Abstract
• Mitochondrial F1FO ATP synthase is the key enzyme for mitochondrial bioenergetics. Dimeric F1FO-ATP synthase, is preferentially located at the edges of the cristae and its oligomerization state determines mitochondrial ultrastructure. The ATP synthase inhibitor protein IF1 modulates not only ATP synthase activity but also regulates both the structure and function of mitochondria. In order to understand this in more detail, we have investigated the effect of IF1 on the spatiotemporal organization of the ATP synthase. Stable cell lines were generated that overexpressed IF1 and constitutively active IF1-H49K. The expression of IF1-H49K induced a change in the localization and mobility of the ATP synthase as analyzed by single molecule tracking and localization microscopy (TALM). In addition, the ultrastructure and function of mitochondria in cells with higher levels of active IF1 displayed a gradual alteration. In state III, cristae structures were significantly altered. The inhibition of the hydrolase activity of the F1FO-ATP synthase by IF1 together with altered inner mitochondrial membrane caused re-localization and altered mobility of the enzyme.
Keywords: F1FO ATP synthase, Inhibitory factor 1, IF1, IF1-H49K, Mitochondria, Superresolution microscopy, Spatiotemporal organization, Tracking and localization microscopy (TALM), Mitochondrial ultrastructure, OXPHOS, ATPase, Opa1
1. Introduction
The mitochondrial F1FO ATP synthase is the last of the complexes known as OXPHOS complexes, which mediate oxidative phosphorylation to generate ATP. It synthesizes most of the ATP under non-fermentative conditions. All OXPHOS complexes are found in the inner membrane (IM) [1–4]. The IM is further divided into sub-compartments: the cristae membranes (CM), which are extrusions of the inner membrane into the matrix and the inner boundary membrane (IBM), which is the part of the IM that parallels the OM. In many cells, CM form disc-like or lamellar structures [5,6]. Dimeric and oligomeric F1FO ATP synthase (CV) bend the cristae membranes and are found at the rims [7–10]. However, the ATP synthase is not a completely immobile protein fixed to the rims of cristae: The localization, mobility and oligomerization state of the ATP synthase depend on its activity mode and influences the ultrastructure [10–20]. Furthermore, ATP synthase is physically and functionally connected with multiple other complexes in and outside of mitochondria [21–24], which also demands some spatio-temporal plasticity. E.g., via mitochondria-associated hexokinases, ATP synthase can dynamically supply ATP for glucose phosphorylation [21]. In a recent study we showed that the localization and mobility of the ATP synthase was restricted to cristae when ATPase activity was inhibited by drug application. In contrast, when the natural inhibitor of ATP synthase, the inhibitory factor IF1 [25–27], was genetically knocked out, the localization and mobility of the ATP synthase changed and the mobility of ATP synthase increased. To deepen our understanding of how the activity and dimerization of ATP synthase affects its spatio-temporal organization in the membrane, we here addressed the impact of constitutively active IF1 on the localization, dynamics and organization of ATP synthase. IF1 binds to the alpha(DP)-beta(DP) interface of the F1FO ATP synthase at the matrix side and contacts the central gamma subunit [28,29] preventing futile hydrolysis of ATP [30]. The inhibitory peptide dimerizes and oligomerizes in a pH-de-pendent manner, for which a protonated histidine residue at position 49 is responsible [31]. Changing the histidine residue to lysine generates IF1-H49K, a pH-independent, constitutively active mutant [31,32]. Binding of IF1 to the F1FO ATP synthase not only inhibits ATPase activity but also promotes interaction between ATP synthase dimers to form tetramers [18,33]. Via ATP synthase activity, IF1 regulates the ultrastructure and function of mitochondria and thus the fate of the cell [25,26,34–38]. To dissect the effect of IF1 on the sub-mitochondrial localization and mobility of ATP synthase, we generated stable cell lines expressing IF1 and IF1-H49K, respectively. The transfected cell lines grew normally with the same proliferation rate. In both cell lines, the cytosolic ATP level was higher, while the mitochondrial membrane potential ΔΨm was decreased as observed before [39]. Cells displayed gradual changes in the inner membrane folding. Finally, we analyzed the impact of IF1 on the spatio-temporal organization of ATP synthase by single particle tracking and localization microscopy in live cells [40,41]. We found a gradual effect of IF1-H49K on the sub-mitochondrial localization of F1FO ATP synthase including a different mobility pattern. This re-organization was beneficial for mitochondrial ATP synthesis and metabolic activity.
2. Material and methods
2.1. Cell culture
HeLa WT, IF1-HA and IF1-H49K-HA cells were cultivated in Minimal Essential Medium Earle’s Salt (5.6 mM glucose + l-Glutamine) supplemented with 10% FCS (Fetal calf serum), 1% HEPES, and 1% NEAA (non-essential amino acids) at 37 °C and 5% CO2. The cells were split when they were 80–100% confluent using Trypsin/EDTA for cell detachment. HeLa cells were purchased from the Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures) and transiently transfected by the calcium transfection method if required. The transfected cells were used 48–72 h after transfection.
2.2. Generation of IF1 and IF1-H49K cell lines
Cells lines stably expressing IF1 and IF1-H49K, respectively, were generated by selection of antibiotic resistant cells (clone picking) after transient transfection with a plasmid encoding for IF1-HA or IF1-H49K-HA. For cloning of IF1-H49K-HA, we used the forward primer GAAG AAGAAATCGTTCATCATAAG with an EcoRV restriction site and the reverse primer CTTGTGTTTTTTCAAAGCTGCCAGTTGTTC with an EcoRI restriction site. The template was integrated into the pSems26 vector after cutting out the sequence encoding for Cox8a-snap-tag purchased from NEB Biosciences™ (formerly Covalys™). The plasmid backbone of pSems26 encodes for ampicillin resistance for bacterial selection and contained the neomycin resistance for selection of stable clones. The promoter was from CMV.
2.3. Generation of a stable mitofilin cell line
For cloning of mitofilin-eGFP, we used the forward primer CGCCT CGAGACCGGTATGCTGCGGGCCTGTCAGTTATCGGGTGTG with an AgeI and XhoI restriction site and the reverse primer CGCGCGGCCGCGATATCCTCTGGCTGCACCTGAGTGGTTCCTATTCCTAC with an EcoRV and NotI restriction site. Mitofilin was N-terminally fused to eGFP in a pSems26 vector. The pSems26 vector had the Cox8a-snap-tag exchanged by eGFP. The vector plasmid was purchased from NEB Biosciences™ (formerly Covalys™). The plasmid backbone of pSems26 encodes for ampicillin resistance for bacterial selection and contained the neomycin resistance for selection of stable clones, as well as CMV promoter.
2.4. Semi- and quantitative real time RT-PCR
The total RNA of HeLa WT and stable IF1-H49K expressing HeLa cells was obtained with an RNA isolation kit (Roti-Prep RNA mini from Roth, # 8485.1), and cDNA was prepared using a cDNA Synthesis Kit (Biozym, # 331470×). The semi-quantitative PCR was performed with suitable primers for IF1, Opa1, GAPDH and Drp1. The following primers were used: IF1-forward: CGCGATATCATGGCAGTGACGGCGTTGG; IF1-reverse: CGCGAATTCATCATCATGTTTTAGCATTTTGATCTTCTG; OPA1-forward: GGCTCTGCAGGCTCGTCTCAAGG; OPA1-reverse: TTCCGCCAGTTGAACGCGTTTACC; GAPDH-forward: CCACCCATGGCAAATTCCATGGCA; GAPDH-reverse: TCTAGACGGCAGGTCAGGTCCACC; Drp1-forward: AACTTGATCTCATGGATGCGGG; Drp1-revers: ATGAACCAGTTCCACACAGCGG. The annealing temperatures were as follows: IF1 (336 bp) 59 °C, OPA1 (562 bp) 54 °C, Drp1 (661 bp) and GAPDH (598 bp) 59 °C. The OPA1 primer bind to transcript variants #1, 4, 6, and 8 (BlastN search), the Drp1 primer to transcript variants #2, 3, 4, 5, 6, 7, 8.
For quantitative real time RT-PCR, total RNA was isolated from cell pellets using the Monarch® Total RNA Miniprep Kit (NEB #T2010) and 1.5 μg of mRNA per sample was transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific #K1621). Quantitaive PCR (qPCR) was carried out on a StepOnePlus™ Real-Time PCR System (Applied Biosystems) using the PowerUP™ SYBR™ Green Master Mix (Applied Biosystems # A25741), between 0.06 and 0.08 μg of cDNA and 0.1 nM of each forward and reverse primer per sample. Samples were run in triplicats. The run consisted of an initial denaturation at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. GAPDH acted as endogenous control. Primers were synthesised by Eurofins Genomics.
GAPDH fwd 5′- CTG GTA AAG TGG ATA TTG TTG CCA T −3′; rev 5′- TGG AAT CAT ATT GGA ACA TGT AAA CC −3′; OPA1 fwd 5′- AGG CAT GGC TCC TGA CAC AAA G-3′; rev 5′- TGC GTT CAG CAT CCA CAG ATC C −3′; IMMT/mitofilin fwd 5′- AGA GGA AGC CAG AAA AGC CCA C −3′; rev 5′- GCT TTG ATG GCC TCA ACT GCA C −3′; Drp1 fwd 5′- GGT GAA GCG GCA AAT CAA ACG −3′; rev 5′- AGC CCA CAA GCA TCA GCA AAG −3′; Mfn1 fwd 5′- CTC CAG CAA CGC CAG ATA ATG C −3′; rev 5′- ACT TGT TGG CAC AGG CGA GC −3′; Mfn2 fwd 5′- GGA TGC TGA TGT GTT TGT GCT GG −3′; rev 5′- AGT CCA TGA TGA GTC GAA CCG C −3′.
2.5. SDS- and blue native PAGE and Immuno-blotting
For SDS-PAGE, cell lysates from samples of confluent T-25 flasks (1 flask would last for 10–15 gels) in sample buffer were heated for 5 min (95 °C) and then separated on 12% Tricine-SDS-PAGEs and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked with 10% nonfat dry milk in TBS-T (200 mM Tris, 1.37 M NaCl, + 0.1% Tween20). SuperSignal West Pico by ThermoScientific (#34080) was used as chemiluminescent substrate to visualize protein bands. A protein ladder was purchased from ThermoScientific (ThermoScientific PageRuler Plus Prestained Protein Ladder #26619). Semi-Dry-Blotting was performed with a Tris-Glycine buffer with 13 mM SDS. The ATP-IF1 Antibody (CD6P1Q) was purchased from Cell Signaling (#13268). The Opa1 antibody was a gift from A. Reichert (Düsseldorf). Subunit β was detected with an anti-ATP5B antibody (Proteintech #17247–1-AP), VDAC with an anti-VDAC antibody (Cell signaling #4661), subunit e with an anti-ATP5I antibody (Abcam #122241), actin with a monoclonal Anti-ß-Actin (Sigma #A1978) and mitofilin with a monoclonal antibody (abcam #ab137057). For the detection of bands, enhanced chemiluminescent substrate consisting of a 1:1 mix of Luminol/Enhancer & Peroxide solution was used.
For blue native PAGE, cell lysates from 2 × T175 flasks were processed as described recently [11]. In short, harvested cells were lysed, mitochondria enriched by differential centrifugation and solubilized using Digitonin 6g/g Mitochondria [42]. ATP synthase was separated by means of high resolution Blue Native (BN) electrophoresis as described in Witting et al. 2007 [43]. After separation on a 3–13% gel, the proteins were electro-blotted onto Hybond-P-polyvinylidene fluoride (PVDF) membranes (GE Healthcare) and immune-blotted with a primary antibody against subunit β of ATP synthase (Abcam 14730, mouse monoclonal) and against IF1 (CD6P1Q, Cell Signaling (#13268)). The secondary antibody was anti-mouse DyLight™ 800 from Rockland (#610145002). The signal was detected using the Odyssey® Infrared Imaging System (Li-COR bioscience). The mean density of the detected bands was determined using the analysis tool measure of ImageJ® after background subtraction.
2.6. Membrane potential measurements
The mitochondrial membrane potential, ΔΨm, was determined using the membrane potential sensitive dye TMRE (tetramethylrhodamine ethyl ester). TMRE is a cell permeant, positively-charged dye with excitation/emission maxima ~549/575 nm, which accumulates in active mitochondria due to its relative negative charge. In addition, mitochondria were stained with MTG (MitoTracker® Green FM) which was used as reference for mitochondrial mass [44]. Cells grown on glass coverslips were incubated in medium with TMRE (7 nM) for 30 min. After 30 min, cells were washed once with PBS and once with medium. Fluorescence was recorded with a cLSM (Leica SP8, equipped with a white light laser, WLL, a 63× water objective, N.A. and special hybrid detectors HyD). Z-stacks (0.36 nm steps, 7 slices) were recorded for TMRE signals (line scanning mode). Cells were kept at 37 °C and 5% CO2 during the measurement. TMRE was excited with laser line 561 nm (WLL) and emission between 620 and 750 nm was recorded. An Otsu filter mask was used as a mask for mitochondria and the background was set to NaN. The mean respective fluorescence intensities (all mitochondria per cell) were determined.
2.7. Cytosolic ATP via fluorescence imaging of Mg2+ levels
For the quantification of cytosolic Mg2+ level in cells, HeLa cells were loaded with Magnesium Green™, AM (Invitrogen). The intensity of the MgGreen signal is inversely related to the ATP concentration. For dye loading, cells were incubated in medium with 5 μM with the addition of 2% Pluronic for 45 min at 37 °C in a humidified atmosphere containing 5% CO2. The cells were washed once with medium before fresh medium was added. Fluorescence was recorded with a cLSM (Leica SP8). Cells were kept at 37 °C and 5% CO2 during the measurement. MgGreen was excited with the laser line 488 nm (WLL) and emission was recorded between 500 and 550 nm. Background subtracted images for intensity analysis were created by using the ImageJ® software (NIH Image). For this, the Otsu filter mask was used as a mask for the cells and the background was set to NaN.
2.8. Mitochondrial ATP via fluorescence imaging of ATP-Red
The BioTracker™ ATP-Red dye (Millipore) targets ATP specifically in the mitochondria of living cells. While the probe is non-fluorescent when forming a closed ring structure in the absence of ATP, the covalent bonds between boron and ribose is broken in the presence of the negatively charged ATP and the ring opens producing fluorescence [45]. For dye loading, cells were incubated in medium with 5 μM ATP-red for 15 min at 37 °C in a humidified atmosphere containing 5% CO2. The cells were washed twice with medium before fresh medium was added. Fluorescence was recorded with a cLSM (Leica SP8). Cells were kept at 37 °C and 5% CO2 during the measurement. ATP-red was excited with the laser line 561 nm (WLL) and emission of Z-stacks (3 slices, stepsize 360 nm) was recorded between 580 and 650 nm. Background subtracted images for intensity analysis were created by using ImageJ® software (NIH Image). For this, the Otsu filter mask was used as a mask for the cells and the background was set to NaN.
2.9. Cell growth
Cell growth was determined with an automatic imaging system (Cytation™, Agilent). Cells were seeded into wells of a 96 well plate and bright field images were taken every 4 h with a 4× objective. 4 wells per condition were analyzed. The overall recording time was 36 h. Only the exponential growth rate was taken into account for calculation of the growth rate. The doubling time (DT) was calculated according to the equation: DT = 0.693*t/ln(N/N0) with t (elapsed time), N0 (starting number of cells), N (number of cells at time point t).
2.10. Preparation of cell monolayers for Cryo-EM
Cells were seeded in culture plates (6 cm in diameter) and fixed with 4% prewarmed (20–37 °C) paraformaldehyde (PFA) + 0.4% glutaraldehyde (GA) in 0.1 M phosphate buffer when 80% confluency was reached. After 5 min, fixation medium was exchanged to 2% PFA + 0.2% GA for 2.5 h at RT in 0.1 M phosphate buffer or 0.1 M HEPES pH 7.4. Then, cells were washed with PBS + 0.1% glycine (e.g. Dulbecco’s without Mg & Ca), then with 0.1% BSA in PBS. Finally, cells were covered with first 2 ml 10% then 12% gelatin in PBS at 37 °C in 0.1 M phosphate puffer and left until the gelatin was dried at RT. For contrasting the cell monolayer, cells were stained as follows: the sample was rinsed once with 0.1% BSA in PBS and a second thin layer of 10% colored gelatin (Methylene blue) was added and dried at 4 °C. The gelatin was then scarified from top to bottom with a scalpel and the sample was infiltrated with 2.3 M sucrose for 24 h (48–72 h) at 4 °C. The gelatin/cell slab was removed with a thin spatula and the blocks were stained for 30 min at 4 °C with 1% Toluidine blue in 2.3 M sucrose. The gelatin block was cut into small pieces (1 mm3), mounted on clean aluminium pins and quickly frozen in liquid nitrogen before cutting into 70 nm slices with a microtome.
2.11. TEM and Electron tomography
Cells were seeded on coated petri dishes and cultivated until confluency. Then, the cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate for 20 min on ice. The cells were rinsed with 0.1 M sodium cacodylate buffer containing 3 μM calcium chloride and incubated in a mixture of 1% OsO4, 0.8% potassium ferrocyanide, 3 μM calcium chloride in 0.1 M sodium cacodylate for 30 min. Then, the cells were washed with double distilled water and stained with 2% uranyl acetate before incubating in increasing EtOH solutions (20%, 50%, 70%, 90%, 100%). Subsequently, the cells were infiltrated with a mixture of 50% EtOH and 50% Durcupan ACM resin (Fluka), and then incubated at least 3 × 2 h in 100% Durcupan and polymerized for at least 48 h at 60 °C in an oven. For TEM, ultrathin sections with a thickness of 70 nm were analyzed on a Zeiss EM902. For electron tomography, semi-thick sections with a thickness of 300–500 nm were poststained in 2% uranyl acetate and Sato lead. As fiducial markers, 20-nm sized gold nanoparticles were deposited on each side of the grid. For each reconstruction, a tilt series of images was recorded at 1-degree tilt increments on a FEI Titan 80–300 (CTWIN) IVEM/STEM. The electron microscopes were operated at 80 kV (Zeiss) and 300 kV (FEI).
2.12. Airyscan-based approach for high resolution imaging of cristae
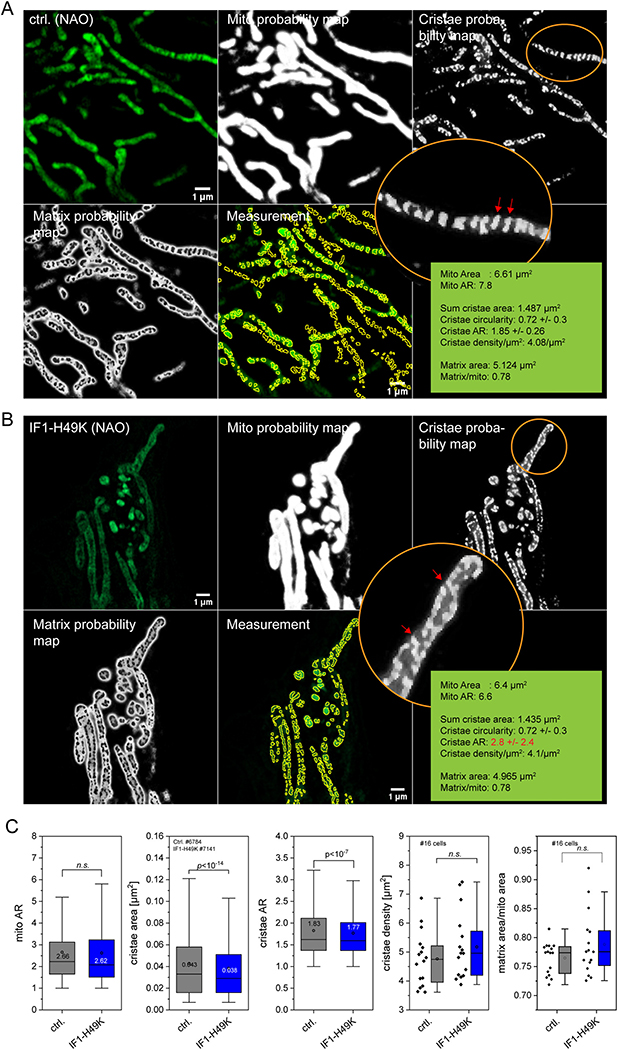
To image cristae with high resolution, a recently presented Airyscan techniques was used [46]. In short, HeLa cells were incubated with 10-N-nonyl acridine orange (NAO, 100 nM). NAO is a fluorescent probe that binds phospholipids in mitochondria, with a high affinity to cardiolipin [47,48]. Mitochondria from living cells were imaged with the Zeiss LSM980 equipped with Airyscan2 technology and with the alpha Plan-Apochromat 63×/1.46 Oil DIC M27 objective. The super-resolution (SR) mode of Airyscan was used. Prior to image analysis, raw .czi files were automatically processed into deconvoluted Airyscan images using the Zen software. For automatic analysis of cristae morphology, a trainable WEKA segmentation (TWS) protocol was used as recently described [49]. In short: The classifier for the WEKA segmentation was trained by creating three different classes: first class for the background signal, second class for mitochondrial matrix and third class for mitochondrial cristae. The classifier was trained on different control images and then used to create probability maps for all three classes for both control cells and IF1-H49K cells. For further analysis of the probability maps, a threshold of 0.65 (> 65% probability of being part of the respective class) was applied and then analyzed with the “Analyze Particles” tool of ImageJ® using a size filter of 4 pixel.
2.13. Single molecule tracking and localization microscopy
The measurements were performed as previously described [40]. In short, HeLa cells stably transfected with CV subunit ɣ fused to the HaloTag™ (SUγ-HaloTag) were labeled with tetramethylrhodamine-HaloTag Ligand (TMRHTL, 0.5–1 nM) for 20 min and imaged with a microscope equipped with a TIRF condenser and with an apochromatic 150× oil immersion objective designed for TIRF microscopy (150 × TIRF objective, N.A. 1.45, Olympus, UAPO). The TMRHTL concentration was low enough to generate distinct single molecule signals due to sub-stoichiometric labeling. A diode pumped solid-state laser (excitation 561 nm, 200 mW, Cobolt Jive 561) was used for TMR excitation and the incident angle was set to achieve a highly inclined optical light sheet [50–52]. Usually, 3000 to 5000 frames were recorded (58 Hz) with a back-illuminated EMCCD camera (Andor iXON 897, pixel size 16 μm2). Assembly and correct intramitochondrial localization were shown earlier with constructs, where similar sized EGFP or pHluorin, were genetically fused to the same position in SUγ as of HaloTag7 [53,54]. The incident angle was set to achieve a highly inclined laminated optical light sheet [55]. Usually, 3000 frames were recorded with a frame rate of 58 Hz (17 fps). Individual signals with a signal to noise ratio above 5 were fitted with a 2D Gaussian fit for localizing molecules. From all frames, a cumulative localization map was generated after movement of mitochondria during the recording time was excluded. For generating trajectory maps, single molecules in subsequent frames were connected when the following criteria were matched: they must have been within a radius of maximal 2 px (pixel size 107 nm) and the intensity distribution profile must have been identical. Only trajectories with a minimum duration of at least 85 ms (5 steps) were analyzed. The tracking algorithm was based on the multi-target tracing (MTT) algorithm described by Sergé et al. [56].
2.14. Theoretical background
Single fluorescent molecules, whose size is below the diffraction limit, project a three-dimensional diffraction pattern onto the image plane, which is defined as the point spread function (PSF). The PSF can be calculated by Abbe’s law:
| (1) |
with d being the diameter of the PSF, λ the wavelength of the emitted light and NA being the numerical aperture of the microscope’s objective. The numerical aperture describes the light gathering capability of an objective and depends on the refractive index n of the medium in which the lens is used and the half angular aperture of the objective α. The “full width at half maximum” (FWHM) criterion is used to describe a PSF, whereby FWHM is the width at 50% of the intensity maximum. For TMR (λem. = 582 nm) and an objective with NA of 1.49, the PSF diameter is approximately 390 nm.
According to the Rayleigh criterion, the resolution of the system is:
| (2) |
where r is the minimal distance between the two objects. n is the refractive index of the medium between the objective and sample, λ is the wavelength of the emitted light, α is the half angular aperture of the objective and NA is the numerical aperture of the microscope objective. Practically, this means that two separate light points can just be distinguished if the intensity maximum of the first point coincides with the first intensity minimum of the second point. Here, the theoretical resolution is ~240 nm. The pixel size was 106.7 nm for an EMCCD camera (512 × 512 px chip) with a pixel area of 16 μm × 16 μm, thus fulfilling the Nyquist-Shannon sampling theorem, which demands a 2-fold oversampling rate for the desired spatial resolution. In order to probe the spatiotemporal behavior of F1FO ATP synthase, the multi-target tracer was used. Details are described in Richter et al. [57]. First, the particle was localized by applying a 2D Gaussian fit. The accuracy of the localization highly depends on the brightness of the signal (corresponding to the number of emitted photons N), the pixel size of the EMCCD/CMOS camera and the background noise:
| (3) |
where σ is the standard error of the mean value of photon distribution, N is the number of gathered photons, a is the pixel size of the EMCCD/CMOS detector, b is the standard deviation of the background and si is the standard deviation in dimension i, with i being either x or y dimension.
2.15. Single molecule localization and tracking
Single molecules were recorded over several frames. The determined PSF was used to localize single emitters by a 2D Gaussian fit [58–60]. The multiple-target tracing algorithm (MTT) (19) was used to generate trajectories. To obtain diffusion data, the mean square displacement (MSD, r2) was determined and the diffusion coefficient was calculated according to:
| (4) |
| (5) |
where r2 is the MSD, D is the lateral diffusion coefficient, Δt, is the lag time, n is the dimension and ε is the localization error. The exponent α is a factor that distinguishes modes of diffusion, with α < 1 for anomalous diffusion. Anomalous diffusion is expected when the movement of particles is restricted by structural constraints such as physical obstacles or inclusion in micro-compartments. Further details are found in Appelhans and Busch (2018) [51].
2.16. Statistics
For statistical analysis, we used ANOVA with post hoc Tukey or Scheffé tests, as indicated, for. At least two (usually three) biological replicates were measured.
3. Results
3.1. IF1-H49K overexpression reduces mitochondrial mass and mildly changes mitochondrial morphology
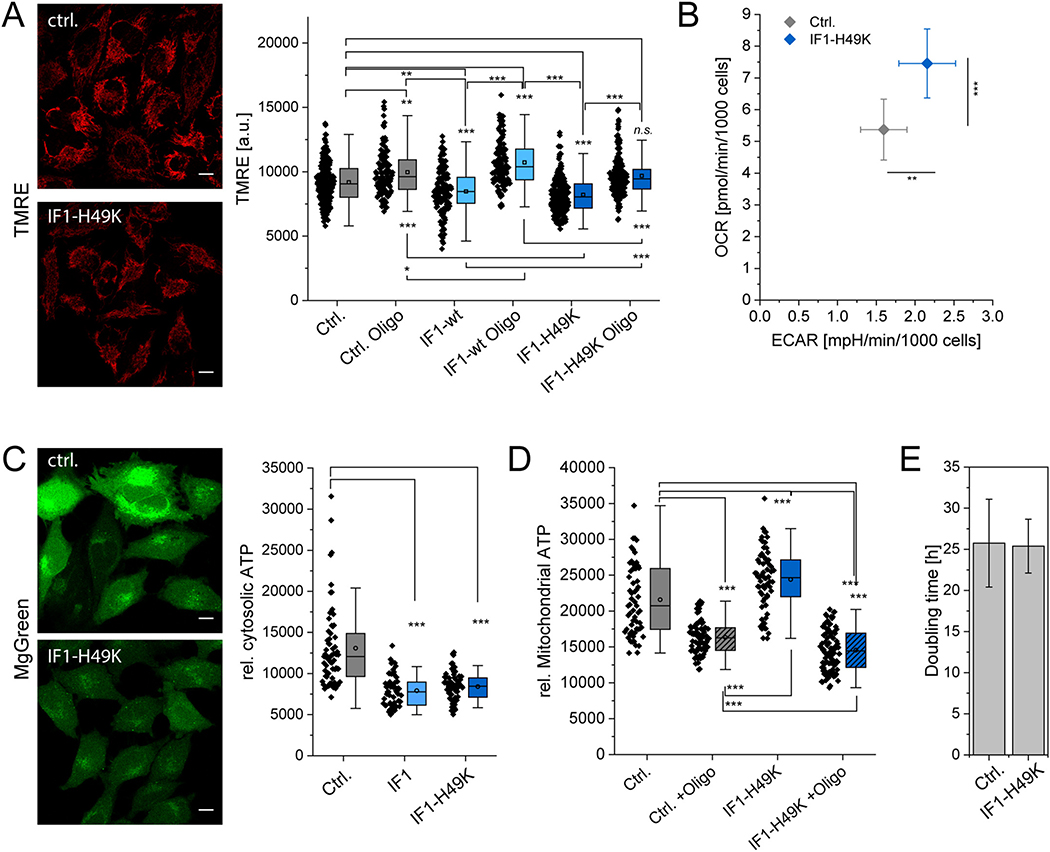
In order to study the effects of IF1 on mitochondrial architecture and the localization of ATP synthase, we first generated stable HeLa cell lines expressing either IF1-HA or the constitutively active IF1-H49K-HA. To check correct mitochondrial localization, IF1-H49K-HA was visualized by immune-staining using an HA antibody and a fluorescent secondary antibody (Cy3). The fluorescence image shows that IF1-H49K-HA clearly was mitochondrially localized since it co-localized with MitoTracker™ Green (Fig. 1A). Immuno-blotting of IF1 in cell extracts from stable cell lines revealed that in cells expressing IF1-HA total IF1 was ~2fold in IF1-HA, and ~3.5 fold in IF1-H49K, when normalized to actin (Fig. 1B).
Fig. 1.
Morphology of cells overexpressing constitutively active IF1-H49K. (A) IF1-H49K is localized in mitochondria. IF1-H49K-HA was visualized by immunostaining using anti-HA antibody in a stable cell line. Second antibody with Cy3 (λexc. 561 nm, λem. 580–620 nm). Cells were stained with MitoTracker™ Green (MTG) before fixation. The overlay of the Cy3 and MTG fluorescence channels clearly shows colocalization. (B) Candidate IF1 overexpression clones of human cervical carcinoma cells were subjected to immunoblot analysis with antibodies against IF1, the mitochondrial marker protein VDAC and cellular actin. Endogenous IF1 and IF1-HA/IF1-H49K-HA levels were determined and normalized to actin levels. Right panel: Total IF1/actin levels in the used cells lines. N = 3. (C) Morphological analysis of mitochondria using the ImageJ® plugin MiNA. Mitochondrial footprint is the total mitochondrial area in one cell. Each symbol corresponds to one analyzed cell. Statistics: N = 3, p ≤ 0.001, ***. p ≤ 0.01, **; n.s.: non significant. Scale bar: 10 μm (A,C).
Then, mitochondrial morphology and area were analyzed in cells expressing IF1-H49K. Cells were transfected with CoxVIIIa-sEcGFP [61], imaged and the mitochondrial morphology quantitatively analyzed with the MiNA (Mitochondrial network analysis) ImageJ plugin. MiNA evaluates the extent of branching within individual networks, thus identifying cells in which mitochondria are normal, hyper-fused or fragmented. We found that IF1-H49K cells have a decreased mitochondrial area (=footprint) and a decreased number of branches per network compared to wildtype (Fig. 1C). The mean branch length was unchanged. Together, these data show that IF1-H49K overexpression induces minor changes to mitochondrial morphology, in particular reduced mass and branching, probably linked to mild alterations in fusion and fission proteins.
3.2. IF1-H49K overexpression is linked to increased respiration, decreased ΔΨm and higher cytosolic ATP levels
Knockdown of IF1 affects cell metabolism and cell growth [30]. We asked, what effect IF1-H49K would have on these parameters. First, we determined the mitochondrial membrane potential, ΔΨm in presence and absence of CV inhibitor oligomycin, the metabolic profile and cellular and mitochondrial ATP levels. To determine ΔΨm, we stained the cells with TMRE (Tetramethylrhodamine ethyl ester), a dye commonly used to determine the mitochondrial membrane potential [62]. The ΔΨm was significantly lower in the presence of constitutively active IF1-H49K (Fig. 2A). This is in accordance to what was found earlier in HeLa cells transiently transfected with IF1 or siRNA for IF1 and stable IF1-KD HeLa cells [25,30]. It indicates that IF1-H49K inhibits ATP hydrolysis and thus proton pumping from the matrix into the ICS. When oligomycin was added, the ΔΨm increased in all cell lines indicating that previously ATP synthesis took place with consumption of the proton gradient. The data strongly supports the model that ATP synthase is at least partially active in the reverse mode under glucose conditions and that IF1-H49K suppresses this reverse activity. This means that ATP production had not been at its maximum before, but only when ATP hydrolysis was inhibited. We coin this a mixed ATP synthase function and suggest that at the same time two subpopulations of ATP synthase exist: a subpopulation that synthesizes ATP and a subpopulation that hydrolyzes ATP. These results also suggest that IF1-H49K was constitutively active and only inhibited the hydrolysis function, but not the ATP synthase activity as described before [26,63,64].
Fig. 2.
Effect of IF1-H49K on mitochondrial function. (A) Mitochondrial membrane potential under the influence of overexpressed IF1-wt and IF1-H49K. HeLa cells were stained with 7 nM TMRE (Scale bars: 10 μm). Right panel: Mean grey values of the TMRE signal per cell (N = 3). (B) Metabolic profiling of control and IF1-H49K cells: both cell lines display a mixed metabolic phenotype with moderate oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) indicating glycolytic activity. The OCR of IF1-H49K cells was corrected for the decreased mitochondrial footprint (N = 2, SEM are shown). (C) Indirect determination of cytosolic ATP levels by MgGreen staining of free Mg2+. The MgGreen fluorescence signal is inversely related to the ATP concentration. N = 2 replicates. Significance levels were determined by One Way ANOVA with post hoc Scheffé test. (D) Determination of mitochondrial ATP levels in mitochondria with active and inhibited ATP synthase via ATP-Red fluorescence. (N = 2 experiments, SD). Every dot is the mean ATP red fluorescence of one cell. (E) Doubling time DT of control cells and cells expressing IF1-H49K. DT is expressed in hours (N = 2 experiments, SD). Scale bars: 10 μm (A, C).
Next, we determined the metabolic profile of control cells and cells expressing IF1-H49K. To do so, oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) of live cells were determined by an automatic flux analyzer (Seahorse/Agilent XF96, 30,000 cells per well). The OCR/ECAR ratio indicates a mixed glycolytic/respiratory metabolism, whereby IF1-H49K expressing cells had a significantly increased respiratory and glycolytic activity. The OCR was corrected for the decreased mitochondrial footprint in the cells (Fig. 2B). It was shown before that in cells with stimulated respiration, the number/area of mitochondria per cell decreases [65].
Ensuing, we checked whether the increased metabolic activity in IF1-H49K cells affected cellular and mitochondrial ATP levels. Cellular ATP levels were indirectly determined by MgGreen™ fluorescence (Fig. 2C). MgGreen™ is a dye that fluoresces in the presence of free Mg2+, but usually most Mg2+ in cells is complexed, mainly by ATP. When ATP levels decrease, Mg2+ is set free and the fluorescence intensity of MgGreen™ increases. Thus, the fluorescence of MgGreen™ is an indirect indicator for ATP content. In IF1-wt and IF1-H49K cells, we found a significant decrease in MgGreen™ fluorescence related to an increase in the cellular ATP content when compared to wildtype cells (Fig. 2C). This might be the result of decreased mitochondrial ATP hydrolysis in IF1-overexpressing cell and more ATP exported to the cytosol. Indeed, mitochondrial ATP levels were increased in IF1-overexpressing cells (Fig. 2D). To determine mitochondrial ATP, BioTracker™ ATP-Red was added to the cells. ATP-Red is targeted to mitochondria of living cells and shows fluorescence proportional to the mitochondrial ATP concentration [45]. In the presence of oligomycin, an inhibitor of ATP synthesis, the ATP-Red signal was significantly lower than in control conditions. In IF1-H49K expressing cells, the dye indicated a significant increase of mitochondrial ATP levels. Interestingly, ATP levels were lowest in IF1-H49K expressing cells that were in treated with oligomycin, indicating a strong ATP synthase activity before inhibition.
However, the increased mitochondrial activity had no influence on the cell growth rate (Fig. 2E). In comparison to wildtype cells, the IF1-H49K-HA cells showed the same doubling time (Ctrl.: 25.75 ± 5.34 h vs. IF1-H49K-HA: 25.38 ± 3.27). Together, these data demonstrate a positive effect of IF1-H49K overexpression on mitochondrial bioenergetics, especially ATP synthesis.
3.3. Inhibition of the ATPase function by IF1-H49K changes the spatio-temporal organization of the ATP synthase
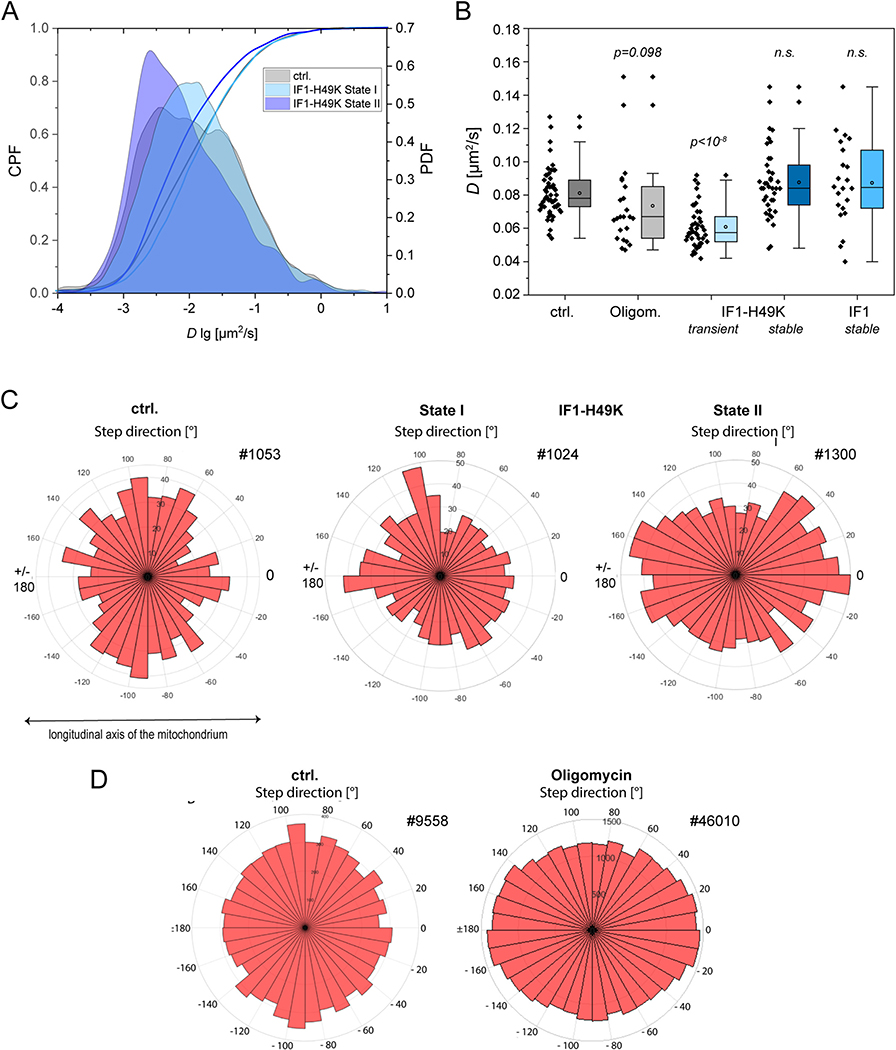
We wondered whether increased levels of IF1 and constitutively active IF1 would influence the spatio-temporal organization of the ATP synthase. IF1 is a known to crosslink dimers of ATP synthase [18,33]. It could be expected that IF1 overexpression would reduce the mobility of ATP synthase in the inner membrane and possibly also its localization. To quantify the mobility of membrane proteins, FRAP and single particle tracking are feasible methods. However, FRAP measurements tend to overestimate the mobility of molecules since only mobile molecules are recorded [66]. We therefore decided to record the mobility of single ATP synthase particles by a technique based on single molecule tracking and localization microscopy (TALM) [67]. Using the multiple-target tracing algorithm (MTT) [56], trajectory maps of F1FO ATP synthase were obtained. Fig. 3 shows typical trajectory maps of the F1FO ATP synthase (CV) in WT HeLa cells and cells stably overexpressing IF1 and IF1-H49K, respectively. Cells were grown in medium supplied with 5.6 mM glucose resulting in a mixed glycolytic/respiratory metabolic profile [68]. As the trajectory maps of F1FO ATP synthase under normal conditions show, the F1FO ATP synthase displays a confined diffusion in cristae as depicted in the orthogonal course of single trajectories (Fig. 3A, inset). Based on the mean step length over 8 steps, three subpopulations of the F1FO ATP synthase with different diffusion behavior could be categorized: mobile, less mobile and immobile molecules with different diffusion coefficients D1 (mobile), D2 (less mobile) and D3 (immobile). This specific mobility behavior was described before and depends on the activity mode of ATP synthase [40,68]. When the F1FO ATP synthase was tracked in cells overexpressing IF1-HA, the pattern only marginally changed (Fig. 3B). However, when CV was localized and tracked in IF1-H49K-HA over-expressing cells, two types of mobility patterns evolved: one similar to the control conditions and one with a clear CV localization and movement at the border of mitochondria and only few trajectories inside mitochondria where cristae are expected (Fig. 3C). These patterns were attributed to a state I and a state II based on ultrastructural data (see Section 3.5). Diffusion coefficients and the directionality of movement in relation to the longitudinal and orthogonal axis of the respective mitochondrion were calculated for the diffusion of the F1FO ATP synthase shown in the trajectory maps in Fig. 3.
Fig. 3.
The spatio-temporal organization of ATP synthase is affected by IF1-H49K. (A) Mobility pattern of F1FO ATP synthase under control conditions. Recording: 59 Hz, 3000 frames, total recording time 51 s (Supplementary material movies S1–S2). The panels show trajectory maps of the F1FO ATP synthase in two individual mitochondria. Inset: selected trajectories from a third mitochondrion showing orthogonal trajectories (arrowheads). Trajectories of the F1FO ATP synthase were generated by using the Multi-Target-Tracing-Tool (MTT). Every single trajectory in one color. (B) Mobility pattern of F1FO ATP synthase in cells expressing IF1-HA, see movie S3 in supplementary information. Recording rate: 59 Hz, 3000 frames. (C) Mobility pattern of F1FO ATP synthase in cells expressing IF1-H49K-HA. Recording: 59 Hz, 3000 frames (51 s) (Supplementary material movies S4–S5). Two types of trajectory maps were distinguishable and classified as state I and state II. Scale bars: 1 μm (A, B, C), 0.5 μm (inset A).
First, the probability density function (PDF) for the diffusion coefficients (D log) is depicted for the F1FO ATP synthase under control conditions and with constitutively active IF1 (IF1-H49K) in Fig. 4A. While the course of the PDF was similar in the control and under IF1-H49K state I, a clear shift towards lower diffusion coefficients is observable for state II conditions. This is also reflected in the cumulative probability function (CPF) for D, which is similar for control and state I conditions, but distinct for state II. As further can be seen in Fig. 4A, the PDF for the control displays three peaks indicating three subpopulations with different mobility. As we described earlier [40,68], this corresponds to a mobile, less mobile and immobile/confined fraction of ATP synthase. The respective diffusion coefficients can be obtained by fitting. Under control conditions, the diffusion coefficients for mobile, less mobile and immobile fractions of ATP synthase shown in Fig. 3 were the following: D1 = 0.080 μm2/s (14%), D2 = 0.018 μm2/s (54%), D3 = 0.006 μm2/s (30%) for control conditions. For the F1FO ATP synthase in stably IF1 expressing cells, the values were similar: D1 = 0.081 μm2/s (17%), D2 = 0.021 μm2/s (48%), D3 = 0.009 μm2/s (36%). When the diffusion coefficients were determined for the F1FO ATP synthase in stably IF1-H49K expressing cells, state II, D1 was lower: D1 = 0.048 μm2/s (28%), D2 = 0.018 μm2/s (28%), D3 = 0.007 μm2/s (19%). This analysis was performed exclusively for the depicted examples from Fig. 3. Next, we asked whether this would also be reflected in a cohort analysis with multiple mitochondria from different cells. We only compared D for the mobile fractions of ATP synthase under different conditions (Fig. 4B). Per condition, > 10,000 trajectories from the F1FO ATP synthase in different cells and from 2 to 3 technical replicates were analyzed. In addition to an IF1-H49K overexpression, which results in the inhibition of reverse ATP synthase (ATP hydrolysis) function, Oligomycin was also tested. Oligomycin inhibits forward and reverse ATP synthase activity. Furthermore, we tested transient transfection with IF1-H49K. Tendentially, Oligomycin resulted in a decrease of the diffusion coefficient D1 of ATP synthase (p = 0.098, ANOVA), while transient transfection of stable CV γ-HaloTag expressing cells with IF1-H49K clearly decreased the D1 of F1FO ATP synthase (p < 10−8). In the cohort measurements, no significant difference in the average mobility was found for cells stably expressing IF1 or IF1-H49K. This can be explained by the similarity of trajectories in control and IF1 expressing cells (state I). For cells stably expressing IF1-H49K, the statistical averaging has apparently averaged out the differences between state I and state II. Finally, we determined the preferential direction of trajectories under the different conditions. As described before, the F1FO ATP synthase is confined to cristae under the glycolytic but non-inhibited condition [67]. This is reflected in a preferential orthogonal movement, which can be quantified. The analysis of four subsequent step directions with respect to the longitudinal axis ( ± 180° and 0°) and the orthogonal axis (from −90° and +90°) is represented on a graduated disk. As the distribution on the graduated disc shows, orthogonal movement of the F1FO ATP synthase dominated for the control (Fig. 4C). Also, the trajectories of the IF1-H49K state I example still show a tendency for preferential orthogonal movement. In IF1-H49K state II, the integrated directionality of four subsequent steps was clearly longitudinal as the graduated disc shows. The directionality analysis thus reveals a different spatiotemporal behavior of the F1FO ATP synthase in this case.
Fig. 4.
Partial inhibition of ATPase function by constitutively active IF1 and complete inhibition of the F1FO ATP synthase by oligomycin mobility in the absence and presence of IF1-H49K-HA. (A) Diffusion coefficients for ATP synthase under control conditions (5.6 mM Glucose, no inhibition), and in the presence of IF1-H49K-HA are shown. The probability density function (PDF) of log D and the cumulative probability function (CPF) are shown. State I and state II refer to the spatio-temporal characteristics of ATP synthase in Fig. 3C. Ctrl.: F1FO ATP synthase mobility without constitutive expression of IF1-H49K. IF1-H49K state I and state II: F1FO ATP synthase mobility in cells expressing IF1-H49K. (B) D1 for the mobile fraction of inhibited (with IF1-H49K) and non-inhibited ATP synthase is shown. Oligom.: Oligomycin (10 μM, 30 min) as inhibitor for ATP synthase forward and reverse function. (C) Analysis of the step directions in relation to the longitudinal axis ( ± 180° and 0°) and the orthogonal axis (from − 90° and + 90°) of the respective mitochondrion. For the determination of the directionality, the direction of 4 successive steps was integrated and displayed on a graduated disc. Trajectories analyzed: ctrl. #1053; IF1-H49K-state I #1024 and IF1-H49K state II #1300 (A, C). (D) Orientation of trajectories of ATP synthase under control and oligomycin inhibited conditions.
3.4. Oligomycin inhibition of the F1FO ATP-synthase affects the directionality of F1FO ATP-synthase movement but not the diffusion coefficients
IF1 only inhibits the reverse function of the ATP synthase. Thus, the observed effects could be related to the elimination of the ATPase function of the ATP synthase. We next tested the effect of oligomycin, a known inhibitor of the forward and reverse functions of the ATP synthase, on its spatiotemporal organization. Trajectory maps were generated and analyzed as before. The diffusion coefficients obtained by fitting step length diagrams were similar as determined before for control conditions: D1 = 0.087 μm2/s (28%), D2 = 0.027 μm2/s (42%), D3 = 0.010 μm2/s (30%). However, when compared to uninhibited conditions, the mean directionality of the ATP synthase changed: In the presence of oligomycin, the analysis of the net directionality of 4 subsequent steps showed a significant increase of the ATP synthase with longitudinal movement similar to the IF1-H49K inhibitory effect in state II. Thus, both inhibitors affected the spatiotemporal organization of the ATP synthase in a similar way.
3.5. Constitutively active IF1 induces changes in the mitochondrial ultrastructure
We next sought to dissect whether the altered spatiotemporal behavior of the ATP synthase in IF1-H49K expressing cells, which we classified as state II, would be related to changes in the ultrastructure. IF1 is known to promote ATP synthase oligomerization [18,69], which is a determining factor of cristae shape [10,13]. Thus, we analyzed the 2D and 3D-ultrastructure of mitochondria in IF1 and IF1-H49K overexpressing cells from transmission electron microscopy (TEM) images and tomograms, which provide a 3D view of selected mitochondria. As the TEM images show, IF1 expressing cells possessed several mitochondria with altered cristae, including arch-like (curved) and shorter cristae (Fig. 5A). These cristae were classified as truncated. While the average mitochondrial length was not altered in IF1-H49K expressing cells, the mitochondrial width was significantly increased. This fits well with the recorded trajectory map shown in Fig. 3C, state II conditions. Stable IF1-H49K expressing cells also had a decreased number of cristae per mitochondrial length and more truncated cristae with shortened length (Fig. 5B).
Fig. 5.
Mitochondrial ultrastructure in cells with constitutively active IF1. (A) TEM of HeLa WT and IF1-H49K-HA cell monolayer. (B) 100 cristae were analyzed for each condition. Determination of mitochondrial length, mitochondrial width, number of cristae per μm mitochondrial length and cristae with shorter length (truncated). Statistical analysis of the parametric data was performed with one-way ANOVA with post hoc Scheffé test. Statistical analysis of the non-parametric data was performed as a Kruskal-Wallis test. Scale bars: 1 μm (A).
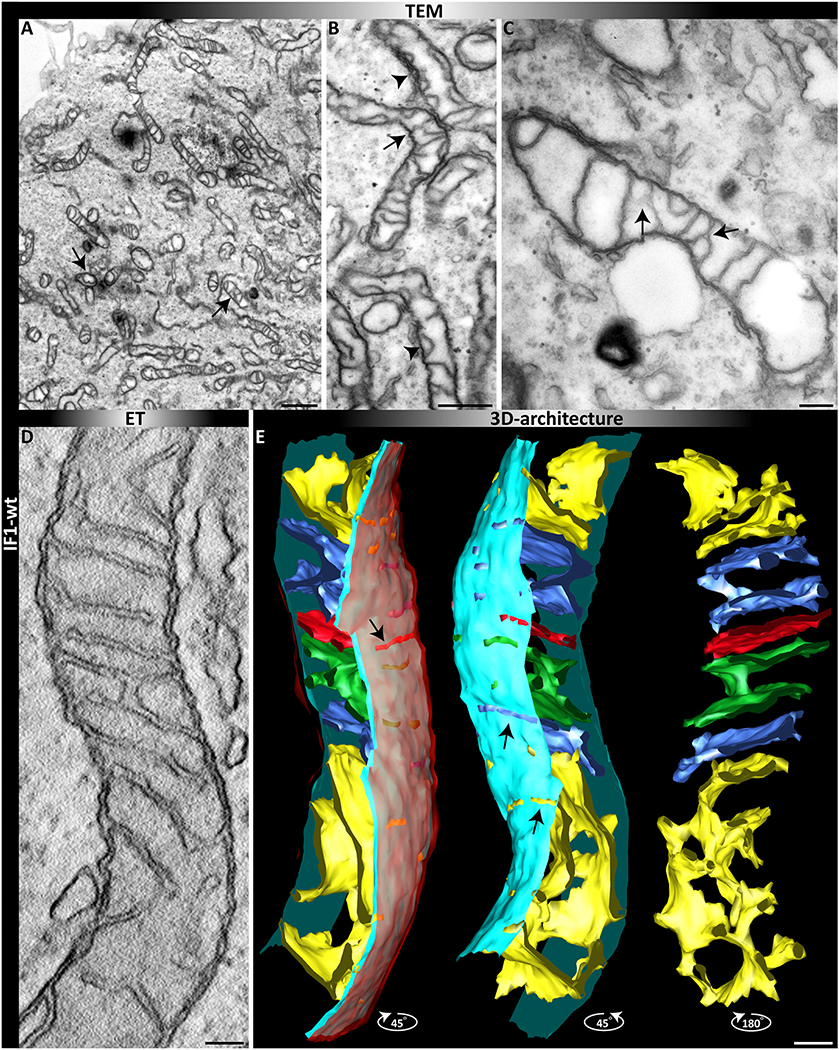
Since TEM images only depict a 2D view and more complex structures might be overseen, we also performed electron tomography (ET). This provides 3D views of single mitochondria with their full cristae architecture. The 3D ET of an individual mitochondrion in cells expressing stable higher IF1 levels shows normally orthogonally arranged mitochondria in the upper part of the mitochondrion, while the lower part contains only one large network-like crista, probably derived from several merged cristae (Fig. 6E). This specific crista is not visible in the single slice view of the same tomogram and it can be assumed that several of these specific cristae are not detected in 2D micrographs of TEMs.
Fig. 6.
2D and 3D view of mitochondria in cells expressing IF1. (A–C) TEM after fixation of monolayers of cells. (A) Overview. (B) Magnified view, arrows indicate altered cristae structures such as arches and branches. (D-E) ET of a single mitochondrion with altered ultrastructure in the presence of IF1. (D) Single tomographic slice. (E) 3D reconstruction after segmentation; different view angles are shown. From left to right: Full segmentation. The outer membrane (OM) is depicted in translucent red, the light blue indicates the inner boundary membrane and cristae are in different colors. Mid: view from a different angle without the OM. Right: only cristae are shown. Scale bars: 1 μM (A); 500 nm (B), 200 nm (C); 100 nm (D, E).
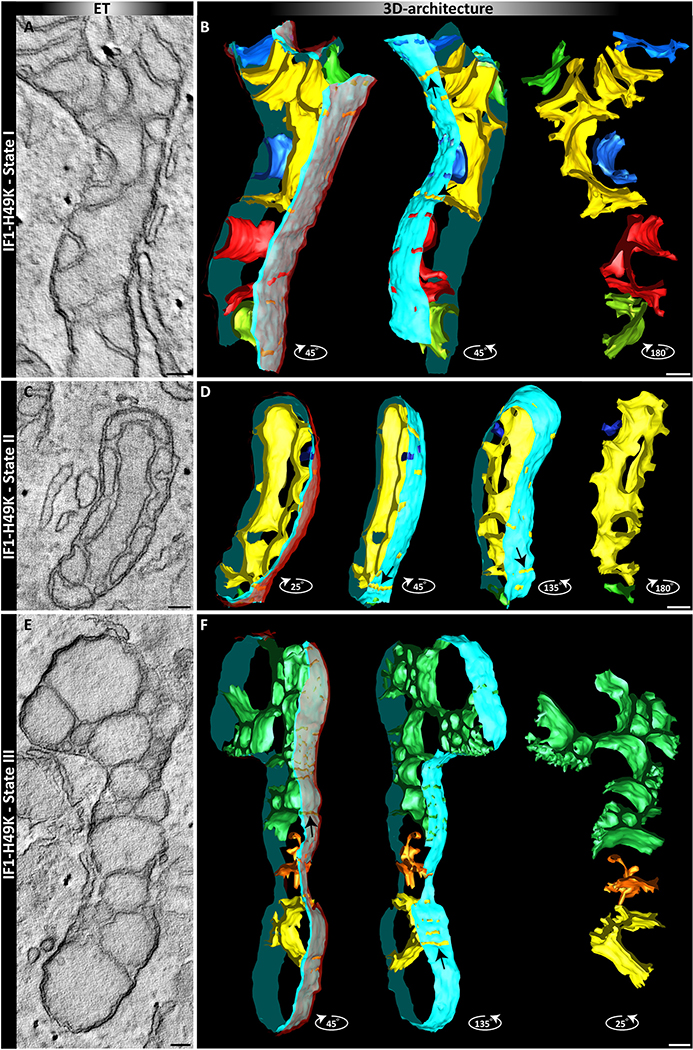
Subsequently, we determined the ultrastructure of mitochondria in IF1-H49K expressing cells. 3D reconstructions using ET revealed protuberant changes in the ultrastructure, already indicated in the single slices. We classified three states (I-III) related to increasing deformation of cristae. While state I resembles the architecture found in IF1(-HA) overexpressing cells, mitochondria in state II exhibit an even more pronounced alteration of the architecture. In state III, cristae display a complete deformation with some cristae having lost any connection to the inner membrane and others with broad slit-like crista junctions. Although the shape of the cristae changed considerably, the area ratio of cristae to OMM remained the same (supplementary Fig. S1). These data show that stable expression of IF1-H49K in glycolytic cells induces a gradual restructuring of the inner membrane (Fig. 7).
Fig. 7.
3D views of mitochondria in cells with constitutively active IF1-H49K. Tomograms of single mitochondria with altered ultrastructure in the presence of IF1-H49K. Three states (I-III) were classified. Left panels: single tomographic slice. Right panels: 3D reconstruction after segmentation, different view angles are shown. From left to right: Full segmentation. The outer membrane is depicted in translucent red, the light blue indicates the inner boundary membrane and cristae are in different colors. Mid: view from a different angle without the OM. Right: only cristae are shown. Scale bars: 100 nm.
3.6. High-throughput quantification of cristae architecture in living cells confirms cristae area reduction in IF1-H49K cells
In order to quantify cristae architecture in an unbiased way, we collected high-resolution images of mitochondria stained with 10-N-nonyl acridine orange (NAO) with a laser scanning confocal microscope equipped with an Airyscan2 detector. NAO labels mitochondrial lipids with high affinity to cardiolipin [70]. These images were then semi-automatically analyzed by using a trainable WEKA segmentation tool, available as an open-source plugin in Fiji. This approach was successfully used to quantify mitochondrial and cristae shape, area and aspect ratio before [49]. In Fig. 8, the processing is shown for a representative image for control and IF1-H49K cells, respectively (Fig. 8A, B). More images are shown in Supplementary Figs. S2 and S3. Then, all recorded images from both conditions were processed using the machine learning approach without any pre-selection. The mitochondria area, aspect ratio, mean cristae area and aspect ratio was calculated for control cells (27 cells), in total 6784 cristae were identified in 666 mitochondria. The same was conducted with images from IF1-H49K cells (51 cells; 1165 mitochondria, 7141 cristae; N = 3). The statistic evaluation resulted in a significant decrease of the cristae area and cristae aspect ratio in IF1-H49K cells, while mitochondrial aspect ratio and cristae density were not changed (Fig. 8C). The obtained data for cristae density (ctrl.: 4.76 ± 0.93 μm−2; IF1-H49K: 5.17 ± 1.15 μm−2) were in the range for what was recently reported for HeLa cells (~5.8 μm−2; [49]), the same for cristae area (ctrl.: 0.043 ± 0.036 μm2 compared to 0.049 taken from SIM images in Segawa et al. [49]). Also, the cristae aspect ratio of control HeLa cells (1.82 ± 0.69) is well in line with the published value for HeLa cells (~1.9, Segawa et al. [49]). Albeit we found a minor decrease of the cristae area, from 0.043 ± 0.036 μm2 in control cells to 0.038 ± 0.032 μm2 in IF1-H49K expressing cells, the matrix per mitochondrion area did not change significantly. However, the data were generated by averaging total matrix per total mitochondrial area in one image, which might cause high variability.
Fig. 8.
Semi-quantitative high through put analysis of cristae morphology in control and IF1-H49K expressing cells. (A, B) Live cell Airyscan images of HeLa mitochondria, control (A) and IF1-H49K (B). Cells were stained with NAO (100 nM), which was present during the measurement. Note, that from left to right original images are shown, followed by probability maps of mitochondria, cristae and of matrix space, and the masks used for cristae measurement. Circled areas show zoomed in regions of cristae probability maps, cristae are marked with red arrowheads. Images were recorded with AiryScan2 (Zeiss) in superresolution (SR) mode equipped with an alpha Plan-Apochromat 63×/1.46 Oil DIC M27 objective on the Zeiss LSM 980. Raw images were automatically processed into deconvolved Airyscan images using the Zen software. (C) Quantification of cristae area between control and IF1-H49K expressing cells, stained with NAO. N = 3 experiments, 16 mitochondria per condition. Data are presented as Box and Whisker Blots. P-values are shown in the panels, n.s.: non-significant. Scale bar: 1 μM (all panels).
Together, the quantitative analysis of the mitochondrial inner membrane from live cell imaging provided sufficient statistics to confirm the reduction in cristae area in IF1-H49K cells suggested from the single tomograms.
3.7. Only the localization but not the mobility of ATP synthase in mitofilin overexpressing cells is changed
In order to test whether an ultrastructural modification of the IMM per se would change the mobility of ATP synthase, we recorded the spatio-temporal behavior of ATP synthase in cells that overexpressed mitofilin (Mic60). Mitofilin is part of the MICOS complex, the mitochondrial contact site. It has membrane-bending activity and thus controls cristae morphology [71–75]. We first generated a stable mitofilin-overexpressing cell line. The analysis of TEM images (Fig. 9A–B) clearly shows an altered inner membrane structure with a significant portion of bent cristae due to mitofilin activity (Fig. 9C). The pattern of the inner membrane was displaying clear irregularity when compared with the cristae pattern in control cells. Therefore, the IMM structure displayed similar features as that in IF1-H49K overexpressing cells, and we used it to determine the influence of the ultrastructure on the spatiotemporal organization of the ATP synthase. The trajectory map of ATP synthase in a single mitochondrion reflected the abnormal IMM microcompartment with no orthogonal trajectory pattern as found in controls (Fig. 9D vs. Fig. 3A). However, the calculated diffusion coefficient Dmob = 0.073 ± 0.015 μm2/s of the mobile F1FO ATP synthase fraction in mitofilin overexpressing cells was similar to the diffusion coefficient in control cell (Dmob = 0.081 ± 0.014 μm2/s) and the fraction of mobile F1FO ATP synthase was also the same. No difference was found when compared to data for F1FO ATP synthase in IF1-H49K expressing cells, either stably or transiently transfected (Fig. 9E–F). This suggests, that the alteration of the ultrastructure of the IMM per se does not affect the mobile fraction of ATP synthase in the IMM. The mitofilin/Mic60 expression level in mitofilin-GFP overexpressing cells was more than 10fold higher than in control cells or IF1-overexpressing cells used in this study (Fig. 9G). Finally, we checked, whether IF1 was upregulated in mitofilin over-expressing cells, but this was not the case (Fig. 9H).
Fig. 9.
Mitofilin overexpression induces ultrastructural changes but does not alter the fraction of mobile ATP synthase. (A) TEM of HeLa cells stably overexpressing mitofilin-GFP. (B) Single mitochondrion with normal and aberrant cristae, lower panel: normal cristae false-colored in blue and aberrant cristae false-colored in red. (C) Determination of aberrant cristae (cell number: ctrl.: > 10 and mitofilin: 5; mitochondria: ctrl.: > 35 and mitofilin: 21; cristae: ctrl.: > 450 and mitofilin: 166). (D) Trajectory map of F1FO ATP synthase (labeled at SU γ-HaloTag with TMRHTL) in one mitochondrion. Cumulative image of 3000 subsequent frames (recording 100 Hz). (E) Diffusion coefficients of F1FO ATP synthase in control cells, cells overexpressing mitofilin and cells expressing IF1-H49K transiently or stably. (F) Percentage of mobile ATP synthase particles under the same condition as in (E). (G) Quantification of mitofilin protein expression levels in ctrl. cells and IF1-H49K-HA expressing cells by quantitative RT-PCR (N = 3 replicates). (H) Immunostaining for IF1 in mitofilin-GFP expressing cells in comparison to other cell lines used in this study. The IF1 levels were not higher when normalized on actin or VDAC. N = 5 replicates. Scale bars: 0.5 μm (A), 300 nm (B, E). Statistics: ANOVA with post hoc Scheffé test, n.s.: non-significant, p ≤ 0.001: ***, error as standard deviation of the mean, SDM. Scale bars: 300 nm (A), 100 nm (B) and 1 μm (E).
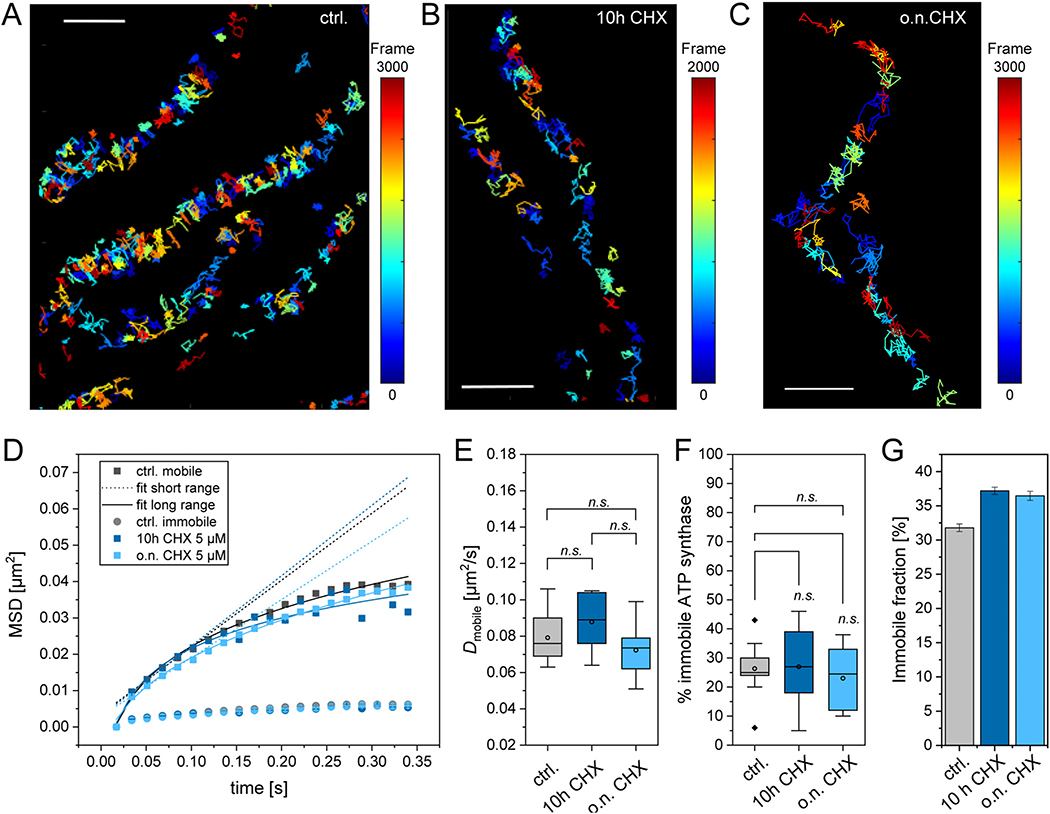
3.8. Blocking de novo protein synthesis has no effect on the mobility of ATP synthase and only assembled ATP synthase is tracked
Finally, we asked whether the mobile fraction measured via fluorescence-labeled subunit γ, might be non-assembled ATP synthase. In human cells, the free F1 domain is anchored to the IMM by binding first to the peripheral stalk [76]. We recorded trajectory maps of ATP synthase labeled at SU γ by TMR-HTL staining of the fused HaloTag. The trajectory map of the control showed confined ATP synthase in accordance with preferential localization in the cristae (Fig. 10A). In order to identify possible distortion of the trajectories by lateral mitochondrial movement, trajectories were depicted in accordance to the time of their appearance. When trajectories from the first frames overlaid with trajectories from the last frames (51 s later), no mitochondria moved significantly and trajectories were evaluated further. The principle pattern of ATP synthase mobility was the same, when cells were inhibited with cycloheximide (CHX) (Fig. 10B, C). CHX interferes with the translocation step in protein synthesis and thus blocks eukaryotic translational elongation. CHX treated cells (5 μM) stopped growing (data not shown). The trajectories of F1FO ATP synthase in the presence of CHX were analyzed to determine the diffusion coefficients and fractions of mobile and immobile ATP synthase. The mean square displacement plots show no conspicuous difference between control conditions and CHX treated cells (application of CHX for 10 h and o.n., respectively) (Fig. 10D), although on the long range, the MSD of ATP in CHX treated cells was tendentially smaller. The diffusion coefficient D was then determined according to Eq. (2) from the MSD. Data from single cells (see e.g. detail in Fig. 9A) were then used to calculate the mean D of the mobile fraction. The diffusion coefficients for the mobile fraction were the same for ATP synthase measured under control conditions and in the presence of CHX (Fig. 10E). Also, the fraction of the immobile ATP synthase particles did not change, when CHX had been applied for more than 10 h (Fig. 10F). Related step length diagrams are shown in Supplementary Fig. S4. When all trajectories of all measurements of one condition were pooled, and the immobile fraction calculated from the pool, the data report a small increase in the presence of CHX (4–5%) (Fig. 10G). However, this variation is less than the standard deviation of the mean value when comparing individual cells (Fig. 10H), and therefore no significant effect of CHX can be read from the data. Since blocking of de novo protein synthesis of subunit γ, which is nuclear encoded, had no effect on the measured mobility, we have no indication that the mobile fraction of ATP synthase is mainly a non-assembled subpopulation of F1. Finally, it should be mentioned that completely detached and thus freely diffusing F1 would diffuse much too fast to be tracked with our recording settings (56 Hz).
Fig. 10.
Blocking of de novo protein synthesis has no impact on the mobility of ATP synthase. (A) Trajectory map of ATP synthase without CHX treatment, no blocking of de novo protein synthesis. A cumulative image of single trajectories from 3000 frames (recording: 56 Hz) is shown. The individual trajectories are color-coded according to their appearance time (frame). (B) Trajectory map of ATP synthase in the presence of CHX (5 μM for 10 h), color indicating time of appearance, the map is a cumulative image of trajectories from 2000 frames (recording: 56 Hz). (C) Trajectory map of ATP synthase in the presence of CHX (5 μM for o.n.), color indicating time of appearance, the map is a cumulative image of trajectories from 3000 frames (recording: 56 Hz). (D) Mean square displacement, MSD, (sliding window, 5 frames) of pooled trajectories from control (black rectangles: mobile fraction, dots: immobile fraction), treatment with CHX for 10 h (dark blue rectangles: mobile fraction, dots: immobile fraction) and treatment with CHX for overnight (o.n., light blue rectangles: mobile, dots: immobile fraction). Dotted lines: linear fit for short range MSD (2–5 frames), solid lines: fit for long range MSD (2–20 frames). (E) Diffusion coefficients for the mobile fraction of ATP synthase. (F) Fraction of immobile ATP synthase under the different conditions, calculated from means of individual cells. (G) Fraction of immobile ATP synthase under the different conditions, calculated after pooling all trajectories of one condition. Statistics: ctrl. 9 cells, 45,390 trajectories; 10 h CHX 6 cells, 14,324 trajectories; o.n. CHX 10 cells, 34,369 trajectories. Significance was determined by One Way ANOVA with post hoc Scheffé test, cut off significance level p = 0.005, SDM shown. Scale bars: 1 μm (A, B).
3.9. The expression level of OPA1 in IF1 overexpressing cells is increased and shifted towards the short form of OPA1
We next asked whether the observed cristae restructuring would be exclusively an IF1-H49K effect or whether other factors may be involved. The protein optic atrophy 1 (OPA1) is a dynamin-related protein associated with the maintenance of mitochondrial function, structure and dynamics. Proteolytic cleavage of inner membrane-anchored long OPA1 (L-OPA1) results in short OPA1 (S-OPA1) and was described to influence the morphology of mitochondria. The ATP synthase and OPA1 in addition are functionally coupled [22,77–80]. First, we analyzed expression of transcripts of IF1 and OPA1 by semiquantitative RT-PCR in control and IF1-H49K cells. The levels of IF1 and OPA1 mRNA were increased compared to control cells (Fig. 11A). We further analyzed the expression levels of the fission proteins Drp1 and hFis, which are also involved in forming mitochondrial shape [81,82]. The expression of the cristae junction protein IMMT was not changed (data not shown, see Fig. 9). The fission protein Drp1 showed increased expressions levels in IF1-overexpressing cells. In addition to semi-quantitative measurements, we also performed quantitative real-time PCR to compare expression levels for OPA1 and Drp1. Furthermore, we tested for the fusion/tethering proteins Mfn1 and Mfn2. In stable IF1 and IF1-H49K cells, OPA1, Drp1, Mfn1 and Mfn2 expression was upregulated (Fig. 11B). OPA1 was tested with two different primer pairs resulting in products of 112 and 562 bp size, both indicating upregulation of OPA1. Next, we determined OPA1 protein levels and processing by immunodetection (Fig. 11C). The immuno-blot shows several distinct bands related to divers long (L-OPA1; a,b) and short (S-OPA1; d,e) OPA1 forms [78]. While the overall protein level of L + S-OPA1 was not significantly changed, we found a trend to higher quota of S-OPA1 forms in IF1 expressing cells (see also Supplementary Fig. S5). As a recent study shows, L-and S-OPA1 affect cristae formation, whereby S-OPA1 dominance maintains oxidative phosphorylation and cristae structure [83]. From these data, we conclude that changes in the overall OPA1 level and a shift towards the short forms of OPA1 primarily contribute to the observed mitochondrial ultrastructure aberrations in IF1 expressing cells.
Fig. 11.
Relative levels of fission and fusion proteins including OPA1 in IF1 overexpressing cells. (A) Semiquantitative RT-PCR blot showing relative mRNA levels of GAPDH, IF1, OPA1, and Drp1 in control cells and cells expressing IF1 (blots: inverse). (C) Immuno-staining of OPA1 in control cells and cells expressing IF1 and IF1-H49K, respectively (antibody α-OPA1: courtesy of A. Reichert lab). OPA1 appears in long and short isoforms due to proteolytic processing of L-OPA1. Line plot profile: relative levels of S- and L-OPA1 protein bands along the cross section indicated with a blue line in the immunoblot. Right panel: Ratio of L-OPA1 and S-OPA1 peak signals indicating the L/S OPA1 relation (peak ratio b/d) for two experiments.
3.10. F1FO ATP synthase oligomerization in IF1H49K expressing cells was not significantly increased
Finally, we checked whether in IF1-H49K overexpressing cells the assembly of ATP synthase monomers to dimers and oligomers has been changed. We determined the relative levels of ATP synthase dimers vs. monomers by quantitative analysis of Immuno-BNGE (Fig. 12). IF1 and Opa1 are known to promote ATP synthase oligomerization [18,22,69]. Our data showed high deviation between different preparations, thus we cannot make conclusive statements whether in IF1-H49K expressing cells the ATP synthase oligomers are stabilized or not (Fig. 12B). Also, the relative amount of IF1 bound to the ATP synthase did not clearly change (Fig. 12C). Next, we determined the relative levels of subunit β (ATP5B) and subunit e (ATP5i), which is involved in dimer formation of the ATP synthase [19,74,84]. The ratio of SU e/SU β and the total level of the ATP synthase apparently did not change (Fig. 12D). Together, these data do not indicate that the overexpression of IF1-H49K in the stable cell line resulted in a shift to more ATP synthase dimers/oligomers or a relative increase of IF1 bound to the ATP synthase.
Fig. 12.
IF1-H49K expressing cells tend to have more ATP synthase dimers and more IF1 bound to ATP synthase. (A) Equal amounts (60 μg per lane) of digitonin-solubilized mitochondrial extracts from HeLa with the indicated IF1 genotype were separated by BNGE and immunoblotted with the indicated antibodies. (B) Quantitative densitometric analysis of ATP synthase dimer vs. monomer (D/M). (C) Quantitative densitometric analysis of IF1 bound to total ATP synthase. (D) Immunoblots after separation of mitochondrial proteins by SDS-PAGE, antibodies as indicated for subunits e (α-ATP5I), β (α-ATP5B) and VDAC as loading control. (E) Ratio of subunit e and subunit β after normalization to VDAC. N = 3 replicates.
4. Discussion
The F1FO ATP synthase is the key enzyme of oxidative phosphorylation. Its main sub-mitochondrial localization is the rims of cristae, which, in many cells, are laminar sheets arranged parallel to each other. Here, we provide new data that inhibition of the ATPase function of the mammalian F1FO ATP synthase affects the sub-mitochondrial localization of the enzyme. ATPase activity was blocked by the expression of the constitutively active IF1-H49K variant [31]. The natural inhibitory factor IF1 is a strong inhibitor of ATP hydrolase function, which is controlled by phosphorylation, Ca2+ levels and pH [85,86]. The mutant IF1-H49K is pH-independent [32,38]. The bioenergetics data suggest that IF1-H49K was constitutively active and only inhibited the hydrolysis function in the investigated cell line, however, ATP synthase inhibition was also described elsewhere [26]. Cytosolic ATP levels augmented, and the mitochondrial membrane potential was decreased. This was expected, since IF1-knockout cells display the contrary [30]. A lower proton motive force (PMF) is comparable with substrate depletion. More recent data suggest, that substrate changes or depletion resulting in different PMF not only affect the activity of the ATP synthase but also the spatiotemporal organization of the enzyme in the inner mitochondrial membrane [12,14,68,87,88]. Indeed, we found that the spatiotemporal organization of ATP synthase was altered in IF1-H49K expressing cells.
In order to determine ATP synthase localization and trajectory maps in normal cells and IF1-H49K expressing cells we recorded single fluorescence-tagged protein complexes. In IF1-H49K expressing cells, we exposed considerable heterogeneity in the spatiotemporal organization of ATP synthase, which we assigned to different states. Those states were characterized by different mobility maps and correlated with gradual changes of the IMM architecture. In state I, the mobility and localization pattern of ATP synthase was similar to control conditions. Three subpopulations of ATP synthase were found: a mobile fraction with D1 = 0.080 μm2/s (14%), a less mobile fraction with D2 = 0.018 μm2/s (54%), and an immobile fraction with D3 = 0.006 μm2/s (30%). The corresponding trajectory map pattern was characterized by a significant amount of ATP synthase trajectories with an orthogonal course following the cristae structure [40,68] and indicating the confinement within this microcompartment. In state 2, however, the majority of F1FO ATP synthases was found at the border of widened mitochondria and only few cristae related trajectories were observed. Quantitative analysis of 2D-TEM micrographs, tomograms and high-resolution live cell imaging of NAO stained mitochondria revealed alterations of cristae such as arches and shortened cristae. But orthogonal cristae often existed in the same mitochondrion. This gradual alteration of the cristae structure might explain why the effects of IF1 on the mitochondrial ultrastructure has been controversially discussed to date [25,30].
The re-translocation of ATP synthase further shows that ATP synthase is not a static enzyme in the membrane as implicated by TEM micrographs and tomograms [3,7,9,15,89]. This, albeit restricted, mobility is responsible for the slow mixing of ATP synthase originating from recently fused mitochondria [90] and is prerequisite for the dynamic physical and functional interaction with other membrane proteins including subunits of the MICOS complex, the inner membrane fusion protein Opa1 and the ADP/ATP carrier [23,91–95], all not explicitly located at cristae rims. Another example of required ATP re-localization is the interaction with the outer membrane translocator protein [96], when forming the mitochondrial transition pore. This is certainly not possible with ATP synthase exclusively localized at the rims of cristae [97]. Thus, a certain mobility of ATP synthase in the inner mitochondrial membrane, as former studies in mammalian and yeast cells indicated earlier [87,98,99] is biophysically and physiologically realistic.
We next aimed to find the reasons for the ultrastructural changes. Opa1 is a known cristae modulator [79,83,100]. Indeed, Opa1 expression levels were increased in IF1-H49K expressing cells, immuno-blotting revealed a shift of long L-Opa1 to short S-Opa1 forms. Sufficiently high ATP levels and a decrease of ΔΨm are known stimulators for Opa1 processing to short forms [77]. It is reasonable to suggest that those factors caused also the processing of Opa1 in this study. While it has been suggested that short Opa1 is sufficient to keep the cristae structure [83,101], this was obviously not the case here. The formation of cristae has been described as being controlled by a stabilization of the ATP-synthase oligomers through Opa1 [22]. Also, IF1 is discussed to promote dimer-dimer interaction and consequently oligomerization dynamics [18]. However, we found no significant alterations in ATP synthase dimer levels. Unfortunately, the conducted BN-PAGE to separate ATP synthase monomers, dimers and oligomers from the different cell lines did not allow for a quantitative analysis of the oligomerization state of ATP synthase, neither the comparison between stably and transiently transfected cell lines. Therefore, we cannot exclude that IF1 promoted the dimerization of ATP synthase dimers. Such stabilization of ATP synthase dimer-dimer interaction by IF1 would probably enhance the membrane bending initiated already by ATP synthase dimers [13] and monomers [20]. Indeed, in transiently transfected cells the mobility of ATP synthase decreased significantly, which might be an indicator for increased oligomerization. In stable cell lines, this decrease in mobility was not observed. We assume that the permanent presence of higher IF1 levels affects ATP synthase differently than short term exposure.
Finally, we checked whether the protein mitofilin/Mic60/IMMT contributes to the changes in IMM structure [75,102]. We generated a mitofilin overexpressing cell line, which displayed similar changes in the IMM architecture as observed in the IF1-H49K cells, but the mitofilin expression level was not changed.
We further used the mitofilin overexpressing cell line to investigate the link between the ultrastructure and the spatiotemporal organization of ATP synthase. In the mitofilin overexpressing condition, the distribution pattern of ATP synthase was changed similar as in IF1-H49K expressing cells with numerous trajectories confined next to the OMM. However, the diffusion coefficient of the mobile fraction of ATP synthase particles was not altered. This suggests that the IMM ultrastructure determines the distribution of the IMM proteins and thus defines the microcompartment in which diffusion is possible, but the structural re-formation does not hamper the diffusion behavior of proteins per se. Other factors such as protein-protein interaction and oligomerization are more likely responsible for the mobility of proteins. Overexpression of ATP synthase subunit g that is responsible for dimerization of ATP synthase [84,103] resulted in a significantly decreased mobility of the F1FO ATP synthase [68].
In summary, we present here data showing that modification of ATP synthase activity, either by inhibition of ATPase activity by IF1 or by complete inhibition with oligomycin, influences the spatiotemporal behavior of ATP synthase. In both cases, the typical inclusion of ATP synthase in cristae was erased, suggesting that the localization of ATP synthase depends on its functional status. This is consistent with recent studies that show that the metabolic state of the mitochondria and thus the activity of the ATP synthase influences the spatiotemporal organization of the enzyme [12,68,87] [12,68]. Further studies are needed to reveal, which impact this re-organization of the key enzyme ATP synthase has on the activity of functionally linked proteins. E.g. juxtaposition of ATP synthase and Hexokinase at the OM offers the possibility to regulate energy fluxes more directly.
5. Conclusions
The mitochondrial F1FO-ATP synthase is an enzyme that has ATP synthesis and ATP hydrolysis function. The specific inhibition of ATPase by constitutively active IF1 leads to a partial relocation of the enzyme towards the OMM related to changes in the mitochondrial ultrastructure in which the inner membrane fusion protein Opa1 is involved. These findings show a close connection between the function of the F1FO-ATP synthase and its spatiotemporal organization. This signifies that the ATP synthase in the inner membrane is more dynamic in terms of localization and mobility than originally assumed.
Supplementary Material
Acknowledgments
We want to thank Christian Richter for his important contribution to the analysis of tracking and localization data. Patrick Duwe and Wladislaw Kohl are acknowledged for their technical assistance in cloning and WESTERN. The study was supported by a grant of the CRC944 (German Research Foundation, GRF) to Karin Busch (INST190/1672), and the z-project. Bettina Rieger was supported by grant of the FOR2848 (Bu2288/3_1, GRF). We thank the Imaging Network of the University of Münster (RI_00497). This work was supported by the German Research Foundation (GRF) through Markus Schwarzländer and the infrastructure grant INST211/903-1 FUGG. Guy Perkins was supported by a NIH grant (5P41 GM103412-29) to National Center for Microscopy and Imaging Research. German Research Foundation: INST190/167-2 (CRC944) and Bu2288/3-1 (FOR 2848/1). NIH 5P41 GM103412-29 to National Center for Microscopy and Imaging Research.
Footnotes
Declaration of competing interest
The authors declare no competing financial interests.
Transparency document
The Transparency document associated with this article can be found in the online version.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbabio.2020.148322.
References
- [1].Gilkerson RW, Selker JM, Capaldi RA, The Cristal membrane of mitochondria is the principal site of oxidative phosphorylation, FEBS Lett. 546 (2003) 355–358. [DOI] [PubMed] [Google Scholar]
- [2].Vogel F, Bornhovd C, Neupert W, Reichert AS, Dynamic subcompartmentalization of the mitochondrial inner membrane, J. Cell Biol. 175 (2006) 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kuhlbrandt W, Macromolecular organization of ATP synthase and complex I in whole mitochondria, Proc. Natl. Acad. Sci. U. S. A. 108 (2011) 14121–14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wilkens V, Kohl W, Busch K, Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution, J. Cell Sci. 126 (2013) 103–116. [DOI] [PubMed] [Google Scholar]
- [5].Frey TG, Mannella CA, The internal structure of mitochondria, Trends Biochem. Sci. 25 (2000) 319–324. [DOI] [PubMed] [Google Scholar]
- [6].Mannella CA, Lederer WJ, Jafri MS, The connection between inner membrane topology and mitochondrial function, J. Mol. Cell. Cardiol. 62 (2013) 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Allen RD, Schroeder CC, Fok AK, An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques, J. Cell Biol. 108 (1989) 2233–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W, Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae, Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 13602–13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W, Dimer ribbons of ATP synthase shape the inner mitochondrial membrane, EMBO J. 27 (2008) 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J, The ATP synthase is involved in generating mitochondrial cristae morphology, EMBO J. 21 (2002) 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Salewskij K, Rieger B, Hager F, Arroum T, Duwe P, Villalta J, Colgiati S, Richter CP, Psathaki OE, Enriquez JA, Dellmann T, Busch KB, The spatio-temporal organization of mitochondrial F1FO ATP synthase in cristae depends on its activity mode, Biochim. Biophys. Acta Bioenerg. 2020 (1861) 148091. [DOI] [PubMed] [Google Scholar]
- [12].Plecita-Hlavata L, Engstova H, Alan L, Spacek T, Dlaskova A, Smolkova K, Spackova J, Tauber J, Stradalova V, Malinsky J, Lessard M, Bewersdorf J, Jezek P, Hypoxic HepG2 cell adaptation decreases ATP synthase dimers and ATP production in inflated cristae by mitofilin down-regulation concomitant to MICOS clustering, FASEB J. 30 (2016) 1941–1957. [DOI] [PubMed] [Google Scholar]
- [13].Blum TB, Hahn A, Meier T, Davies KM, Kuhlbrandt W, Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 4250–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dlaskova A, Spacek T, Engstova H, Spackova J, Schrofel A, Holendova B, Smolkova K, Plecita-Hlavata L, Jezek P, Mitochondrial cristae narrowing upon higher 2-oxoglutarate load, Biochim. Biophys. Acta Bioenerg. 1860 (2019) 659–678. [DOI] [PubMed] [Google Scholar]
- [15].Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP, Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane, FEBS Lett. 579 (2005) 5769–5772. [DOI] [PubMed] [Google Scholar]
- [16].Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ, Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria, FEBS Lett. 580 (2006) 3427–3432. [DOI] [PubMed] [Google Scholar]
- [17].Giraud MF, Paumard P, Soubannier V, Vaillier J, Arselin G, Salin B, Schaeffer J, Brethes D, di Rago JP, Velours J, Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim. Biophys. Acta 1555 (2002) 174–180. [DOI] [PubMed] [Google Scholar]
- [18].Gu J, Zhang L, Zong S, Guo R, Liu T, Yi J, Wang P, Zhuo W, Yang M, Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1, Science 364 (2019) 1068–1075. [DOI] [PubMed] [Google Scholar]
- [19].Habersetzer J, Larrieu I, Priault M, Salin B, Rossignol R, Brethes D, Paumard P, Human F1F0 ATP synthase, mitochondrial ultrastructure and OXPHOS impairment: a (super-)complex matter? PLoS One 8 (2013) e75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jiko C, Davies KM, Shinzawa-Itoh K, Tani K, Maeda S, Mills DJ, Tsukihara T, Fujiyoshi Y, Kuhlbrandt W, Gerle C, Bovine F1Fo ATP synthase monomers bend the lipid bilayer in 2D membrane crystals, Elife 4 (2015) e06119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Depaoli MR, Karsten F, Madreiter-Sokolowski CT, Klec C, Gottschalk B, Bischof H, Eroglu E, Waldeck-Weiermair M, Simmen T, Graier WF, Malli R, Real-time imaging of mitochondrial ATP dynamics reveals the metabolic setting of single cells, Cell Rep. 25 (2018) 501–512 (e503). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Quintana-Cabrera R, Quirin C, Glytsou C, Corrado M, Urbani A, Pellattiero A, Calvo E, Vazquez J, Enriquez JA, Gerle C, Soriano ME, Bernardi P, Scorrano L, The cristae modulator optic atrophy 1 requires mitochondrial ATP synthase oligomers to safeguard mitochondrial function, Nat. Commun. 9 (2018) 3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beutner G, Alanzalon RE, Porter GA Jr., Cyclophilin D regulates the dynamic assembly of mitochondrial ATP synthase into synthasomes, Sci. Rep 7 (2017) 14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gerle C, Mitochondrial F-ATP synthase as the permeability transition pore, Pharmacol. Res 160 (2020) 105081. [DOI] [PubMed] [Google Scholar]
- [25].Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, Duchen MR, Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1, Cell Metab. 8 (2008) 13–25. [DOI] [PubMed] [Google Scholar]
- [26].Garcia-Bermudez J, Cuezva JM, The ATPase inhibitory factor 1 (IF1): a master regulator of energy metabolism and of cell survival, Biochim. Biophys. Acta 1857 (2016) 1167–1182. [DOI] [PubMed] [Google Scholar]
- [27].Willers IM, Cuezva JM, Post-transcriptional regulation of the mitochondrial H (+)-ATP synthase: a key regulator of the metabolic phenotype in cancer, Biochim. Biophys. Acta 1807 (2011) 543–551. [DOI] [PubMed] [Google Scholar]
- [28].Cabezon E, Montgomery MG, Leslie AG, Walker JE, The structure of bovine F1-ATPase in complex with its regulatory protein IF1, Nat. Struct. Biol. 10 (2003) 744–750. [DOI] [PubMed] [Google Scholar]
- [29].Bason JV, Montgomery MG, Leslie AG, Walker JE, Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 11305–11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fujikawa M, Imamura H, Nakamura J, Yoshida M, Assessing actual contribution of IF1, inhibitor of mitochondrial FoF1, to ATP homeostasis, cell growth, mitochondrial morphology, and cell viability, J. Biol. Chem. 287 (2012) 18781–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cabezon E, Butler PJ, Runswick MJ, Walker JE, Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH, J. Biol. Chem. 275 (2000) 25460–25464. [DOI] [PubMed] [Google Scholar]
- [32].Schnizer R, Van Heeke G, Amaturo D, Schuster SM, Histidine-49 is necessary for the pH-dependent transition between active and inactive states of the bovine F1-ATPase inhibitor protein, Biochim. Biophys. Acta 1292 (1996) 241–248. [DOI] [PubMed] [Google Scholar]
- [33].Cabezon E, Arechaga I, Jonathan P, Butler G, Walker JE, Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1, J. Biol. Chem. 275 (2000) 28353–28355. [DOI] [PubMed] [Google Scholar]
- [34].Faccenda D, Campanella M, Molecular regulation of the mitochondrial F(1)F(o)-ATPsynthase: physiological and pathological significance of the inhibitory factor 1 (IF(1)), Int J Cell Biol 2012 (2012) 367934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Faccenda D, Tan CH, Seraphim A, Duchen MR, Campanella M, IF1 limits the apoptotic-signalling cascade by preventing mitochondrial remodelling, Cell Death Differ. 20 (2013) 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Campanella M, Seraphim A, Abeti R, Casswell E, Echave P, Duchen MR, IF1, the endogenous regulator of the F(1)F(o)-ATPsynthase, defines mitochondrial volume fraction in HeLa cells by regulating autophagy, Biochim. Biophys. Acta 1787 (2009) 393–401. [DOI] [PubMed] [Google Scholar]
- [37].Formentini L, Pereira MP, Sanchez-Cenizo L, Santacatterina F, Lucas JJ, Navarro C, Martinez-Serrano A, Cuezva JM, In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning, EMBO J. 33 (2014) 762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Garcia-Aguilar A, Cuezva JM, A review of the inhibition of the mitochondrial ATP synthase by IF1 in vivo: reprogramming energy metabolism and inducing mitohormesis, Front. Physiol 9 (2018) 1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Faccenda D, Nakamura J, Gorini G, Dhoot GK, Piacentini M, Yoshida M, Campanella M, Control of mitochondrial remodeling by the ATPase inhibitory factor 1 unveils a pro-survival relay via OPA1, Cell Rep. 18 (2017) 1869–1883. [DOI] [PubMed] [Google Scholar]
- [40].Appelhans T, Busch K, Single molecule tracking and localization of mitochondrial protein complexes in live cells, Methods Mol. Biol. 1567 (2017) 273–291. [DOI] [PubMed] [Google Scholar]
- [41].Appelhans T, Richter CP, Wilkens V, Hess ST, Piehler J, Busch KB, Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy, Nano Lett. 12 (2012) 610–616. [DOI] [PubMed] [Google Scholar]
- [42].Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA, Respiratory active mitochondrial supercomplexes, Mol. Cell 32 (2008) 529–539. [DOI] [PubMed] [Google Scholar]
- [43].Wittig I, Karas M, Schagger H, High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes, Mol. Cell. Proteomics 6 (2007) 1215–1225. [DOI] [PubMed] [Google Scholar]
- [44].Wikstrom JD, Katzman SM, Mohamed H, Twig G, Graf SA, Heart E, Molina AJ, Corkey BE, de Vargas LM, Danial NN, Collins S, Shirihai OS, Beta-cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels, Diabetes 56 (2007) 2569–2578. [DOI] [PubMed] [Google Scholar]
- [45].Wang L, Yuan L, Zeng X, Peng J, Ni Y, Er JC, Xu W, Agrawalla BK, Su D, Kim B, Chang YT, A multisite-binding switchable fluorescent probe for monitoring mitochondrial ATP level fluctuation in live cells, Angew Chem Int Ed Engl 55 (2016) 1773–1776. [DOI] [PubMed] [Google Scholar]
- [46].Wolf DM, Segawa M, Kondadi AK, Anand R, Bailey ST, Reichert AS, van der Bliek AM, Shackelford DB, Liesa M, Shirihai OS, Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent, Embo J (2019) e101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH, Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces, FEBS Lett. 507 (2001) 187–190. [DOI] [PubMed] [Google Scholar]
- [48].Leung CW, Hong Y, Hanske J, Zhao E, Chen S, Pletneva EV, Tang BZ, Superior fluorescent probe for detection of cardiolipin, Anal. Chem. 86 (2014) 1263–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Segawa M, Wolf DM, Hultgren NW, Williams DS, van der Bliek AM, Shackelford DB, Liesa M, Shirihai OS, Quantification of Cristae Architecture Reveals Time-dependent Characteristics of Individual Mitochondria, Life Sci Alliance, 3, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Vignolini T, Gardini V, Curcio V, Capitanio M, Pavone FS, Optimization of Highly Inclined Optical Sheet Illumination for Super-Resolution Microscopy, Biophys. J. 114 (2018) 14a. [DOI] [PubMed] [Google Scholar]
- [51].Appelhans T, Beinlich FR, Richter CP, Kurre R, Busch KB, Multi-color localization microscopy of single membrane proteins in organelles of live mammalian cells, JoVE (Journal of Visualized Experiments) (2018) e57690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tokunaga M, Imamoto N, Sakata-Sogawa K, Highly inclined thin illumination enables clear single-molecule imaging in cells, Nat. Methods 5 (2008) 455–461. [DOI] [PubMed] [Google Scholar]
- [53].Muster B, Kohl W, Wittig I, Strecker V, Joos F, Haase W, Bereiter-Hahn J, Busch K, Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells, PLoS One 5 (2010) e11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rosselin M, Santo-Domingo J, Bermont F, Giacomello M, Demaurex N, L-OPA1 regulates mitoflash biogenesis independently from membrane fusion, EMBO Rep. 18 (2017) 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tokunaga M, Imamoto N, Sakata-Sogawa K, Highly inclined thin illumination enables clear single-molecule imaging in cells, Nat. Methods 5 (2008) 159–161. [DOI] [PubMed] [Google Scholar]
- [56].Serge A, Bertaux N, Rigneault H, Marguet D, Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes, Nat. Methods 5 (2008) 687–694. [DOI] [PubMed] [Google Scholar]
- [57].Richter D, Moraga I, Winkelmann H, Birkholz O, Wilmes S, Schulte M, Kraich M, Kenneweg H, Beutel O, Selenschick P, Paterok D, Gavutis M, Schmidt T, Garcia KC, Muller TD, Piehler J, Ligand-induced type II interleukin-4 receptor dimers are sustained by rapid re-association within plasma membrane microcompartments, Nat. Commun 8 (2017) 15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF, Imaging intracellular fluorescent proteins at nanometer resolution, Science 313 (2006) 1642–1645. [DOI] [PubMed] [Google Scholar]
- [59].Gould TJ, Verkhusha VV, Hess ST, Imaging biological structures with fluorescence photoactivation localization microscopy, Nat. Protoc. 4 (2009) 291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hess ST, Girirajan TP, Mason MD, Ultra-high resolution imaging by fluorescence photoactivation localization microscopy, Biophys. J. 91 (2006) 4258–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rieger B, Junge W, Busch KB, Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP-synthase in folded mitochondrial membranes, Nat. Commun 5 (2014) 1–6. [DOI] [PubMed] [Google Scholar]
- [62].Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA, Mitochondrial membrane potential probes and the proton gradient: a practical usage guide, Biotechniques 50 (2011) 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sanchez-Cenizo L, Formentini L, Aldea M, Ortega AD, Garcia-Huerta P, Sanchez-Arago M, Cuezva JM, Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype, J. Biol. Chem. 285 (2010) 25308–25313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zanotti F, Gnoni A, Mangiullo R, Papa S, Effect of the ATPase inhibitor protein IF1 on H+ translocation in the mitochondrial ATP synthase complex, Biochem. Biophys. Res. Commun. 384 (2009) 43–48. [DOI] [PubMed] [Google Scholar]
- [65].Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA, Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells, Cancer Res. 64 (2004) 985–993. [DOI] [PubMed] [Google Scholar]
- [66].Appelhans T, Busch KB, Dynamic imaging of mitochondrial membrane proteins in specific sub-organelle membrane locations, Biophys. Rev 9 (2017) 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J, High-density mapping of single-molecule trajectories with photoactivated localization microscopy, Nat. Methods 5 (2008) 155–157. [DOI] [PubMed] [Google Scholar]
- [68].Salewskij K, Rieger B, Hager F, Arroum T, Duwe P, Villalta J, Colgiati S, Richter CP, Psathaki OE, Enriquez JA, Dellmann T, Busch KB, The spatio-temporal organization of mitochondrial F1FO ATP synthase in cristae depends on its activity mode, Biochim. Biophys. Acta Bioenerg. 148091 (2019). [DOI] [PubMed] [Google Scholar]
- [69].Garcia JJ, Morales-Rios E, Cortes-Hernandez P, Rodriguez-Zavala JS, The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase, Biochemistry 45 (2006) 12695–12703. [DOI] [PubMed] [Google Scholar]
- [70].Jacobson J, Duchen MR, Heales SJ, Intracellular distribution of the fluorescent dye nonyl acridine orange responds to the mitochondrial membrane potential: implications for assays of cardiolipin and mitochondrial mass, J. Neurochem. 82 (2002) 224–233. [DOI] [PubMed] [Google Scholar]
- [71].Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W, The mitochondrial contact site complex, a determinant of mitochondrial architecture, EMBO J. 30 (2011) 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bohnert M, Wenz LS, Zerbes RM, Horvath SE, Stroud DA, von der Malsburg K, Muller JM, Oeljeklaus S, Perschil I, Warscheid B, Chacinska A, Veenhuis M, van der Klei IJ, Daum G, Wiedemann N, Becker T, Pfanner N, van der Laan M, Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane, Mol. Biol. Cell 23 (2012) 3948–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J, The mitochondrial inner membrane protein mitofilin controls cristae morphology, Mol. Biol. Cell 16 (2005) 1543–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS, Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g, J. Cell Biol. 185 (2009) 1047–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tarasenko D, Barbot M, Jans DC, Kroppen B, Sadowski B, Heim G, Mobius W, Jakobs S, Meinecke M, The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology, J. Cell Biol. 216 (2017) 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].He J, Ford HC, Carroll J, Douglas C, Gonzales E, Ding S, Fearnley IM, Walker JE, Assembly of the membrane domain of ATP synthase in human mitochondria, Proc. Natl. Acad. Sci. U. S. A. 115 (2018) 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Del Dotto V, Fogazza M, Carelli V, Rugolo M, Zanna C, Eight human OPA1 isoforms, long and short: what are they for? Biochim. Biophys. Acta Bioenerg. 1859 (2018) 263–269. [DOI] [PubMed] [Google Scholar]
- [78].Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, Chomyn A, Bauer MF, Attardi G, Larsson NG, Neupert W, Reichert AS, Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology, J. Biol. Chem. 281 (2006) 37972–37979. [DOI] [PubMed] [Google Scholar]
- [79].Ishihara N, Fujita Y, Oka T, Mihara K, Regulation of mitochondrial morphology through proteolytic cleavage of OPA1, EMBO J. 25 (2006) 2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Santo-Domingo J, Giacomello M, Poburko D, Scorrano L, Demaurex N, OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange, EMBO J. 32 (2013) 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yu T, Fox RJ, Burwell LS, Yoon Y, Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1, J. Cell Sci. 118 (2005) 4141–4151. [DOI] [PubMed] [Google Scholar]
- [82].Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ, Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis, J. Cell Biol. 159 (2002) 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lee H, Smith SB, Yoon Y, The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure, J. Biol. Chem. 292 (2017) 7115–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wagner K, Rehling P, Sanjuan Szklarz LK, Taylor RD, Pfanner N, van der Laan M, Mitochondrial F1Fo-ATP synthase: the small subunits e and g associate with monomeric complexes to trigger dimerization, J. Mol. Biol. 392 (2009) 855–861. [DOI] [PubMed] [Google Scholar]
- [85].Boreikaite V, Wicky BIM, Watt IN, Clarke J, Walker JE, Extrinsic conditions influence the self-association and structure of IF1, the regulatory protein of mitochondrial ATP synthase, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 10354–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Garcia-Bermudez J, Sanchez-Arago M, Soldevilla B, Del Arco A, Nuevo-Tapioles C, Cuezva JM, PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H(+)-ATP synthase, Cell Rep. 12 (2015) 2143–2155. [DOI] [PubMed] [Google Scholar]
- [87].Jimenez L, Laporte D, Duvezin-Caubet S, Courtout F, Sagot I, Mitochondrial ATP synthases cluster as discrete domains that reorganize with the cellular demand for oxidative phosphorylation, J. Cell Sci. 127 (2014) 719–726. [DOI] [PubMed] [Google Scholar]
- [88].Dlaskova A, Engstova H, Spacek T, Kahancova A, Pavluch V, Smolkova K, Spackova J, Bartos M, Hlavata LP, Jezek P, 3D super-resolution microscopy reflects mitochondrial cristae alternations and mtDNA nucleoid size and distribution, Biochim. Biophys. Acta Bioenerg. 1859 (2018) 829–844. [DOI] [PubMed] [Google Scholar]
- [89].Fernandez Moran H, Oda T, Blair PV, Green DE, A macromolecular repeating unit of mitochondrial structure and function. Correlated electron microscopic and biochemical studies of isolated mitochondria and submitochondrial particles of beef Heart muscle, J. Cell Biol. 22 (1964) 63–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wilkens V, Appelhans T, Kohl W, Piehler J, Busch K, Mitochondrial ultrastructure and complex formation are determining factors for the mobility of OXPHOS complexes in the IMM, BBA-Bioenergetics S72 (2012). [Google Scholar]
- [91].Eydt K, Davies KM, Behrendt C, Wittig I, Reichert AS, Cristae architecture is determined by an interplay of the MICOS complex and the F1FO ATP synthase via Mic27 and Mic10, Microb Cell 4 (2017) 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Clemencon B, Yeast mitochondrial interactosome model: metabolon membrane proteins complex involved in the channeling of ADP/ATP, Int. J. Mol. Sci 13 (2012) 1858–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wittig I, Schagger H, Structural organization of mitochondrial ATP synthase, Biochim. Biophys. Acta 1777 (2008) 592–598. [DOI] [PubMed] [Google Scholar]
- [94].Wittig I, Schagger H, Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes, Biochim. Biophys. Acta 1787 (2009) 672–680. [DOI] [PubMed] [Google Scholar]
- [95].Alavian KN, Li H, Collis L, Bonanni L, Zeng L, Sacchetti S, Lazrove E, Nabili P, Flaherty B, Graham M, Chen Y, Messerli SM, Mariggio MA, Rahner C, McNay E, Shore GC, Smith PJ, Hardwick JM, Jonas EA, Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase, Nat. Cell Biol. 13 (2011) 1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sileikyte J, Petronilli V, Zulian A, Dabbeni-Sala F, Tognon G, Nikolov P, Bernardi P, Ricchelli F, Regulation of the inner membrane mitochondrial permeability transition by the outer membrane translocator protein (peripheral benzodiazepine receptor), J. Biol. Chem. 286 (2011) 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ricchelli F, Sileikyte J, Bernardi P, Shedding light on the mitochondrial permeability transition, Biochim. Biophys. Acta 1807 (2011) 482–490. [DOI] [PubMed] [Google Scholar]
- [98].Sukhorukov V, Busch K, Bereiter-Hahn J, Accurate Analysis of CLSM FRAP Data for Mitochondrial Proteins, Annual Meeting of the German Society for Cell Biology, Elsevier GMBH, Urban & Fischer Verlag Office Jena, PO Box 100537, 07705 JENA, (2007), p. 21–21. [Google Scholar]
- [99].Sukhorukov VM, Dikov D, Busch K, Strecker V, Wittig I, Bereiter-Hahn J, Determination of protein mobility in mitochondrial membranes of living cells, Biochim. Biophys. Acta 1798 (2010) 2022–2032. [DOI] [PubMed] [Google Scholar]
- [100].Faelber K, Dietrich L, Noel JK, Wollweber F, Pfitzner AK, Muhleip A, Sanchez R, Kudryashev M, Chiaruttini N, Lilie H, Schlegel J, Rosenbaum E, Hessenberger M, Matthaeus C, Kunz S, von der Malsburg A, Noe F, Roux A, van der Laan M, Kuhlbrandt W, Daumke O, Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1, Nature 571 (2019) 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Herlan M, Bornhovd C, Hell K, Neupert W, Reichert AS, Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor, J. Cell Biol. 165 (2004) 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Colina-Tenorio L, Horten P, Pfanner N, Rampelt H, Shaping the mitochondrial inner membrane in health and disease, J. Intern. Med. 6 (2020) 645–669. [DOI] [PubMed] [Google Scholar]
- [103].Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schagger H, Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits, EMBO J. 17 (1998) 7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.