal brain compared to the control GW21 brain (Figure 5C; Figures S5A–E), but also showed an aberrant neuronal distribution. In the ZikV infected brain, both Satb2+ upper layer neurons (Figures 5D, E; Figures S5F, G) and Ctip2+ lower layer neurons (Figures 5F, G; Figures S5H, I), were distributed throughout the CP. Moreover, cortical neurons ectopically localized in the intermediate zone in the ZikV infected brain (Figures S5C, D), was similar to cortical abnormalities caused by cilia deficiencies such as in Arl13b mutant radial glial cells (Higginbotham et al., 2013).
Figure 5.-. Microcephaly in ZikV infected Human foetal brain is accompanied by a severe disorganization of the cortical plate.
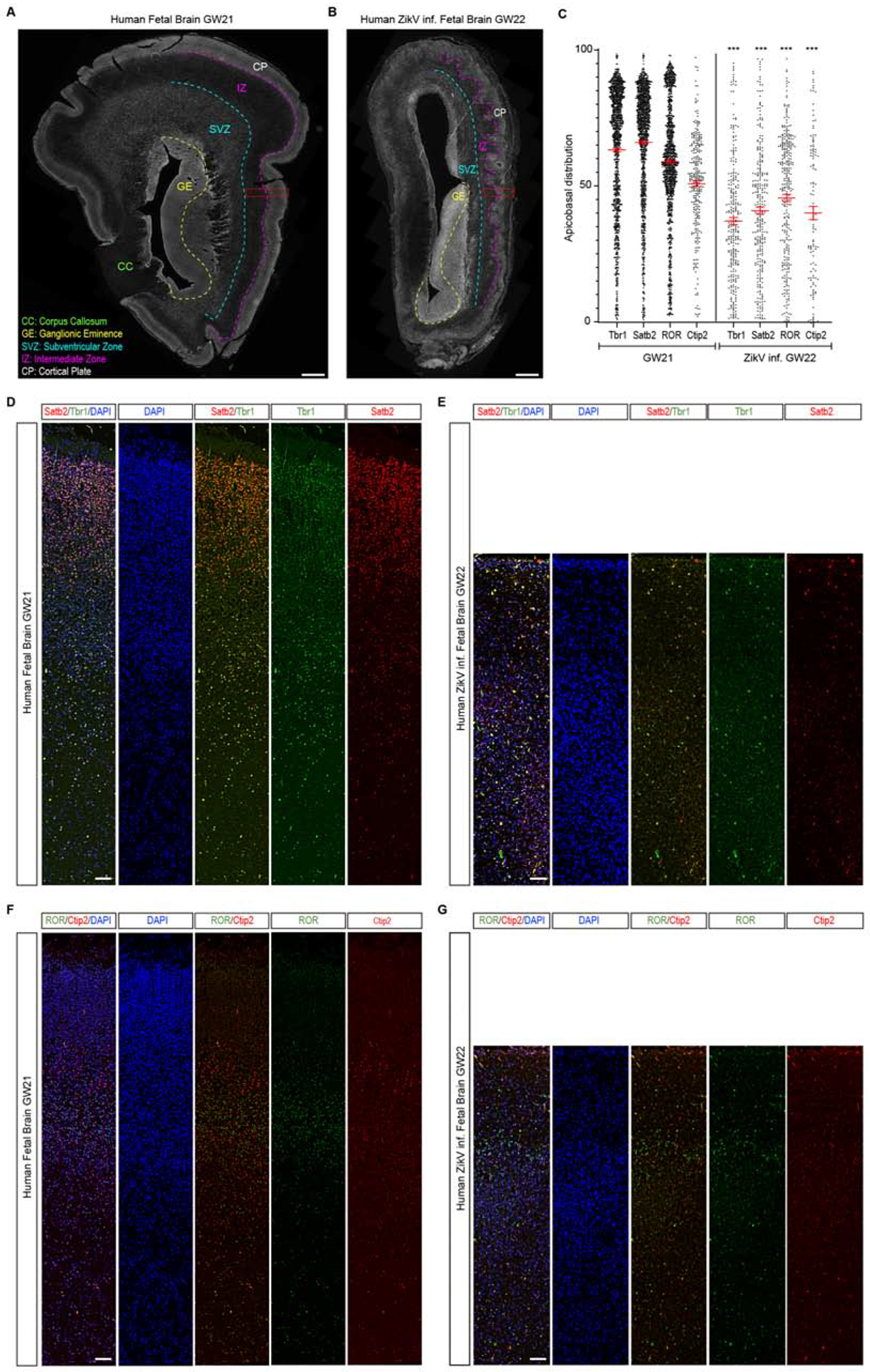
(A) Representative image of control GW21 forebrain coronal sections indicating the different forebrain areas. Red inset highlights the area quantified in (C) and the images shown in (D) and (F).
(B) Representative image of ZikV infected GW22 forebrain coronal sections. Red box highlights the area quantified in (C) and the images shown in (E) and (G).
(C) Relative apicobasal position of Tbr1+, Satb2+, ROR+, or Ctip2+ neurons at GW21 and ZikV infected GW22. Values represent median ± s.e.m. Total number of neurons counted, GW21: Tbr1+ n = 1.304, Satb2+ n = 1.303, ROR+ n = 1.329, Ctip2+ n = 293; ZIKV inf. GW22: Tbr1+ n = 283, Satb2+ n = 290, ROR + n = 320, Ctip2+ n = 120.
(D, E) Representative image of the generating cortical plate and intermediate zone at GW21 (D) and ZikV infected GW2 (E), immunostained for marker of post-mitotic neurons Tbr1+ and upper-layer neurons, Satb2.
(F-G) Representative image of the generating cortical plate and intermediate zone at GW21 (F) and ZikV infected GW2 (G), immunostained for markers of deep-layer neurons, ROR and Ctip2.
All figures were digitally stitched by the appropriate image software as described in STAR Methods.
***p<0.001, **p<0.01, *P<0.05. C, Mann-Whitney U test. Scale bars 1.5 mm (A, B), 100 μm (D-G).
See also Figure S5.
During development and upon termination of primary neurogenesis, monociliated NPCs differentiate into multiciliated ependymal cells (ECs) (Spassky et al., 2005). To test whether the ciliopathy described in the chick embryo NPCs might persist in the ECs of ZikV infected human foetal brain, we first assessed the distribution of the viral NS5 protein (Retallack et al., 2016) in the ZikV infected brain. Consistent with the viral infection early in gestation, the NS5 protein spread from the ependymal layer, to the SVZ and CP (Figure 6A). Moreover, the viral NS5 protein was detected in subependymal cells (Figure 6B) and in ECs, co-localizing with the acetylated α-tubulin-labelled cilia shaft (Figures 6C, D), cilia that are much shorter than in the control GW 21 ECs (Figure 6E).
Figure 6.-. Microcephaly in ZikV infected Human foetal brain is accompanied by ciliopathy in ependymal cells.
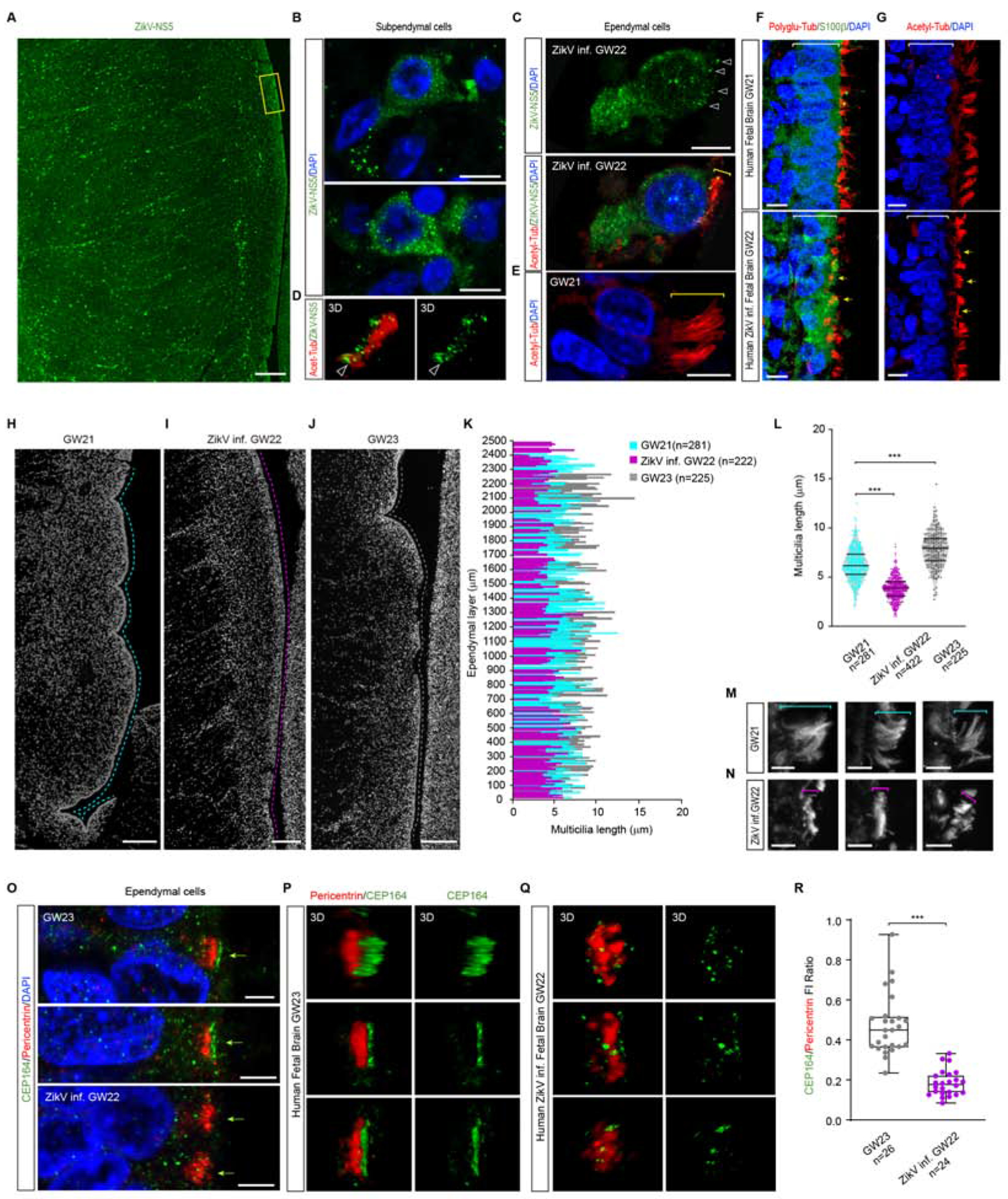
(A) Immunostaining for the viral NS5 protein in ZikV infected GW22 coronal forebrain sections.
(B) Selected images from C (yellow inset) showing the viral ZikV-NS5 protein in subependymal cells with DAPI stained nuclei.
(C) Images show acetylated-tubulin labelled multicilia in ZikV-NS5 infected ependymal cells with DAPI stained nuclei. Arrowheads point to the ZikV-NS5 protein and the bracket indicates the cilia length.
(D) 3D image of ZikV-NS5 (arrowhead) localized to the basal acetylated-tubulin labelled multicilia. The arrowheads clearly show the green dots at the base of cilia.
(E) Images showing acetylated-tubulin labelled multicilia in control GW21 ependymal cells with DAPI stained nuclei, in which the bracket indicates the cilia length.
(F) Images of poly-glutamylated-tubulin stained multicilia in S100β+ ependymal cells of control GW21 foetal brain, and ZikV GW22 foetal brain.
(G) Images of acetylated-tubulin stained multicilia in control GW21 and ZikV GW22 ependymal cells in which the yellow arrows indicate multicilia shortening.
(H-J) Images depicting the 2.5 mm ependymal areas (dotted lines) used to measure cilia length in control GW21, ZikV infected GW22 and control GW23 forebrain sections.
(K) Plots of the cilia length along the ependymal layer at the developmental stages indicated.
(L) Plots of the cilia length in ependymal cells of control GW21, ZikV infected GW22 and control GW23 forebrain sections (upper and lower lines indicate the interquartile range, the middle line the median)
(M, N) Images of acetylated-tubulin stained multicilia in control GW21 (M) and in ZikV infected GW22 ependymal cells (N, brackets indicate cilia length).
(O) Images of PCNT stained multiple centrosomes and CEP164 stained multi cilia base in control GW21 and ZikV infected GW22 ependymal cells.
(P, Q) 3D image showing PCNT and CEP164 localization to the base of multicilia in control GW23 (P) and ZikV infected GW22 (Q) ependymal cells.
(R) Box plot of CEP164/PCNT F. I. Ratio, showing reduced expression of CEP164 in the multiple centrosomes of ZikV infected ependymal cells.
Figures A, H-J were digitally stitched by the appropriate image software as described in STAR Methods.
***p<0.001, **p<0.01, *P<0.05. N, Kruskal-Wallis test, R, Mann-Whitney U test. Scale bars 200 μm (A, H-J). 6 μm (B, C, E-G, M, N), 3 μm (O).
In addition, we observed a severe disorganization of the S100β+ ECs lining the ventricle (Figures 6F,G), as reported in ZikV infected macaque tissues (Adams Waldorf et al., 2018), with differentiating ECs expanding their apical surface and assembling multiple motile cilia (Mahuzier et al., 2018). To next investigate whether ZikV infection affected ciliogenesis, the length of the acetylated α-tubulin cilia shafts protruding from the VZ surface was measured in the ~2.5 mm ependymal layer facing the Ganglionic Eminence (GE) in ZikV infected ECs, comparing these values to those in control ECs in the 21st and 23rd GW (Figures 6H–K). While control tissue showed an increase in cilia length associated with EC maturation (GW21 median=6.3 ± 1.57 μm, n=281; GW23 median=7.8 ± 1.77 μm, n=225), the EC multicilia were significantly shorter and more disorganized in ZikV infected tissue (median=3.86 ± 1.14 μm, n=422: Figures 6L–N).
Multiciliated EC differentiation involves the assembly of a large number of centrioles that migrate apically and anchor to the plasma membrane. In the case of these cilia, Cep164 localizes to the distal appendages and Rootletin extends from the proximal end of the centrioles to their rootlets (Mahuzier et al., 2018) (Figure 6O). Indeed, 3D imaging highlighted the disturbed CEP164 distribution at the apical surface of ZikV infect ECs relative to the controls (Figures 6P,Q). To search for the mechanisms underlying the ZikV mediated ciliopathy in human tissue, we quantified the endogenous Cep164 protein in ECs, relative to the centriole marker Pericentrin. ZikV infected ECs had significantly less CEP164 protein than control ECs, which is itself required for motile cilia assembly (Siller et al., 2017) (control mean=0.47 ± 0.15, n=26; ZikV infected mean=0.18± 0.06, n=24: Figure 6R). Together our analysis revealed that ZikV infection of human foetal brain tissue not only compromises primary neurogenesis, resulting in a severe microcephalic brain but also, it disrupts EC differentiation and provokes a ciliopathy in these cells. This ciliopathy is likely to impair the flux of CSF elements necessary for brain homeostasis in these cells, as well as for toxin wash-out, further contributing to the developmental malformations associated with congenital Zika syndrome, such as microcephaly and ventriculomegaly.
DISCUSSION
ZikV has evolved multiple mechanisms to exploit or perturb fundamental cellular processes in NPCs (Scaturro et al., 2018). Previous data showed that NS4AB interferes with the mTOR pathway in NPCs, reducing proliferation and inducing autophagy (Liang et al., 2016), and that NS2A interacts with AJs, while disrupting the integrity of the developing neuroepithelium and again, perturbing NPC proliferation (Yoon et al., 2017). The NS2B3 heterodimer was also shown to disturb the cell-cycle as it affects the host protein Septin-2 that is involved in NPCs cytokinesis (Li et al., 2019). To date, NS5 has only been shown to counteract host antiviral mechanisms by targeting the interferon pathway (Grant et al., 2016a). Here we describe a toxic effect of NS5 on NPCs by showing its capacity to interact with multiple host proteins at the cilium base, consequently promoting a ciliopathy and a premature NPC differentiation.
Primary and motile cilia differ in their structure, composition and function. In the developing brain, primary cilia are non-motile signaling organelles present on NPCs that help integrate growth signals. Multiple motile cilia are found on the surface of the ECs lining all the brain ventricles, where they contribute to the flow of CSF. During brain development, monociliated NPCs differentiate into multiciliated ECs, providing an ideal system to study the impact of pathogen infection on these two developmental stages. The primary cilium has a microtubule-based core and an axoneme that extends from a specialized centriole at the base of the cilium, the site where Cep164, Rootletin and Bart are located (Figure 1E). Differentiation of ECs involves the assembly of a large number of centrioles that migrate apically and anchor to the plasma membrane (Mahuzier et al., 2018). Here we show that in NPCs, ZikV-NS5 binds directly to the ciliary basal body, impairing the normal development of this organelle and resulting in cilia shortening (Figures 1–2). We hypothesize that NS5 interaction with CEP164, Rootletin and BART, might target these proteins for ubiquitination and degradation (Grant et al., 2016b), at the cilia basal body, therefore perturbing cilia assembly. Failure to elongate a primary cilium after mitosis diminishes the reponses to external stimuli like Shh, and to other growth factors that promote symmetric NPC division and CNS growth (Goetz and Anderson, 2010; Saade et al., 2018). Remarkably, reduced levels of CEP164 and centriole damage were previously associated to ZikV infection (Gabriel et al., 2017). In addition, dysregulation of neurogenesis through the Notch pathway, a modulator of NPCs response to Shh ligand, was also reported in ZikV infections (Ferraris et al., 2019). Hence, the primary ciliopathy provoked by ZikV-NS5 seems to be responsible for the microcephaly induced by ZikV. Interestingly, we found that in human post-mortem tissue of a presumed ZikV-infected microcephalic fetus from a ZikV infected mother, ECs exhibited a severe ciliopathy that might prevent the flow of the CSF necessary to maintain brain homoeostasis and toxin washout (Figures 5–6). The distinction between a motile ciliopathy and a primary ciliopathy seems to be unclear, as hydrocephalus may also be observed in a number of primary cilia disorders (Valente et al., 2014). Indeed, here we show that a single pathogen can cause both types of ciliopathy, depending on the cell targeted.
We have also demonstrated that both ciliopathy and the imbalance in the mode of division produced by ZikV-NS5 are strongly dependent of its quaternary structure. The recently reported X-ray structures of the full-length ZikV NS5 revealed a dimer based helicoidal arrangement of the protein that is mediated by MTase-MTase interactions, involving residues Y25, K28 and K29 (Figure 4H) (Ferrero et al., 2019). Here we show that the monomeric ZikV-NS5 Y25A/K28S/K28A variant is retained within the cell nucleus forming a diffuse pattern. This data suggests that nuclear multimerization of NS5 is required to escape from this cellular compartment and travel to the basal body. Indeed, our results show that the capacity of ZikV-NS5 to form multimers contributes to its interference with components of the cilium base, a fundamental mechanism underlying the distinct developmental malformations attributed to congenital Zika syndrome. These findings suggest alternative routes that could be followed in order to discover potential antiviral agents that target the intermolecular interfaces involved in dimer/oligomer formation.
Limitations of Study
In this study, we recognize the limitations associated to the ZikV-NS5 strategy for passing the nuclear envelop barrier. The key mechanisms featuring at the nuclear envelop and how ZikV-NS5 multimerization capacity favors nuclear egress is still debated. Testing nuclear export inhibitors such as Leptomycin B on infected NPCs could help deciphering whether active export via the nuclear pore complexes pathway is involved in this process. We should also consider that the exit of ZikV-NS5 from the nucleus is clearly accompanied with Lamin B blebbing and disorganization (Figure 4M). This let us suggest that ZikV-NS5 nuclear egression could involve structural and mechanical aberrations of the nuclear lamina, a mechanism already reported in several viral infections (Stiekema et al., 2020).
STAR METHODS
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elisa Marti (emgbmc@ibmb.csic.es).
Materials Availability
All plasmids generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The macro generated during this study to measure the length and positioning of the multiciliated patches along the ependymal cell layer in human brain is available at MolecularImagingPlatformIBMB’s Github (https://github.com/MolecularImagingPlatformIBMB/MeasureMultiCilia).
Experimental Model and Subject Details
Tissue Procurement
All work was performed according to EU guidelines for the acquisition and distribution of human tissue for bio-medical research purposes and with approval by the human Investigation Committees and Institutional Ethics Committees of CSIC-007/2019 and VHIR. De-identified postmortem human brain specimens were obtained from tissue collection at the Neuropathology laboratory, Pathology Department, H. U. Vall d’Hebron-PR(AG)129/201. Appropriate informed consent was obtained and all available non-identifying information was recorded for each specimen. Tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the WMA Declaration of Helsinki.
Chick embryos
Eggs from white Leghorn chickens were staged according to the method of Hamburger and Hamilton (Hamburger, 1951). In ovo electroporation was performed at stage HH10-12 (36 hours post fertilization -hpf-, 12 somite stage) and the embryos were recovered at 24 hpf. Embryos were electroporated with Clontech purified plasmid DNA (0,051μg/ml in H2O) with Fast Green (50 ng/ml). Briefly, the plasmid DNA was injected into the NT lumen and electrodes were placed either side of the embryo to perform electroporation using an Intracel Dual Pulse (TSS10) electroporator, delivering five 50 millisecond pulses of 20-30V.
Human NPC culture
Human neural progenitor cells (hNPCs, WiCell) were obtained as previously described (Akizu et al 2013). Briefly, embryoid bodies (EBs) were formed by mechanical dissociation of H9 human embryonic stem cell clusters, plated in suspension in neural induction media (DMEM F12, 1x N2, 1x B27, 1μM LDN and 1μM SB431542) for 9 days with daily media replacement. Resultant EBs were then plated on polyornithine/laminin coated dishes in NBF medium (DMEM F12, 1x N2, 1x B27, 20 ng/ml bFGF, 1x Penicillin-Streptomycin). Neural rosettes, visible after 3-5 days, were dissociated with Accutase (ThermoFisher) to obtain hNPCs. hNPCs were expanded and all experiments performed between passage 3 and 6.
hNPCs were cultured in 24-well plates (1.8x105 cells/well) and transfected with 1μg of pCIG or pCIEGO-ZikVNS5-GFP using lipofectamine stem (ThermoFisher) in NBF media following manufacturer’s instructions. 48h after transfection cells were washed once with PBS and fixed with cold methanol for 10 minutes at −20C. Fixed cells were permeabilized/blocked with blocking solution (2% fetal serum bovine plus 0.1% Tween 20 in PBS) for 2h. Cells were incubated with primary antibodies in blocking solution overnight at 4°C. After 3 washes with 0.1% Tween 20-PBS, cells were incubated with fluorescently labelled secondary antibodies for 2 h at room temperature. Fluorescence signal was detected using Leica confocal microscope and images were processed with image J (ImageJ).
Method Details
DNA Constructs
All oligonucleotides designed to generate the following DNA constructs are listed in Table S3. DNA constructs used for control conditions are the pCAGGS without florescent marker (pCIEGO) and pCAGGS_ires_H2B: GFP (pCIG). Tagged ZikV proteins were expressed with pCIEGO-EGFP and/or its modified version pCIEGO-FLAG, constructed ligating a short dsDNA sequence codifying for FLAG followed by a stop codon in EcoRI multiple cloning site of pCIEGO-EGFP vector. The dsDNA was formed annealing an equimolar mixture of two oligos that generate cohesive ends compatible with EcoRI site. Polyprotein ORF codifying for ZikV structural proteins (C, Pr-M, E) and ZikV non-structural proteins (NS1-NS5) were chemically synthetized (GeneArt, ThermoFisher) based on the Suriname strain, human isolate sequence Z1106033 (accession number: KU312312.1) introducing some silence mutations to eliminate restriction sites naturally present. Polyprotein ORF codifying for ZikV-NS5s were chemically synthetized (GeneArt, ThermoFisher) based on the African strain MR766 (GenBank accession code: DQ859059) and the Asian H/PF/2013 strain (GenBank accession code: KJ776791). This sequence was employed as a template to obtain ZikV-NS proteins ORFs by PCR using Phusion DNA polymerase (Thermo Fisher, F530S) and specific DNA primers with restriction sites and following the standard protocol provided by manufacturers.
MTDead ZikV-NS5-D146A and MultimDead Y25A/K28S/K29A punctual mutants in pCIEGO-FLAG vector were generated by PCR using pCIEGO-ZikV-NS5-FLAG vector as template and QuickChange site directed mutagenesis kit (Agilent) according to manufacturer’s instructions. DNA fragments codifying for ZikV-NS5 residues ranging from 1-262 and 267-903 were obtained by PCR using the proper primers and Phusion DNA polymerase (ThermoFisher), subsequently digested with restriction enzymes, purified from agarose gel and cloned in pCIEGO-FLAG as described above.
Human Cep164 cDNA was kindly received from Pr. Eric Nigg (Graser et al., 2007) (Biozentrum, University of Basel, Switzerland) cloned into pCJW206 vector to produce a C-terminal myc tagged CEP164 protein. Full length Rootletin in pEGFP vector was purchased from Addgene. R1463 codon was replaced by stop codon to express a more soluble protein with the construct pEGFP-Rootletin (1-1462) as previously reported (Potter et al., 2017). Punctual mutagenesis was introduced using specific primers and QuickChange mutagenesis kit following the manufacturer’s protocol (Agilent). BART-1 gene was kindly provided by Pr. Michael Cheetham (UCL, London). The ORF was amplified by PCR using specific primers flanked by restriction sites and Phusion DNA polymerase following the standard protocol provided by manufacturers (ThermoFisher). The DNA fragment was digested with proper enzymes, purified and cloned into previously digested pEGFP-C1 vector (Clontech) to generate pEGFP-BART-1 construct. pEGFP-C1 Emerin (637) construct was obtained from Addgene and was transfected to produce the fusion protein EGFP-Emerin (637).
pCS2_H2B-GFP was electroporated to label the nucleus. DsRedex (kindley provided by Pr. Roger Tsien, UCSD, USA) encodes DsRedexpress, a red fluorescent protein that diffuses throughout neuroepithelial cells and thereby reveals their morphology (Wang et al., 2009). CEP152-GFP (50 ng/ul: Origene), encoding a centriolar component required for centriole duplication, was electroporated to label the centrosome (Dzhindzhev et al., 2010; Saade et al., 2017). Arl13b-RFP and Arl13b-GFP (Kindly provided by Pr. Magdalena Götz, LMU, Germany), encoding the ciliary membrane component Arl13b (Caspary et al., 2007; Saade et al., 2017) (50ng/ul) was electroporated to label the primary cilia membrane. The pSox2:GFP and pTis21:RFP reporters used to assess the modes of divisions undergone by NPCs were previously described in details (Saade et al., 2013). For luciferase assays, The pSox2:luc and pTis21: RFP were derived from the pSox2:GFP and pTis21: RFP, respectively (Saade et al., 2017; Saade et al., 2013). The pNeuroD:luc was kindly provided by Pr. François Guillemot (The Francis Crick Institute, UK) (Castro et al., 2006). The pTubb3:luc reporter was obtained by subcloning the Tubb3 enhancer region present in the pTubb3enh:GFP plasmid (Bergsland et al., 2011) (kindly provided by Jonas Muhr, Karolinska Institute, Sweden) into the pGL3:luc vector (Promega).
Yeast Two-Hybrid Analysis
Yeast two-hybrid screening was performed by Hybrigenics Services, S.A.S., Paris, France (http://www.hybrigenics-services.com). The coding sequence of ZikV-NS5 protein from Zika virus was PCR-amplified and cloned into pB29 as an N-terminal fusion to LexA (ZikV-NS5-LexA). The construct was checked by sequencing the entire insert and used as a bait to screen a random-primed human Fetal Brain cDNA library constructed into pP6. pB29 and pP6 derive from the original pBTM116 (Beranger et al., 1997; Vojtek and Hollenberg, 1995) and pGADGH (Bartel, 1993) plasmids, respectively.
99 million clones (9-fold the complexity of the library) were screened using a mating approach with YHGX13 (Y187 ade2-101::loxP-kanMX-loxP, MATα) and L40ΔGal4 (MATa) yeast strains as previously described (Fromont-Racine et al., 1997). 141 His+ colonies were selected on a medium lacking tryptophan, leucine and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5’ and 3’ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (NCBI) using a fully automated procedure.
The exact codes of the algorithm used to analyse the Y2H data and to calculate the confidence score for each interaction, PBS (predicted biological score), are described in detail in the following publications (Formstecher et al., 2005; Rain et al., 2001). Briefly, the description of the confidence score, the Predicted Biological Score (PBS) relies on two different levels of analysis. Firstly, a local score takes into account the redundancy and independency of prey fragments, as well as the distribution of reading frames and stop codons in overlapping fragments. Secondly, a global score takes into account the interactions found in all the screens performed at Hybrigenics using the same library. The scores were divided into four categories, from A (highest confidence) to D (lowest confidence). A fifth category (E) specifically flags interactions involving highly connected prey domains previously found several times in screens performed on libraries derived from the same organism. Finally, several of these highly connected domains have been confirmed as false-positives of the technique and are now tagged as F. The PBS scores have been shown to positively correlate with the biological significance of interactions (Rain et al., 2001; Wojcik et al., 2002).
Cell culture transfection and Pull-down assays
293T cells (ATCC) were cultured in 100 mm dishes (Corning) with standard cell culture conditions at 37°C in DMEM suplemented with 10% FBS (SIGMA), 1% Non-essential Amino acids (SIGMA), 1% penicilin and streptomicin (SIGMA), 0.5 μg/ml Amphotericin B (GIBCO). Plasmids containing human ZikV-NS5 interacting proteins (pEGFP-BART-1, pEGFP-Rootletin, pCJW206-CEP164 and pEGFP-Emerin) and empty pEGFP-C1 (to express GFP) vector were purified by miniprep kit (Macherey-Nagel) and were independently transfected into 293T cells in 10 mm culture dishes using Lipofectamine 2000 reagent (Invitrogen) following manufacturer’s instructions. 48 hs post transfection cells were washed twice with PBS, harvested and centrifuged at 1500 rpm for 5 min. The cellular pellets were individually re-suspended in 1 ml lysis buffer (20 mM Tris pH8, 120 mM NaCl, 1% Triton X-100, 0.05% SDS, 10% glycerol) supplemented with protease inhibitors (Complete Mini EDTA Free, Roche). Cell extracts were clarified by centrifugation at 13.000 rpm for 10 min. Recombinant purified c-terminal His tagged ZikV-NS5 protein (Ferrero et al., 2019) (5 μg) was added to each supernatant tube and incubated for 4 hs at 4°C. 10 μl of His-trap TALON resin (Takara) was added to each tube and incubated for additional 2 hs. Resin was pull-down by centrifugation at 1500 rpm at 4°C during 5 min and washed twice with lysis buffer and 1 mM imidazole. Proteins were finally eluted washing TALON resin with 500 mM imidazole in PBS. Comparative pull-down assay between WT ZikV-NS5 and MultimDead Y25A/K28S/K29A mutant was performed identically using 2.5 μg of each protein.
For Pull-down assays of endogenous proteins, hNPC were cultured in 100 mm dishes, harvested, centrifuged and cell pellet stored in dry ice. The cellular pellets from 2 dishes were re-suspended in 600 μl lysis buffer (20 mM Tris pH8, 120 mM NaCl, 1% Triton X-100, 0.05% SDS, 10% glycerol) supplemented with protease inhibitors (Complete Mini EDTA Free, Roche). Cell extracts were clarified by centrifugation at 13.000 rpm for 10 min. Supernatant was recovered and equally divided in two Eppendorf. Recombinant purified c-terminal His tagged ZikV-NS5 protein (Ferrero et al., 2019) (5 μg) was added only to one supernatant tube and incubated for 4 hs at 4°C. After that, 10 μl of His-trap TALON resin (Takara) was added to each tube and incubated for additional 2 hs. Resin was pull-down by centrifugation at 1500 rpm at 4°C during 5 min and washed twice with lysis buffer and 1 mM imidazole. Proteins were finally eluted washing TALON resin with 500 mM imidazole in PBS.
Western Blot
Proteins from pull down eluted samples were separated in a 10% SDS-PAGE and transferred onto nitrocellulose membranes (Amersham) by wet electroblotting (50V, 150 mA, ON) in 25 mM Tris-HCl pH 8.3, 192 mM glycine, 20% (v:v) methanol buffer. Membranes were blocked with 5% non-fat milk in PBST (0.5% Tween 20 in PBS) during 2 hours at RT, followed by incubation with primary antibody anti-GFP, (Saade et al., 2017) or anti-His (Qiagen) during 1 hours at RT in blocking buffer, two washes with PBST and a final incubation with secondary antibody anti-Mouse IgG-HRP, diluted 1:2000 (Thermo Fisher Scientific). Immunoreactive signals were detected with ECL kit (BIO-RAD) and visualized by Odyssey Fc imaging system (LI-COR). Western Blot quantifications were performed using Image J/ Fiji softwares.
Western Blot from pull down of endogenous hNPC proteins, were performed identically. The primary antibodies diluted in blocking buffer were anti-Cep164 (1:500, provided by Dr. Cyaran Morisson-NUI), anti-Rootletin (1:1000,Abcam) and anti-Bart (1:2000, provided by Richard Khan-EMORY). Secondary antibody was replaced for anti-Rabbit-HRP antibody (1:2000) for Rootletin and BART detection.
Immunohistochemistry and microscopy
Human tissues were fixed in 4% PFA or Bouins solution overnight, paraffin embedded and sectioned following standard procedures. For markers of cortical layers and ZikV-NS5 stainings, heat-induced antigen retrieval was performed in 10 mM sodium citrate buffer (pH 6) at 95 °C for 20 min after deparafination, as described before (Retallack et al., 2016). Chick embryos were fixed in 4% PFA for 2 h at 4 °C, and immunostaining was performed on vibratome sections (40 μm) following standard procedures. For centrosome staining, Chick embryos were fixed in pre-chilled (−20 °C) 100% methanol overnight at −20 °C. For en-face imaging, the neural tube of (HH16-18) chick embryos was open sagittally (dorsoventrally) at the lumen along the ventricle. Notochord and somites were discarded to allow a better whole mounting of the neural tube. After washing in PBS-0.1% Triton, the sections were incubated overnight at 4C with the appropriate primary antibodies diluted in a solution of PBS-0.1% Triton supplemented with 10% bovine serum albumin or sheep serum. After washing in PBS-0.1% Triton, sections were incubated for 2 hr at room temperature with the appropriate secondary antibodies diluted in a solution of PBS-0.1% Triton supplemented with 10% bovine serum albumin or sheep serum. Alexa488-, Alexa555- and Alexa633- antibodies were obtained from Thermo Fisher Scientific. Sections were finally stained with 1 μg/ml DAPI (Sigma) and mounted in Mowiol (Sigma). Images were acquired at room temperature on a Zeiss LSM780 confocal microscope system using 25× (Plan-Apochromat 25x/0.8 Imm Korr DIC), 40× (Plan-Apochromat 40x/1.3 Oil DIC M27) or 63× (Plan-Apochromat 63x/1.4 Oil DIC M27) lenses. Human sections were acquired using the Thunder Imager 3D Live Cell from Leica microsystems, equipped with the objectives Plan Fluotar 10x/0.32, Plan Fluotar 25x/0.8 MultiImmersion and Plan-Apochromat 100x/1.4 Oil. Human forebrain coronal sections, cortical and ependymal layers were built with mosaic merge using the Thunder Imager. Tiled images were digitally stitched by Leica LAS X software (Leica) to generate full scan images shown. For image processing and data analysis, we used the ImageJ /FIJI. Three-dimensional (3D) reconstruction of whole neuroepithelial cells from an en-face view were built from z-stacks using the Volocity v.6.2 software (PerkinElmer). This software was also used to 3D reconstruct NP mitotic cells and the multi-centrosome/cilia of human ependymal cells.
Flow cytometry
Chicken embryos were recovered 24 hr after co-electroporation of the pSox2:eGFP and pTis21:RFP reporters together with the indicated encoding plasmids. Cell suspensions were obtained from pools of 6–8 dissected neural tubes after digestion with Trypsin-EDTA (Sigma) for 10–15 min, and further processed on a FACS Aria III cell sorter (BD Biosciences) for measurement of eGFP and RFP fluorescences. At least 5000 cells for each progenitor population (PP, PN and NN) were analyzed per sample.
Luciferase reporter assay
The activity of pTis21, pNeuroD, Tubb3enh, and pSox2 were assessed in the neural tube using the corresponding Luciferase vectors. Reporters were co-electroporated with a Renilla luciferase reporter construct carrying the cytomegalovirus immediate early enhancer promoter for normalization (Promega). NTs obtained at 24 hpe were processed following the Dual Luciferase Reporter Assay protocol (Promega), as described previously (Saade et al., 2013). The data are presented as the mean ± s.e.m. from 6-8 embryos per experimental condition (biological replicates).
Quantification of ciliary length
In NPCs, primary cilia length was assessed from Arl13b and polyglutamylated tubulin signals. Pictures were taken at the confocal microscope and the maximum intensity projection in the z-axis (0.5 μm spaced optical sections) was used for the measurement of cilia length and assessment of morphology using ImageJ /Fiji software. Arl13b signal was used to measure the length of the cilia in μm from the cilia tip to the basal body as stained with FOP antibody (provided by Dr. Olivier Rosnet, IBDM). Since microtubules constitute the core structure of primary cilia, the fluorescence average pixel intensity of tubulin polyglutamylation (Enzo Life Sciences) has been measured. To measure the length of multicilia in human forebrain ependymal cells, we used the 3D Live Cell Thunder Imager that allowed for fast and precise multidimensional image acquisition of a large (~ 2.5 mm) ependymal layer that faces the ganglionic eminence in coronal sections; a Plan-Apochromat 100x/1.4 Oil lens was used to avoid resolution loss. The whole area of interest was acquired as a mosaic of Z-stacks (0.5 μm spacing). The image data set was first Z-projected, using as criterion the maximum intensity of acetylated-tubulin signal (Sigma), and then merged to a single mosaic image. The length and positioning of the multiciliated patches was measured along the ependymal cell layer using the following macro (https://github.com/MolecularImagingPlatformIBMB/MeasureMultiCilia).
Measurement of fluorescence intensities and area
All measurements of fluorescent intensities were carried out using image j/ FIJI softwares. The fluorescence average pixel intensity of CEP164 and Rootletin was measured at the centrosome level (FOP+) in electroporated NPCs showing a cilia base localization of ZikV-NS5 and ciliopathy (Arl13b+). For controls, the fluorescence average pixel intensity of centrosomal CEP164 and Rootletin was measured in the surrounding electroporated NPCs where ZikV-NS5 is not localized in the cilia base and where the cilia structure is normal. The fluorescence average pixel intensity of BART was measured at the centrosome level in en-face view of NPCs showing a ZikV-NS5 centrosomal localization. The control was defined by measuring the fluorescence average pixel intensity of centrosomal BART in the apical area of the non-transfected close neighbour cells. BART distribution in the apical cell border and Rootletin linking the rootlet fiber were excluded from these quantifications. Centrosomal proteins mean intensity were measured in the sum projection of the 6 z-stacks encompassing CEP164, Rootletin and BART stainings (0.5 μm spaced optical sections). In human ependymal cells, the fluorescence average pixel intensity of CEP164 and Pericentrin (Abcam, ) was measured and presented as a ratio. N-cadherin staining was used to define the size of apical area. The apical area ratio was obtained by dividing the apical area of an electroporated NPC by the mean apical area of four of its non-electroporated neighbour cells (spaced by one cell row from the electroporated cell). To characterize the differentiation state of NPCs according to the subcellular localization of the ZikV-NS5, the level of electroporation was first defined by measuring the mean intensity of ZikV-NS5-flag or the control vector H2B-GFP in the nucleus. NPCs with the same level of electroporation (same nuclear mean intensity of FLAG or GFP staining) were considered for pTis21-RFP quantification. In human forebrain, the average number of cells positive for Tbr1 (Abcam), Satb2 (Abcam, ), ROR (R and D Systems, ) and Ctip2 (Abcam,) per field and along the apicobasal axis was counted using image j/ FIJI softwares ImageJ.
Apical foot length measurement
For cell 3D reconstruction from en-face neural tube, electroporated NPCs showing apical end-foot restriction (ratio of apical area < 0.5μm) have been acquired with a z-stack encompassing ~ 70μm for the control dsRedex staining from the most apical end-foot to the nucleus (0.5μm spaced optical sections). ZikV-NS5+ NPCs have been acquired with a z-stack encompassing ZikV-NS5 distribution from the most apical end-foot to the nucleus with a ranging from 6 to 36 μm. 3D reconstructions were built from z-stacks using the Volocity v.6.2 software (PerkinElmer) and image J/Fiji softwares. The reconstruction and measurement of the apical foot length was performed using the Simple Neural Tracer framework from image J (Frangi, 1998; Longair et al., 2011; Pool et al., 2008). Briefly, a semi-automatic tracing was performed from the end-foot to the nucleus through 3D image stacks taking into consideration the tube-like structure of the apical foot. Details of the traces were exported as CSV and SWC files for a 3D Python plotting (7.04) using the Matplotlib library (Python).
GO Analysis
The ZikV-NS5 target proteins were in silico analysed to identify protein-protein interactions experimentally reported among them using the Agile Protein Interactomes DataServer (APID) (Alonso-Lopez et al., 2016). Moreover, in order to identify the main cell compartments where ZikV-NS5 belongs, a Gene Ontology (GO) analysis was performed using the Protein ANalysis THrough Evolutionary Relationships (PANTHER) classification system version 14.0 (Mi et al., 2013).
Sequence alignments
Human Cep164, Rootletin and BART protein sequences were obtained from Sanger sequenced constructs that are identical to Genebank NP_055771.4, NP_055490.4, AAH03087.1 sequences, respectively. Chicken (Gallus gallus) CEP164, Rootletin and BART-1 protein sequences were obtained from NCBI (Genebank accesion numbers: XP_024999041.1; XP_015152501.1; NP_001007971.1, respectively). Protein sequences alignments were performed using Clustal Omega program (Madeira et al., 2019) and colored using ESPript 3.0 program (Robert and Gouet, 2014). For CEP164 and Rootletin, only ZikV-NS5 interacting domain sequences identified by Y2H assay were employed for sequence alignment.
ZikV-NS5 Structure Cartoons
ZikV-NS5 structure cartoons were generated by Chimera software (UCSF Chimera), a visualization system for exploratory research and analysis (Pettersen et al., 2004) using the crystal structure coordinates deposited at PDB (pdb code: 6I7P) and specifically colored to enhance protein features like domains or residues.
Quantification and Statistical Analysis
No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments or outcome assessment. Statistical analyses were performed using the GraphPad Prism 8.0.2 software (GraphPad Software, Inc.). For apical foot length measurement, Kernel density plots have been applied to represent the data set distribution corresponding to the ratio of the apical area versus the apical foot length of electroporated NPCs
Quantitative data are expressed as mean ± sem or mean ± stdev except for violin plots that present median with quartile range. The n values are indicated in figures or the corresponding legends. The normal distribution of the values was assessed by the Shapiro-Wilk normality test. Significance was then assessed with a two-sided unpaired t-test, one-way ANOVA + Tukey’s test or two-way ANOVA + Sidak’s or +Dunnett’s test for data presenting a normal distribution, or alternatively with non-parametric Mann–Whitney or Kruskal-Wallis +Dunn’s multiple comparisons’ tests for non-normally distributed data. n.s: non-significant; *: p<0.05 or less, as indicated in individual figures.
Supplementary Material
Video 1. (Related to Figure 2)
Premature basal migration of the centrosome in ZikV-NS5 electroporated NPCs
3D reconstruction of a ZikV-NS5-Flag (shown in Green) electroporated NPC showing a premature basal migration of the Fop+ centrosome (shown in Red). The Apical area of the electroporated NPC is delimitated with N-Cadherin (shown in Magenta)
Video 2. (Related to Figure 2)
Premature apical end-foot restriction in ZikV-NS5 electroporated NPCs
3D reconstruction of a ZikV-NS5-Flag (shown in Green) electroporated NPCs showing a short apical foot with a restricted apical-end foot area (Yellow Arrow) delimitated with N-Cadherin (shown in Magenta, asterisk points the nucleus)
Table S1. (Related to Figure 1)
ZikV-NS5-host interactome determined by Yeast two hybrid and subdivided based on their subcellular localization according to the Gene Ontology (GO) analysis
Italic letters correspond to genes.
Table S2. (Related to Figure 1)
Comparative analysis of the ZikV-NS5 yeast two hybrid data with two published independent interactome datasets
Italic letters correspond to genes and bold letters correspond to common hits.
Table S3. (Related to Star Methods)
List of oligonucleotides for the cloning, truncation and mutation of the corresponding DNA constructs
Video 3. (Related to Figure 2)
Premature apical end-foot restriction in ZikV-NS5 electroporated NPCs
3D reconstruction of a ZikV-NS5-Flag (shown in Green) electroporated two neighbour NPCs showing a short apical foot with a restricted apical-end foot area (Yellow and blue Arrows) delimitated with N-Cadherin (shown in Magenta, Asterisks with the same colour code than the arrows point the corresponding nucleus)
Video 4. (Related to Figure 2)
Apical end-foot restriction in control NPCs
3D reconstruction of a DsRedEx (shown in Green) electroporatde NPCs showing a long apical foot with a restricted apical end-foot area (Yellow arrow) delimitated with N-Cadherin (shown in Magenta, asterisk points the nucleus).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BART | Dr. Richard Khan (EMORY) | N/A |
| CEP164 | Dr. Cyaran Morisson (NUI) | N/A |
| FOP | Dr. Olivier Rosnet (IBDM) | N/A |
| Rootletin | Abcam | Cat#ab121653, RRID:AB_11129547 |
| Pericentrin | Abcam | Cat#ab4448, RRID:AB_304461 |
| Flag | Our Lab; Saade et al., 2017 | N/A |
| GFP | Our Lab; Saade et al., 2017 | N/A |
| Penta.HIS | Qiagen | Cat#34660, RRID:AB_2619735 |
| Myc | ProSci | Cat# 51-118, RRID:AB_1947601 |
| Anti-Rabbit IgG-Peroxidase | Sigma | Cat# A0545, RRID:AB_257896 |
| Anti-Mouse IgG-Peroxidase | Thermo Fisher Scientific | Cat# A16011, RRID:AB_2534685 |
| RFP | Our Lab; Saade et al., 2017 | N/A |
| Acetylated tubulin | Sigma | Cat# T6793, RRID:AB_477585 |
| Polyglutamylated-tubulin | Enzo Life Sciences | Cat# AG-20B-0020, RRID:AB_2335608 |
| N-Cadherin | Thermo Fisher Scientific | Cat# 13-2100, RRID:AB_2533007 |
| Lamin B1 | Thermo Fisher Scientific | Cat# 33-2000, RRID:AB_2533106 |
| PH3 | Millipore | Cat# Ser-10, RRID:AB_2315135 |
| ROR | R and D Systems | Cat# PP-H3925-00, RRID:AB_2254092 |
| Ctip2 | Abcam | Cat# ab18465, RRID:AB_2064130 |
| Satb2 | Abcam | Cat# ab51502, RRID:AB_882455 |
| Tbr1 | Abcam | Cat# ab183032 |
| Caspase 3 | BD Biosciences | Cat# 550821, RRID:AB_393906 |
| HUC/D | Molecular Probes | Cat# A-21271, RRID:AB_221448 |
| NS5 | Novus Biologicals | Cat# NBP2-42900 |
| S100β | Agilent | Cat# Z0311, RRID:AB_10013383 |
| Alexa Fluor 488 donkey anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat #: A-21206; RRID: AB_2535792 |
| Alexa Fluor 488 donkey anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat #: A-21202; RRID: AB_141607 |
| Alexa Fluor 555 donkey anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat #: A-31572; RRID: AB_162543 |
| Alexa Fluor 555 donkey anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat #: A-31570 RRID:AB_2536180 |
| Alexa Fluor 633 donkey anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat #: A-21050 RRID:AB_141431 |
| Alexa Fluor 633 donkey anti-rat IgG (H+L) | Thermo Fisher Scientific | Cat #: A-21094 RRID:AB_141553 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| C-terminal His tagged ZikV-NS5 protein | Our Lab; Ferrero et al., 2019 | N/A |
| Dulbecco’s Modification of Eagle’s Medium (DMEM) | Sigma-Aldrich | Cat# D5796 |
| Fetal Bovine Serum (FBS) | Sigma-Aldrich | Cat# 12103C |
| Non-essential Amino acids | Sigma-Aldrich | Cat# M7145 |
| penicilin and streptomicin | Sigma-Aldrich | Cat# P4333 |
| Amphotericin B | GIBCO | Cat# 15290-18 |
| Lipofectamine 2000 reagent | Invitrogen | Cat# 11668019 |
| Phusion DNA polymerase | Thermo Fisher Scientific | Cat# F530S |
| Complete™ULTRA Tablets, Mini, EDTA-free, EASYpack Protease Inhibitor Cocktail | Roche | Cat# 05892791001 |
| His-trap TALON resin | Takara | Cat# 635503 |
| ECL kit | BIO-RAD | Cat# 170-5061 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Mowiol | Sigma-Aldrich | Cat# 81381 |
| Trypsin-EDTA | Sigma-Aldrich | Cat# T3924 |
| Accutase | ThermoFisher | Cat# 1110501 |
| Lipofectamine Stem | ThermoFisher | Cat# STEM00001 |
| Biological Samples | ||
| Postmortem GW21 human brain sections | Neuropathology laboratory, Pathology Department, H. U. Vall d’Hebron-PR(AG)129/201, Barcelona, Spain. | N/A |
| Postmortem GW23 human brain sections | Neuropathology laboratory, Pathology Department, H. U. Vall d’Hebron-PR(AG)129/201, Barcelona, Spain. | N/A |
| ZIKV Infected Postmortem GW22 human brain sections | Neuropathology laboratory, Pathology Department, H. U. Vall d’Hebron-PR(AG)129/201, Barcelona, Spain. | N/A |
| Critical Commercial Assays | ||
| GeneArt™ Site-Directed Mutagenesis PLUS System | Agilent | Cat# A14604 |
| Dual Luciferase Reporter Assay System | Promega | Cat# E1910 |
| NucleoSpin Plasmid, Mini kit for plasmid DNA | Macherey-Nagel | Cat# 740588.50 |
| Deposited Data | ||
| ZikV-NS5 structure | Ferrero et al., 2019 | N/A |
| Experimental Models: Cell Lines | ||
| Human cell line 293T (or HEK 293T) | ATCC | Cat# CRL-3216, RRID:CVCL_0063 |
| H9 Human Embryonic Stem Cell ( WA09) | WiCell | RRID:CVCL_9773 |
| Experimental Models: Organisms/Strains | ||
| White Leghor chick embryos (Stages 12 to 24 somites) | Granja Gibert | N/A |
| ZikV Suriname strain, human isolate sequence Z1106033 | Enfissi et al., 2016 | GeneBank accession code: KU312312.1 |
| ZikV African strain MR766 | Grard et al., 2010 | GeneBank accession code: DQ859059 |
| ZikV Asian strain H/PF/2013 | Baronti et al., 2014 | GenBank accession code: KJ776791 |
| Oligonucleotides | ||
| Primers for pCIEGO-Flag, See Table S3 | Our Lab; Saade et al., 2017 | N/A |
| Primers for pCIEGO- ZikV Structural genes (C, Pre-M, E) from Suriname strain, See Table S3 | This paper | N/A |
| Primers for pCIEGO- ZikV Non Structural (NS1-NS5) genes from Suriname strain, See Table S3 | This paper | N/A |
| Primers for pCIEGO- ZikV-NS5 truncations and mutations, See Table S3 | This paper | N/A |
| Primers for pCIEGO- ZikV-NS5 African strain, See Table S3 | This paper | N/A |
| Primers for pCIEGO- ZikV-NS5 Asian strain, See Table S3 | This paper | N/A |
| Primers for pEGFP Rootletin (1-1462), See Table S3 | This paper | N/A |
| Primers for pEGFP-BART1, See Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pCAGGS without florescent marker (pCIEGO) | Our Lab; Saade et al., 2017 | N/A |
| pCAGGS_ires_H2B: GFP(pCIG) | Our Lab; Saade et al., 2017 | N/A |
| pCS2_H2B-GFP | Our Lab; Saade et al., 2017 | N/A |
| pEGFP-C1 | Clontech | Cat #6084-1 |
| HumanCep164cDNA | Dr. Eric Nigg (Biozentrum), | RRID:Addgene_41149 |
| Full length Rootletin in pEGFP | Addgene | RRID:Addgene_41166 |
| BART-1 cDNA | Dr. Michael Cheetham (UCL) | |
| pEGFP-C1 Emerin | Addgene | RRID:Addgene_61993 |
| DsRedex | Dr. Roger Tsien (UCSD), | N/A |
| CEP152-GFP | OriGene | CAT#: RG211581 |
| Arl13b-RFP, Arl13b-GFP | Dr. Magdalena Götz (LMU) | N/A |
| pSox2:GFP | Our Lab; Saade et al., 2013 | N/A |
| pTis21:RFP | Our Lab; Saade et al., 2013 | N/A |
| pSox2:luc | Our Lab; Saade et al., 2017 | N/A |
| pTis21:luc | Our Lab; Saade et al., 2017 | N/A |
| pNeuroD:luc | Dr. François Guillemot (The Francis Crick Institute) | N/A |
| pTubb3:luc | Our Lab; Le Dreau et al., 2018 | N/A |
| Software and Algorithms | ||
| Fiji/Image J (2.0) | ImageJ; Schneider et al., 2012 | https://imagej.nih.gov/ij/ RRID:SCR_003070 |
| Agile Protein Interactomes DataServer (APID) | Alonso-Lopez et al., 2016 | http://apid.dep.usal.es RRID:SCR_008871 |
| (PANTHER) classification system version 14.0 | Mi et al., 2013 | http://pantherdb.org RRID:SCR_004869 |
| Clustal Omega | https://www.ebi.ac.uk/Tools/msa/clustalo/ RRID:SCR_001591 | |
| ESPript 3.0. Structure protein | http://espript.ibcp.fr/ESPript/ESPript/ RRID:SCR_006587 | |
| Chimera software | UCSF Chimera; Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ RRID:Addgene_15601 |
| BD FACSDiva™ Software v. 6.1.3 | BD Biosciences | RRID:SCR_001456 |
| Flowjo (10.2) software | FlowJo | RRID:SCR_008520 |
| Volocity v.6.2 software | Perkinelmer | RRID:SCR_002668 |
| LSM Software ZEN 2.1 | Zeiss | RRID:SCR_013672 |
| Leica LAS X software -Thunder imaging system software | Leica Microsystems | RRID:SCR_013673 |
| GenBank public database | NCBI | https://www.ncbi.nlm.nih.gov/genbank/ RRID:SCR_002760 |
| Ensembl public database | Ensembl | https://www.ensembl.org/info/data/index.html RRID:SCR_002344 |
| GraphPad Prism (8.02) | GraphPad | http://cicblade.dep.usal.es:8080/APID/init.action RRID:SCR_002798 |
| Python (7.04) | Python | https://wingware.com/news/2019-07-11 RRID:SCR_008394 |
| Macro for measuring Multicilia length in Human Ependymal cells | This paper; Github | https://github.com/MolecularImagingPlatformIBMB/MeasureMultiCilia |
| Other | ||
HIGHLIGHTS.
ZikV-NS5 interacts with and depletes centrosome proteins at the primary cilia in NPCs
ZikV-NS5 at the centrosome causes ciliopathy and promotes neuron delamination
Multimeric arrangement of ZikV-NS5 is requiered for nuclear exit and for ciliopathy
In human fetal brain, ZikV affects motile cilia integrity in Ependymal cells
ACKNOWLEDGEMENTS
We thankDr Elena Rebollo and Jaume Boix for their technical assistance at the AFMU Facility (IBMB); Leica Microsystems for supporting and collaborating with the AFMU Facility. We are grateful to researchers that kindly provided DNAs and antibodies. The work in EM’s laboratory was supported by grants BFU2016-77498-P, BFU2016-77498-P and La Maratò de TV3 foundation 201833-10. The work in NV’s laboratory was supported by grants BIO2017-83906-P, Maria de Maeztu Unit of Excellence MDM-2014-0435 (MCIU) and La Maratò de TV3 foundation 201833-10. The work in NA laboratory was supported by NIH/NINDS R00NS089859 and IDDRC-NPDA (CHOP/Penn).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abbott A (2011). Tissue-bank shortage: Brain child. Nature 478, 442–443. [DOI] [PubMed] [Google Scholar]
- Adams Waldorf KM, Nelson BR, Stencel-Baerenwald JE, Studholme C, Kapur RP, Armistead B, Walker CL, Merillat S, Vornhagen J, Tisoncik-Go J, et al. (2018). Congenital Zika virus infection as a silent pathology with loss of neurogenic output in the fetal brain. Nat Med 24, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N, Cantagrel V, Schroth J, Cai N, Vaux K, McCloskey D, Naviaux RK, Van Vleet J, Fenstermaker AG, Silhavy JL, et al. (2013). AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 154, 505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Lopez D, Gutierrez MA, Lopes KP, Prieto C, Santamaria R, and De Las Rivas J (2016). APID interactomes: providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res 44, W529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek C, Freem L, Goiame R, Sang H, Morin X, and Tozer S (2018). Mib1 prevents Notch Cis-inhibition to defer differentiation and preserve neuroepithelial integrity during neural delamination. PLoS Biol 16, e2004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Chien C-T, Sternglanz R, Fields S (1993). Using the twohybrid system to detect protein-protein interactions Cellular Interactions in Development: A Practical Approach (Hartley DA, ed), Oxford University Press, Oxford, 153–179. [Google Scholar]
- Beranger F, Aresta S, de Gunzburg J, and Camonis J (1997). Getting more from the two-hybrid system: N-terminal fusions to LexA are efficient and sensitive baits for two-hybrid studies. Nucleic Acids Res 25, 2035–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, and Muhr J (2011). Sequentially acting Sox transcription factors in neural lineage development. Genes Dev 25, 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, and Anderson KV (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 12, 767–778. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, et al. (2006). Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell 11, 831–844. [DOI] [PubMed] [Google Scholar]
- Coyaud E, Ranadheera C, Cheng D, Goncalves J, Dyakov BJA, Laurent EMN, St-Germain J, Pelletier L, Gingras AC, Brumell JH, et al. (2018). Global Interactomics Uncovers Extensive Organellar Targeting by Zika Virus. Mol Cell Proteomics 17, 2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, and Rana TM (2016). Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell 19, 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AE, Schwarz N, Zelinger L, Stern-Schneider G, Shoemark A, Spitzbarth B, Gross M, Laxer U, Sosna J, Sergouniotis PI, et al. (2013). Mutations in ARL2BP, encoding ADP-ribosylation-factor-like 2 binding protein, cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet 93, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Song H, Wang H, Chai Y, Su C, Qi J, Shi Y, and Gao GF (2017). The crystal structure of Zika virus NS5 reveals conserved drug targets. Embo j 36, 919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, and Glover DM (2010). Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718. [DOI] [PubMed] [Google Scholar]
- Ferraris P, Cochet M, Hamel R, Gladwyn-Ng I, Alfano C, Diop F, Garcia D, Talignani L, Montero-Menei CN, Nougairede A, et al. (2019). Zika virus differentially infects human neural progenitor cells according to their state of differentiation and dysregulates neurogenesis through the Notch pathway. Emerg Microbes Infect 8, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DS, Ruiz-Arroyo VM, Soler N, Uson I, Guarne A, and Verdaguer N (2019). Supramolecular arrangement of the full-length Zika virus NS5. PLoS Pathog 15, e1007656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. (2005). Protein interaction mapping: a Drosophila case study. Genome Res 15, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangi AF, Niessen WJ, Vincken KL , and Viergever MA (1998). Multiscale vessel enhancement filtering In Wells WM, Colchester A, and Delp S, editors, Medical Image Computing and Computer-Assisted Intervention-MICCAI’98 1496 of Lecture Notes in Computer Science, 130–137. [Google Scholar]
- Fromont-Racine M, Rain JC, and Legrain P (1997). Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16, 277–282 [DOI] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, et al. (2017). Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 20, 397–406.e395. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, and Rehen SK (2016). Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818 [DOI] [PubMed] [Google Scholar]
- Goetz SC, and Anderson KV (2010). The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, et al. (2016a). Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 19, 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, et al. (2016b). Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 19, 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, and Nigg EA (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V.a.H., L. H (1951). A series of normal stages in the development of the chick embryo. J Morphol 88, 49–92 [PubMed] [Google Scholar]
- Hatch E, and Hetzer M (2014). Breaching the nuclear envelope in development and disease. J Cell Biol 205, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Guo J, Yokota Y, Umberger NL, Su CY, Li J, Verma N, Hirt J, Ghukasyan V, Caspary T, et al. (2013). Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat Neurosci 16, 1000–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, and Huttner WB (1999). Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc Natl Acad Sci U S A 96, 4639–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasioulis I, Das RM, and Storey KG (2017). Inter-dependent apical microtubule and actin dynamics orchestrate centrosome retention and neuronal delamination. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, and Dynlacht BD (2013). Assembling a primary cilium. Curr Opin Cell Biol 25, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dreau G, Saade M, Gutierrez-Vallejo I, and Marti E (2014). The strength of SMAD1/5 activity determines the mode of stem cell division in the developing spinal cord. J Cell Biol 204, 591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, Terskikh AV, Shresta S, and Gleeson JG (2016). Zika Virus Infects Neural Progenitors in the Adult Mouse Brain and Alters Proliferation. Cell Stem Cell 19, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Saucedo-Cuevas L, Yuan L, Ross D, Johansen A, Sands D, Stanley V, Guemez-Gamboa A, Gregor A, Evans T, et al. (2019). Zika Virus Protease Cleavage of Host Protein Septin-2 Mediates Mitotic Defects in Neural Progenitors. Neuron 101, 1089–1098.e1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, et al. (2016). Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 19, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, and Armstrong JD (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahuzier A, Shihavuddin A, Fournier C, Lansade P, Faucourt M, Menezes N, Meunier A, Garfa-Traore M, Carlier MF, Voituriez R, et al. (2018). Ependymal cilia beating induces an actin network to protect centrioles against shear stress. Nat Commun 9, 2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, and Thomas PD (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41, D377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, and Kriegstein AR (2016). Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell 18, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, et al. (2016). Zika Virus Disrupts Phospho-TBK1 Localization and Mitosis in Human Neuroepithelial Stem Cells and Radial Glia. Cell Rep 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Wilsch-Brauninger M, and Huttner WB (2013). Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell 155, 333–344. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pierson TC, and Kielian M (2013). Flaviviruses: braking the entering. Curr Opin Virol 3, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool M, Thiemann J, Bar-Or A, and Fournier AE (2008). NeuriteTracer: a novel ImageJ plugin for automated quantification of neurite outgrowth. J Neurosci Methods 168, 134–139. [DOI] [PubMed] [Google Scholar]
- Potter C, Zhu W, Razafsky D, Ruzycki P, Kolesnikov AV, Doggett T, Kefalov VJ, Betleja E, Mahjoub MR, and Hodzic D (2017). Multiple Isoforms of Nesprin1 Are Integral Components of Ciliary Rootlets. Curr Biol 27, 2014–2022.e2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. (2016). Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, et al. (2001). The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215. [DOI] [PubMed] [Google Scholar]
- Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, et al. (2016). Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci U S A 113, 14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saade M, Blanco-Ameijeiras J, Gonzalez-Gobartt E, and Marti E (2018). A centrosomal view of CNS growth. Development 145. [DOI] [PubMed] [Google Scholar]
- Saade M, Gonzalez-Gobartt E, Escalona R, Usieto S, and Marti E (2017). Shh-mediated centrosomal recruitment of PKA promotes symmetric proliferative neuroepithelial cell division. Nat Cell Biol 19, 493–503. [DOI] [PubMed] [Google Scholar]
- Saade M, Gutierrez-Vallejo I, Le Dreau G, Rabadan MA, Miguez DG, Buceta J, and Marti E (2013). Sonic hedgehog signaling switches the mode of division in the developing nervous system. Cell Rep 4, 492–503. [DOI] [PubMed] [Google Scholar]
- Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Plaszczyca A, Bartenschlager R, Gotz M, and Pichlmair A (2018). An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561, 253–257. [DOI] [PubMed] [Google Scholar]
- Siller SS, Sharma H, Li S, Yang J, Zhang Y, Holtzman MJ, Winuthayanon W, Colognato H, Holdener BC, Li FQ, et al. (2017). Conditional knockout mice for the distal appendage protein CEP164 reveal its essential roles in airway multiciliated cell differentiation. PLoS Genet 13, e1007128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Merkle FT, Flames N, Tramontin AD, Garcia-Verdugo JM, and Alvarez-Buylla A (2005). Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci 25, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiekema M, van Zandvoort M, Ramaekers FCS, and Broers JLV (2020). Structural and Mechanical Aberrations of the Nuclear Lamina in Disease. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. (2016). Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 18, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Rosti RO, Gibbs E, and Gleeson JG (2014). Primary cilia in neurodevelopmental disorders. Nat Rev Neurol 10, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen JA, and Vartiainen MK (2017). Diverse functions for different forms of nuclear actin. Curr Opin Cell Biol 46, 33–38. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, and Hollenberg SM (1995). Ras-Raf interaction: two-hybrid analysis. Methods Enzymol 255, 331–342. [DOI] [PubMed] [Google Scholar]
- Wang B, Tan XF, Thurmond S, Zhang ZM, Lin A, Hai R, and Song J (2017). The structure of Zika virus NS5 reveals a conserved domain conformation. Nat Commun 8, 14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, and Shi SH (2009). Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsch-Brauninger M, Peters J, Paridaen JT, and Huttner WB (2012). Basolateral rather than apical primary cilia on neuroepithelial cells committed to delamination. Development 139, 95–105. [DOI] [PubMed] [Google Scholar]
- Wilson KL (2000). The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol 10, 125–129. [DOI] [PubMed] [Google Scholar]
- Wojcik J, Boneca IG, and Legrain P (2002). Prediction, assessment and validation of protein interaction maps in bacteria. J Mol Biol 323, 763–770. [DOI] [PubMed] [Google Scholar]
- Yan X, Habedanck R, and Nigg EA (2006). A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol Biol Cell 17, 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Adamian M, and Li T (2006). Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell 17, 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu X, Yue G, Adamian M, Bulgakov O, and Li T (2002). Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol 159, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, et al. (2017). Zika-Virus-Encoded NS2A Disrupts Mammalian Cortical Neurogenesis by Degrading Adherens Junction Proteins. Cell Stem Cell 21, 349–358.e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Huang XY, Liu ZY, Zhang F, Zhu XL, Yu JY, Ji X, Xu YP, Li G, Li C, et al. (2017). A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 358, 933–936. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, and Li H (2007). Structure and function of flavivirus NS5 methyltransferase. J Virol 81, 3891–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1. (Related to Figure 2)
Premature basal migration of the centrosome in ZikV-NS5 electroporated NPCs
3D reconstruction of a ZikV-NS5-Flag (shown in Green) electroporated NPC showing a premature basal migration of the Fop+ centrosome (shown in Red). The Apical area of the electroporated NPC is delimitated with N-Cadherin (shown in Magenta)
Video 2. (Related to Figure 2)
Premature apical end-foot restriction in ZikV-NS5 electroporated NPCs
3D reconstruction of a ZikV-NS5-Flag (shown in Green) electroporated NPCs showing a short apical foot with a restricted apical-end foot area (Yellow Arrow) delimitated with N-Cadherin (shown in Magenta, asterisk points the nucleus)
Table S1. (Related to Figure 1)
ZikV-NS5-host interactome determined by Yeast two hybrid and subdivided based on their subcellular localization according to the Gene Ontology (GO) analysis
Italic letters correspond to genes.
Table S2. (Related to Figure 1)
Comparative analysis of the ZikV-NS5 yeast two hybrid data with two published independent interactome datasets
Italic letters correspond to genes and bold letters correspond to common hits.
Table S3. (Related to Star Methods)
List of oligonucleotides for the cloning, truncation and mutation of the corresponding DNA constructs
Video 3. (Related to Figure 2)
Premature apical end-foot restriction in ZikV-NS5 electroporated NPCs
3D reconstruction of a ZikV-NS5-Flag (shown in Green) electroporated two neighbour NPCs showing a short apical foot with a restricted apical-end foot area (Yellow and blue Arrows) delimitated with N-Cadherin (shown in Magenta, Asterisks with the same colour code than the arrows point the corresponding nucleus)
Video 4. (Related to Figure 2)
Apical end-foot restriction in control NPCs
3D reconstruction of a DsRedEx (shown in Green) electroporatde NPCs showing a long apical foot with a restricted apical end-foot area (Yellow arrow) delimitated with N-Cadherin (shown in Magenta, asterisk points the nucleus).
Data Availability Statement
The macro generated during this study to measure the length and positioning of the multiciliated patches along the ependymal cell layer in human brain is available at MolecularImagingPlatformIBMB’s Github (https://github.com/MolecularImagingPlatformIBMB/MeasureMultiCilia).


