Abstract
The current pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in the approval of numerous molecular diagnostic assays with various performance and technical capacities. There are limited data comparing performance among assays. We conducted a retrospective analysis of >10,000 test results among three widely used RT-PCR assays for coronavirus disease 2019 (CDC, Simplexa Direct, and TaqPath) to assess performance characteristics. We also retested remnant weakly positive specimens to assess analytical sensitivity. All assays had strong linear correlation and little bias among CT values for PCR targets. In patients with first-test negative results (n = 811), most (795, 98.0%) remained negative for all subsequent testing. Retesting of weakly positive specimens (CT > 30) showed sensitivities as follows: TaqPath (97.8%), CDC (91%), Simplexa (75.3%). Our analysis showed no performance difference among PCR targets within the same assay, suggesting a single target is sufficient for SARS-CoV-2 detection. Lower respiratory tract specimens had a higher negative predictive value (100%) than upper respiratory tract specimens (98%), highlighting the utility of testing lower respiratory tract specimens when clinically indicated. Negative predictive value did not increase on further repeated testing, providing strong evidence for discouraging unnecessary repeated testing for SARS-CoV-2.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which emerged in December 2019 in Wuhan, China, has resulted in the currently ongoing pandemic of coronavirus disease 2019 (COVID-19).1, 2, 3, 4 The first molecular diagnostic assays were developed5 shortly after the characterization of the viral genome. In February 2020, the CDC received an Emergency Use Authorization for the COVID-19 Real-Time Reverse Transcriptase PCR assay. Following that, numerous commercial molecular diagnostic assays were approved with Emergency Use Authorization for COVID-19, with various performance, throughput, speed, and technical complexity factors. Since then, numerous reports comparing analytical performance between and among assays have been published.6, 7, 8, 9, 10, 11, 12, 13 However, these data were obtained on the basis of small sample sizes, making robust conclusions difficult.
Herein, we report retrospective data collected from March 9 to April 29 on >10,000 clinical specimens from 8948 unique patients tested using three assays: CDC COVID-19 RT-PCR, Simplexa COVID-19 Direct Real-Time RT-PCR (DiaSorin Molecular, Cypress, CA), and TaqPath COVID-19 RT-PCR (Thermo Fisher Scientific, Waltham, MA). We measured negative predictive values (NPVs) among assays and bias among gene targets within each assay. We also report data for >900 patients with multiple test results to assess the NPV of the first result. Finally, we retested >100 previously weakly positive specimens by all three assays to compare analytical sensitivities.
Materials and Methods
Study Overview and Ethics
Three commercially available assays approved under the Emergency Use Authorization were used for the qualitative detection of SARS-CoV-2 as part of standard of care testing at the University of California, Los Angeles Health System. A retrospective analysis of PCR results obtained from these assays between March 9, 2020, and April 29, 2020 was performed. During this period, testing was mostly performed on patients with symptoms or exposure history. This study is exempt from Institutional Review Board review.
Clinical Samples
Tests were performed on various specimen types: nasopharyngeal (NP) swab (n = 10,215), bronchoalveolar lavage (n = 121), expectorated sputum (n = 22), and miscellaneous sample types (n = 35). Of the 630 positive tests, 613 were NP swabs, 11 were bronchoalveolar lavage, 2 were expectorated sputum, and 4 were other sample types.
Description of Assays
The CDC COVID-19 RT-PCR assay targets two regions of the SARS-CoV-2 N gene (ie, N1 and N2). Nucleic acid extraction was performed using EZ1 Advanced XL (Qiagen, Hilden, Germany) or NUCLISENS easyMAG (bioMérieux, Hazelwood, MO). RT-PCR was performed on the Applied Biosystems 7500 Fast Real-Time PCR instrument (Thermo Fisher Scientific, Waltham, MA). Detection of both gene targets was considered positive; detection of one of the two targets was deemed inconclusive. This assay was mainly performed on bronchioalveolar lavage and sputum because it was the only assay approved for the lower respiratory tract (LRT) sample types in the early phase of the COVID-19 outbreak. Other respiratory specimens, such as tracheal aspirate, were tested and resulted with a disclaimer stating a nonvalidated specimen source was used.
The Simplexa COVID-19 Direct Real-Time RT-PCR assay was performed on the LIASON MDX instrument (DiaSorin Molecular). This assay targets the SARS-CoV-2 S and ORF1ab genes. Detection of one or both targets was deemed positive. This assay was performed on NP swabs only. This test was primarily used on inpatients and high-risk health care workers because of its faster turnaround time.
The TaqPath COVID-19 RT-PCR assay targets the SARS-CoV-2 N, S, and ORF1ab genes. Nucleic acid extraction was performed with the MagMax Viral/Pathogen Nucleic Acid Isolation Kit using the automated KingFisher Flex Purification System (Thermo Fisher Scientific). RT-PCR was performed on the Applied Biosystems 7500 Fast Real-Time PCR instrument. Detection of two or all targets was deemed positive; detection of only one target was deemed inconclusive. This assay was performed on NP swabs, NP aspirates, and bronchoalveolar lavage specimens that are not viscous. This test was primarily used on ambulatory patients and low-risk health care workers because of its highest throughput.
Comparison Study of Weakly Positive Specimens
A total of 107 weakly positive (defined as any target regardless of assay, with a CT value of ≥30) NP swab specimens were selected. These frozen specimens (stored at −80°C for 3 to 8 weeks) were thawed and retested by all three assays within 8 hours.
Statistical Analysis
Categorical variables were reported as frequencies and percentages; continuous variables were reported as means with SD. Pearson χ2 or Fisher exact tests and t-tests or analysis of variance were used to compare categorical and continuous results, respectively.
NPV was calculated for the retrospective analysis of within-patient data. NPV was defined as the true negatives over the true negatives plus the false negatives. True negatives were defined as patients with repeatedly negative test results. False negatives were defined as an initial negative test result followed by a subsequent positive test result.
Results
Overview of PCR Results
From March 9, 2020, to April 29, 2020, 10,393 tests were performed on 10,165 unique specimens from 8948 patients. A total of 630 (6.1%) positive results from 527 (5.9%) patients were obtained. A total of 25 (0.2%) inconclusive and 13 (0.1%) indeterminate test results were obtained, and all were repeated.
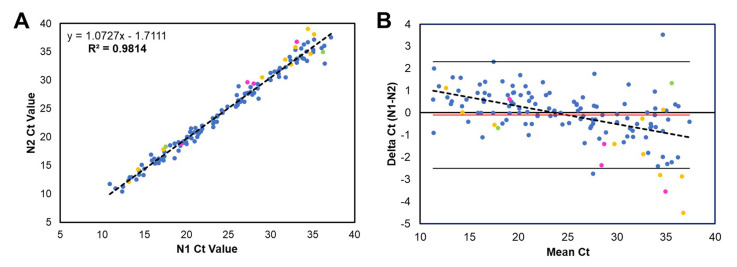
The CDC assay had a positivity rate of 7.3% (132/1820), with CT values ranging from 10.9 to 37.2 (24.7 ± 7.1) for N1 and from 10.4 to 39.0 (24.8 ± 7.7) for N2. Strong linear correlation (Supplemental Figure 1A) and little bias (Supplemental Figure 1B) between target CT values were observed. Ten (0.6%, 10/1820) specimens were inconclusive, with CT ranges of 31.0 to 40.7. On repeated testing by the same assay, one specimen was positive, five were negative, and four remained inconclusive with the same single target detected.
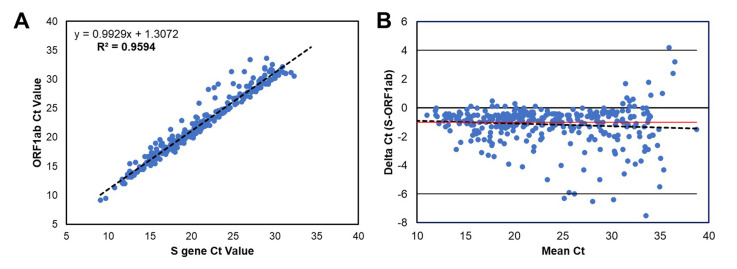
The Simplexa Direct assay had a positivity rate of 6.7% (358/5356), with CT values ranging from 9.1 to 38.7 (23.2 ± 6.8) for S and from 9.2 to 39.5 (24.2 ± 6.7) for ORF1ab. A total of 5.6% (20/358) of positive tests had only one target detected (S gene, 12/20; ORF1ab, 8/20). Strong linear correlation (Supplemental Figure 2A) between target CT values was seen. A slight bias toward higher CT values (0.34) for the ORF1ab target was observed, but it was not statistically significant (Supplemental Figure 2B). These results suggested comparable performance of the two PCR targets (S and ORF1ab) in this assay.
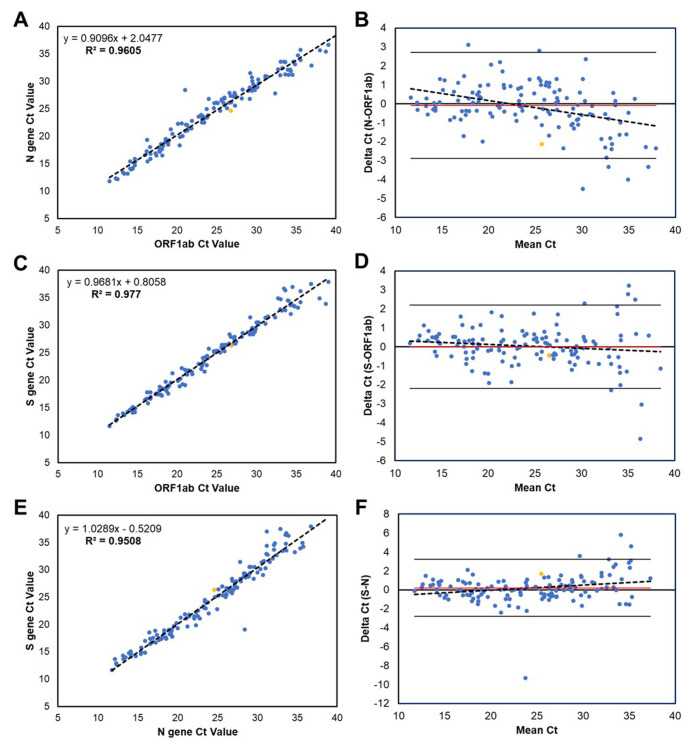
The TaqPath assay had a positivity rate of 4.4% (140/3217), with CT values ranging from 11.5 to 39.8 (24.8 ± 7.3) for ORF1ab, from 11.8 to 36.7 (24.8 ± 6.8) for N, and from 11.7 to 39.4 (24.7 ± 7.1) for S. A total of 6.4% (9/140) of positive tests had only two targets detected. All target comparisons showed strong linear correlation and little bias between CT values (Supplemental Figure 3). Eleven specimens were inconclusive (ie, only one of three targets detected), with CT ranges of 34.0 to 39.9. On repeated testing using other assays (Simplexa Direct = 10 and CDC = 1), 10 were tested negative (9 by Simplexa and 8 by CDC) and 1 was tested positive by Simplexa.
Estimation of NPVs
Of the 8949 patients, 907 had greater than one unique specimen tested, 702 had two specimens, 132 had three specimens, and 73 had greater than four specimens. Most (795, 87.7%) of these patients were repeatedly negative; 112 (12.3%) had at least one positive result. A false-negative initial test result was defined as a patient with an initial negative PCR test result followed by a subsequent positive test at any time. Of 811 patients with an initial negative test result, most of them (795, 98.0%) remained negative (ie, true negatives) for all subsequent tests (636 were tested two times, 107 were tested three times, and 52 were tested four or more times). NPVs were calculated by dividing the true negatives by the true negatives plus the false negatives. The overall NPV for patients with an initial negative test result was 98.0% (95% CI, 97.0%–98.7%) (Table 1 ). The NPV was also calculated on the basis of patients with second (NPV = 98.8%) and third test negatives (NPV = 96.3%). These results indicate that NPVs do not significantly change with additional testing.
Table 1.
NPV for Initial Negative Test Results
| Variable | Total | True negative | False negative | NPV, % (95% CI) |
|---|---|---|---|---|
| Result 1: negative | 811 | 795 | 16 | 98.0 (97.0–98.7) |
| Results 1–2: negative | 161 | 159 | 2 | 98.8 (95.5–99.7) |
| Results 1–3: negative | 54 | 52 | 2 | 96.3 (88.4–98.9) |
True negative was defined as all subsequent patient tests (regardless of specimen type) were negative/not detected. False negative was defined as at least one subsequent patient test (regardless of specimen type) was positive/detected.
NPV, negative predictive value.
NPV calculation was also performed based on results from the three different assays and it was found that NPVs did not differ significantly among assays (Table 2 ). In the 16 cases (2.0%, 16/811) that met our false-negative definition (ie, initial test negative and then became positive subsequently), 15 of them were tested positive by the same assay subsequently. Only in one case, an initial negative result by Simplexa was followed by a positive result using CDC. In this case, the negative result was an NP swab and the positive result was a tracheal suction; the specimens were collected on the same day. These data show that false-negative results are unrelated to the specific assay performed in our institution.
Table 2.
NPV for Initial Negative Test Results by Assay or Specimen Type
| Comparison by | Group | Initial negative (n = 811) | True negative (n = 795) | False negative (n = 16) | NPV, % (95% CI) |
|---|---|---|---|---|---|
| Assay | CDC RT-PCR | 168 | 166 | 2 | 98.8 (95.7–99.7) |
| Simplexa Direct | 518 | 506 | 12 | 97.7 (96.2–98.6) | |
| TaqPath RT-PCR | 125 | 123 | 2 | 98.4 (94.6–99.5) | |
| Specimen type | Initial URT | 785 | 769 | 16 | 98.0 (96.9–98.7) |
| Initial LRT | 26 | 26 | 0 | 100 |
LRT, lower respiratory tract; NPV, negative predictive value; URT, upper respiratory tract.
NPVs differed slightly by initial specimen type [ie, upper respiratory tract (URT) or LRT] (Table 2). Patients with initially negative URT specimens (n = 785) had an NPV of 98.0%, whereas patients with initially negative LRT specimens (n = 26) had no false negatives, giving an NPV of 100%. Twenty-nine patients had a URT and LRT specimen collected and tested on the same day. The same-day specimens were usually both negative (86.2%, 25/29); one pair (3.4%) was both positive, and three pairs (10.4%) had conflicting results. Among those with conflicting results, all were URT-/LRT+.
Comparison Study of Weakly Positive Samples
A total of 107 weakly positive (any target CT ≥ 30) NP specimens were selected for retesting on all three assays. These samples consisted of 17 (15.9%), 64 (59.8%), and 26 (24.3%) samples originally tested by CDC, Simplexa, and TaqPath, respectively. Retesting was performed on the frozen specimen, and all assays were performed within 8 hours to ensure the same sample stability to allow an unbiased comparison among the three assays. 61.7% (66/107) of specimens tested positive on all three assays, whereas 3.7% (4/107) were negative on all three. The remaining 34.6% (37/107) tested positive on only one (9/37) or two assays (28/37).
TaqPath was shown to be most sensitive (92.5%, 99/107), followed by CDC (90.7%, 97/107) and Simplexa (62.6%, 67/107). The freeze-thaw cycle negatively affected the performance of Simplexa the most, with only 49 of the 64 (76.6%) specimens retested positive on the same assay. However, the freeze-thaw cycle seemed to have less impact on the retested positivity rate on the CDC assay (82.4%, 14/17) and the TaqPath assay (92.3%, 24/26). Because freeze-thaw cycles were shown to affect the assays differently, specimens that were not reproducibly positive by the same assay (n = 18) were excluded to remove this confounding factor. The recalculated sensitivities of the three assays were as follows: 97.8% (TaqPath), 91.0% (CDC), and 75.3% (Simplexa) (Table 3 ). The 15 specimens that were not repeat positive on Simplexa had a significantly higher (P < 0.01) average CT value (33.6 ± 2.3) than the 49 that were repeat positive (32.0 ± 1.9). These data show that Simplexa had lower sensitivity for the weakly positive samples. Similar comparisons for the TaqPath and CDC assays were not statistically significant.
Table 3.
Results for Retesting Weakly Positive Specimens among Assays
| Original test | CDC RT-PCR |
Simplexa Direct |
TaqPath RT-PCR |
|||||
|---|---|---|---|---|---|---|---|---|
| D | ND | I | D | ND | D | ND | I | |
| Detected (n = 89) | 81 (91.0) | 4 (4.5) | 4 (4.5) | 67 (75.3) | 22 (27.4) | 87 (97.8) | 2 (2.2) | 0 |
| CDC RT-PCR (n = 16) | 14 | 0 | 2 | 8 | 8 | 14 | 2 | 0 |
| Simplexa Direct (n = 49) | 49 | 0 | 0 | 49 | 0 | 49 | 0 | 0 |
| TaqPath RT-PCR (n = 24) | 18 | 4 | 2 | 10 | 14 | 24 | 0 | 0 |
Data are given as n or n (%).
D, detected; I, inconclusive; ND, not detected.
Discussion
Over 10,000 test results were retrospectively evaluated for the detection of SARS-CoV-2 in patient specimens from three RT-PCR assays: CDC, Simplexa Direct, and TaqPath. CT values were plotted for every pair of targets for each assay and linear regression was used to show that values were strongly correlative; this was consistent among assays and specimen types. Bland-Altman plots were used to show that the mean bias of CT values between targets was near 0 for all assays. Considering the large sample size used to generate these data, these findings strongly support the claim that testing of a single target is appropriate for SARS-CoV-2 detection. Multiple targets per assay generates the opportunity for inconclusive results, which could lead to increased complexity in result interpretation. Most of the inconclusive results obtained herein tested negative on repeat testing (15/21).
The NPV for patients was calculated with initial negative test results. The overall NPV was high (98%) and did not differ significantly by number of repeated testing or assay used in our institution. These data show that retesting negative patients is unlikely to yield a subsequent positive result. These findings are consistent with other reports14 and serve as important considerations for clinical and epidemiologic decision-making regarding suspected COVID-19 patients. The NPV of LRT specimens (100%) was significantly higher than that of URT specimens (98%). Considering the pathogenesis of the virus, these data likely relate to the stage of disease. Several studies have shown that URT specimens are positive earlier in infection, whereas LRT specimens have prolonged positivity for SARS-CoV-2.15, 16, 17 By assessing the 29 patients with both URT and LRT specimens collected on the same day, most test results (26/29) matched between paired specimens. However, in three cases, the URT specimens were negative, whereas the LRT samples were positive, highlighting the importance of testing the LRT samples in a setting of negative URT sample but still clinically suspected for COVID-19.
A comparison study was performed using weakly positive samples to assess the sensitivity of the three SARS-CoV-2 RT-PCR assays, and the results showed Simplexa had a lower sensitivity than TaqPath or CDC. Notably, the sensitivity of the Simplexa assay is greatly reduced for extremely weak positive specimens with CT values >33.6. However, this is not a significant concern because only 2.1% (7/338) of the Simplexa-positive specimens had both CT values >33.6. In addition, its ease of use, its fast speed, and our findings that its NPV was not statistically different from the other two assays justify the use of Simplexa for clinical diagnostic testing of symptomatic patients. Ultimately, we believe preanalytical parameters, including specimen types, sample collection quality, and timing, are more important factors for the clinical performance of the PCR assays for SARS-CoV-2 detection.
Limitations of our study included the lack of clinical chart review. This would be particularly important to determine the stage of disease of positive patients and could be used to determine clinical false-negative results. Variation in specimen collection techniques and collection time relative to symptoms was not considered in this analysis; however, the large sample size likely overcomes this limitation. In addition, our definition of false-negative results is not perfect because of lack of clinical information or further data, such as serologic test results. Finally, the weakly positive specimens used for the comparison study were previously frozen and may have degradation, which reduced the reproducibility. Our data showed that the freeze-thaw cycle likely affected PCR assay sensitivity, especially on Simplexa.
In summary, this is one of the first studies that assessed >10,000 patient test results to compare SARS-CoV-2 PCR assay performance. Our analysis showed no performance difference among different PCR targets within the same assay, suggesting only one target is sufficient for SARS-CoV-2 detection. The NPV of the initial PCR test was high (approximately 98%) and did not differ among different assays, despite a noticeable difference in sensitivity toward weakly positive samples. We also demonstrated that the NPV did not increase on further repeated testing, providing strong evidence for discouraging unnecessary repeated testing for SARS-CoV-2.
Footnotes
Supported by the University of California Los Angeles (UCLA) Department of Pathology and Laboratory Medicine.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.11.008.
Supplemental Data
Supplemental Figure S1.
Correlation and bias between CT values of the CDC COVID-19 RT-PCR assay for the N1 and N2 regions of the SARS-CoV-2 N gene. CT values for positive specimens are shown for the N1 and N2 targets. Specimen type is shown by color: blue (nasopharyngeal swab), yellow (bronchoalveolar lavage), green (expectorated sputum), and pink (other). A: Linear regression line (dotted line) was deduced from the CT values; the equation is shown. Correlation coefficient is R = 0.9814 (95% CI, 1.047–1.098; P < 0.001). B: Bland-Altman plot shows difference between N1 and N2 CT values versus the mean of the two measurements. The mean bias is −0.1 (red line), and the SD is 1.2 (two times SD is solid black lines). n = 132 positive specimens (A and B).
Supplemental Figure S2.
Correlation and bias between CT values of the Simplexa COVID-19 Direct assay for the SARS-CoV-2 S gene and ORF1ab. CT values for positive (both targets detected) specimens are shown for the S gene and ORF1ab targets. Specimen type is shown by color: blue (nasopharyngeal swab). A: Linear regression line (dotted line) was deduced from the CT values; the equation is shown. Correlation coefficient is R = 0.9594 (95% CI, 0.913–0.996; P < 0.001). B: Bland-Altman plot shows difference between S and ORF1ab CT values versus the mean of the two measurements. The mean bias is −1.0 (red line), and the SD is 1.3 (two times SD is solid black lines). n = 338 positive specimens (A and B).
Supplemental Figure S3.
Correlations among CT values of the TaqPath COVID-19 RT-PCR assay for the SARS-CoV-2 N gene, S gene, and ORF1ab. A–F: CT values for positive (all three targets detected) specimens are shown for the N and ORF1ab targets (A and B), S and ORF1ab targets (C and D), and S and N targets (E and F). Specimen type is shown by color: blue (nasopharyngeal swab) and yellow (bronchoalveolar lavage). A, C, and E: Linear regression lines (dotted lines) were deduced from the CT values; equations are shown. A: Correlation coefficient is R = 0.9605 (95% CI, 0.877–0.942; P < 0.001). C: Correlation coefficient is R = 0.9770 (95% CI, 0.942–0.994; P < 0.001). E: Correlation coefficient is R = 0.9508 (95% CI, 0.988–1.070; P < 0.001). B, D, and F: Bland-Altman plots show difference between pairs of CT values versus the mean of the two measurements. B: Difference between N and ORF1ab; mean bias is −0.1 (red line), and the SD is 1.4 (two times SD is solid black lines). D: Difference between S and ORF1ab; mean bias is 0.0 (red line), and the SD is 1.1 (two times SD is solid black lines). F: Difference between S and N; mean bias is 0.2 (red line), and the SD is 1.5 (two times SD is solid black lines). n = 131 positive specimens (A–F).
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Guizhen W., Gao G., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Joanna Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P. Drosten CDetection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00743-20. e00743–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., Love N. The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00772-20. e00772–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00760-20. e00760–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P., Maggiore J., Kahn S. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00798-20. e00798–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00821-20. e00821–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D., Deconinck L., Lescure F.X., Lucet J.C., Bouadma L., Timsit J.F., Descamps D., Yazdanpanah Y., Casalino E., Houhou-Fidouh N. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00630-20. e00630–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poljak M., Korva M., Knap Gasper N., Fujs Komlos K., Sagadin M., Ursic T., Zupanc T.A., Petrovec M. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00599-20. e00599–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smithgall M., Scherberkova I., Whittier S., Green D. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the rapid detection of SARS-CoV-2. J Clin Viol. 2020;128:e104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Testing for SARS-CoV-2: can we stop at two? Clin Infect Dis. 2020;71:2246–2248. doi: 10.1093/cid/ciaa459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee T.H., Lin R.J., Lin R.T.P., Barkham T., Rao P., Leo Y.S., Lye D.C., Young B. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]