Alzheimer’s disease is the most common neurodegenerative disease with complex genetic architecture (Liu et al., 2014). Recently, large-scale genome-wide association studies (GWAS) have been performed and successfully identified more than 40 novel Alzheimer’s disease genetic variants (Lambert et al., 2013; Cuyvers et al., 2015; Kunkle et al., 2019). Meanwhile, large-scale GWAS of Alzheimer’s disease endophenotypes have also been reported (DemiNg et al., 2017, 2018; Chung et al., 2018; Dumitrescu et al., 2019; Moreno-Grau et al., 2019), and identified sex differences (Deming et al., 2018; Dumitrescu et al., 2019). In 2018, Deming and colleagues conducted sex-specific GWAS of CSF levels of amyloid-β42 and tau from 1527 males and 1509 females (Deming et al., 2018). They identified rs316341 (sex-interaction P = 0.04) and rs13115400 (sex-interaction P = 0.002) to show stronger association with amyloid-β42 in females than males (Deming et al., 2018). In 2019, Dumitrescu and colleagues analysed a GWAS dataset with 2701 males and 3275 females (Dumitrescu et al., 2019). They identified variant rs34331204, which showed significant sex-specific association with P = 2.90 × 10−4 (Dumitrescu et al., 2019). The rs34331204 variant minor allele C was associated with a lower risk of neurofibrillary tangles (NFT) in males (P = 2.50 × 10−8) but not females (P = 0.85). Interestingly, rs34331204 was also associated with increased hippocampal volume and executive function only in males (Dumitrescu et al., 2019). Hence, their findings provide a male-specific protective genetic variant against tau pathology.
There are still four main concerns to be mentioned, although these are important and interesting findings. First, Dumitrescu and colleagues established the association between rs34331204 variant C allele and reduced NFT burden. It is known that increased NFT burden is a key Alzheimer’s disease neuropathology. However, it remains unclear whether the rs34331204 variant is associated with Alzheimer’s disease risk, especially in males. Second, the rs34331204 variant is a non-coding variant. Dumitrescu and colleagues conducted an expression quantitative trait loci (eQTL) analysis to identify the candidate genes within the rs34331204 variant, and further evaluate the association between the tau load and the expression of target genes in the prefrontal cortex (PFC) (Dumitrescu et al., 2019). Using the Braineac data, eQTL analyses—including the exon-specific level and the transcript level—were carried out. However, they only evaluated the association between target gene expression and tau load at the transcript level. Importantly, these eQTL analyses are based on the average expression profile across all 10 brain tissues in Braineac (Dumitrescu et al., 2019). It remains unclear whether these target genes have different expression in these different brain tissues. Third, ours and other studies have clearly indicated that eQTL analyses vary considerably in different tissue/cell types, and disease statuses (Liu et al., 2016, 2017a, 2018, 2019a, b, 2020; Peters et al., 2016; Soldner et al., 2016; Hu et al., 2017b). Hence, a tissue-specific eQTL analysis should be performed, especially in the PFC. Fourth, if one gene is the target gene of the rs34331204 variant in PFC, and its expression shows sex-specific association with tau pathology in PFC, it remains unclear whether there is significant difference regarding differential expression (Alzheimer’s disease versus controls) in males and females. These concerns prompted us to further evaluate their findings.
In stage 1, we conducted a candidate variant study to evaluate the potential association between the rs34331204 variant and Alzheimer’s disease risk using three large-scale Alzheimer’s disease GWAS datasets. The first dataset was from the International Genomics of Alzheimer’s Project (IGAP) (Kunkle et al., 2019). The IGAP stage 1 dataset included 21 982 Alzheimer’s disease patients and 41 944 cognitively normal control subjects of European descent from four consortia including the Alzheimer Disease Genetics Consortium (ADGC), Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE), The European Alzheimer’s Disease Initiative (EADI), and the Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES) (Kunkle et al., 2019). All Alzheimer’s disease patients were diagnosed using the NINCDS-ADRDA criteria or DSM-IV guidelines (Kunkle et al., 2019). Meanwhile, we selected two sex-specific Alzheimer’s disease GWAS datasets including one Alzheimer’s disease GWAS dataset in males diagnosed by paternal history of Alzheimer’s disease (14 338 cases and 245 941 controls), and one Alzheimer’s disease GWAS dataset in females diagnosed by maternal history of Alzheimer’s disease (27 696 cases and 260 980 controls), both of which were from the UK Biobank (Marioni et al., 2018). Here, we defined the significance threshold to be P < 0.05. Using the Alzheimer’s disease GWAS dataset in IGAP, we found no significant association between the rs34331204 variant and Alzheimer’s disease risk. Interestingly, the sex-specific analysis indicated that rs34331204 variant C allele was associated with reduced Alzheimer’s disease risk in males (P = 0.046), but not in females (P = 0.365), as provided in Supplementary Table 1.
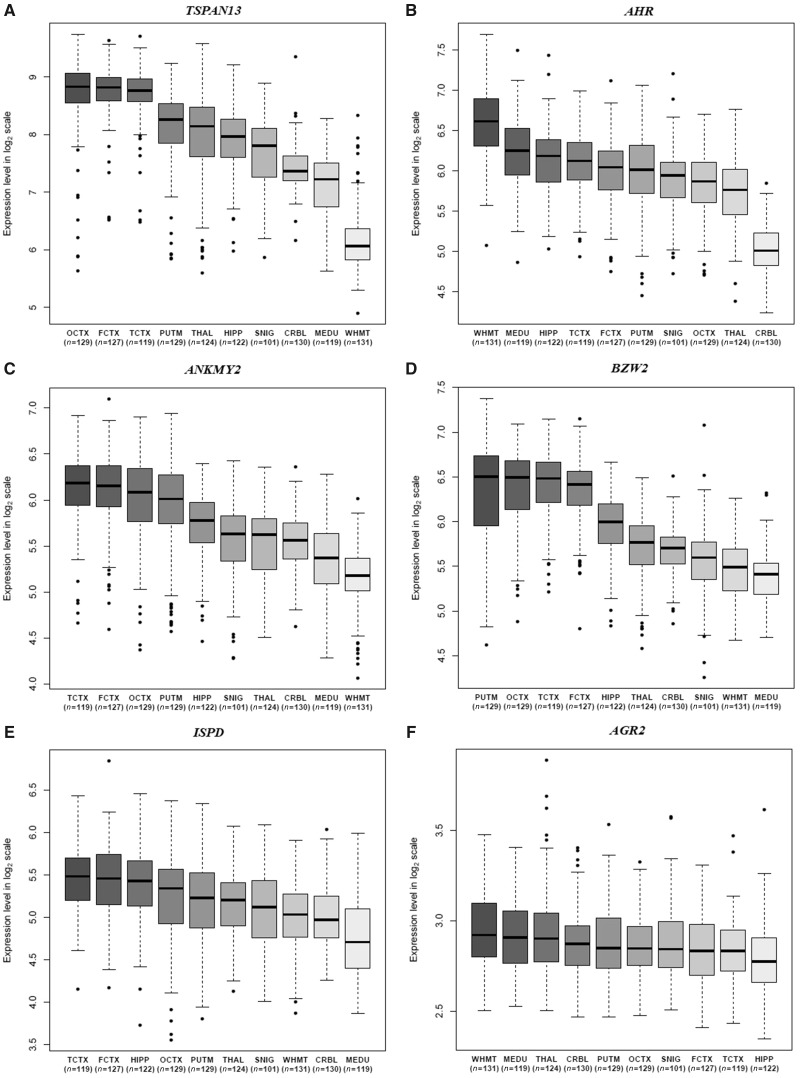
In stage 2, we conducted a gene expression analysis to demonstrate the significant expression difference of the target genes of rs34331204 across the 10 brain tissues in Braineac. Using the Braineac dataset, Dumitrescu and colleagues (2019) identified eight target genes for rs34331204 variant: BZW2, TSPAN13, AGR3, ANKMY2, LRRC72, AGR2, ISPD, and AHR, and evaluated the association of six genes with tau pathology in PFC by excluding AGR3 and LRRC72. Using the online Braineac database (http://www.braineac.org/), we found that all of these six genes showed significant expression difference across the 10 brain tissues including TSPAN13 (maximum fold change = 5.8, P = 2.20 × 10−64), AHR (maximum fold change = 3, P = 2.30 × 10−68), ANKMY2 (maximum fold change = 2, P = 8.80 × 10−44), BZW2 (maximum fold change = 2, P = 1.00 × 10−37), ISPD (maximum fold change = 1.6, P = 3.50 × 10−22), and AGR2 (maximum fold change = 1.1, P = 5.00 × 10−8). A box plot showing the expression of these six genes across the 10 brain tissues in Braineac is provided in Fig. 1. All of these findings indicate that a tissue-specific eQTL analysis is needed, especially in PFC.
Figure 1.
A box plot showing the expression of the six genes across the 10 brain tissues in Braineac. (A) Fold change between OCTX and WHMT = 5.8 (P = 2.20 × 10−64) for TSPAN13; (B) fold change between WHMT and CRBL = 3 (P = 2.30 × 10−68) for AHR; (C) fold change between TCTX and WHMT = 2 (P = 8.80 × 10−44) for ANKMY2; (D) fold change between PUTM and MEDU = 2 (P = 1.00 × 10−37) for BZW2; (E) fold change between TCTX and MEDU = 1.6 (P = 3.50 × 10−22) for ISPD; (F) fold change between WHMT and HIPP = 1.1 (P = 5.00 × 10−8) for AGR2. CRBL = cerebellar cortex; FCTX = frontal cortex; HIPP = hippocampus; MEDU = medulla; OCTX = occipital cortex; PUTM = putamen; SNIG = substantia nigra; TCTX = temporal cortex; THAL = thalamus; WHMT = intralobular white matter.
In stage 3, we performed an eQTL analysis of the rs34331204 variant in PFC using two independent datasets. The first dataset was from the Religious Orders Study and Memory and Aging Project (ROSMAP), which included 494 human PFC samples (Ng et al., 2017). Ninety-seven per cent of these samples were diagnosed with pathological Alzheimer’s disease and clinical Alzheimer’s disease (Ng et al., 2017). In ROSMAP, a Spearman’s rank correlation was used to perform the eQTL analysis (Ng et al., 2017). The second dataset was from the PsychENCODE Consortium (Wang et al., 2018). There were a total of 1866 PFC individuals including 1039 control individuals [113 from Genotype-Tissue Expression Consortium (GTEx version 7) and 926 from PsychENCODE], and 827 disease samples (558 schizophrenia, 217 bipolar disorder, 44 autism spectrum disorder, and eight affective disorder from PsychENCODE) (Wang et al., 2018). In PsychENCODE, a liner regression analysis was used to conduct the eQTL analysis (Wang et al., 2018). The statistical significance was a Bonferroni-corrected threshold of P < 0.05/31 = 1.61 × 10−3. The results showed that the rs34331204 variant C allele was associated with increased TSPAN13 expression only in the ROSMAP dataset (P = 1.23 × 10−3) (Table 1).
Table 1.
The rs2293871 variant and HTRA1 expression in human brain tissues
| SNP | EA | NEA | Gene symbol | Gencode ID | Beta | P-value | Dataset |
|---|---|---|---|---|---|---|---|
| rs34331204 | C | A | SOSTDC1 | ENSG00000171243.7 | –0.0257 | 5.69 × 10−1 | ROSMAP |
| rs34331204 | C | A | ANKMY2 | ENSG00000106524.4 | 0.0372 | 4.09 × 10−1 | ROSMAP |
| rs34331204 | C | A | TSPAN13 | ENSG00000106537.7 | 0.1450 | 1.23 × 10−3 | ROSMAP |
| rs34331204 | C | A | BZW2 | ENSG00000136261.10 | –0.0213 | 6.37 × 10−1 | ROSMAP |
| rs34331204 | C | A | AC006041.1 | ENSG00000229379.1 | 0.0097 | 7.45 × 10−1 | PsychENCODE |
| rs34331204 | C | A | RP11-196O16.1 | ENSG00000273477.1 | 0.0002 | 9.95 × 10−1 | PsychENCODE |
| rs34331204 | C | A | RPL36AP29 | ENSG00000224683.1 | –0.0111 | 8.41 × 10−1 | PsychENCODE |
| rs34331204 | C | A | CRPPA-AS1 | ENSG00000229688.3 | 0.0218 | 4.56 × 10−1 | PsychENCODE |
| rs34331204 | C | A | ISPD | ENSG00000214960.5 | 0.0132 | 5.26 × 10−1 | PsychENCODE |
| rs34331204 | C | A | SOSTDC1 | ENSG00000171243.7 | 0.0394 | 1.55 × 10−1 | PsychENCODE |
| rs34331204 | C | A | GS1-166A23.1 | ENSG00000272537.1 | 0.0032 | 9.32 × 10−1 | PsychENCODE |
| rs34331204 | C | A | LRRC72 | ENSG00000205858.5 | –0.0335 | 4.16 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC005014.5 | ENSG00000224280.1 | 0.0265 | 3.92 × 10−1 | PsychENCODE |
| rs34331204 | C | A | GS1-166A23.2 | ENSG00000272361.1 | 0.0176 | 6.90 × 10−1 | PsychENCODE |
| rs34331204 | C | A | ANKMY2 | ENSG00000106524.4 | 0.0060 | 6.92 × 10−1 | PsychENCODE |
| rs34331204 | C | A | BZW2 | ENSG00000136261.10 | 0.0160 | 3.55 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC073333.8 | ENSG00000235837.1 | 0.0662 | 2.29 × 10−1 | PsychENCODE |
| rs34331204 | C | A | TSPAN13 | ENSG00000106537.7 | 0.0012 | 9.54 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC073333.1 | ENSG00000267906.1 | 0.0356 | 4.31 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AGR2 | ENSG00000106541.7 | –0.0370 | 4.04 × 10−1 | PsychENCODE |
| rs34331204 | C | A | RP11-455J15.1 | ENSG00000270593.1 | –0.0329 | 2.63 × 10−1 | PsychENCODE |
| rs34331204 | C | A | RAD17P1 | ENSG00000232400.1 | 0.1042 | 2.24 × 10−2 | PsychENCODE |
| rs34331204 | C | A | AGR3 | ENSG00000173467.4 | 0.0305 | 4.95 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC098592.2 | ENSG00000227965.1 | –0.0135 | 4.72 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC098592.1 | ENSG00000223867.1 | –0.0466 | 1.51 × 10−1 | PsychENCODE |
| rs34331204 | C | A | BRWD1P3 | ENSG00000232841.1 | 0.0200 | 6.50 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC073332.1 | ENSG00000237773.1 | 0.0418 | 3.41 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AHR | ENSG00000106546.8 | 0.0229 | 3.23 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC019117.1 | ENSG00000236318.1 | 0.0019 | 9.71 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC019117.1 | ENSG00000236039.1 | –0.0421 | 1.93 × 10−1 | PsychENCODE |
| rs34331204 | C | A | AC017060.1 | ENSG00000226598.1 | –0.0209 | 2.54 × 10−1 | PsychENCODE |
EA = effect allele; EAF = effect allele frequency; NEA = non-effect allele. Beta is the regression coefficient based on the effect allele. Beta > 0 and Beta < 0 means that this effect allele increase and reduce disease or phenotype, respectively. The statistical significance for eQTL analysis was a Bonferroni-corrected threshold of P < 0.05/31 = 1.61 × 10−3.
However, this association was not replicated in the PsychENCODE dataset (Table 1). We consider that multiple disease statuses in PsychENCODE may have caused this negative finding, although there was larger sample size compared with ROSMAP. Here, we carried out a further subgroup eQTL analysis of the rs34331204 variant in PFC using the neuropathologically normal samples from two independent datasets. The first dataset was from the Braineac, including 134 PFC samples (Ramasamy et al., 2014). The second dataset was from the GTEx (version 8), including 175 PFC samples (Battle et al., 2017). We first evaluated the association between the rs34331204 variant and TSPAN13 expression using a liner regression analysis in both datasets. We then extracted the corresponding summary statistics of the rs34331204 variant in both datasets, and determined the heterogeneity of the rs34331204 variant using Cochran’s Q test. Finally, we conducted a meta-analysis to evaluate the association between the rs34331204 variant and TSPAN13 expression using R Package (meta: General Package for Meta-Analysis) (Hu et al., 2017a). The overall odds ratio (OR) was calculated by the fixed effect model (Mantel-Haenszel) or random-effect model (DerSimonian-Laird), which is determined by the heterogeneity (Hu et al., 2017a; Liu et al., 2017b). The statistical significance was 0.05. Interestingly, we found no significant heterogeneity with P = 0.7425. A meta-analysis using the fixed effect model highlighted a significant association between the rs34331204 variant C allele and reduced TSPAN13 expression (beta = −0.15, P = 0.0107). Hence, all of these findings indicate that the directions regarding the effect of rs34331204 variant C allele on TSPAN13 expression are different in neuropathologically normal samples and neurodegenerative disease individuals.
In stage 4, we performed an Alzheimer’s disease control gene expression analysis of TSPAN13 in males and females, respectively. We selected the gene expression dataset from the Harvard Brain Tissue Resource Center (HBTRC), including 129 (62 males and 67 females) Alzheimer’s disease samples and 101 (82 males and 19 females) non-demented healthy control samples in human PFC (Zhang et al., 2013). Here, we selected P < 0.05 to define the differential expression of TSPAN13 in Alzheimer’s disease and healthy control subjects. Using the online Bioconductor R package GEO2R, we found that TSPAN13 indicated stronger differential expression in males (fold change = 0.81 for Alzheimer’s disease versus control, P = 2.90 × 10−16) than females (fold change = 0.82 for Alzheimer’s disease versus control, P = 3.20 × 10−4).
Taken together, Dumitrescu and colleagues identified the rs34331204 variant C allele to be significantly associated with the reduced NFT in males (Dumitrescu et al., 2019). However, four main concerns remained unclear. Here, we performed a multi-stage analysis to answer these questions. In stage 1, we identified the rs34331204 variant C allele to be associated with reduced Alzheimer’s disease risk only in males. In stage 2, we demonstrated the different expression of the target genes of rs34331204 across the 10 brain tissues in Braineac. In stage 3, we found that the rs34331204 variant only regulated TSPAN13 expression in PFC, and in stage 4, we identified stronger differential expression of TSPAN13 in males than females in the PFC. We believe that these findings provide important supplementary information regarding the role of the rs34331204 variant in Alzheimer’s disease.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Supplementary Material
Acknowledgements
We thank the International Genomics of Alzheimer's Project (IGAP) and UK Biobank for the AD GWAS summary statistics. We thank the Braineac, GTEx, ROSMAP and PsychENCODE for the eQTL datasets, and HBTRC for the gene expression dataset. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families.
Funding
The i-Select chips were funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer's Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC-10-196728. This work was partially supported by funding from the Science and technology Beijing one hundred leading talent training project (Z141107001514006), Beijing Municipal Administration of Hospitals’ Mission Plan (SML20150802), and the National Natural Science Foundation of China (81620108011 and 6180010993).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature 2017; 550: 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Wang X, Maruyama T, Ma Y, Zhang X, Mez J, et al. Genome-wide association study of Alzheimer's disease endophenotypes at prediagnosis stages. Alzheimers Dement 2018; 14: 623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyvers E, De Roeck A, Van den Bossche T, Van Cauwenberghe C, Bettens K, Vermeulen S, et al. Mutations in ABCA7 in a Belgian cohort of Alzheimer's disease patients: a targeted resequencing study. Lancet Neurol 2015; 14: 814–22. [DOI] [PubMed] [Google Scholar]
- Deming Y, Dumitrescu L, Barnes LL, Thambisetty M, Kunkle B, Gifford KA, et al. Sex-specific genetic predictors of Alzheimer's disease biomarkers. Acta Neuropathol 2018; 136: 857–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K, et al. Genome-wide association study identifies four novel loci associated with Alzheimer's endophenotypes and disease modifiers. Acta Neuropathol 2017; 133: 839–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, Bush WS, et al. Sex differences in the genetic predictors of Alzheimer's pathology. Brain 2019; 142: 2581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Cheng L, Zhang Y, Bai W, Zhou W, Wang T, et al. Rs4878104 contributes to Alzheimer's disease risk and regulates DAPK1 gene expression. Neurol Sci 2017. a; 38: 1255–62. [DOI] [PubMed] [Google Scholar]
- Hu Y, Jin S, Cheng L, Liu G, Jiang Q. Autoimmune disease variants regulate GSDMB gene expression in human immune cells and whole blood. Proc Natl Acad Sci USA 2017. b; 114: E7860–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019; 51: 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Hu Y, Han Z, Jin S, Jiang Q. Genetic variant rs17185536 regulates SIM1 gene expression in human brain hypothalamus. Proc Natl Acad Sci USA 2019. a; 116: 3347–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Hu Y, Jin S, Jiang Q. Genetic variant rs763361 regulates multiple sclerosis CD226 gene expression. Proc Natl Acad Sci USA 2017. a; 114: E906–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Hu Y, Jin S, Zhang F, Jiang Q, Hao J. Cis-eQTLs regulate reduced LST1 gene and NCR3 gene expression and contribute to increased autoimmune disease risk. Proc Natl Acad Sci USA 2016; 113: E6321–E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Jin S, Hu Y, Jiang Q. Disease status affects the association between rs4813620 and the expression of Alzheimer's disease susceptibility gene TRIB3. Proc Natl Acad Sci USA 2018; 115: E10519–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Xu Y, Jiang Y, Zhang L, Feng R, Jiang Q. PICALM rs3851179 variant confers susceptibility to Alzheimer’s disease in Chinese population. Mol Neurobiol 2017. b; 54: 3131–6. [DOI] [PubMed] [Google Scholar]
- Liu G, Yao L, Liu J, Jiang Y, Ma G, Chen Z, et al. Cardiovascular disease contributes to Alzheimer's disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 2014; 35: 786–92. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhang H, Liu B, Ji X. Rs2293871 regulates HTRA1 expression and affects cerebral small vessel stroke and Alzheimer's disease. Brain 2019. b; 142: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang H, Liu B, Wang T, Han Z, Ji X. rs4147929 variant minor allele increases ABCA7 gene expression and ABCA7 shows increased gene expression in Alzheimer's disease patients compared with controls. Acta Neuropathol 2020; 139: 937–40. [DOI] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, et al. GWAS on family history of Alzheimer's disease. Transl Psychiatry 2018; 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Grau S, de Rojas I, Hernandez I, Quintela I, Montrreal L, Alegret M, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: the GR@ACE project. Alzheimers Dement 2019; 15: 1333–47. [DOI] [PubMed] [Google Scholar]
- Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, et al. An xQTL map integrates the genetic architecture of the human brain's transcriptome and epigenome. Nat Neurosci 2017; 20: 1418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Lyons PA, Lee JC, Richard AC, Fortune MD, Newcombe PJ, et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet 2016; 12: e1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17: 1418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, et al. Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature 2016; 533: 95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 2018; 362: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 2013; 153: 707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.