Abstract
Access to safe drinking water is a global problem, and methods to reliably and easily detect contaminants could be transformative. We report the development of a cell-free in vitro transcription system that uses RNA output sensors activated by ligand induction (ROSALIND) to detect contaminants in water. A combination of highly processive RNA polymerases, allosteric protein transcription factors and synthetic DNA transcription templates regulates the synthesis of a fluorescence-activating RNA aptamer. The presence of a target contaminant induces the transcription of the aptamer, and a fluorescent signal is produced. We apply ROSALIND to detect a range of water contaminants, including antibiotics, small molecules and metals. We also show that adding RNA circuitry can invert responses, reduce crosstalk and improve sensitivity without protein engineering. The ROSALIND system can be freeze-dried for easy storage and distribution, and we apply it in the field to test municipal water supplies, demonstrating its potential utility for monitoring water quality.
INTRODUCTION
Synthetic biology is creating new capabilities for engineered biological systems to address pressing global challenges through repurposing, evolving and rewiring the functions of biological components for a range of applications [1]. Among these functions, the ability of cells to use molecular biosensors and genetic networks to sense and respond to changing conditions is being harnessed to create new diagnostic technologies [2]. In particular, cell-free nucleic-acid diagnostics are being developed to detect pathogens and human health biomarkers in complex matrices. These systems use RNA genetic switch design [3] or CRISPR-mediated interactions [4, 5] to detect specific target DNA or RNA sequences and trigger gene expression, fluorophore release, or other biochemical reactions to produce visual outputs when target molecules are present. The ability to freeze-dry cell-free reaction components for simple storage and distribution has enabled field-deployable technologies for profiling the gut microbiome [6] and detecting pathogens such as Ebola and Zika [3].
It is important to extend the capability of cell-free synthetic biology diagnostics to sense chemical targets other than nucleic acids. This is especially true in the case of water quality monitoring. Nearly 80% of the world’s population is at risk of water insecurity, partly due to a lack of access to clean and safely managed water [7]. Among the many contaminants present in water, some chemical species pose serious threats since they cannot be seen or easily detected. In fact, reliable methods to test for toxic species remain largely constrained to sophisticated, centralized laboratories with expensive instrumentation that requires significant technical expertise. These limitations preclude implementation of water quality testing in the field, most notably in low-resource settings where they are most needed.
To address this need, synthetic biologists have engineered microorganisms to serve as whole cell biosensors (WCBs) [8]. For example, cellular reporter genes can be regulated by allosteric transcription factors (aTFs) that respond to specific chemical contaminants to produce a detectable signal. Changing the aTF allows cellular systems to be engineered to sense a range of contaminants including metals, aromatic compounds and antibiotics [8]. Advantages of this approach include signal amplification through gene expression and selectivity afforded by processes that affect compound bioavailability [9]. However, there are challenges associated with WCBs, including maintenance of viable cells, difficulties with sensing toxic contaminants, membrane transport limitations and the need to address biocontainment and regulatory concerns for deploying living organisms in the field [10].
Here, we sought to preserve the modularity and signal amplification advantages of WCBs, while addressing some of their limitations. We developed cell-free diagnostics using RNA output sensors activated by ligand induction (ROSALIND) to detect contaminants in water (Fig. 1). ROSALIND comprises a highly processive phage RNA polymerase (RNAP), aTFs and engineered DNA transcription templates, that together produce visible outputs upon exposure to specific ligands. The use of RNA-level outputs allows signals to be observed within minutes and removes the need for complex and resource-intensive protein translation to detect responses. In addition, the use of in vitro transcription (IVT) reactions removes the need for cells to be metabolically active and overcomes biocontainment concerns. ROSALIND represents a new capability for simple and rapid field monitoring of chemical species to address growing global health crises in water quality.
Fig. 1 |. The ROSALIND system.
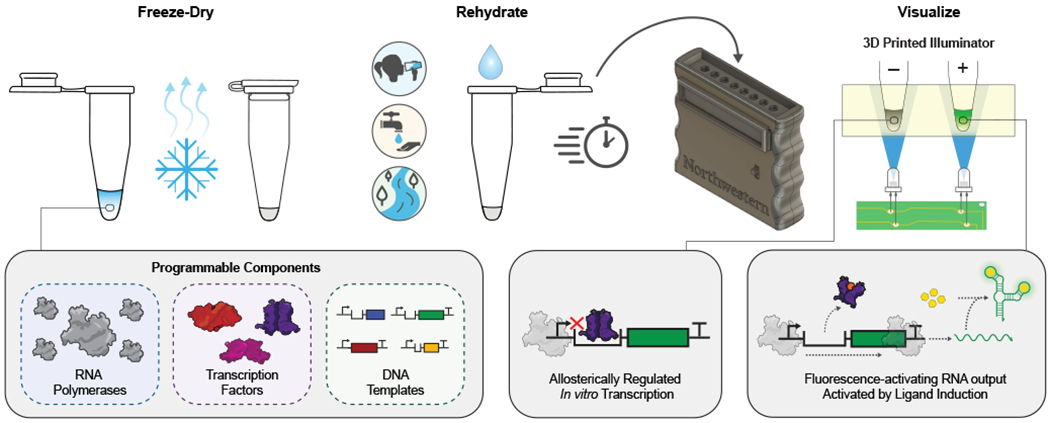
The RNA Output Sensors Activated by Ligand Induction (ROSALIND) system consists of three programmable components: highly-processive RNA polymerases, allosteric transcription factors and synthetic DNA transcription templates. Together, these components allosterically regulate the in vitro transcription of a fluorescence-activating RNA aptamer: in the absence of an analyte of interest, transcription is blocked, while in its presence, a fluorescent signal is produced. This design is compatible with downstream RNA circuitry that can tune the specificity and sensitivity of transcription factor outputs. ROSALIND can be freeze-dried for field deployment, thereby creating water quality sensors that are activated upon rehydration with the sample of interest. A low-cost, 3D-printed handheld device provides easy visualization of the sensor’s RNA output.
RESULTS
Fluorescent aptamers are rapid visible transcription reporters
We reasoned that fluorescent RNA-level outputs generated through IVT would allow biosensing reactions to be simplified, thereby resulting in faster signal generation than protein-level outputs that must be translated in more complex reaction environments [11]. To demonstrate the viability of fluorescent RNA outputs, we chose the three-way junction dimeric Broccoli (3WJdB) aptamer [12] and the fast T7 phage RNAP capable of transcribing 300 nucleotides (nt)/s in vitro [13]. DNA transcription templates encoding 3WJdB behind a T7 RNAP promoter were applied to a commercially available transcription kit supplemented with the Broccoli-binding dye DFHBI-1T (Fig. 2a). Fluorescence activation was monitored in real-time and standardized to known amounts of fluorescein (FITC) (Supplementary Fig. 1). The IVT reactions generated fluorescence signal comparable to micromolar equivalent fluorescein (MEF) across a range of input DNA template concentrations within minutes (Fig. 2b, Supplementary Video 1). Furthermore, the fluorescence activation of 3WJdB was easily visualized at 0.1’s MEF (µM FITC) with low-cost LEDs and a stage-lighting filter (Fig. 2c, Supplementary Data File 1). Additionally, we found that including a T7 terminator in the transcription templates resulted in improved fluorescence activation (Supplementary Fig. 2).
Fig. 2 |. In vitro transcription of a fluorescence-activating aptamer rapidly generates a visible RNA output that can be allosterically regulated.
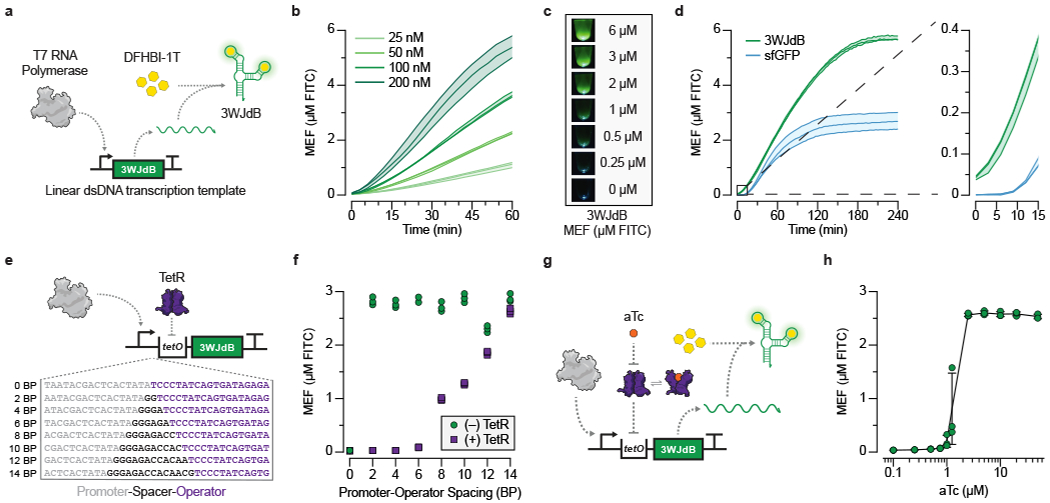
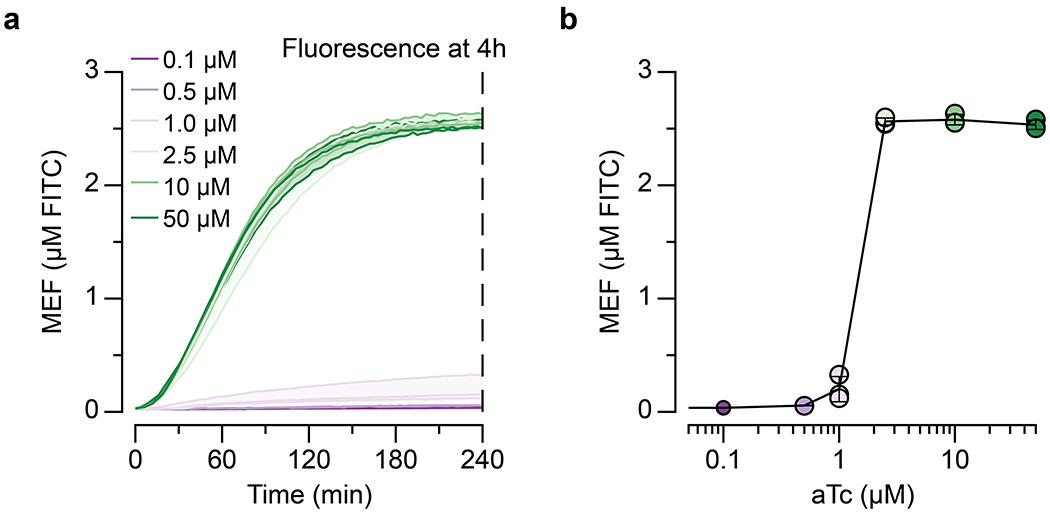
a, T7 RNA polymerase transcription of the three-way junction dimeric Broccoli (3WJdB) aptamer. Binding of DFHBI-1T by 3WJdB activates its fluorescence. b, Titration of the linear dsDNA template in a commercial transcription kit generates fluorescence signal comparable to micromolar equivalent fluorescein (MEF) when standardized to soluble fluorescein (µM FITC). c, Visual detection of 3WJdB:DFHBI-1T fluorescence, with values corresponding to MEF of soluble FITC. Results are shown for one representative of n=3 independent biological replicates. d, Comparison of fluorescence kinetics of 3WJdB synthesized from in vitro transcription, and sfGFP synthesized from in vitro transcription-translation, using an equimolar DNA template (50 nM). e, In vitro transcriptions (IVT) can be allosterically regulated with a template configured to bind a purified transcription factor (TetR) via the operator sequence (tetO) placed downstream of the T7 promoter. A series of spacers in 2 base pair (BP) intervals was constructed to evaluate the impact of spacer length on the ability to regulate transcription. f, Four-hour end point data shown for promoter-operator spacer variants regulated (with 2.5 µM TetR dimer, 25 nM DNA template) and unregulated (without TetR). g, Induction of a TetR-regulated IVT reaction occurs in the presence of the cognate ligand, anhydrotetracycline (aTc), which binds to TetR and prevents its binding to tetO. This allows transcription to proceed, leading to fluorescence activation. h, Dose response with aTc, measured at 240 minutes with 25 nM DNA template and 1.25 µM TetR dimer. All data shown for n=3 independent biological replicates as lines (b, d) or points (f, h) with raw fluorescence values standardized to MEF (µM FITC). Shading (b, d) and error bars (h) indicate the average value of 3 independent biological replicates ± standard deviation. The original, uncropped image shown in c can be found in Supplementary Data File 1.
We next compared the fluorescence intensity and kinetics of the RNA output to that of superfolder GFP (sfGFP) [14] produced using a commercial kit based on the PURE system [15]. 3WJdB-expressing IVT reactions and sfGFP-expressing PURE reactions were run using equimolar amounts of DNA templates. In this setup, the RNA output immediately began to increase in fluorescence intensity, while sfGFP required at least nine minutes before a detectable signal was observed (Fig. 2d, Supplementary Fig. 3). Additionally, over the four-hour time course, 3WJdB transcription produced substantially more fluorescence than sfGFP (Fig. 2d, Supplementary Fig. 3), and the signal generated with a homemade transcription reaction was comparable to that of the commercial kit (Extended Data Fig. 1).
Together, these results demonstrate that RNA-level outputs represent a viable and advantageous approach to biosensing signal generation by reducing the complexity associated with producing a visibly fluorescent output.
Allosteric transcription factors enable ligand-responsiveness
We next sought to create a ligand-responsive biosensor by using an aTF to regulate T7 RNAP IVT. We reasoned that the binding of an aTF to its cognate operator sequence would prevent T7 RNAP from promoter escape in the absence of ligand, and ligand-mediated de-repression of an aTF would result in a fluorescence signal (Fig. 1).
We first investigated how aTF-operator interactions can block T7 RNAP transcription in the absence of ligand. We chose TetR, a well-characterized model aTF that binds the 19 base-pair (BP) palindromic tet operator sequence (tetO) in the absence of its cognate ligand tetracycline [16]. We designed a series of transcription templates to explore the effect of spacing between the minimal 17 BP T7 promoter and the tetO sequence (Fig. 2e). We utilized the native sequence that follows the canonical T7 RNAP promoter [17] as a spacer in 2 BP increments from 0-14 BP, immediately followed by tetO, 3WJdB and T7 terminator sequences. IVT reactions were setup as before using homemade reactions, with the addition of purified recombinant TetR protein in 100-fold excess of the DNA template. We observed that the absence of any spacing resulted in no fluorescence activation in either the presence (regulated) or absence (unregulated) of TetR (Fig. 2f). However, robust fluorescence activation was observed without TetR when using a 2-6 BP spacer, which was reduced to baseline levels when regulated by TetR. Spacers that were 8 BP or longer resulted in fluorescence activation in the regulated reactions. We therefore chose the shorter functional spacers for subsequent promoter-operator designs.
We next sought to determine if TetR can be de-repressed with a cognate ligand to permit IVT and therefore act as a biosensor (Fig. 2g). We first performed IVT reactions over a range of TetR concentrations using the transcription template with a 4 BP spacer (Extended Data Fig. 2). We found that a 25-fold excess of TetR dimer to transcription template was the minimal amount of repressor needed to ensure tight regulation of transcription, while a 50-fold excess of TetR dimer performed similarly to the 100-fold excess (Extended Data Fig. 2). We then setup IVT reactions using these TetR and DNA template concentrations and included a range of anhydrotetracycline (aTc) concentrations. We observed strong repression over four hours through high nanomolar amounts of aTc, with half-maximal induction between 1 and 2.5 µM aTc (Fig. 2h, Extended Data Fig. 3). Importantly, when induced with 2.5 µM aTc, the kinetics of induction follow those of the unregulated transcription reaction (Extended Data Fig. 2).
Together, these results show that aTF-regulated IVT of fluorescence-activating RNA aptamers can serve as biosensors, establishing a proof-of-concept basis for the ROSALIND system.
Modularity allows sensing of organic molecules and metals
We next sought to expand the range of molecules that can be detected with ROSALIND by leveraging the natural diversity of aTFs [18]. Inspired by earlier work with WCBs [8], we reasoned that ROSALIND could sense a wide range of important targets by changing the operator sequence within the transcription templates and including the corresponding aTF in the reaction (Extended Data Fig. 2, Supplementary Data File 2).
We began with antibiotics whose overuse is resulting in the contamination of freshwater supplies and driving antimicrobial resistance [19]. We first tested our TetR-based sensor to detect tetracycline (Fig. 3a, Extended Data Fig. 4a) and the semi-synthetic antibiotic doxycycline (Fig. 3b, Extended Data Fig. 4b), both of which are on the World Health Organization’s (WHO) list of essential medicines. Reactions with TetR-regulated transcription templates and purified TetR were run using either lab-grade water (−), or similar water with added antibiotic (+). In both cases, we observed generation of fluorescence with the antibiotic present and no fluorescence in the water control.
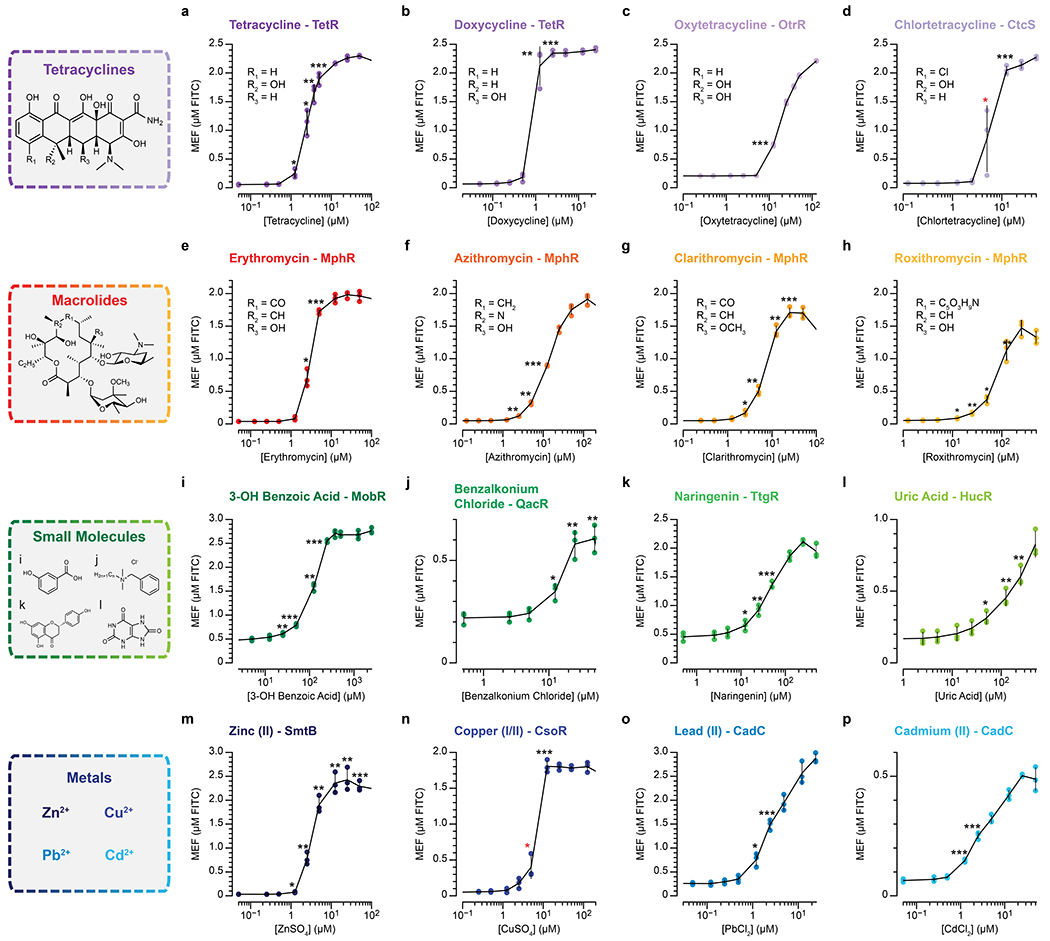
Fig. 3 |. ROSALIND is modular and can be used to sense complex molecules, small molecules and metal ions.
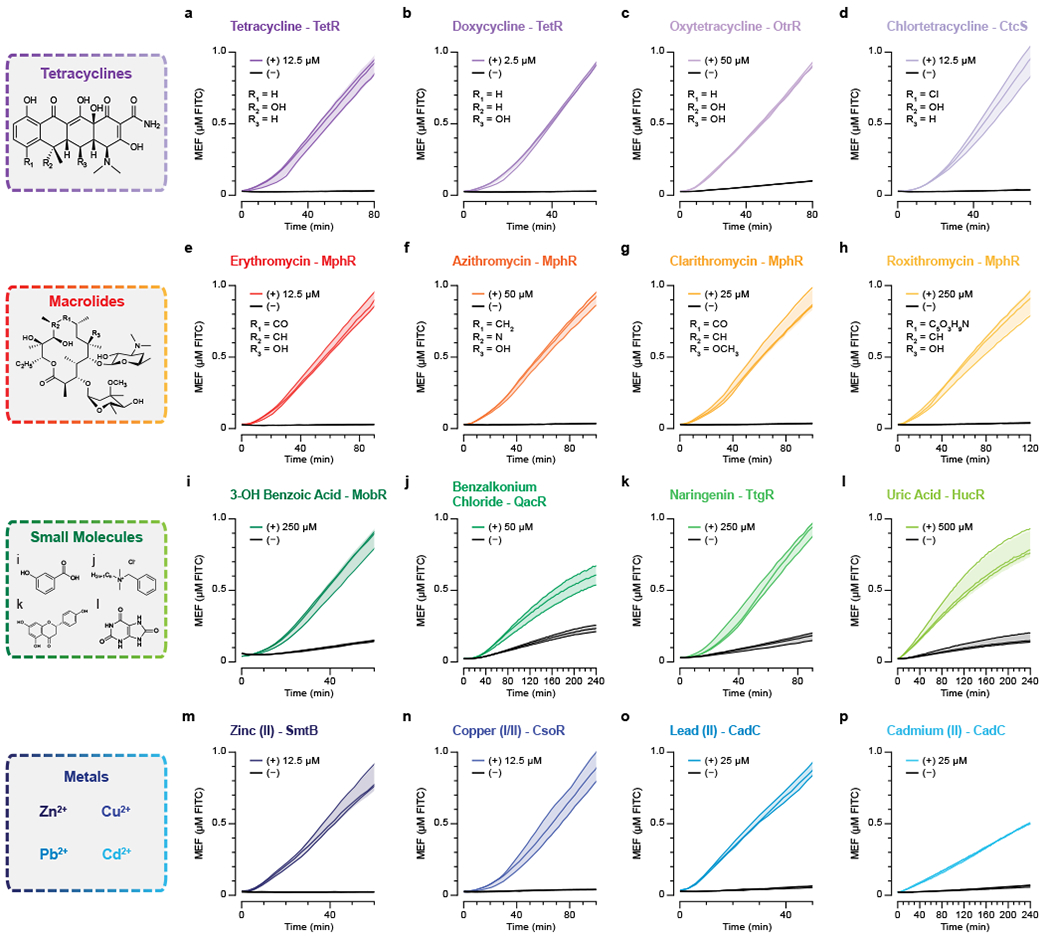
TetR can be used to sense a, tetracycline, b, doxycycline and other tetracycline family antibiotics. c, Oxytetracycline sensing with OtrR. d, Chlortetracycline sensing with CtcS. MphR senses the macrolides e, erythromycin, f, azithromycin, g, clarithromycin and h, roxithromycin. i, 3-hydroxy benzoic acid sensing with MobR. j, Benzalkonium chloride sensing with QacR. k, Naringenin sensing with TtgR. l, Uric acid sensing with HucR. m, Zn2+ sensing from ZnSO4 with SmtB. n, Cu2+/Cu+ sensing from CuSO4 with CsoR from Bacillus subtilis. o, Pb2+ sensing from PbCl2 and p, Cd2+ sensing from CdCl2 with CadC. Each reaction contains the indicated amount of ligand (+) dissolved in lab-grade water, ethanol, Tris-base buffer, dimethyl sulfoxide, or diluted sodium hydroxide, or a lab-grade water control (−) (Supplementary Table 2). All data shown for n=3 independent biological replicates as lines with raw fluorescence values standardized to MEF (µM FITC). Shading indicates the average value of 3 independent biological replicates ± standard deviation. DNA template and aTF concentrations used in each reaction can be found in Supplementary Table 2.
We then leveraged OtrR [20], a member of the multiple antibiotic resistance repressor (MarR) family of aTFs [21], to serve as a biosensor for oxytetracycline (Fig. 3c, Extended Data Fig. 4c). To expand the range of tetracycline antibiotic sensing further, we identified a putative MarR-family aTF analogous to OtrR, CtcS [22] and the divergent operon regulated by CtcS, and designed a minimal operator sequence to enable its use in ROSALIND for sensing chlortetracycline (Fig. 3d, Extended Data Fig. 4d).
We next sought to expand sensing to the macrolide class of antibiotics that are also widely used in human health. We chose a TetR-family aTF, MphR [23] to detect erythromycin, a drinking water contaminant of emerging concern [24], azithromycin, clarithromycin and roxithromycin (Fig. 3e–h, Extended Data Fig. 4e–h). We note that this observed broad range of MphR specificity is expected based on its known promiscuity and is likely a common feature of xenobiotic-sensing aTFs [25].
Having demonstrated the ability of ROSALIND to detect complex antibiotic molecules, we sought to leverage the system for the detection of other organic molecules, focusing on those with public health relevance. In particular, there has been a growing concern that unregulated contaminants, such as those generated from the large use of pharmaceuticals and personal care products (PPCPs), interfere with or are unaffected by water treatment and are causing widespread health and environmental concerns [26]. We started with the perfume additive 3-hydroxy benzoic acid as a model PPCP that can de-repress the Comamonas testosteroni KH122-3S MarR-family aTF, MobR [27]. When ROSALIND reactions were configured with MobR and its corresponding DNA template, we observed strong activation of fluorescence only in the presence of 3-hydroxy benzoic acid (Fig. 3i, Extended Data Fig. 4i). We further demonstrated that ROSALIND could be used to sense benzalkonium chloride, a widely used disinfectant that can promote antimicrobial resistance [28] by leveraging the Staphylococcus aureus TetR-family aTF, QacR [29] (Fig. 3j, Extended Data Fig. 4j).
Some organic molecules such as naringenin [30] and uric acid can serve as important markers for human health and nutrition. Using the Pseudomonas putida TetR-family aTF, TtgR [31] and an optimized ttgO variant sequence [32], we designed ROSALIND reactions to sense naringenin in the tens of micromolar range (Fig. 3k, Extended Data Fig. 4k). Additionally, we sought to detect uric acid, an early indicator of gout when present at 6 mg/dL or above within bodily fluids [33]. By incorporating HucR, a MarR-family aTF from Deinococcus radiodurans [34], and the corresponding DNA template, we were able to detect uric acid within water samples at approximately 50 µM (~0.84 mg/dL) or higher (Fig. 3l, Extended Data Fig. 4l).
Among water contaminants, metals have become a major issue due to aging infrastructure, mining operations, industrial wastes disposal and from natural abundance [35–37]. To enable metal detection using ROSALIND, we first identified the canonical zinc sensing aTF, SmtB, from the ArsR/SmtB family of regulators [38]. Using SmtB and the corresponding DNA template, we were able to sense Zn(II) at 2.5 µM (163 ppb) and above, which is below the US EPA secondary drinking guideline of 5 ppm (Fig. 3m, Extended Data Fig. 4m, Supplementary Table 1).
While many metals are toxic at high concentrations, of particular concern are copper and lead [36, 37] due to toxic effects such as psychiatric disorders, impaired neural development, and organ damage. As such, their levels are regulated in drinking water by the US EPA’s Lead and Copper Rule (LCR). Among the many copper-sensing aTFs is CsoR, a Cu(I) sensing aTF first identified in Mycobacterium tuberculosis that forms a homotetramer to bind four copper ions [39]. When configured into ROSALIND with the appropriate DNA template, our sensor was able to detect copper at 5 µM (318 ppb) and above (Fig. 3n, Extended Data Fig. 4n). Interestingly, while the cognate ligand for CsoR is Cu(I), we observed that the reaction senses Cu(II) from added CuSO4, possibly due to the presence of 10 mM DTT in IVT reactions that can reduce Cu(II) to Cu(I). Another ArsR/SmtB family aTF, CadC, is found in pI258 of Staphylococcus aureus and known to bind a number of metals, including lead and cadmium [40]. When implemented in ROSALIND with appropriately configured DNA templates, we were able to robustly detect both lead and cadmium at 1.25 µM (259 ppb for lead and 141 ppb for cadmium) and above (Fig. 3o,p, Extended Data Fig. 4o,p).
In total, we employed 11 different aTFs to sense 16 different ligands that are structurally and functionally diverse. An investigation into their limits of detection (Extended Data Fig. 4, Supplementary Data File 3) showed that the majority detect their respective ligands in the micromolar range. Additionally, characterization of the inhibitory effects of the ligands and the solvents used to prepare them (Supplementary Fig. 4, Supplementary Table 2) suggests that ROSALIND can tolerate much higher concentrations of the ligands than their respective WHO/EPA regulated limits (Supplementary Table 1).
RNA circuitry can invert logic and fix crosstalk
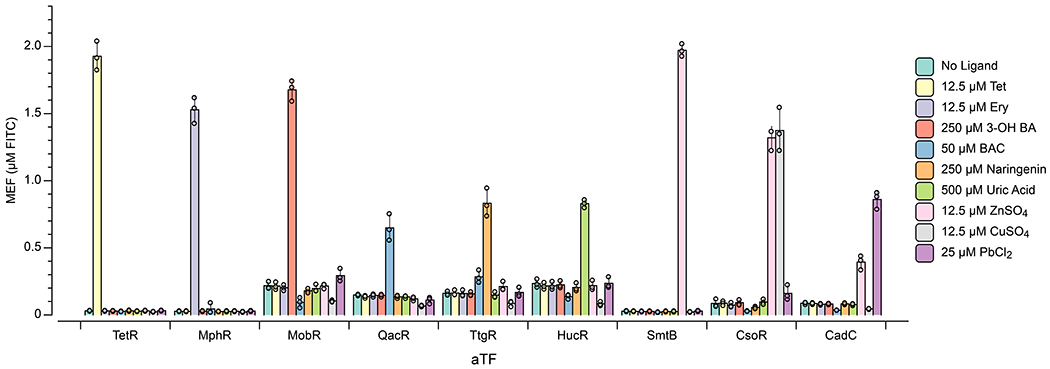
Specificity is a key metric for the performance of biosensors. In some cases, a single aTF is well-known to sense multiple ligands, as is the case with TetR sensing tetracyclines, MphR sensing macrolides and CadC sensing both lead and cadmium [18, 25, 40]. In these cases, ROSALIND is best viewed as reporting on the presence of a specified family of stereochemically related ligands. To investigate whether ROSALIND reactions are specific for the indicated compounds outside of these known cases, we tested ROSALIND reactions configured with TetR, MphR, MobR, QacR, TtgR, HucR, SmtB, CsoR, or CadC, along with their corresponding DNA templates, with a panel of cognate ligands. Tetracycline, erythromycin and lead were chosen as representative members of each class of compounds for TetR, MphR and CadC, respectively. In general, we found strong signals from cognate ligands with minimal crosstalk, indicating that ROSALIND reactions are generally specific for their intended ligands when DNA template and aTF concentrations are optimized (Fig. 4a, Extended Data Fig. 5).
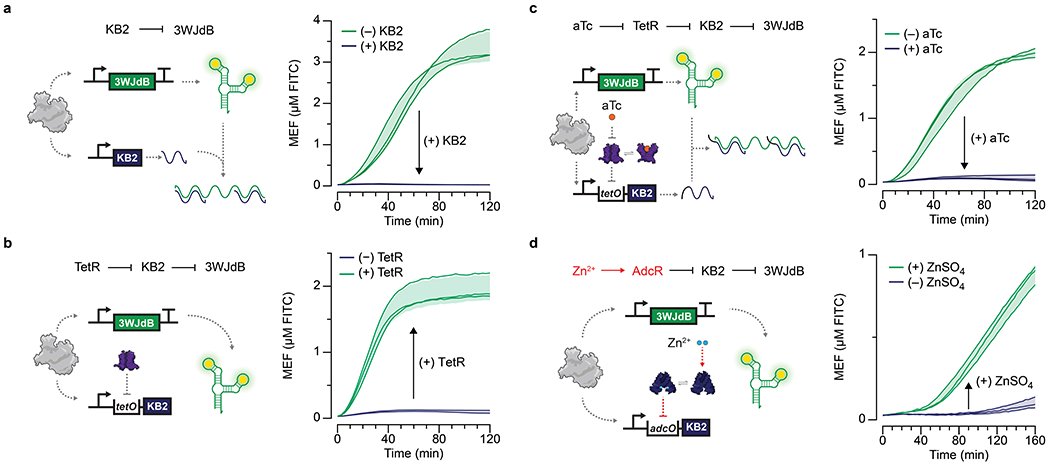
Fig. 4 |. Cross-reactivity of ROSALIND reactions can be corrected using an RNA logic circuit.
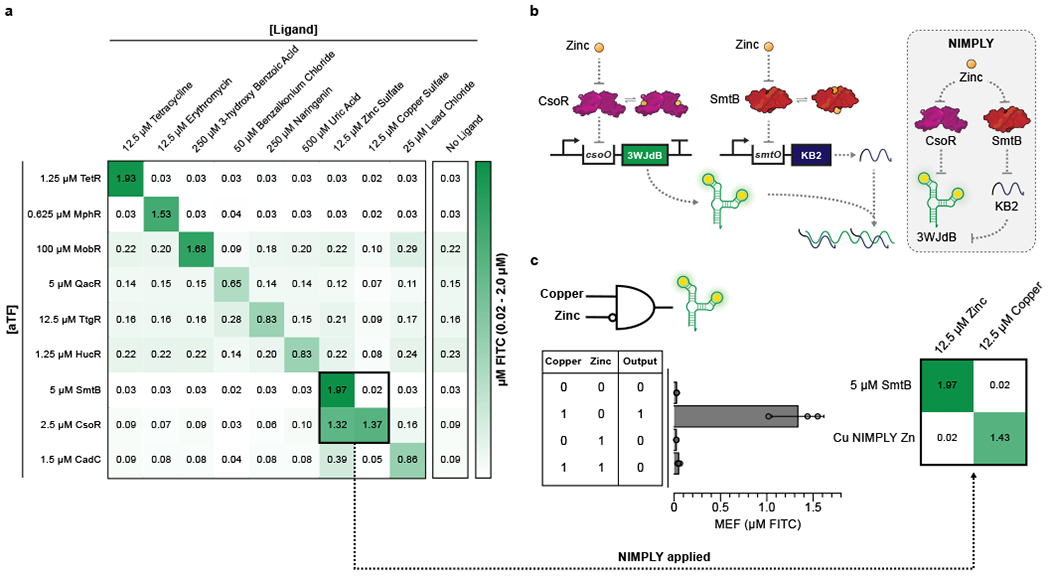
a, A subset of ROSALIND reactions were tested to evaluate cross-reactivity with different ligands. Significant crosstalk was observed from the copper sensing aTF, CsoR, where the sensor detects copper as well as zinc. The values on the heat map represent the average MEF (µM FITC) of n=3 independent biological replicates after 240 minutes (see Extended Data Fig. 5 for all data). 25 nM of DNA templates were used for reactions with TetR, MphR, SmtB and CsoR, 20 nM for QacR, 12.5 nM for HucR, 10 nM for CadC and 5 nM for MobR and TtgR. b, To fix the cross-reactivity of CsoR, a Cu NIMPLY Zn (Cu AND NOT Zn) logic gate was implemented consisting of the CsoR-controlled 3WJdB template and a SmtB-controlled template encoding a 3WJdB-interfering kleptamer (KB2). In the presence of zinc, the fluorescence of 3WJdB is inhibited by the presence of KB2, while in the presence of copper alone, only 3WJdB is produced. c, When implemented in ROSALIND, the Cu NIMPLY Zn circuit fixes the cross-reactivity of the copper sensor. Each reaction contained 25 nM of the 3WJdB template, 150 nM of the KB2 template, 2.5 µM of CsoR tetramer and 5 µM of SmtB dimer. The concentrations of copper and zinc tested in the bar plot are 10 µM each. The bar plot in c shows data for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of the replicates. Error bars indicate the average value of 3 independent biological replicates ± standard deviation. The matrix in c shows the average MEF (µM FITC) of n=3 independent biological replicates after 240 minutes. The aTF concentrations indicated in a and c are dimer concentrations except for CsoR, which is a tetramer.
We did however find significant crosstalk from the CsoR containing copper sensor when induced with either copper or zinc (Fig. 4a, Extended Data Fig. 5). Notably there was no corresponding crosstalk between SmtB, the zinc sensor, and copper. This created the opportunity to construct a new sensor by combining the activity profiles of CsoR and SmtB in a single reaction to fix the crosstalk of CsoR. Specifically, this new sensor was configured based on a copper NIMPLY zinc circuit that would only respond to copper AND NOT zinc. Implementation of this circuit requires the presence of zinc to inhibit the 3WJdB signal produced by CsoR.
To achieve this zinc-mediated inhibition, we leveraged the recent development of RNA kleptamers—short antisense RNAs designed to inhibit Broccoli by disrupting the aptamer folding that enables fluorescence activation [41]. We found that fluorescence activation of 3WJdB could be inhibited through unregulated IVT of the KB2 kleptamer with T7 RNAP (Extended Data Fig. 6a), and that KB2 transcription could be regulated by TetR by including the tetO sequence in the KB2 template (Extended Data Fig. 6b). Addition of its cognate ligand, aTc, effectively de-repressed TetR from the KB2 template, thereby allowing KB2 transcription and inverting the output of the TetR sensor (Extended Data Fig. 6c). We also demonstrated the generality of the kleptamer inversion strategy using the aporepressor AdcR from Streptococcus pneumoniae [21, 42], which binds its operator in the presence of Zn(II), and inverting it into a zinc-activated sensor (Extended Data Fig. 6d).
We used the kleptamer strategy within the copper NIMPLY zinc circuit. In this circuit design, the transcription of 3WJdB is regulated by CsoR, while the transcription of KB2 is regulated by SmtB (Fig. 4b). In the presence of copper only, CsoR is de-repressed from the 3WJdB transcription template, generating 3WJdB. In the presence of zinc only, both CsoR and SmtB are de-repressed from their respective transcription templates, generating 3WJdB and KB2, which is then able to prevent 3WJdB from binding its dye, shutting off the fluorescence signal. When implemented in ROSALIND, the copper NIMPLY zinc circuit performed as expected, correcting the crosstalk of CsoR with zinc (Fig. 4c). Furthermore, the circuit was able to fix the crosstalk across a wide range of zinc concentrations, while keeping the dose response curves against copper unaffected (Supplementary Fig. 5). Overall these results show that RNA genetic circuitry can be designed to address issues with aTF specificity without having to utilize protein engineering strategies.
Sensitivity can be tuned with transcriptional circuitry
A shortcoming of our approach is that it is limited by the natural sensitivity of the aTF employed in the ROSALIND reaction. For example, our TetR sensor was activated by doxycycline at 1.25 µM (556 ppb), whereas the typical concentrations of antibiotic pollution found in environmental water samples are in the range of tens of ppb [43]. We therefore sought to tune the sensitivity of ROSALIND reactions using a generalizable strategy that did not require engineering of the aTF.
As proof-of-concept demonstrations, we first sought to tune the copper and lead sensing reactions containing CsoR and CadC, respectively. The dose response curves of the copper and lead sensors indicate that the limit of detection for copper (0.3 ppm of Cu(II)) is below the threshold set by the US EPA’s LCR (1.3 ppm), while that of lead (259 ppb) is an order of magnitude above the desired threshold (15 ppb) (Extended Data Fig. 7, Supplementary Table 1). To desensitize the copper sensor, we increased the CsoR concentration to allow excess amount to act as a copper chelator and shifted the dose response curve to activate near the desired LCR threshold of 1.3 ppm of copper from added CuSO4 (Extended Data Fig. 7a). To sensitize the lead sensor, we thus employed the converse of our CsoR strategy. By lowering CadC dimer concentration to 0.5 µM, we were able to shift the dose response curve to be more sensitive to detect tens of ppb Pb(II) (Extended Data Fig. 7b). However, this strategy resulted in significant increase of signal in the absence of lead.
To address this limitation, we sought to implement a different method to improve sensor sensitivity, based on the previous development of a TetR-binding RNA aptamer (“antiTetR”) which de-represses TetR by mimicking its operator binding site tetO [44]. Taking advantage of this feature of antiTetR, we implemented an RNA feedback loop designed to enhance the expression of 3WJdB at low concentrations of doxycycline by regulating antiTetR transcription by TetR (Fig. 5a). As configured, when doxycycline is present, the circuit is designed to de-repress TetR and initiate the expression of 3WJdB as well as antiTetR. Once transcribed, antiTetR is then able to further de-repress any TetR that is blocking the transcription of 3WJdB. When implemented in ROSALIND, the feedback circuit was able to improve the sensitivity of the doxycycline sensor by an order-of-magnitude to the ~0.2 µM (89 ppb) range (Fig. 5b, Supplementary Data File 3). These results demonstrate that TetR-based sensors can be sensitized to the ranges of antibiotic concentrations typically found in the environment, though pre-concentration strategies would further enhance this. Moreover, this demonstrates that ROSALIND is compatible with RNA feedback circuits that can improve the sensitivity of aTFs without any protein engineering.
Fig. 5 |. Sensitivity of ROSALIND reactions can be tuned using an RNA feedback circuit.
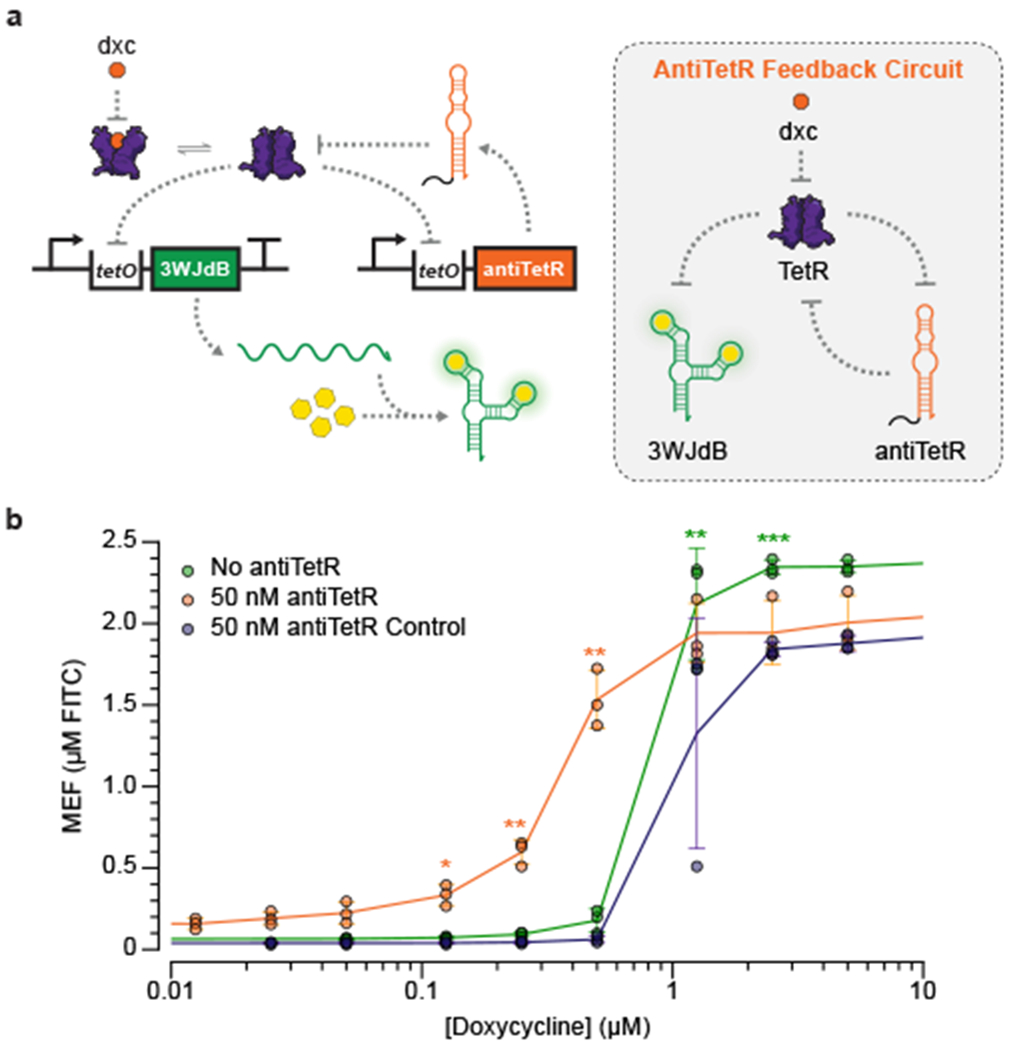
a, A TetR binding RNA, antiTetR, can be used to improve the limit of detection of the TetR sensor. In this feedback circuit, TetR regulates both the expression of 3WJdB and antiTetR. In the presence of the cognate ligand, doxycycline (dxc), antiTetR is transcribed and further de-represses TetR to enhance 3WJdB expression. b, When implemented in ROSALIND, the antiTetR feedback circuit improves the sensitivity of TetR by an order of magnitude. Each reaction contains 25 nM of the 3WJdB template, 50 nM of the antiTetR template and 1.25 µM of TetR dimer. A transcription template encoding the reverse sequence of antiTetR was used as a control to demonstrate that sensitization required the antiTetR interaction. All data shown for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC). Error bars indicate the average value of 3 independent biological replicates ± standard deviation. The ligand concentrations at which the signal is distinguishable from the background were determined using two-tailed heteroscedastic t-test against the no ligand condition, and their p-value ranges are indicated with asterisks (*** < 0.001, ** = 0.001 – 0.01, * = 0.01 – 0.05). Exact p-values along with degrees of freedom can be found in Supplementary Data File 3. Data for no ligand condition were excluded in b since the x-axis is on the log scale and are presented in Supplementary Data File 3.
Application to municipal water samples
We next examined whether ROSALIND can be formulated for distribution in the field. Many of the advances in synthetic biology diagnostic technologies have been enabled through freeze-drying cell-free systems for convenient storage and distribution before rehydration at the point-of-use [3, 45]. To test whether the ROSALIND system can be freeze-dried, we explored a range of known lyoprotectants and determined that nonreducing disaccharides, such as sucrose and trehalose, in combination with a sugar alcohol, mannitol, provided substantial lyoprotection. In addition, we developed a freeze-drying and packaging procedure (Supplementary Video 2) that enabled freeze-dried ROSALIND reactions to be stored at ambient temperature for at least 2.5 months and remain functional, although with some impact to measured fluorescence (Extended Data Fig. 8).
In addition to freeze-drying, field deployment also requires a convenient approach to detecting the fluorescence signals generated by ROSALIND. To this end, we developed a 3D-printed handheld device that could visualize fluorescent output using the naked eye (Extended Data Fig. 9, Supplementary Data File 4). The use of off-the-shelf components assembled onto a printed circuit board allowed us to optimize the device for visualization of 3WJdB fluorescence using blue light for fluorescence-excitation and yellow stage light film as a long-pass optical filter.
With these results, we next sought to demonstrate a field application of ROSALIND to address a water quality issue of growing concern–the contamination of municipal water supplies with metals. We studied the influence that the 2018 Camp Fire in Paradise, California had on its water distribution system. Preliminary field sampling of unused Paradise municipal water fountains during the cleanup efforts in the spring of 2019 suggested elevated levels of both copper and zinc. We therefore sought to field test our copper (CsoR), zinc (SmtB) and copper NIMPLY zinc sensors in the context of Paradise municipal water.
We first confirmed that this set of ROSALIND reactions functioned correctly when municipal drinking water collected in Evanston, Illinois, spiked with either copper or zinc, was used for rehydration (Supplementary Fig. 6). Sensors were then manufactured, arrayed in strip tubes, freeze-dried, packaged in Evanston and overnight-shipped to Chico, California at ambient temperature (Fig. 6a). We included control reactions for rehydration with lab-grade, metal-free water to confirm reactions performed as intended and to provide a convenient visual comparison against sampled water by eye (Fig. 6b). Upon receipt in California, we sampled and filtered water from four different municipal locations in Paradise [46]. The shipped sensors were rehydrated with these samples or lab-grade, metal-free water and placed in a portable incubator at 37° C for four hours. Photographs were taken of the reactions placed in our handheld illuminator after two hours and overnight (Fig. 6c–f). Rehydrated reactions were then shipped back to Evanston for plate reader quantification (Fig. 6c–f), which was possible due to the observed stability of fluorescence signals from ROSALIND reactions once complete (Extended Data Fig. 8a). Of the four sites sampled, two contained zinc only and two contained both zinc and copper, as verified using Flame Atomic Absorption Spectroscopy (FAAS) (Supplementary Fig. 7). In all cases, the freeze-dried, shipped and applied reactions behaved as expected (Fig. 6c–f). To further demonstrate ROSALIND functionality in complex matrices, additional laboratory tests of freeze-dried copper sensors were performed against Lake Michigan water spiked with copper as well as surface waters from Chile, known to be highly enriched with copper from a combination of geogenic and anthropogenic processes [47]. We confirmed that the sensors can function with environmental water samples as well, though with some inhibition of fluorescence signal (Extended Data Fig. 10).
Fig. 6 |. Field-deployment of ROSALIND.
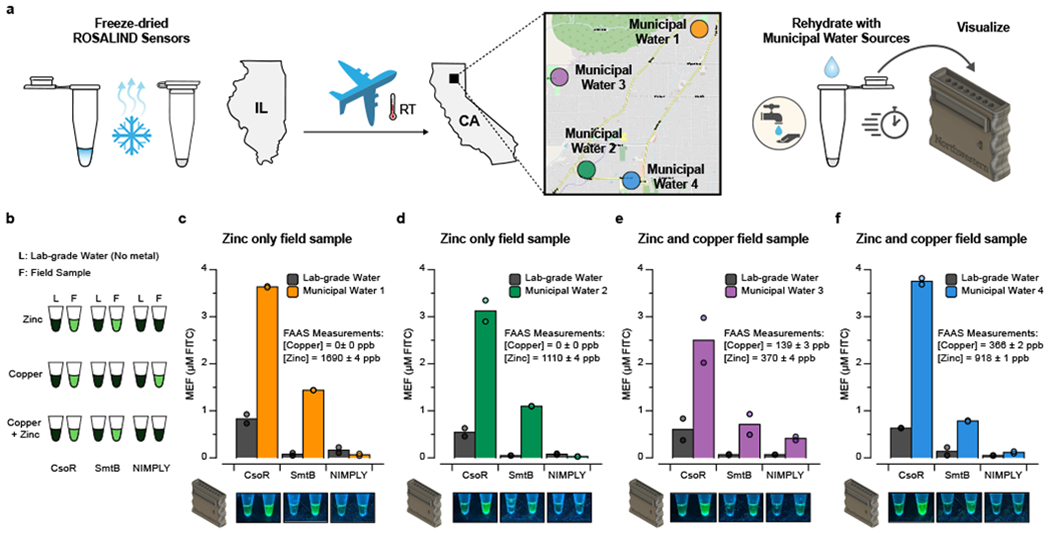
a, ROSALIND reactions were freeze-dried in Evanston, Illinois, shipped at ambient conditions and applied to municipal water from Paradise, California where the water quality was affected by the destructive Camp Fire. Four different municipal water samples were collected, filtered and used to rehydrate the sensors, which were then incubated in a portable incubator. A 3D-printed handheld illuminator was used to visualize signals from the rehydrated sensors within a day. b, Three different types of sensors were shipped and tested in strips of tubes: zinc sensing SmtB, copper sensing CsoR and the Cu NIMPLY Zn sensor. Negative controls were included where each sensor was rehydrated with lab-grade, metal-free water. Expected results for different metal contaminations are shown. c – f, Rehydrated ROSALIND reactions were shipped back to IL for plate reader quantification (top), aligned with handheld illuminator images taken in CA (bottom). The collected field samples were tested with flame atomic absorption spectroscopy (FAAS) for metal concentration validation. All sensors shipped and tested resulted in expected signal patterns. Data from n=2 technical replicates are shown as points with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of 2 technical replicates. Images are shown for one replicate with other replicate images included in Supplementary Data File 1. The concentrations of DNA template(s) and aTF(s) in each sensor are the same as the ones used in Fig. 4c. Geographical data in a © OpenStreetMap contributors [46].
Overall, these results demonstrate that ROSALIND can function in the field to detect water contaminants of interest and, as configured, are best suited to indications of presence/absence at specific thresholds.
DISCUSSION
Here we report the development of a new cell-free biosensor platform, ROSALIND, for detection of a wide range of chemical compounds and elements in water. ROSALIND consists of simple and defined aTF-regulated IVT reactions that produce RNA outputs as a function of ligand concentration. Inspired by previous WCB approaches [8], this modular architecture allowed us to develop 16 different sensors by changing the aTF-operator pair (Fig. 3). The use of fast phage polymerases generated visible signals to the naked eye in less than an hour. In addition, the reactions and handheld illuminators could be manufactured inexpensively (Supplementary Table 3, Supplementary Data File 4).
A key advantage of transcriptional RNA outputs was our ability to design RNA-level circuits to address well-known shortcomings of aTFs without performing any protein engineering. In particular, we were able to demonstrate that aTF-ligand crosstalk can be eliminated with RNA-level logic gates (Fig. 4), and aTF sensitivity can be improved through a new RNA-mediated feedback circuit (Fig. 5). We anticipate that these RNA-level circuit strategies will be applicable to optimizing broad classes of aTFs.
The simple and defined nature of ROSALIND makes it amenable to extensions. For example, multiplexing can be added with additional fluorescent aptamers [48], and further engineering of the aTF or operator sequences can be used to enhance sensitivity, modify specificity, and otherwise tune system performance [49]. In addition, other uses besides water contaminant detection are possible, including the high-throughput screening of poorly-characterized aTFs to uncover their cognate ligands and putative operator sequences, as we showed with our validation of the putative aTF, CtcS (Fig. 3d).
A key limitation of ROSALIND is that it is currently limited to aTFs that block RNAP elongation unless bound to a ligand. In addition, while the sensors are easy to operate from a user perspective, purifying aTFs that are functional in vitro can be non-trivial. Finally, signal detection by eye can be difficult due to variations in lighting and reaction conditions. While the inclusion of control reactions helps with this, more sophisticated illuminator design that incorporates quantification would improve this aspect of the system.
A main feature of our study is demonstrating that freeze-dried ROSALIND reactions can be manufactured and shipped across the US at ambient temperature and applied to municipal water samples in the field (Fig. 6). We also found that ROSALIND reactions could be applied to other environmental water sources, though with some inhibition of the observed fluorescence levels (Extended Data Fig. 10). This can potentially be due to molecular components in the water samples that can inhibit transcriptional activity or quench fluorescence. Nonetheless, we did find that a practical workflow of testing serial dilutions could be used to detect copper, which we anticipate dilutes the interfering components in the reactions (Extended Data Fig. 10). In addition, different test formats may be used to include calibration curves that control for matrix effects [50].
ROSALIND builds off of significant prior work in developing WCBs [8] and cell extract-based biosensors [2]. Compared to WCBs, the defined in vitro environment of ROSALIND enables rapid optimization of sensor performance (Extended Data Fig. 2) and implementation of RNA-level circuitry to address key limitations of aTF-based sensors [8]. However, this flexibility comes at the cost of purifying functional aTFs and optimizing reaction conditions, and one advantage of WCBs is the simplicity of creating engineered strains. WCBs may take advantage of active transports to accumulate and detect only bio-available ligand, i.e., the fraction that can cross the cell membrane [9], though more systematic comparisons are needed to precisely define these benefits. Finally, WCBs require additional equipment or procedures to maintain cells in physiologically active states, encapsulation to manage biocontainment and normalization of signals to account for different numbers of cells in each reaction – challenges which are significantly mitigated by cell-free biosensing approaches such as ROSALIND.
Taken together, WCBs and cell-free synthetic biology approaches like ROSALIND help fill a gap in existing water quality monitoring technology, which currently consists mainly of centralized and expensive instrumentation which cannot always be deployed in the field (Supplementary Table 4). We anticipate ROSALIND to be a part of a synthetic biology-based solution to enable communities and individuals to rapidly and economically monitor their water quality to meet their needs, to inform when and where more accurate measurements are needed and to facilitate the gathering of critical information necessary for achieving the UN global sustainable development goal of safe and clean water.
ONLINE METHODS
Strains and growth medium
E. coli strain K12 (NEB Turbo Competent E. coli, New England Biolabs #C2984) was used for routine cloning. E. coli strain Rosetta 2(DE3)pLysS (Novagen #71401) was used for recombinant protein expression. LB supplemented with the appropriate antibiotic(s) (100 µg/mL carbenicillin, 100 µg/mL kanamycin, and/or 34 µg/mL chloramphenicol) was used as the growth media.
Plasmids and genetic parts assembly
DNA oligonucleotides for cloning and sequencing were synthesized by Integrated DNA Technologies. Genes encoding aTFs were synthesized either as gBlocks (Integrated DNA Technologies) or gene fragments (Twist Bioscience). Expression plasmids were cloned using Gibson Assembly (NEB Gibson Assembly Master Mix, New England Biolabs #E2611) into a pET-28c plasmid backbone and were designed to overexpress recombinant proteins as either C-terminus or N-terminus His-tagged fusions. Certain constructs additionally incorporated a recognition sequence for cleavage and removal of the His-tag using TEV protease (Supplementary Table 5). Gibson assembled constructs were transformed into NEB Turbo cells, and isolated colonies were purified for plasmid DNA (QIAprep Spin Miniprep Kit, Qiagen #27106). Plasmid sequences were verified with Sanger DNA sequencing (Quintara Biosciences) using the primers listed in Supplementary Data File 5.
Plasmids encoding transcription templates were constructed using pUC19-T7-3WJdB-T (Addgene plasmid #87308) as a backbone. These plasmids included a T7 RNAP promoter, an aTF operator site, the 3WJdB coding sequence [51], and a T7 terminator. Here, we define the T7 RNAP promoter as a minimal 17 BP sequence (TAATACGACTCACTATA) excluding the first G that is transcribed. Plasmids were cloned using either Gibson Assembly or inverse PCR followed by blunt-end ligation, and then transformed into NEB Turbo cells for clonal isolation and sequence verification using the primer listed in Supplementary Data File 5. Once sequence verified, transcription templates were generated by PCR amplification (Phusion High-Fidelity PCR Kit, New England Biolabs #E0553) of the plasmids to include a 5’ region upstream of the T7 promoter and a 3’ region ending with either 3WJdB or the T7 terminator with the primers listed in Supplementary Data File 5. Amplified templates were then purified (QIAquick PCR purification kit, Qiagen #28106), verified for the presence of a single DNA band of expected size on a 2% TAE-Agarose gel, and concentrations were determined using the Qubit dsDNA BR Assay Kit (Invitrogen #Q32853).
The KB2 DNA transcription templates were generated by denaturing two complementary oligonucleotides (Supplementary Data File 5) at 95° C for 3 minutes and slow cooling (-0.1° C/s) to room temperature in 1X annealing buffer (100 mM potassium acetate and 30 mM HEPES, pH 8.0). Annealed oligonucleotides where then purified by resolving them on a 20% native PAGE-TBE gels, isolating the band of expected size, and eluting at room temperature overnight in 1X annealing buffer. The eluted DNA template was then ethanol precipitated, resuspended in MilliQ H2O, and quantified using the Qubit dsDNA BR Assay Kit. The KB2 DNA template for the copper NIMPLY zinc circuit was constructed by placing the smtO sequence 5 BP downstream of T7 promoter and immediately upstream of the KB2 coding sequence [52]. The NIMPLY control circuit was designed by reversing the KB2 sequence, while keeping the other parts consistent. These DNA templates were generated using the same method described above.
The antiTetR DNA transcription template was constructed by placing the tetO sequence 4 BP downstream of T7 promoter and immediately upstream of the antiTetR coding sequence [53]. This template was generated by three sequential PCR amplifications using primers listed in Supplementary Data File 5. The first PCR reaction was performed using a single-stranded DNA oligonucleotide as its template, and the resulting PCR product was used as the template for the second PCR reaction. After the second PCR amplification, the expected DNA band was extracted from a 2% agarose gel (QIAquick Gel Extraction Kit, Qiagen #28706) for the final PCR amplification. Amplified templates were then purified, verified for the presence of a single DNA band of expected size on a 2% TAE-Agarose gel, and concentrations were determined using the Qubit dsDNA BR Assay Kit. The antiTetR control circuit was designed by reversing the antiTetR sequence, and its DNA template was generated using the same method described above.
A table listing the sequences and Addgene accession numbers of all plasmids generated in this study are listed in Supplementary Data File 5.
aTF expression and purification
Supplementary Table 5 provides a listing of the exact expression and purification process for each aTF. In general, with the exception of CadC and AdcR, each protein was expressed and purified as follows. Sequence-verified pET-28c plasmids were transformed into the Rosetta 2(DE3)pLysS E. coli strain for protein expression. A single transformed colony from each expression plasmid transformation was then used to seed a small overnight culture grown at 37° C. Overnight cultures were then seeded into larger expression cultures (10 mL starter culture / 1 L expression culture). Cultures were induced for overexpression at an optical density (600 nm) of ~0.5 with 100 to 500 µM of IPTG and grown for 4 additional hours at 37° C or overnight at 30° C. Cultures were then pelleted by centrifuging at 4,000 RCF for 20 minutes at 4 °C in a Thermo Scientific Sorvall Lynx 4000, and then either were immediately stored at −80° C or resuspended in lysis buffer (10 mM Tris-HCl pH 7.5 – 8.5, 500 mM NaCl, 1 mM TCEP, and protease inhibitor (cOmplete EDTA-free Protease Inhibitor Cocktail, Roche)). Resuspended cells were then lysed on ice through four rounds of ultrasonication (50% duty cycle) for 1 min, with 2 min of rest on ice in between rounds. Lysates were then centrifuged at 13,000 RCF for 30 minutes to remove insoluble material. Clarified supernatants were then purified using His-tag affinity chromatography with a Ni-NTA column (HisTrap FF 5mL column, GE Healthcare Life Sciences) followed by size exclusion chromatography (Superdex HiLoad 26/600 200 pg column, GE Healthcare Life Sciences) using an AKTAxpress fast protein liquid chromatography (FPLC) system. The collected fractions from the FPLC were concentrated and buffer exchanged (25 mM Tris-HCl, 100 mM NaCl, 1mM TCEP, 50% glycerol v/v) using centrifugal filtration (Amicon Ultra-0.5, Millipore Sigma). Protein concentrations were determined using a Qubit Protein Assay Kit (Invitrogen #Q33212). The purity and size of the proteins were validated on SDS-PAGE gel (Mini-PROTEAN TGX and Mini-TETRA cell, Bio-Rad). Purified proteins were stored at −20° C. CadC and AdcR, whose genes were cloned into a pET-3d and pET-3a plasmid backbone respectively, were purified with the protocol previously described [54]. For CadC purification, the protocol was modified to have a higher concentration of TCEP (5 mM) in its buffers.
In vitro transcription (IVT) reactions
NEB HiScribe™ T7 Quick High Yield RNA Synthesis Kit (#E2050S, New England Biolabs) was used for data shown in Fig. 2b,d, Supplementary Fig. 2, 3, and Extended Data Fig. 1 according to the manufacturers protocol with the addition of 2.25 mM DFHBI-1T (Tocris #5610) in the reaction mix. Homemade IVT reactions were set up by adding the following components listed at their final concentration: IVT buffer (40 mM Tris-HCL pH 8, 8 mM MgCl2, 10 mM DTT, 20 mM NaCl, and 2 mM spermidine), 2.25 mM DFHBI-1T, 11.4 mM Tris-buffered NTPs pH 7.5, 0.3U thermostable inorganic pyrophosphatase (#M0296S, New England Biolabs), DNA transcription template(s), and MilliQ H2O to a total volume of 20 µL. Regulated IVT reactions additionally included a purified aTF at the indicated concentration and were equilibrated at 37° C for 15 minutes. Immediately prior to plate reader measurements, 0.2 ng of T7 RNA polymerase and, optionally, a ligand at the indicated concentration (Supplementary Table 2) were added to the reaction. Reactions were then characterized on a plate reader as described below (Plate reader quantification and micromolar equivalent fluorescein (MEF) standardization). Supplementary Data File 2 contains a master experimental planning sheet that describes how a typical ROSALIND reaction is setup.
NEB PURExpress® In Vitro Protein Synthesis Kit (#E6800, New England Biolabs) was used for data shown in Fig. 2d and Supplementary Fig. 3 according to the manufacturer’s protocol. Linear DNA template encoding a T7 promoter followed by sfGFP and a T7 terminator was PCR amplified (Phusion High-Fidelity PCR Kit, New England Biolabs #E0553) from the plasmid ADS004 (Addgene plasmid #69496) using primers listed in Supplementary Data File 5. Amplified template was then purified, quantified using the Qubit dsDNA BR Assay Kit, and verified for the presence of a single band of expected size on a 1% TAE-Agarose Gel.
Freeze-drying
Prior to lyophilization, PCR tube caps were punctured with a pin to create three holes. Lyophilization of ROSALIND reactions was then performed by assembling the components of regulated in vitro transcription (see above) with the addition of 50 mM sucrose and 250 mM D-mannitol. Assembled reaction tubes were immediately transferred into a pre-chilled aluminum block and placed in a −80°C freezer for 10 minutes to allow slow-freezing. Following the slow-freezing, reaction tubes were wrapped in Kimwipes and aluminum foil, submerged in liquid nitrogen, and then transferred to a FreeZone 2.5 L Bench Top Freeze Dry System (Labconco) for overnight freeze-drying with a condenser temperature of −85° C and 0.04 millibar pressure. Unless rehydrated immediately, freeze-dried reactions were packaged as follows. The reactions were placed in a light-protective bag (Mylar open-ended food bags, Uline #S-11661) with a desiccant (Dri-Card Desiccants, Uline #S-19582), purged with Argon using an Argon canister (ArT Wine Preserver, Amazon #8541977939), and immediately impulse heat-sealed (Metronic 8 inch Impulse Bag Sealer, Amazon #8541949845) (Supplementary Video 2).
Plate reader quantification and micromolar equivalent fluorescein (MEF) standardization
A NIST traceable standard (Invitrogen #F36915) was used to convert arbitrary fluorescence measurements to micromolar equivalent fluorescein (MEF). Serial dilutions from a 50 µM stock were prepared in 100 mM sodium borate buffer at pH 9.5, including a 100 mM sodium borate buffer blank (total of 12 samples). The samples were prepared in technical and experimental triplicate (12 samples X 9 replicates = 108 samples total), and fluorescence values were read at an excitation wavelength of 472 nm and emission wavelength of 507 nm for 3WJdB-activated fluorescence, or 485 nm excitation and 515 nm emission for sfGFP fluorescence on a plate reader (Synergy H1, BioTek). Fluorescence values for a concentration in which a single replicate saturated the plate reader were excluded from analysis. The remaining replicates (9 per sample) were then averaged at each FITC concentration, and the average fluorescence value of the blank was subtracted from all values. Linear regression was then performed for concentrations within the linear range of fluorescence (0-6.25 µM FITC) between the measured fluorescence values in arbitrary units and the concentration of FITC to identify the conversion factor. For each plate reader, excitation, emission, and gain setting, we found a linear conversion factor that was used to correlate arbitrary fluorescence values to MEF (µM FITC).
To characterize ROSALIND reactions, 19 µL of ROSALIND reactions were loaded onto a 384-well, black, optically-clear, flat-bottom plate using a multichannel pipette and measured on a plate reader (Synergy H1, BioTek). Kinetic analysis was performed by reading the plate at 3 min intervals with excitation and emission wavelengths of 472 nm and 507 nm, respectively for four hours at 37oC. PURExpress reactions were quantified and standardized to MEF (µM FITC) as described for ROSALIND reactions, but with excitation and emission wavelengths set to 485 nm and 515 nm, respectively.
We note that no background subtraction was performed when analyzing outputs from any ROSALIND reaction. An example of this standardization procedure is shown in Supplementary Fig. 1.
Municipal and Surface Water Sampling
Municipal water was collected from water fountains in public parks of Paradise, California in October 2019, as the town recovered from the most destructive wildfire in California’s history (Camp Fire, November 2018). This urban fire severely damaged the municipal water system in Paradise, resulting in widespread contamination of drinking water pipes. After the fire, a Water Quality Advisory (WQA) was issued, asking the public not to drink municipal water. With the WQA still in effect at the time of sampling, public water fountains had likely remained unflushed for ~11 months. Water samples were collected from self-standing water spigots (brass or stainless steel) in metal free plastic tubes and filtered (0.45 µm nylon membranes) in the field using acid-washed plastic syringes. 50-mL filtered subsamples were immediately acidified to pH ~2 with HNO3 for metal analysis (FAAS) and kept cold (4 °C) with 50-mL filtered, unacidified subsamples until their return to California State University, Chico, where ROSALIND tests were initiated within four hours of collection using filtered, unacidified subsamples.
Tests were initiated by applying 20 µL of filtered unacidified samples to reaction tubes, or lab-grade, metal-free water (ultrapure Type 1 water, resistivity > 18 MΩ-cm) as indicated. Reactions were incubated in a portable incubator at 37oC and periodically monitored. Photographs were taken of reaction tubes after two hours and overnight using the handheld illuminator.
Reaction strips and water samples were shipped back to Northwestern University for fluorescence measurements by plate reader (Plate reader quantification and micromolar equivalent fluorescein (MEF) standardization, above) and metals analysis (Flame atomic absorption spectroscopy measurement, below).
For Chilean environmental samples, river water samples were taken from the Yerba Loca Creek in the upper Mapocho-Maipo watershed, and the Toro River in the upper Elqui River watersheds. Both streams are metal-polluted tributaries to rivers that provide ecosystem services (e.g., drinking water sources, habitat for ecosystems, recreation) to major Chilean cities. Acid-washed HDPP bottles were used to collect 200 mL of filtered (0.45 µm nylon membranes) samples and shipped in a cooler. Given the low pH (2.9 < pH < 4) and the objective of the experiments, pH was not adjusted.
For copper / zinc spiked municipal water from Evanston, IL, two bottles of approximately 250 mL of the water samples were collected from a drinking fountain. One of the bottles was then filtered at 0.45 µm using 47 mm cellulose acetate filter membranes (Thermo Fisher). The other was spiked with a known amount of CuSO4 / ZnSO4 and then filtered using the same method. Then, the copper / zinc spiked fountain water was diluted with the filtered non-spiked fountain water to create 40 mL of solutions in the range of 0.125 – 4 ppm for copper and 0.05 – 3 ppm for zinc. 10 mL of these water samples were then acidified with HNO3 and measured using FAAS for concentration validation (Flame atomic absorption spectroscopy measurement, below). 20 µL of the water samples were used to rehydrate freeze-dried CsoR, SmtB, and copper NIMPLY zinc sensors. Upon rehydration, fluorescence measurements of the reactions were performed by plate reader (Plate reader quantification and micromolar equivalent fluorescein (MEF) standardization, above). For copper spiked Michigan water sample, the same method described above was used with the Lake Michigan water sample.
Flame atomic absorption spectroscopy measurements
A Flame Atomic Absorption Spectroscopy instrument (FAAS, PerkinElmer PinAAcle 500) equipped with PerkinElmer Lumina™ hallow-cathode lamps was used to determine copper and zinc concentrations in the drinking water spiked with copper and zinc (Supplementary Fig. 6), lake water spiked with copper, the Chilean surface water samples (Extended Data Fig. 10) and the Paradise municipal water samples (Fig. 6, Supplementary Fig. 7). The spectral lines were 324.8 nm for copper and 213.9 nm for zinc. FAAS was calibrated for Cu(II) using CuNO3 reference solution as standard (Fisher Scientific, CAS10031-43-3) and for Zn(II) using certified ISO Guide 34 standard solution (Fluka Analytical, #18562), with calibration curves comprised between 0.05 and 2 ppm for copper and 0.05 and 1 ppm for zinc. Samples were diluted with 18 MΩ-cm deionized water (Millipore), when necessary, and acidified with ultra-pure HNO3 (Fisher Chemical, CAS7697-37-2). Three replicates were performed and averaged, and error propagations were taken into account to estimate uncertainties.
Photographs of reactions
Photographs of reaction tubes in Fig. 2 and Fig. 6 were taken on an Apple iPhone X and Google Pixel 3a using their default camera applications, respectively. Photographs in Extended Data Fig. 8 were taken on an Apple iPhone 6S using the default Apple iOS Camera application. The resulting images were only cropped. Source image files are available as Supplementary Data File 1.
Statistics and reproducibility
The number of replicates and types of replicates performed are described in the legend to each figure. Individual data points are shown, and where relevant, the average ± standard deviation is shown; this information is provided in each figure legend. The type of statistical analysis performed in Fig. 5 and Extended Data Fig. 4 is described in the legend to each figure. Exact p-values along with degrees of freedom computed from the statistical analysis can be found in Supplementary Data File 3.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
DATA AVAILABILITY
All data presented in this manuscript are available as supplementary data files. All plasmids used in this manuscript are available in Addgene with the identifiers 140371-140399.
CODE AVAILABILITY
No custom software code was used in this manuscript.
Extended Data
Extended Data Fig. 1 |.
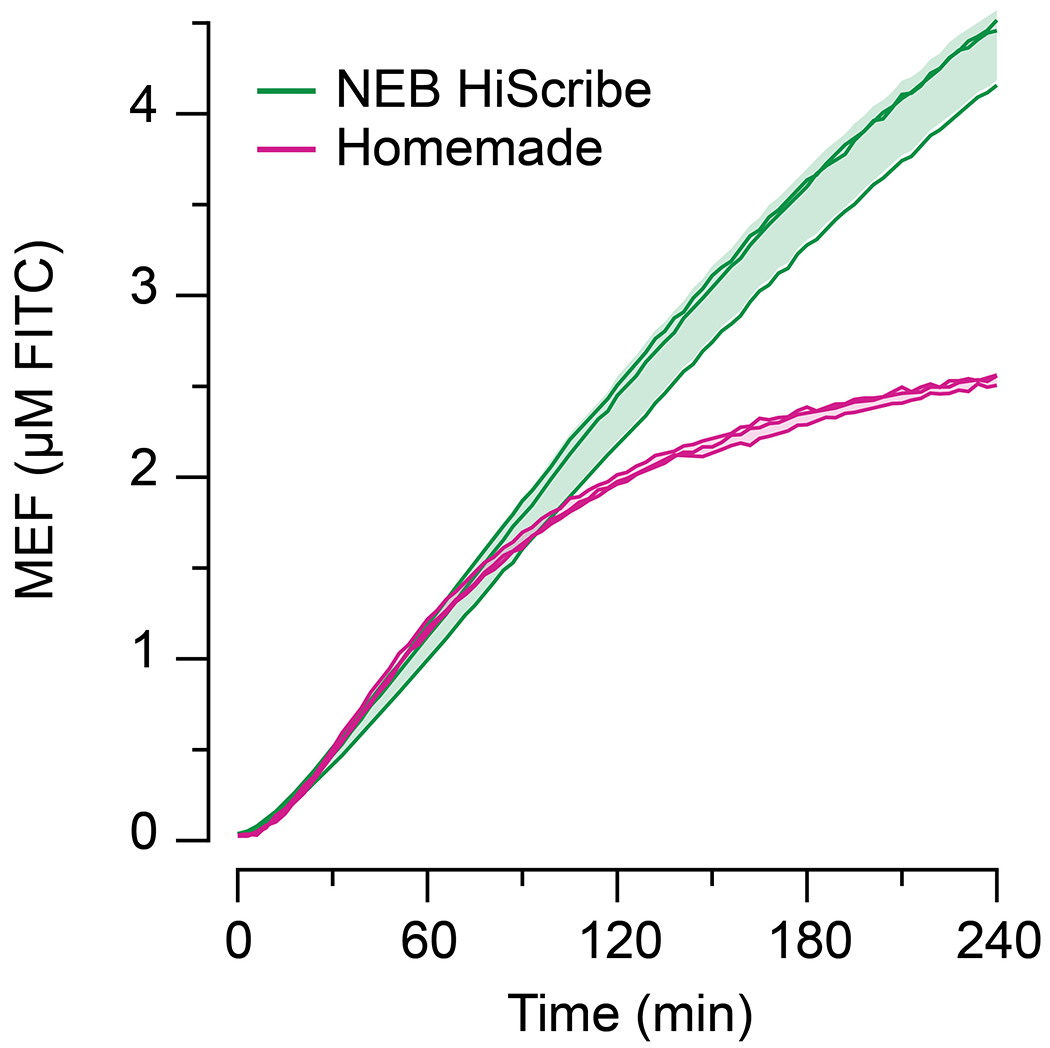
Homemade in vitro transcription reactions were compared to a commercially available high yield transcription kit (NEB HiScribe™ T7 Quick High Yield RNA Synthesis Kit). 25 nM DNA encoding T7-3WJdB-T was added to each reaction in a total reaction volume of 20 µL. Over the course of 1 hour, the data show similar fluorescence activation. However, the homemade reaction begins to saturate after 1 hour, likely due to the exhaustion of NTPs in the homemade reaction (11.4 mM) when compared to the commercial kit (40 mM). All data shown for n=3 independent biological replicates as lines with raw fluorescence values standardized to MEF (µM FITC). Shading indicates the average value of 3 independent biological replicates ± standard deviation.
Extended Data Fig. 2 |.
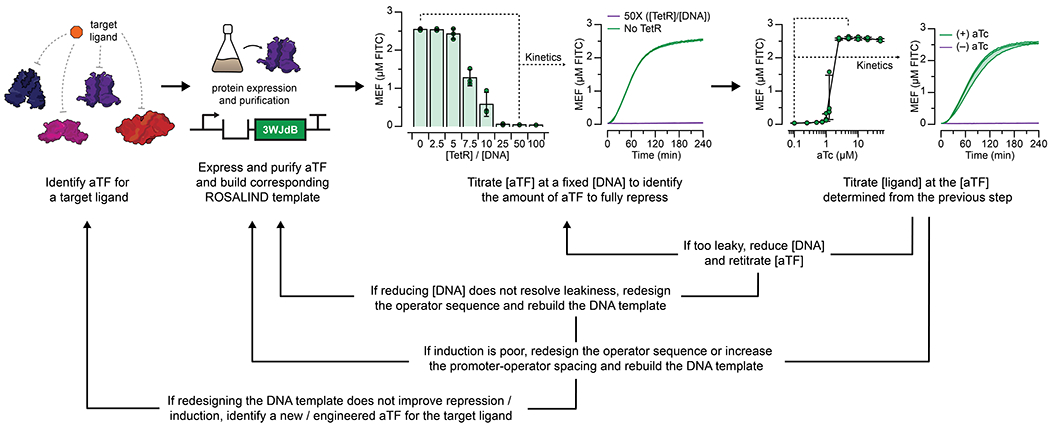
Constructing a ROSALIND reaction begins with identifying a ligand of interest, an aTF capable of binding or unbinding a DNA operator sequence as a function of the ligand, and the aTF’s cognate operator sequence. Once identified, two DNA constructs are designed, one to separately express and purify the aTF, and the other to generate a linear transcription template encoding the promoter, spacer, operator, fluorescent RNA aptamer and optional terminator. The first step in optimizing the sensor is performing a titration of the purified aTF at a fixed DNA template concentration to determine the amount of aTF needed to fully repress the expression of 3WJdB. Then, the target ligand is titrated (at the concentration of aTF previously determined) to test for induction. If the sensor needs improvement, components of ROSALIND can be redesigned and retested. The data shown here are for the TetR-based ROSALIND reaction described in Fig. 2f,h. The bar graphs shown are data taken at four hours after initiating reactions with T7 RNAP and 25 nM DNA template, and the kinetics data are shown for 0 µM (green lines) and 1.25 µM TetR dimer (purple lines). Fluorescence-activation is substantially repressed at 25-fold excess or greater of TetR dimer over DNA template. The TetR-aTc dose response curve shown are the data presented in Fig. 2h, and the kinetics data are shown for 0 µM (purple lines) and 2.5 µM aTc (green lines). All data shown for n=3 independent biological replicates as points or lines with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of the replicates. Shading and error bars indicate the average value of 3 independent biological replicates ± standard deviation.
Extended Data Fig. 3 |.
A dose response curve of a TetR-regulated ROSALIND reaction is plotted from the measured and calibrated kinetics traces, using the four-hour end point fluorescence values at different ligand concentrations. a, kinetic traces of TetR–aTc induction at different aTc concentrations, and b, the corresponding four-hour fluorescence values plotted against the µM of aTc added. Reactions are generated using 25 nM of DNA and 1.25 µM of TetR dimer. All data shown for n=3 independent biological replicates as lines (a) or points (b) with raw fluorescence values standardized to MEF (µM FITC). Shading (a) and error bars (b) indicate the average value of 3 independent biological replicates ± standard deviation.
Extended Data Fig. 4 |.
The dose response curves of ROSALIND reactions shown in Fig. 3 are presented. The amount of DNA templates and aTF used in each panel are configured as described in Supplementary Table 2. All data shown for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC). Error bars indicate the average value of 3 independent biological replicates ± standard deviation. The ligand concentrations at which the signal is distinguishable from the background were determined using two-tailed heteroscedastic t-test against the no ligand condition, and their p-value ranges are indicated with black asterisks (*** < 0.001, ** = 0.001 – 0.01, * = 0.01 – 0.05). The asterisks indicated in red have p-values in the range between 0.05 – 0.15 due to variability between replicates, although the average signals were clearly above background. Exact p-values along with degrees of freedom can be found in Supplementary Data File 3. Data for no ligand conditions were excluded in the plots since the x-axis is on the log scale, but they can be found in Supplementary Data File 3.
Extended Data Fig. 5 |.
The corresponding bar graph data of the orthogonality matrix shown in Fig. 4a are presented. All data shown for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of replicates. Error bars indicate the average value of 3 independent biological replicates ± standard deviation.
Extended Data Fig. 6 |.
a, A kleptamer RNA (KB2) antisense to the dye-binding region of the broccoli aptamer can disrupt folding of 3WJdB and lead to the loss of fluorescence. Addition of a KB2-expressing template in a 4:1 ratio with the 3WJdB template inhibits signal. b, KB2 transcription can be regulated by TetR by placing the tetO site in between the T7 promoter and KB2 coding sequence. In the presence of 1.25 μM TetR dimer, the KB2 transcription is fully repressed, and the signal from 3WJdB is restored. c, Kleptamers can be used to invert the response of transcription factors when KB2 is regulated by TetR (1.25 μM dimer) and induced by aTc (2.5 μM shown). d, This scheme was used to create a ROSALIND zinc sensor with the aporepressor AdcR. When bound to Zn2+ (30 μM), AdcR (1.5 μM dimer) binds to its cognate operator sequence, adcO, placed upstream of the KB2 coding sequence, preventing KB2 expression and thereby activating fluorescence from 3WJdB. Arrows inside of the plots represent direction of regulation when indicated species are added. All data shown for n=3 independent biological replicates as lines with raw fluorescence values standardized to MEF (µM FITC). Shading indicates the average value of 3 independent biological replicates ± standard deviation. 3WJdB template concentrations used are: 25 nM for a-c and 7.5 nM for d. KB2 template concentrations used are: 100 nM for a, and 150 nM for b-d.
Extended Data Fig. 7 |.
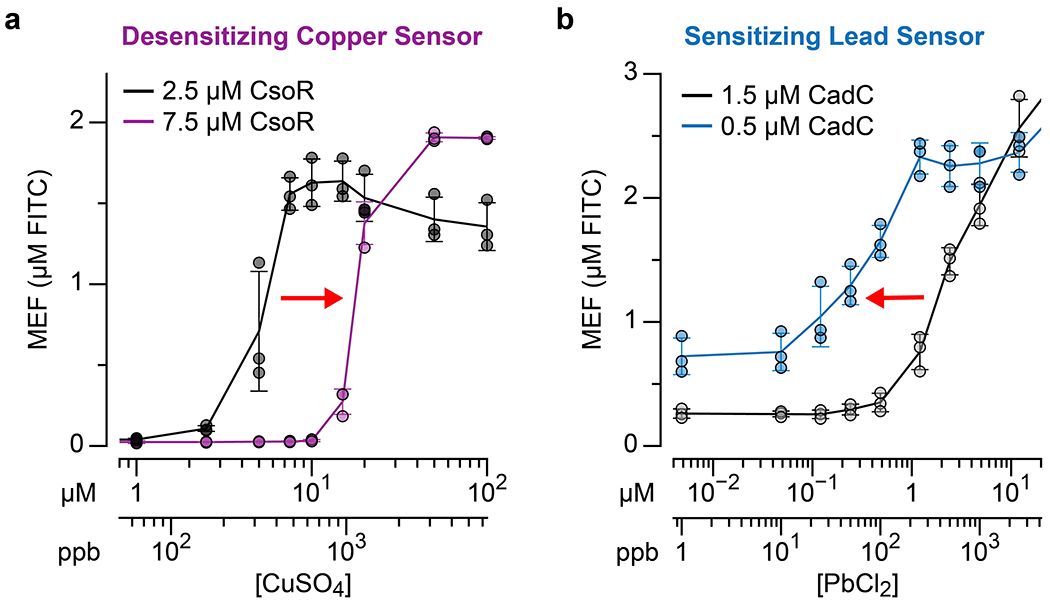
a, increasing the concentration of CsoR desensitizes the copper sensor (dose response shift to higher concentrations), while b, decreasing the concentrations of CadC sensitizes the lead sensor (dose response shift to lower concentrations). The red arrows in the plots indicate the direction of dose response shift. All data shown for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC). Error bars indicate the average value of 3 independent biological replicates ± standard deviation. 3WJdB DNA template concentrations used are: 25 nM for a and 1.5 µM CadC condition in b, and 10 nM for 0.5 µM CadC condition in b.
Extended Data Fig. 8 |.
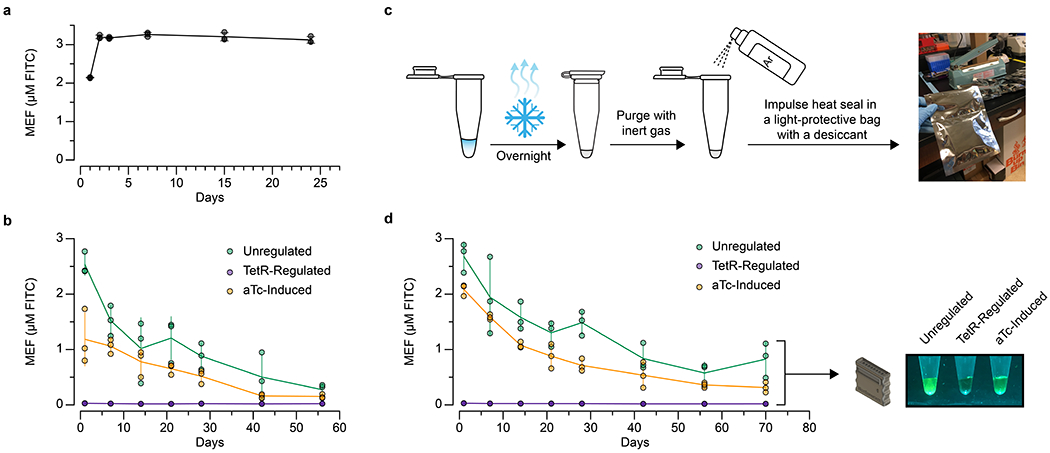
a, The fluorescence signal generated from unregulated ROSALIND reactions are stable over weeks. The increase in fluorescence from Day 1 is likely due to a concentration increase caused by evaporation when the plate was taken out of the incubator after the first measurements. b, The shelf-stability of freeze-dried ROSALIND reactions (unregulated, TetR-regulated, and aTc-induced) decay over the course of a month without proper packaging. c, Packaging of freeze-dried ROSALIND: 1) reactions are lyophilized overnight, 2) the overnight lyophilized reactions are purged with inert gas such as argon, and 3) the reactions are placed into a light-protective bag with a desiccant and immediately impulse heat sealed (Supplementary Video 2). d, When this packaging method is implemented, lyophilized reactions are functional out to 2.5 months. Though we observed signal decay, the signal from rehydrated reactions after 2.5 months is clearly visible. Images are shown for one replicate with other replicate images included in Supplementary Data File 1. Unregulated reactions were lyophilized with 25 nM of the 3WJdB template, and TetR-regulated reactions with additional 1.25 µM TetR dimer along with the components of IVT specified in the In vitro transcription reactions method section. Unregulated and TetR-regulated reactions were then rehydrated with laboratory-grade water, and aTc-induced reactions were rehydrated with 10 µM of aTc. All data shown for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and center values representing averages of the replicates. Error bars indicate the average value of 3 independent biological replicates ± standard deviation. The original, uncropped images shown in c and d can be found in Supplementary Data File 1.
Extended Data Fig. 9 |.
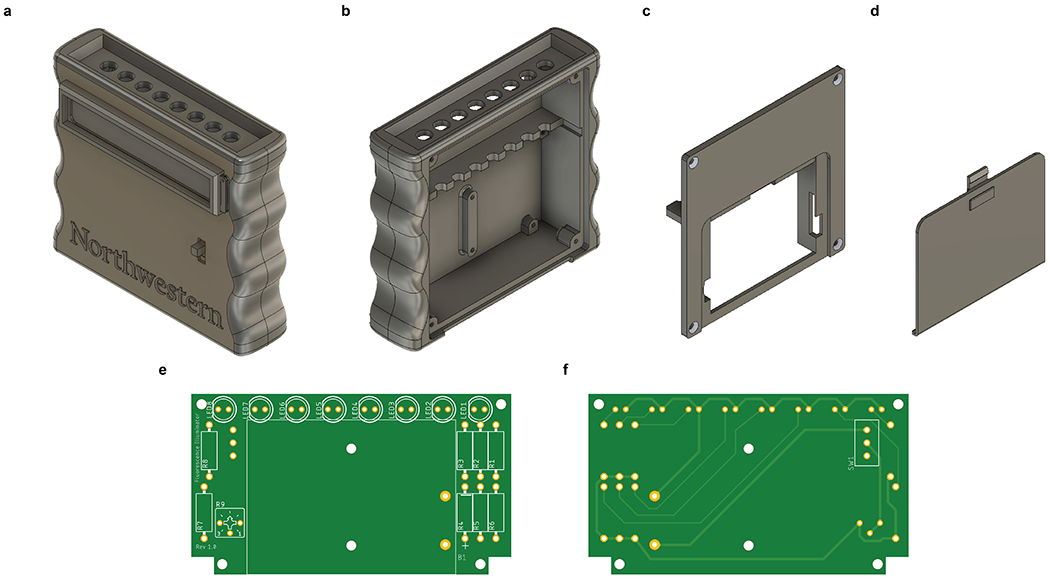
a, A low-cost handheld fluorescence illuminator enables point-of-use functionality for ROSALIND. 3D printed components include a, b, front case, c, back case and d, battery cover. A printed circuit board (e, front view facing back case, f, rear view facing front case) mounts in the front case and connects LEDs, resistors, a trimmer potentiometer, a power switch and a battery holder. Full CAD files, 3D print files (.STL), PCB design files, and assembly instructions are provided in the Supplementary Data File 4. The estimated cost of building one device is $8.69 USD.
Extended Data Fig. 10 |.
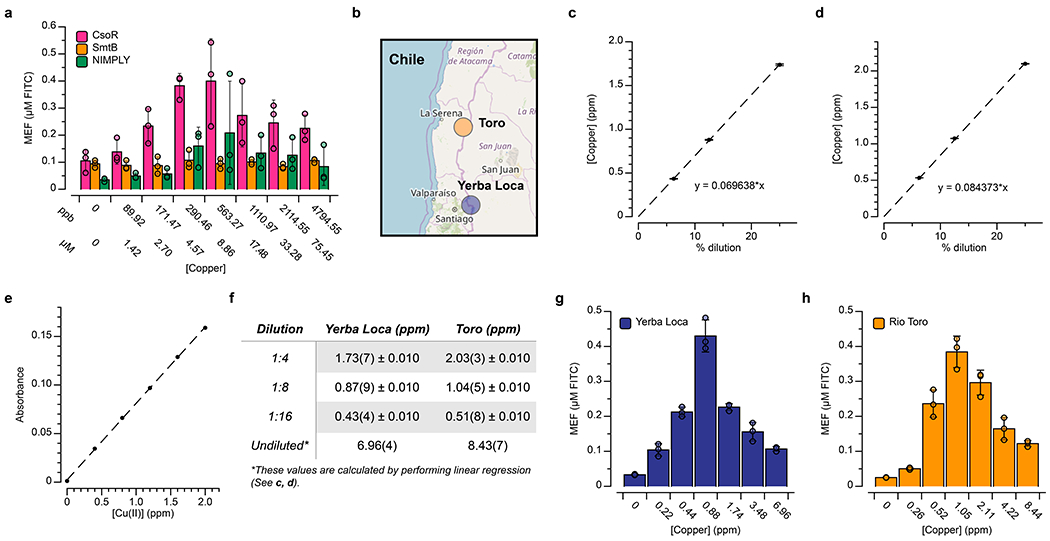
a, The ROSALIND reactions shown in Fig. 6 were characterized for response to CuSO4 as in Supplementary Fig. 6, but using water from Lake Michigan (collected in Evanston, Illinois). We observed lower signals from these spiked environmental samples than with the lab-grade water samples due to uncharacterized matrix effects. b, To test ROSALIND functionality on real-world environmental samples, surface freshwater samples were obtained from two streams known to contain elevated levels of copper: (1) the Yerba Loca Creek, upstream from Santiago, in the Maipo watershed of Central Chile, and (2) the Toro River, upstream from La Serena, in the Elqui watershed of Northern Chile. c, d, Copper concentrations were determined using Flame Atomic Absorption Spectroscopy (FAAS) and found to be in the range of 6.9 – 8.5 ppm. FAAS was calibrated with a Cu(NO3)2 standard solution as shown in e. Serial dilution was performed on each sample to create three separate diluted samples that are in the operating ppm range of FAAS, and their ppm values were measured and reported in Table f as the average value of 3 independent replicates ± standard deviations. Using these measurements, linear regression on the averages of each dilution was performed to calculate the ppm value of the undiluted sample. g, h, In order to generate a series of tests that cover our detectable copper range, we serially diluted each sample with lab-grade, metal-free water (undiluted, 1:2, 1:4, 1:8, 1:16 and 1:32 dilutions) and measured their concentrations by FAAS before using this series to rehydrate freeze-dried copper sensors. The copper concentrations indicated were taken from c, d, or calculated from the extrapolated ppm value of the undiluted sample in Table f. When rehydrated with the 1:8 dilution, we observed clear visible signals, corresponding to copper concentrations of 0.88 ppm and 1.05 ppm from the Yerba Loca Creek and Toro River samples, respectively. Four-hour end-point data are shown in a, g, h for n=3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of the replicates. Error bars indicate the average value of 3 independent biological replicates ± standard deviation. Geographical data in b © OpenStreetMap contributors46
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Zhaoxun Yang (Northwestern University) for assistance with FAAS measurements; Nina Donghia (Wyss Institute) for helpful discussions on lyophilization of cell-free reactions; Sergii Pshenychny (Recombinant Protein Production Core at Northwestern University) for assistance in protein purification; Jackson Webster, Robert Gruenberg, John Machado and Erica Plasencia-Campos (CSU, Chico) for assisting field work; and John Bussan, Frank Lantz and Bob Golenia (Northwestern University Research Shop) for assistance in the development of the handheld illuminator. J.K.J. was supported in part by Northwestern University’s Graduate School Cluster in Biotechnology, System, and Synthetic Biology, which is affiliated with the Biotechnology Training Program. P.R.C. was supported in part by the NSF Synthetic Biology REU (SynBREU) (DBI-1757973). This work was also supported by funding from the Pew Charitable Trusts (to D.A.C.), NIH R35 GM118157 (to D.P.G.), CONICYT/FONDECYT 1161337 (to P.A.P.), ANID/FONDAP/15110020 (to P.A.P.), NSF CAREER (1452441 to J.B.L.), NSF MCB RAPID (1929912 to J.B.L. and J.-F.G), support from the Crown Family Center for Jewish and Israel Studies at Northwestern University (to J. B. L.), and Searle Funds at The Chicago Community Trust (to J.B.L.).
Footnotes
COMPETING INTERESTS STATEMENT
K.K.A., J.K.J. & J.B.L. have submitted a US provisional patent application (No. 62/758,242) relating to regulated in vitro transcription reactions. K.K.A., J.K.J., M.S.V., P.R.C., J.W.L., J.J.C. & J.B.L. have submitted a US provisional patent application (No. 62/838,852) relating to the preservation and stabilization of in vitro transcription reactions. K.K.A. & J.B.L. are founders and have financial interest in Stemloop, Inc. The latter interests are reviewed and managed by Northwestern University in accordance with their conflict of interest policies. All other authors declare no conflicts of interest.
REFERENCES
- 1.French KE, Harnessing synthetic biology for sustainable development. Nature Sustainability, 2019. 2(4): p. 250–252. [Google Scholar]
- 2.Slomovic S, Pardee K, and Collins JJ, Synthetic biology devices for in vitro and in vivo diagnostics. Proc Natl Acad Sci U S A, 2015. 112(47): p. 14429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardee K, et al. , Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell, 2016. 165(5): p. 1255–1266. [DOI] [PubMed] [Google Scholar]
- 4.Gootenberg JS, et al. , Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, 2018. 360(6387): p. 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JS, et al. , CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 2018. 360(6387): p. 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi MK, et al. , A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat Commun, 2018. 9(1): p. 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vorosmarty CJ, et al. , Global threats to human water security and river biodiversity. Nature, 2010. 467(7315): p. 555–61. [DOI] [PubMed] [Google Scholar]
- 8.van der Meer JR and Belkin S, Where microbiology meets microengineering: design and applications of reporter bacteria. Nature Reviews Microbiology, 2010. 8(7): p. 511–522. [DOI] [PubMed] [Google Scholar]
- 9.McNerney MP, et al. , Active Analyte Import Improves the Dynamic Range and Sensitivity of a Vitamin B12 Biosensor. ACS Synth Biol, 2020. 9(2): p. 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belkin S, et al. , Remote detection of buried landmines using a bacterial sensor. Nat Biotechnol, 2017. 35(4): p. 308–310. [DOI] [PubMed] [Google Scholar]
- 11.Siegal-Gaskins D, et al. , Gene circuit performance characterization and resource usage in a cell-free “breadboard”. ACS Synth Biol, 2014. 3(6): p. 416–25. [DOI] [PubMed] [Google Scholar]
- 12.Alam KK, et al. , A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly In Vivo. ACS Synth Biol, 2017. 6(9): p. 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, et al. , Single-molecule tracking of the transcription cycle by sub-second RNA detection. Elife, 2014. 3: p. e01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedelacq JD, et al. , Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol, 2006. 24(1): p. 79–88. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu Y, et al. , Cell-free translation reconstituted with purified components. Nat Biotechnol, 2001. 19(8): p. 751–5. [DOI] [PubMed] [Google Scholar]
- 16.Cuthbertson L and Nodwell JR, The TetR family of regulators. Microbiol Mol Biol Rev, 2013. 77(3): p. 440–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn JJ and Studier FW, Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol, 1983. 166(4): p. 477–535. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Lopez R, et al. , Transcription factor-based biosensors enlightened by the analyte. Front Microbiol, 2015. 6: p. 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baquero F, Martinez JL, and Canton R, Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol, 2008. 19(3): p. 260–5. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, et al. , Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol, 2006. 72(4): p. 2573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deochand DK and Grove A, MarR family transcription factors: dynamic variations on a common scaffold. Crit Rev Biochem Mol Biol, 2017. 52(6): p. 595–613. [DOI] [PubMed] [Google Scholar]
- 22.Zhu T, et al. , Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab Eng, 2013. 19: p. 69–78. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi N, et al. , Regulation of transcription of the mph(A) gene for macrolide 2'-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(A). J Bacteriol, 2000. 182(18): p. 5052–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson SD and Ternes TA, Water Analysis: Emerging Contaminants and Current Issues. Anal Chem, 2018. 90(1): p. 398–428. [DOI] [PubMed] [Google Scholar]
- 25.Kasey CM, et al. , Development of Transcription Factor-Based Designer Macrolide Biosensors for Metabolic Engineering and Synthetic Biology. ACS Synth Biol, 2018. 7(1): p. 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carballa M, et al. , Behavior of pharmaceuticals, cosmetics and hormones in a sewage treatment plant. Water Res, 2004. 38(12): p. 2918–26. [DOI] [PubMed] [Google Scholar]
- 27.Hiromoto T, et al. , Characterization of MobR, the 3-hydroxybenzoate-responsive transcriptional regulator for the 3-hydroxybenzoate hydroxylase gene of Comamonas testosteroni KH122–3s. J Mol Biol, 2006. 364(5): p. 863–77. [DOI] [PubMed] [Google Scholar]
- 28.Kim M, et al. , Widely Used Benzalkonium Chloride Disinfectants Can Promote Antibiotic Resistance. Applied and Environmental Microbiology, 2018. 84(17): p. e01201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grkovic S, et al. , QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem, 1998. 273(29): p. 18665–73. [DOI] [PubMed] [Google Scholar]
- 30.Lu WJ, et al. , Enantiomers of naringenin as pleiotropic, stereoselective inhibitors of cytochrome P450 isoforms. Chirality, 2011. 23(10): p. 891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers JK, et al. , Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res, 2015. 43(15): p. 7648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, et al. , De novo design of programmable inducible promoters. Nucleic Acids Research, 2019. 47(19): p. 10452–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juraschek SP, et al. , Gout, Urate-Lowering Therapy, and Uric Acid Levels Among Adults in the United States. Arthritis Care & Research, 2015. 67(4): p. 588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson SP and Grove A, HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J Biol Chem, 2004. 279(49): p. 51442–50. [DOI] [PubMed] [Google Scholar]
- 35.Abraham J, Dowling K, and Florentine S, Risk of post-fire metal mobilization into surface water resources: A review. Science of the Total Environment, 2017. 599: p. 1740–1755. [DOI] [PubMed] [Google Scholar]
- 36.Hanna-Attisha M, et al. , Elevated Blood Lead Levels in Children Associated With the Flint Drinking Water Crisis: A Spatial Analysis of Risk and Public Health Response. American Journal of Public Health, 2016. 106(2): p. 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calle GR, et al. , Enhanced copper release from pipes by alternating stagnation and flow events. Environ Sci Technol, 2007. 41(21): p. 7430–6. [DOI] [PubMed] [Google Scholar]
- 38.VanZile ML, Chen X, and Giedroc DP, Allosteric negative regulation of smt O/P binding of the zinc sensor, SmtB, by metal ions: a coupled equilibrium analysis. Biochemistry, 2002. 41(31): p. 9776–86. [DOI] [PubMed] [Google Scholar]
- 39.Chang FM, et al. , Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J Biol Chem, 2014. 289(27): p. 19204–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busenlehner LS, et al. , Spectroscopic properties of the metalloregulatory Cd(II) and Pb(II) sites of S. aureus pI258 CadC. Biochemistry, 2001. 40(14): p. 4426–36. [DOI] [PubMed] [Google Scholar]
- 41.Lloyd J, et al. , Dynamic Control of Aptamer-Ligand Activity Using Strand Displacement Reactions. ACS Synth Biol, 2018. 7(1): p. 30–37. [DOI] [PubMed] [Google Scholar]
- 42.Reyes-Caballero H, et al. , The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol, 2010. 403(2): p. 197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danner MC, et al. , Antibiotic pollution in surface fresh waters: Occurrence and effects. Sci Total Environ, 2019. 664: p. 793–804. [DOI] [PubMed] [Google Scholar]
- 44.Steber M, et al. , Mechanistic basis for RNA aptamer-based induction of TetR. Chembiochem, 2011. 12(17): p. 2608–14. [DOI] [PubMed] [Google Scholar]
- 45.Pardee K, et al. , Paper-based synthetic gene networks. Cell, 2014. 159(4): p. 940–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haklay M.a.W. P., Openstreetmap: UserGenerated Street Maps. IEEE Pervasive Computing, 2008. 7(4): p. 12–18. [Google Scholar]
- 47.Vega AS, Lizama K, and Pastén PA, Water Quality: Trends and Challenges, in Water Policy in Chile, Donoso G, Editor. 2018, Springer International Publishing: Cham: p. 25–51. [Google Scholar]
- 48.Chen X, et al. , Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat Biotechnol, 2019. 37(11): p. 1287–1293. [DOI] [PubMed] [Google Scholar]
- 49.Ellefson JW, Ledbetter MP, and Ellington AD, Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nat Chem Biol, 2018. 14(4): p. 361–367. [DOI] [PubMed] [Google Scholar]
- 50.McNerney MP, et al. , Point-of-care biomarker quantification enabled by sample-specific calibration. Sci Adv, 2019. 5(9): p. eaax4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 51.Alam KK, et al. , A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly In Vivo. ACS Synth Biol, 2017. 6(9): p. 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lloyd J, et al. , Dynamic Control of Aptamer-Ligand Activity Using Strand Displacement Reactions. ACS Synth Biol, 2018. 7(1): p. 30–37. [DOI] [PubMed] [Google Scholar]
- 53.Steber M, et al. , Mechanistic basis for RNA aptamer-based induction of TetR. Chembiochem, 2011. 12(17): p. 2608–14. [DOI] [PubMed] [Google Scholar]
- 54.Capdevila DA, et al. , Tuning site-specific dynamics to drive allosteric activation in a pneumococcal zinc uptake regulator. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this manuscript are available as supplementary data files. All plasmids used in this manuscript are available in Addgene with the identifiers 140371-140399.


