Extended Data Figure 7. Genetic targeting of osmotic and hypovolemic thirst activated cell populations in the SFO and OVLT.
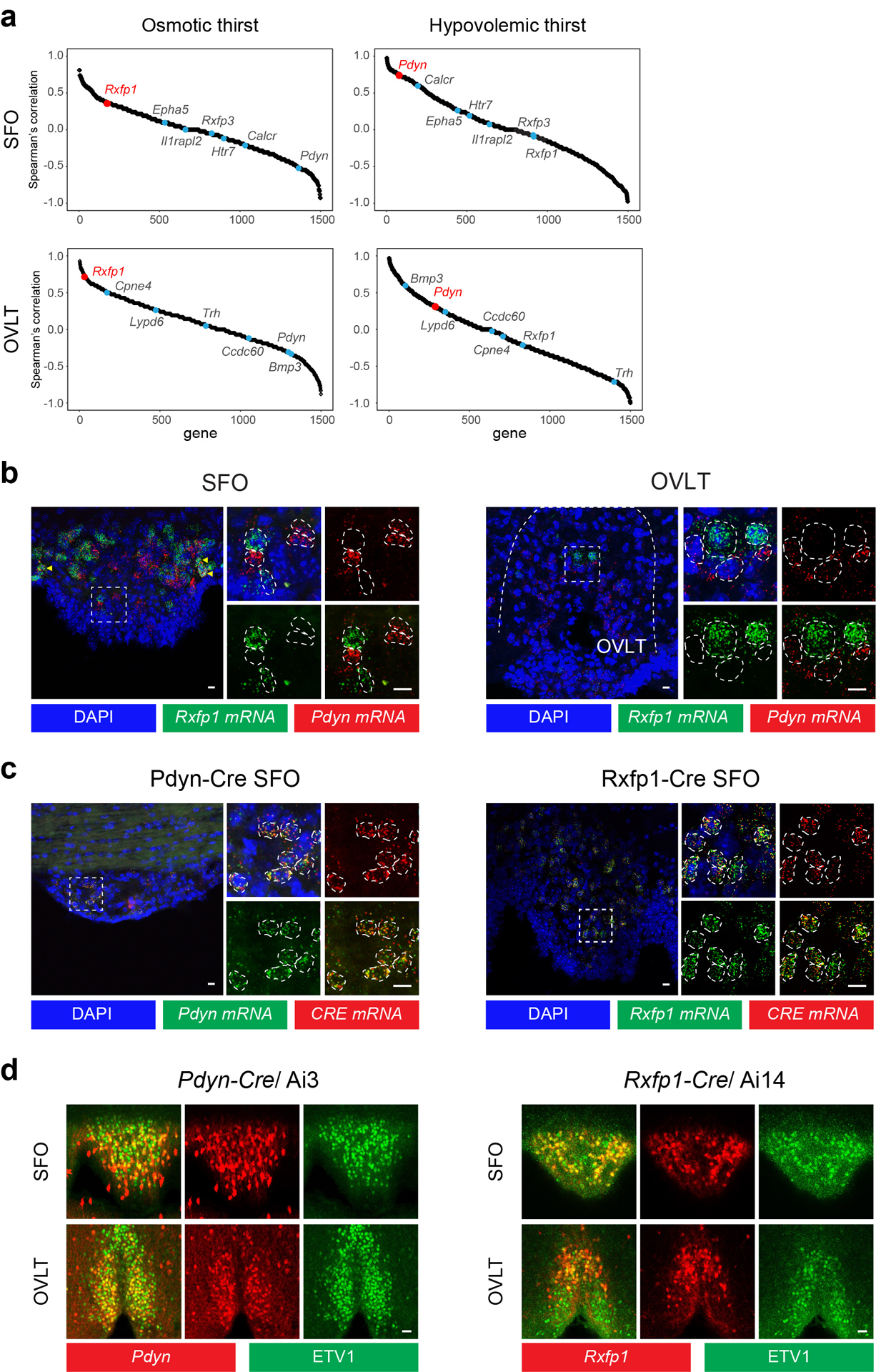
a, Spearman correlation between c-Fos expression under distinct thirst states and cell-type-specific and thirst-state-specific marker genes. Thirst-state-specific marker genes (Rxfp1 and Pdyn) show higher correlation with c-Fos expression compared to cell-type-specific genes. b, Two-color in situ hybridization of Pdyn and Rxfp1. These gene expression patterns are mostly distinct with minor overlap (arrowhead). Representative images from 8 and 2 slices from 2 independent experiments for SFO (left) and OVLT (right) respectively. Scale bar 10 μm. c, Validation of Cre expression in Pdyn-Cre and Rxfp1-Cre lines. 95.5% of Pdyn-Cre and 100% of Rxfp1-Cre expression matched endogenous gene expression. Representative images from 3 slices from 2 independent experiments for both Pdyn/Cre and Rxfp1/Cre stains. Scale bar 10 μm. d, Immunostaining of the SFO (top) and OVLT (bottom). Shown are Pdyn-positive neurons in Pdyn-Cre/Ai3 animals (representative images out of 8 slices from 4 mice for both SFO and OVLT, left) and Rxfp1-positive neurons in Rxfp1-Cre/Ai14 animals (representative images out of 6 slices from 3 mice for both SFO and OVLT, right). Pdyn- and Rxfp1-positive neurons (red) are a partial population of Etv1-positive excitatory neurons (green). Almost all (>90%) Pdyn- and Rxfp1-positive neurons expressed Etv1. Rxfp1 and Pdyn data are from Fig. 4d. Scale bar 10 μm.
