Abstract
High-dose chemotherapy with melphalan followed by autologous transplantation is a first-line treatment for multiple myeloma. Here, we present pre-clinical evidence that this treatment may be significantly improved by the addition of exportin 1 inhibitors (XPO1i). The XPO1i selinexor, eltanexor, and KOS-2464 sensitized human MM cells to melphalan. Human 8226 and U266 MM cell lines and melphalan-resistant cell lines (8226-LR5 and U266-LR6) were highly sensitized to melphalan by XPO1i. MM cells from newly diagnosed and relapsed/refractory MM patients were also sensitized by XPO1i to melphalan. In NOD/SCID-γ mice challenged with either parental 8226 or U266 MM and melphalan-resistant MM tumors, XPO1i/melphalan combination treatments demonstrated stronger synergistic anti-tumor effects than single-agent melphalan with minimal toxicity. Synergistic cell death resulted from increased XPO1i/melphalan-induced DNA damage in a dose-dependent manner and decreased DNA repair. In addition, repair of melphalan-induced DNA damage was inhibited by selinexor, which decreased melphalan-induced monoubiquitination of FANCD2 in MM cells. Knockdown of FANCD2 was found to replicate the effect of selinexor when used with melphalan, increasing DNA damage (γH2AX) by inhibiting DNA repair. Thus, combination therapies that include selinexor or eltanexor with melphalan may have the potential to improve treatment outcomes of MM in melphalan-resistant and newly diagnosed patients. The combination of selinexor and melphalan is currently being investigated in the context of high-dose chemotherapy and autologous transplant (NCT02780609).
Keywords: Exportin 1 inhibitors, melphalan, preclinical studies, multiple myeloma, drug resistance
Introduction
Multiple myeloma (MM) accounts for about 10% of all hematologic malignancies (1). Significant improvements in response/survival have been seen over the past several years, with the current 5-year survival rate at approximately 50% (2). The US Food and Drug Administration has approved 6 novel therapies for the treatment of relapsed/refractory MM since 2015. These include the nuclear export inhibitor selinexor (SEL) (3), proteasome inhibitors carfilzomib (4) and ixazomib (5), monoclonal antibodies daratumumab (6) and elotuzumab (7), and the histone deacetylase inhibitor panobinostat (8). The introduction of these new agents may further increase response/survival rates. Improvements seen with these new agents notwithstanding, MM remains incurable and patients ultimately die from progressive disease refractory to anti-myeloma therapy. A common treatment approach for patients with newly diagnosed MM consists of induction chemotherapy for 4 to 6 cycles with some combination of an immunomodulator/proteasome inhibitor/dexamethasone (DEX)/alkylating agent, followed by high-dose melphalan (HD MEL) and autologous hematopoietic stem cell transplant (HSCT). Many patients do well with this treatment approach and remain progression free for several years. However, a subset of patients (often defined by specific high-risk characteristics) (9) relapse soon after HD chemotherapy. The achievement of complete response (CR), before or after HD MEL, has been associated with improved outcomes; thus, increasing the rate of CR post-HSCT (among patients who have not had a CR pre-HSCT) by adding a new agent is an appropriate goal.
MEL is a bifunctional alkylating agent that produces DNA interstrand crosslinks (ICLs) and is a widely used and effective treatment for MM (10). HD MEL is the most commonly used preparative regimen before autologous HSCT. MEL-induced ICLs are repaired in non-replicating cells by the nucleotide excision repair machinery and by the DNA translocase FANCM (11). FANCM, together with additional Fanconi anemia (FA) proteins, senses the ICL lesion and binds to the chromatin where it acts as a landing platform for the FA core complex (12). This FA core complex is recruited to the sites of DNA damage and consists of 14 proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, FANCM, FANCT, FAAP100, MHF1, MHF2, FAAP20 and FAAP24). The FA core complex physically catalyzes the conjugation of an ubiquitin moiety to FANCD2 and FANCI proteins which dimerize, thereby activating their DNA repair function. Monoubiquitination of FANCD2 and FANCI is catalyzed by the core complex through its ubiquitin ligase subunit FANCL. The ubiquitinated FANCD2-FANCI complex is loaded on to the chromatin at the site of the ICL and produces a nucleolytic incision at the converged replication forks to release the ICL from one of the two parental strands, (referred to as “unhooking”) thus initiating the repair event. Although there are several DNA repair pathways (normal and deregulated) that are operational in human MM, including base excision repair, nucleotide excision repair, mismatch repair, non-homologous end joining, homologous recombination, and single-strand annealing, the FA pathway is critical to MEL-induced ICL repair and in the resistance of MM cells to MEL (11–14).
Exportin 1 (XPO1), a nuclear protein that exports >250 proteins with specific nuclear export signals from the nucleus to the cytoplasm, is overexpressed 2- to 4-fold in several human malignancies, including MM (15). Overexpression frequently correlates with poor prognosis and/or reduced survival, suggesting that XPO1 could have a direct role in the etiology of the malignant phenotype (15). A new class of XPO1 inhibitors (XPO1i) has been introduced by Karyopharm Therapeutics and includes selinexor and eltanexor (Figure S1). Investigators have demonstrated pre-clinical activity with these agents in solid tumors (16–24) and hematologic malignancies (25–28), including MM (29–31). KOS-2464 is a semisynthetic derivative of leptomycin B and has been shown to be a very potent inhibitor of XPO1 (in the nanomolar range). This compound was used in our studies to show that structurally unrelated XPO1i produced similar anti-myeloma results when used in combination with MEL (32).
In recent years several clinical trials combining SEL or eltanexor (ELT) with other MM drugs have been conducted. Clinical trials in our research group include an ongoing Phase 1/2 study (NCT02186834) with SEL and pegylated liposomal doxorubicin (PLD) (33) and an ongoing Phase 1/2 trial (NCT02780609) combining SEL and MEL in the context of high-dose chemotherapy and autologous transplant (34). The SEL/PLD trial showed a 20% response rate in penta-refractory patients. The SEL/MEL trial with HSCT treatment was found to be well-tolerated and engraftment kinetics were not altered. The phase 2 arm of this study with SEL (80 mg) with high-dose MEL and autologous HSCT is currently ongoing. Additional combination studies with SEL/DEX/carfilzomib (35), SEL/DEX/bortezomib (36) and SEL/DEX/pomalidomide (37) have each shown promising effects using these drug combinations in relapsed/refractory MM (RRMM). The most important trial to date in terms of clinical relevance was the STORM clinical trial which led to accelerated FDA approval of SEL in RRMM patients (3). A total of 122 patients that were triple-class refractory to MM treatments were enrolled. Patients were treated with 80 mg SEL and 20 mg dexamethasone twice weekly. A partial response or better was observed in 26% of patients. In addition to the SEL clinical studies, a phase 1/2 study using the second generation XPO1i ELT was performed in combination with DEX in patients with quad- or penta-refractory MM (38). This combination was given to patients five times weekly without reaching a maximum tolerated dose, thus indicating enhanced tolerability in patients treated with eltanexor.
In previous studies, we have shown that XPO1i significantly sensitized parental and drug-resistant MM cells to doxorubicin and proteasome inhibitors (31, 39–44). In the current study we have demonstrated that XPO1i sensitize human MM cells to MEL, re-sensitize MEL-resistant 8226/LR5 and U266/LR6 cells to MEL, and that this sensitization to MEL likely involves changes in expression of proteins in the FA pathway. Our central hypothesis is that molecules that disrupt the binding of XPO1 to the nuclear export signals (NES) on proteins in human MM significantly sensitize MM cells to MEL by altering the formation/repair of MEL-induced DNA ICLs.
Materials and Methods
For human MM sample acquisition, written informed consent approved by an Institutional Review Board was obtained from all patients, in accordance with the Declaration of Helsinki. Patient samples were de-identified and obtained through the Institutional Review Board–approved Total Cancer Care protocol at the H. Lee Moffitt Cancer Center and Research Institute.
Cell lines
RPMI8226, U266, and NCI-H929 human MM cell lines were obtained from the American Type Culture Collection (ATCC). MEL-resistant 8226-LR5 and U266-LR6 cells were provided by Dr. William Dalton at the Moffitt Cancer Center (13, 45). These drug-resistant cells were produced by incrementally increasing their exposure to MEL; cells were passaged routinely in media containing 5 μM (8226-LR5) or 6 μM (U266-LR6) MEL to maintain resistance. All cell lines were grown in RPMI1640 media (Corning/Cellgro) containing 10% fetal bovine serum (Sigma) and kept at 37°C in 5% CO2. H929 cell media required the addition of 0.025% β-mercaptoethanol (Sigma). All cell lines were authenticated and assayed for mycoplasma contamination by the Moffitt Genomics Core Facility in accordance with ATCC guidelines (46). Peripheral blood mononuclear cells (PBMC) were obtained from normal donors (Florida Blood Services).
MM cell lines treated with XPO1i and MEL
Human parental MM 8226, H929, and U226 and MEL-resistant 8226-LR5 and U266-LR6 cells were treated with 300 nM SEL (Karyopharm Therapeutics), 300 nM ELT (Karyopharm Therapeutics), or 10 nM KOS-2464 (Bristol-Myers Squibb) concurrently with various concentrations (5, 10, 15, or 20 μM) MEL (Sigma) for 20 hours (see figure S1 for chemical structures of XPO1i). Optimal drug concentrations were determined by titration experiments. Cells were fixed and permeabilized, and apoptosis was measured by using anti-activated caspase 3/A488 (Cell Signal) staining, in accordance with the manufacturer’s standard protocol. Apoptosis percentages were determined by flow cytometry (FACS) on an LSRII (Becton-Dickinson) bench-top analyzer. Data analyses were performed by Flowjo version 9.9.6 software (Tree Star). All experiments were repeated 3–5 times.
Automated in vitro cell viability assay
Combinatorial Index (CI) values were determined in cells treated with MEL combined with SEL and ELT. Both parental U266/8226 cells and U266-LR6/8226-LR5 cells were evaluated by a high-throughput CellTiter-Blue (Promega) cell viability assay, as previously described (47). The CI method developed by Chou and Talalay (48) was used to analyze cell viability assay results for synergistic, additive, and antagonistic effects.
Ex vivo MM patient studies
BD Vacutainer 15 mL CPT tubes (BD Biosciences) were used to isolate mononuclear cells from bone marrow aspirates collected from newly diagnosed (n = 20) and relapsed/refractory (n = 11) MM patients. Mononuclear cells were incubated at 4–8×106 cells/mL in 200 μL of RPMI1640 media (Fisher) containing 10% fetal bovine serum and treated with either SEL (300 nM), ELT (300 nM), or KOS-2464 (10 nM) +/− MEL (10 μM), in 96-well plates. Incubation lasted for 20 hours in a 5% CO2 humidified incubator. The cells were then fixed, and the MM cells identified and assayed for apoptosis by FACS, in accordance with the methods outlined in Turner et al (43).
NSG mouse studies with SEL and MEL
All mouse studies were reviewed and approved by the University of South Florida Institutional Animal Care and Use Committee. Drug-resistant U266-LR6 (5×106) human myeloma cells or parental U266 cells (107) were injected subcutaneously into the flanks of NSG mice, and the resulting tumors were allowed to grow for 14 days before the start of treatment. Each experimental group consisted of 6 mice, 3 males and 3 females. Tumor volumes were plotted against time. Mice were treated (1) once weekly with MEL (3 mg/kg) by intraperitoneal injection, (2) twice weekly (Tuesday and Friday) with SEL (10 mg/kg) or ELT (10 mg/kg) by oral gavage, or (3) the SEL or ELT were given in combination with MEL, in which case, XPO1i administration was followed 1 to 2 hours later by MEL injection. Tumors were measured by calipers, and tumor volumes (mm3) were calculated by measuring length×width2/2. Mice were euthanized when tumor volume exceeded 2000 mm3 or when they lost > 10% of their bodyweight; these criteria were used to define survival. Drug toxicity was assayed by mouse weights, with a decrease of ≥ 10% considered to be an indication of drug regimen toxicity.
For the NSG mouse studies comparing sequential and concurrent drug treatment, NSG mice were challenged with subcutaneous flank MM tumors (5×106 cells/injection) and treated with either single-agent SEL (10 mg/kg) or MEL (3 mg/kg) as above, or they received either sequential treatment with SEL followed 2 hours later by MEL or concurrent treatment with SEL and MEL. Each experimental group consisted of 6 mice, 3 males and 3 females.
For the studies of NSG mice challenged with human MM tumors and treated with SEL, MEL, and DEX, NSG mice were challenged with 8226 and U266 human MM tumors as described above and treated with a triple-drug combination of SEL/MEL/DEX, which is similar to the combination used in our current clinical trial (NCT02780609). The 3-drug combination was compared to single-agent SEL (5 mg/kg), MEL (1 mg/kg), DEX (5 mg/kg), or combinations of MEL/DEX, SEL/MEL, or SEL/DEX. Each experimental group consisted of 6 mice, 3 males and 3 females. All drug combinations were given currently twice weekly on days 1 and 4.. Mice were assayed for tumor growth or survival as described previously (31).
Alkaline comet assay
H929 MM cells (3×106/mL) were exposed to SEL (100 nM) for 20 hours followed by MEL (1, 2.5, 5, and 10 μM) for 2 hours. Drugs were removed, and cells were incubated for another 3 hours to allow for crosslink formation. Cells were exposed to 900 rads (X-Rad 160 X-ray biological irradiator), and alkaline comet assays were performed immediately (Trevigen Inc) in the manner described by Chen et al (13). Experiments were repeated 5 times and data were combined and analyzed. These experiments were repeated at lower density cell concentrations (2×105), and incubation times extended for 4, 8, 16, 24, and 48 hours to see how the crosslinks were repaired over time (n = 4).
γH2AX assay and DNA damage repair time course
Human H929 MM cells were treated with SEL (300 nM), ELT (300 nM), or KOS-2464 (10 nM) +/− MEL (2 μM, 5 μM, or 10 μM) and assayed for γH2AX expression (JBW301/ FITC, Millipore) by FACS. 8226, 8226-LR5, U266 and U266-LR6 MM cells were treated for 2 hours with 50 μM MEL or untreated controls. Cells were then pelleted, resuspended in fresh media, and incubated with SEL (300 nM) for 4, 8, 16, 24, and 48 hours. Cells were fixed, stained with anti-γH2AX and analyzed by FACS for γH2AX.
Western blot
Human U266, 8226, 8226-LR5 and U266-LR6 MM cells (2×105/mL) were treated for 2 hours with 50 μM MEL, pelleted, resuspended in fresh media and incubated with SEL (300 nM) for 8, 16, 24, and 48 hours. Cell protein lysates (100 μg) were loaded on a 3–8% NuPAGE Tris-Acetate gel (Thermo/Fisher) and separated for 4 hours at 90 V. Protein was transferred overnight (30 V at 4ºC) and proteins identified with anti-FANCD2 (FI17/Santa Cruz) or anti-γ-tubulin antibody (GTU-88/Sigma). Mono-ubiquitinated FANCD2 was initially identified as the “long-form” and is a 162 kDa band (7 kDa larger than the 155 kDa non-ubiquitinated “short form”) on the Western blots presented in this paper (49). H929 MM cells (3×106 cells/mL) were treated for 6 hours with 10 μM MEL +/− 300 nM SEL (n=2). Western blots were assayed for FANCF (Novus), FANCL (Santa Cruz), NFĸB (Millipore) and IKKα (Novus). U266 and U266-LR6 cells were incubated with 50 μM MEL for 2 hours, washed and incubated with 0, 50, 100, 500 or 1000 nM SEL for 20 hours and assayed for FANCD2, ubFANCD2 (FI17/Santa Cruz), BRCA1 (NB100–599/Novus Bio), BRAC2 (ab123491/Abcam), RAD51 (D4B10/Cell Signal) and vinculin (4650/Cell Signal) (n=3). Data for Western blots were digitized using iBright Analysis software.
Proximity ligation assay
Log phase 8226 and 8226-LR5 human MM cells were either treated with 50 μM MEL for 2 hours or left untreated. Cells were then removed, incubated with fresh media +/− 300 nM SEL for 24 hours, and assayed for proximity co-localization of FANCD2 and ubiquitin. After treatment, the cells were washed with PBS, and cytoslides were made, with 8×104 cells used for each treatment. Slides were fixed with 4% paraformaldehyde, washed, and stored overnight in PBS at 4°C. The assay was performed by using Duolink in situ proximity ligation assay (Olink Bioscience), as previously described (31). Images were taken with a Leica TCS SP8 acousto-optical beam-splitter laser scanning confocal microscope, through a Plan-Apochromat 63X/1.4NA oil-immersion objective lens (Leica Microsystems). A minimum of 700 cells were assayed for each experimental condition (n=3).
FANCD2 small interfering RNA knockdown
Small interfering RNA (siRNA) duplexes for FANCD2 (cat#SR301519) and universal scrambled negative control duplexes (cat#SR30004/517–220063241) were obtained from OriGene (Rockville, MD). Three sets of 27-mer siRNA duplexes were used to perform knockdown of FANCD2 gene expression. Briefly, human U266 and U266-LR6 MM cells (5×106) were transfected in 600 μl of Opti-MEM media (ThermoFisher) premixed with 9 μL of Lipofectamine RNAiMAX reagent (ThermoFisher) and 3 μL of each siRNA duplex (10 μM). After being incubated for 48 hours, the cells were treated with 50 μM MEL for 2 hours, washed, and then incubated for a further 48 hours. At the 24- and 48-hour time points, DNA damage was assessed by measuring γH2AX protein expression via FACS analysis.
Statistical analyses
All experiments were performed 3–5 times, and the mean and standard error of the means are shown for each experiment where appropriate. GraphPad Prism 7 and SAS version 9.4 software were used to produce Kaplan-Meier survival plots of animal data and analyses. The difference between survival curves was log-rank test evaluated. Depending on the datasets being analyzed, data were analyzed by using either paired or Welch-Satterthwaite t tests, ANOVA, Dunnett test, or P values adjusted by the Bonferoni method. The pairwise comparisons for the experiments with ≥3 groups were made by applying Tukey’s method. The difference in linear trend between groups is assessed by the linear mixed effect model. IC50 values were calculated using a sigmoidal equilibrium model regression with XLfit version 5.2 (ID Business Solutions Ltd.).
Results
In vitro, ex vivo, and in vivo MM studies
Inhibitors of XPO1 sensitize human MM and MEL-resistant cell lines to MEL
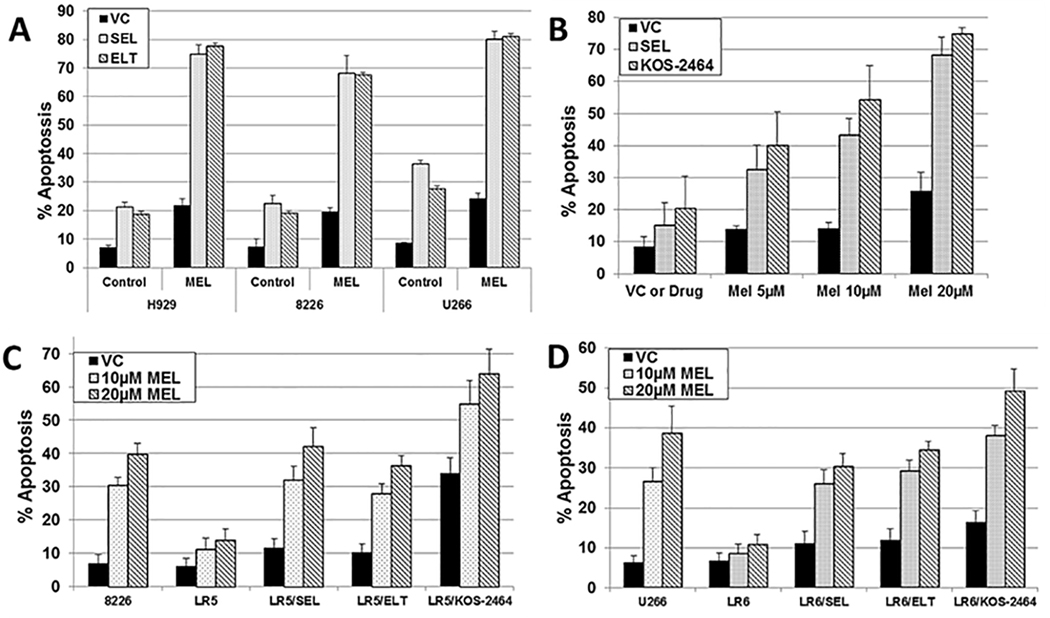
We found that H929, 8226, and U266 human MM cells, treated concurrently with SEL/MEL or ELT/MEL synergistically increased apoptosis (activated caspase 3) (P ≤ .00032 and P ≤ .00031, respectively) in all human MM cell lines tested (Figure 1A). This finding was evidenced by comparisons with the same cell lines treated with single-agent MEL, SEL, or ELT (Figure 1A). 8226 MM cells were also sensitized to MEL by SEL or KOS-2464 in a dose-dependent manner (P ≤ .009 and P ≤ .0001, respectively), as shown by comparative rates of apoptosis (Figure 1B). Normal PBMCs were not affected by in vitro XPO1i/MEL treatment (P ≥ .212) (n = 4). Human 8226/U266 and 8226-LR5/U266-LR6 MM cell lines were 3.6- to 9.5-fold more resistant to single-agent MEL than parental cells. The addition of SEL, ELT, or KOS-2464 significantly sensitized 8226-LR5 cells and U266-LR6 cells to MEL (P < .0001; n=5) (Figure 1C/D).
Fig. 1. Inhibitors of XPO1 sensitize human parental MM cell lines and MEL-resistant cell lines to MEL.
(A) H929 (3 × 106 cells/mL), 8226 (2 × 106 cells/mL), and U266 (4 × 106 cells/mL) human MM cells were treated for 20 hours with SEL (300 nM) or ELT (300 nM) as single agents (control) or the cells were treated with SEL or ELT combined with MEL (15 μM) (n=3). (B) Human 8226 MM cells were treated with SEL (300 nM) or KOS-2464 (10 nM) +/− MEL and assayed for apoptosis (n=3). (C/D) Human 8226 and U266 drug-resistant (8226-LR5 and U266-LR6) and parental MM cell lines were treated for 20 hours with MEL alone (VC) or with SEL (300 nM), ELT (300 nM), or KOS-2464 (10 nM) concurrent with 10 or 20 μM MEL and assayed for apoptosis by flow cytometry (using activated caspase 3) (n = 5).
XPO1 inhibitors sensitize MM cells obtained from newly diagnosed and relapsed/refractory MM patients to MEL
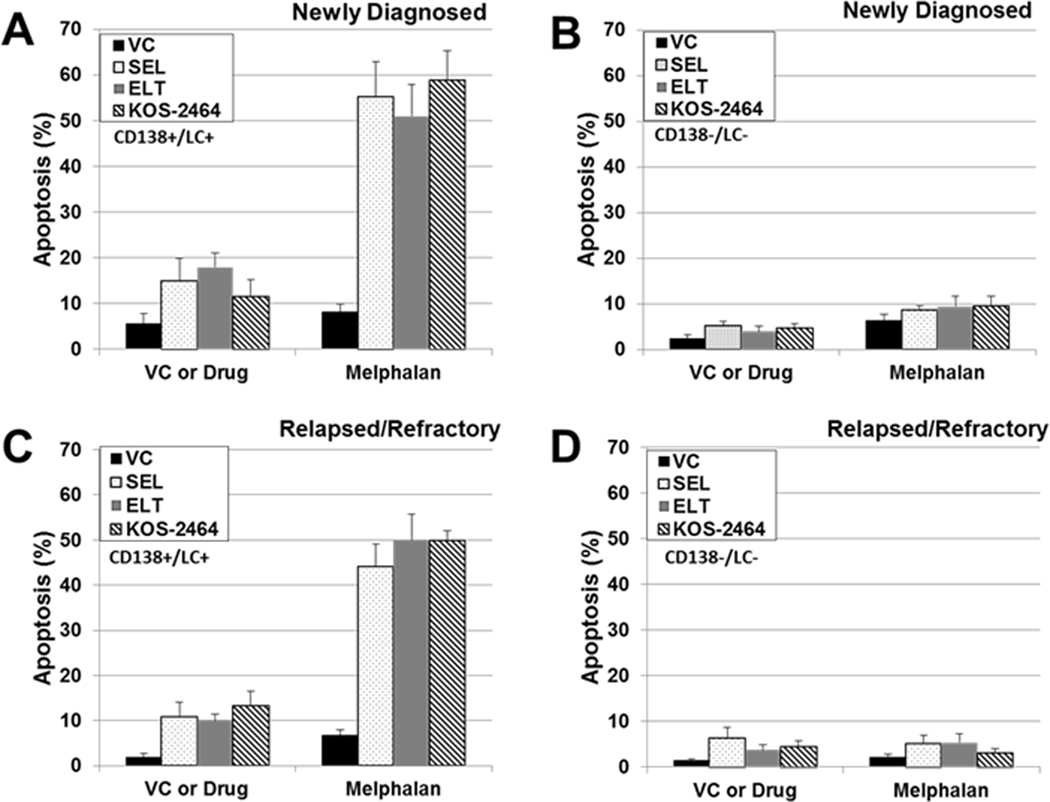
Bone marrow mononuclear cells were isolated from patients with MM and treated with XPO1i +/− MEL. CD138+/LC+ MM cells were gated and assayed for apoptosis (activated caspase 3) by FACS. Newly diagnosed (Figure 2A) (n = 20) and relapsed/refractory (Figure 2C) (n = 11) MM patient samples (CD138+/LC+) were sensitized by SEL, ELT, and KOS to MEL when compared to single-agent MEL, as shown by increased apoptosis in newly diagnosed (SEL, P < .0001; ELT, P < .0001; and KOS, P < .0001) and relapsed/refractory (SEL, P = .001; ELT, P < .0001; and KOS, P < .0001) patient aspirates. This increase indicates greater sensitization to MEL with the combination treatment. Non-myeloma CD138-/LC- patient cells were not sensitized by XPO1i (Figure 2, B and D). Table S1 shows the last treatments that the relapsed/refractory patients received before bone marrow aspirates were obtained. Seventy-three percent of relapsed/refractory patients had received HD chemotherapy with MEL and autologous stem cell rescue during the course of their treatments.
Fig. 2. SEL, ELT, and KOS sensitize newly diagnosed and relapsed/refractory patient MM cells to MEL.
Bone marrow mononuclear cells from newly diagnosed (n = 20) and relapsed/refractory (n = 11) patients: (A) newly diagnosed CD138+/light chain+ cells; (B) CD138−/light chain− cells; (C) relapsed/refractory CD138+/light chain+ cells; and (D) relapsed/refractory CD138−/light chain− cells. Patient bone marrow mononuclear cells were isolated and treated with SEL (300 nM) or ELT (300 nM) ± MEL (10 μM) for 20 hours. Cells were fluorescently labeled with antibodies against activated caspase 3, CD138, and light chain antigen (LC) (κ or λ) and analyzed by flow cytometry. Supplemental table S2 shows the last treatments that the relapsed/refractory patients received before the bone marrow aspirate was obtained.
Combination index values from human MM cell lines and PBMCs treated with MEL and XPO1i
CI values (Table S2) were determined by CT-blue assays for parental U266 and 8226 human MM cells and 8226-LR5 and U266-LR6 cells treated with SEL or ELT, and MEL concurrently. These CI values show that concurrent administration of both SEL/MEL and ELT/MEL acted synergistically to reduce viability in both parental and MEL-resistant MM cell lines.
Parental and drug-resistant MM tumors treated with MEL in combination with SEL or ELT have reduced tumor growth and increased survival in vivo in NOD/SCID-γ (NSG) mouse studies
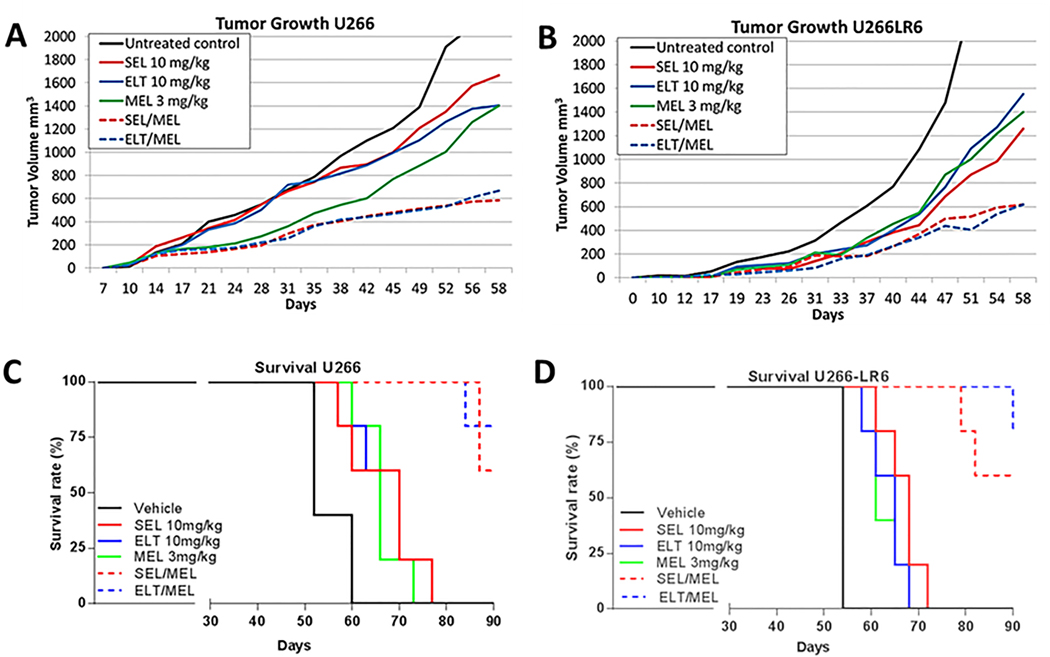
In mice implanted with U266 tumors, tumor growth was significantly decreased in SEL/MEL- and ELT/MEL-treated tumors (P = .00206 and P = .00207, respectively) than in those treated with MEL alone (Figure 3A). SEL/MEL and ELT/MEL combination treatments significantly decreased tumor growth (P = .0056 and P = .0033, respectively) in U266-LR6 tumors when compared to tumor growth following treatment with MEL alone (Figure 3B). We found in both parental U266 and MEL-resistant U266-LR6 cells that XPO1i/MEL combination treatments significantly improved survival (P < .0001) (Figure 3, C and D) when compared to survival with single-agent SEL, ELT, or MEL treatments.
Fig. 3. NSG mice in vivo studies: tumor growth in the presence of XPO1i and MEL.
NSG mice were challenged with human U266 and MEL-resistant U266-LR6 MM cells (subcutaneous flank injection) and treated with single agent SEL or ELT ± MEL. Mice were assayed for (A) U266 tumor growth, (B) MEL-resistant U266-LR6 tumor growth, (C) U266 tumor survival, and (D) MEL-resistant U266-LR6 tumor survival. Mice were euthanized when tumor volumes reached 2000 mm3. No toxicity (defined as weight loss > 10%) was observed.
NSG mouse studies (Figure S2) were also performed to compare sequential and concurrent drug treatments in mice challenged with subcutaneous tumors. Mice were treated with either single-agent SEL or MEL or SEL/MEL administered sequentially or concurrently. There was no statistically significant difference in tumor growth between concurrent and sequentially treated 8226 (Figure S2A) and 8226-LR5 (Figure S2B) or U266 (Figure S2C) and U266-LR6 (Figure S2D) MM tumors.
NSG mice challenged with 8226 and U266 human MM tumors were treated with a triple drug regimen of SEL/MEL/DEX (Figure S3) or control regimens. The control regimens were single-agents or combinations of MEL/DEX, SEL/MEL, or SEL/DEX. Mice were challenged with 8226 MM tumors (Figure S3A/B) and U266 MM tumors (Figure S3C/D). Mice challenged with U266 but not 8226 tumors and treated with the SEL/MEL/DEX had less tumor growth (P = .0037) and greater survival (P = .0001) than those treated with the control regimens.
XPO1 inhibitor/MEL mechanistic studies
The addition of SEL to MEL increases DNA damage and prevents DNA repair in human MM cells
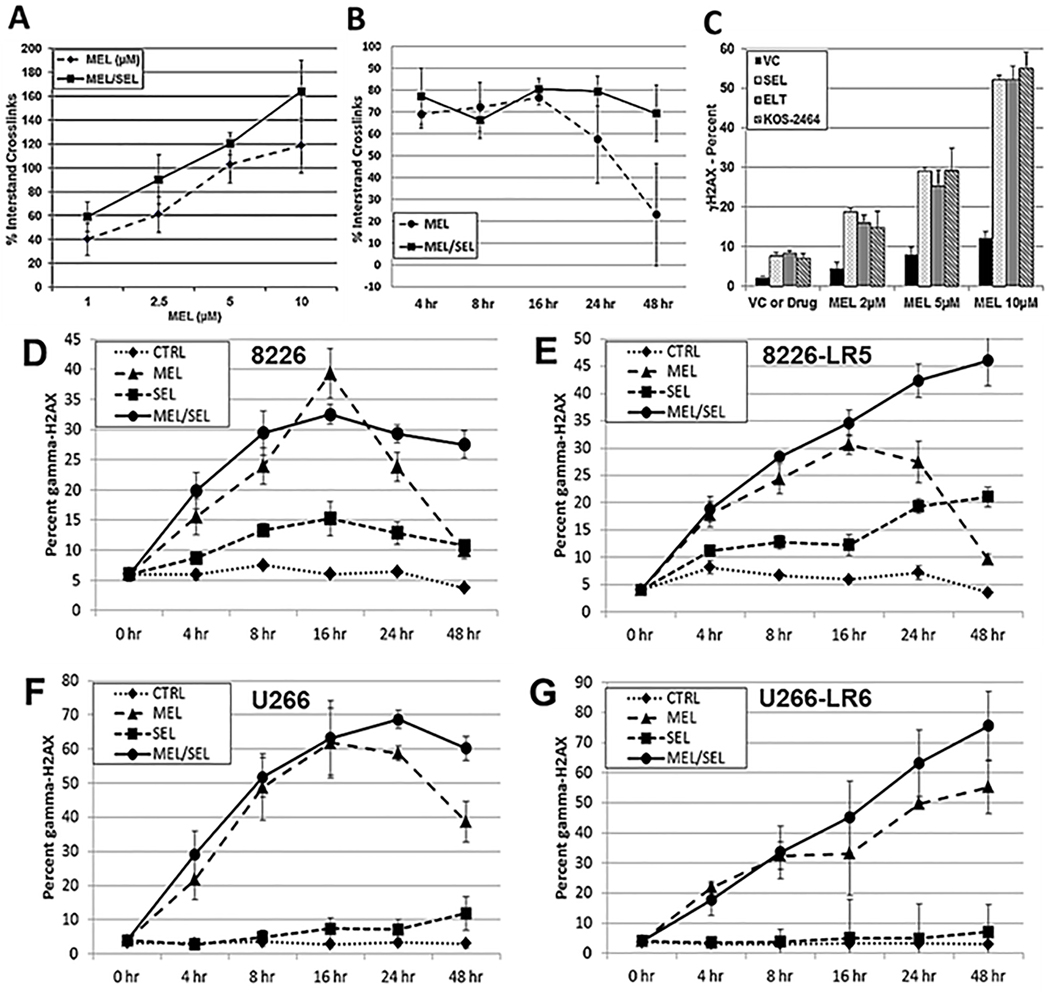
We found that the addition of SEL increased MEL-induced DNA ICLs when human MM cells were treated with SEL for 20 hours, followed by MEL for 2 hours (Figure 4A); the MEL/SEL combination produced significantly more DNA ICLs than single-agent MEL at all concentrations tested (P ≤ .0011). Additionally, alkaline comet assays were performed over a time course of 0 to 48 hours (Figure 4B). H929 human MM cells were treated with MEL (50 μM) for 2 hours and incubated with 300 nM SEL and showed decreased repair of DNA crosslinks at 24 and 48 hours (P ≤ .026) (Figure 4B). DNA damage (γH2AX) increased in tandem with greater concentrations of MEL combined with XPO1i, showing that DNA damage is dose-dependent (P < .0001) (Figure 4C). To further examine whether SEL induced DNA damage or inhibited DNA repair (Figures 4D, E, F and G), human 8226, U266, 8226-LR5 and U266-LR6 MM cells were treated for 2 hours with MEL, washed, and then incubated with SEL for 48 hours. We found that there was significantly less DNA repair in both MEL-resistant and parental cells treated with combined SEL/MEL than in those cells treated with MEL alone, showing that the addition of SEL prevented DNA repair.
Fig. 4. SEL increases DNA damage and prevents DNA repair.
(A) Alkaline comet assay experiments were used to assay MEL-induced DNA ICL formation. H929 human myeloma cells were exposed to SEL (100 nM) for 20 hours followed by MEL (1, 2.5, 5, and 10 μM) for 2 hours or they were treated with doses of MEL alone at the same increments. Drugs were removed and cells were incubated for 3 hours to allow for ICL formation. Cells were exposed to 900 rads (X-Rad 160 X-ray biological irradiator), and the alkaline comet assay was performed immediately (Trevigen Inc). Image and data analyses were performed by using the Loats Comet Analysis System (Loats Associates) (n=3). (B) Alkaline comet assay outcomes over a 0- to 48-hour time course. H929 human MM cells were treated with MEL (50 μM) for 2 hours to produce DNA damage. MEL was removed and either fresh media or media including SEL (100 nM) were added. Cells were incubated and aliquots were taken at 4, 8, 16, 24, and 48 hours and ICL measured (n=5). (C) Human H929 MM cells were incubated for 20 hours with varying concentrations of MEL (2, 5, and 10 μM) ± SEL (300 nM), ELT (300 nM), or KOS-2464 (100 nM) and then analyzed by FACS for γH2AX (n=3). (D/E) Human 8226 and MEL-resistant 8226-LR5, and (F/G) U266 and MEL-resistant U266-LR6 MM cells were treated for 2 hours with 50 μM MEL, washed, and then incubated with SEL (300 nM) for 4, 8, 16, 24, and 48 hours. Cells were analyzed by FACS for γ-H2AX (n=3).
FANCD2 mono-ubiquitination is decreased when MEL-treated MM cells are incubated with SEL
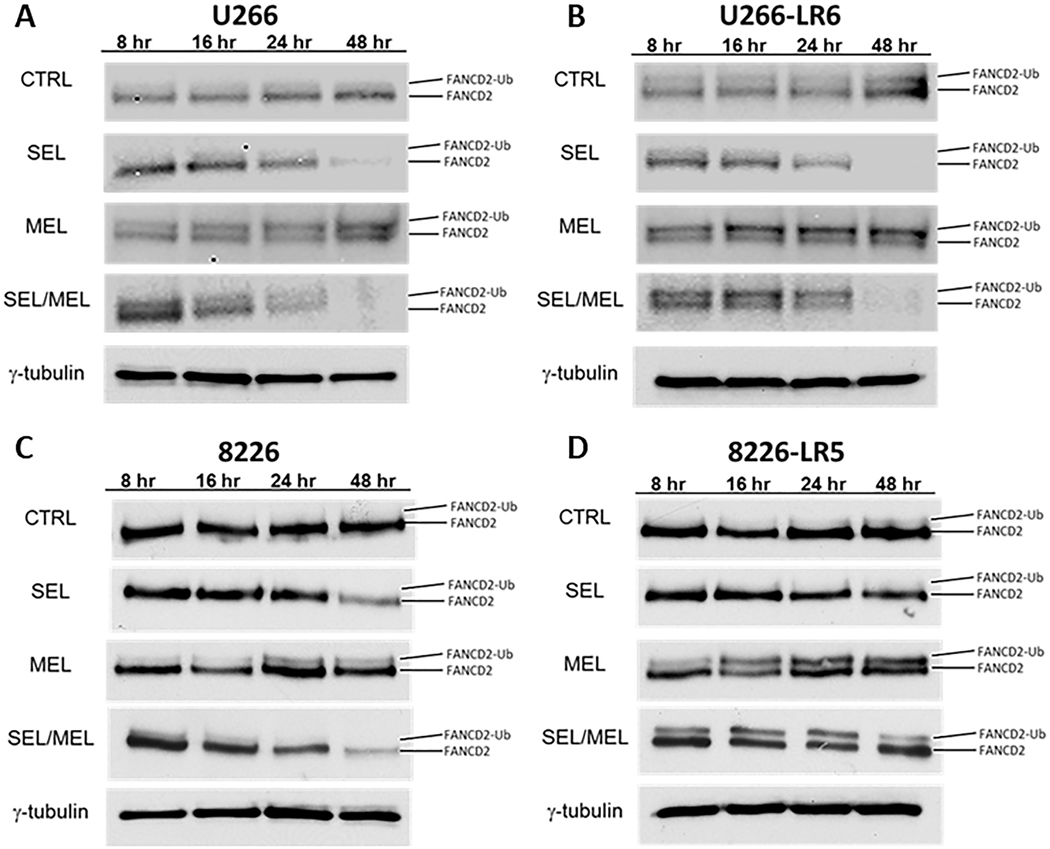
FANCD2 mono-ubiquitination is the pivotal FA DNA repair event of DNA ICLs. To investigate whether SEL would inhibit the formation of ubFANCD2, we induced ICL DNA damage by treating cells for 2 hours with 50 μM MEL. Cells were then placed in fresh media or 300 nM SEL and incubated for 48 hours. Western blot analyses showed that MEL increased FANCD2 mono-ubiquitination. However, the addition of SEL decreased FANCD2 mono-ubiquitination and FANCD2 protein over the 48-hour time course in all cells when compared to untreated cells or cells treated with MEL alone (Figure 5A, B, C, D), thus indicating that DNA repair was inhibited by the combination of MEL/SEL. These data parallel the γH2AX time-course study shown in Figure 4D, E, F and G.
Fig. 5. SEL decreases FANCD2 and mono-ubiquitination FANCD2.
Human U266 (A), 8226 (C) and MEL-resistant U266-LR6 (B) 8226-LR5 (D) MM cells were treated for 2 hours with 50 μM MEL, washed, and placed in fresh media with or without control or SEL. Protein (100 μg) was separated by gradient acrylamide gel electrophoresis for 4 hours to separate FANCD2 from mono-ubiquitinated-FANCD2. SEL was found to decrease both ubiquitinated and total FANCD2 as shown in representative Western blots from both parental U266 (A) and 8226 (B) and MEL-resistant U266-LR6 (C) and 8226-LR5 (D) cells(n=4).
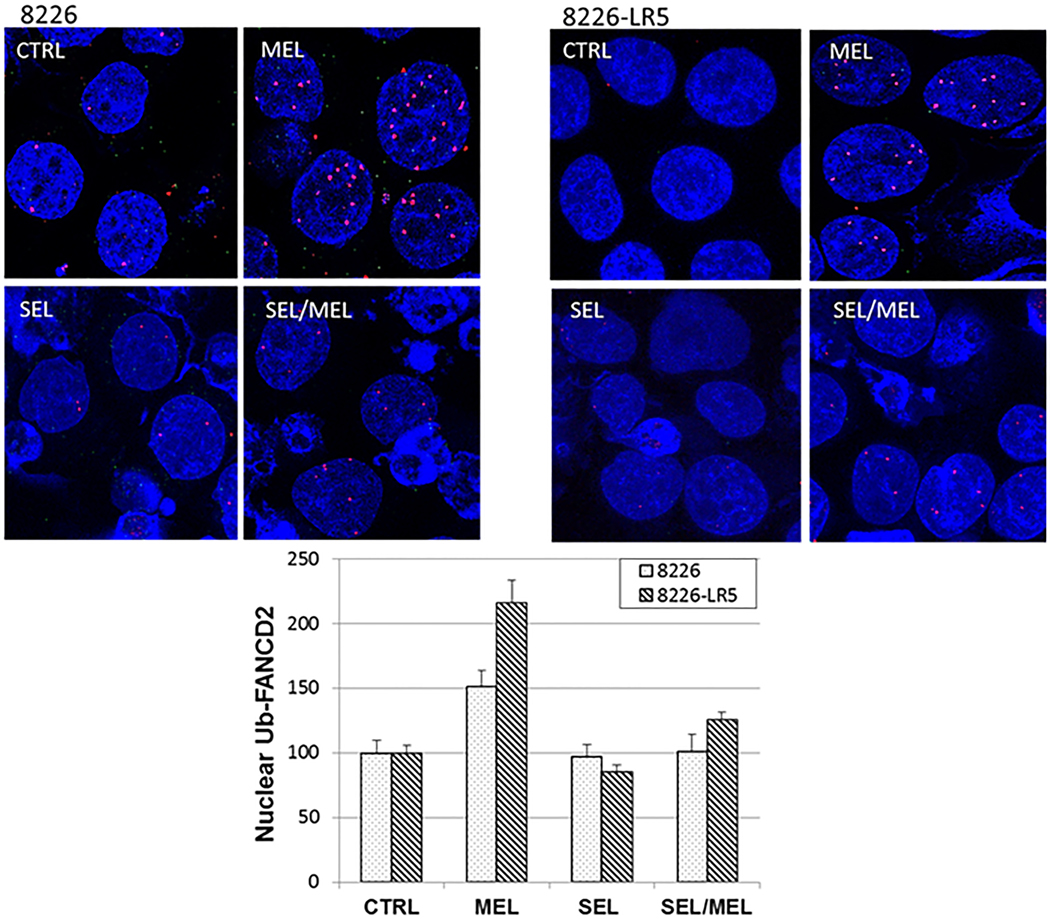
SEL inhibits MEL-induced mono-ubiquitination of FANCD2, as shown by proximity ligation assay
To confirm the Western blot analyses, 8226 and 8226-LR5 human MM cells treated with MEL followed by 300 nM SEL were assayed by proximity ligation assay, using separate antibodies for FANCD2 and ubiquitin (Figure 6). MEL treatment increased the number of ubiquitin-FANCD2 foci by 51.2% in 8226 and by 115.8% in 8226-LR5, when compared to untreated controls (P = .002 and P < .0001, respectively), indicating that MEL significantly increased FANCD2 mono-ubiquitination. The addition of SEL to MEL-treated cells significantly decreased FANCD2 mono-ubiquitination in both 8226 (50.3%, P = .0066) and 8226-LR5 MM cells (90.2%, P = .00038). This experiment was repeated 3 times and the images are representative of the results obtained.
Fig. 6. SEL inhibits MEL-induced mono-ubiquitination of FANCD2 in both parental and drug-resistant 8226 human MM cells.
8226 and 8226-LR5 human MM cells were either untreated or treated with 50 μM MEL for 2 hours. The cells were then collected by centrifugation and incubated for 24 hours with fresh media with or without 300 nM SEL. The cells were assayed for proximity co-localization of FANCD2 and ubiquitin. More than 700 cells were assayed for each experimental treatment group. The proximity ligation assay generates a red fluorescent signal when the two proteins tested (FANCD2 and ubiquitin) are within 40 nM of each other. The values on the y-axis represent the change in ubiquitin-FANCD2 nuclear foci percentages compared to untreated controls (whose values were set at 100%).
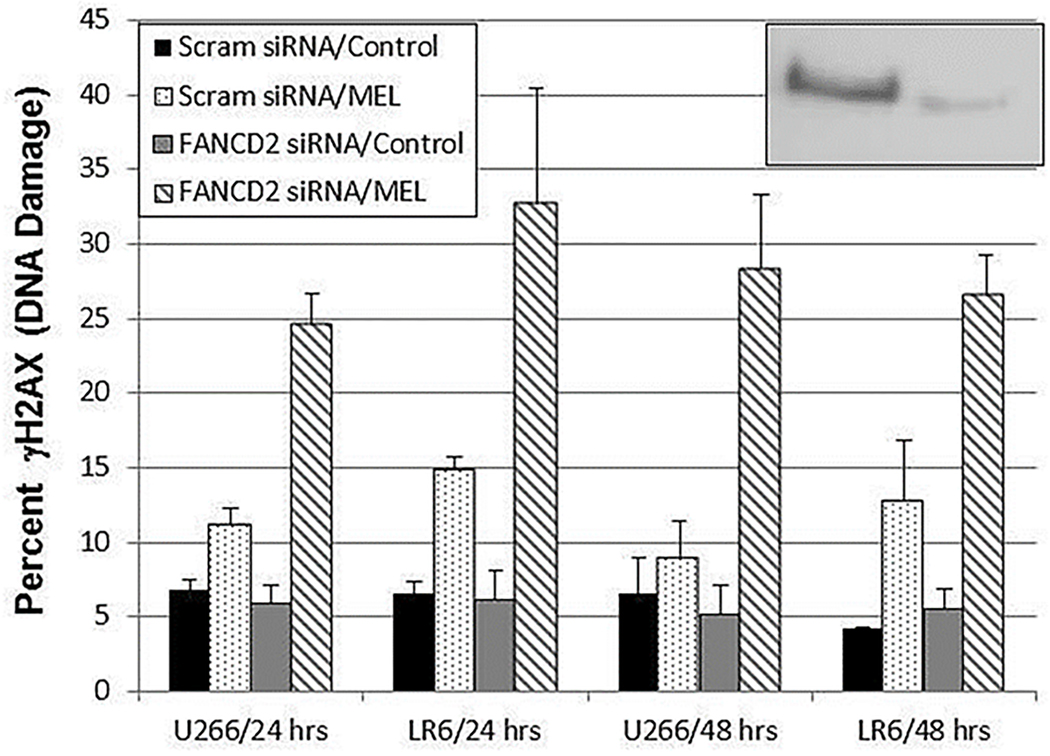
FANCD2 siRNA knockdown prevents DNA repair and increases DNA damage in both parental and MEL-resistant human MM cells
To approximate the effect of the SEL-induced decrease in ubFANCD2 and total FANCD2, U266 and U266-LR6 MM cells were transfected with control siRNA or a FANCD2-specific siRNA (Figure 7). Two days after siRNA knockdown, the cells were treated with MEL, and DNA damage was assessed by γH2AX protein expression. FANCD2 knockdown increased MEL-induced DNA damage; thereby approximating the effect of SEL on FANCD2 expression and DNA damage (see also Figures 4, 5, and 6). DNA damage was significantly increased in both cell lines at the 24- and 48-hour time points by FANCD2 siRNA and MEL treatment, as compared to DNA damage in scram/control (P ≤ .0002), scram/MEL (P ≤ .039), and FANCD2 siRNA only (P ≤ .0005). This may demonstrate, at least in part, how SEL and MEL worked synergistically to increase DNA damage by inhibiting FANCD2-mediated DNA repair.
Fig. 7. FANCD2 siRNA knockdown prevents DNA repair and increases DNA damage (γH2AX protein expression) in the presence of MEL in both parental and MEL-resistant human MM cells (n = 4).
Human MM U266 and MEL-resistant U266-LR6 cells were transfected with control (scram) siRNA and FANCD2 siRNA. After 48 hours, cells were treated with 50 μM MEL for 2 hours, washed, and then incubated for a further 48 hours. At the 24- and 48-hour time points, DNA damage was assessed by measuring γH2AX protein expression by FACS analysis. FANCD2 knockdown was > 82% (U266 cells inset).
SEL decreased MEL-induced expression of NFĸB, IκB kinase α (IKKα), FANCF, and FANCL
H929 MM cells were treated for 6 hours with MEL (10 μM), SEL (300 nM), or a combination of both. Whole cell lysates were assayed by Western blot for FANCF, FANCL, IKKα, and NFĸB. MEL was found to increase DNA repair enzymes FANCF and FANCL and cell proliferation proteins NFĸB and IKKα. However, FANCF, FANCL, IKKα, and NFĸB were decreased by the addition of SEL (Figure S4) (n = 2).
SEL decreases total FANCD2, ubiquitinated FANCD2, BRCA1, BRCA2 and RAD51 proteins in a dose-dependent manner.
U266 and U266-LR6 cells were incubated with MEL for 2 hours to activate DNA repair, cells were then incubated with 0, 50, 100, 500 or 1000 nM SEL for an additional 24 hours. Cell lysates were assayed by Western blot for DNA repair proteins FANCD2, ubFANCD2, BRCA1, BRAC2 and RAD51 (figure S5). Protein bands were assayed for percent change versus untreated controls. We found that SEL caused a decrease in all DNA repair proteins assayed in a dose-dependent manner. In addition we found that U266 cells demonstrated a greater inhibition of FANCD2, ubFANCD2, BRCA1, BRCA2 and RAD51 expression than MEL-resistant U266-LR6 cells. (n=2).
Discussion
Nearly all MM patients who receive HD chemotherapy before autologous HSCTs are treated with HD MEL. Depending on the progression-free interval between a first autologous HSCT and relapse, HD MEL is also often used as the preparative regimen for a second transplant. No additions to this MEL backbone have resulted in significant improvements. In this study we investigated whether adding SEL to the MEL regimen may improve outcomes for MM patients.
MEL is a bifunctional alkylating agent; a nitrogen mustard (4-[bis(2-chloroethyl)amino]-1-phenylalanine) that induces mono-adducts in DNA (alkylation of N-7 in guanine or N-3 in adenine) and DNA ICLs between guanine/guanine or guanine/adenine (50). Ninety-five percent of the lesions are mono-adducts and 5% are ICLs. Although ICLs are infrequent, they are highly toxic to cells because they impair DNA transcription and replication (50). The repair of MEL-induced DNA ICLs largely depends on the Fanconi anemia (FA) ICL repair pathway. Nineteen FA genes have been identified to date, and their corresponding proteins cooperate with other proteins in nucleotide excision repair to resolve ICLs (12). FA proteins also assist in other DNA repair pathways including non-homologous end joining, translesion synthesis, and homologous recombination (11–14, 50). The mono-ubiquitinated FANCD2/FANCI complex is the key regulator of nucleotide excision (ICL) repair resulting from MEL-induced DNA damage (50). Identification of the ICL is made by FANCM. FANCM recruits 14-FA core complex proteins including an ubiquitin ligase. Monoubiquitination of FANCD2 and FANCI is then catalyzed by the FA protein core. The ubiquitinated FANCD2-FANCI dimer is loaded on to the chromatin at the site of the ICL by the FA core complex and produces a nucleolytic incision (unhooking event) at the converged replication forks to release the ICL from the parental strands, and thus initiates the repair event. We show that both FANCD2 and Ub-FANCD2 are downregulated by the addition of SEL in a dose-dependent manner, resulting in a decrease in repair of MEL induced ICL DNA damage as shown by both the Comet assay and γ-H2AX protein expression. This was demonstrated in both parental MM cell lines and MEL-resistant cell lines. Even though SEL treatment of MM cells was shown to decrease the expression of other DNA-repair related proteins such as IKKα, RAD51, BRCA1 and BRCA2, it is the ubiquitinated FANCD2-FANCI dimer that initiates ICL DNA repair and is the pivotal event controlling the ICL DNA damage response (13, 45). The SEL mediated decrease in the expression of additional FA proteins, FANCF and FANCL, may also result in decreased DNA damage repair. For example a decrease in FANCL, the FA complex E3 ubiquitin ligase, would result in a decrease in FANCD2/FANCI ubiquitination and prevent DNA repair. A decrease in FANCF would prevent formation of the FA core complex, again resulting in a decrease in the FANCD2/FANCI ubiquitination and subsequent ICL repair (51).
In summary, the combination of MEL and an XPO1i results in synergistic MM cell kill. Mechanistically, this appears to result from increased DNA damage (γH2AX and comet assay) and a decreased rate of DNA repair (Ub-FANCD2). Preliminary data demonstrate that the increase in FA mediated DNA repair proteins observed with MEL treatment (ubiquitinated FANCD2/FANCI) is blocked by SEL, possibly contributing to the synergistic cell kill observed with the combined SEL and MEL treatment. These data were confirmed in this study by siRNA knockdown of FANCD2 which resulted in decreased DNA repair.
Drug resistance is the greatest obstacle to the treatment of MM and many other cancers. In this study, we demonstrated the possibility that acquired MM drug resistance may be overcome by combinatorial therapy of SEL or ELT with MEL. We explored the combination of a new class of agents, XPO1i, with MEL in the treatment of sensitive and MEL-resistant MM and have begun an early-phase trial combining SEL and HD MEL (NCT02780609). This project is the first effort to examine the synergistic interaction between a bifunctional alkylating agent and an XPO1i in several models of MM and to use this combination to overcome MEL drug resistance.
Supplementary Material
Statement of significance:
Inhibition of exportin 1 with selinexor synergistically sensitizes human multiple myeloma to melphalan by inhibiting Fanconi anemia pathway-mediated DNA repair.
Acknowledgments:
We thank Paul Fletcher and Daley Drucker (Moffitt Cancer Center) for editorial assistance. This work has been supported in part by Jonathan Nguyen at the Moffitt Analytic Microscopy Core Facility, Ashley Blanchard at the Translational Research Core, John Robinson at the Flow Cytometry Core, and the Tissue Core Facility at the H. Lee Moffitt Cancer Center and Research Institute. The H. Lee Moffitt Cancer Center and Research Institute is supported in part by NCI Cancer Center Support Grant P30 CA076292. Additional support was provided by NIH grant CA194051.
Footnotes
Data and materials availability: Selinexor and eltanexor were provided by Karyopharm Therapeutics; KOS-2464 was provided by Bristol-Myers Squibb.
Conflicting interests: The authors declare that there are no conflicts of interest regarding the publication of this article.
References
- 1.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Kosary CL, et al. American Cancer Society Cancer Facts & Figures 2018. Atlanta: American Cancer Society; 2018. [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Chari A, Vogl DT, Gavriatopoulou M, Nooka AK, Yee AJ, Huff CA, et al. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N Engl J Med. 2019;381:727–38. [DOI] [PubMed] [Google Scholar]
- 4.Alsina M, Trudel S, Furman RR, Rosen PJ, O’Connor OA, Comenzo RL, et al. A phase I single-agent study of twice-weekly consecutive-day dosing of the proteasome inhibitor carfilzomib in patients with relapsed or refractory multiple myeloma or lymphoma. Clin Cancer Res. 2012;18:4830–40. [DOI] [PubMed] [Google Scholar]
- 5.Mateos MV, Masszi T, Grzasko N, Hansson M, Sandhu I, Pour L, et al. Impact of prior therapy on the efficacy and safety of oral ixazomib-lenalidomide-dexamethasone vs. placebo-lenalidomide-dexamethasone in patients with relapsed/refractory multiple myeloma in TOURMALINE-MM1. Haematologica. 2017;102:1767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonial S, Weiss BM, Usmani SZ, Singhal S, Chari A, Bahlis NJ, et al. Daratumumab monotherapy in patients with treatment-refractory multiple myeloma (SIRIUS): an open-label, randomised, phase 2 trial. Lancet. 2016;387:1551–60. [DOI] [PubMed] [Google Scholar]
- 7.Lonial S, Dimopoulos M, Palumbo A, White D, Grosicki S, Spicka I, et al. Elotuzumab therapy for relapsed or refractory multiple myeloma. N Engl J Med. 2015;373:621–31. [DOI] [PubMed] [Google Scholar]
- 8.San-Miguel JF, Hungria VT, Yoon SS, Beksac M, Dimopoulos MA, Elghandour A, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15:1195–206. [DOI] [PubMed] [Google Scholar]
- 9.Usmani SZ, Rodriguez-Otero P, Bhutani M, Mateos MV, Miguel JS. Defining and treating high-risk multiple myeloma. Leukemia. 2015;29:2119–25. [DOI] [PubMed] [Google Scholar]
- 10.Tew K, Colvin O, Jones R. Clinical and high-dose alkylating agents In: Chabner B, Longo D, editors. Cancer Chemotherapy and Biotherapy: Principles and Practice. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2006. p. 283–309. [Google Scholar]
- 11.Gourzones-Dmitriev C, Kassambara A, Sahota S, Reme T, Moreaux J, Bourquard P, et al. DNA repair pathways in human multiple myeloma: role in oncogenesis and potential targets for treatment. Cell Cycle. 2013;12:2760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Van der Sluis PC, Boulware D, Hazlehurst LA, Dalton WS. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood. 2005;106:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo F, Li J, Du W, Zhang S, O’Connor M, Thomas G, et al. mTOR regulates DNA damage response through NF-kappaB-mediated FANCD2 pathway in hematopoietic cells. Leukemia. 2013;27:2040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizawa J, Kojima K, Hail N Jr., Tabe Y, Andreeff M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther. 2015;153:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue H, Kauffman M, Shacham S, Landesman Y, Yang J, Evans CP, et al. CRM1 blockade by selective inhibitors of nuclear export attenuates kidney cancer growth. J Urol. 2013;189:2317–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salas Fragomeni RA, Chung HW, Landesman Y, Senapedis W, Saint-Martin JR, Tsao H, et al. CRM1 and BRAF inhibition synergize and induce tumor regression in BRAF-mutant melanoma. Mol Cancer Ther. 2013;12:1171–9. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y, Gery S, Sun H, Shacham S, Kauffman M, Koeffler HP. KPT-330 inhibitor of XPO1-mediated nuclear export has anti-proliferative activity in hepatocellular carcinoma. Cancer Chemother Pharmacol. 2014;74:487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun H, Hattori N, Chien W, Sun Q, Sudo M, GL EL, et al. KPT-330 has antitumour activity against non-small cell lung cancer. Br J Cancer. 2014;111:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gravina GL, Tortoreto M, Mancini A, Addis A, Di Cesare E, Lenzi A, et al. XPO1/CRM1-selective inhibitors of nuclear export (SINE) reduce tumor spreading and improve overall survival in preclinical models of prostate cancer (PCa). J Hematol Oncol. 2014;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendonca J, Sharma A, Kim HS, Hammers H, Meeker A, De Marzo A, et al. Selective inhibitors of nuclear export (SINE) as novel therapeutics for prostate cancer. Oncotarget. 2014;5:6102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez-Canales J, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538:114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Camacho SC, Silvers TR, Razak AR, Gabrail NY, Gerecitano JF, et al. Inhibition of the Nuclear Export Receptor XPO1 as a Therapeutic Target for Platinum-Resistant Ovarian Cancer. Clin Cancer Res. 2017;23:1552–63. [DOI] [PubMed] [Google Scholar]
- 24.Garg M, Kanojia D, Mayakonda A, Said JW, Doan NB, Chien W, et al. Molecular mechanism and therapeutic implications of selinexor (KPT-330) in liposarcoma. Oncotarget. 2017;8:7521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT, et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol. 2013;161:117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120:1765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muqbil I, Aboukameel A, Elloul S, Carlson R, Senapedis W, Baloglu E, et al. Anti-tumor activity of selective inhibitor of nuclear export (SINE) compounds, is enhanced in non-Hodgkin lymphoma through combination with mTOR inhibitor and dexamethasone. Cancer Lett. 2016;383:309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranganathan P, Kashyap T, Yu X, Meng X, Lai TH, McNeil B, et al. XPO1 inhibition using selinexor synergizes with chemotherapy in acute myeloid leukemia by targeting DNA repair and restoring topoisomerase IIalpha to the nucleus. Clin Cancer Res. 2016;22:6142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt J, Braggio E, Kortuem KM, Egan JB, Zhu YX, Xin CS, et al. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia. 2013;27:2357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai YT, Landesman Y, Acharya C, Calle Y, Zhong MY, Cea M, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2014;28:155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner JG, Dawson JL, Grant S, Shain KH, Dalton WS, Dai Y, et al. Treatment of acquired drug resistance in multiple myeloma by combination therapy with XPO1 and topoisomerase II inhibitors. J Hematol Oncol. 2016;9:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PB, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69:510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baz RC, Shain KH, Alsina M, Brayer J, Rashal T, Cooksey JL, et al. Phase I trial of the combination of selinexor (SEL), liposomal doxorubicin (DOX) and dexamethasone (Dex) for relapsed and refractory multiple myeloma (RRMM). Journal of Clinical Oncology, Abstract. 2016;34:8013. [Google Scholar]
- 34.Nishihori T, Alsina M, Ochoa J, Puglianini O, Baz R, Shain K, Brayer JB, Kim J, Dumala M, Turner J, and Sullivan D. The Result of a Phase 1 Study of Selinexor in Combination with High-Dose Melphalan and Autologous Hematopoietic Cell Transplantation for Multiple Myeloma. Blood Abstract 3314 2019. [Google Scholar]
- 35.Jakubowiak A, Jasielec J, Rosenbaum CA, Cole CE, Chari A, Nam J, et al. Final results of phase 1 MMRC trial of selinexor, carfilzomib, and dexamethasone in relapsed/refractory multiple myeloma (RRMM). Am Soc Hematology, Abstract; 2016. [Google Scholar]
- 36.Bahlis NJ, Sutherland H, White D, Sebag M, Lentzsch S, Kotb R, et al. Selinexor plus low-dose bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma. Blood. 2018;132:2546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CI, Bahlis N, Gasparetto C, Tuchman SA, Lipe BC, Baljevic M, et al. Selinexor, pomalidomide, and dexamethasone (SPd) in patients with relapsed or refractory multiple myeloma. American Society of Hematology, Abstract #141; 2019. [Google Scholar]
- 38.Cornell R, Rossi A, Baz R, Hofmeister C, Shustik C, Richter J, Chen C, Vogl D, Baloglu E, Senapedis W, Ellis J, Williams T, Shacham S, Kauffman M. Eltanexor (KPT-8602), a Second-Generation Selective Inhibitor of Nuclear Export (SINE) Compound, in Patients with Refractory Multiple Myeloma. Blood, Abstract 130:3134 2017. [Google Scholar]
- 39.Turner JG, Dawson J, Emmons MF, Cubitt CL, Kauffman M, Shacham S, et al. CRM1 Inhibition Sensitizes Drug Resistant Human Myeloma Cells to Topoisomerase II and Proteasome Inhibitors both In Vitro and Ex Vivo. J Cancer. 2013;4:614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner JG, Dawson JL, Cubitt CL, Kauffman M, Shacham S, Sullivan D. Combination therapy of human multiple myeloma using proteosome and CRM1 inhibitors. Cancer Res, Abstract. 2013;73. [Google Scholar]
- 41.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83:1021–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner JG, Engel R, Derderian JA, Jove R, Sullivan DM. Human topoisomerase IIalpha nuclear export is mediated by two CRM-1-dependent nuclear export signals. J Cell Sci. 2004;117:3061–71. [DOI] [PubMed] [Google Scholar]
- 43.Turner JG, Marchion DC, Dawson JL, Emmons MF, Hazlehurst LA, Washausen P, et al. Human multiple myeloma cells are sensitized to topoisomerase II inhibitors by CRM1 inhibition. Cancer Res. 2009;69:6899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner JG, Sullivan DM. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr Med Chem. 2008;15:2648–55. [DOI] [PubMed] [Google Scholar]
- 45.Bellamy WT, Dalton WS, Gleason MC, Grogan TM, Trent JM. Development and characterization of a melphalan-resistant human multiple myeloma cell line. Cancer Res. 1991;51:995–1002. [PubMed] [Google Scholar]
- 46.Marx V. Cell-line authentication demystified. Nat Methods. 2014;11:483–8. [DOI] [PubMed] [Google Scholar]
- 47.Gray J, Cubitt CL, Zhang S, Chiappori A. Combination of HDAC and topoisomerase inhibitors in small cell lung cancer. Cancer Biol Ther. 2012;13:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–62. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Martinez D, Liang CC, Cohn MA. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell Mol Life Sci. 2016;73:3097–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Léveillé F, Blom E, Medhurst AL, Bier P, Laghmani EH, Johnson M, et al. The Fanconi anemia gene product FANCF is a flexible adaptor protein. Journal of Biological Chemistry. 2004;279:39421–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.