Abstract
Dental caries is the most prevalent oral disease affecting nearly 70% of children in India and elsewhere. Micro-ecological niche based acidification due to dysbiosis in oral microbiome are crucial for caries onset and progression. Here we report the tooth bacteriome diversity compared in Indian children with caries free (CF), severe early childhood caries (SC) and recurrent caries (RC). High quality V3–V4 amplicon sequencing revealed that SC exhibited high bacterial diversity with unique combination and interrelationship. Gracillibacteria_GN02 and TM7 were unique in CF and SC respectively, while Bacteroidetes, Fusobacteria were significantly high in RC. Interestingly, we found Streptococcus oralis subsp. tigurinus clade 071 in all groups with significant abundance in SC and RC. Positive correlation between low and high abundant bacteria as well as with TCS, PTS and ABC transporters were seen from co-occurrence network analysis. This could lead to persistence of SC niche resulting in RC. Comparative in vitro assessment of biofilm formation showed that the standard culture of S. oralis and its phylogenetically similar clinical isolates showed profound biofilm formation and augmented the growth and enhanced biofilm formation in S. mutans in both dual and multispecies cultures.
Subject terms: Next-generation sequencing, Bioinformatics, Biofilms, Microbial communities, Infectious-disease diagnostics, Biochemical reaction networks, Classification and taxonomy
Introduction
Interaction among more than 700 species of microbiota under different micro-ecological niches of the human oral cavity1,2 acts as a primary defense against various pathogens. This has been observed to play a significant role in child’s oral and general health. Dysbiosis among these microbes due to excessive and frequent intake of carbohydrates results in acidic niche, thereby lowering the buffering provided by healthy microbiome. This condition leads to demineralization of tooth surface causing caries in children3,4. Once lesions advance beyond the white spot stage and the enamel surface is damaged, they cannot be biologically reversed resulting in Severe Early Childhood Caries (SC) among the children with the age group of 3–6 years5. Considerable change in persistent oral biota in response to the functional molecules even after the treatment of SC facilitates recurrent caries (RC) in children, affecting both primary and permanent dentition6.
Moreover, dysbiosis of oral microbes also results in acute to chronic disease conditions either directly or indirectly by producing metabolically active compounds that interrupt host immune system. Various studies have shown that harmful oral microbiome may hold a significant impact beyond the oral cavity that is related to systemic diseases7, including elevated cardiovascular risk8,9, rheumatoid arthritis10, adverse pregnancy outcome11 and digestive diseases12. Such factors make it imperative to know the colonization patterns of oral commensals occurring during childhood and their benign impact in oral and systemic diseases and health conditions.
Metagenomics studies with the help of high-throughput sequencing technologies revolutionized the human microbiome research13, providing an opportunity to tap and focus on the unexplored complex microbial systems that are difficult to cultivate in-vitro. Increased understanding about the functional activity of microbes within the complex microbial community with that of host ecosystems was possible with the advancement of computational analysis tools for these sequences14. According to Keystone Pathogen Hypothesis15, understanding and identifying the complex interactions between high and low abundant microbes and its functional potential that resist any therapeutic agents under the micro-ecological niche. Such analysis may prevent an adverse effect on the human system and can become a new target for treatment and preventive care for caries as well as other related diseases.
Here, we report the analysis of both over-represented and under-represented caries causing microbiota, its predictive functional traits, co-aggregation with each other and their susceptibility towards secondary caries in SC and RC of Indian children. Importance was given to the exploration of biomarkers in-order to identify the key pathogen and its functional properties that resist the micro ecological stress. Interaction network analysis among the oral microbiota and functional traits would enhance the understanding about the strong network of oral bacterial species especially in SC and RC that could paves way for early detection and management of dental caries.
Results
Demography of caries status
In the current report, study subjects were categorized into three study groups and two gender groups. Caries status of the participants, their age and gender are presented in Table 1. Sequencing were performed for 30 samples, Caries Free (CF) (n = 10), Severe Early Childhood Caries (SC) (n = 10), Recurrent Early Childhood Caries (RC) (n = 10), from the 55 children enrolled because of low yield and quality of DNA. Gender proportion (Female/Male) of the study subjects CF, SC and RC were 5/5, 4/6, 6/4 respectively. Significant differences were not observed in the mean age and gender proportion among three study groups.
Table 1.
Patient demographic data and QC of NGS data.
| Sample ID | Study group | Gender | Age | DMFT | No. of sequences | Unique reads | Duplicate reads | % Trimmed | % of top overrepresented sequence | % of the sum of remaining overrepresented sequences |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | Normal | Female | 3 | 0 | 324,653 | 30,867 | 293,786 | 0.029 | 13.8 | 58.08 |
| P2 | Normal | Female | 6 | 0 | 368,052 | 34,579 | 333,473 | 0.03 | 8.2 | 64.01 |
| P3 | Normal | Female | 5 | 0 | 500,131 | 51,239 | 448,892 | 0.025 | 11.7 | 56.07 |
| P8 | Normal | Female | 3 | 0 | 843,584 | 154,048 | 689,536 | 0.032 | 17.4 | 51.24 |
| P9 | Normal | Female | 4 | 0 | 426,340 | 39,929 | 386,411 | 0.029 | 12.4 | 57.52 |
| P10 | Normal | Male | 5 | 0 | 791,013 | 69,365 | 721,648 | 0.035 | 1.6 | 65.50 |
| P11 | Normal | Male | 4 | 0 | 1,044,615 | 87,922 | 956,693 | 0.033 | 2.1 | 57.34 |
| P15 | Normal | Male | 4 | 0 | 989,960 | 91,168 | 898,792 | 0.035 | 2.7 | 61.17 |
| P17 | Normal | Male | 5 | 0 | 959,351 | 83,279 | 876,072 | 0.034 | 2.1 | 63.31 |
| P4 | Disease | Male | 5 | 5 | 581,388 | 62,484 | 518,904 | 0.031 | 8.3 | 58.69 |
| P5 | Disease | Female | 4 | 4 | 541,995 | 46,244 | 495,751 | 0.033 | 7.1 | 64.15 |
| P6 | Disease | Female | 5 | 4 | 763,526 | 53,837 | 709,689 | 0.033 | 10.8 | 65.19 |
| P7 | Disease | Male | 4 | 4 | 519,897 | 44,592 | 475,305 | 0.03 | 17.0 | 59.49 |
| P13 | Disease | Female | 5 | 4 | 928,077 | 67,115 | 860,962 | 0.036 | 4.4 | 68.04 |
| P14 | Disease | Male | 6 | 5 | 790,177 | 38,068 | 752,109 | 0.038 | 16.4 | 64.44 |
| P33 | Disease | Male | 5 | 5 | 423,672 | 37,885 | 385,787 | 0.031 | 13.0 | 58.97 |
| P35 | Disease | Male | 6 | 5 | 976,071 | 65,141 | 910,930 | 0.034 | 2.1 | 61.96 |
| P39 | Disease | Male | 6 | 5 | 289,569 | 30,562 | 259,007 | 0.031 | 6.2 | 65.03 |
| P40 | Disease | Female | 5 | 4 | 400,603 | 43,049 | 357,554 | 0.03 | 6.9 | 61.39 |
| P19 | Recurrent | Male | 6 | 2 | 1,197,632 | 76,828 | 1,120,804 | 0.037 | 19.5 | 60.19 |
| P21 | Recurrent | Female | 7 | 2 | 963,211 | 69,289 | 893,922 | 0.035 | 3.8 | 59.44 |
| P23 | Recurrent | Male | 6 | 2 | 946,556 | 91,744 | 854,812 | 0.033 | 2.4 | 58.57 |
| P25 | Recurrent | Female | 7 | 3 | 916,099 | 85,611 | 830,488 | 0.033 | 3.8 | 57.66 |
| P31 | Recurrent | Female | 7 | 3 | 980,648 | 69,794 | 910,854 | 0.036 | 3.7 | 66.27 |
| P32 | Recurrent | Female | 6 | 4 | 1,018,812 | 56,302 | 962,510 | 0.037 | 9.6 | 73.01 |
| P36 | Recurrent | Female | 7 | 3 | 423,917 | 39,608 | 384,309 | 0.03 | 8.0 | 63.27 |
| P37 | Recurrent | Female | 7 | 2 | 332,321 | 44,845 | 287,476 | 0.031 | 9.1 | 58.15 |
| P38 | Recurrent | Male | 6 | 3 | 343,591 | 38,805 | 304,786 | 0.029 | 7.3 | 61.88 |
Biodiversity of microbiota among SC, RC and CF micro ecosystem through high quality sequencing
A total of 15,270,361 high quality reads were generated from 28 samples with an average of 391,573 per sample ranging between 110,367 and 772,880 after trimming out low quality reads (Table 1). High richness in species diversity using Shannon, Chao and ACE index with 52,628 unique OTU’s after trimming out the low-quality reads were observed. RC exhibits significant difference in Chao 1 index with CF (P = 0.039), SC (P = 0.043) and in ACE with CF (0.012) and SC (P = 0.009) (Fig. 1A). Out of 52,628 OTU’s obtained in the analysis SC, CF and RC are characterized by 19,984, 12,473 and 8306 unique OTU’s respectively. SC and CF shared 7314 OTU, CF and RC shared 4556, RC and SC shared 5320 OTU’s. In common 3682 OTU’s were shared by all three study groups. Representative Venn diagram illustrates the number of unique and shared OTUs in each study group (Fig. 1B). To investigate the relatedness of microbiome composition, separation of beta diversity among CF, SC and RC samples based on PCoA (Principle Co-ordinates) axes 1 and 2 was given in the Bray Curtis Plot (Fig. 1C). The microbiota shows clear segregation between CF and RC but between CF–SC and SC–RC the microbiota overlaps considerably. Boxplot analysis of Weighted UniFrac distance showed significant difference between all groups and within group comparison except the distance between SC vs SC to RC vs RC and SC vs SC to all within the study group and between study groups (Fig. 1D). Similarly, significant differences were observed for all with in gender to male vs male and female vs female (Fig. 1D).
Figure 1.
Diversity comparison of SC, CF and RC microbiota. (A) Boxplot of ACE and Chao 1 diversity in the three groups. Outliers are represented by dots. Significant differences between groups are shown by lines and the following notation. P < 0.05 (*), P < 0.01 (**), P < 0.001 (***). (B)Venn diagram illustrated the number of shared OTUs between SC, CF and RC at 97% similarity. Coloured circles represented each group and intersection part between circles represented the number of shared OTUs. (C) Principal coordinates analysis (PCoA) based on weighted UniFrac distance of community structure among all the individuals of three treatment groups. Each color circle represents one sample: pink for SC group (n = 10); blue for CF group (n = 8); violet for RC group (n = 10). (D) Box plot comparison of beta diversity analysis across treatment groups at genera level. The X-axis denotes the population studied while Y-axis denotes the corresponding Shannon–Weaver index representing the beta diversity of treatment and gender group.
Taxonomical analysis of bacterial diversity and its phylogenetic relationship in SC, RC and CF group of Indian children
Upon aligning the 52,628 unique OTU’s with the HOMD database at a 97% similarity level a total of 780 species from 10 phyla were classified. Percentage distribution of relative abundance of bacterial diversity at genus level was given in Fig. 2A. Among the 5 most abundant phyla Firmicutes dominated in all the three groups (CF—46.56%, SC—3.25%, RC—42.02%) followed by Proteobacteria (CF—36.99%, SC—32.98%, RC—29.55%). The next dominant group was Actinobacteria followed by Fusobacteria in SC, whereas CF was dominated by unassigned (9.3%) and RC was dominated by Fusobacteria (12.43%) and Bacteroidetes (6.43%). The abundance level of Actinobacteria was diminished in CF with 2.6% and it was slightly higher in RC with 5.1%. The Candidate Phyla Radiation (CPR) phyla TM7 was found to occur in all three groups with low abundance whereas SR1 and GN02 were represented only in the CF group. All 10 phyla constituted around 93.3% of aligned taxonomy while the remaining 6.63% constitute unassigned bacteria (Fig. S1). Coloured heat map depicts the phylogenetic relatedness based on Euclidean distance between samples and their constituent microbial taxa that are represented by vertical and horizontal dendrogram (Fig. 2B). Horizontal clustering clearly exhibits the grouping of taxon based on their relative abundance in every individual with major clusters of Neisseria, Streptococcus, Haemophilus, Veillonella and Leptotrichia.
Figure 2.
(A) Broad and Fine detail compositional differences of microbiota at genus level. (B) Coloured heatmap represent the hierarchical clustering of microbiota data at a bacterial family level. Abundances are coloured according to the colour key on the top left with blue representing a value of zero. Euclidean distance and complete linkage were used to cluster the rows and columns of the heatmap. All taxa present at less than 1% in all three groups were excluded from the heatmap. Heatmap was annotated using plotly_4.8.0.
Over-represented and under-represented bacteria in SC and RC
To understand the over and underrepresented bacteria in the tooth biofilm of each study group, top 100 taxa based on their relative abundance was subjected to Two-way ANOVA with multiple comparisons among the study groups. High relative abundance of Streptococcaceae, Neisseriaceae, Pasteurellaceae, Veillonellaceae, Leptotrichiaceae and low relative abundance of Corynebacteriaceae was observed to be common in all three study groups. In SC samples Micrococcaceae, Selenomonadaceae, Bifidobacteriaceae, Lactobacillales were overrepresented and Absconditabacteria_(SR1)_[F1], Fusobacteriaceae, Porphyromonadaceae, Campylobacteraceae were represented with low relative abundance. The CF group is overrepresented by Gemellaceae, Flavobacteriaceae with a high relative abundance and Gracilibacteria_(GN02), Absconditabacteria_(SR1), Campylobacteraceae, Prevotellaceae, with low abundancy level. Families like Fusobacteriaceae, Campylobacteraceae, are expressed at high abundance in RC, whilst it exhibited low to very low abundance in SC and CF group. RC group show relatively low abundance in families like Gemellaceae, Absconditabacteria_(SR1)_[F1], Prevotellaceae, Lactobacillaceae, Corynebacterium, Selenomonadaceae. Significance of high abundant bacteria at genus level between the study groups was depicted in (Fig. S2). Streptococcus oralis subsp. tigurinus clade 071 and other non-mutan Streptococcus exhibit a highly significant difference in SC and RC group from that of the CF group. Fusobacterium was found to be significantly very low in the CF group than in the SC and RC groups. Veillonella parvula and Veillonella dispar were reported in the CF group with a highly significant difference from that of RC and SC group. Of Neisseria and Haemophilus that occur high in CF, Neisseria was found to be highly significant than that of Haemophilus from RC and SC group. Other caries causing bacteria like Leptotrichia, Aggregatibacter, Capnocytophaga leadbetteri, Selenomonas, Prevotella, Rothia found in high occurrence in SC and RC group when compared to the CF group with no significant difference among them.
Diversified bacteria that represents potential biomarkers in caries and caries-free conditions
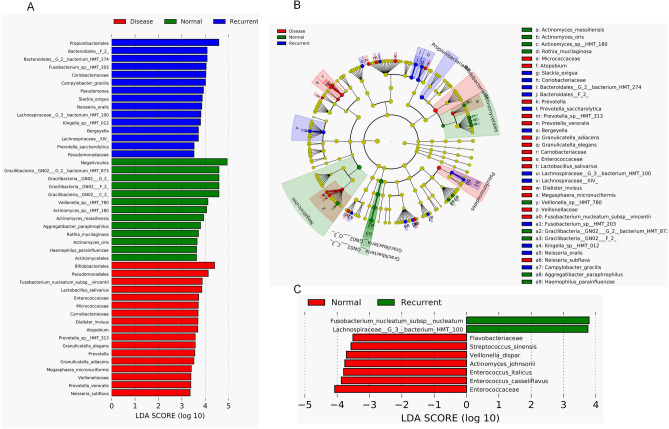
Unique bacterial community composition associated with the tooth biofilm was examined by comparing the relative abundance of the taxa among SC, RC and CF groups by LeFse analysis. The SC group is sturdily represented by 17 differentially abundant taxa that include Bifidobacteriales, Pseudomonadales, Fusobacterium nucleatum subsp. vincentii, Lactobacillus salivarius, Enterococcaceae, Micrococcaceae, Carnobacteriaceae, Dialister invisus, Atopobium, Prevotella sp_HMT_313, Granulicatella adiacens, Granulicatella elegans, Megasphaera micronuciformis, Veillonellaceae, Prevotella and Neisseria subflava. In recurrent groups the differences were attributable to the enrichment of Propionibacteriales, Bacteroidales_G_2_bacterium_HMT_274, Fusobacterium sp._HMT_203, Coriobacteriaceae, Campylobacter gracilis, Pseudomonas, Slackia exigua, Neisseria oralis, Lachnospiraceae_G_3_bacterium_HMT_100, Kingella sp._HMT_100, Prevotella saccharolytica and Bergeyella. Interestingly the tooth biofilm of the CF group was dominated by Negativicutes followed by four different Gracilibacteria_GN02, three different Actinomyces sp., Veillonella sp._HMT_780, Rothia mucilaginosa and Haemophilus parainfluenzae (Fig. 3A). Thus, the significant differences observed in the abundance of Bifidobacteriales, Granulicatella, Micrococcaceae, Carnobacteriacea, Enterococcaceae and Lactobacillus between the CF and SC groups may be related to the disease condition in affected children. Additionally, the significant differences observed in the abundance of Propionibacteriales, Pseudomonadales, Lachnospiraceae, Bergeyella, Prevotella, Slackia could be related to the secondary caries formation in RC groups (Fig. 3B). It has been observed that Fusobacterium nucleatum and Lachnospiraceae_G_3_bacterium HMT 100 might be the key organism in the severity of the disease conditions (Fig. 3C).
Figure 3.
The potential biomarkers defined by LeFSe analysis and LDA (A–C). Cladogram for the taxonomic representation of significant differences between CF, SC and RC groups. The coloured nodes from the inner to the outer circles represent taxa from the phylum to genus level. Histogram of the LDA scores for differentially abundant features among groups. The threshold on the logarithmic LDA score for discriminative features was set to 3.0. (A) Histogram of the LDA scores (log 10) computed for features that were differentially abundant in children with SC, RC and CF. (B) Taxonomic representation of statistically and biologically consistent differences in children with SC, RC and CF. (C) Histogram of the possible biomarkers between RC and CF. https://huttenhower.sph.harvard.edu/galaxy/.
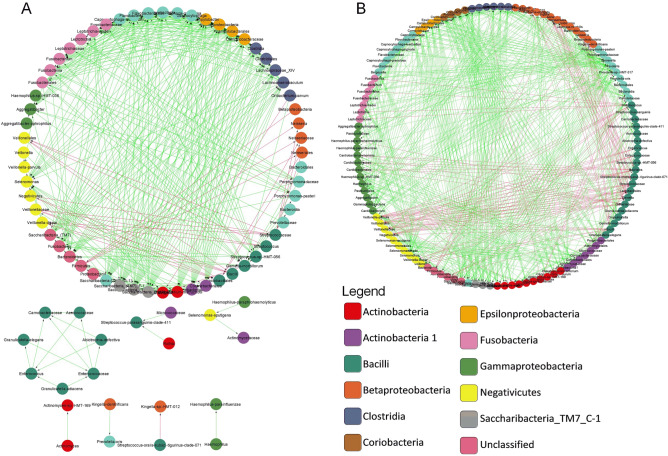
Mutual relationship among biofilm forming caries microbiota exhibited in SC and RC ecological niche
Co-occurrence network among the core microbiome was constructed with the organisms that occur in 90% of the study group which consists of 80 nodes and 357 edges. Both positive and negative correlation among the interacting groups were shown as edges with green and red colour respectively. This network was characterized with two major clusters one with early colonizers and other with middle and late colonizers. All the late colonizers were found to interact mutually with each other. Separate clusters among the bacteria Carnobacteriaceae, Aerococcaceae, Granulicatella elegans, Enterococcus and Abiotrophia defective that belongs to the class Bacilli were observed. (Fig. 4A). The network of disease conditions constitutes 127 nodes with 2115 interactions indicating a strong association of complex networks among the bacterial community. Analysis on interacting microbes reveals that the Coriobacteria, Micrococceae was found to occur exclusively in caries condition exhibiting positive interaction with most of the early and late colonizers. S. oralis subsp. tigurinus clade 071 that shows high abundance in all three study groups had negative interaction with Kingella_sp._HMT_012 in the core microbiome. However, in the disease network S. oralis subsp. tigurinus clade 071 exhibited positive interaction with Streptococcus, Carnobacteriaceae, Bacillales, Gemella, S. parasanguinis and Granulicatella (Fig. 4B). Central interaction mainly occurs among Corynebacterium durum, Haemophilus parainfluenzae, Oribacterium, Enterococcaceae, Micrococcaceae, Capnocytophaga sputigena, Capnocytophaga leadbetteri, Prevotella and Lachnospiraceae. The interaction that occurs among Pasteurellaceae, Corynebacterium matruchotii, Eikenella, Fusobacterium, Veillonella tobetsuensis and Bifidobacterium dentium shows positive co-occurrence and Veillonella parvula and Gemella shows mutual exclusion with Bifidobacterium dentium (Fig. S3). Under disease condition three different strains of Saccharibacteria_TM7_(G-1,C-1,F-1) found to interact with most of the middle and late colonizers like Selenomonas, Tannerella, Prevotella, Eikenella, Lachnoanaerbaculum, Clostridia, Camphylobacter, Epsilonproteobacteria, Capnocytophaga and Aggregatibacter positively while there seems to be no interaction among the three strains of Saccharibacteria_TM7 (Fig. S4). Gracillibacteria GN02 was found to have mutual interaction with the majority of biofilm forming microbes in caries free condition.
Figure 4.
Co-occurrence interaction networks of oral microbial communities is shown in the above figure. (A) The complete network of core microbiome that represented in 95% of the all three study group. (B) The significant central interaction in recurrent caries. The solid green line represents co-presence and dotted red line indicates mutual exclusion. Arrows at the end of each line represent the direction of interaction among the different microbes. All the interacting microbes are represented with different colour according to their Class.
Functional prediction of the predominant taxa among three study groups
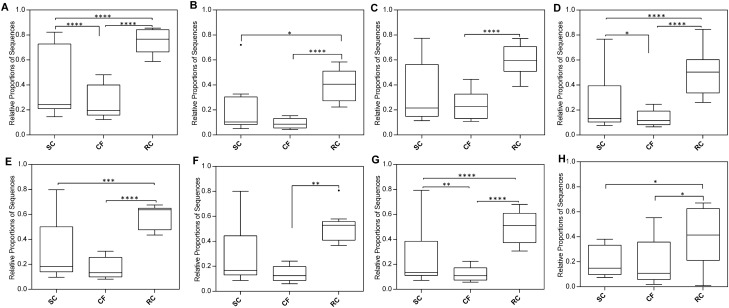
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to understand and infer the metabolic interactions between the microbiota of CF, SC and RC groups. Essential carbohydrate metabolisms that are related to caries like starch and sucrose metabolism, amino sugar metabolism, TCA cycle, fructose and mannose metabolism, galactose metabolism, glycolysis, pentose phosphate pathway in SC and RC exhibit highly significant difference from CF group (Fig. S5A). A distinct difference was not observed in pyruvate and inositol phosphate metabolism. Analysis on cell motility related pathway shows an increased level of bacterial toxins, bacterial chemotaxis, bacterial motility protein and flagellar assembly in RC and SC groups. The major transport systems related to quorum sensing Phosphotransferase System (PTS), ABC transport system show distinct differences between RC with that of the CF group. Glycan biosynthesis which is the prime function of oral bacterial taxon towards plaque formation found to be high in the RC group (Fig. 5).
Figure 5.
Box plot showing the significantly different KEGG pathways among the three treatment groups CF, SC, and RC. (A) ABC transporters, (B) phosphotransferase system (PTS), (C) bacterial secretion, (D) bacterial toxin, (E) sugar metabolism, (F) starch and sucrose metabolism, (G) galactose metabolism, (H) N-glycan biosynthesis. Statistical significance was evaluated using Tukey’s multiple comparisons test. P-values: * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Outliers are plotted as circles.
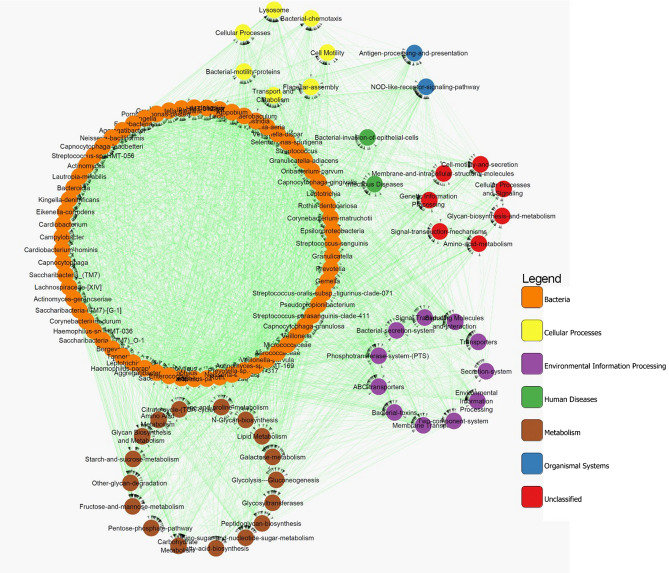
Network analysis of bacterial community with functional traits
Inter and intra relationship among the biofilm forming microbes and its predictive potential functional molecule was constructed to reveal the role of functional molecules for the ecological interactions among the bacteria in all three study groups. A total of 113 nodes with 2717 mutual interactions was obtained using Spearman correlation and P values were adjusted with Benjamini Hochberg multiple test correction to 0.05. More number of positive interactions was observed among different biofilm forming bacteria with that of the predicted functional traits. The network association was clustered based on the interaction among bacteria, cellular processes, environmental information processing, metabolism and organismal systems (Fig. 6). In the case of starch and sucrose metabolism we could observe a positive interaction among major bacteria like Streptococcus, Cardiobacterium hominis, Lachnospiraceae, Selenomonas, Leptotrichia, Atopobium, Prevotella oris, Campylobacter, Coriobacteria, Rothia aeria, Actinobacteria and Corynebacterium. Starch and sucrose metabolism is also positively correlated with other major functional traits like bacterial chemotaxis, pentose phosphate pathway, TCA cycle, bacterial motility proteins, flagellar assembly, N-Glycan biosynthesis, PTS, Two component system and other transport systems (Fig. S7). Of the three transport mechanism ABC transport mechanism exhibited positive interaction with almost all the carbohydrate metabolising bacteria. Along with the above mentioned bacteria in starch and sucrose metabolism Kingella sp. HMT-012, Veillonella parvula, Betaproteobacteria, S. oralis subsp. tigurinus clade 071, Haemphilus shows positive correlation with the TCS (Fig. S8). The PTS shows positive association with Saccharibacteria-TM7, Fusobacteria, Lautropia mirabilis, Tannerella, Neisseria bacilliformis, Clostridia, Corynebacterium matruchotii, Epsilonproteobacteria, Lachnospiraceae, Gemella morbillorum, S. parasanguinis, Aggregatibacter aphophilus, Capnocytophaga (Fig. S9).
Figure 6.
Mutual interaction networks of oral microbial communities with functional traits is shown in the above figure. The significant clustered interaction were given in different colours in recurrent caries. The solid green line represents edges with positive interactions. Arrows at the end of each line represent the direction of interaction among the interacting nodes.
Comparative in vitro characterization of biofilm formation by clinical isolates and standard culture
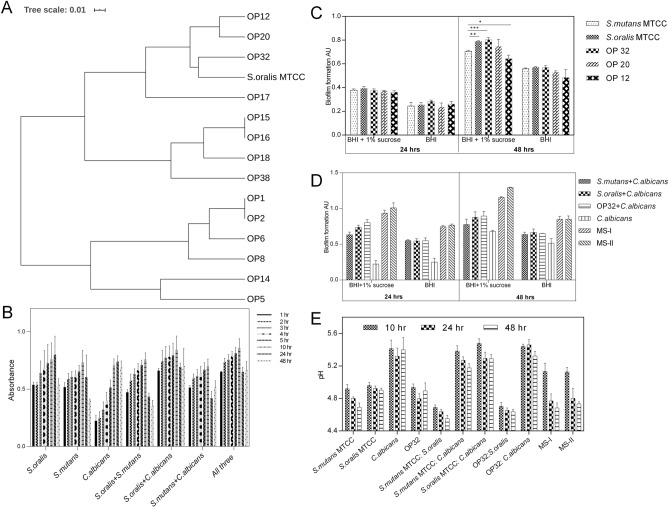
Biofilm formation is a major step in caries plaque pathology. Colonies isolated from the tooth swab of SC showed varied levels of biofilm forming capabilities. Isolated clinical cultures grown in congo red agar showed various colour determinations ranging from dark red, red, light brown, black and dry crystalline black. In CRA, 15 isolated clinical cultures (OP 1, OP 2, OP 5, OP 6, OP 8, OP 10, OP 12, OP 14, OP 15, OP 16, OP 17, OP 18, OP 20, OP 32, OP 38) and all standard cultures showed positive results by producing dark black colonies in the agar plate indicating the production of exo-polysaccharide which is a main factor for biofilm formation. Representative CRA assay was given in (Fig. S10A). Phylogenetic comparison between the biofilm forming clinical isolates and S. oralis (MTCC) by BOX PCR showed 5 OPs closer to S. oralis grouped in the same clade. OP32 was the closest to S. oralis (MTCC) while OP12 and OP 29 showed branches near to the standard culture (Fig. 7A). OP32 was used for further analysis. Co-culture of CRA positive (OP32) with CRA negative along with standard colonies showed no antagonistic interaction. This indicated OP32 did not show negative growth impact on non-biofilm forming bacteria and vice-versa (Fig. S10B).
Figure 7.
(A) Similarity UPGAMA tree of BOX-PCR patterns of 5 bacterial strains belonging to S. oralis isolated from SECC patients along with the standard S. oralis MTCC culture. (B) Biomass production assay for standard MTCC cultures both planktonic and multispecies in BHI broth for various time interval. (C) Analysis of Biofilm formation by crystal violet assay for standard and isolated culture in the presence and absence of 1% sucrose. (D) Comparative analysis of biofilm formation by crystal violet assay of standard culture and isolated culture OP32 in the presence or absence of C. albicans. MS-I (S. oralis MTCC + S. mutans MTCC + C. albicans), MS-II (OP32 + S. mutans MTCC + C. albicans) (E) Comparative analysis on effect of pH upon addition of sucrose in planktonic, dual and mixed species at 10 h, 24 h, and 48 h for both standard cultures and OP32. Statistical significance was evaluated using Tukey’s multiple comparisons test. P-values: * < 0.05, ** < 0.01, *** < 0.001.
Effect of sucrose on biofilm formation on dual and multispecies biofilm growth over 24 h and 48 h
Biofilm forming potential of the standard culture as planktonic, dual and mixed species at various time intervals were performed. In all the cultures, exponential growth was observed from 1 hr to 10 h and declined growth in 24 and 48 h culture which could have happened due to deficit in nutrient availability (Fig. 7B). When compared to the planktonic cells, the co-cultures (dual cultures) showed increased biofilm formation especially for C. albicans (MTCC) and S. mutans (MTCC). Both exhibited increased production of biomass in the presence of S. oralis (MTCC). Similarly, significant increase in growth of biomass in mixed species was observed at all time intervals (Fig. 7B). Of the three clinical isolates that show phylogenetic similarity to S. oralis (MTCC) and biofilm forming capacity, OP32 was found to be significantly different from that of the standard cultures and other planktonic cultures at 48 h, however no significant difference was observed in 24 h culture in the presence of sucrose. Hence for dual and multispecies analysis we took OP32 as the reference clinical isolate that showed genetic similarity with S. oralis (MTCC) (Fig. 7C). After 24 h and 48 h of biofilm growth, both S. oralis (MTCC): C. albicans (MTCC) and OP32: C. albicans (MTCC) dual species were observed to have an increased biofilm formation compared to mono-species biofilms and S. mutans (MTCC): C. albicans (MTCC) in the presence of sucrose in both 24 h and 48 h. Significance difference in biofilm formation in multispecies at 48 h was observed. Especially at 48 h the biofilm formed by MSII (OP 32: S. mutans (MTCC): C. albicans (MTCC)) significantly differ from MSI (S. oralis (MTCC): S. mutans (MTCC) : C. albicans (MTCC)) with P < 0.001 (Fig. 7D).
The pH of the biofilm was observed to decline over 48 h of growth period ranging below 5 in all the planktonic and multispecies cultures in the presence of sucrose when compared to the control media. S. mutans (MTCC): S. oralis (MTCC) and OP32: S. mutans (MTCC) biofilms had the largest decrease in supernatant pH. In the case of C. albicans (MTCC) dual culture we could observe slightly elevated pH above 5 (Fig. 7E).
Discussion
Severe early childhood caries (SC) and its recurrence (RC) is a critical health concern affecting both physical and psychological health of a child. The oral microbiome dysbiosis and formation of conducive cariogenic niche results in severe caries16,17. Poor oral hygiene and generalized treatment methods do not completely eradicate the caries forming bacteria that might lead to persistence of these bacteria causing recurrence in Indian children. High-throughput sequencing methods were used in this study to untap the hidden diversity13 and its varied abundance among CF, SC and RC children in India. In general, we observed varied diversity of bacteria among the study groups. We could observe high bacterial diversity in SC as compared to CF group contradicting earlier reports4,18–20 (Fig. 1B) while RC group was found to exhibit low diversity of bacteria enriched with late colonizers. Notably we also encountered very low diversity of bacteria in sample P19 from RC that was found to be enriched with Firmicutes (98.30%). The low diversity in RC is in corroboration with previous studies18,20. However, differences in beta diversity analysis revealed there could be varied proportion of taxa in RC form that of CF and SC (Fig. 1B). This might be the consequence of significantly higher metabolic activities of microbiota that contribute to host-microbiome interaction when compared with SC and CF, triggering its survival in the unique ecological niche in RC. However, significant correlation was not found between gender and any other oral demographic status with caries occurrence, similar to earlier finding21.
Most of the caries microbiome related studies are based on saliva samples22 because it is a patient friendly non-invasive approach and is easy to collect. However, this may not address the complete details of potential microbial community composition, as it varies at different intraoral sites due to the diversified ecological niche present in oral cavity23. In this study, the samples collected from the lesions of carious dentine gives an insight into the variation in diversity of bacteria as well as its notable community interaction in both SC and RC condition. To the best of our knowledge, the current study provides a preliminary snapshot of the tooth biofilm microbiome variation of Indian children affected by severe early childhood caries and its recurrence. About 80% of OTU based taxonomical identification constitutes to the core microbiome (Fig. 5A) of the study group which includes the representation from almost all the reported phyla except for Fusobacteria found abundance in RC and SC groups. This clearly indicates that presence of Fusobacteira plays a vital role in caries formation and progression. The decrease in abundance of dominant phyla like Firmicutes, Proteobacteria in such a way CF > SC > RC corroborated with the reported low diversity of bacteria occurring upon caries progression19,20. The observed correlation of interspecies between the low abundant heterogenic oral microbiome and the resultant niche pH might play a determining role in caries formation and its recurrence. Our study showed significant variations in low abundant species among all the study groups which may act as a potential biomarker. Late colonizers like Prevotella saccharolytica, Campylobacter gracilis, Pseudomonas, Neisseria oralis, Coriobacteriaceae, Atopobium, Bergeylla, Fusobacterium HMT 203, Scardovia wiggsiae, Selenomonas flueggei, Stomatobaculum, Oribacterium parvum, Parvimonas and Peptostreptococcaceae in SC and its maintenance in RC (Fig. S6) shows its capability of being key pathogens in the severity of the disease. This augments the keystone hypothesis, that relative abundance of prevalent taxon may not be strongly attributed to the progression of caries15. Co-occurrence network analysis exhibits positive association among these microbes and also with that of the early colonizers emphasizing their role in severity of caries, predicting an incomplete elimination of these microbes from the caries niche with current therapeutic methods20,24.
Among the oral commensals the dysbiosis of Streptococcus plays a pivotal role in caries formation and progression. However, the relative abundance of S. mutans was very low in our observation on Indian children, though it has been reported as a pioneer in caries forming bacteria25,26. We could find only 5% of S. mutans in the SC microbiome and 2% in RC (Fig. S1B,C) which clearly indicates the scarcity of S. mutans in advanced caries, as seen before6,19,27. Whereas the non-mutan Streptococcus and S. oralis subsp. tigurinus clade 071 was found to be abundant in all the study groups subjected in this report, particularly high in RC (Fig. S2). The specific biochemical activities in Streptococcus oralis subsp. tigurinus clade 071 is not clearly understood, however, its significant abundance in caries depicts a possible overlap in biochemical properties of acting as a flux and AI24 activity that helps in quorum sensing similar to S. oralis.
In vitro analysis of the clinical isolates on biofilm formation supports this. The clinical isolates OP32, OP12 and OP20 were found closely related to S. oralis (MTCC) by genetic fingerprinting analysis using BOX PCR that helps to identify the bacterial strains at species level28,29. Clinical isolates closer to S. oralis MTCC might be S. oralis subsp. tigurinus clade 071 as seen from the NGS data analysis and closer phylogenetic clade. These oral isolates are supposed to be the subgroup of S. oralis as seen earlier30. The positive growth interaction between the OP32 with the non-biofilm forming clinical isolates show that the interspecies interaction is significant in the SECC microbiome. Further, the increased biofilm formation in co-culture of S. oralis and OP32 with the major cariogen S. mutans under high sucrose environment show the cariogenic interrelation between the species, as observed earlier31,32. Also high pH reduction in the dual culture S. mutans: S. oralis and S. mutans: OP32 co-culture suggests its role in acidification of oral niche.
Additionally the interaction of interkingdom microbe C. albicans enhances the biofilm forming capability in both standard and clinical isolates. Recent report states that the glucans produced by gtfR mediates cross-kingdom interaction between C. albicans and S. oralis which in turn have positive effect on stronger biofilm matrix formation31. Similarly, we observed increased biofilm formation by S. oralis MTCC and OP32 in the presence of C. albicans both in dual and multispecies especially in the presence of sucrose and positively correlated with incubation time.
Increased level of starch and sucrose metabolism along with metabolites like ABC transporters, Phosphotransferase system, Two Component system in functional prediction analysis and its positive interaction with S. oralis subsp. tigurinus clade 071 (Fig. 6) supports our in vitro findings. These processes support an acidic environment that might allow growth of low abundant bacteria like Micrococceae, Abiotrophia defectiva, Granulicaterlla elegans, Aerococcaceae and Carnobacteriaceae. Moreover, the interaction network analysis on Two Component System which is one of the key player of biofilm formation (Fig. S8) clearly shows that S. oralis subsp. tigurinus clade 071 positively correlates with TCS and anaerobic late colonizers like Capnocytphaga leadbetteri, Capnocytphaga granulosa and Aggrgatibacter. These in turn may exhibit positive interaction with signal transduction mechanisms inviting other bacteria to interact with the biofilm more firmly and might cause the severity of the conditions.
However high abundance of other Streptococcus genera in SC and RC groups and its co-occurrence with other disease causing microbes and metabolic pathways exemplifies that the untapped species level diversity of Streptococcus still exists in the oral cavity that contributes to a greater extent in caries formation and progression. Based on the observations we envisage that Streptococcus oralis subsp. igurinus clade 071 initially colonize the tooth surface, with its carbohydrate metabolism with EPS production resulting in increased quorum sensing signals that invite other unidentified Streptococcus making the environment more acidic and further allows all the other middle and late colonizers to form a strong network that leads to demineralization and rupturing of teeth which may cause Severe early childhood caries.
Leptotrichia the third largest significant bacteria in RC group (Fig. S2) could be one of the key pathogens in RC because of its reported property to metabolize sucrose and its isomers with the help of PTS to lactic acid in the absence of S. mutans33. Significantly high PTS in RC group (Fig. 6) and its positive interaction with Leptotrichia in the network analysis (Fig. 7) in the current report gave a piece of clear evidence that Leptotrichia could create acidic niche, thereby inviting other acid tolerant bacteria such as Capnocytophaga, Aggregatibacter, Fusobacterium, Campylobacter, Granulicatella, Gemella and Prophyromonas (Fig. S9). Association of these bacteria develops a strong network of complex microbial associations that may resist any treatment methods resulting in recurrence.
Unusually, among other abundant species, we found a potent cariogen V. parvula34–37 high in CF individuals compared to RC and SC (Figs. 3, S2). The inability of V. parvula to enhance caries formation in these CF individuals could be linked with the high abundance of non-cariogenic arginolytic bacteria Neisseria and V. dispar38 in the CF oral microbiome that might neutralize the acidification of the niche through arginine metabolism19. The PICRUST analysis showed high arginine metabolism in the CF group, possibly from the profound Neisseria and V. dispar in CF (Fig. S5B). Occurrence of Micrococcaceae and Bifidobacteriaceae in SC and RC groups, and not in CF shows its intrinsic role in caries formation and especially recurrence condition. We found that Micrococcaceae interacts positively with most of the reported facultative anaerobes and with the major predicted carbohydrate metabolism like starch and sucrose, fructose and mannose and amino acid metabolism. This could play a critical role in metabolism of carbohydrate even in the anaerobic environment of SC children that could maintain the acidic niche. The abundance of Bifidobacterium in RC corroborated with its fluoride resistance property with the help of bifid shunt, a bypass pathway, under fluoride inhibition of enolase to reduce the pH39 that could lead to recurrent caries.
The co-aggregation of microbes is of prime importance among oral commensals dysbiosis that leads to complex network of biofilm formation resulting in caries manifestation24. Most of the core microbiome exhibited positive interaction with each other, indicting the microbes are mutually benefiting from each other’s metabolites. Co-occurrence network analysis showed that the interaction between microbes resulting in recurrence is not a single large hub, instead, several inter-related hubs of co-occurrence and mutual exclusion. Significant hubs like Pseudomonas, Bergeyella show mutual exclusion and Neisseria shows positive interaction with most of the bacteria. Prominent bacteria like Actinomyces, Neisseria, Fusobacterium, Corynebacterium, Prevotella, Bifidobacteria, Olesenella, Selenomonas, Oribacterium and Gracillibacter form a complete network of early, middle and late colonizers both in high and low abundant with different kinds of interactions depicting their role of heterogeneity in recurrence as well as severe caries (Fig. S3A). V. tobetuensis, one of the early colonizers possess very high autoinducer 2 activity, a strong quorum sensor40, co-aggregated to Fusobacterium in recurrent groups which could be a potent quorum sensing bacteria in the microbiome of Indian children especially in RC. This is supported by the increased sugar metabolism that resulted in increased Glycan biosynthesis in RC (Fig. 6) forming a strong matrix of biofilm with the help of bridging bacteria like Fusobacteria and Corneybacerium to link the early and late colonizers in the biofilm (Fig. S3B)1,41. A different bridging bacteria Corynebacterium matruchotii and Corynebacterium durum which produce acid from mannitol and galactose42 family were found high in RC. This has a long filamentous structure that acts as an anchoring site for other microbes1,2. Early colonizers like Capnocytophaga leadbetteri, Haemophilus, Streptococcus and late colonizers like Oribacterium, Prevotella, Capnocytophaga sputigena were interacting with Corynebacterium rather than Fusobacterium, while the abundance of Fusobacterium was more. This might add a demographic effect in the Indian population, as well as synergistic or competitive effect between Fusobacteria and Corynebacteria (Fig. S3).
As most of the earlier studies on dental caries have focused on the dysbiosis in oral microbiome20,36, wherein functional properties of these species in causing caries and its recurrence is contentious6. Positive interactions among the predicted carbohydrate metabolism and quorum sensing systems like ABC transport system, TCS, PTS with that of middle and late colonizers like Prevotella, Cardiobacterium, Carnobacteriaceae, Selenomonas, Prophyromonas, Micrococcaceae, Rothia, Bifidobacterium, Corynebacterium, Oribacterium, Lachnoanaerobaculum, Actinomyces and Saccharibacteria (TM07) could be correlated to the pathway of energy flow among these microbes that could enhances the severity of the condition by enriching more number of aciduric or acidogenic bacteria and evading the treatment method due to complexity of the bacteria present at that particular ecological niche. Significant difference in flagellar assembly between caries free and caries condition could be related to the abundance of late colonizer Selenomonas and also other flagellated organisms. Positive associations of three members of CPR phyla members Gracillibacteria GN02, Absconditabacteria SR1 and Saccharibacteria TM7 with that of three study groups CF and SC, RC respectively was observed. These findings are in line with the studies that proved the role of SR1 with H2S production43 and TM7 as obligate symbiont with Actinomyces species in oral disease conditions44. From this study we could infer that GN02 could be considered as a potential biomarker for the healthy status and TM7 for caries status of oral cavity (Fig. 4).
Thus, from the results, we observed the significant interaction among overrepresented and underrepresented bacteria and the functional metabolic interrelationship is essential in SC as well as its recurrence. In Indian children, we observed different combinations of middle and late colonizers in the polymicrobial system with the presence or absence of the so far reported early colonizers. This tends to develop a strong network of microbial association in caries condition that could develop resistance against most of the treatment methods and also nullifies the effect of resident bacteria that are trying to regain the acid environment which leads to the recurrent condition.
Conclusion
Severe Early Childhood Caries is a chronic and prevalent disease that affects the overall growth of the children, and recurrence after treatment persists also in adulthood. This study reports the nature and heterogeneity of microbiome in the dentine micro-ecological niche among the healthy and caries affected children in Indian sub-populations. This will help to develop some simple and cost effective solutions to treat this disease. Increased depth of high throughput sequencing analysis allowed us to identify significant high and low abundant bacteria and its co-occurrence towards the aetiology of caries formation and its recurrence in Indian children. From our study, we understood that S. oralis subsp. tigurinus clade 071 is the prevalent and significant among caries affected Indian children instead of S. mutans and is a potent cariogen. Detailed investigation of S. oralis subsp. tigurinus clade 071 towards caries etiology and its inter-kingdom ecological interactions would deepen the knowledge about its persistence towards treatment method and how it evades the host immune system. The biomarker and co-occurrence analysis have given a better insight on the contribution of low abundant bacteria towards the severity of the disease. Our findings significantly mitigate the understanding of changes in bacterial profile in response to interaction with the predictive functional traits especially quorum sensing signals at different stages of childhood caries. This has opened up new dimensions into the oral microbiome at the community level under different micro-ecological niche to support future metabolomics and transcriptomic studies, coupled with functional assays, thereby contributing for the development of novel strategies to identify and manage the risk of caries in children.
Materials and methods
Study subjects and sample collection
The complete study was performed under informed consent from the parents concerned about the experimental and control group children following the World Medical Association Declaration of Helsinki guidelines. The Ethical Committee of K.S.R Institute of Dental Science and Research endorsed the design, protocol and informed written consent (132/KSRIDSR/EC/2016). Medically healthy children between the age group of 3–7 years who were reported between November 2016 and April 2018 at the Department of Pediatric Dentistry at K.S.R Institute of Dental Science and Research were recruited for this study. There was no genetic relationship among the individuals of both genders and they do not share a homogeneous living environment. Children treated with antibiotics within 3 months of the study visit were excluded. The dental health and hygiene of every subject in the study was assessed by a team of professional dentists satisfying the DMFT index standards45. A questionnaire on child’s food habits, brushing habit, caries prevalence in their family, especially intake frequency of fermentable carbohydrates excluding their regular interval of meals like breakfast, lunch and dinner were also collected.
A total of 55 children were divided into three groups, 1. No clinical evidence of caries experience with zero DMFT was considered as Caries-Free (CF): n = 15, 2. Children with lesion and decayed teeth with DMFT ≥ 4 were considered as Severe Early Childhood Caries (SC): n = 20. 3. Children with secondary caries and filled teeth for primary caries were considered as Recurrent Early Childhood Caries (RC): n = 20. After thorough dental examination of the children by the dentist, the teeth were gently wiped with cotton rolls followed by gentle air stream to avoid saliva contamination and biofilm was obtained using soft ended swab (Himedia, India) according to the prescribed manual of procedures for Human Microbiome (http://hmpdacc.org/resources/tools_pot.cols.php) with minor modifications and transported to lab within 4 h of sample collection46.
DNA extraction from swab samples for metagenomic studies
Bacterial DNA was extracted using QIAamp DNA microbiome Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions with minor modification in the pathogen lysis step to enhance better lysis Gram’s positive and Gram’s negative organisms. The enriched microbial DNAs were purified by ethanol precipitation. The DNA concentration was estimated spectrophotometrically using a NanoDrop spectrophotometer (Thermo Electron Corporation, USA), and molecular size was estimated by agarose gel electrophoresis. DNA samples that had passed the QC with NanoDrop concentration > 10 ng/µL was used for further processing. DNA samples were then stored at − 20 °C until further use.
PCR amplification of the 16S rRNA gene and illumina sequencing
The V3 and V4 regions of 16S rRNA were amplified using PCR with primers Pro341F-5′ CCTACGGGNBGCASCAG; Pro805R-5′-GACTACNVGGGTATCTAATCC47. PCR was carried out in 50 μL reaction volume, including 32.5 μL of ddH2O, 10 μL of 10× Taq buffer, 1 μL of 10 mM dNTPs, 0.5 μL of 50 mM MgCl2, 2 μL each of 10 μM forward and reverse primers and 1 unit of Taq DNA polymerase (Thermo Scientific, USA) and 20 ng of DNA at the following PCR conditions: initial denaturation at 98 °C for 30 s followed by 30 cycles of 98 °C for 15 s, 66 °C for 25 s and 72 °C for 30 s and an final extension at 72 °C for 10 min. Thus, amplified products were subjected to next generation sequencing (2 × 250 bp) at Scigenome Research Foundation using Illumina Miseq platform according to the manufacturer’s instructions.
Sequence analysis
The reads obtained from Illumina platform were filtered to discard sequences with an average Phred score > 30 and sequences containing incorrect barcodes and/or lacking primer sequences. Filtered reads were de-replicated, followed by subsequent removal of singletons and identified chimeras and subjected to further analysis using QIIME pipeline (version 1.9.1)48. The processed reads were then clustered at 97% similarity using UCLUST to identify the species-level Operational Taxonomic Units (OTUs) and the reads were mapped to the filtered OTUs to determine the exact count of each OTU in each sample49,50. To determine the bacterial genera the OTUs were BLAST against the HOMD database for human oral bacteria51. The species or phylotypes were identified by their HOMD identity. OTUs with less than 10 sequences were excluded from the analysis37. The sequences which could not be assigned to a genus in HOMD (at 97% identity) were considered as the probable candidates that might be unique or novel in the oral cavity of the Indian populations.
Biomarker and network analysis
In order to identify the potential biomarker, LDA effect size (LEFSe) was performed to find out the differentially enriched taxa among the groups, the threshold for discriminative features was set to 3.052 and the results were displayed in a cladogram and a bar graph53. Krona plot was performed to find out the percentage of enriched taxa between groups54. The functional prediction of microbiota was done with PICRUST. Only reads identified in closed reference picking were used for PICRUST analysis and aligned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) data base47. Co-occurrence network analysis was performed to understand the inter and intrarelationship of oral microbiome with its predicted functional potentials obtained from PICRUST analysis among the study groups using CoNet55 and the model was visualized using Cytoscape56 tool version 3.7.2.
Bacterial strains, growth conditions, Biofilm formation and quantification
The standard bacterial strains Streptococcus mutans 497T, Streptococcus oralis 2696, Candida albicans 1367T used in this study were obtained from the Microbial Type Culture Collection (MTCC), Chandigarh, India. All the microbial strains were cultured and maintained using culturing conditions given by MTCC. Clinical isolates were obtained from swab samples of caries affected children involved in this study in K.S.R. Dental Science and Research, Tiruchengode. The swab samples were cultured on blood agar (Himedia pvt Ltd, India) in order to isolate the alpha haemolytic Streptococcal strains. Thus isolated 40 strains were named as OP1-OP40 were cultured and maintained in Brain Heart Infusion (BHI) (Himedia pvt Ltd, India) media and stored as glycerol stock at − 80 °C for further use. To identify the biofilm forming isolates among the clinical cultures all the isolates were subjected to Congo Red Agar test as described earlier57. Co-culture of the clinical isolates and standard colonies were performed in BHI agar. The co-culture growth was observed at 37 °C growth for 24 h58. Biofilm was formed by pipetting out 10 µL of OD cultures for planktonic cells and equal concentration for dual and mixed cells on the wells of a flat-bottom tissue-culture treated 96-well microtiter plate containing 100μL of BHI broth. For some experiments the media were supplemented with 1% W/V sucrose. The plates were incubated statically for 24 h at 37 °C for biofilm formation. After incubation, the biofilm biomass by crystal violet assay and pH assay was performed as described previously59.
BOX PCR
The 22-mer BOXAIR oligonucleotide was used to generate BOX-PCR profiles. Amplification reactions were performed in volume of 25 μL, containing 2 μM of BOX primer (5′-CTACGGCAAGGCGACGCTGACG-3′), 200 μM dNTP, PCR reaction buffer (10 mM Tris Hcl, 50 mM Kcl, 1.5 mM Mgcl 2) 1.5 units of Taq DNA polymerase and template DNA 5 μL of bacterial cell at 108 CFU/mL. Amplification was performed using Lark thermocycler with the following PCR condition: initial denaturation step of 5 min at 94 °C followed by 40 cycles of 1 min at 94 °C, 1 min at 60 °C, and 1 min at 72 °C, and a final elongation step of 10 min at 72 °C followed by the analysis of BOX PCR fingerprint was done according to previous protocol28 and the tree was visualized using iTOL (Tree Of Life v1.0).
Statistical analysis
OTU table that was created by QIIME analysis contains raw counts were normalized to a relative abundance OTU table. Using this relative abundance, similar types of taxa were aggregated at the phylum, class, order, family, genus and species level48. Biodiversity between classified groups were examined using non-parametric Mann–Whitney and/chi-squared, Fisher’s exact test. Significant differences in the alpha diversity indexes between the different groups (P < 0.05) were identified by Student’s t-test46. Weighted Unifrac distance of OTU samples were used to perform PCoA to analyze the differences among groups (beta diversity). Differences in the Unifrac distances for pairwise comparisons among groups were determined using Multiple t-test and visualized by the construction of a box and whiskers plot. The correlation was evaluated by the Spearman Correlation. The graphical representation of the results was performed by using GraphPad Prism version 6.01 (GraphPad Software, La Jolla California USA).
Supplementary information
Acknowledgements
We extend our heart filled thanks to the Management and Principal of K S Rangasamy College of Technology for providing infrastructure to carry out this research work. The corresponding author acknowledges DST-FIST, DBT-STAR scheme for providing infrastructure facilities. The authors are grateful for the genomics grant awarded by SciGenom Research Foundation (SGRF), Chennai to support the DNA sequencing in this work. This study used the Nephele platform from the National Institute of Allergy and Infectious Diseases (NIAID) Office of Cyber Infrastructure and Computational Biology (OCICB) in Bethesda, MD. We thank all the previous researchers and their findings which has led us to this output.
Author contributions
B. K. conceptualized the study, designed, performed the experiments, analysed, interpreted the data and drafted the manuscript. P.P., A.H.B. and A.S., performed part of the bioinformatics analysis, S.A., designed the study, collected the sample from children and interpreted the data. R.A., interpreted the data and drafted the manuscript, R.S.V. and S.S.G conceptualized the study and critically revised the manuscript.
Data availability
The raw reads generated in this study were deposited into the NCBI Sequence Read Archive (SRA) database under Bioproject Accession Number PRJNA454811.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-78057-z.
References
- 1.Ferrer MD, Mira A. Oral biofilm architecture at the microbial scale. Trends Microbiol. 2016;24:246–248. doi: 10.1016/j.tim.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fakhruddin KS, Ngo HC, Samaranayake LP. Cariogenic microbiome and microbiota of the early primary dentition: a contemporary overview. Oral Dis. 2019;25:982–995. doi: 10.1111/odi.12932. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J. Clin. Microbiol. 2007;45:81–87. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kothia NR, et al. Assessment of the status of national oral health policy in India. Int. J. Health Policy Manage. 2015;4:575–581. doi: 10.15171/ijhpm.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu C, et al. The predictive potentiality of salivary microbiome for the recurrence of early childhood caries. Front. Cell. Infect. Microbiol. 2018;8:1–11. doi: 10.3389/fcimb.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graves DT, Corrêa JD, Silva TA. The oral microbiota is modified by systemic diseases. J. Dent. Res. 2018;98:148–156. doi: 10.1177/0022034518805739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan NS, Tribble G, Angelov N. Oral microbiome and nitric oxide: the missing link in the management of blood pressure. Curr. Hypertens. Rep. 2017;19:33. doi: 10.1007/s11906-017-0725-2. [DOI] [PubMed] [Google Scholar]
- 9.Demmer RT, Desvarieux M. Periodontal infections and cardiovascular disease: the heart of the matter. J. Am. Dent. Assoc. 2006;137:S14–S20. doi: 10.14219/jada.archive.2006.0402. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, et al. Variations in oral microbiome profiles in rheumatoid arthritis and osteoarthritis with potential biomarkers for arthritis screening. Sci. Rep. 2018;8:17126. doi: 10.1038/s41598-018-35473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobb CM, et al. The oral microbiome and adverse pregnancy outcomes. Int. J. Womens Health. 2017;9:551–559. doi: 10.2147/IJWH.S142730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 2019;11:1586422. doi: 10.1080/20002297.2019.1586422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr. Rev. 2012;70(Suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilian M, et al. The oral microbiome—an update for oral healthcare professionals. Br. Dent. J. 2016;221:657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- 18.Ledder RG, Kampoo K, Teanpaisan R, McBain AJ. Oral microbiota in severe early childhood caries in Thai children and their families: a pilot study. Front. Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross EL, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurley E, et al. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. 2019;19:13. doi: 10.1186/s12903-018-0693-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez A, Nelson KE. The oral microbiome of children: development, disease, and implications beyond oral health. Microb. Ecol. 2017;73:492–503. doi: 10.1007/s00248-016-0854-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, et al. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 2012;6:1–10. doi: 10.1038/ismej.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bik EM, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bramhachari PV, Ahmed VKS, Selvin J, Hassan S. Quorum Sensing and Biofilm Formation by Oral Pathogenic Microbes in the Dental Plaques: Implication for Health and Disease BT—Implication of Quorum Sensing and Biofilm Formation in Medicine, Agriculture and Food Industry. Singapore: Springer; 2019. pp. 129–140. [Google Scholar]
- 25.Banas JA, Drake DR. Are the mutans streptococci still considered relevant to understanding the microbial etiology of dental caries? BMC Oral Health. 2018;18:129. doi: 10.1186/s12903-018-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson AC, et al. In-vivo shift of the microbiota in oral biofilm in response to frequent sucrose consumption. Sci. Rep. 2018;8:1–13. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grover V. Discriminating Life Forms in Oral Biofilms. New York: IntechOpen; 2018. [Google Scholar]
- 28.Marques A, Marchaison A, Gardan L, Samson R. BOX-PCR-based identification of bacterial species belonging to Pseudomonas syringae-P. viridiflava group. Genet. Mol. Biol. Genet. Mol. Biol. 2008 doi: 10.1590/S1415-47572008000100019. [DOI] [Google Scholar]
- 29.Poonchareon K, Pulsrikarn C, Nuanmuang N, Khamai P. Effectiveness of BOX-PCR in differentiating genetic relatedness among Salmonella enterica serotype 4,[5],12:i:- isolates from hospitalized patients and minced pork samples in northern Thailand. Int. J. Microbiol. 2019;2019:5086240. doi: 10.1155/2019/5086240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen A, Scholz CFP, Kilian M. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int. J. Syst. Evol. Microbiol. 2016;66:4803–4820. doi: 10.1099/ijsem.0.001433. [DOI] [PubMed] [Google Scholar]
- 31.Souza JGS, et al. Role of glucosyltransferase R in biofilm interactions between Streptococcus oralis and Candida albicans. ISME J. 2020;14:1207–1222. doi: 10.1038/s41396-020-0608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, van der Mei HC, Busscher HJ, Ren Y. Streptococcus mutans adhesion force sensing in multi-species oral biofilms. NPJ Biofilms Microbiomes. 2020;6:25. doi: 10.1038/s41522-020-0135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J, Pikis A. Metabolism of sugars by genetically diverse species of oral Leptotrichia. Mol. Oral Microbiol. 2012;27:34–44. doi: 10.1111/j.2041-1014.2011.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanasi E, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanner ACR, et al. Cultivable anaerobic microbiota of severe early childhood caries. J. Clin. Microbiol. 2011;49:1464–1474. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, et al. Dynamic alterations in salivary microbiota related to dental caries and age in preschool children with deciduous dentition: a 2-year follow-up study. Front. Physiol. 2018;9:1–12. doi: 10.3389/fphys.2018.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koopman JE, et al. Stability and resilience of oral microcosms toward acidification and candida outgrowth by arginine supplementation. Microb. Ecol. 2015;69:422–433. doi: 10.1007/s00248-014-0535-x. [DOI] [PubMed] [Google Scholar]
- 38.Arif N, Sheehy EC, Do T, Beighton D. Diversity of Veillonella spp. from sound and carious sites in children. J. Dent. Res. 2008;87:278–282. doi: 10.1177/154405910808700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur R, Gilbert SC, Sheehy EC, Beighton D. Salivary levels of Bifidobacteria in caries-free and caries-active children. Int. J. Paediatr. Dent. 2013;23:32–38. doi: 10.1111/j.1365-263X.2011.01220.x. [DOI] [PubMed] [Google Scholar]
- 40.Mashima I, Nakazawa F. The interaction between Streptococcus spp. and Veillonella tobetsuensis in the early stages of oral biofilm formation. J. Bacteriol. 2015;197:2104–2111. doi: 10.1128/JB.02512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lif Holgerson P, Öhman C, Rönnlund A, Johansson I. Maturation of oral microbiota in children with or without dental caries. PLoS ONE. 2015;10:e0128534. doi: 10.1371/journal.pone.0128534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett SL, Cookson BT, Carlson LC, Bernard KA, Coyle MB. Diversity within reference strains of Corynebacterium matruchotii includes Corynebacterium durum and a novel organism. J. Clin. Microbiol. 2001;39:943–948. doi: 10.1128/JCM.39.3.943-948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aas JA, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker JL, Bor B, Agnello M, Shi W, He X. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ismail AI, et al. The international caries detection and assessment system (ICDAS): an integrated system for measuring dental caries. Community Dent. Oral Epidemiol. 2007;35:170–178. doi: 10.1111/j.1600-0528.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 46.Xiao C, Ran S, Huang Z, Liang J. Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S pyrosequencing. Front. Microbiol. 2016;7:1145. doi: 10.3389/fmicb.2016.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-García A, et al. Comparison of mothur and QIIME for the Analysis of rumen microbiota composition based on 16S rRNA amplicon sequences. Front. Microbiol. 2018;9:3010. doi: 10.3389/fmicb.2018.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allali I, et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017;17:194. doi: 10.1186/s12866-017-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole JR, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen T, et al. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42–R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Research. 2016;5:1519. doi: 10.12688/f1000research.9050.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Castro Melo P, et al. Comparison of methods for the detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Braz. J. Microbiol. 2013;44:119–124. doi: 10.1590/S1517-83822013005000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heersema LA, Smyth HDC. A multispecies biofilm in vitro screening model of dental caries for high-throughput susceptibility testing. High-throughput. 2019;8:14. doi: 10.3390/ht8020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads generated in this study were deposited into the NCBI Sequence Read Archive (SRA) database under Bioproject Accession Number PRJNA454811.