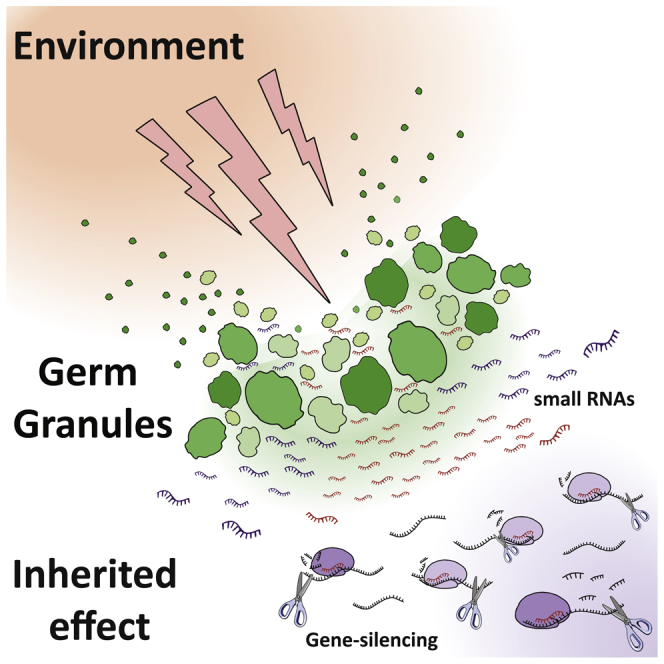
Summary
In the recent decade small RNA-based inheritance has been implicated in a variety of transmitted physiological responses to the environment. In Caenorhabditis elegans, heritable small RNAs rely on RNA-dependent RNA polymerases, RNA-processing machinery, chromatin modifiers, and argonauts for their biogenesis and gene-regulatory effects. Importantly, many of these factors reside in evolutionary conserved germ granules that are required for maintaining germ cell identity and gene expression. Recent literature demonstrated that transient disturbance to the stability of the germ granules leads to changes in the pools of heritable small RNAs and the physiology of the progeny. In this piece, we discuss the heritable consequences of transient destabilization of germ granules and elaborate on the various small RNA-related processes that act in the germ granules. We further propose that germ granules may serve as environment sensors that translate environmental changes to inheritable small RNA-based responses.
Subject Areas: Biological Sciences, Molecular Biology, Cell Biology
Graphical Abstract
Biological Sciences; Molecular Biology; Cell Biology
Introduction
More than a century ago August Weismann postulated that acquired traits are not inherited, and that “germ plasm,” a nuclear substance, is passed from parental germ cells to the progeny's germ cells to ensure the heredity of the species qualities (Weismann, 1893). Weismann's followers “evolved” the “germ plasm” term and instead used it to describe maternally contributed cytoplasmic substance, consisting of germ granules, that is required for the formation of germ cells in the next generation (Eddy, 1976; Hegner, 1914; Ritter, 1890). Ironically, recent studies demonstrate that these germ granules play a crucial role in the inheritance of regulatory small RNAs that can defy Weismann's law and allow acquired traits to be inherited. In the recent decade regulatory small RNAs were implicated in the transmission of various phenotypes in worms (Ewe et al., 2020; Gammon et al., 2017; Moore et al., 2019; Rechavi et al, 2011, 2014), flies (Casier et al., 2019; Hermant et al., 2015), plants (Ito et al, 2011, 2016; Liu et al., 2019; Zhong et al., 2013), and mice (Chen et al., 2016; Conine et al., 2018; Gapp et al., 2014; Grandjean et al., 2015; Rodgers et al., 2015; Zhang et al., 2018b). Notably, transgenerationally heritable small RNAs seem to differ from DNA-based genetics: their inheritance obeys a different set of rules (Houri-Ze'evi et al., 2016; Houri-Zeevi et al., 2020), their biogenesis can be induced in response to the environment (Ni et al., 2016; Rechavi et al, 2011, 2014), and their cellular pools change across generations in a continuous (rather than discrete) manner that seems to dilute (Vastenhouw et al., 2003; Alcazar et al., 2008; Houri-Ze'evi et al., 2016) or concentrate (Dodson and Kennedy, 2019; Houri-Ze'evi et al., 2016; Lev et al., 2017; 2019b; Ouyang et al., 2019; Simon et al., 2014). This review covers the role of the germ granules in regulating transgenerational small RNA responses and focuses on knowledge gained from studies in the nematode Caenorhabditis elegans, which plays an instrumental role (Perez and Lehner, 2019) in uncovering the principles and rules of transgenerational epigenetic inheritance.
In C. elegans exposure to synthetic exogenous double-stranded RNA leads to the production of small RNAs (Aoki et al., 2007; Pak and Fire, 2007; Vasale et al., 2010). These small RNAs silence the complementary gene and are inherited to the progeny (Buckley et al., 2012; Burton et al., 2011; Fire et al., 1998; Vastenhouw et al., 2006). Likewise propagation of RNA viruses leads to the production of antiviral small RNAs that silence the viral genes across multiple generations and protects the progeny (Gammon et al., 2017; Rechavi et al., 2011). Beyond the two examples above, heritable small RNAs were implicated in a variety of inherited responses to different environmental conditions. Inheritance of small RNAs results, for example, in enhanced longevity in the progeny of starved worms (Rechavi et al., 2014), altered endoderm development following parental diapause (Ewe et al., 2020), competent chemotactic behavior (Posner and Toker, 2019), inherited bacterial avoidance (Kaletsky et al., 2020; Moore et al., 2019; Palominos et al., 2017; Pereira et al., 2020), transgenerational loss of fertility in specific mutants (Mortal germline phenotype) (Barucci et al., 2020; Buckley et al., 2012; Lev et al., 2017; Simon et al., 2014), and heritable gene-expression responses to temperature changes (Ni et al., 2016; Schott et al., 2014).
The transgenerational inheritance of small RNAs in the germline relies on small RNA-binding argonauts (Buckley et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012), RNA-dependent RNA polymerases (RdRPs) (Rechavi et al., 2011), additional biogenesis factors (Grishok et al., 2000), and in some cases (but not always) also chromatin modifiers (Ashe et al., 2012; Kalinava et al, 2017, 2018; Lev et al, 2017, 2019a; Mao et al., 2015; Shirayama et al., 2012). The argonaute proteins act as effectors guided by the bound small RNAs to regulate the expression of the targeted gene. The RdRPs use the gene's mRNA as template to produce additional “secondary” small interfering RNAs (siRNAs) in every generation to maintain the response. This inheritance requires nuclear RNAi factors (nrde genes) (Buckley et al., 2012; Burton et al., 2011; Mao et al., 2015). However, many of the factors involved, including multiple argonauts and the RdRPs, reside in cytoplasmic germ granules (see Table 1).
Table 1.
Germ Granule-Localized Small RNA Factors
| Protein Name(s) | Function | Germ Granule Localization | Temperature-dependent Fertility Defects |
|---|---|---|---|
| WAGO-1 | Argonaute | P granules (Gu et al., 2009) | Unknown |
| CSR-1 | Argonaute | P granules (Claycomb et al., 2009) and Z granules (Charlesworth et al., 2020) | General sterility in hermaphrodites (Yigit et al., 2006), temperature-sensitive sterility in males (Conine et al., 2013) |
| WAGO-4 | Argonaute | Z granules (Wan et al., 2018) | Yes (Wan et al., 2018; Xu et al., 2018) |
| PRG-1 | Argonaute | P granules (Batista et al., 2008; Wang and Reinke, 2008) | Yes (Batista et al., 2008; Simon et al., 2014; Yigit et al., 2006) |
| ALG-3/4 | Argonautes | P granules (Conine et al., 2010) | Yes (Conine et al., 2010) |
| ALG-5 | Argonaute (microRNAs) | P granules (Brown et al., 2017) | General (not temperature-dependent) defective fertility (Brown et al., 2017) |
| EGO-1 | RdRP, small RNA biogenesis | P granules (Claycomb et al., 2009) | General sterility (Qiao et al., 1995; Smardon et al., 2000) |
| DRH-3 | Helicase, small RNA biogenesis | P granules (Claycomb et al., 2009) | Yes (Claycomb et al., 2009; Nakamura et al., 2007) |
| RRF-1 | RdRP, small RNA biogenesis | M granules (Phillips et al., 2012) | Unknown |
| DCR-1 | Ribonuclease, small RNA biogenesis | Interacting with P granules (Barucci et al., 2020; Beshore et al., 2011) | Yes (Pavelec et al., 2009; Welker et al., 2010) |
| RDE-3(MUT-2) | Poly-UG polymerase, small RNA biogenesis | M granules (Phillips et al., 2012) | Yes (Chen et al., 2005) |
| RDE-12 | Small RNA biogenesis | P granules (Shirayama et al., 2014) | Unknown |
| CDE-1(CID-1 or PUP-1) | Poly(A) polymerase, implicated in sorting of small RNAs | P granules (van Wolfswinkel et al., 2009) | Yes (Spracklin et al., 2017) |
| ZNFX-1 | Small RNA biogenesis | Z granules (Ishidate et al., 2018; Wan et al., 2018) | Yes (Wan et al., 2018) |
| RDE-8 | Small RNA biogenesis, mRNA cleavage. | M granules (Tsai et al., 2015) | Unknown |
| MUT-16, MUT-15, MUT-8 (RDE-2) | Small RNA biogenesis. | M granules (Phillips et al., 2012) | Yes (Vastenhouw et al., 2003) |
| MUT-14 | RNA helicase, small RNA biogenesis | M granules (Phillips et al., 2012) | Yes (Vastenhouw et al., 2003) |
| MUT-7 | Exonuclease, small RNA biogenesis | M granules (Phillips et al., 2012) | Yes (Ketting et al., 1999) |
Germ granules are cytoplasmic RNA-rich membrane-less condensates found in the germline (Eddy, 1976; Krieg et al., 1978; Wolf et al., 1983). Germ granules are evolutionary conserved and have been described in dozens of organisms, including worms, flies, and mice (Eddy, 1976). In C. elegans, the best studied class of germ granules are the P granules. The P granules are transmitted from the mother's germline to the embryo and segregate together with the P lineage, which ultimately forms the adult germline in the progeny (Hird et al., 1996). In contrast to the germ granules of Drosophila melanogaster, the P granules in C. elegans are not required for specification of germ cell identity (Gallo et al., 2010). Instead, the P granules are required for maintaining germline identity and proper germline gene-expression patterns. The proteins PGL-1 and PGL-3 are required for germ granule nucleation (Hanazawa et al., 2011; Kawasaki et al., 2004; Updike et al., 2011) while the GLH1 and GLH-4 proteins are promoting the anchoring of germ granules to nuclear periphery (Updike et al., 2011). Simultaneous knockdown of these four key germ granule components leads to ectopic expression of sperm genes in the germline (Campbell and Updike, 2015), and in later adulthood, to additional ectopic expression of somatic transcripts (Knutson et al., 2017; Updike et al., 2014). How germ granules maintain germline gene expression is an ongoing field of research; however, recent studies shed new light on these mechanisms (see Box 1 for outstanding questions).
Box 1. Outstanding Questions:
-
1.
What is the role of the organization of small RNA factors in the different sub-classes of germ granule?
-
2.
How the developmental changes in germ granule composition affect small RNA biology?
-
3.
What is the role of germ granules in sorting of amplified siRNAs to their partner argonauts?
-
4.
Which environmental cues impact germ granule structure and composition in vivo, and how?
Germ Granules Survey mRNAs as They Leave the Nuclear Pores
In the cytoplasm, the germ granules are strategically located adjacent to the nuclear pores, where they survey mRNAs as they are released into the cytoplasm. In agreement with this model, studies using fluorescence in situ hybridization (FISH) demonstrated that mRNAs transiently move through the germ granules when exiting the nuclear pore (Schisa et al., 2001; Sheth et al., 2010). Although the germ granules are largely perinuclear, they transiently lose their perinuclear localization during specific developmental stages. For example, in mature oocytes the germ granules are released into the cytosol and after fertilization migrate to the posterior part of the zygote to segregate with the germline blastomeres (Strome and Wood, 1983). Later during embryonic development, toward the 24-cell stage, the germ granules regain their perinuclear localization (Strome and Wood, 1983).
Germ granules can be subclassified into more specific entities of physically adjacent condensates, some of which stereotypically divide into separate entities at defined developmental stages (Wan et al., 2018). So far three classes of germ granules have been identified: the P (Hird et al., 1996), Z (Wan et al., 2018), and M (or Mutator) (Phillips et al., 2012) granules. Each subclass harbors different small RNA-related proteins. For example, the RdRP EGO-1 is localized in the P granules, whereas the argonaute WAGO-4 is localized in the Z granules (Claycomb et al., 2009; Wan et al., 2018) (for a comprehensive list see Table 1). The P and the Z granules separate to form two distinct condensates specifically in the primordial germ cells in the embryo and also later during the pachytene stage of the adult germline (Wan et al., 2018). This separation leads to the formation of a tri-condensate a “PZM” structure, where the Z granule is hypothesized to bridge between the P and M granules. Accordingly, the PZM assemblage's structure was hypothesized to play a role in organizing and coordinating small RNA pathways (Wan et al., 2018).
Germ Granules Regulate Gene Expression via the Small RNA Machinery
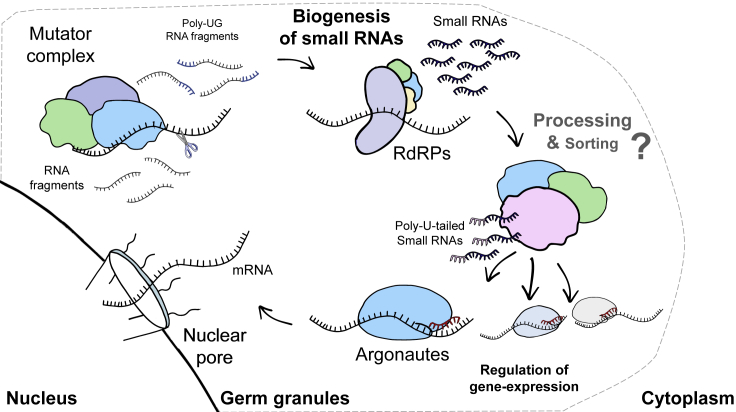
Initial genetic studies showed that mutants in structural germ granule proteins such as pgl-1 (Robert et al., 2005; Spike et al., 2008b) and deps-1 (Houri-Ze'evi et al., 2016; Spike et al., 2008b) and later also glh-1 (Spracklin et al., 2017) and meg-3;4 (Wan et al., 2018) have defective small RNA-mediated RNAi responses. Conversely, mutating or knocking down small RNA-related factors such as the argonaute CSR-1, the RdRP EGO-1, and the helicase DRH-3 disrupts germ granule structures (Claycomb et al., 2009; Updike and Strome, 2009; Vought et al., 2005). In addition, mutants in many small RNA-related factors exhibit fertility defects that are commonly temperature sensitive (see Table 1), similar to mutants in germ granule proteins. For example, the M granule protein MUT-16 is required for biogenesis of endogenous (Mutator-class) small RNAs (Phillips et al., 2012). When grown at elevated temperatures, mut-16 mutants exhibit reduced fertility and ectopic expression of somatic and sperm genes in the germline, thus phenocopying P granule mutants and the outcome of P granule knockdown (Campbell and Updike, 2015; Kawasaki et al., 1998, 2004; Knutson et al., 2017; Rogers and Phillips, 2020a; Spike et al., 2008a; Updike et al., 2014). Similarly, knockdown of the argonaute CSR-1 results in ectopic expression of sperm genes (Campbell and Updike, 2015). Overall, evidence from co-localization and genetic analyses strongly suggest a mechanistic link between small RNAs and germ granules. The presence of the ribonuclease RDE-8 in the M granules also supports this idea. This ribonuclease was shown to be recruited by the small RNA machinery and to be required for the formation of cleaved RNA fragments and small RNAs from the targeted mRNA (Tsai et al., 2015). A recent study provided further evidence for germ granule-mediated regulation of gene expression, which is mediated by the recruitment of the small RNA machinery. In this study, artificial tethering of a germline expressed transcript to the P granules resulted in robust silencing of the transcript (Aoki et al., 2018). Importantly, silencing was dependent on both the integrity of the P granules and the P granule argonaute WAGO-1. Altogether, the mechanism of germ granules' regulation of gene expression seems to be, at least in part, mediated by the small RNA machinery (summarized in Figure 1).
Figure 1.
Conceptual Scheme Depicting the Different Small RNA-Related Processes that Occur Inside the Germ Granules
When exiting the nucleus, the mRNA molecule passes through the germ granule. Guided by small RNA molecules different argonauts regulate the expression of the mRNA. RNA-dependent-RNA polymerases (RdRPs) amplify and maintain the small RNA response across generations by creating additional small RNAs. The amplification process involves a feedback between poly-UG RNA fragments derived from the targeted mRNA and small RNA production. The small RNAs are then processed and sorted to the different argonaute proteins. The sorting process is hypothesized to involve untemplated poly-urydilation of the small RNAs, which direct the small RNAs to specific argonauts.
Amplification of Small RNAs in Germ Granules
Different primary small RNA pathways such as dsRNA-derived small RNAs, ERGO-1-associated small RNAs, and piRNAs converge by inducing the production of similar secondary siRNAs (reviewed by Houri-Zeevi and Rechavi, 2016). These secondary siRNAs are ∼22 base pair long with a strong 5′ guanine (22G) bias (Gu et al., 2009; Pak and Fire, 2007; Aoki et al., 2007). The secondary siRNAs are antisense to the mature mRNA, covering mainly the exons. The amplification step, which is central to siRNA-mediated silencing and siRNA inheritance seems to take place in the germ granules. The RdRPs that produce the amplified siRNAs, EGO-1 and RRF-1, are localized in the germ granules (Claycomb et al., 2009; Phillips et al., 2012). In addition, mutants lacking core germ granule components misexpress different populations of amplified siRNAs (Dodson and Kennedy, 2019; Lev et al., 2019b; Ouyang et al., 2019; Suen et al., 2020) (discussed further below). Additionally, a Z granule component, the helicase-domain protein ZNFX-1, contributes to the alignment pattern of amplified small RNAs aligning along the targeted gene. In its absence, siRNAs are produced preferentially from the 5′ of the transcript (Ishidate et al., 2018). A recent study showed that the amplification step requires the presence of germ granule-localized RNA fragments with untemplated UG tails (19–75 bp long) (Shukla et al., 2020). This study demonstrated that targeting of mRNAs by silencing siRNAs leads to the production of poly-UG-tagged RNA fragments that derive from the targeted mRNAs. The poly-UG fragments then facilitate the production of additional amplified siRNAs. Together the poly-UG RNAs and amplified siRNAs form a feedforward loop that maintains the production of siRNAs and the silencing of the targeted gene. Both RDE-3, a poly-UG ribo-nucleotidyl-transferase, and the poly-UG RNAs are localized in the germ granules (specifically the M granules) (Shukla et al., 2020). In conclusion, accumulating evidence shows that germ granules serve as centers of small RNA amplification (summarized in Figure 1).
Sorting of Small RNAs in Germ Granules
Another vital step in small RNA-mediated regulation of gene expression is their assignment to different argonauts. It is generally thought that the identity of the binding argonaute determines, to some extent, the effect of the amplified small RNA on its targeted mRNA. Despite their structural similarity (same length and 5′ nucleotide bias), the different amplified 22G siRNAs are bound by different argonauts (such as WAGO-1, WAGO-4, CSR-1, and HRDE-1). How the amplified siRNAs find their correct partner argonaute is still unclear. Untemplated poly-urydilation of siRNAs is thought to be involved in this process. The siRNAs that bind with the CSR-1 argonaute (specifically the CSR-1b isoform, Charlesworth et al., 2020; van Wolfswinkel et al., 2009), tend to have longer untemplated poly-U tails when compared with the siRNAs that bind with the WAGO-1 and HRDE-1 argonauts (de Albuquerque et al., 2015; Houri-Ze'evi et al., 2016; Lev et al., 2019b; Phillips et al., 2015; van Wolfswinkel et al., 2009). Poly-uridylation of siRNAs is partially dependent on CDE-1, a germ granule-localized poly uracil polymerase (van Wolfswinkel et al., 2009; Xu et al., 2018). Accordingly, it was hypothesized that CDE-1-dependent uridylation targets the modified siRNAs for loading to specific argonauts such as CSR-1 (van Wolfswinkel et al., 2009). WAGO-4-bound siRNAs are also poly-urydilated (Xu et al., 2018). In addition, the binding of WAGO-4 to these siRNAs is defective in cde-1 mutants (Xu et al., 2018). RNAi inheritance is defective in both cde-1 and wago-4 mutants (Lev et al., 2019b; Spracklin et al., 2017; Wan et al., 2018; Xu et al., 2018). Altogether, these results lead the authors to suggest that CDE-1-dependent uridylation of small RNAs promotes the transition of siRNAs from the CSR-1 argonaute to WAGO-4 to facilitate their inheritance (Xu et al., 2018). The recent finding that like WAGO-4, the CSR-1b argonaute localizes to the Z germ granule is in accordance with this view (Charlesworth et al., 2020; Wan et al., 2018). The argonaute HRDE-1 is crucial for RNAi inheritance (Ashe et al., 2012; Buckley et al., 2012; Kalinava et al., 2017; Lev et al., 2017; Luteijn et al., 2012; Pak et al., 2012; Shirayama et al., 2012). A recent study showed that mutants of core germ granule factors MEG-3/4 exhibit ectopic uridylation of HRDE-1-class siRNAs and unexpected HRDE-1-independent inheritance of RNAi (Lev et al., 2019b). These findings further support the idea that germ granules play a role in sorting of small RNAs to specific pathways. Although poly-uridylation of siRNAs seem to distinguish between siRNA associated with different argonauts, it is still unclear whether it plays a role in their sorting. For example, it is possible that the poly-uridylation occurs only after the loading of the siRNA to the argonaute. Future studies will clarify the roles of poly-uridylation in sorting of siRNAs, and how this process is regulated by the germ granules (see Box 1 for outstanding questions).
Germ Granules Regulate the Inheritance of Small RNAs
Initial studies reported that worms with disrupted germ granule structures (defective in their organization or morphology) cannot initiate RNAi-derived siRNA responses (Robert et al., 2005; Spike et al., 2008b). The presence of key siRNA inheritance factors in the germ granules and their intricate organization further suggested that the structure of the germ granule structures will be required for inheritance of small RNAs. Recent studies on fertile mutants defective in germ granule structures revealed a more complicated picture. The proteins PGL-1, MEG-3 and MEG-4 are core constituents of germ granules (Kawasaki et al., 1998; Wang et al., 2014). The MEG-3 and MEG-4 proteins function redundantly to scaffold the germ granules (Wang et al., 2014). The stability of the germ granules is further regulated by the PP2A phosphatase and its regulatory subunit PPTR-1 (Gallo et al., 2010) (reviewed in detail by Seydoux, 2018). Accordingly meg-3;meg-4 and pptr-1 mutants have small germ granules that do not segregate well with the germ cell lineage in the embryo (Gallo et al., 2010; Wang et al., 2014). Post embryogenesis, however, the germ granules are normally formed in meg-3;meg-4 mutants (Wang et al., 2014). Despite having largely disrupted embryonic germ granules, these mutants were recently found to display potent small RNA inheritance (Lev et al., 2019b). Moreover, pptr-1 and meg-3;meg-4 exhibit enhanced transgenerational RNAi-mediated silencing that lasts for tens of generations (Lev et al., 2019b). Therefore, the integrity of the germ granules in the embryo is not essential for the inheritance of RNAi-derived small RNAs. Instead, the structure of the germ granules seems to play a role in limiting the duration of the heritable response.
Homozygous mutants lacking functional meg-3;meg-4 or pgl-1 genes gradually lose their ability to initiate RNAi responses across generations (Dodson and Kennedy, 2019; Lev et al., 2019b; Ouyang et al., 2019). This explains why previous studies found pgl-1 and meg-3;meg-4 mutants to have defective RNAi responses (Robert et al., 2005; Spike et al., 2008b; Wan et al., 2018). Further investigations showed that in the absence of meg-3;meg-4, ectopic siRNAs are transmitted to the progeny (Dodson and Kennedy, 2019; Lev et al., 2019b). These siRNAs accumulate across generations and ultimately render the mutants refractive to RNAi (Dodson and Kennedy, 2019; Lev et al., 2019b; Ouyang et al., 2019). Removal and subsequent rescue of the HRDE-1 argonaute “resets” the ability of the meg-3;4 mutants to initiate an RNAi response (Lev et al., 2019b), showing that HRDE-1-dependent siRNAs are the heritable agents that mediate this transgenerational phenomenon. Among the accumulating ectopic siRNAs in the meg-3;meg-4 mutants are siRNAs that target the sid-1 and rde-11 genes (Dodson and Kennedy, 2019; Lev et al., 2019b; Ouyang et al., 2019). These genes encode for proteins required for the RNAi responses, and specifically for the inter-tissue spreading and amplification steps of RNAi (Feinberg and Hunter, 2003; Yang et al., 2012; Zhang et al., 2012). These siRNAs silence the sid-1 and rde-11 mRNAs in meg-3;meg-4 mutants, and their inheritance is dependent on HRDE-1 (Dodson and Kennedy, 2019; Ouyang et al., 2019). Elegant RNA-FISH experiments revealed that the germ granules encapsulate the sid-1 and rde-11 mRNAs and that in meg-3;meg-4 mutants this compartmentalization is impaired (Ouyang et al., 2019). The sid-1 and rde-11 genes are regularly targeted by piRNAs, but in the wild type this targeting does not lead to the production of amplified siRNAs and silencing of these genes. The authors suggested that the germ granules protect the mRNAs of sid-1 and rde-11 from piRNA targeting (Ouyang et al., 2019). Altogether, heritable siRNA-mediated silencing of the sid-1 and rde-11 accounts for the observed heritable RNAi defects found in the meg-3;meg-4 mutants. Notably, this mechanism cannot explain the enhanced transgenerational inheritance of RNAi responses of pptr-1 mutants and early generation (following homozygosity) of meg-3;meg-4 mutants. Instead, the genome-wide disruption of competing heritable small RNA species found in these mutants seems to contribute to these inheritance dynamics (Lev et al., 2019b). For example, pptr-1 mutants exhibit a reduction in the total amounts of endogenous siRNAs (endo-siRNAs, Lev et al., 2019b). The endo-siRNAs compete over shared machinery (such as RdRPs) with exogenous dsRNA-derived siRNAs (exo-siRNAs). Therefore, in the absence of endo-siRNAs, exogenous siRNA production is potentiated. Indeed, pptr-1 mutants were shown to synthesize higher levels of exo-siRNAs (Lev et al., 2019b). Similar mechanism was described for other conditions (or mutants) where worms exhibit an enhanced RNAi phenotype (Duchaine et al., 2006; Houri-Ze'evi et al., 2016; Lev et al., 2017). Recently an eri-6/7-based feedback between MUT-16 and ERGO-1 small RNA pathways (Rogers and Phillips, 2020b) was described. It is possible that similar feedback mechanism between different small RNA pathways may also be involved in these inheritance patterns. In summary, recent studies revealed that germ granules maintain proper endogenous small RNA production and regulate the animal's capacity to produce and inherit RNAi-derived siRNAs.
Heritable Small RNAs Maintain the Effects of Germ Granule Disruption across Generations
As germ granules regulate heritable small RNAs, even transient changes to the germ granule assembly or composition can have transgenerational consequences. In accordance with this idea, wild-type progenies that are isolated from a cross with meg-3;meg-4 mutants exhibit defects in both initiation and inheritance of RNAi responses (Dodson and Kennedy, 2019; Lev et al., 2019b). These defects are maintained in the wild-type lineage for multiple generations, and their inheritance depends on the argonaute HRDE-1 (Dodson and Kennedy, 2019; Lev et al., 2019b). This heritable phenotype was accompanied by inheritance of many HRDE-1-dependent endogenous siRNAs to the wild-type progeny (Dodson and Kennedy, 2019; Lev et al., 2019b). Recently, heritable phenotypes have also been described for mutants of the germ granule-residing argonauts ERGO-1 and PRG-1. In both cases, the heritable phenotypes are mediated by small RNAs. The F1 wild-type progeny of mutants of the P granule argonaute ERGO-1 are more sensitive to RNAi (Almeida et al., 2019). Similarly, wild-type progeny of mutants in the P granules-residing argonaute PRG-1 have reduced fertility for multiple generations (Barucci et al., 2020). Importantly, other examined germ granule components were unaltered in prg-1 mutants. Nevertheless, both studies highlight that transient changes to germ granule-residing small RNA factors can lead to heritable phenotypes. In conclusion, changes to germ granule integrity or composition can affect the physiology of ensuing generations via their effect on regulatory small RNAs.
Interestingly, the meg-3;meg-4-defective RNAi initiation phenotype is transmitted more efficiently through the maternal lineage, as wild-type progeny of meg-3;meg-4 males have less severe RNAi defects than progeny of meg-3;meg-4 hermaphrodites (Lev et al., 2019b). This maternal effect is not due to inheritance of the defective granules (Lev et al., 2019b). Intriguingly, the earliest report on the M granule protein MUT-7, which plays a key role in small RNA biogenesis, described a similar phenomenon (Ketting et al., 1999). Heterozygotus mut-7 (+/−) mutants had less severe RNAi defects if they were segregated from a cross with a mutant male (Ketting et al., 1999). Recently, a similar maternal parent-of-origin effect was reported for small RNA-based transgenerational inheritance of an endoderm development phenotype (Ewe et al., 2020). Further research would reveal if this is a general theme for small RNA-based heritable responses.
Do Germ Granules Make Heritable Small RNAs Sensitive to the Environment?
As discussed above, various environmental challenges induce heritable physiological responses. Heritable small RNAs have been implicated in the inheritance of some of these effects. It is, however, still unclear how abiotic environmental changes induce the production of specific small RNA species. The ability of germ granules to affect heritable small RNA populations mark them as a likely candidate mediator (Dodson and Kennedy, 2019; Lev et al., 2019b; Ouyang et al., 2019). Germ granules possess liquid-like properties such as fusion, dripping, and wetting (Brangwynne et al., 2009). As biophysical entities, the P and M germ granules are sensitive to temperature and tend to disperse as temperatures rise (Putnam et al., 2019; Uebel et al., 2018). Transient growth at higher temperatures elevates the expression of transgenes and repetitive elements (Klosin et al., 2017). These gene-expression changes are inherited for multiple generations. Growth at high temperatures also interferes with piRNA production (Belicard et al., 2018) and temperature shifts impact heritable small RNAs and the expression of some of their endogenous targets (Manage et al., 2020; Ni et al., 2016; Schott et al., 2014). As discussed above, mutants in structural germ granule components and germ granule-residing small RNA factors exhibit temperature-dependent sterility. It is therefore tempting to speculate that the intrinsic biophysical sensitivity of germ granules to temperature allows them to translate transient temperature shifts into changes in the pools of heritable small RNAs. As germ granules are highly conserved, it is possible that they will play a similar role in other organisms. Future studies will uncover the involvement of germ granules in rendering small RNAs responsive to the environment.
Acknowledgments
We thank Itai Antoine Toker, Shahar Bracha, and Nitzan Aframian for reading the paper and for their insightful comments. I.L. thanks the EMBO funding agency for the personal long-term fellowship (ALTF 1037-2019). O.R. is supported by the Adelis Foundation (grant 0604916191) and the Israel Science Foundation (grant #1339/17).
Author Contributions
I.L and O.R. conceptualized and wrote the paper.
Contributor Information
Itamar Lev, Email: itamai.et@gmail.com.
Oded Rechavi, Email: odedrechavi@gmail.com.
References
- Alcazar R.M., Lin R., Fire A.Z. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.V., de Jesus Domingues A.M., Ketting R.F. Maternal and zygotic gene regulatory effects of endogenous RNAi pathways. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1007784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S.T., Lynch T., Crittenden S., Bingman C., Wickens M., Kimble J. Liquid droplet germ granules require assembly and localized regulators for mRNA repression. bioRxiv. 2018:382838. doi: 10.1101/382838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J. PiRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barucci G., Cornes E., Singh M., Li B., Ugolini M., Samolygo A., Didier C., Dingli F., Loew D., Quarato P., Cecere G. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 2020;22:235–245. doi: 10.1038/s41556-020-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P.J. PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Molecular Cell. 2008 doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belicard T., Jareosettasin P., Sarkies P. The piRNA pathway responds to environmental signals to establish intergenerational adaptation to stress. BMC Biol. 2018;16:103. doi: 10.1186/s12915-018-0571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshore E.L. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Developmental Biology. 2011 doi: 10.1016/j.ydbio.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/SCIENCE.1172046. [DOI] [PubMed] [Google Scholar]
- Brown K.C. ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Research. 2017 doi: 10.1093/nar/gkx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton N.O., Burkhart K.B., Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A.C., Updike D.L. CSR-1 and P granules suppress sperm-specific transcription in the C. elegans germline. Development. 2015;142:1745–1755. doi: 10.1242/dev.121434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casier K., Delmarre V., Gueguen N., Hermant C., Viodé E., Vaury C., Ronsseray S., Brasset E., Teysset L., Boivin A. Environmentally-induced epigenetic conversion of a piRNA cluster. Elife. 2019;8 doi: 10.7554/eLife.39842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A.G., Lehrbach N.J., Seroussi U., Renaud M.S., Molnar R.I., Woock J.R., Aber M.J., Diao A.J., Ruvkun G., Claycomb J.M. Two isoforms of the essential C. elegans Argonaute CSR-1 differentially regulate sperm and oocyte fertility through distinct small RNA classes. bioRxiv. 2020 doi: 10.1101/2020.07.20.212050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Chun Chieh G., Simard Martin J, Tabara Hiroaki, Brownell Daniel R, McCollough Jennifer A, Mello Craig C. A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Current Biology. 2005 doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F. The argonaute CSR-1 and its 22g-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine Colin C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine Colin C. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. Elegans. Cell. 2013 doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C., Sun F., Song L., Rivera-Pérez J., Rando O. Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. bioRxiv. 2018:311670. doi: 10.1101/311670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque B.F.M., Placentino M., Ketting R.F. Maternal piRNAs are essential for germline development following de novo establishment of endo-siRNAs in Caenorhabditis elegans. Dev. Cell. 2015;34:448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Dodson A.E., Kennedy S. Germ granules coordinate RNA-based epigenetic inheritance pathways. Dev. Cell. 2019;50:704–715.e4. doi: 10.1016/j.devcel.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Eddy E.M.M. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 1976;43:229–280. doi: 10.1016/S0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Ewe C.K., Torres Cleuren Y.N., Flowers S.E., Alok G., Snell R.G., Rothman J.H. Natural cryptic variation in epigenetic modulation of an embryonic gene regulatory network. Proc. Natl. Acad. Sci. U S A. 2020;117:13637–13646. doi: 10.1073/pnas.1920343117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg E.H., Hunter C.P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gallo C.M., Wang J.T., Motegi F., Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/SCIENCE.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon D.B., Ishidate T., Li L., Gu W., Silverman N., Mello C.C. The antiviral RNA interference response provides resistance to lethal arbovirus infection and vertical transmission in Caenorhabditis elegans. Curr. Biol. 2017;27:795–806. doi: 10.1016/j.cub.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E., Mansuy I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V., Fourré S., De Abreu D.A.F., Derieppe M.A., Remy J.J., Rassoulzadegan M. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci. Rep. 2015;5:18193. doi: 10.1038/srep18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Tabara H., Mello C.C. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J. Distinct argonaute-mediated 22g-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa M., Yonetani M., Sugimoto A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 2011 doi: 10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegner R.W. Studies on germ cells. I. The history of the germ cells in insects with special reference to the Keimbahn-determinants. II. The origin and significance of the Keimbahn-determinants in animals. J. Morphol. 1914;25:375–509. doi: 10.1002/jmor.1050250302. [DOI] [Google Scholar]
- Hermant C., Boivin A., Teysset L., Delmarre V., Asif-Laidin A., Van Den Beek M., Antoniewski C., Ronsseray S. Paramutation in drosophila requires both nuclear and cytoplasmic actors of the piRNA pathway and induces cis-spreading of piRNA production. Genetics. 2015;201:1381–1396. doi: 10.1534/genetics.115.180307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird S.N., Paulsen J.E., Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- Houri-Ze’evi L., Korem Y., Sheftel H., Faigenbloom L., Toker I.A., Dagan Y., Awad L., Degani L., Alon U., Rechavi O. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- Houri-Zeevi L., Korem Kohanim Y., Antonova O., Rechavi O. Three rules explain transgenerational small RNA inheritance in C. elegans. Cell. 2020;182:1186–1197.e12. doi: 10.1016/j.cell.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houri-Zeevi L., Rechavi O. A matter of time: small RNAs regulate the duration of epigenetic inheritance. Trends Genet. 2016;xx:1–12. doi: 10.1016/j.tig.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Ishidate T., Ozturk A.R., Durning D.J., Sharma R., Shen E., Chen H., Seth M., Shirayama M., Mello C.C. ZNFX-1 functions within perinuclear nuage to balance epigenetic signals. Mol. Cell. 2018;70:639–649.e6. doi: 10.1016/J.MOLCEL.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–120. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- Ito H., Kim J.M., Matsunaga W., Saze H., Matsui A., Endo T.A., Harukawa Y., Takagi H., Yaegashi H., Masuta Y. A stress-activated transposon in arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R., Moore R.S., Vrla G.D., Parsons L.R., Gitai Z., Murphy C.T. C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance. Nature. 2020:1–7. doi: 10.1038/s41586-020-2699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinava N., Ni J.Z., Gajic Z., Kim M., Ushakov H., Gu S.G. C. elegans heterochromatin factor SET-32 plays an essential role in transgenerational establishment of nuclear RNAi-mediated epigenetic silencing. Cell Rep. 2018;25:2273–2284.e3. doi: 10.1016/J.CELREP.2018.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinava N., Ni J.Z., Peterman K., Chen E., Gu S.G. Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenetics Chromatin. 2017;10:6. doi: 10.1186/s13072-017-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Amiri A., Fan Y., Meyer N., Dunkelbarger S., Motohashi T., Karashima T., Bossinger O., Strome S. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167:645–661. doi: 10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/S0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp T.H.A., Van Luenen H.G.A.M., Plasterk R.H.A. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/S0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- Knutson A.K., Egelhofer T., Rechtsteiner A., Strome S. Germ granules prevent accumulation of somatic transcripts in the adult Caenorhabditis elegans germline. Genetics. 2017;206:163–178. doi: 10.1534/genetics.116.198549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg C., Cole T., Deppe U., Schierenberg E., Schmitt D., Yoder B., von Ehrenstein G. The cellular anatomy of embryos of the nematode Caenorhabditis elegans. Analysis and reconstruction of serial section electron micrographs. Dev. Biol. 1978;65:193–215. doi: 10.1016/0012-1606(78)90190-2. [DOI] [PubMed] [Google Scholar]
- Lev I., Seroussi U., Gingold H., Bril R., Anava S., Rechavi O. MET-2-Dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Lev I., Gingold H., Rechavi O. H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes. Elife. 2019;8:338582. doi: 10.7554/elife.40448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev I., Toker I.A., Mor Y., Nitzan A., Weintraub G., Antonova O., Bhonkar O., Ben Shushan I., Seroussi U., Claycomb J.M. Germ granules govern small RNA inheritance. Curr. Biol. 2019;29:2880–2891.e4. doi: 10.1016/j.cub.2019.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Feng L., Gu X., Deng X., Qiu Q., Li Q., Zhang Y., Wang M., Deng Y., Wang E. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019;29:379–390. doi: 10.1038/s41422-019-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M.J., van Bergeijk P., Kaaij L.J.T., Almeida M.V., Roovers E.F., Berezikov E., Ketting R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manage K.I., Rogers A.K., Wallis D.C., Uebel C.J., Anderson D.C., Nguyen D.A.H., Arca K., Brown K.C., Rodrigues R.J.C., de Albuquerque B.F.M. A tudor domain protein, SIMR-1, promotes sirna production at pirna-targeted mrnas in C. Elegans. Elife. 2020;9 doi: 10.7554/eLife.56731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Zhu C., Zong D., Weng C., Yang X., Huang H., Liu D., Feng X., Guang S. The nrde pathway mediates small-RNA-directed histone H3 lysine 27 trimethylation in Caenorhabditis elegans. Curr. Biol. 2015;25:2398–2403. doi: 10.1016/j.cub.2015.07.051. [DOI] [PubMed] [Google Scholar]
- Moore R.S., Kaletsky R., Murphy C.T. Piwi/PRG-1 argonaute and TGF-β mediate transgenerational learned pathogenic avoidance. Cell. 2019;177:1827–1841.e12. doi: 10.1016/j.cell.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes to Cells. 2007 doi: 10.1111/j.1365-2443.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Kalinava N., Chen E., Huang A., Trinh T., Gu S.G. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin. 2016;9:3. doi: 10.1186/s13072-016-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J.P.T., Folkmann A., Bernard L., Lee C.Y., Seroussi U., Charlesworth A.G., Claycomb J.M., Seydoux G. P granules protect RNA interference genes from silencing by piRNAs. Dev. Cell. 2019;50:716–728.e6. doi: 10.1016/j.devcel.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007 doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pak J., Maniar J.M., Mello C.C., Fire A. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell. 2012;151:885–899. doi: 10.1016/j.cell.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palominos M.F., Verdugo L., Gabaldon C., Pollak B., Ortíz-Severín J., Varas M.A., Chávez F.P., Calixto A. Transgenerational diapause as an avoidance strategy against bacterial pathogens in Caenorhabditis elegans. MBio. 2017;8 doi: 10.1128/mBio.01234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec D.M. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009 doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A.G., Gracida X., Kagias K., Zhang Y. C. elegans aversive olfactory learning generates diverse intergenerational effects. J. Neurogenet. 2020:1–11. doi: 10.1101/2020.02.07.939017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M.F., Lehner B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 2019;21:143–151. doi: 10.1038/s41556-018-0242-9. [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Brown K.C., Montgomery B.E., Ruvkun G., Montgomery T.A. PiRNAs and piRNA-dependent siRNAs protect conserved and essential C. elegans genes from misrouting into the RNAi pathway. Dev. Cell. 2015;34:457–465. doi: 10.1016/j.devcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C.M., Montgomery T.A., Breen P.C., Ruvkun G. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012;26:1433–1444. doi: 10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner R., Toker I.A. Neuronal Small RNAs Control Behavior Transgenerationally. Cell. 2019 doi: 10.1016/j.cell.2019.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam A., Cassani M., Smith J., Seydoux G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019;26:220–226. doi: 10.1038/s41594-019-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics. 1995 doi: 10.1093/genetics/141.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O., Houri-Ze’evi L., Anava S., Goh W.S.S., Kerk S.Y., Hannon G.J., Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O., Minevich G., Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter R. Die Entwicklung der Geschlechtsorgane nnd des Darmes bei Ghironomns. Von. Z. für wiss. Zool. 1890;50:408. [Google Scholar]
- Robert V.J.P., Sijen T., Van Wolfswinkel J., Plasterk R.H.A. Chromatin and RNAi factors protect the C. elegans germline against repetaitive sequences. 2005. [DOI] [PMC free article] [PubMed]
- Rodgers A.B., Morgan C.P., Leu N.A., Bale T.L. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. U S A. 2015;112:13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A.K., Phillips C.M. RNAi pathways repress reprogramming of C. elegans germ cells during heat stress. Nucleic Acids Res. 2020;48:4256–4273. doi: 10.1093/nar/gkaa174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A.K., Phillips C.M. A small-RNA-mediated feedback loop maintains proper levels of 22g-RNAs in C. elegans. Cell Rep. 2020;33:108279. doi: 10.1016/j.celrep.2020.108279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa J.A., Pitt J.N., Priess J.R. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development. 2001;128:1287–1298. doi: 10.1242/dev.128.8.1287. [DOI] [PubMed] [Google Scholar]
- Schott D., Yanai I., Hunter C.P. Natural RNA interference directs a heritable response to the environment. Sci. Rep. 2014;4:7387. doi: 10.1038/srep07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G. The P granules of C. elegans: a genetic model for the study of RNA–protein condensates. J. Mol. Biol. 2018 doi: 10.1016/j.jmb.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Pitt J., Dennis S., Priess J.R. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M. The Vasa homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Current Biology. 2014 doi: 10.1016/j.cub.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.C., Gu W., Ishidate T., Conte D., Mello C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Yan J., Pagano D.J., Dodson A.E., Fei Y., Gorham J., Seidman J.G., Wickens M., Kennedy S. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature. 2020;582:283–288. doi: 10.1038/s41586-020-2323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Sarkies P., Ikegami K., Doebley A.-L., Goldstein L.D., Mitchell J., Sakaguchi A., Miska E.A., Ahmed S. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans piwi mutants. Cell Rep. 2014;7:762–773. doi: 10.1016/j.celrep.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C., Meyer N., Racen E., Orsborn A., Kirchner J., Kuznicki K., Yee C., Bennett K., Strome S. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics. 2008;178:1973–1987. doi: 10.1534/genetics.107.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Current Biology. 2000 doi: 10.1016/S0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Spike C.A., Bader J., Reinke V., Strome S., Kohara Y., Rhoads R.E., Strome S. DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development. 2008;135:983–993. doi: 10.1242/dev.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G., Fields B., Wan G., Vijayendran D., Wallig A., Shukla A., Kennedy S. Identification and characterization of C. elegans RNAi inheritance machinery. Genetics. 2017:1–19. doi: 10.20944/preprints201702.0096.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W.B. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Suen K.M., Braukmann F., Butler R., Bensaddek D., Akay A., Lin C.C., Milonaitytė D., Doshi N., Sapetschnig A., Lamond A. DEPS-1 is required for piRNA-dependent silencing and PIWI condensate organisation in Caenorhabditis elegans. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-18089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.Y., Chen C.C.G., Conte D., Moresco J.J., Chaves D.A., Mitani S., Yates J.R., Tsai M.D., Mello C.C. A ribonuclease coordinates siRNA amplification and mRNA Cleavage during NAi. Cell. 2015;160:407–419. doi: 10.1016/j.cell.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebel C.J., Anderson D.C., Mandarino L.M., Manage K.I., Aynaszyan S., Phillips C.M. Distinct regions of the intrinsically disordered protein MUT-16 mediate assembly of a small RNA amplification complex and promote phase separation of Mutator foci. PLoS Genet. 2018;14:e1007542. doi: 10.1371/journal.pgen.1007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D.L., Hachey S.J., Kreher J., Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 2011;192:939–948. doi: 10.1083/JCB.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D.L., Knutson A.K.A.K., Egelhofer T.A., Campbell A.C., Strome S. Germ-granule components prevent somatic development in the C. Elegans germline. Curr. Biol. 2014;24:970–975. doi: 10.1016/j.cub.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D.L., Strome S. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics. 2009;183:1397–1419. doi: 10.1534/genetics.109.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel J.C., Claycomb J.M., Batista P.J., Mello C.C., Berezikov E., Ketting R.F. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Vasale J.J., Gu W., Thivierge C., Batista P.J., Claycomb J.M., Youngman E.M., Duchaine T.F., Mello C.C., Conte D. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. U S A. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N.L. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 2003 doi: 10.1016/S0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- Vastenhouw N.L., Brunschwig K., Okihara K.L., Müller F., Tijsterman M., Plasterk R.H.A. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Vought V.E., Ohmachi M., Lee M.H., Maine E.M. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/Notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics. 2005;170:1121–1132. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G., Fields B.D., Spracklin G., Shukla A., Phillips C.M., Kennedy S. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature. 2018;557:679–683. doi: 10.1038/s41586-018-0132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Reinke V. A C. elegans Piwi, PRG-1, Regulates 21U-RNAs during Spermatogenesis. Curr. Biol. 2008 doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3 doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A. Charles Scribner’s Sons; 1893. The Germ-Plasm: A Theory of Heredity. [Google Scholar]
- Welker N.C. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA. 2010 doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N., Priess J., Hirsh D. Segregation of germline granules in early embryos of Caenorhabditis elegans: an electron microscopic analysis. J. Embryol. Exp. Morphol. 1983;73:297–306. [PubMed] [Google Scholar]
- Xu F., Feng X., Chen X., Weng C., Yan Q., Xu T., Hong M., Guang S. A cytoplasmic argonaute protein promotes the inheritance of RNAi. Cell Rep. 2018;23:2482–2494. doi: 10.1016/j.celrep.2018.04.072. [DOI] [PubMed] [Google Scholar]
- Yang H., Zhang Y., Vallandingham J., Li H., Florens L., Mak H.Y. The RDE-10/RDE-11 complex triggers RNAi-induced mRNA degradation by association with target mRNA in C. elegans. Genes Dev. 2012;26:846–856. doi: 10.1101/gad.180679.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E. Analysis of the C. elegans Argonaute Family Reveals that Distinct Argonautes Act Sequentially during RNAi. Cell. 2006 doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zhang C., Montgomery T.A., Fischer S.E.J., Garcia S.M.D.A., Riedel C.G., Fahlgren N., Sullivan C.M., Carrington J.C., Ruvkun G. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 2012;22:881–890. doi: 10.1016/j.cub.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Shi J., Tuorto F., Li X., Liu Y., Liebers R., Zhang L., Qu Y., Qian J. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 2018;20:535–540. doi: 10.1038/s41556-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S.H., Liu J.Z., Jin H., Lin L., Li Q., Chen Y., Yuan Y.X., Wang Z.Y., Huang H., Qi Y.J. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2013;110:9171–9176. doi: 10.1073/pnas.1219655110. [DOI] [PMC free article] [PubMed] [Google Scholar]