Abstract
An ongoing pandemic Coronavirus disease (COVID-19), caused by a newly emerged Coronavirus, SARS-CoV-2 has affected millions of people globally. One of the most crucial structural proteins of SARS-CoV-2 is the Spike glycoprotein (S-glycoprotein), for which the first de novo modelling was envisaged by our group in early 2020, and was superimposed to its predecessor SARS-CoV S-glycoprotein, to determine structural divergence, glycosylation and antigenic variation between SARS-CoV-2 and SARS-CoV. S-glycoprotein is involved in binding with the cellular receptor, membrane fusion, internalization via angiotensin-converting enzyme 2 (ACE2) receptor, and tissue tropism. Upon internalization into the target host cells, the viral genome encodes two precursor polypeptides which get processed into 16 mature nonstructural proteins that play a crucial role in replication and transcription of SARS-CoV-2. Currently S-glycoprotein is one of the most vital targets for vaccine and therapeutics development for COVID-19.
Keywords: SARS-CoV-2, COVID-19, Spike glycoprotein, Virus-host interaction, ACE-2 receptor
Introduction
Numerous cases of pneumonia with novel etiological factors have emerged in Wuhan, Hubei region of China in December 2019 [1–4]. Consequently, the metagenomic sequencing analysis of bronchoalveolar lavage fluid samples taken from the patient suggested the novel coronavirus (nCoV) outbreak in Wuhan. On January 31, 2020, the World Health Organization (WHO) confirmed this nCoV as a public health emergency of international concern [2, 5]. Considering the high sequence similarity with SARS-CoV, the International Committee on Taxonomy of Viruses (ICTV) has renamed this nCoV as the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) [6]. So far, at least 50 million confirmed cases and ~1.2 million deaths have been reported globally [7]. Clinically, a person infected with SARS-COV-2 shows respiratory illness, which is related to the other respiratory viral infections. Understanding of viral dynamics and host responses are necessary for development of effective vaccination strategies, antiviral treatment, and the epidemiological control of COVID-19 [8]. So far, the SARS-CoV-2 is 7th coronavirus which has infected humans [9, 10]. SARS-CoV-2 morphology is elliptic or round or pleomorphic with an approximate diameter of 60–140 nm in size [11]. Coronaviruses encodes for 16 nonstructural proteins (nsp1-nsp16) and four structural proteins spike glycoprotein (S-glycoprotein), membrane (M), envelope (E), nucleocapsid (N) and hemagglutinin (HE) protein as exhibited in Fig. 1a [12, 13]. However, SARS-CoV-2 is devoid of the hemagglutinin gene. SARS-CoV-2 is a positive sense single-stranded (+ ss) RNA virus of 29.8–29.9 kb genome size encoding ~9860 amino acids long polyprotein [14]. The infection of coronaviruses (CoVs) commence with S-glycoprotein mediated virus attachment to the host cellular receptor. The first de novo modelling of S-glycoprotein was envisaged by our group in early 2020 before the WHO has announced the public health emergency [12]. Receptor recognition is crucial for cellular tropism of a CoVs. Among the seven identified human coronaviruses (hCoVs), HKU1 and HCoV-OC43 are able to exceptionally connect to sugar moieties for cellular attachment, while the other hCoVs identifies proteinaceous peptidases as their potential target receptors. The HCoV-229E virus interacts with aminopeptidase N (hAPN) whereas, human dipeptidyl peptidase 4 (hDPP4 or hCD26) is required for MERS-CoV interaction for their internalization. Whereas, SARS-CoV and hCoV-NL63 attach to the hACE2 for entry [15–18]. Similarly, SARS-CoV-2 uses hACE2 receptor for its attachment and internalization into the host cell. S-glycoprotein embedded at the surface of the SARS-CoV-2 exhibits receptor recognition and membrane fusion activity. Upon S protein binding of the hACE2 receptor, conformational transformation occurs in S protein that results in the integration of E protein with the host membrane. The process of viral attachment to ACE2 receptors is a crucial step during SARS-CoV-2 infection therefore, understanding this phenomenon is important for identification of potential attachment inhibitors [6, 19]. Here, we highlights the genome structure of the CoV and discuss the mechanism of S-glycoprotein mediated SARS-CoV-2 attachment to target host cell receptor ACE2.
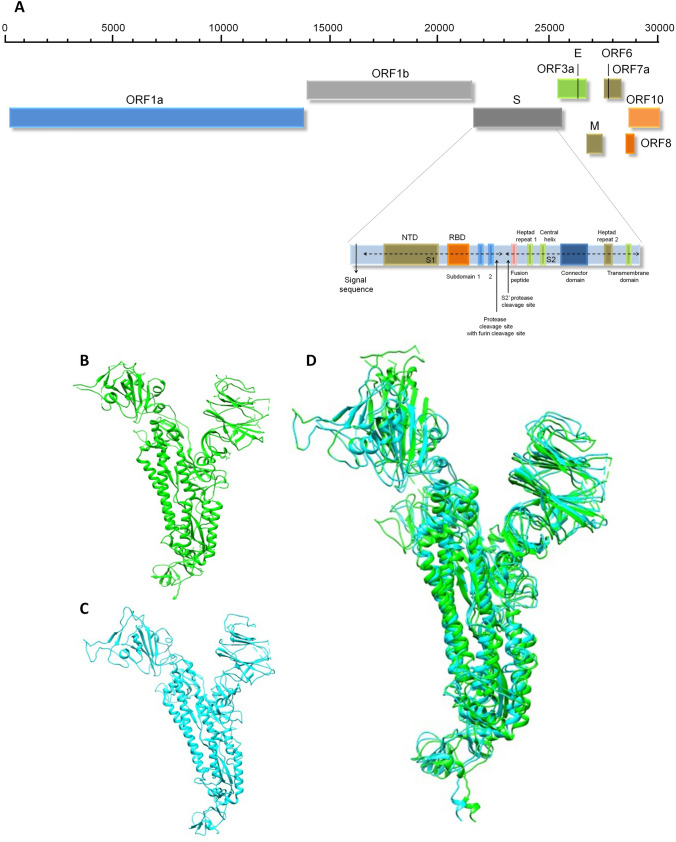
Fig. 1.
Representation of the genomic organization of the SARS-CoV-2 and domain structure of Spike glycoprotein (A). S-glycoprotein contains S1 and S2 subunits where S1 subunit consists of signal sequence, N-terminal domain (NTD) and receptor binding domain (RBD). The S2 subunit consists of heptad-repeat (HR) regions as HR-N and HR-C involves the formation of coiled coil structure within the protein ectodomain. At the interface of S1 and S2 subunit various cleavage sites are present including protease, furin and S2′ protease. S- Spike glycoprotein gene, E-Envelope protein gene, N- nucleocapsid protein gene, M- membrane protein gene, RBD- receptor-binding domain. Structural comparison of Spike glycoprotein of SARS-CoV-2 (B), with SARS-CoV (C), suggesting minimal structural difference in superimposed spike glycoproteins structures (D)
SARS-CoV-2 genome organization
Upon interaction of S-glycoprotein with hACE2 receptor, SARS-CoV-2 gets internalized via an endosomal pathway that results in release of viral RNA. The SARS-CoV-2 genome is 29.8–29.9 kb positive sense RNA with 5′ UTR region of 265 and 3′UTR region of 358 nucleotides. The 5′ end comprises of orf1ab which is more than two third of the genome that encodes for orf1ab polyproteins whereas 3′end comprises of genes encoding structural and accessory proteins (3a, 6, 7a, 7b, 8, and 10). Following internalization, viral RNA gets translated into long polyproteins (pp1a and pp1b). These polyproteins get processed via proteolytic cleavage of polyproteins into nsps. The negative strand of viral RNA acts as a template strand for generation of genomic RNA (gRNA) of positive sense as well as subgenomic RNA (sgRNA). sgRNA further encodes for structural proteins which involves in the packaging of gRNA for assembling of progeny virions [20].
Spike glycoprotein of CoVs
Entry of CoV is mediated by S-glycoprotein which protrudes from the viral surface as homotrimeric transmembrane protein. S-glycoprotein assembled into trimeric conformation on the surface of the virus to form the typical “corona”, or crown-like appearance [21]. S-glycoprotein is essential for membrane fusion, receptor binding, internalization, tissue tropism, and host range. S-glycoprotein consists of two functional subunits S1, required for the attachment to cellular receptors and S2, involved in viral and cell membrane fusion. The S-glycoprotein is cleaved from the interface of S1 and S2 subunits that remains non-covalently linked in prefusion conformation. S1 subunit is required for the stability of prefusion conformation of membrane linked S2 subunit. To activate the S-glycoprotein, host proteases are involved in the processing of spike protein specifically at the S2′ site present at the upstream fusion peptide which ultimately requires for its membrane fusion activity.
Spike glycoprotein and ACE-2 mediated SARS-CoV-2 entry
The functional aspect of ACE2 receptor in coronavirus infection has been primarily identified during SARS-CoV outbreak. The general physiological function of the receptor ACE2 is the conversion of angiotensin I and II into angiotensin (1–9) and angiotensin (1–7) respectively [22, 23]. Angiotensin (1–9) binds with Mas receptor that leads to anti-inflammatory effects and vasodilatation which is antagonistic to the ACE function. Two functional regions of ACE2 have been identified as N-terminal peptidase domain M2 and the C-terminal collectrin domain. Considering the higher sequence similarity among the S-glycoproteins, a similar function of ACE2 receptor was observed during SARS-CoV-2. S1 subunit of S-glycoprotein comprises a receptor binding domain (RBD) (aa residues 333–527) which is responsible for the virus attachment with ACE2 receptor [23]. Due to the presence of four residues (Gly-Val-Glu-Gly) at 482–485 named as receptor binding motif (RBM), the affinity of S-glycoprotein ectodomain towards ACE2 receptor is 10–20 folds higher in comparison with SARS-CoV [24]. The critical point of attachment for S-glycoprotein is the presence of two lysine residues in ACE2 receptor that interacts with Gln493 and Leu455 in the SARS-CoV-2 S-glycoprotein RBM for substantial binding stabilization with receptor [23].
SARS-CoV-2 spike glycoprotein structure
Like other CoVs, SARS-CoV-2 internalization in the target host cell is exhibited by S-glycoprotein. A prominent sequence variation in the S-glycoprotein of SARS-CoV-2 was determined as compared with SARS-CoV and other CoVs. Specifically, we have observed, 12.8% of sequence variation in S-glycoprotein and 23.6% differences in minimal RBD among SARS-CoV and SARS-CoV-2 [12]. However, length of encoded proteins in both the coronaviruses is found almost the same [25] with the introduction of four amino acids at the interface of S1 and S2 that acts as a furin cleavage site necessary for priming of SARS-CoV-2 S-glycoproteins. Around 3.8 Å RMSD of structural divergence was observed when crystal structures of S-glycoprotein of SARS-CoV-2 with SARS-CoV were compared [26]. Our first de novo modelling of S-glycoprotein [12] has exhibited the structural divergence of S-glycoprotein of SARS-CoV-2 (Fig. 1b) and SARS-CoV (Fig. 1c) suggests structural resemblance and raises hope towards the treatment of COVID-19 (Fig. 1d). Considering this, the earlier trialed attachment inhibitor used for SARS-CoV might be effective as the current option of treatment of COVID-19. Interestingly, SARS-CoV-2 S-glycoprotein exhibits larger differences in RBDs with 10 times more binding affinity towards ACE2. Similar to other CoVs, S-glycoprotein of SARS-CoV-2 comprises two subunits S1 and S2 (Fig. 1a). S1 subunit consists of signal peptide, N-terminal domain (NTD) and RBD. Whereas, S2 subunit consists of two heptad-repeat (HR) regions both at N and C terminal (HR-N and HR-C) that forms coiled coil structure within protein ectodomain that assist in membrane fusion [6, 20, 25, 27].
Proteolytic cleavage and activation of spike glycoprotein
SARS-CoV-2 entry into host cell follows several steps that start with binding of S-glycoprotein with host cell surface receptor ACE2 via RBD (Fig. 2), changes in S-glycoprotein conformation, its proteolysis and the release of subunit S2, which further exhibits virus fusion and endocytosis [23, 28]. The S-glycoprotein cleaved from the site of cleavage that makes S1 and S2 subunits. The RBD is present in S1 subunit that mediates S-glycoprotein binding to the host cell receptor [29]. Upon binding to the ACE2 receptor, a proteolytic cleavage site gets exposed on S2 subunit, where transmembrane protease serine 2 (TMPSSR2), a host cell protease start the cellular entry process by priming the S-glycoprotein [30]. During this process, the RBD ties up to the carboxypeptidase area of ACE2. The RBM present inside the RBD facilitates the initial interaction with ACE2 peptidase domain. The processing of the S-glycoprotein occurs in two steps, first ‘priming’ and second ‘activation’ of cleavage at the boundary of S1 and S2 subunits that comprises multi-basic arginine residues. In addition, a furin cleavage site (R-R-A-R) is present at the interface of S1 and S2 subunits of S-glycoprotein, which is absent in SARS-CoV [30]. Elimination of this motif affects the cellular attachment and internalization into the target host cell. The furin site extends the flexibility of SARS-CoV-2 for processing by cellular proteases and possibly the tropism and transmissibility attributable to the wide cell articulation of furin proteases especially in the respiratory system. This implies that recently synthesized virions may be emitted in a 'pre-activated' form ready to infect diverse cell types without the requirement to bind with a cell receptor, for example, ACE2. In the two-step mechanism, the cleavage site at the S2′ is required for activation of the S-glycoprotein for fusion with cell membrane. Hindrance of TMPRSS2 utilizing camostat mesylate somewhat obstructs the entry of SARS-CoV-2 [32]. Despite TMPRSS2, S-glycoprotein of SARS-CoV-2 can also get activated proteolytically by a number of other proteases such as endosomal cysteine proteases; cathepsin B and L. Some other proteases like furin, factor X, trypsin and elastase are also able to perform this ‘priming’ proteolysis that begins the cellular entry process [23]. Using computer aided drug designing; we have recently shown several of the potential antiviral molecules that may interfere with the interaction of spike with ACE2 receptors [33, 34].
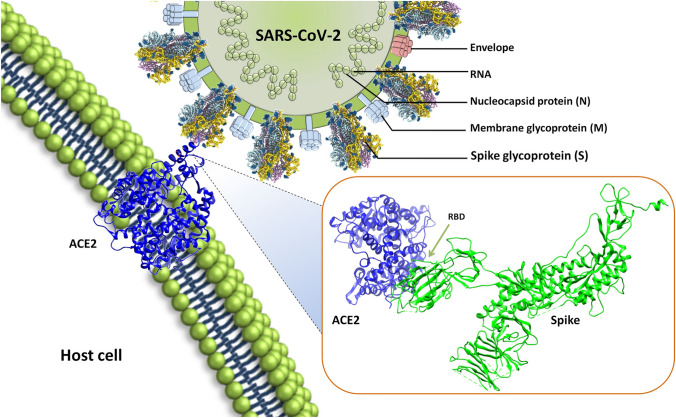
Fig. 2.
Structure of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and interaction with host cellular receptor ACE2. SARS-CoV-2 encodes for four structural proteins, spike glycoprotein, membrane glycoprotein (M), Envelope (E), nucleocapsid (N). Spike glycoprotein is embedded in the host derived membrane which binds with the host cell receptor ACE2. The inset is showing the protein–protein interaction of SARS-CoV-2 S-glycoprotein with host cell receptor ACE2 mediated via receptor binding domain (RBD) present in spike glycoprotein and peptidase domain of ACE2
Spike glycoprotein mediated translation inhibition in coronavirus infected cell
Upon SARS-CoV-2 infection, host cells mediate various host genes expression including chemokines and proinflammatory cytokines up-regulation at both transcriptional and translational levels [35]. More specifically, IL-8, IL-6, TNF-a and IFN, have been considered as the major mediators of the SARS-CoV-2 pathogenesis. These over-produced cytokines and chemokines elicit inflammasome response by the induction of acute injury in the cell and the tissues [36]. S-glycoprotein of SARS-CoV interacts with the eIF-3f, a subunit of the eukaryotic initiation factor 3, which results in inhibition of protein synthesis [37]. Since the inhibition takes place at the later stages of the virus replication cycle, virus-induced transcripts including proinflammatory cytokines and chemokines translational process is primarily affected. The inhibition of translation exhibits a crucial role in the pathogenesis of CoVs. The inhibitory effect seems to be general, as both the viral and cellular protein synthesis gets affected. Conversely, aggregation of S-glycoprotein up to a certain amount in the host cell is a signal for translational inhibition, it would affect only at the final stage of viral infection. This can be majorly advantageous to the virus. Upto this, the viral replication and translation in the host cell are almost completed, inhibition of viral protein synthesis would be less affected causing sufficient viral production and following contamination of neighboring cells. In contrast, inhibition of infected cell protein synthesis, specifically, the virus-induced transcripts translation which accumulated at high concentration would have reduced the protein product production dramatically from these genes. Most of the synthesized proteins are proinflammatory in nature, and thereby production and secretion of these cytokines/chemokine causes inflammatory response, induces the adjacent cells and may cause cell death [38]. The direct outcome of this cell/tissue reaction could be the restraint and elimination of viral infection.
Post translational modifications of S-glycoprotein and other structural, non-structural proteins
Numerous CoV proteins undergo post translational modifications (PTMs) which include glycosylation, palmitoylation of the E and S-glycoprotein. More specifically, glycosylation at Asn residues (N linked) or hydroxyl group of Ser and Thr (O-linked) of the M-protein, ADP-ribosylation of N-protein and other PTMs on NSPs and accessory protein have been observed in CoVs [39]. In addition, incorporation of disulfide bonds in S-glycoprotein and conserved cysteine residues into its cytosolic domain has been reported with palmitoylation.
N-linked glycosylation
The N-linked glycosylation in S-glycoprotein of CoVs was first reported for mouse hepatitis virus. S-glycoprotein of SARS-CoV has exhibits oligosaccharides containing high mannose and trimers within 30 min after the entry into endoplasmic reticulum, former to the acquirement of multifaceted glycans present in Golgi apparatus [40]. Monitoring of SARS-CoV S-glycoprotein susceptibility to endoglycosidase H (endo H), shows its maturation status [39]. Using molecular cloning techniques, the S-glycoprotein coding sequences of various CoVs were cloned, following determination of N-linked glycosylation sites. However, all the putative glycosylation sites were not functional. In SARS-CoV S-glycoprotein, 12 sites were found glycosylated out of the 23 putative sites [41]. N-linked glycosylation is associated with protein conformation, and hence intensely affects the antigenicity of S-glycoprotein and receptor binding. Similarly, S-glycoprotein of SARS-CoV-2 has been found to exhibit 22 N-linked glycan structure detected using mass spectrometry which showed complex and hybrid glycans likely to play an important role in protein folding and immune evasion [42].
Palmitoylation
Palmitoylation is a process of the covalent attachment of fatty acids like palmitic acid, to cysteine etc. Four cysteine rich clusters are present in the cytoplasmic ends of SARS-CoV S-glycoprotein. Mutational study revealed palmitoylation of cysteine clusters I and II. Even though the cellular expression of SARS-CoV S-glycoprotein was unaffected considerably by cysteine clusters I and II mutations, S mediated fusion of cells was clearly reduced as compared with the wild type protein. This suggests the requirement of palmitoylation in the endodomain for the SARS-CoV S-glycoprotein fusogenic activity.
However, post translational modifications have also been observed in following other structural and accessory proteins of SARS-CoV [39].
Envelope protein
N-linked glycosylation and palmitoylation
In SARS-CoV, N-linked glycosylation depends on the membrane topologies
Membrane protein
O-linked glycosylation in some beta-coronaviruses while in other PTMs occur by N-linked glycosylation
Nucleocapsid protein
Modification by proteolytic cleavage, phosphorylation, ADP-ribosylation and sumoylation.
During coronavirus induced programmed cell death, caspase cleaved the N protein of coronavirus.
Nonstructural proteins and accessory proteins
CoV nsp3 and nsp4 are multi-spanning transmembrane proteins which get modified via N-linked glycosylation. This may have a crucial role during viral replication and vesicle formation.
In some beta-coronaviruses the accessory proteins hemagglutinin-esterase 3a and 8ab of SARS-CoV modified by glycosylation.
Possible remedial approaches to deal with COVID-19
Development of vaccine based on S-glycoprotein
S-glycoprotein plays a major role in attachment of virus particles. The similarity in the antigenicity of the SARS-CoV-2 with SARS-CoV may be allied with the similar type of antigenic response and thus this may be used for the designing peptide-based vaccine based on S-glycoprotein [12, 43].
Restraining the activity of TMPRSS2 and protease activity
The initial priming of the S-glycoprotein is a crucial step for the entry and infection of SARS-CoV-2 via ACE2 receptor. Inhibition of the proteases (TMPRSS2, endosomal cysteine proteases; cathepsin B and L.) may prevent the viral entry into the host cell. It is reported that camostat mesylate, a serine protease inhibitor can partially block TMPRSS2 activity [23, 32. 44, 45].
ACE2 receptor blockage
It is well reported that SARS-CoV-2 enters into the host cell via ACE2 receptor. The use of some other molecule which targets this binding site can prohibit the entry of virus in the host cell [44, 46]. We have also shown the use of convalescent plasma therapy in order to prevent the infection [47].
Novel epitope-based vaccine
The variation found in the RBD domain of S-glycoprotein suggests that SARS-CoV-2 may experience modification in virus binding capacity and internalization into the host cell. While comparing the antigenic sites of S-glycoprotein of SARS-CoV-2, we have found novel CTL epitopes exhibited by SARS-CoV-2 that may cause distinctive antigenic responses as compared to SARS-CoV. Our novel CTL epitopes in S-glycoprotein suggested various opportunities for the designing peptide based vaccine for preventing COVID-19 [12]. Consequently, a recombinant adenovirus type-5 (Ad5) vectored COVID-19 vaccine has been designed that express S-glycoprotein of SARS-CoV-2 which found to elicit rapid specific T-cell responses [48], suggesting the importance of S-glycoprotein associated vaccines for further investigations.
Conclusions
In the middle of the worldwide public health emergency i.e. COVID-19, it is logical to question why the origin of this pandemic matters. Thorough understanding of cross species viral transmission will support the prevention of upcoming zoonotic events. SARS-CoV-2 shows many similarities and variations with SARS-CoV on sequence alignment, structural, and antigen basis. SARS-CoV-2 utilizes the receptor ACE2 for the entry into the host cell. S-glycoproteins present in the virus particle help in attachment and activation of the process. Still, many areas are unexplored and the mechanism is unknown. Efficient SARS-CoV-2 vaccines are crucial to reduce the severity and transmission of the disease. Developing a vaccine takes time, till then it is important to follow the authority’s instructions and maintain social distancing. Restricted information on the SARS-CoV-2 results in some limitations to explain the entire role of S-glycoprotein and antigenicity.
Acknowledgements
The authors are grateful to the Vice Chancellor, King George’s Medical University (KGMU), Lucknow, India for the encouragement for this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Abbreviations
- ACE2
Angiotensin-Converting Enzyme 2
- APN
Aminopeptidase N
- BALF
Broncho-Alveolar Fluid
- CoV
Coronavirus
- CTL
Cytotoxic T Lymphocyte
- MERS
Middle East Respiratory Syndrome
- NSP
Non-structural protein
- ORF
Open Reading Frame
- RBD
Receptor-Binding Domain
- RBM
Receptor Binding Motif
- RMSD
Root Mean Square Deviation
- SARS
Severe Acute Respiratory Syndrome
- ssRNA
Single-Stranded Ribonucleic Acid
- TM
Transmembrane
- TMPRSS2
Transmembrane Protease Serine 2
Authors’ Contributions
SKS conceived the idea. SKS, PB, SK, NS, BP and RKR collected the data, devised the initial draft, reviewed the final draft and finalized the draft for submission. All authors read and approved the final version of the manuscript.
Funding
None.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest. The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pneumonia of unknown cause – China, World health Organization 2020. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/. Accessed 10 Nov 2020.
- 2.Yang P, Wang X. COVID-19: a new challenge for human beings. Cell Mol Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yadav T, Saxena SK. Transmission Cycle of SARS-CoV and SARS-CoV-2. Coronavirus Disease 2019 (COVID-19) 2020 doi: 10.1007/978-981-15-4814-7_4. [DOI] [Google Scholar]
- 4.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava N, Baxi P, Ratho RK, Saxena SK. Global trends in epidemiology of coronavirus disease 2019 (COVID-19) Coronavirus Disease 2019 (COVID-19) 2020 doi: 10.1007/978-981-15-4814-7_2. [DOI] [Google Scholar]
- 6.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181(894–904):e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coronavirus disease (COVID-2019) situation reports (2020). World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-report Accessed 10 Nov 2020.
- 8.Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020;55:105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Napoli RD. Features, evaluation and treatment coronavirus (COVID-19) NCBI Bookshelf. https://www.ncbi.nlm.nih.gov/books/NBK554776/ Accessed 10 Nov 2020.
- 12.Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim YX, Ng YL, Tam JP, Liu DX. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005;24:S223–S237. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 18.Hulswit RJG, Lang Y, Bakkers MJG, Li W, Li Z, Schouten A, et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Nyodu R, Maurya VK, Saxena SK. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Coronavirus Disease 2019 (COVID-19) 2020 doi: 10.1007/978-981-15-4814-7_3. [DOI] [Google Scholar]
- 21.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillay TS. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J Clin Pathol. 2020;73:366–369. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- 24.Baig AM, Khaleeq A, Ali U, Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 25.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020;11:576622. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(271–280):e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M, Schroeder S, Kleine-Weber H, Müller MA, Drosten C, Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob Agents Chemother. 2020;64:e00754–e820. doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurya VK, Kumar S, Bhatt MLB, Saxena SK. Antiviral activity of traditional medicinal plants from ayurveda against SARS-CoV-2 infection. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1832577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurya VK, Kumar S, Prasad AK, Bhatt MLB, Saxena SK. Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virusdisease. 2020;31:179–193. doi: 10.1007/s13337-020-00598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ratajczak MZ, Kucia M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine "storm" and risk factor for damage of hematopoietic stem cells. Leukemia. 2020;34:1726–1729. doi: 10.1038/s41375-020-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao H, Xu LH, Yamada Y, Liu DX. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS ONE. 2008;3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song P, Li W, Xie J, Hou Y, You C. Cytokine storm induced by SARS-CoV-2. Clin Chim Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fung TS, Liu DX. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen S, Tan TH, Tan YJ. Expression, glycosylation, and modification of the spike (S) glycoprotein of SARS CoV. Methods Mol Biol. 2007;379:127–135. doi: 10.1007/978-1-59745-393-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krokhin O, Li Y, Andonov A, Feldmann H, Flick R, Jones S, et al. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol Cell Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava N, Saxena SK. Prevention and Control Strategies for SARS-CoV-2 Infection. Coronavirus Disease 2019 (COVID-19) 2020 doi: 10.1007/978-981-15-4814-7_11. [DOI] [Google Scholar]
- 44.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxena SK, Kumar S, Maurya VK, Sharma R, Dandu HR, Bhatt MLB. Current Insight into the novel coronavirus disease 2019 (COVID-19) Coronavirus Disease 2019 (COVID-19) 2020 doi: 10.1007/978-981-15-4814-7_1. [DOI] [Google Scholar]
- 46.Adedeji AO, Severson W, Jonsson C, Singh K, Weiss SR, Sarafianos SG. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J Virol. 2013;87:8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta A, Karki R, Dandu HR, Dhama K, Bhatt ML, Saxena SK. COVID-19: benefits and risks of passive immunotherapeutics. Hum Vaccin Immunother. 2020 doi: 10.1080/21645515.2020.1808410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]