Abstract
Purpose
Magnetic resonance-guided online adaptive radiation therapy (MRgOART) requires accurate and efficient segmentation. However, the performance of current autosegmentation tools is generally poor for magnetic resonance imaging (MRI) owing to day-to-day variations in image intensity and patient anatomy. In this study, we propose a patient-specific autosegmentation strategy using multiple-input deformable image registration (DIR; PASSMID) to improve segmentation accuracy and efficiency for MRgOART.
Methods and materials
Longitudinal MRI scans acquired on a 1.5T MRI-Linac for 10 patients with abdominal cancer were used. The proposed PASSMID includes 2 steps: applying a patient-specific image processing pipeline to longitudinal MRI scans, and populating all contours from previous sessions/fractions to a new fractional MRI using multiple DIRs and combining the resulted contours using simultaneous truth and performance level estimation (STAPLE) to obtain the final consensus segmentation. Five contour propagation strategies were compared: planning computed tomography to fractional MRI scans through rigid body registration (RDR), pretreatment MRI to fractional MRI scans through RDR and DIR, and the proposed multi-input DIR/STAPLE without preprocessing, and the PASSMID. Dice similarity coefficient (DSC) and mean distance to agreement (MDA) with ground truth contours were calculated slice by slice to quantify the contour accuracy. A quantitative index, defined as the ratio of acceptable slices, was introduced using a criterion of DSC > 0.8 and MDA < 2 mm.
Results
The proposed PASSMID performed well with an average 2-dimensional DSC/MDA of 0.94/1.78 mm, 0.93/1.04 mm, 0.93/1.06 mm, 0.93/1.14 mm, 0.92/0.83 mm, 0.84/1.53 mm, 0.86/2.39 mm, 0.81/2.49 mm, 0.72/5.48 mm, and 0.70/5.03 mm for the liver, left kidney, right kidney, spleen, aorta, pancreas, stomach, duodenum, small bowel, and colon, respectively. Starting from the third fractions, the contour accuracy was significantly improved with PASSMID compared with the single-DIR strategy (P < .05). The mean ratio of acceptable slices were 13.9%, 17.5%, 60.8%, 70.6%, and 71.8% for the 5 strategies, respectively.
Conclusions
The proposed PASSMID solution, by combining image processing, multi-input DIRs, and STAPLE, can significantly improve the accuracy of autosegmentation for intrapatient MRI scans, reducing the time required for further contour editing, thereby facilitating the routine practice of MRgOART.
Introduction
With the incorporation of a magnetic resonance imaging (MRI) scanner into a traditional linac machine (eg, MRI_Linac),1 MRI-guided (MRg) online adaptive radiation therapy (OART) is being introduced into the clinic. For each fraction, MRI with superior soft tissue contrast and possibly functional information is acquired before treatment delivery to provide patient anatomy of the day without additional radiation exposure. Thus, an adaptive treatment plan can be developed using the fractional MRI. MRgOART has to be efficient because the plan adaptation must be accomplished online while the patient is on the couch awaiting treatment.2 Currently, the bottleneck in the process is the efficiency and accuracy of segmenting the patient’s daily anatomy because the segmentation affects all subsequent processes, such as plan optimization, evaluation, and accumulation.
Manual segmentation is tedious and time-consuming, and can take >30 minutes,3 presenting a barrier to widespread implementation for OART. Registration-based contour propagation is a way to accelerate the contouring process, whereby organ contours from a reference image are mapped to the fractional image through image registration, either rigidly or deformably. The performance of the rigid registration is usually limited by patient anatomic changes, such as weight loss, tumor shrinkage, and organ motion. To account for these changes, deformable image registration (DIR)–based contour propagation is performed by establishing voxel-to-voxel spatial correspondence through an iterative optimization process that maximizes the similarity between the fractional and reference images.4 With the availability of several commercial systems, DIR has gained widespread acceptance for the application of automatic contouring for specific treatment sites.5 For instance, clinically acceptable DIR-based autosegmentations have been reported in regions that experience minimal deformation, such as the head and neck region.6 However, current DIR algorithms for organs that may exhibit large deformations, such as the abdominal organs, frequently do not yield acceptable results.5
In current use of the DIR-based contour propagation method, only 1 single reference image set is used.7 The reference is typically chosen to be a precontoured image set from the same subject (ie, previously acquired images) or from other subjects.8 McLaren et al proposed population-based average contour templates for DIR using multiple trainning images/contours to better accomodate subject variations.9 Intrapatient fractional MRI scans acquired during MRgOART share inherently higher anatomic and image similarity over interpatient images. In current MRgOART practice, target(s) and critical structures are usually autocontoured and then manually modified for plan adaptation for each fraction.10 Longitudinal images acquired from a patient’s previous fractions contain the anatomy variation for the patient. Therefore, these images are potentially useful to provide patient-specific information that would aid in the autosegmentation of new fractional images. However, these intrapatient image characteristics have been ignored in general autosegmentation solutions.
Unlike standardized computed tomography (CT) images, MRI intensities do not possess tissue-specific numeric meanings.11 Significant intensity variations have been observed among images across subjects, protocols, and scanners. Intensity inhomogeneities exist even for the same patient using the same protocol on the same scanner.12 This inherent feature of MRI scans can adversely impact the accuracy and precision of any intensity-based image analysis procedure, such as registration, segmentation, and quantitative radiomics studies.13,14 Image processing methods that can improve the image quality or similarity between the subject and reference images may potentially improve the performance of DIR and DIR-based autosegmentation.
In this study, we proposed a novel patient-specific autosegmentation strategy using multi-input DIR (PASSMID) that combines an image processing pipeline and multi-input DIRs to improve the accuracy and efficiency of autosegmentation. The feasibility and effectiveness of the proposed approach was demonstrated using fractional MRI scans from patients who underwent MRgOART with a 1.5T MRI-Linac (Unity, Elekta AB) at our institution. To the best of our knowledge, there is no similar approach reported in the literature.
Methods and Materials
Images and contours
Fractional MRI scans acquired before treatment delivery from 10 patients with abdominal cancer (4 pancreas, 5 liver, and 1 adrenal) treated with MRgOART on the MRI-Linac were used to demonstrate the proposed method. For each patient, 7 longitudinal MRI scans from the MRI-Linac, including a pretreatment MRI scan (for imaging optimization) taken before the first fraction and 6 fractional MRI scans (except for 1 patient with liver cancer with only 5 fractional MRI scans) were used. MRI acquisition parameters, including protocol time, echo time, repetition time, and flip angle, are summarized in Table 1 for each patient. Each MRI scan consisted of 63 to 100 slices with a spatial resolution of 1.6 mm in each 2-dimensional (2D) transversal slice and 5 mm in slice thickness. For each patient, an optimal scanning protocol for better tumor/organs-at-risk visualization was determined in the imaging optimization session before the first treatment. In our current clinical practice of MRgOART, a set of free-breathing 4-dimensional MRI scans15 were acquired to reconstruct a motion-averaged image set. Contours of target and organs at risk were first rigidly propagated from planning CT to the fractional MRI scans and then edited manually by the MRI-Linac team members (ie, physicians, physicists, and therapists) using a parallel recontouring workflow.10 For this retrospective study, these clinical contours were manually checked and modified by 2 experienced researchers and then used as the ground truth. Ten abdominal organs at risk were included: liver, left kidney, right kidney, spleen, aorta, pancreas, stomach, duodenum, small bowel, and colon.
Table 1.
Magnetic resonance imaging acquisition parameters for each patient
| Patient no. | Tumor site | No. of images | Protocol | Echo Time/msec | Repetition time in ms | Flip angle in degrees |
|---|---|---|---|---|---|---|
| 1 | Pancreas | 7 | FFE | 1.44 | 4.11 | 4 |
| 3.30 | 6.80 | 25 | ||||
| 2 | Liver | 7 | FFE | 1.44 | 4.11 | 4 |
| 3.30 | 6.80 | 25 | ||||
| 3 | Liver | 7 | FFE | 1.44 | 4.11 | 4 |
| 3.30 | 6.80 | 25 | ||||
| 4 | Pancreas | 7 | FFE | 3.30 | 6.80 | 25 |
| 5 | Liver | 6 | tFE2 | 1.85 | 4.60 | 25 |
| 6 | Liver | 7 | tFE | 1.85 | 4.60 | 25 |
| 7 | Pancreas | 7 | btFE3 | 2.07 | 4.30 | 50 |
| 8 | Left adrenal | 7 | btFE | 2.07 | 4.30 | 50 |
| 9 | Liver | 7 | btFE_f4 | 2.07 | 4.30 | 50 |
| 10 | Pancreas | 7 | btFE | 2.07 | 4.30 | 50 |
Abbreviations: btFE = balanced turbo field echo; btFE_f = fat suppressed balanced fast field echo; FFE = fast-field echo; tFE = turbo-field echo.
Proposed solution
The proposed PASSMID, as illustrated in Figure 1, includes 2 steps. First, a patient-specific MRI processing pipeline is applied to the longitudinal MRI scans of the patient to improve the image quality and similarity. Second, all images along with the contours from previous sessions/fractions are populated onto the MRI of a subsequent fraction by multiple DIRs, and the resulting deformed contours are then combined using a statistical label fusing algorithm (ie, simultaneous truth and performance level estimation [STAPLE]15) to obtain the final consensus segmentation for the fractional MRI. The STAPLE method was originally proposed for the estimation of the unknown ground truth from multiple manual segmentations of the same image.15 In this study, the STAPLE method was applied to the fusion of contours from multiple intrapatient DIR-based segmentations.
Figure 1.
Proposed patient-specific autosegmentation strategy using multiple-input deformable image registration.
Preprocessing pipeline
The proposed image processing pipeline in PASSMID consists of 4 steps. The first step is bias correction, whereby all MRI scans are corrected for the radio-frequency coil inhomogeneity via an intensity-based nonparametric bias correction method (ie, N4 algorithm).16 For the second step, denoising, the anisotropic diffusion filter17 was used for each image set to smooth noisy pixels while preserving the intensity of most edges. The third step involves intensity clipping. To clip off the air/noisy voxels outside the body and high-intensity outlier voxels, each image is thresholded using an intensity threshold, determined as the first peak and the 99.5 percentile from the histogram (bin width 100). The fourth and final step is intensity normalization. All images are first linearly normalized to a maximum intensity of 10240. Compared with the pretreatment MRI from the same patient, if the Pearson correlation coefficient is <0.99, a landmark-based histogram matching was performed to improve the image similarity.18
Bias correction and denoising were performed using commercial software (MIM Software, Cleveland, OH), and the succeeding steps were done automatically using an in-house Matlab program.
Automatic contour propagation strategies
Five autogenerated contour sets on each fractional MRI scan were generated using the following strategies: S1, CT-MRI-rigid (rigidly transferring the ground truth contours from the planning CT to the fractional MRI using rigid registration); S2, MRI-MRI-rigid (rigidly transferring the ground truth contours from the pretreatment MRI to each fractional MRI); S3, original-single (propagating the ground truth contours from the pretreatment MRI to a fractional MRI using DIR without first processing the MRI image); S4, original STAPLE (populating the contours from all previous MRI scans to a new fractional MRI using DIR without image processing, then fusing all contours using the STAPLE algorithm); and S5, PASSMID (repeating the multiple-input DIR method described in S4, but using the processed MRI scans).
The rigid registration was performed manually using the MIM software to ensure that gross tumor volume contours were aligned as accurately as possible. For DIR and STAPLE fusion, a research software (ADMIRE, research 3.7-beta1, Elekta AB, Stockholm, Sweden) was used. The general image registration framework in ADMIRE consists of 3 major steps: global mutual information-based linear registration, block-matching-based 3-dimensional nonlinear image registration, and nonparametric full free-form deformable registration with a hybrid image matching metric.19, 20, 21
Contour quality evaluation
For every autogenerated contour from different strategies, 2 similarity indices (ie, Dice similarity coefficient [DSC] and mean distance to agreement [MDA]) with the corresponding ground truth contours were calculated to quantify the contouring accuracy. For a large or serial organ, such as the liver and bowels, different ranges were delineated for different fractions. As a result, the autogenerated contours (both rigid and DIR contours) may have different numbers of slices compared with the ground truth, which deteriorates the 3-dimensional volume-overlapping index (ie, 3-dimensional DSC). To avoid this, both the DSC and MDA were calculated on a slice-by-slice basis, and the mean value across all slices were reported. A pair-wise t test was used to evaluate the statistical difference between any pairs of contours from different strategies, and P < .05 was considered statistically significant.
In addition, the impact of using different numbers of prior images/contours in the proposed PASSMID method was also evaluated. For simplification purposes, only results from the single-input DIR-based method (S3) and PASSMID method (S5) were used for this comparison. For S3, only the pretreatment MRI scan was used for all fractional MRI scans, but for PASSMID, the number of prior images/contours was equal to the corresponding fraction number. The difference between 2D DSC for each fraction and the trends of mean value with the increased number of inputs was analyzed and compared between the 2 methods.
To evaluate the improvement on the contouring efficiency, a quantitative index, defined as the ratio of acceptable slices (ROA) was introduced for each strategy using a criterion of DSC > 0.8 and MDA < 2 mm based on the recommendation by the AAPM Task Group report 132.22 A hgher ROA indicates less time needed for further manual contour editing.
Results
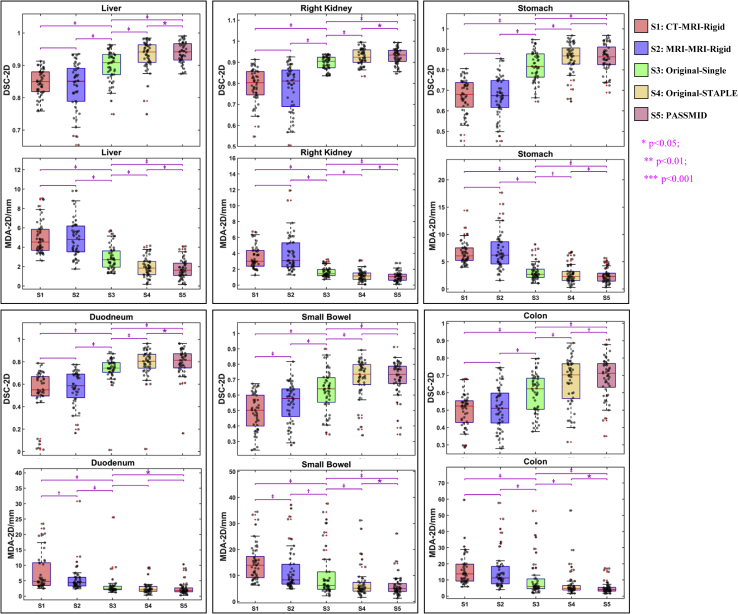
In general, the contour quality improves gradually from strategy 1 to 5. The accuracy of DIR-based contours (S3, S4, and S5) is higher than those using rigid registration (S1 and S2). The proposed PASSMID method had the best performance, with an average 2D DSC/MDA of 0.94/1.78 mm, 0.93/1.04 mm, 0.93/1.06 mm, 0.93/1.14 mm, 0.92/0.83 mm, 0.84/1.53 mm, 0.86/2.39 mm, 0.81/2.49 mm, 0.72/5.48 mm, and 0.70/5.03 mm for the liver, left kidney, right kidney, spleen, aorta, pancreas, stomach, duodenum, small bowel, and colon, respectively. Figure 2 shows the contour accuracy comparisons of 2D DSC and MDA for 6 representative organs (liver, right kidney, stomach, duodenum, small bowel, and colon) generated from different strategies. For the multi-input DIR strategies (S4 and S5), 2D DSCs are all significantly higher (P < .001) and 2D MDAs are significantly smaller (P < .01) compared with those from the commonly used rigid propagation (S1 and S2) or single-reference DIR-based contours (S3). Moreover, the contour accuracy is further improved with the proposed PASSMID method with significant difference (P < .05) compared with the results using original images (S4). The contour accuracy comparison for the remaining organs (aorta, left kidney, pancreas, and spleen) is provided in the Figure E2.
Figure 2.
Box-and-whisker plots of 2-dimensional Dice similarity coefficient and mean distance to agreement for autogenerated contours (liver, right kidney, stomach, duodenum, small bowel, and colon) using different strategies. The sample distribution is also shown with black dots. ∗P < .05 based on the paired t test of the linked 2 boxes; †P < .01 based on the paired t test of the linked 2 boxes; ‡P < .001 based on the paired t test of the linked 2 boxes.
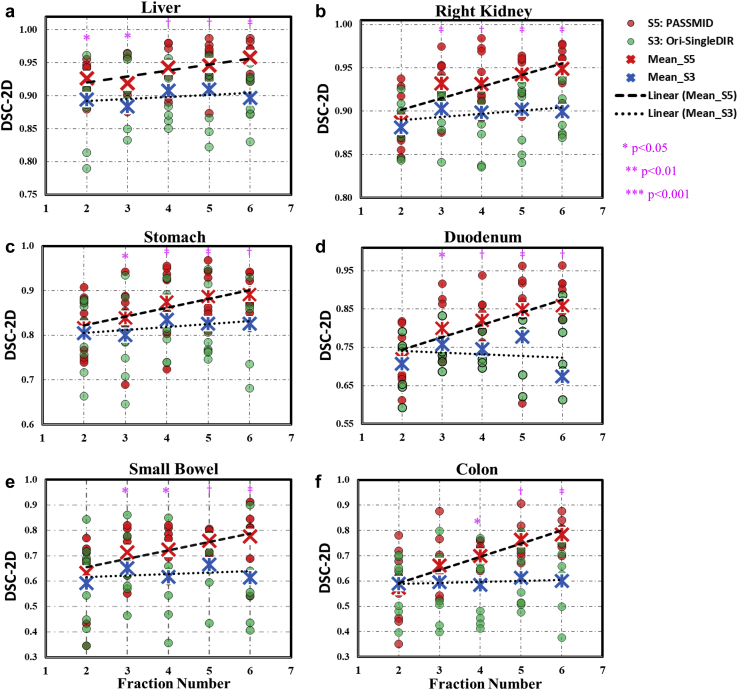
Figure 3 shows the 2D DSC comparison for different fractional contours (liver, right kidney, stomach, duodenum, small bowel, and colon) from the single-input DIR strategy (S3) and proposed PASSMID method (S5). The number of inputs for PASSMID was equal to the corresponding fraction number; thus, the latter was used in Figure 3 to better present the results. For S3, the mean 2-dimensional DSC of each fraction (blue x samples and dotted trendlines) shows no or minor variations across different fractions. In contrast, for PASSMID, the mean 2D DSC of each fraction (red x samples and dash trendlines) increases with the increased number of additional prior images/contours. As indicated by the purple star samples on top of each column, starting from the third fractions (ie, with >3 prior images/contours), significant improvement (P < .05) was observed for PASSMID compared with S3.
Figure 3.
Comparison of 2-dimensional Dice similarity coefficient for fractional contours of the (a) liver, (b) right kidney, (c) stomach, (d) duodenum, (e) small bowel, and (f) colon, using strategy S3 (green) and S5 (red). The mean values of each fraction are highlighted with red and blue x symbols, and the corresponding linear trendlines are presented in dash and dotted lines for S5 and S3. Statistically significant difference between S5 and S3 for certain fractions are illustrated in pink. ∗P < .05; †P < .01; ‡P < .001.
Representative examples of the autogenerated contours using the proposed PASSMID method are depicted in Figure 4. The STAPLE algorithm was applied for 5 or 6 DIR-based contours from preprocessed MRI scans of previous fractions to get consensus structures for the fifth and sixth fractional MRI scans. The corresponding 2-dimensional DSC and MDA for abdominal organs are reported in the tables below Figure 4. Autogenerated contours for most organs, including the liver, spleen, spinal cord, and kidneys, have 2D DSC > 0.8 and 2D MDA < 2 mm for both fractions, which are acceptable based on the recommendations by the AAPM Task Group report 132.22 Even for challenging gastrointestinal organs (eg, stomach, duodenum, colon, and small bowel), the mean 2D DSCs are in the range of 0.72 to 0.93 for fraction 5 and 0.81 to 0.94 for fraction 6.
Figure 4.
Autogenerated contours using the proposed patient-specific autosegmentation strategy using multiple-input deformable image registration method (S5) for the fifth and sixth fractional images from 1 patient. The 2-dimensional Dice similarity index and mean distance to agreement are listed in the table below each image.
Table 2 shows the ROA values for different organ contours from different strategies. The mean ROAs for the selected organs contours are 13.9%, 17.5%, 60.8%, 70.6%, and 71.8% for the 5 strategies. Only a very small percentage of slices are acceptable for the rigid registration-based strategies (S1 and S2), which is expected considering the large daily anatomy change. The ROA of the multi-input DIR strategy (S4 and S5) are largely increased, indicating a reduced time for manual contour editing. The ROAs of the small bowel and colon (not shown in Table 2) are quite low (<20%) for all strategies.
Table 2.
Comparison of ROA for different strategies using a quantitative criterion of Dice similarity coefficient > 0.8 and mean distance to agreement < 2 mm based on the recommendation by the AAPM Task Group report 132
| ROA/%∗ |
Mean | SD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Liver | Left Kidney | Right Kidney | Spleen | Pancreas | Stomach | Duodenum | Aorta | |||
| S1: CT-MRI-rigid | 4.2 | 25.9 | 19.3 | 10.0 | 4.9 | 1.2 | 5.9 | 39.4 | 13.9 | 13.3 |
| S2: MRI-MRI-rigid | 9.7 | 24.2 | 22.0 | 16.5 | 12.6 | 4.3 | 6.2 | 44.3 | 17.5 | 12.9 |
| S3: Original-single | 50.2 | 78.8 | 75.5 | 70.8 | 50.4 | 33.2 | 36.8 | 90.3 | 60.8 | 20.9 |
| S4: Original-STAPLE | 66.2 | 85.1 | 81.7 | 80.0 | 64.1 | 48.3 | 47.6 | 92.2 | 70.6 | 16.8 |
| S5: PASSMID | 67.6 | 86.2 | 82.4 | 81.5 | 65.2 | 50.1 | 49.2 | 92.3 | 71.8 | 16.4 |
Abbreviations: MRI = magnetic resonance imaging; PASSMID = patient-specific autosegmentation strategy using multiple-input deformable image registration; ROA = ratio of acceptable slices; STAPLE = simultaneous truth and performance level estimation; SD = standard deviation.
The small bowel and colon are excluded because the ROAs are quite low (<20%) for all strategies.
Discussion
The results demonstrate that the proposed image preprocessing pipeline can effectively improve the image quality and similarity for intrapatient MRI scans and therefore help achieve better DIR-based autosegmentation. The pipeline is designed for patient-specific intensity normalization of fractional MRI scans acquired during MRgOART, which are usually acquired with the same scanning protocol and supposed to share a similar image histogram for a given patient. The pipeline is not expected to solve the MRI intensity standardization issue, which is a more complex problem and has been investigated in other studies.23,24
We compared the results from 5 registration-based contour propagation strategies. Two rigid registration strategies (rigid propagation from planning CT to fractional MRI and from pretreatment MRI to fractional MRI) were included to compare the performance of the newly proposed method with current clinical MRgOART practice. Our results indicate that contour accuracy increased significantly using the multi-input DIR strategy, confirming the benefit of including the patient’s prior longitudinal images and contours into the autosegmentation process. As expected, the accuracy increased with the increasing number of prior image/contour sets used. Significant improvement was observed for most critical organ contours using >3 prior image sets (ie, starting from the third fraction). Currently, at our clinic, only 1 pretreatment image is acquired. However, acquiring >1 set of pretreatment images and use them for the multi-input contour propagation strategy to increase the contour accuracy is possible, especially for the first and second fractions, which are less accurate in this study.
For the sake of efficiency, only target and adjacent organs at risk would be carefully edited in our current adaptive replanning practice. However, accurate contours are always desired for dose accumulation and plan evaluation. As indicated by the increased ratio of acceptable contour slices for different organs, the proposed method can reduce the time required for further manual editing during MRgOART. With a fast autosegmentation, the online daily replanning process can be practiced routinely.
In this study, we have demonstrated the feasibility of the proposed PASSMID method for abdominal organs using MRI scans from a specific MRI-Linac system. This approach can be easily extended for other organs or tumor sites for other MRI-guided systems, where multiple fractional images are available.
In recent years, various machine learning-based autosegmentation methods have been developed to help the image segmentation task. Good organ segmentation results have been reported,23, 25 and some commercial products have been implemented at the clinic for CT-based autosegmentations. However, as mentioned earlier, image intensity in MRI scans can vary remarkably for different acquisition protocols or different scanners, indicating that a specific autosegmentation model may need to be trained for each scenario. To train a robust, widely applicable model, a large data set, including images and high-quality contours, would be needed. At this early stage of MRgOART, such a large data set is not yet available. The proposed patient-specific autosegmentation approach, which does not require a large training data set, demonstrates comparable results in terms of contour accuracy with state-of-the-art machine learning-based autosegmentation methods for abdominal MRI scans. A detailed comparison is provided in the Table E1. Only images and contours acquired from previous sessions/fractions for the same patient are required; thus, the proposed PASSMID method can be a practical alternative for the emerging MRgOART.
Even with significantly increased contour accuracy, manual review and editing are still needed. As a part of an underdevelopment autosegmentation tool for OART in our group, a contour quality assurance method will be used to identify the inaccurate slices from the autogenerated contours using quantitative MRI texture and shape features. The identified inaccurate slices will then be automatically corrected using a texture map-based active contour method. Preliminary results show that the 2D contour quality assurance method yielded an average sensitivity of 91% and specificity of 89% for a set of autogenerated contours from T1-weighted noncontrast MRI scans of patients with pancreatic cancer,26 and the contour-correction method can quickly and significantly improve the quality of the inaccurate contours.27 Our focus of future research is to integrate all pieces into a fully automatic and robust autosegmentation tool, saving substantial time for the segmentation and evaluation process and thereby facilitating the routine practice of MRgOART.
Conclusions
The proposed PASSMID method that integrates MRI preprocessing and multi-DIR with the STAPLE algorithm can significantly improve the accuracy of contour generation on longitudinal MRI scans of a patient. The PASSMID performance is improved with an increasing number of previous longitudinal, fractional image sets. With further development, this approach can improve the efficiency and accuracy of segmentation on fractional MRI scans, thereby facilitating the routine practice of MRgOART.
Acknowledgments
The authors thank Tia E. Plautz, PhD, Laura Buchanan, PhD, and Asma Amjad, PhD, for their inputs.
Footnotes
Sources of support: The research was supported in part by the Medical College of Wisconsin (MCW) Cancer Center and Froedtert Hospital Foundation, the MCW Meinerz and Fotsch Foundations, Manteia Med., Elekta AB, and the National Cancer Institute of the National Institutes of Health (award number R01CA247960).
Disclosures: Medical College of Wisconsin received research support from Elekta AB, Siemens Healthineers, Philips Health Care and Manteia Med. Dr Li received an honorarium from Elekta AB for educational presentations. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Research data are stored in an institutional repository and will be shared upon request to the corresponding author.
Supplementary material for this article can be found at https://doi.org/10.1016/j.adro.2020.04.027.
Supplementary data
References
- 1.Tijssen R.H.N., Philippens M.E.P., Paulson E.S. MRI commissioning of 1.5T MR-linac systems - A multi-institutional study. Radiother Oncol. 2019;132:114–120. doi: 10.1016/j.radonc.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Plautz T.E., Hao Y., Kinchen C., Li X.A. Texture-based, automatic contour validation for online adaptive replanning: A feasibility study on abdominal organs. Med Phys. 2019;46:4010–4020. doi: 10.1002/mp.13697. [DOI] [PubMed] [Google Scholar]
- 3.Lamb J., Cao M., Kishan A. Online adaptive radiation therapy: Implementation of a new process of care. Cureus. 2017;9:e1618. doi: 10.7759/cureus.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao X., Yang J., Zhang J. Deformable image registration based on similarity-steered CNN regression. Med Image Comput Comput Assist Interv. 2017;10433:300–308. doi: 10.1007/978-3-319-66182-7_35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loi G., Fusella M., Lanzi E. Performance of commercially available deformable image registration platforms for contour propagation using patient-based computational phantoms: A multi-institutional study. Med Phys. 2018;45:748–757. doi: 10.1002/mp.12737. [DOI] [PubMed] [Google Scholar]
- 6.Lee C., Langen K.M., Lu W. Evaluation of geometric changes of parotid glands during head and neck cancer radiotherapy using daily MVCT and automatic deformable registration. Radiother Oncol. 2008;89:81–88. doi: 10.1016/j.radonc.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Cabezas M., Oliver A., Llado X., Freixenet J., Cuadra M.B. A review of atlas-based segmentation for magnetic resonance brain images. Comput Methods Programs Biomed. 2011;104:e158–e177. doi: 10.1016/j.cmpb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Rohlfing T. Handbook of Biomedical Image Analysis. Springer; New York City, NY: 2005. Quo vadis, atlas-based segmentation? pp. 435–486. [Google Scholar]
- 9.McLaren D.G., Kosmatka K.J., Oakes T.R. A population-average MRI-based atlas collection of the rhesus macaque. NeuroImage. 2009;45:52–59. doi: 10.1016/j.neuroimage.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulson E.S., Ahunbay E.E., Chen X., Erickson B.A., Hall W.A., Li A. First clinical use of a parallel workflow to reduce recontouring time and errors in MR-fuided online adaptive radiotherapy. Int J Radiat Oncol Biol Phys. 2019;105:E788. [Google Scholar]
- 11.De Nunzio G., Cataldo R., Carlà A. Robust intensity standardization in brain magnetic resonance images. J Digit Imaging. 2015;28:727–737. doi: 10.1007/s10278-015-9782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vovk U., Pernus F., Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging. 2007;26:405–421. doi: 10.1109/TMI.2006.891486. [DOI] [PubMed] [Google Scholar]
- 13.Bağcı U., Udupa J.K., Bai L. The role of intensity standardization in medical image registration. Pattern Recog Letters. 2010;31:315–323. [Google Scholar]
- 14.Park S., Plishker W., Quon H., Wong J., Shekhar R., Lee J. Deformable registration of CT and cone-beam CT with local intensity matching. Phys Med Biol. 2017;62:927–947. doi: 10.1088/1361-6560/aa4f6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warfield S.K., Zou K.H., Wells W.M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tustison N.J., Avants B.B., Cook P.A. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perona P., Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Analysis Machine Intellig. 1990;12:629–639. [Google Scholar]
- 18.Nyúl L.G., Udupa J.K., Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging. 2000;19:143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 19.CMS Software. Atlas-based auto-segmentation of CT images for radiotherapy planning – A White Paper.
- 20.Han X., Hoogeman M.S., Levendag P.C. Atlas-based auto-segmentation of head and neck CT images. Med Image Comput Assist Interv. 2008;11:434–441. doi: 10.1007/978-3-540-85990-1_52. [DOI] [PubMed] [Google Scholar]
- 21.Xiao H, Hibbard LS, Willcut V. GPU-accelerated, gradient-free MI deformable registration for atlas-based MR brain image segmentation. Paper presented at: 2009 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops. June 20-25, 2009; Miami, FL.
- 22.Brock K.K., Mutic S., McNutt T.R., Li H., Kessler M.L. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys. 2017;44:e43–e76. doi: 10.1002/mp.12256. [DOI] [PubMed] [Google Scholar]
- 23.Robitaille N., Mouiha A., Crepeault B., Valdivia F., Duchesne S. The Alzheimer’s Disease Neuroimaging Initiative. Tissue-based MRI intensity standardization: Application to multicentric datasets. Int J Biomed Imaging. 2012;2012:11. doi: 10.1155/2012/347120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gholizadeh N., Fuangrod T., Greer P.B., Lau P., Ramadan S., Simpson J. An inter-centre statistical scale standardisation for quantitatively evaluating prostate tissue on T2-weighted MRI. Australas Phys Eng Sci Med. 2019;42:137–147. doi: 10.1007/s13246-019-00720-1. [DOI] [PubMed] [Google Scholar]
- 25.Roth H.R., Lu L., Lay N. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localiation and segmentation. Med Image Anal. 2018;45:94–107. doi: 10.1016/j.media.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Ceballos F., Ahunbay E., Li X.A. A framework of automatic contour quality validation for MRI-guided online adaptive radiation therapy. Med Phys. 2019;46:500. [Google Scholar]
- 27.Zhang Y., Schott D., Ahunbay E., Li X.A. Automatic contour correction based on image texture for MRI-guided online adaptive replanning. Med Phys. 2018;45:4010–4020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.