Abstract
Background
Urinary tract infections (UTIs) are the most common bacterial infections and are often caused by uropathogenic Escherichia coli (UPEC). We investigated the distribution of phylogenetic groups, adhesin genes, antimicrobial resistance, and biofilm formation in E. coli isolated from patients with UTIs.
Methods
In the present study, 208 UPEC isolated from Thai patients were classified into phylogenetic groups and adhesin genes were detected using multiplex PCR. Antimicrobial susceptibility testing was performed using agar disk diffusion. The Congo red agar method was used to determine the ability of the UPEC to form biofilm.
Results
The most prevalent UPEC strains in this study belonged to phylogenetic group B2 (58.7%), followed by group C (12.5%), group E (12.0%), and the other groups (16.8%). Among adhesin genes, the prevalence of fimH (91.8%) was highest, followed by pap (79.3%), sfa (12.0%), and afa (7.7%). The rates of resistance to fluoroquinolones, trimethoprim-sulfamethoxazole, and amoxicillin-clavulanate were 65%, 54.3%, and 36.5%, respectively. The presence of adhesin genes and antibiotic resistance were more frequent in groups B2 and C compared to the other groups. Of the 129 multidrug-resistant UPEC strains, 54% were biofilm producers. Our findings further indicated that biofilm production was significantly correlated with the pap adhesin gene (p ≤ 0.05).
Conclusion
These findings provide molecular epidemiologic data, antibiotic resistance profiles, and the potential for biofilm formation among UPEC strains that can inform further development of the appropriate prevention and control strategies for UTIs in this region.
Keywords: Uropathogenic Escherichia coli, Phylogenetic group, Adhesin genes, Biofilm, Antimicrobial resistance
Introduction
Urinary tract infections (UTIs) are a common bacterial infection, with 150 million UTI cases observed annually worldwide (Stamm & Norrby, 2001). Uropathogenic Escherichia coli (UPEC) is the most common causative agent of both uncomplicated and complicated UTIs, accounting for 75% and 65% of cases, respectively (Flores-Mireles et al., 2015). Clermont and colleagues developed a new polymerase chain reaction (PCR)-based method to classify the eight phylogenetic groups of E. coli, of which seven are clustered in E. coli sensu stricto (A, B1, B2, C, D, E, and F) and one belongs to Escherichia Clade 1 (Clermont et al., 2013). Several studies have reported that phylogenetic groups B2 and D are associated with extraintestinal infection, while the other groups are more prevalent among diarrheagenic and commensal bacteria (Picard et al., 1999; Kumar, Nahid & Zahra, 2017; Ahumada-Santos et al., 2020).
Adherence and colonization are the crucial steps in UTI pathogenesis. UPEC generally use various adhesins to recognize uroepithelium cells and mediate colonization (Flores-Mireles et al., 2015). Type 1 fimbriae consist of a major protein, FimA, that is associated with the ancillary proteins FimF, FimG, and the adhesin FimH, all of which are encoded by the fim gene cluster (Orndorff & Falkow, 1984). The P fimbriae are encoded by the pap gene cluster, which contains 11 genes (papA to papK) (Fernández & Berenguer, 2000). P fimbriae promote early colonization of the epithelial cells lining the tubules, while type 1 fimbriae appear to play a role in inter-bacterial binding and biofilm formation (Melican et al., 2011). The S fimbriae are expressed by the sfa operon, which was reported to be most often found in E. coli strains implicated in human meningitis and septicemia (Antao, Wieler & Ewers, 2009). The P-independent, X-binding fimbrial adhesin encoded by the afa1 operon mediates specific binding to uroepithelial cells and human erythrocyte receptors (Labigne-Roussel & Falkow, 1988). Different studies have investigated the presence of the adhesion-encoding genes pap (P fimbriae), sfa (S fimbriae), afa (afimbrial adhesin), and fimH (type 1 fimbriae) across UPEC strains using multiplex PCR (Rahdar et al., 2015; Dadi et al., 2020; Tarchouna et al., 2013; Shetty et al., 2014).
Currently, the empirical treatment of UTIs is an issue of concern due to the increasing rates of antibiotic resistance. The resistance to trimethoprim-sulfamethoxazole (TMP-SMZ), ciprofloxacin, and amoxicillin-clavulanate (AMC) among UPEC isolates is higher in developing countries (ranging from ∼50% to 85%) than in developed countries (ranging from 3% to 40%) (Kot, 2019). Routine standard antimicrobial susceptibility testing must be performed in order to reduce the rates of inappropriate empirical antibiotic therapy of UTIs and thereby decrease the occurrence of multidrug-resistant (MDR) UPEC (Adamus-Białek et al., 2018).
Biofilms are microbial communities that adhere to various surfaces, and the cells within a biofilm are encased in self-produced extracellular polymeric matrix (Hall & Mah, 2017). The ability of UPEC to form biofilms is important, as biofilms increase antimicrobial agent tolerance and facilitate evasion of the urinary tract host defense, contributing to the evolution of MDR strains and the recurrence of UTIs (Mittal, Sharma & Chaudhary, 2015).
A study of virulence genes and antimicrobial susceptibility patterns of UPEC in southern Thailand was previously reported (Themphachanal et al., 2015), but there is no information on the new classification of phylogenetic groups or the biofilm-forming ability of UPEC. Therefore, the aim of the present study was to determine the phylogenetic groups, adhesin gene distribution, antimicrobial resistance profiles, and biofilm formation ability of UPEC isolated from patients with UTIs in central Thailand. We also investigated the possible correlation between adhesin genes and the ability to form biofilm.
Materials and Methods
Ethical approval
E. coli strains were isolated from patients with UTI then identified and collected at the Nopparat Rajathanee Hospital as part of the routine microbiological laboratory. The study protocol was approved by the Ethics Review Board (ERB) of the Research Institute of Rangsit University (DPE.No.RSUERB2018-002). All the bacterial strains were acquired with permission from the Director of Nopparat Rajathanee Hospital.
Bacterial strains
The 208 non-repetitive E. coli strains isolated from urine specimens of UTI patients between February and May 2018 were used from the current study. E. coli strains were isolated from pure cultures and identified in the department of microbiological laboratory in the Nopparat Rajathanee Hospital. The bacteria were confirmed as E. coli by considering Gram’s staining morphology, colony characteristic on MacConkey agar (Oxoid, UK), and biochemical properties (Bergey & Holt, 1994). The oxidase test, catalase test, sugar fermentation, motility test, indole production, methyl red test, Voges-proskauer reaction, urease production, citrate utilization, and ornithine and lysine decarboxylase test were used as the standard biochemical testing in our laboratory. The only one isolate from each patient was investigated.
Characterization of phylogenetic groups and adhesin genes
Bacterial DNA was extracted using the optimized boiling method (Dashti et al., 2009). The phylogenetic groups of E. coli were characterized using multiplex PCR according to the protocol previously published (Clermont et al., 2013). Table S1 shows the primer sequences and the size of amplicons. In addition, four adhesin genes, pap, sfa, afa, and fimH, were detected in all isolates using multiplex PCR (Yamamoto et al., 1995; Le Bouguenec, Archambaud & Labigne, 1992; Struve & Krogfelt, 1999). The details of the primers and sizes of PCR products are listed in Table S2. The PCR reaction volume contained 15 µl of 2X AmpMaster™ HS-Taq (GeneAll®, Korea), 10 pmol/µl of each primer, 3 µl of DNA template, and DNase-free H2O to a final volume of 30 µl. Amplification was carried out in the Mastercycler® nexus (Eppendorf, Germany) under the following conditions: initial denaturation at 95 °C for 3 min, 45 cycles of 45 s denaturation at 95 °C, 45 s of primer annealing at 55 °C (to characterize the phylogenetic groups) and 54 °C (to amplify the adhesin genes), 60 s of extension at 72 °C, and further extension for 5 min at 72 °C. PCR products were separated on a 2% agarose gel with a 100-bp DNA ladder (Fermentas, US) and visualized on a UV trans-illuminator.
Antimicrobial susceptibility testing
Antimicrobial susceptibility tests were performed using the agar disk diffusion method according to Clinical and Laboratory Standards Institute guidelines (CLSI, 2018). The antibiotic disks (Oxoid, UK) ampicillin (10 µg), amoxicillin-clavulanate (20/10 µg), piperacillin-tazobactam (100/10 µg), cefoperazone-sulbactam (75/30 µg), cefazolin (30 µg), cefotaxime (30 µg), ceftriaxone (30 µg), ceftazidime (30 µg), imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), gentamicin (10 µg), amikacin (30 µg), netilmicin (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), norfloxacin (10 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), and fosfomycin (200 µg) were used. Escherichia coli ATCC 25922 was used as a control in all antibiogram tests. Whether a strain was MDR was determined on the basis of acquired non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012).
Detection of biofilm formation
The biofilm production of all E. coli strains was determined using the Congo red agar (CRA) method, as previously published (Neupane et al., 2016; Sm, Jayakumar & Aravazhi, 2016; Tajbakhsh et al., 2016). The medium contains brain heart infusion agar (52 gm/L); sucrose (36 gm/L) and Congo red dye (0.8 gm/L). The tested organisms were cultured on CRA and incubated under the aerobic condition at 37 °C for 24 to 48 h. The six color tones of colonies were categorized as follows: very black, black, almost black, which were interpreted as strong, moderate, and weak biofilm producers, respectively, and bordeaux, red, and very red, reported as non-biofilm producers.
Statistical analysis
Chi-square test was used for comparisons of proportions the demographic characteristics of patients. The correlations between phylogenetic group, the presence of adhesin genes, biofilm production, and antimicrobial resistance were determined by performing Pearson’s chi-square tests. SPSS version 21 software was used for data analysis (IBM SPSS Inc., Armonk, NY, USA). Results were considered statistically significant if the p-value was ≤ 0.05.
Results
Among 1,926 patients with symptoms of UTI, a total of 208 isolates were identified as E. coli. The demographic characteristics of patients infected with UPEC are shown in Table 1. Among the patients, 154 (74%) were female and 54 (26%) were male. Patients were stratified into five different age groups, and those over 65 years represented 63.9% of all patients. The highest number of UPEC samples was isolated from catheter urine samples (150, 72.1%). The highest proportion of UPEC isolates came from the internal medicine ward (80, 38.5%), followed by the emergency room (45, 21.6%), intensive care unit (34, 16.3%), and outpatients (22, 10.6%).
Table 1. Demographic characteristics of patients infected with uropathogenic E. coli (N = 208).
| Parameter | No. of isolates (%) | Chi-square | Degree of freedom | p-value | |
|---|---|---|---|---|---|
| Gender | |||||
| Female | 154 (74.0) | 48.08a | 1 | <0.0001 | |
| Male | 54 (26.0) | ||||
| Age (years) | |||||
| <14 | 11 (5.3) | 265.02b | 4 | <0.0001 | |
| 15–24 | 6 (2.9) | ||||
| 25–44 | 15 (7.2) | ||||
| 45–64 | 43 (20.7) | ||||
| ≥65 | 133 (63.9) | ||||
| Type of samples | |||||
| Midstream urine | 58 (27.9) | 40.69a | 1 | <0.0001 | |
| catheter urine | 150 (72.1) | ||||
| Hospital Unit | |||||
| Out-patient | 22 (10.6) | 188.23c | 7 | <0.0001 | |
| In-patient | |||||
| Internal medicine | 80 (38.5) | ||||
| ER | 45 (21.6) | ||||
| ICU | 34 (16.3) | ||||
| Surgery | 12 (5.8) | ||||
| Pediatrics | 8 (3.8) | ||||
| Stroke | 5 (2.4) | ||||
| Burn | 2 (1.0) | ||||
| MDR stains | |||||
| MDR | 129 (62.0) | 12.02a | 1 | 0.001 | |
| Non-MDR | 79 (38.0) |
Notes.
0 cells (.0%) have expected frequencies less than 5. The minimum expected cell frequency is 104.0.
0 cells (.0%) have expected frequencies less than 5. The minimum expected cell frequency is 41.6.
0 cells (.0%) have expected frequencies less than 5. The minimum expected cell frequency is 26.0.
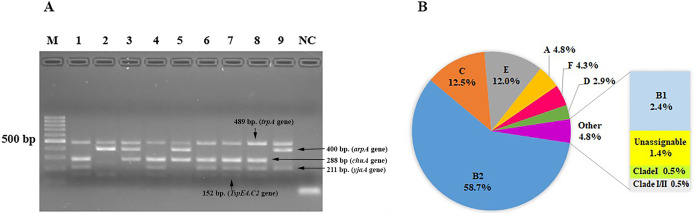
We characterized the phylogenetic groups of E. coli from urine specimens by detecting the arpA (400 bp), chuA (288 bp), yjaA (211 bp), and TspE4.C2 (152 bp) genes using multiplex PCR (Fig. 1A). Primers specific for the trpA (489 bp) gene were added to all PCR reactions to provide an internal control. Groups C and E were classified by amplification of the trpA (219 bp) and arpA (301 bp) genes using specific primers. The majority of the 208 E. coli isolates were group B2 (122, 58.7%), followed by group C (26, 12.5%), group E (25, 12%), group A (10, 4.8%), group F (9, 4.3%), group D (6, 2.9%), group B1 (5, 2.4%), unassignable (3, 1.4%), and clade I or clade II (2, 1.0%; Fig. 1B).
Figure 1. The distribution of phylogenetic groups among uropathogenic Escherichia coli isolates by the new Clermont phylo-typing method.
(A) Multiplex PCR profiles for specific uropathogenic Escherichia coli isolates by detecting the arpA (400 bp), chuA (288 bp), yjaA (211 bp), and TspE4.C2 (152 bp) genes. Lane M, 100-base pair ladder (Fermantas); Lane 1, group B2 (-, +, +, +); Lane 2, group B1 (+, -, -, +); Lane 3, group D or E (+, +, -,-); Lane 4, group B2 (-, +, +, +); Lane 5, group D or E (+, +, -,-); Lane 6, group B2 (-, +, +, +); Lane 7, group B2 (-, +, +, +); Lane 8, group B2 (-, +, +, +); Lane 9, group A or C (+, -, +, -); Lane NC, negative control. The trpA (489 bp) internal control gene appeared in all samples except the negative control. Distilled water without any DNA as negative controls was used in PCR experiments. (B) The percentage of phylogenetic groups among uropathogenic Escherichia coli isolates.
Adhesin-encoding genes were successfully amplified by multiplex PCR. The most frequent UPEC adhesin gene was fimH (191, 91.8%), followed by pap (165, 79.3%), sfa (25, 12.0%), and afa (16, 7.7%). We also investigated the adhesin gene patterns of the strains (Table 2). Among the isolates, 30 (14.4%), 167 (80.3%), and 11 (5.3%) possessed 1, 2, and 3 adhesin genes, respectively. A high prevalence of combined fimH and pap genes was significantly found (69.2%, p < 0.0001). Moreover, the fimH gene has significant association with UPEC phylogenetic groups B2 (p = 0.041). There were significant associations between phylogenetic group E and two adhesin genes namely pap and afa (p = 0.002 and p < 0.0001, respectively). Similarly, there were significant associations between phylogenetic group F and adhesin genes fimH and sfa (p = 0.005 and p = 0.044, respectively) (See Table 3).
Table 2. Profiles of adhesin genes in uropathogenic Escherichia coli strains.
| No. of genes | Adhesin genes patterns | No. of isolates (%) | Chi-square | Degree of freedom | P-value |
|---|---|---|---|---|---|
| 1 gene, n = 30 (14.4%) | |||||
| fimH | 19 (9.1) | ||||
| pap | 4 (1.9) | ||||
| sfa | 5 (2.4) | ||||
| afa | 2 (1.0) | ||||
| 2 genes, n = 167 (80.3%) | |||||
| fimH, pap | 144 (69.2) | 922.88a | 10 | <0.0001 | |
| fimH, sfa | 11 (5.3) | ||||
| fimH, afa | 6 (2.9) | ||||
| pap, sfa | 2 (1.0) | ||||
| pap, afa | 4 (1.9) | ||||
| 3 genes, n = 11 (5.3%) | |||||
| fimH, pap, sfa | 7 (3.4) | ||||
| fimH, pap, afa | 4 (1.9) | ||||
Notes.
0 cells (.0%) have expected frequencies less than 5. The minimum expected cell frequency is 18.9.
Table 3. The association between phylogenetic groups and adhesin genes of uropathogenic Escherichia coli isolates.
| Adhesin genes | Phylogenetic group | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B2 (n = 122) | C (n = 26) | E (n = 25) | A (n = 10) | F (n = 9) | |||||||||||||||||
| B2 | Non-B2 | χ2 | P-value | C | Non-C | χ2 | P-value | E | Non-E | χ2 | P-value | A | Non-A | χ2 | P-value | F | Non-F | χ2 | P-value | ||
| fimH | Present | 116 | 75 | 4.166 | 0.041 | 24 | 167 | 0.009 | 0.924 | 22 | 169 | 0.554 | 0.456 | 10 | 181 | 0.935 | 0.334 | 6 | 185 | 7.935 | 0.005 |
| Absent | 6 | 11 | 2 | 15 | 3 | 14 | 0 | 17 | 3 | 14 | |||||||||||
| pap | Present | 100 | 65 | 1.254 | 0.263 | 20 | 145 | 0.105 | 0.746 | 14 | 151 | 9.428 | 0.002 | 10 | 155 | 2.738 | 0.098 | 7 | 158 | 0.014 | 0.907 |
| Absent | 22 | 21 | 6 | 37 | 11 | 32 | 0 | 43 | 2 | 41 | |||||||||||
| sfa | Present | 19 | 6 | 3.526 | 0.060 | 1 | 24 | 1.877 | 0.171 | 1 | 24 | 1.728 | 0.189 | 0 | 25 | 1.435 | 0.231 | 3 | 22 | 4.041 | 0.044 |
| Absent | 103 | 80 | 25 | 158 | 24 | 159 | 10 | 173 | 6 | 177 | |||||||||||
| afa | Present | 6 | 10 | 3.198 | 0.074 | 0 | 16 | 2.476 | 0.116 | 8 | 8 | 23.645 | 0.000 | 0 | 16 | 0.875 | 0.349 | 1 | 15 | 0.155 | 0.694 |
| Absent | 116 | 76 | 26 | 166 | 17 | 175 | 10 | 182 | 8 | 184 | |||||||||||
| Adhesin genes | Phylogenetic group | ||||||||||||||||||||
| D (n=6) | B1 (n=5) | Unassignable (n=3) | CladeI (n=1) | Clade I or II (n=1) | |||||||||||||||||
| D | Non-D | χ 2 | P-value | B1 | Non-B1 | χ 2 | P-value | Unassign | Non-unassign | χ 2 | P-value | CladeI | Non-cladeI | χ 2 | P-value | CladeI/II | Non-cladeI/II | χ 2 | P-value | ||
| fimH | Present | 5 | 186 | 0.594 | 0.441 | 5 | 186 | 0.456 | 0.500 | 2 | 189 | 2.567 | 0.109 | 1 | 190 | 0.089 | 0.765 | 0 | 191 | 11.290 | 0.001 |
| Absent | 1 | 16 | 0 | 17 | 1 | 16 | 0 | 17 | 1 | 16 | |||||||||||
| pap | Present | 5 | 160 | 0.060 | 0.806 | 4 | 161 | 0.001 | 0.970 | 3 | 162 | 0.739 | 0.373 | 1 | 164 | 0.262 | 0.609 | 1 | 164 | 0.262 | 0.609 |
| Absent | 1 | 42 | 1 | 42 | 0 | 43 | 0 | 43 | 0 | 43 | |||||||||||
| sfa | Present | 0 | 25 | 0.844 | 0.358 | 1 | 24 | 0.309 | 0.579 | 0 | 25 | 0.416 | 0.519 | 0 | 25 | 0.137 | 0.711 | 0 | 25 | 0.137 | 0.711 |
| Absent | 6 | 177 | 4 | 179 | 3 | 180 | 1 | 182 | 1 | 182 | |||||||||||
| afa | Present | 0 | 16 | 0.515 | 0.473 | 0 | 16 | 0.427 | 0.513 | 1 | 15 | 2.818 | 0.093 | 0 | 16 | 0.084 | 0.772 | 0 | 16 | 0.084 | 0.772 |
| Absent | 6 | 186 | 5 | 187 | 2 | 190 | 1 | 191 | 1 | 191 | |||||||||||
We performed antimicrobial susceptibility tests on E. coli strains using different categories of antibiotics. There were significant associations between E. coli phylogenetic groups and resistance rates of antibiotics (p < 0.05) except ampicillin, gentamicin and trimethoprim-sulfamethoxazole (Table 4). All isolates showed high rates of resistance to ampicillin (84.1%), ciprofloxacin (65.4%), norfloxacin (65.4%), levofloxacin (64.9%), trimethoprim-sulfamethoxazole (54.3%), cefazolin (44.7%), cefotaxime (43.8%), ceftriaxone (43.8%), ceftazidime (43.8%), amoxicillin-clavulanate (36.5%), and gentamicin (33.7%). The rates of resistance to other antibiotics were between ∼1% and 6%. E. coli phylogenetic group C had the highest rates of resistance to all antibiotics (p <0.05) except ampicillin, gentamicin, amikacin, netilmicin, and fosfomycin (Table S3). Three isolates (1.4%) in group C were carbapenems-resistant. Interestingly, most of the 129 isolates (62.0%) that were MDR and belonged to group B2 (59.7%; 77 of 129). However, the lower resistance rates to piperacillin-tazobactam and carbapenems were observed in group B2 (p = 0.005 and p = 0.0038, respectively) (Table S3). The lowest rates of resistance to cephalosporin were observed in group A (p = 0.02), while group D was more susceptible to fluoroquinolones than the other groups (p = 0.01). The only one isolate of group A was resistant to fosfomycin (p < 0.0001).
Table 4. Chi-square test for comparisons of resistance rates to antimicrobial agents among various phylogenetic groups of uropathogenic Escherichia coli isolates.
| Antimicrobial resistance rates | Phylogenetic group | Chi- square | P-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B2 | C | E | A | F | D | B1 | Unassignable | Clade I and I or II | Total | |||
| n = 122(%) | n = 26(%) | n = 25(%) | n = 10(%) | n = 9(%) | n = 6(%) | n = 5(%) | n = 3(%) | n = 2(%) | n = 208(%) | |||
| Penicillins | ||||||||||||
| AMP | 101 (82.8) | 25 (96.2) | 21 (84) | 8 (80) | 6 (66.7) | 6 (100) | 4 (80) | 3 (100) | 1 (50) | 175 (84.1) | 16.707 | 0.054 |
| β-lactam/ β-lactamase inhibitor combinations | ||||||||||||
| AMC | 39 (32) | 17 (65.4) | 9 (36) | 3 (30) | 4 (44.4) | 1 (16.7) | 3 (60) | 0 | 0 | 76 (36.5) | 16.906 | 0.050 |
| TZP | 2 (1.6) | 7 (26.9) | 1 (4) | 0 | 1 (11.1) | 0 | 0 | 0 | 0 | 11 (5.3) | 29.961 | 0.000 |
| SCF | 5 (4.1) | 7 (26.9) | 1 (4) | 0 | 0 | 0 | 0 | 0 | 0 | 13 (6.3) | 22.477 | 0.007 |
| Cephalosporins | ||||||||||||
| KZ | 54 (44.3) | 18 (69.2) | 11 (44) | 1 (10) | 5 (55.6) | 2 (33.3) | 1 (20) | 1 (33.3) | 0 | 93 (44.7) | 15.248 | 0.084 |
| CTX | 53 (43.4) | 18 (69.2) | 11 (44) | 1 (10) | 5 (55.6) | 1 (16.7) | 1 (20) | 1 (33.3) | 0 | 91 (43.8) | 16.977 | 0.049 |
| CRO | 53 (43.4) | 18 (69.2) | 11 (44) | 1 (10) | 5 (55.6) | 1 (16.7) | 1 (20) | 1 (33.3) | 0 | 91 (43.8) | 16.977 | 0.049 |
| CAZ | 52 (42.6) | 18 (69.2) | 11 (44) | 1 (10) | 5 (55.6) | 1 (16.7) | 2 (40) | 1 (33.3) | 0 | 91 (43.8) | 16.977 | 0.049 |
| Carbapenems | ||||||||||||
| IPM | 0 | 3 (11.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (1.4) | 21.307 | 0.011 |
| MEM | 0 | 3 (11.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (1.4) | 21.307 | 0.011 |
| ERT | 0 | 3 (11.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (1.4) | 21.307 | 0.011 |
| Aminoglycosides | ||||||||||||
| CN | 43 (33.6) | 12 (46.2) | 9 (36) | 1(10) | 4 (44.4) | 1 (16.7) | 1 (20) | 0 | 0 | 70 (33.7) | 10.759 | 0.293 |
| AK | 0 | 0 | 0 | 0 | 1 (11.1) | 0 | 0 | 0 | 0 | 1 (0.5) | 25.121 | 0.003 |
| NET | 0 | 0 | 0 | 0 | 1 (11.1) | 0 | 0 | 0 | 0 | 1 (0.5) | 25.121 | 0.003 |
| Fluoroquinolones | ||||||||||||
| CIP | 87 (71.3) | 25 (96.2) | 10 (40) | 4 (40) | 5 (55.6) | 1 (16.7) | 2 (40) | 2 (66.7) | 0 | 136 (65.4) | 36.148 | 0.000 |
| NOR | 88 (72.1) | 25 (96.2) | 10 (40) | 4 (40) | 5 (55.6) | 1 (16.7) | 1 (20) | 2 (66.7) | 0 | 136 (65.4) | 36.148 | 0.000 |
| LEV | 86 (70.5) | 25 (96.2) | 10 (40) | 4 (40) | 5 (55.6) | 1 (16.7) | 2 (40) | 2 (66.7) | 0 | 135 (64.9) | 35.411 | 0.000 |
| Folate pathway inhibitors | ||||||||||||
| SXT | 61 (50) | 20 (76.9) | 16 (64) | 5 (50) | 5 (55.6) | 4 (66.7) | 0 | 1 (33.3) | 0 | 113 (54.3) | 10.853 | 0.286 |
| Fosfomycins | ||||||||||||
| FOS | 0 | 0 | 0 | 1 (10) | 0 | 0 | 0 | 0 | 0 | 1 (0.5) | 19.896 | 0.019 |
Notes.
- Amp
- ampicillin
- AMC
- amoxicillin-clavulanic acid
- TZP
- piperacillin-tazobactam
- SCF
- cefoperazone-sulbactam
- KZ
- cefazolin
- CTX
- cefotaxime
- CRO
- ceftriaxone
- CAZ
- ceftazidime
- CN
- gentamicin
- CIP
- ciprofloxacin
- NOR
- norfloxacin
- LEV
- levofloxacin
- SXT
- trimethoprim-sulfamethoxazole
- IPM
- Imipenem
- MEM
- meropenem
- ERT
- ertapenem
- CN
- gentamicin
- AK
- amikacin
- NET
- netilmicin
- FOS
- fosfomycin
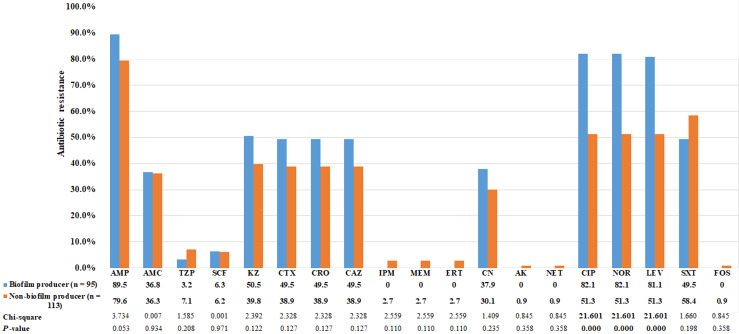
Using the CRA method, the abilities of bacteria to form biofilm were categorized into four groups based on the color tones of colonies. Among the 95 E. coli strains that could form biofilm, 4 (4.2%) showed strong biofilm-forming ability, 38 (40.0%) showed moderate ability, and 53 (55.8%) showed weak ability. The biofilm-producing strains were predominantly clustered in phylogenetic group B2 (Table 5). Biofilm- and non-biofilm-producing UPEC showed different antimicrobial resistance profiles. Among the biofilm producers, the rate of resistance was highest for ampicillin (90%), followed by fluoroquinolones (82%), cephalosporins (50%), and gentamicin (38%). No biofilm producer was resistant to carbapenems. In contrast, the non-biofilm producers were more resistant to TMP-SMZ (58%), followed by piperacillin-tazobactam (7%) and carbapenems (3%). The frequency distribution is presented in Fig. 2. The resistance rates to ciprofloxacin, norfloxacin and levofloxacin among biofilm producers were significantly higher than non-biofilm producers (p < 0.0001; Fig. 2). Of the 129 MDR E. coli isolates, 54% were biofilm producers.
Table 5. Biofilm forming ability among various phylogenetic groups of uropathogenic Escherichia coli isolates.
| Phylogenetic group | Prevalence of biofilm formation ability | |||
|---|---|---|---|---|
| Strong (n = 4), % | Moderate (n = 38), % | Weak (n = 53), % | Absent (n = 113), % | |
| B2 (n = 122) | 3 (2.5) | 36 (29.5) | 46 (37.7) | 37 (30.3) |
| C (n = 26) | 0 | 0 | 3 (11.5) | 23 (88.5) |
| E (n = 25) | 0 | 0 | 1 (4) | 24 (96) |
| A (n = 10) | 0 | 0 | 0 | 10 (100) |
| F (n = 9) | 0 | 1 (11.1) | 1 (11.1) | 7 (77.8) |
| D (n = 6) | 0 | 0 | 0 | 6 (100) |
| B1 (n = 5) | 1 (20) | 0 | 1 (20) | 3 (60) |
| Unassignable (n = 3) | 0 | 1 (33.3) | 0 | 2 (66.7) |
| Clade I (n = 1) | 0 | 0 | 0 | 1 (100) |
| Clade I or II (n = 1) | 0 | 0 | 1 (100) | 0 |
Figure 2. Comparison of antibiotic resistance between biofilm producers and non-biofilm producers.
Uropathogenic E. coli strains were evaluated for in vitro susceptibility to nineteen antibiotics: Amp, ampicillin; AMC, amoxicillin-clavulanic acid; TZP, piperacillin-tazobactam; SCF, cefoperazone-sulbactam; KZ, cefazolin; CTX, cefotaxime; CRO, ceftriaxone; CAZ, ceftazidime; CN, gentamicin; CIP, ciprofloxacin; NOR, norfloxacin; LEV, levofloxacin; SXT, trimethoprim-sulfamethoxazole; IPM, Imipenem; MEM, meropenem; ERT, ertapenem; CN, gentamicin; AK, amikacin; NET, netilmicin; FOS, fosfomycin. Bar graphs show the percentage of antibiotic resistance among biofilm producers in blue and non-biofilm producers in orange.
We also investigated the association between the presence or absence of the four adhesin genes and biofilm formation ability. The results demonstrated that biofilm production was significantly correlated with the presence of pap adhesin gene (p ≤ 0.05; Table 6). Among the biofilm producer group, we found the prevalence of pap gene was lower in strong biofilm formers than in weak and moderate.
Table 6. Prevalence of virulence genes among various groups of different biofilm formation ability.
| Virulence | Percentage of biofilm formation ability | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| genes |
Strong (n = 4), % |
Moderate (n = 38), % |
Weak (n = 53), % |
Total (n = 95), % |
Absent (n = 113), % |
Pearson Chi-square |
p-value | ||
| fimH | 4 (100) | 37 (97.4) | 49 (92.5) | 90 (94.7) | 101 (89.4) | 1.97a | 0.16 | ||
| pap | 2 (50) | 35 (92.1) | 44 (83.0) | 81 (85.3) | 84 (74.3) | 3.76b | 0.05 | ||
| sfa | 0 | 1 (2.6) | 6 (11.3) | 7 (7.4) | 18 (15.9) | 3.58c | 0.06 | ||
| afa | 0 | 2 (5.3) | 3 (5.7) | 5 (5.3) | 11 (9.7) | 1.45d | 0.23 | ||
Notes.
P-values were calculated using the Pearson Chi-squared test. P-values ≤ 0.05 are indicated in bold.
0 cells (.0%) have expected count less than 5. The minimum expected count is 7.76.
0 cells (.0%) have expected count less than 5. The minimum expected count is 19.64.
0 cells (.0%) have expected count less than 5. The minimum expected count is 11.42.
0 cells (.0%) have expected count less than 5. The minimum expected count is 7.31.
Discussion
The higher proportion of UTIs in female (74%) than male (26%) patients in this study was observed. This is most likely to the anatomical structure of the female urethra, which is shorter, wider, and closer to the anus than that of males. E. coli is common in the gastrointestinal tract flora and can be easily moved from the anus to the urinary tract, leading to UTIs (Dadi et al., 2020). Half of the UTI cases in this study (50%) were observed in female patients over 65 years of age. In postmenopausal women, the low level of estrogen and high intravaginal pH are associated with increased bacterial adherence to the uroepithelium cell, which causes UTIs (Johansson et al., 1996; Beyer et al., 2001). Our study included a large number of catheter urine specimens, which was correlated with the high percentage of infections in the over-65 age group. The low immunity level in the elderly puts those of advanced age at a high risk of bacterial infection and is responsible for the high prevalence in catheterized cases (Themphachanal et al., 2015).
Phylogenetic groups B2 and D are common strains implicated in UTIs (Ejrnæs et al., 2011). In contrast to the results of studies from Uruguay and Southern Thailand, where high prevalences of phylogenetic group D were found (Themphachanal et al., 2015; Robino et al., 2014), we observed that group B2 was the most prevalent UPEC (58.7%), followed by group C (12.5%). Our results are in accordance with several studies in which the dominant strain was found to be group B2. These studies were conducted in North America (45% prevalence of group B2) (Johnson et al., 2003), Denmark (67%) (Ejrnæs et al., 2011), Poland (35%) (Kot et al., 2016), South Korea (79%) (Lee et al., 2016), and Ethiopia (30%) (Dadi et al., 2020). Using a novel PCR-based method (Clermont et al., 2013), we could classify UPEC into groups C, E, and F and clade I, resulting in a lower percentage of strains in groups A, B1, and D than in earlier studies. This finding indicates that the triplex method of phylo-grouping misidentifies groups C, E, and F and clade I as belonging to group A, B1, B2, or D (Kumar, Nahid & Zahra, 2017). It had been reveal that some strains (1.4%) could not be assigned to a phylogenetic group due to simply relying upon PCR of a few small number of genes. As stated by Clermont et al. (2013), the unassignable strains are more likely the result of large-scale recombination events from two different groups or genome plasticity driven by loss and gain of genes. In this study, 1% of UPEC belonged to cryptic clade I/II. This is a much lower percentage than in a study conducted in Mexico (9%) (Kumar, Nahid & Zahra, 2017). The cryptic clades are primarily associated with environmental E. coli; thus, the observed results may be related to a lack of good hygiene practices. The different distributions of phylogenetic groups may depend on the geographic area, health status of the host, use of antibiotics, and/or variations in research design and sample size of the studies (Derakhshandeh et al., 2013).
The most prevalent adhesin gene was fimH, followed by pap, sfa, and afa. In agreement with studies conducted in Ethiopia (Dadi et al., 2020) and Iran (Tajbakhsh et al., 2016), phylogenetic group B2 strains showed the highest frequency of the adhesin genes in our study. We found a coexistence of fimH and pap genes (69.2%), indicating a high presence of virulence genes among UPEC isolated from UTI patients in Thailand. This outcome was different from that of a study in Iran, in which the combination of pap and afa virulence genes was more common (Rahdar et al., 2015). The ability of UPEC to form biofilm is a crucial virulence property. We found that 45.7% of UPEC were biofilm producers and that most of these classified into phylogenetic group B2. This finding demonstrates that biofilm formation may be associated with phylogenetic group B2. The association between biofilm-forming ability and some adhesin genes among UPEC was previously reported (Rahdar et al., 2015; Tajbakhsh et al., 2016; Naves et al., 2008). Consistently, the most significant correlation observed in our study was the correlation between the pap gene and biofilm production. The negative correlation found closely to significance between sfa gene and biofilm formation ( p = 0.06), as the prevalence of this gene was lower in biofilm producer. In contrast, no significant correlation was seen between the fimH, or afa genes and biofilm production in the strains evaluated in this study. This finding is in agreement with other studies that did not find significant correlations in clinical isolates of pathogenic E. coli (Reisner et al., 2006; Hancock, Ferrie‘res & Klemm, 2007). The discrepant results imply that these genes are not the only determinants of biofilm production in UPEC strains; rather, environmental and genetic factors may also be involved (Reisner et al., 2006). Adhesin genes such as fimH are under strict control by phase variation in many strains. The presence of adhesin genes certainly does not imply their expression. It would have been far more informative if the further study had been able to correlate expression of these genes rather than just their presence or absence by PCR.
It is important to perform antimicrobial susceptibility testing to select the appropriate empiric antibiotic therapy for UTIs. Our findings showed that the rate of resistance to ampicillin (84.1%) was higher than rates of resistance to other antibiotics. In general, fluoroquinolones are recommended for oral antimicrobial therapy in uncomplicated pyelonephritis. TMP-SMZ is commonly used in the treatment of uncomplicated cystitis, while AMC was a first-line therapy for complicated UTIs (Bonkat et al., 2019). However, our results revealed that rates of resistance to fluoroquinolones, TMP-SMZ, and AMC were 65%, 54%, and 37%, respectively. This result is consistent with a previous mini-review reporting increases in resistance rates of those drugs among UPEC isolates in developing countries (Kot, 2019). This likely emerged due to the widespread use of fluoroquinolones for uncomplicated UTIs or the inappropriate use of TMP-SMZ for empiric UTI treatment (Bartoletti et al., 2016). In this study, the strains in phylogenetic group C showed the highest rates of antibiotic resistance. In recent decades, the increasing rate of MDR in UPEC has become a public health threat. A high prevalence of MDR UPEC of approximately 62% was observed in the current study, similar to the findings reported in Iran (60.2%) (Tajbakhsh et al., 2016) and Nepal (63.2%) (Ganesh et al., 2019). The majority of MDR UPEC belonged to phylogenetic group B2, consistent with the outcomes reported in South Korea (73%) (Lee et al., 2016).
The present study found that biofilm producer strains were more resistant to ciprofloxacin, norfloxacin and levofloxacin than non-biofilm producers. These results were in agreement with previous studies indicating that the sessile bacterial cells are much less susceptible to antimicrobial agents than nonattached (planktonic) cells (Costerton, Stewart & Greenberg, 1999). A higher rate of resistance to TMP-SMZ was found among the non-biofilm producers than among the biofilm producers. One explanation for this finding is that these strains may carry the dhfr or dhps gene mutation on chromosomal DNA, which are common causes of resistance to this drug (Huovinen et al., 1995).
In conclusion, the majority of UPEC among patients with UTIs in this geographical area belonged to phylogenetic group B2. UPEC in this group also showed the highest prevalence of adhesin genes and biofilm formation. The analysis of the antimicrobial resistance of strains tested in this study showed a high level of resistance to cephalosporins, fluoroquinolones, TMP-SMZ, and AMC among strains belonging to groups B2 and C. Therefore, further study of the molecular epidemiology of UPEC and their antibiotic susceptibility patterns will improve our understanding of the organism and lead to a better management of UTIs.
Supplemental Information
R, red; AB, almost black; VB, very black. The color tones of colonies were categorized as follows: very black, and almost black, which were interpreted as strong, and weak biofilm producers, respectively, and red reported as non-biofilm producers.
Acknowledgments
We would like to acknowledge the staff of the Center of Excellence in Clinical Virology, Faculty of Medicine, Chulalongkorn University, for their excellent technical assistance. We also thank all of the medical technicians in the hospital for helping with bacteria collection and Ms. Naraumon Beakee, Ms. Pacharida Pattum, Ms. Sakonwan Thanoochan, and Ms. Thanyaporn Sidafong for assistance with the laboratory process.
Funding Statement
This work was supported by the Research Institute of Rangsit University (Grant No. 77/2017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Nipaporn Tewawong conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Siriporn Kowaboot analyzed the data, prepared figures and/or tables, and approved the final draft.
Yaowaluk Pimainog and Yong Poovorawan conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Naiyana Watanagul and Thanunrat Thongmee performed the experiments, prepared figures and/or tables, and approved the final draft.
Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The study protocol was approved by the Ethics Review Board (ERB) of the Research Institute of Rangsit University (DPE.No. RSUERB2018-002).
Data Availability
The following information was supplied regarding data availability:
Raw data are available in the Supplementary Files.
References
- Adamus-Białek et al. (2018).Adamus-Białek W, Baraniak A, Wawszczak M, Głuszek S, Gad B, Wróbel K, Bator P, Majchrzak M, Parniewski P. The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Molecular Biology Reports. 2018;45(5):1055–1065. doi: 10.1007/s11033-018-4254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahumada-Santos et al. (2020).Ahumada-Santos YP, Báez-Flores ME, Díaz-Camacho SP, Uribe-Beltrán MJ, Eslava Campos CA, Parra-Unda JR, Delgado-Vargas F. Association of phylogenetic distribution and presence of integrons with multidrug resistance in Escherichia coli clinical isolates from children with diarrhoea. Journal of Infection and Public Health. 2020;13(5):767–772. doi: 10.1016/j.jiph.2019.11.019. [DOI] [PubMed] [Google Scholar]
- Antao, Wieler & Ewers (2009).Antao EM, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut pathogens. 2009;1:22. doi: 10.1186/1757-4749-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoletti et al. (2016).Bartoletti R, Cai T, Wagenlehner FM, Naber K, Bjerklund Johansen TE. Treatment of urinary tract infections and antibiotic stewardship. European Urology Supplements. 2016;15(4):81–87. doi: 10.1016/j.eursup.2016.04.003. [DOI] [Google Scholar]
- Bergey & Holt (1994).Bergey DH, Holt JG. Bergey’s manual of determinative bacteriology. 9th edition Williams & Wilkins; Baltimore: 1994. [Google Scholar]
- Beyer et al. (2001).Beyer I, Mergam A, Benoit F, Theunissen C, Pepersack T. Management of urinary tract infections in the elderly. Zeitschrift fur Gerontologie und Geriatrie. 2001;34(2):153–157. doi: 10.1007/s003910170080. [DOI] [PubMed] [Google Scholar]
- Bonkat et al. (2019).Bonkat G, Bartoletti RR, Bruyère F, Cai T, Geerlings SE, Köves B, Schubert S, Wagenlehner F. EAU Guidelines on Urological infection [Internet]. Arnhem (The Netherlands): European Association of Urology. 2019. https://uroweb.org/wp-content/uploads/EAU-Guidelines-on-Urological-infections-2019.pdf. [20 June 2020]. https://uroweb.org/wp-content/uploads/EAU-Guidelines-on-Urological-infections-2019.pdf
- Clermont et al. (2013).Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental Microbiology Reports. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2018).Clinical Laboratory Standards Institute (CLSI) CLSI supplement M100. 28th ed. Clinical and Laboratory Standards Institute; Wayne: 2018. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- Costerton, Stewart & Greenberg (1999).Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dadi et al. (2020).Dadi BR, Abebe T, Zhang L, Mihret A, Abebe W, Amogne W. Distribution of virulence genes and phylogenetics of uropathogenic Escherichia coli among urinary tract infection patients in Addis Ababa, Ethiopia. BMC Infectious Diseases. 2020;20:108. doi: 10.1186/s12879-020-4844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti et al. (2009).Dashti AA, Jadaon MM, Abdulsamad AM, Dashti HM. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. The Journal of the Kuwait Medical Association. 2009;41(2):117–122. [Google Scholar]
- Derakhshandeh et al. (2013).Derakhshandeh A, Firouzi R, Moatamedifar M, Motamedi A, Bahadori M, Naziri Z. Phylogenetic analysis of Escherichia coli strains isolated from human samples. Molecular Biology Research Communications. 2013;2(4):143–149. [Google Scholar]
- Ejrnæs et al. (2011).Ejrnæs K, Stegger M, Reisner A, Ferry S, Monsen T, Holm SE, Lundgren B, Frimodt-Møller N. Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence. 2011;2(6):528–537. doi: 10.4161/viru.2.6.18189. [DOI] [PubMed] [Google Scholar]
- Fernández & Berenguer (2000).Fernández LA, Berenguer J. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiology Reviews. 2000;24(1):21–44. doi: 10.1111/j.15746976.2000.tb00531.x. [DOI] [PubMed] [Google Scholar]
- Flores-Mireles et al. (2015).Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature Reviews. Microbiology. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh et al. (2019).Ganesh R, Shrestha D, Bhattachan B, Rai G. Epidemiology of urinary tract infection and antimicrobial resistance in a pediatric hospital in Nepal. BMC Infectious Diseases. 2019;19(1):420. doi: 10.1186/s12879-019-3997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall & Mah (2017).Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiology Reviews. 2017;41(3):276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- Hancock, Ferrie‘res & Klemm (2007).Hancock V, Ferrie‘res L, Klemm P. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiology Letters. 2007;267(1):30–37. doi: 10.1111/j.1574-6968.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- Huovinen et al. (1995).Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrobial Agents and Chemotherapy. 1995;39(2):279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson et al. (1996).Johansson C, Molander U, Milsom I, Ekelund P. Association between urinary incontinence and urinary tract infections, and fractures in postmenopausal women. Maturitas. 1996;23(3):265–271. doi: 10.1016/0378-5122(95)00982-5. [DOI] [PubMed] [Google Scholar]
- Johnson et al. (2003).Johnson JR, Kuskowski MA, Owens K, Gajewski A, Winokur PL. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. The Journal of Infectious Diseases. 2003;188(5):759–768. doi: 10.1086/377455. [DOI] [PubMed] [Google Scholar]
- Kot (2019).Kot B. Antibiotic resistance among uropathogenic Escherichia coli. Polish Journal of Microbiology. 2019;68(4):403–415. doi: 10.33073/pjm-2019-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kot et al. (2016).Kot B, Wicha J, Gruzewska A, Piechota M, Wolska K, Obrebska M. Virulence factors, biofilm-forming ability, and antimicrobial resistance of urinary Escherichia coli strains isolated from hospitalized patients. Turkish Journal of Medical Sciences. 2016;46(6):1908–1914. doi: 10.3906/sag-1508-105. [DOI] [PubMed] [Google Scholar]
- Kumar, Nahid & Zahra (2017).Kumar N, Nahid F, Zahra R. Association of virulence factors, phylogenetic groups and antimicrobial resistance markers in Escherichia coli from Badin city, Pakistan. Journal of Chemotherapy. 2017;29(1):8–13. doi: 10.1080/1120009X.2016.1154682. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel & Falkow (1988).Labigne-Roussel A, Falkow S. Distribution and degree of heterogeneity of the afimbrial-adhesin-encoding operon (afa) among uropathogenic Escherichia coli isolates. Infection and Immunity. 1988;56(3):640–648. doi: 10.1128/IAI.56.3.640-648.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouguenec, Archambaud & Labigne (1992).Le Bouguenec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. Journal of Clinical Microbiology. 1992;30(5):1189–1193. doi: 10.1128/JCM.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2016).Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, Koo SH, Oh MH, Kim HJ, Choi CH. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Letters in Applied Microbiology. 2016;62(1):84–90. doi: 10.1111/lam.12517. [DOI] [PubMed] [Google Scholar]
- Magiorakos et al. (2012).Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Melican et al. (2011).Melican K, Sandoval RM, Kader A, Josefsson L, Tanner GA, Molitoris BA, Richter-Dahlfors A. Uropathogenic Escherichia coli P and Type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLOS Pathogens. 2011;7(2):e1001298. doi: 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, Sharma & Chaudhary (2015).Mittal S, Sharma M, Chaudhary U. Biofilm and multidrug resistance in uropathogenic Escherichia coli. Pathogens And Global Health. 2015;109(1):26–29. doi: 10.1179/2047773215Y.0000000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naves et al. (2008).Naves P, Del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Dahbi G, Blanco M, Ponte Mdel C, Soriano F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microbial Pathogenesis. 2008;45(2):86–91. doi: 10.1016/j.micpath.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Neupane et al. (2016).Neupane S, Pant ND, Khatiwada S, Chaudhary R, Banjara MR. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrobial Resistance and Infection Control. 2016;5:5. doi: 10.1186/s13756-016-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orndorff & Falkow (1984).Orndorff PE, Falkow S. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. Journal of Bacteriology. 1984;159(2):736–744. doi: 10.1128/JB.159.2.736-744.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard et al. (1999).Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infection and Immunity. 1999;67(2):546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar et al. (2015).Rahdar M, Rashki A, Miri HR, Rashki Ghalehnoo M. Detection of pap, sfa, afa, foc, and fim adhesin-encoding operons in uropathogenic Escherichia coli isolates collected from patients with urinary tract infection. Jundishapur Journal of Microbiology. 2015;8(8):e22647. doi: 10.5812/jjm.22647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner et al. (2006).Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. Journal of Bacteriology. 2006;188(10):3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robino et al. (2014).Robino L, García-Fulgueiras V, Araujo L, Algorta G, Pírez MC, Vignoli R. Urinary tract infection in Uruguayan children: aetiology, antimicrobial resistance and uropathogenic Escherichia coli virulotyping. Journal of Global Antimicrobial Resistance. 2014;2(4):293–298. doi: 10.1016/j.jgar.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Shetty et al. (2014).Shetty AV, Kumar SH, Shekar M, Shetty AK, Karunasagar I, Karunasagar I. Prevalence of adhesive genes among uropathogenic Escherichia coli strains isolated from patients with urinary tract infection in Mangalore. Indian Journal of Medical Microbiology. 2014;32(2):175–178. doi: 10.4103/0255-0857.129812. [DOI] [PubMed] [Google Scholar]
- Sm, Jayakumar & Aravazhi (2016).Sm N, Jayakumar K, Aravazhi AN. The effectiveness of antibiotics against a major uropathogen-Escherichia coli and its biofilm assay by phenotypic methods. International Journal of Research in Medical Sciences. 2016;4:4820–4828. [Google Scholar]
- Stamm & Norrby (2001).Stamm WE, Norrby SR. Urinary tract infections: disease panorama and challenges. The Journal of Infectious Diseases. 2001;183(Suppl 1):S1–S4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- Struve & Krogfelt (1999).Struve C, Krogfelt KA. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology. 1999;145(Pt 10):2683–2690. doi: 10.1099/00221287-145-10-2683. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh et al. (2016).Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrobial Resistance and Infection Control. 2016;5:11. doi: 10.1186/s13756-016-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchouna et al. (2013).Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. International Journal of Infectious Diseases. 2013;17(6):e450-e453. doi: 10.1016/j.ijid.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Themphachanal et al. (2015).Themphachanal M, Kongpheng S, Rattanachuay P, Khianngam S, Singkhamanan K, Sukhumungoon P. Molecular characterization of virulence and antimicrobial susceptibility profiles of uropathogenic Escherichia coli from patients in a tertiary hospital, southern Thailand. The Southeast Asian Journal of Tropical Medicine and Public Health. 2015;46(6):1021–1030. [PubMed] [Google Scholar]
- Yamamoto et al. (1995).Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunology and Medical Microbiology. 1995;12(2):85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R, red; AB, almost black; VB, very black. The color tones of colonies were categorized as follows: very black, and almost black, which were interpreted as strong, and weak biofilm producers, respectively, and red reported as non-biofilm producers.
Data Availability Statement
The following information was supplied regarding data availability:
Raw data are available in the Supplementary Files.