Abstract
Although many researchers talk about a “patient database,” they typically are not referring to a database at all, but instead to a spreadsheet of curated facts about a cohort of patients. This article describes relational database systems and how they differ from spreadsheets. At their core, spreadsheets are only capable of describing one-to-one (1:1) relationships. However, this article demonstrates that clinical medical data encapsulate numerous one-to-many relationships. Consequently, spreadsheets are very inefficient relative to relational database systems, which gracefully manage such data. Databases provide other advantages, in that the data fields are “typed” (that is, they contain specific kinds of data). This prevents users from entering spurious data during data import. Because each record contains a “key,” it becomes impossible to add duplicate information (ie, add the same patient twice). Databases store data in very efficient ways, minimizing space and memory requirements on the host system. Likewise, databases can be queried or manipulated using a highly complex language called SQL. Consequently, it becomes trivial to cull large amounts of data from a vast number of data fields on very precise subsets of patients. Databases can be quite large (terabytes or more in size), yet still are highly efficient to query. Consequently, with the explosion of data available in electronic health records and other data sources, databases become increasingly important to contain or order these data. Ultimately, this will enable the clinical researcher to perform artificial intelligence analyses across vast amounts of clinical data in a way heretofore impossible. This article provides initial guidance in terms of creating a relational database system.
Introduction
This issue of Advances in Radiation Oncology presents a series of articles around applications of artificial intelligence (AI) in our field. One of the potential benefits of AI is that it can pore through large amounts of data to discover patterns not evident to clinicians. However, this vast volume of data cannot be accommodated within a single spreadsheet (which is how most clinical researchers work when conducting standard multivariable analyses). In fact, many researchers erroneously describe spreadsheets as databases. This article will demonstrate both what a relational database system is and how it is superior to a spreadsheet. It will also discuss considerations when implementing a relational database system (RDBS) for your own research purposes, using an actual lung cancer radiation therapy database as an example. I have also provided some excellent Wikipedia references that contain abundant additional information, beyond what can be encapsulated in a single article. (These, in turn, reference computer science literature for the very intrepid reader, but such references might extend beyond the level of understanding of all but the most technically inclined.)
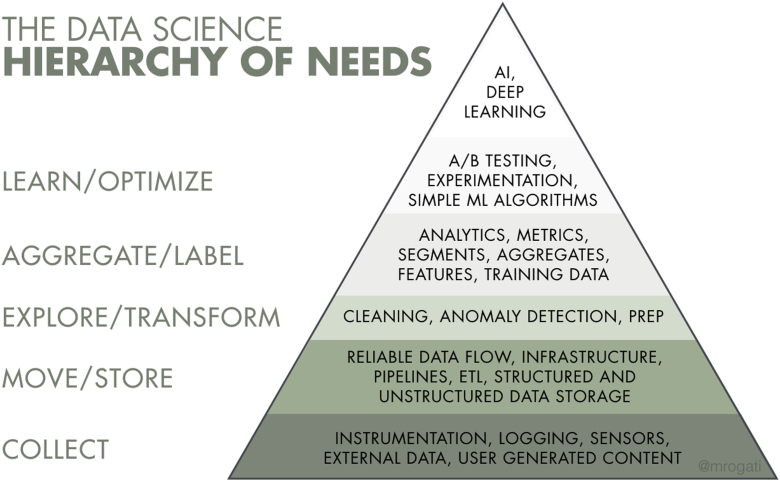
One might question why a database system is necessary for AI research. This article will demonstrate that a database enables creation of a multidimensional structure to cleanly and accurately contain these data. To perform AI analysis requires efficient storage of hundreds or thousands of data points on a single patient or even on a single course of radiation therapy. There is a famous illustration of the “data science hierarchy of needs” (Fig 1). To perform an AI analysis, one must first create an RDBS to serve as the storage mechanism. This creation of a system to store structured data entails a major part of the bottom row of the pyramid. To create a database, then, will set the reader down the path toward conducting AI research at their own institution.
Figure 1.
The data science hierarchy of needs. Used with permission of Monica Rogati (aipyramid.com). For details, see text. Abbreviation: AI = artificial intelligence.
Origin of Relational Databases
The concept of a RDBS was first described in a seminal article in 1970.1 The theoretic construct was that all data could be defined or represented as a series of relations with or to other data. The article was quantitative in that it used relational algebra and tuple relational calculus to prove its points.2 IBM used this theoretic framework to design what became the initial SQL (pronounced “see-quell” or “ess-cue-ell”) language, which they used to manipulate and retrieve data from IBM’s original RDBS.3 Since that time, the American National Standards Institute and the International Standards Organization have deemed SQL to be the standard language in relational database communication.2 Today, there are a wide variety of commercial and open-source relational database systems available for use. These vary in their features and relative strengths or weaknesses, but, fundamentally, they all operate using the principles defined in the Codd article.1 The SQL language is well defined and is used to write code to query (or update) the data within an RDBS.
Fundamental Disadvantage of Spreadsheets
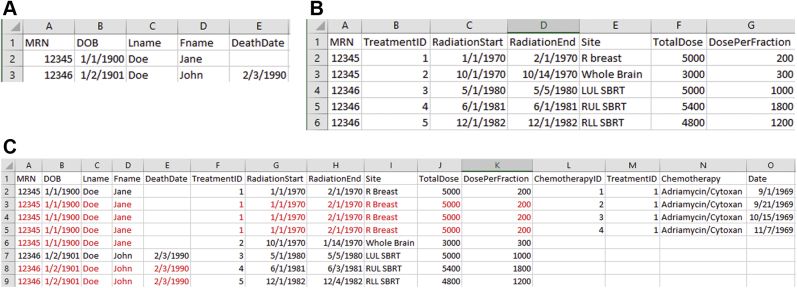
Spreadsheets are designed to incorporate and analyze one-to-one (1:1) relationships (Fig 2a). Each patient has a single birth date and a single death date. However, medical records are rife with “one-to-many” relationships (Fig 2b). A patient might receive multiple different courses of radiation therapy treatment, as in the example provided, or might have multiple chemotherapy administrations. To accommodate these data, a spreadsheet quickly balloons in size (Fig 2c). Not only is this inefficient (duplication of data), but it also makes maintenance of the spreadsheet extremely cumbersome and prone to error. For instance, in this example, when patient “12345” passes away, the “DeathDate” needs to be updated in 5 rows of the spreadsheet (because she had 2 courses of radiation therapy and 4 cycles of chemotherapy). It is not difficult to imagine that a researcher could neglect to update the “DeathDate” in each place, introducing errors. To further expound upon the issue, imagine a patient who takes numerous medications or has variable numbers of comorbid illnesses; the rows required to encapsulate 1 patient explode. To use a data science term, the dimensionality of the data balloons. But, to reiterate the point, spreadsheets are only designed to encapsulate 1:1 relationships (2-dimensional data). But patient data are multidimensional.
Figure 2.
(a) Spreadsheets are useful where there is a one-to-one correspondence of data. For instance, each unique medical record number (MRN) represents a single patient, with a single birth/death date and a single first and last name. (b) Spreadsheets “break down” when describing 1-to-many correspondences. In this example, 2 patients have a total of 5 courses of radiation therapy treatment between them. (c) To accommodate all the data in our simple example, a spreadsheet needs to store redundant data (colored in red). The data storage requirements quickly balloon. Furthermore, as additional traits and factors are added to the spreadsheet, it becomes impossible to follow, as one patient will require untold numbers of rows to capture all relevant data concepts. Stated another way, the data are multidimensional. Maintenance and updating of fields become error-prone (see text). Abbreviations: DOB = date of birth; Lname = last name; Fname = first name; LUL = upper lobe; MRN = medical record number; RLL, right lower lobe; RUL = right upper lobe; SBRT = stereotactic body radiotherapy.
Fundamental Advantage of Relational Databases
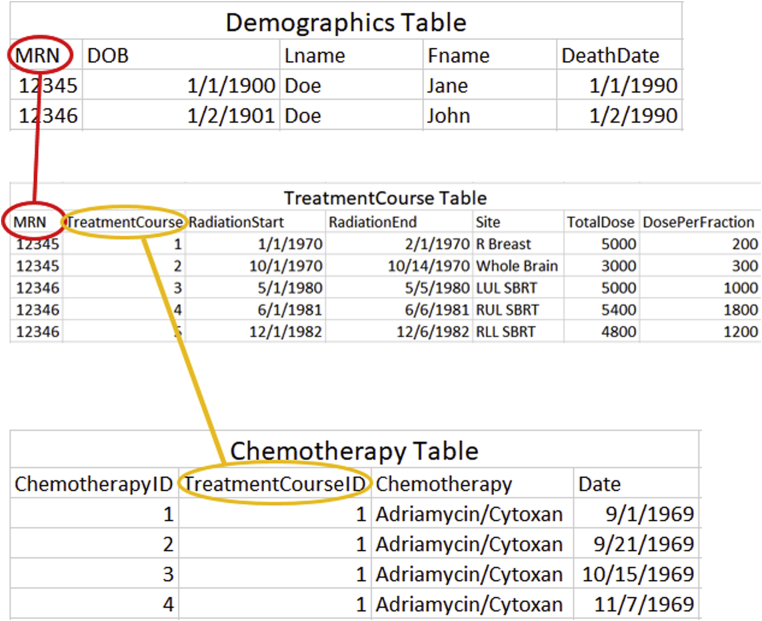
RDBS gracefully manage one-to-many relationships. They can do so because a database is created of numerous different tables, which are explicitly linked (Fig 3). Every table must also contain a key, which is a unique, required identifier for each row of data. Relationships between the tables are defined when creating the database tables or fields. In the “demographics” table, medical record number, “MRN,” is the key. For the “TreatmentCourse” and “Chemotherapy” tables, the keys are “TreatmentCourse” and “ChemotherapyID,” respectively. Note that “TreatmentCourse 1” in the “TreatmentCourse” table pertains to breast radiation therapy treatment. This, in turn, is linked to 4 cycles of chemotherapy in the “Chemotherapy” table, each of which is uniquely identified in that table, in turn.
Figure 3.
In a relational database, data are stored in multiple tables, which are joined via defined variables. In this fictitious example, note that the patient only has one “DeathDate” to update. Furthermore, each course of treatment (“TreatmentCourse”) can have multiple chemotherapy cycles associated with it. Note, too, that medical record number (MRN) only appears in 2 of the 3 tables (it is not needed in the “Chemotherapy” table). If the researcher wishes to retrieve the MRN, it can be obtained via a SQL query, linking back to one of the tables that contains it. Abbreviations: DOB = date of birth.
When comparing Fig 3 (a database) to Fig 2c (a spreadsheet), note that Fig 3 contains the same information as Fig 2c without the addition of repetitious information (colored red in Fig 2c). Alternately, in a spreadsheet, the researcher could manually aggregate and summarize the chemotherapy delivered into a single cell in a single row of the spreadsheet (ie, “flatten” the data, to use a data science term), but then some data would be lost. Using the chemotherapy administrations as an example, if one were to “flatten” the data down to a single spreadsheet cell stating “4 cycles of Adriamycin/Cytoxan,” one loses the dates of administration. If one summarizes the data as “4 cycles of Adriamycin/Cytoxan: <date1>, <date2>, <date3>, <date4>,” the dates and the chemotherapy occupy the same cell and the data are retained but are no longer discrete; one loses the ability to filter the spreadsheet by chemotherapy kind or by date.
Conversely, a SQL database cleanly encapsulates these multidimensional data (Fig 3). Each table is 2-dimensional in structure. But because it can contain multiple rows of data on 1 patient (chemotherapy administrations, to follow the same example), a multidimensional structure is created, as 4 chemotherapy cycles link to one of 2 courses of radiation therapy (“TreatmentCourse” table) for 1 patient (“Demographics” table). Now, imagine a clinical database with millions of rows of data spread across hundreds of tables, as in the real-life example described below. Clearly, a spreadsheet would not be adequate.
Additional Advantages of Relational Databases
-
1.
Each row of data in a table has a unique identifier (a key). Consequently, one cannot accidentally add a row of data into a database table twice.
-
2.
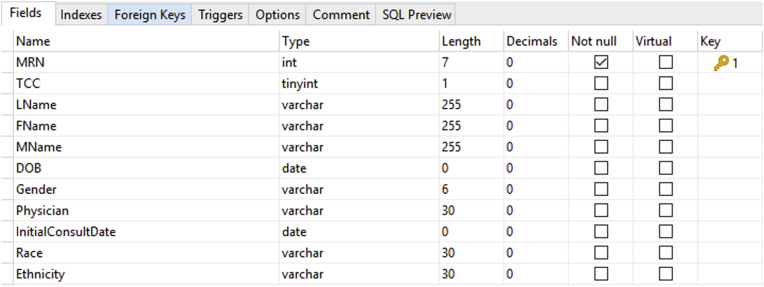
Each field in the database is specifically defined as to what it contains (Fig 4). Consequently, if data are imported from some external source, the data types must correspond or the import will fail. For example, if an entry contained an integer where a date was indicated, the database would signal that the variable contained erroneous data. This feature helps maintain consistency of data and minimize data errors.
-
3.
Not only must the data types correspond, but the data lengths must be observed. If the database design states that a field is a decimal with 3 places to the right of the decimal place, then a fourth decimal place would be truncated at import. Alternately, the database could declare an error, which might also imply that the field contains erroneous data.
-
4.
A key from one table can be linked “backward” to a key from another table (termed a “foreign key”). As an example (Fig 3), the database can be designed such that the MRN from “TreatmentCourse” must refer to an MRN already contained within the “Demographics” table. If one tried to import data into “TreatmentCourse” and it used an MRN not listed in “Demographics,” the import would fail. Such a situation might imply, for instance, that the MRN was incorrect in the external data source (or in the database). Or perhaps it relates to a patient who received prostate radiation therapy (but you have a breast cancer database).
-
5.
Foreign key relationships also work in the opposite direction: If one realizes that a patient is represented in the database who should not be, one can delete the patient from the “Demographics” table and the database will delete all data about that patient from all the other data tables automatically.
-
6.
RDBS are specifically optimized to manage vast amounts of data. Large spreadsheets (containing thousands of rows and hundreds of columns) are extremely slow and memory intensive. However, one can query across or manipulate many gigabytes of data in fractions of a second in many RDBS, as the data stores are highly optimized and efficient from both a computational and memory utilization perspective.
-
7.
RDBS are much more secure than spreadsheets. An institution’s IT team might allow one to access some tables within an institution’s data warehouse, but not others. One’s access could be restricted to defined subsets of patients. One might have “read” access to these data, but not “write” access (or “write” access to only some subset of fields). Databases might likewise be set up such that only users from specific IP addresses or computers may access them. The login systems set up by IT departments for these purposes typically use state of the art encryption algorithms, 2-factor authentication, and the like. In contrast, an Excel spreadsheet can be “locked” such that only some fields are editable. But it is not possible to restrict data access by user. Furthermore, this restriction is to “write” access only, not to “read” access. It is true that one can “hide” columns in a spreadsheet and then lock it, to prevent a given user from viewing them, but the spreadsheet maintainer must do this manually before distributing the spreadsheet (time-consuming and prone to error).
Figure 4.
Note that each field in this database table is specifically designed. It has a “type” (kind) and a “size” (length). When importing data from numerous external sources, these definitions can prevent erroneous imports (for details, see text). Note that the field “MRN” is the key for this table. All the data in this table refer back to “MRN” via a one-to-one relationship. MRN can be used as the key because no 2 patients have the same MRN. Abbreviations: DOB = date of birth; MRN = medical record number.
Benefits of SQL
As described earlier, SQL is a defined, standardized language for composing queries within an RDBS, or to manipulate and update these data. Some database systems provide “extensions” to the SQL standard, to provide some additional and specific functionality (details available in the vendors’ literature). It is beyond the scope of this article to teach SQL coding. However, many excellent online resources are available for the interested reader. Functionally, SQL allows one to search for any number of variables or combinations of variables across any number of tables, simultaneously. This can be extremely powerful and useful, both for retrieving and for manipulating and updating data. Queries can be saved for reuse or modification later. As stated above, these queries typically produce output in fractions of a second, even across vast pools of data.
Our institution has a database of patients who have received radiation therapy to the lung, whether for primary cancer or metastatic disease to the lung.4 The database and some of its details of implementation are described below, but first, some “real-life” examples of what such an RDBS system can do (not possible when using a spreadsheet):
-
•
Real-life example 1: Find patients who might be candidates for a certain lung cancer clinical trial. For this particular study, they must have previously received lung SBRT, have nonmetastatic disease, no evidence of recurrence, be alive (obviously), be at least 2 years out from the end of the prior SBRT treatment, and must have been seen in follow-up within the past 1.5 years. By constructing an appropriate SQL query, 135 patients were found (out of more than 4600 in the database) to pass along to the PI for closer inspection.
-
•
Real-life example 2: It takes only a few minutes to set up very complex queries. If one has a basic facility with SQL, one can design a query such as: “Find all patients with stage II or III lung cancer treated with concurrent chemoradiotherapy who developed neutropenia during treatment, who are female, 70 years of age or older, and who take any antihypertensive medication (defined in a certain list).” Ultimately, such queries are only limited by one’s imagination (and the richness or completeness of the data coming from the source systems).
It is true that one can “filter” data in Excel to rapidly find subsets. However, this filtering is limited to “true or false” matching. In this example, it would be impossible to discover the patients who developed neutropenia while undergoing radiation therapy unless one had a “neutropenia (yes or no)” column. But one cannot perform the arithmetic “where date of neutropenia > RadiationStartDate and < RadiationEndDate” to filter the data without writing code in Visual Basic, which is likely beyond the ability of most.
Disadvantage of SQL
With SQL, it is possible to create highly complex queries; it is a rich and powerful language. However, these can be quite complicated and obtuse to a nontechnical person. Some systems do provide graphical tools to help build SQL queries, but, even so, there are some users for whom all but the simplest SQL queries will be beyond their technical skills.
Database Implementation
Databases may, and often do, contain many thousands of tables and millions of rows of data. (In other words, they can contain data far in excess of the requirements of any one radiation oncologist or even any one radiation oncology department). In fact, some systems allow even single tables to contain terabytes or even petabytes of data.5 Consequently, there are numerous systems available to accommodate any researcher’s needs. Some of the very best are open source (free). Software is available across a wide variety of operating systems. Wikipedia provides an outstanding overview of the topic.5
To implement a database system, it is first necessary to have a discussion with the IT Department at your institution. There is no single solution for creating a data repository that holds true for researchers across all institutions. The solution can vary, depending on the resources at your institution and the level of access the IT Department has into the underlying patient data source systems (often defined in the institutional contracts signed with the individual vendors). Some large centers have elaborate data warehouse systems. Smaller centers might provide access to data from individual source systems but might not have compiled them into a single data warehouse. Some IT departments might have adequate resources to provide output data from their data warehouse to individual researchers, when needed. Others might not. Some might provide a dedicated research server on which the researcher can construct a database. Other researchers might need to rely instead upon existing servers within their department. I do not recommend that one set up a database system on a free-standing laptop or desktop machine, as there are Health Insurance Portability and Accountability Act concerns (the computer could be stolen). Data should be backed up across a secured network electronically.
Creation of a Lung Cancer Radiation Therapy Database
I began my own database several years ago. My need grew out of a sense of frustration regarding lack of access to clinical data. At the time, at my institution, it was a difficult (and somewhat mysterious) process to procure data from the data warehouse. However, data from Mosaiq (Elekta AB, Stockholm, Sweden), which is our department’s record or verify system, were available. These data formed the nucleus of the original database. Basic demographic information and radiation therapy prescriptions, dates of treatment, dose delivered, tumor stage, and the like, were exported, using custom software. Research IT provided a Linux server, on which I implemented the database. I chose to use MariaDB (MariaDB Foundation, DE), as it is a powerful, well-regarded, commercially supported, free, open-source database system whose SQL functions are congruent with those of Oracle (which is a database system I had used previously). Because my institution could not support an implementation of an Oracle, MariaDB was an excellent alternate option. MariaDB does include a Windows GUI database administration tool for administering its databases (creating tables, writing SQL code, importing or exporting data, and the like). I had previously used a similar commercial database administration product called Navicat (Premium Cybertech Ltd, Hong Kong), which provides similar functionality, so I elected to purchase that. Similarly, I imported all data I had captured in spreadsheets for previous research projects. More recently, I have gained access to data from our data warehouse and so have created numerous additional tables to store the information. At present, the database contains more than 3 million rows of data on approximately 4800 patients, spread across more than 170 tables.
Incorporation of Data from Other Institutional Data Systems
To import data from outside source systems requires a multistep process, referred to as “ETL” (“Extraction, Transformation, and Loading”) in the data science literature.”6 The issues go far beyond the physical importation of data into the database; importing spreadsheets of data are a trivial task. There are numerous issues in ETL, which are critical to consider when designing a database and importing data into it. Furthermore, many of these issues are not inherently obvious. In fact, a large proportion of the time required to create a database and fill it with clinically useable data derives from the ETL involved. The oft-reproduced “Data Science Hierarchy of Needs” illustrates this fact (Fig 1). Most of the discussion in this article addressed aspects of the bottom-most layer of the pyramid. ETL comprises the majority of the next 2 layers of the pyramid and is the topic of another article.
Footnotes
Research data are not available at this time.
Sources of support: none.
Disclosures: Dr Dilling reports personal fees and nonfinancial support from NCCN, personal fees from Varian, personal fees and nonfinancial support from Harborside Press, nonfinancial support from Astra Zeneca, all outside the submitted work.
References
- 1.Codd E.R. A relational model of data for large shared data banks. Commmunications of the ACM. 1970;13:377–387. [Google Scholar]
- 2.Wikipedia. SQL. https://en.wikipedia.org/wiki/SQL Available at:
- 3.Wikipedia IBM System R. https://en.wikipedia.org/wiki/IBM_System_R Available at:
- 4.Dilling T.J. Artificial Intelligence (AI) for clinical analysis: Design and implementation of a suitable database resource. Int J Radiat Oncol Biol Phys. 2019;105:E131–E132. [Google Scholar]
- 5.Wikipedia Comparison of relational database managment systems. https://en.wikipedia.org/wiki/Comparison_of_relational_database_management_systems Available at:
- 6.Wikipedia Extract, Transform, Load. https://en.wikipedia.org/wiki/Extract,_transform,_load Available at: