Abstract
Expansions of trinucleotide or hexanucleotide repeats lead to several neurodegenerative disorders, including Huntington disease [caused by expanded CAG repeats (CAGr) in the HTT gene], and amyotrophic lateral sclerosis [ALS, possibly caused by expanded GGGGCC repeats (G4C2r) in the C9ORF72 gene], of which the molecular mechanisms remain unclear. Here, we demonstrated that lowering the Drosophila homologue of tau protein (dtau) significantly rescued in vivo neurodegeneration, motor performance impairments, and the shortened life-span in Drosophila expressing expanded CAGr or expanded G4C2r. Expression of human tau (htau4R) restored the disease-related phenotypes that had been mitigated by the loss of dtau, suggesting an evolutionarily-conserved role of tau in neurodegeneration. We further revealed that G4C2r expression increased tau accumulation by inhibiting autophagosome–lysosome fusion, possibly due to lowering the level of BAG3, a regulator of autophagy and tau. Taken together, our results reveal a novel mechanism by which expanded G4C2r causes neurodegeneration via an evolutionarily-conserved mechanism. Our findings provide novel autophagy-related mechanistic insights into C9ORF72-ALS and possible entry points to disease treatment.
Electronic supplementary material
The online version of this article (10.1007/s12264-020-00518-2) contains supplementary material, which is available to authorized users.
Keywords: ALS, C9orf72, G4C2, Huntington disease
Introduction
An expansion of the GGGGCC repeat (G4C2r) in intron 1 of C9ORF72 is a common mutation associated with sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [1, 2]. ALS is a fatal neurodegenerative disorder primarily affecting motor neurons, whereas FTD is caused by neurodegeneration primarily in the frontal, insular, and anterior temporal cortex. The aggregation-prone dipeptides synthesized from the expanded G4C2 via repeat-associated non-ATG translation and/or G4C2 RNA foci/membraneless granules originating via phase separation are likely the major cause of neurodegeneration, but the downstream molecular mechanisms remain unclear [1–7]. Abnormalities of the microtubule-binding protein tau play a central role in several neurodegenerative diseases termed tauopathies, including Alzheimer’s disease (AD), progressive supranuclear palsy (PSP), and tau-positive FTD with parkinsonism (FTD with parkinsonism-17). Among these, FTD with parkinsonism-17 is caused by aberrant splicing of tau, and similar mechanisms may contribute to the pathology in Huntington’s disease (HD) [8], another disease caused by nucleotide repeat expansion (CAG repeat expansion in exon 1 of the HTT gene) [9]. Abnormal phosphorylation and aggregation of tau has also been reported in patients with C9ORF72-ALS [10]. Thus, we investigated whether tau mediates expanded G4C2r-induced neurotoxicity, which may share molecular commonalities in the pathological mechanisms with CAG repeat expansion diseases.
Tau is the key protein in several neurodegenerative diseases such as AD and other tauopathies. The tau-related toxicity in neurodegenerative disorders may be a mixture of both loss-of-function of normal tau and gain-of-function of aggregation-prone tau caused by abnormal splicing and post-translational modification under pathological conditions. Loss of normal tau may have an impact on axonal transport and thus contribute to synaptic dysfunction and neurodegeneration [11]. Meanwhile, aggregated tau can be generated by hyperphosphorylation [12, 13] and/or an altered isoform ratio [14]. Hyperphosphorylated tau detaching from microtubules usually misfolds and sequesters normal tau and other related proteins to produce large insoluble tau aggregates, which may impair the cellular clearance machinery [15, 16], axonal transport [17], mitochondrial function [18, 19], and other cellular processes.
Drosophila models have been widely used to study genetic neurodegenerative disorders, especially CAG expansion and G4C2 expansion diseases, probably because of their monogenetic nature [20, 21]. Many of the genetic and chemical modifiers identified in these models have been validated in patient cells and mammalian models [22–25]. In this study, we first characterized the pathophysiological action of the Drosophila homologue of tau (dtau) in a model of HD. We confirmed an evolutionarily-conserved role of dtau in HD neurotoxicity by showing that the loss of dtau significantly rescued phenotypes in the Drosophila HD model expressing an exon 1 fragment of human mutant HTT protein, consistent with the report in a mouse model of HD expressing this fragment [8]. We then investigated the potential role of dtau in the expanded G4C2r-induced neurotoxicity in the Drosophila model, and explored tau-related pathogenic mechanisms, in which autophagy plays a key role.
Materials and Methods
Fly Stocks and Genetics
Fly cultures and crosses were maintained on standard food according to standard procedures and raised at 25 °C. The elav(c115)-GAL4 (elav(I)-GAL4) (no. 458), elav(II)-GAL4 (no. 8765), elav(III)-GAL4 (no. 8760), OK371-GAL4 (no. 26160), and UAS-GFP-mCherry-atg8a (no. 37749) were from the Bloomington Drosophila Stock Center at Indiana University (Bloomington, Indiana). The dtau−/− stock was as described previously [26]. GMR61G12-GAL4 was from the FlyLight GAL4 collection organization (http://flweb.janelia.org/cgi-bin/flew.cgi). The UAS-(G4C2)30-EGFP and UAS-(G4C2)3-EGFP lines were as described previously [27]. The transgenic Drosophila line expressing human wild-type was as described previously [28]. The expression of (G4C2)30 in all cells of the peripheral and central nervous systems using elav(I)-GAL4 caused lethality in early development as described previously [27]. As the pupal lethality of (G4C2)30 driven by elav(I)-GAL4 precluded studies in mature neurons of the adult brain, we used elav(II)-GAL4 or elav(III)-GAL4 as a substitute. UAS-HTT.ex1.Q25 and UAS-HTT.ex1.Q72 were generated by injecting pUAST- HTT.ex1.Q25 or pUAST- HTT.ex1.Q72 plasmid with helper-plasmids named Δ2-3 into w1118.
Behavioral and Lifespan Experiments
In behavioral experiments (climbing assay), we placed 15 age-matched virgin female flies in an empty vial and tapped them down. The percentage of flies that climbed past a 9-cm line after 15 s was recorded. The mean of five observations was plotted for each vial on each day, and data from multiple vials containing different batches of flies were plotted and analyzed using two-way ANOVA. Flies were randomly placed into each tube. For lifespan measurements, we placed ≥ 60 age-matched virgin females in an empty plastic vial and recorded the survival in each vial on each day. In both behavioral and lifespan measurement experiments, the investigator who performed the experiments was blind to the genotypes until data analysis. The survival distribution of the two genotypic groups were compared using the log-rank (Mantel-Cox) test.
Plasmids Used for Fly Strain Generation and Mammalian Cell Transfection
The HTT.ex1.Q25 and HTT.ex1.Q72 cDNA were obtained by PCR amplification from pcDNA- HTT.ex1.Q25 and pcDNA- HTT.ex1.Q72 plasmid, and they were cloned into the pUAST vector. The tau-snap plasmid was generated by inserting the cDNA expressing one transcript of human tau (0N4R, cloned by PCR amplification from the UAS-htau4R Drosophila strain) into a snap-tag vector (P9312S, New England Biolabs, CA). (G4C2)3 and (G4C2)25 plasmids were generated by inserting (G4C2)3 or (G4C2)25 repeats into the pTT-sfGFP vector. The (G4C2)3 repeats were generated by the primer-annealing process and (G4C2)25 repeats were generated using an enzyme to cut the plasmid named pHR-Tre3G-29xGGGGCC-12xMS2 (cat.no. #99149, Addgene, MA). The BAG3 (BAG family molecular chaperone regulator 3) vector was constructed by cloning the human BAG3 cDNA sequence from PUC57_BAG3wt-eGFP (cat.no. #98182, Addgene, MA) into the pTT-sfGFP vector. As the end of the BAG3 gene PCR product was a termination codon, GFP protein in the vector was not expressed with BAG3 protein. The mRFP-GFP-LC3B vector was from Addgene (cat.no. #21074, Addgene, MA). The Lamp1-mcherry vector was a gift from Prof. Shouqing Luo (Peninsula School of Medicine and Dentistry, University of Plymouth, UK).
SiRNA Treatment
The siRNAs were reverse-transfected into HEK293T cells with Lipofectamine 2000 (11668027, ThermoFisher, MA). All transfections were performed according to the manufacturer’s protocol. Cells were collected 3 days after siRNA transfection for western blots. The siRNAs were validated by qPCR and/or western blots for target knockdown. The siRNA target sequence was 5′-AAGGUUCAGACCAUCUUGGAA-3′.
Immunostaining
For immunofluorescence studies (Figs. 2D, 5A and 6C), cultured HeLa cells were fixed in 4% paraformaldehyde (PFA) at room temperature for 10 min after washing three times with 1× PBS (20 min each), and then washed again and permeabilized in 0.5% (v/v) TritonX-100 for 10 min. The cells were then blocked in blocking buffer [4% bovine serum albumin + 0.1% (v/v) Triton X-100 in 1× PBS] for 30 min, incubated overnight at 4 °C with the primary antibody (clone 5A6, P10636, Developmental Studies Hybridoma Bank, IA), and then washed three times with blocking buffer and incubated with secondary antibody at room temperature for 1 h. The cells on coverslips were then washed three times, stained with 0.5 mg/mL DAPI for 5 min at room temperature, and then mounted in Vectashield mounting medium (H-1002, Vector Laboratories, CA). Images were captured on a Zeiss Axio Vert A1 confocal microscope (H1009664-1, Zeiss, Oberkochen, Germany) and the number of htau puncta in each cell was analyzed blindly using ImageJ (ImageJ 1.52a, Wayne Rasband National Institures of Health, CA).
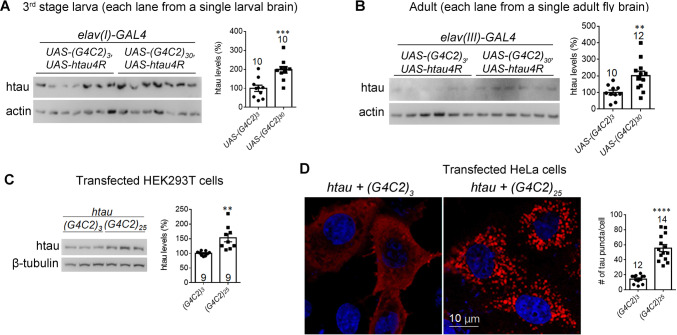
Fig. 2.
Expanded G4C2r expression increases tau protein levels or aggregation in Drosophila and in human cells. A, B Representative western blots and quantification of htau in brain lysates of the indicated genotypes showing that the total tau levels were increased by (G4C2)30 expression in the third instar larva (A) and adult (B) flies, compared to the controls expressing (G4C2)3 (numbers above bars, numbers of flies tested; statistical analysis, two-tailed unpaired t test). C Representative western blots and quantification showing that total htau levels were increased by (G4C2)25 compared to (G4C2)3 controls in transfected HEK293T cells (statistical analysis, two-tailed unpaired t test). D Representative immunofluorescence images and quantification (numbers of tau puncta per cell) showing that (G4C2)25 expression increased tau aggregates in HeLa cells compared to controls expressing (G4C2)3 (statistical analysis, two-tailed unpaired t tests). Note: aggregates were tested in HeLa cells because tau aggregates were hardly visible in HEK293T cells. In all plots, bars indicate the mean ± SEM; **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
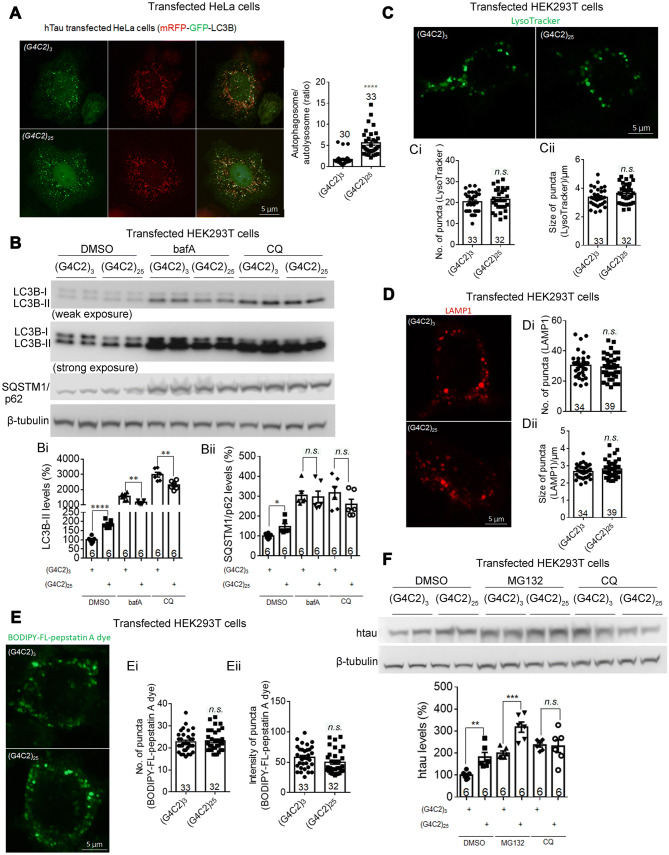
Fig. 5.
Expanded G4C2r expression impairs autophagic flux by inhibiting autophagosome–lysosome fusion in human cells. A Representative immunofluorescence images of mRFP-GFP-LC3B puncta and quantification of the autophagosome /autolysosome ratio (red + green + puncta : red + green – puncta). (G4C2)25 expression significantly decreased autophagosome–lysosome fusion in HeLa cells compared to controls expressing (G4C2)3 (n, number of individual flies; statistical analysis, two-tailed unpaired t test). B Representative Western blots and quantification showing that (G4C2)25 expression significantly increased LC3B-II and SQSTM1/p62 protein levels in HEK293T cells, compared to controls expressing (G4C2)3. Bi, Bii After the treatment with lysosome inhibitors bafilomycin A1 (bafA, 100 nmol/L) or chloroquine (CQ, 10 μmol/L), (G4C2)25 expression mildly decreased LC3B-II levels and did not change the SQSTM1/p62 levels in HEK293T cells, compared to controls expressing (G4C2)3 (n, number of biological replicates; statistical analysis, two-tailed unpaired t test). C Representative immunofluorescence images labeled by LysoTracker and quantifications of lysosomes (Ci, Cii) showing that (G4C2)25 expression did not change the number or size of lysosomes in HEK293T cells, compared to controls expressing (G4C2)3 (n, number of cells; statistical analysis, two-tailed unpaired t test). D As in C, but measured by the number and the averaged size of mCherry-LAMP1 puncta per cell in transfected HEK293T cells. E As in C, but measured by BODIPY-FL-pepstatin A, which specifically binds to and inhibits active CTSD (a key protease in the lysosome) at low pH (number, active lysosomes; intensity, level of active CTSD). F Representative western-blots and quantification of htau protein levels in HEK293T cells transfected with the indicated plasmids and treated with the indicated compounds (2 μmol/L MG132; 10 μmol/L CQ; n, number of independently plated and treated wells; statistical analysis, two-tailed unpaired t test). In all plots, bars indicate the mean ± SEM; n.s. P > 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
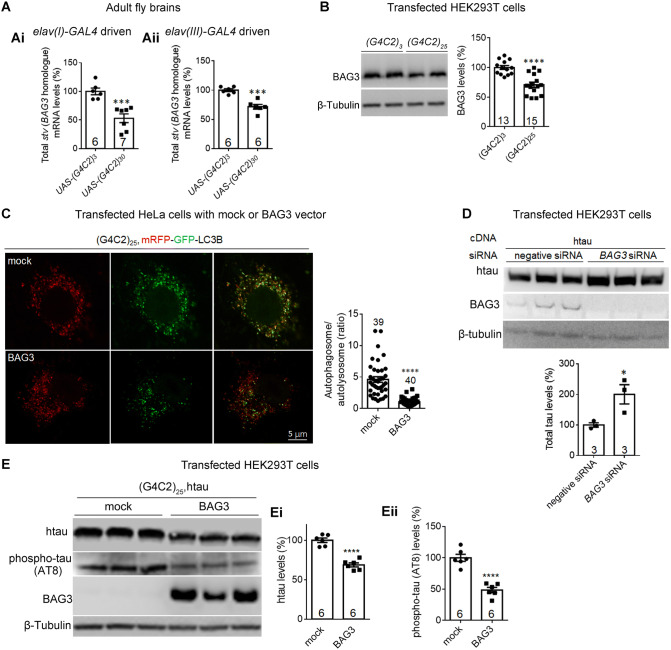
Fig. 6.
Expanded G4C2r expression decreases BAG3 (starvin) levels, contributing to the decreased autophagosome–lysosome fusion and increased tau accumulation. Ai, Aii qPCR quantification of the starvin (stv, the BAG3 homologue in Drosophila) mRNA levels in adult fly brains expressing (G4C2)30 versus (G4C2)3. B Representative western blots and quantification of lysates from HEK293T cells transfected with the indicated plasmids showing decreased endogenous BAG3 protein levels with (G4C2)25 expression. C Representative immunofluorescence images of mRFP-GFP-LC3B puncta and quantification of autophagosome/autolysosome ratio (red+green+ puncta : red+green- puncta) in HeLa cells transfected with the indicated plasmids (n, number of cells; statistical analysis, two-tailed unpaired t test). D, E Representative western blots and quantification of lysates from HEK293T cells transfected with the indicated cDNA plasmids and/or siRNAs. Ei, Eii Total tau and phospho-tau levels decreased with the expression of BAG3 protein in HEK293T cells expressing (G4C2)25 compared to mock controls (n, number of biological replicates; statistical analysis, two-tailed unpaired t test). In all plots, bars indicate the mean ± SEM; *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
For immunofluorescence of cultured HEK293T cells (Fig. 5C–E), all the procedures for immunostaining were the same as for HeLa cells, except for LysoTracker and cathepsin D (CTSD) immunostaining, for which LysoTracker dye (1:1000; L7526, ThermoFisher, MA) or BODIPY-FL-pepstatin A dye (1:500; P12271, ThermoFisher, MA) was added to live cells and incubated for 1 h; after which the cells were immediately sealed on slides with nail polish. Images were captured on a Zeiss Axio Vert A1 confocal microscope and the number of puncta per cell was analyzed blindly using ImageJ.
To image neurodegeneration in the fly brain (Figs. 1A, S1A, B, and S2), the whole brains of adult flies at the indicated ages were dissected on ice and then fixed in 4% PFA on ice for 20 min. Immunostaining was then performed in the same way as for cultured HeLa cells. The primary antibody used was anti-GFP (50430-2-AP, ProteinTech, Wuhan, China). The red fluorescence signals of DenMarker (Fig. S2) were imaged directly without antibody staining.
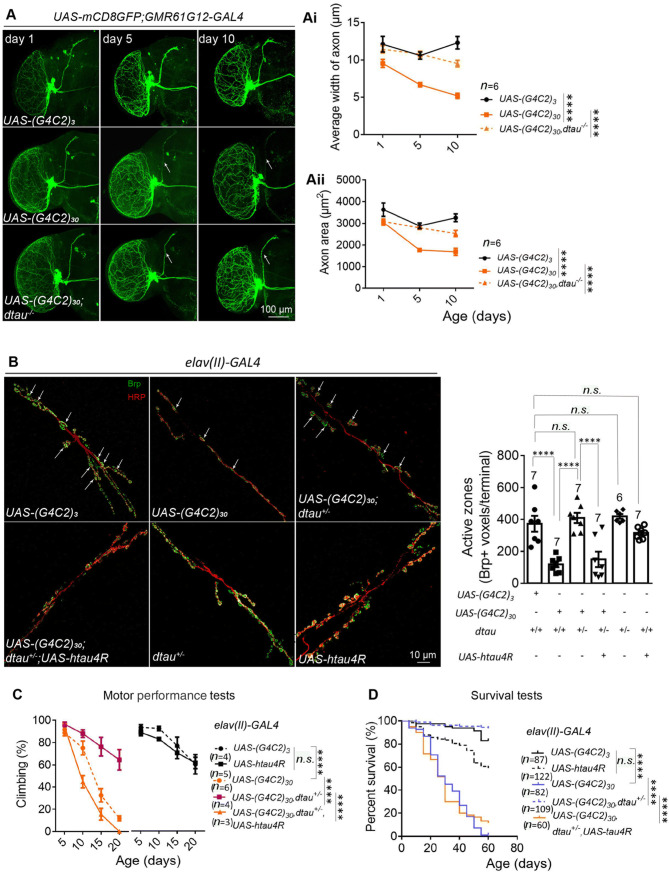
Fig. 1.
Homozygous (dtau−/−) and heterozygous (dtau+/−) knockout rescue the neurodegeneration, motor deficits, and shortened lifespan in flies expressing (G4C2)30. A Representative immunofluorescence images of whole-mount brains from flies of the indicated genotypes at the indicated ages showing neuronal degeneration. The small ventral lateral (sLNv) clock neurons were labeled by mCD8GFP protein, which was driven by GMR61G12-GAL4 (scale bar, 100 μm). Dtau−/− significantly rescued the neuronal degeneration induced by (G4C2)30 expression, using (G4C2)3 as the control (Ai, Aii; n, number of flies tested; statistical analysis, two-way ANOVA with Turkey’s post-hoc test). B Representative images and quantification of neuronal degeneration in the neuromuscular junctions (NMJs) of flies with the indicated genotypes. NMJs of muscle 6/7 in abdominal segments A2 and A3 of third instar larvae were stained with anti-HRP (neuronal axon marker, red) and anti-Bruchpilot (Brp) (active zone marker, green) (arrows, boutons of NMJs; scale bar, 10 μm). The Brp-positive active zones were reduced by (G4C2)30 expression, and rescued by dtau+/− (numbers above bars, numbers of flies tested; statistical analysis, one-way ANOVA with Turkey’s post-hoc test). C Motor performance (climbing) of flies with the indicated genotypes and ages. Dtau+/− rescued the motor deficits induced by (G4C2)30 expression (n, number of tested vials, each containing 15 virgin females; statistical analysis, two-way ANOVA with Dunnett’s post-hoc test). D Lifespans of flies with indicated genotypes. Dtau+/− rescued the shortened life-span induced by (G4C2)30 expression (n, number of flies; statistical analysis, log-rank test). For all plots in A–D, error bars indicate mean ± SEM; n.s. P > 0.05, ****P ≤ 0.0001).
To immunostain neuromuscular junctions (NMJs; Fig. 1B), muscle tissue was isolated by dissection of third stage larvae. The dissected samples were fixed in 4% PFA at room temperature for 30 min, and then washed 3 times in 0.5% PBST (~ 20 min each time). Immunostaining was then performed as for cells. The primary anti-Brp antibody nc82 (AB 2314866, Developmental Studies Hybridoma Bank, IA) was then added to the samples at 1:20 for incubation at 4 °C overnight. The samples were washed 3 times (20 min each), then incubated with the fluorophore-labeled primary anti-HRP antibody Cy3-HRP (1:200; 123-165-021, Jackson ImmunoResearch, PA) and the secondary antibody (goat anti-mouse, 633 nm) for 1 h at room temperature. The samples were then washed three times and mounted in Vectashield mounting medium (H-1002, Vector Laboratories, CA). Images were captured on the Zeiss Axio Vert A1 confocal microscope and analyzed blindly using ImageJ. The NMJs in muscles 6/7 of segment A2 or A3 were chosen for analysis.
The immunostaining process for neurons driven by OK371-GAL4 (Fig. 4D) was the same as for NMJ immunostaining but without the antibody staining. Images of neurons in the ventral nerve cord of each larva were captured and analyzed blindly using ImageJ.
Fig. 4.
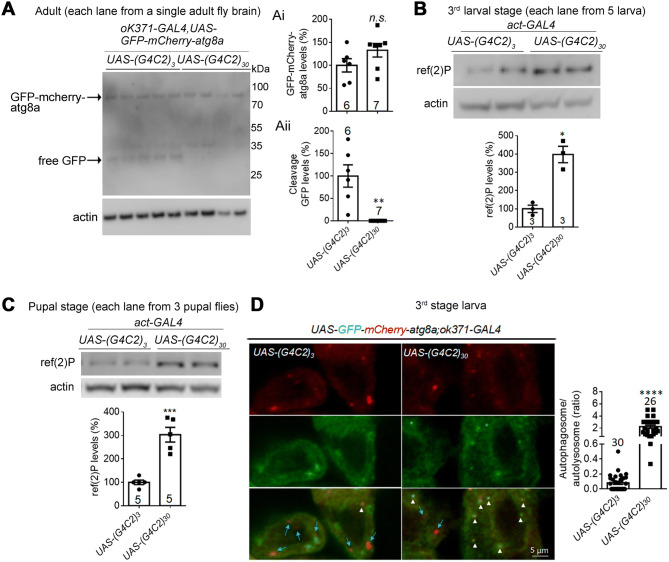
Expanded G4C2r expression impairs autophagic flux by inhibiting autophagosome–lysosome fusion in flies. A Representative western blots and quantification of fly brain lysates of indicated genotypes showing that the cleaved product band (free GFP cleaved from GFP-mCherry-atg8) is largely missing from motor neurons expressing (G4C2)30 driven by OK371-GAL4 (i, ii), suggesting an inhibition of autophagy activity in these neurons (numbers above bars, numbers of flies; statistical analysis, two-tailed unpaired t test). Note: each lane is loaded with the lysate from a single fly brain, and the cleaved product may be too low to detect in samples expressing (G4C2)30. B, C Representative western blots of the ref(2)P protein (homologue of human SQSTM1/p62) showing an increased level in larvae expressing (G4C2)30 (B) and pupae (C) (numbers in bars, numbers of flies; statistical analysis, two-tailed unpaired t test). D Representative immunofluorescence images of GFP-mCherry-atg8 puncta and quantification of the autophagosome/autolysosome ratio (red + green + puncta : red + green – puncta) in fly larvae (arrows, putative autolysosomes; arrowheads, putative autophagosomes; n, number of individual cells in >20 flies per group; statistical analysis, two-tailed unpaired t test). In all plots, bars indicate the mean ± SEM; n.s. P > 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Western Blot Experiments
To extract proteins from fly tissues, samples (brains or whole bodies) were dissected on ice and homogenized with a tissue grinder for 5 min at 60 Hz and lysed on ice for 60 min in 1× RIPA buffer (P0013B, Beyotime, Shanghai, China) and 1× complete protease inhibitor (4693159001, Sigma, MO). The samples were then sonicated for 10 cycles of 15 s on and 20 s off. The whole lysates were then loaded onto a 4%–12% bis-tris gradient gel for western blots. For western blots of ref(2)P (Fig. 4B, C), the lysates were centrifuged at > 20,000 g at 4 °C for 30 min. The precipitates were then collected and loaded for western blots. To extract proteins from cells, the cell pellets were lysed on ice for 30 min in 2% SDS [in 1× PBS + 1× complete protease inhibitor (4693159001, Sigma, MO)] and sonicated for 10 cycles, 15 s on and 20 s off. The whole lysates were then loaded for western blots.
Antibodies for Western Blots and Immunostaining
The antibodies used were: anti-Brp (ab2314866, Developmental Studies Hybridoma Bank, IA); anti-htau (clone 5A6, P10636, Developmental Studies Hybridoma Bank, IA); anti-actin (Millipore, cat. no. 92590); anti-β-tubulin (ab6046, Abcam, Cambridge, UK); anti-ref(2)P (ab178440, Abcam, Cambridge, UK); anti-GFP (50430-2-AP, ProteinTech, Wuhan, China); anti-BAG3 (10599-1-AP, ProteinTech, Wuhan, China); AT8 [detects phosphorylated tau (phospho-tau), MN1020, ThermoFisher, MA]; pS262 (detects Ser262 phospho-tau, OPA1-03142, ThermoFisher, MA); and pS199 (detects Ser199 phospho-tau, 701054, ThermoFisher, MA). The specificity of each antibody has been validated in this study (Fig. S4) or in the literature (e.g., as cited or indexed in Antibodypedia).
Bafilomycin A1 (bafA), chloroquine (CQ), and MG132 Treatment
To assess the LC3B-II protein levels in HEK293T cells expressing (G4C2)25 (Fig. 5B), after the cells were transfected with (G4C2)3 or (G4C2)25 for 48 h, they were incubated with the autophagy inhibitor bafA (100 mmol/L; S1413, Selleck, TX) or CQ (10 μmol/L; C6628, Sigma, MO) for another 24 h, then collected for western blots.
To determine the htau protein levels in HEK293T cells expressing (G4C2)25 (Fig. 5F), after they were transfected with (G4C2)3 or (G4C2)25 plus snap-tau vectors for 24 h, they were incubated with CQ (10 μmol/L; C6628, Sigma, MO) for another 48 h. Since MG132 (S2619, Selleck, TX) is highly toxic to HEK293T cells, we used a concentration of 2 μmol/L and the treatment time was ~15 h. Then cells were collected for western blots.
RNA Extraction and RT-qPCR
RNA was extracted from fly tissues or cells using an RNAprep kit (DP419, Tiangen, Beijing, China) followed by purification using an RNA-clean kit (DP412, Tiangen, Beijing, China) to remove proteins, and RNase-free DNase I (RT411, Tiangen) to break down the genomic DNA. cDNA was obtained by reverse transcription using the FastQuant RT Kit with the oligo (dT) primer (RR047A, Takara, Japan). qPCR was then performed using SYBR Green Realtime PCR Master Mix (QPK-201, Toyobo, Japan). All the primers have been published, validated, and further tested using standard and melting curves. Amplification efficiency was 95%–105% and the R2 for a linear relationship was >0.999 for all primers. No reverse-transcriptase controls were used to ensure the specificity of the signals. The qPCR primer sequences were as follows:
3R + 4R-F 5′-GGCGGCGAGAAGAAGATA-3′, 3R + 4R-R 5′-GCGAACCGATTTTGGACTT-3′; 4R-F 5′-TGGGCTCGACGGCCAATGTGAAACA-3′, 4R-R 5′-CCGCCACCGGGCTTGTGCTTTACA-3′; 3R-PI-F 5′-AAGGACAAGGCCAAGCCGAAGGTG-3′, 3R-PI-R 5′-GGTGCCTTCCAATACTTGATGTCTCCGC-3′; 3R-PJ-F 5′-AAGGACAAGGCCAAGCCGAAGGTG-3′, 3R-PJ-R 5′-TGGACTCTTGATGTCTCCGCCACCC-3′; EGFP-F 5′-TATATCATGGCCGACAAGCA-3′, EGFP-R 5′-GTTGTGGCGGATCTTGAAGT-3′; Upstream-F 5′-TCAATTAAAAGTAACCAGCAACCA-3′, and Upstream-R 5′-TCCCTATTCAGAGTTCTCTTCTTGTA-3′.
The process for testing standard curves was as follows. The fly or cellular cDNA was diluted 5 times step-by-step until 5 different concentration gradients were achieved. Then the samples, including the 5 concentration gradients, were subjected to qPCR. The results of qPCR for the samples had amplification efficiency 95%–105% and an R2 for linear relationship > 0.999.
Statistical Analysis
Statistical comparisons between two groups were assessed with unpaired two-tailed t tests; for comparisons among multiple groups with one-way ANOVA and post-hoc tests (Turkey’s tests); and for comparisons of series of data collected at different times with two-way ANOVA tests. The similarity of variances between groups to be compared was tested using GraphPad Prism 7 and Microsoft Excel 2016. Normality of data sets was assumed for ANOVA and t tests, and was assessed by Shapiro-Wilk tests. When the data were significantly different from a normal distribution, nonparametric tests were used. All statistical tests were unpaired and two-tailed.
Results
Homozygous or Heterozygous dtau-Knockout Rescues HD-Related Phenotypes in the Drosophila Model of HD
In order to capture the neuronal morphology in the Drosophila brain clearly, we used a simple GAL4-UAS system to express a membrane GFP marker (mCD8GFP) in a very small group of neurons using GMR61G12-GAL4, so that their morphology could be clearly observed. We assessed the HD-related neurodegeneration by observing the loss of major axon bundles from neurons labeled with GFP induced by the expression of the human mHTT but not the wild-type HTT exon1 fragment (HTT.ex1.Q72 versus HTT.ex1.Q25; Fig. S1A). The neurodegeneration in both dendrites and axons was detected by fluorescent protein-tagged marker proteins (Fig. S2).
At the whole-animal level, flies expressing mutant HTTexon1 (HTT.ex1.Q72) by the pan-neural driver elav-GAL4 caused motor impairments and shortened the life-span compared to flies expressing the wild-type version (HTT.ex1.Q25, Fig. S1C, D), recapitulating certain aspects of the HD-related phenotypes in patients.
We then investigated the potential role of tau in HD by testing knockout of the Drosophila tau gene (dtau, the homologue of human MAPT) to eliminate the expression of dtau proteins. The dtau knockout line we used was previously reported to present a specific deletion of the genetic region spanning exon 2 to exon 6, which codes for the microtubule-binding region of dtau [26]. Interestingly, homozygous knockout (dtau−/−) or heterozygous knockout (dtau+/−) significantly rescued the HD-related neurodegeneration, behavioral deficits, and shortened life-span phenotypes, suggesting that dtau mediates mHTT neurotoxicity, at least partially (Fig. S1B–D). These findings are consistent with a study using the mouse R6/2 HD model, confirming an evolutionarily-conserved role of dtau in neurodegeneration. To further demonstrate this, we expressed human tau (htau4R) in these flies and found that it restored the HD-related phenotypes in dtau+/− Drosophila (Fig. S1C, D).
Homozygous or Heterozygous dtau-Knockout Rescues the Expanded G4C2r-Induced ALS-Related Neurodegeneration, Behavioral Deficits, and Shortened Life-Span
We next investigated the potential functions of dtau in the expanded G4C2r-induced neurotoxicity in a well-characterized and widely-used Drosophila model expressing 30 repeats of G4C2 [(G4C2)30] [27]. Similar to the Drosophila model of HD, the GMR61G12-GAL4-driven UAS-mCD8GFP and UAS-(G4C2)30 expression led to neurodegeneration (Fig. 1A). In addition, (G4C2)30 expression in the neurons driven by elav-GAL4 led to a significantly reduced number of active zones in NMJs, as detected by the NMJ marker Brp and the axon marker HRP (Fig. 1B). At the animal level, neuronal expression of (G4C2)30 driven by elav-GAL4 led to motor performance impairments and a shortened life-span (Fig. 1C, D). Homozygous dtau-knockout (dtau−/−) or heterozygous knockout (dtau+/−) significantly mitigated all of these disease-related phenotypes (Fig. 1A–D), suggesting that dtau mediates the neurotoxicity induced by expanded G4C2r as well.
We then determined whether dtau and human tau are evolutionarily-conserved in mediating the expanded G4C2r-induced toxicity by expressing htau4R in the neurons driven by elav-GAL4. The expression of htau4R restored the (G4C2)30-induced neurodegeneration, deficient motor performance, and the shortened life-span in the dtau+/− flies (Fig. 1C, D), confirming that dtau plays a role in the expanded G4C2r-induced neurodegeneration.
Expanded G4C2r Increases Total Tau and Phospho-Tau Protein Levels in Drosophila and Human Cells
We then explored potential mechanisms through which dtau influenced the expanded G4C2r-induced toxicity, since the mechanisms had not yet been revealed. There were two major possibilities: (1) dtau is an upstream regulator of expanded G4C2r expression; and (2) dtau is a downstream factor that is modulated by the expanded G4C2r and mediates its neurotoxicity. To distinguish these possibilities, we first determined whether dtau-knockout lowered the expanded G4C2r RNA, which is likely the fundamental source of neurotoxicity in this model [27]. Since it is extremely difficult to amplify the G4C2 repeat region per se by quantitative PCR (qPCR), we chose the upstream and downstream sequences for qPCR measurements (Fig. S3A). We found no significant changes in the expanded G4C2r mRNA levels in the dtau+/− flies, suggesting that dtau is not an upstream regulator of expanded G4C2r expression (Fig. S3B).
The above evidence suggested that dtau is likely a downstream mediator of the expanded G4C2r-induced toxicity. We thus investigated the potential mis-splicing of tau transcripts, which is known to cause neurodegeneration [29–31]. Alternative splicing of exon 10 of the MAPT gene results in tau isoforms containing either three or four microtubule-binding repeats (3R and 4R) [30, 32]. Similar to human tau, dtau proteins expressed from different tau transcripts (Fig. S3C) contained either three or four putative microtubule-binding domains (MTBDs) that are homologous with those of mammals [32, 33]. In addition, all dtau proteins had an extra MTBD that is homologous with Caenorhabditis elegans but not mammals. Thus, dtau had the 4R versus the 3R isoform due to alternative splicing (or 5R versus 4R if considering the extra MTBD), corresponding to the 4R versus 3R isoforms in human tau. We thus measured the different dtau transcript levels and the 4R/3R ratio in fly brains with (G4C2)30 expression versus the control [(G4C2)3], and found no significant changes in either the transcript levels or the 4R/3R ratio (Fig. S3D), suggesting that the expanded G4C2r does not induce dtau mis-splicing.
An increased level of tau protein and/or phospho-tau is another possible driver of neurodegeneration [34–45]. We thus investigated whether the tau or phospho-tau protein levels are increased by expressing the expanded G4C2r in Drosophila. Expression of (G4C2)30 led to increased total tau protein levels in tissue samples from third-stage larvae and adult Drosophila (Fig. 2A, B), suggesting that the expanded G4C2r expression increases tau accumulation. Consistent with the finding in Drosophila, the exogenous expression of expanded G4C2r [(G4C2)25] in HEK293T cells also resulted in tau protein accumulation (Fig. 2C), as suggested by western blots using a tau-specific antibody (Fig. S4). We further investigated the levels of tau aggregates, which are likely to be a biomarker associated with toxic tau accumulation [46]. While tau aggregates were hardly observable in Drosophila and HEK293T cells with htau transgene expression [consistent with previous reports (not shown)] [28, 47], HeLa cells expressing (G4C2)25 showed a drastic increase of tau aggregates by both kinds of imaging (Fig. 2D), suggesting that expanded G4C2r is capable of enhancing the aggregated form of tau as well.
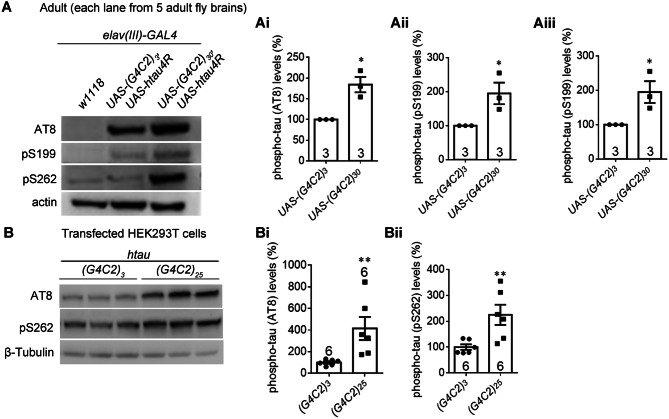
We further investigated the levels of phospho-tau, which is considered to be a major toxic species that causes neurodegeneration [37, 48]. Western blots of the brains of adult Drosophila expressing (G4C2)30 showed an increase of certain forms of phospho-tau as detected by the phospho-tau antibody AT8, pS199, or pS262 (Fig. 3A, Ai–Aiii). Consistent with this, the phospho-tau increase by (G4C2)25 expression was also found in HEK293T cells (Fig. 3B, Bi, Bii). The phospho-tau increase is likely a consequence of increased tau accumulation (Fig. 2A–C), and may also contribute to the neurotoxicity induced by expanded G4C2r expression.
Fig. 3.
Expanded G4C2r expression increases the levels of specific forms of phosphorylated tau protein in Drosophila and in human cells. A Representative western blots and quantification of brain lysate samples from indicated genotypes showing that the phosphorylated tau levels detected by the antibodies AT8, pS262, and pS199 were increased by (G4C2)30 expression in the adult fly head, compared to controls expressing (G4C2)3 (i, ii, iii) (numbers in bars, numbers of biological replicates; statistical analysis, two-tailed unpaired t test). B As in A, but in transfected HEK293T cells. In all plots, bars indicate the mean ± SEM; *P ≤ 0.05, **P ≤ 0.01.
Expanded G4C2r Expression Impairs Autophagic Flux by Inhibiting Autophagosome–Autolysosome Fusion in Flies
The tau protein is known to be degraded via autophagy in cells [35], so we hypothesized that the expanded G4C2r expression may impair autophagy, resulting in tau accumulation. We assayed the autophagic flux by assessing the cleavage of GFP-mCherry-atg8a (autophagy-related protein 8a). Atg8a in Drosophila is the homologue of the mammalian protein LC3, which is the key autophagosome protein that is cleaved in lysosomes. Thus, GFP-atg8a or GFP-LC3 cleavage is a widely used assay for measurements of autophagic flux [49, 50]. By expressing GFP-mCherry-atg8a driven by OK371-GAL4 in Drosophila, we measured the autophagic flux in motor neurons by the levels of free GFP [51], the cleaved product of the GFP-mCherry-atg8a protein. The free GFP band essentially disappeared in all the (G4C2)30-expressing Drosophila brains tested compared to those expressing (G4C2)3 (Fig. 4A, Ai, Aii), indicating an impairment of autophagic flux. Consistent with this, the ref(2)P protein (the Drosophila homologue of SQSTM1/p62 [52]) was significantly increased by (G4C2)30 expression at larval and pupal stages, compared to those expressing (G4C2)3 (Fig. 4B, C), further demonstrating that autophagic flux was impaired by the expanded G4C2r expression.
The major causes of impaired autophagic flux include decreased autophagosome formation, deficient autophagosome–autolysosome fusion, and defects of lysosome functions. To distinguish these possibilities, we measured the autophagosome–autolysosome fusion using GFP-mCherry-atg8a, which labels autophagosomes with both red and green fluorescence and autolysosomes with only red fluorescence because of the quenching of green fluorescence by the low pH in autolysosomes. By expressing GFP-mCherry-atg8a driven by OK371-GAL4 in Drosophila larvae, we found significantly more yellow puncta in Drosophila brains expressing (G4C2)30 than in those expressing (G4C2)3 (Fig. 4D), suggesting a reduction of autolysosome numbers and a likely inhibition of autophagosome–autolysosome fusion.
Expanded G4C2r Expression Impairs Autophagic Flux by Inhibiting Autophagosome–Autolysosome Fusion in Human Cells
Consistent with the results in Drosophila, in HeLa cells transfected with mRFP-GFP-LC3B, (G4C2)25 expression increased the autophagosome/autolysosome ratio compared to (G4C2)3 (Fig. 5A), further demonstrating deficient autophagosome–autolysosome fusion induced by expanded G4C2r expression. Consistent with this, the LC3B-II and SQSTM1/p62 protein levels were significantly higher in HEK293T cells expressing (G4C2)25 than in controls (Fig. 5B, Bi, Bii). The increased LC3-II levels could be an indication of increased autophagosome formation, decreased autophagosome–lysosome fusion, or decreased lysosome functions. To distinguish among these possibilities, we treated the cells with the lysosome inhibitors bafA and CQ, which block autophagosome–lysosome fusion and lysosome functions. If (G4C2)25 expression increased the LC3-II levels by activating autophagosome formation, the increase would persist in the presence of these inhibitors. Contradicting this prediction, the (G4C2)25-induced increase of LC3B-II and SQSTM1/p62 protein levels was ameliorated or even reversed in cells treated with bafA or CQ (Fig. 5B, Bi, Bii), suggesting defective autophagosome–lysosome fusion or lysosome functions in HEK293T cells expressing (G4C2)25. To measure potential changes in lysosomes, we labeled them with LysoTracker or LAMP1-mCherry and quantified the numbers and sizes of puncta. We found no significant difference between HEK293T cells expressing (G4C2)25 and (G4C2)3 in these parameters (Fig. 5C, Ci, Cii, D, Di, Dii), suggesting that the lysosomes were not influenced. We further measured lysosome functions using BODIPY-FL-pepstatin A, which specifically binds to active CTSD under low pH conditions [53]. The functional lysosomes were labeled by this dye and when we quantified the number and intensity of puncta, they showed no significant difference between HEK293T cells expressing (G4C2)25 and (G4C2)3 (Fig. 5E, Ei, Eii), suggesting that the lysosomes were not influenced. Taken together, the expanded G4C2r most likely impairs autophagosome–lysosome fusion.
Since tau protein may be degraded by proteasomes in addition to autophagy, we tested the effects of the proteasome inhibitor MG132 versus the autophagy inhibitor CQ on (G4C2)25-induced tau accumulation. The effects were not significantly influenced by MG132, but were largely abolished by CQ (Fig. 5F), confirming that the expanded G4C2r-induced tau accumulation is largely due to deficient autophagy.
Taken together, expanded G4C2r expression impaired autophagic flux by inhibiting autophagosome–autolysosome fusion, leading to tau accumulation.
Expanded G4C2r Expression Decreases BAG3 (starvin) mRNA Levels, Contributing to Deficient Autophagosome–Autolysosome Fusion and Tau Accumulation
We then explored the potential molecular mechanisms of autophagy impairment. Studies have shown that BAG3 (BAG family molecular chaperone regulator 3), a key modulator of autophagy [54, 55], is negatively associated with tau protein levels [56, 57]. We thus hypothesized that the expanded G4C2r expression may reduce BAG3 levels, leading to impaired autophagy and tau accumulation. Consistent with this hypothesis, (G4C2)30 expression in Drosophila neurons significantly reduced the mRNA levels of starvin (stv), the Drosophila homologue of BAG3 [58], compared to controls expressing (G4C2)3 (Fig. 6A, Ai, Aii). Due to a lack of anti-starvin antibody, we examined potential BAG3 protein level changes in HEK293T cells and found consistent BAG3 lowering by expanded G4C2r expression (Fig. 6B).
To confirm that deficient autophagosome–autolysosome fusion is due to the decreased BAG3 level, we over-expressed BAG3 in HeLa cells expressing (G4C2)25 and investigated fusion using the mRFP-GFP-LC3 marker. Compared to controls with (G4C2)25 expression alone, BAG3 overexpression significantly rescued the deficient autophagosome–autolysosome fusion (Fig. 6C), suggesting a role of reduced BAG3 levels.
Given that reduced BAG3 levels contribute to the impairment of autophagy and that tau is degraded by autophagy, it is likely that reduced BAG3 leads to tau accumulation. This is also consistent with the previously-reported negative association between BAG3 and tau protein levels [56, 57]. We thus investigated whether BAG3 functionally modulates tau protein levels by knocking down BAG3 in HEK293T cells co-expressing (G4C2)25 and tau, and found that lowering BAG3 expression increased the tau level (Fig. 6D). In the reverse direction, overexpressing BAG3 reduced the tau level (Fig. 6E, Ei, Eii).
Taken together, expanded G4C2r expression decreased BAG3 levels, leading to reduced autophagic flux and tau accumulation.
Discussion
Our study revealed a functional role of tau in the expanded G4C2r-mediated neurotoxicity. While some of the previous studies have suggested a potential link between tau and G4C2r expansion [10, 59], including a recent report showing synergistic neurotoxicity induced by htau and (G4C2)30 over-expression in Drosophila [60], there have been no loss-of-function experiments to demonstrate tau as a potential mediator of neurotoxicity in these flies. Our data revealed a novel pathogenic role of tau in mediating the expanded G4C2r-induced neurotoxicity by showing that dtau+/− and dtau−/− mitigated the neuronal degeneration, motor performance deficits, and a shortened lifespan in the Drosophila (G4C2)30 model (Fig. 1).
While our study mainly used the Drosophila model, the pathogenic action of tau in mediating the expanded G4C2r-induced neurotoxicity is likely to be evolutionarily conserved between humans and flies. Dtau has been cloned and demonstrated to have microtubule-binding properties [33], but its potential role in neuronal degeneration has been controversial. Studies using either Drosophila RNAi lines or tau hypomorphic/deficient lines led to contradictory results regarding the potential detrimental or beneficial effects of dtau removal [61], possibly because of off-target effects or incomplete removal of dtau, and there were no disease phenotype “restoration” experiments expressing human tau (htau) to test the functional similarity between dtau and htau. Meanwhile, dtau knockout did not trigger major effects on fly survival and climbing ability in the wild-type background (Figs 1 and S1), and it did not influence the toxic effects on fly survival associated with the expression of human Aβ [26], suggesting that dtau mediates neurodegeneration in a specific context, not generally.
On the other hand, our results demonstrated that expression of htau restored most of the phenotypes that were ameliorated by dtau+/− or dtau−/− (Figs. 1B–D and S1C–D). In addition, the expanded G4C2r expression increased the tau levels in both Drosophila and mammalian cells (Fig. 2A–C). Finally, dtau+/− and dtau−/− rescued the HD-related phenotypes in Drosophila expressing mHTT protein (Fig. S1), consistent with the report in a mouse model [8]. These pieces of cross-species evidence suggest an evolutionarily conserved role of dtau in mediating neuronal degeneration, and justify using dtau for mechanistic studies of tauopathies, providing a much faster and more convenient model for research in this field compared to mouse models.
In neurodegenerative mouse and cellular models, different tau protein species including non-phosphorylated tau and phosphorylated tau have different turnover rates [62–64]. The short-lived and misfolded tau protein species are degraded through the ubiquitination–proteasome system [65, 66], while the long-lived and aggregated proteins are preferentially degraded through the autophagy pathway [67, 68]. Consistent with this, the level of tau without expanded G4C2r co-transfection was increased more by MG132 treatment than by CQ treatment, suggesting that the proteasome system is more involved. The likely explanation is that the transient transfection leads to comparatively higher levels of proteasome-sensitive short-lived tau species than autophagy-sensitive long-lived species, because of the limited expression time. Meanwhile, this does not affect our conclusion that the expanded G4C2r-induced tau increase is mediated by inhibition of autophagy, because this increase was insensitive to MG132 treatment.
BAG3-mediated autophagy may play a key role in maintaining cellular homeostasis under stress as well as during aging (reviewed in [69]), when the increased demand for protein degradation enhances BAG3 expression, followed by a functional switch from the BAG1-mediated proteasomal to BAG3-mediated autophagic pathway. Meanwhile, the potential changes or functional roles of BAG3 in G4C2r models were previously unknown, and it was unclear which specific process in autophagy is influenced by BAG3. Our study revealed that BAG3 influences autophagosome–autolysosome fusion, at least in cells expressing expanded G4C2r (Fig. 6C). In addition, we demonstrated a potential role of the BAG3–autophagy pathway in the neurotoxicity induced by expanded G4C2r. However, the molecular mechanism by which expanded G4C2r expression causes BAG3 reduction is yet to be elucidated.
In summary, our study reveals a possible functional role of tau and BAG3-mediated autophagy in the neuronal degeneration caused by expanded G4C2r, providing entry points for targeting C9ORF72-ALS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Prof. Peng Jin and Ranhui Duan for providing the G4C2 Drosophila, Prof. L. Partridge, Haihuai He, and Peng Lei for providing the dtau-knockout Drosophila, Prof. Yongqin Zhang for providing related tool Drosophila stocks and UAS-htau4R [28], and Prof. Shouqing Luo for providing the Lamp1-mcherry vector. This work was supported by the National Natural Science Foundation of China (81925012 and 31961130379) and a Newton Advanced Fellowship (NAF_R1_191045).
Conflict of interest
The authors claim no conflict of interest.
Footnotes
Xue Wen and Ping An have contributed equally to this work.
Contributor Information
Jian Wang, Email: wangjian_hs@fudan.edu.cn.
Lixiang Ma, Email: lxma@fudan.edu.cn.
Boxun Lu, Email: luboxun@fudan.edu.cn.
References
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 5.Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173(958–971):e917. doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fay MM, Anderson PJ, Ivanov P. ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017;21:3573–3584. doi: 10.1016/j.celrep.2017.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Nogales M, Cabrera JR, Santos-Galindo M, Hoozemans JJ, Ferrer I, Rozemuller AJ, et al. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat Med. 2014;20:881–885. doi: 10.1038/nm.3617. [DOI] [PubMed] [Google Scholar]
- 9.TheHuntington’sDiseaseCollaborativeResearchGroup. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72: 971–983. [DOI] [PubMed]
- 10.King A, Al-Sarraj S, Troakes C, Smith BN, Maekawa S, Iovino M, et al. Mixed tau, TDP-43 and p62 pathology in FTLD associated with a C9ORF72 repeat expansion and p.Ala239Thr MAPT (tau) variant. Acta Neuropathol 2013, 125: 303–310. [DOI] [PubMed]
- 11.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 12.Andorfer C, Kress Y, Espinoza M, de Silva R, Tucker KL, Barde YA, et al. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J Neurochem. 2003;86:582–590. doi: 10.1046/j.1471-4159.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Gotz J. Tau-based therapies in neurodegeneration: opportunities and challenges. Nat Rev Drug Discov. 2017;16:863–883. doi: 10.1038/nrd.2017.155. [DOI] [PubMed] [Google Scholar]
- 14.Schoch KM, DeVos SL, Miller RL, Chun SJ, Norrbom M, Wozniak DF, et al. Increased 4R-Tau induces pathological changes in a Human-Tau mouse model. Neuron. 2016;90:941–947. doi: 10.1016/j.neuron.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo JL, Buist A, Soares A, Callaerts K, Calafate S, Stevenaert F, et al. The dynamics and turnover of Tau aggregates in cultured cells: Insights into therapies for tauopathies. J Biol Chem. 2016;291:13175–13193. doi: 10.1074/jbc.M115.712083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jo C, Gundemir S, Pritchard S, Jin YN, Rahman I, Johnson GV. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun. 2014;5:3496. doi: 10.1038/ncomms4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ittner LM, Fath T, Ke YD, Bi M, van Eersel J, Li KM, et al. Parkinsonism and impaired axonal transport in a mouse model of frontotemporal dementia. Proc Natl Acad Sci U S A. 2008;105:15997–16002. doi: 10.1073/pnas.0808084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy PH. Abnormal tau, mitochondrial dysfunction, impaired axonal transport of mitochondria, and synaptic deprivation in Alzheimer’s disease. Brain Res. 2011;1415:136–148. doi: 10.1016/j.brainres.2011.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu MH, Zhao XY, Yao PP, Xu DE, Ma QH. The mitochondrion: A potential therapeutic target for Alzheimer’s disease. Neurosci Bull. 2018;34:1127–1130. doi: 10.1007/s12264-018-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito K, Sass H, Urban J, Hofbauer A, Schneuwly S. GAL4-responsive UAS-tau as a tool for studying the anatomy and development of the Drosophila central nervous system. Cell Tissue Res. 1997;290:1–10. doi: 10.1007/s004410050901. [DOI] [PubMed] [Google Scholar]
- 21.Guo C, Pan Y, Gong Z. Correction to: Recent advances in the genetic dissection of neural circuits in Drosophila. Neurosci Bull. 2019;35:1138. doi: 10.1007/s12264-019-00398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu B, Al-Ramahi I, Valencia A, Wang Q, Berenshteyn F, Yang H, et al. Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat Neurosci. 2013;16:562–570. doi: 10.1038/nn.3367. [DOI] [PubMed] [Google Scholar]
- 23.Yao Y, Cui X, Al-Ramahi I, Sun X, Li B, Hou J, et al. A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity. Elife 2015, 4. [DOI] [PMC free article] [PubMed]
- 24.Yu M, Fu Y, Liang Y, Song H, Yao Y, Wu P, et al. Suppression of MAPK11 or HIPK3 reduces mutant Huntingtin levels in Huntington’s disease models. Cell Res. 2017;27:1441–1465. doi: 10.1038/cr.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Ramahi I, Giridharan SSP, Chen YC, Patnaik S, Safren N, Hasegawa J, et al. Inhibition of PIP4Kgamma ameliorates the pathological effects of mutant huntingtin protein. Elife 2017, 6. [DOI] [PMC free article] [PubMed]
- 26.Burnouf S, Gronke S, Augustin H, Dols J, Gorsky MK, Werner J, et al. Deletion of endogenous Tau proteins is not detrimental in Drosophila. Sci Rep. 2016;6:23102. doi: 10.1038/srep23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, et al. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- 29.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himmler A. Structure of the bovine tau gene: alternatively spliced transcripts generate a protein family. Mol Cell Biol. 1989;9:1389–1396. doi: 10.1128/mcb.9.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himmler A, Drechsel D, Kirschner MW, Martin DW., Jr Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Mol Cell Biol. 1989;9:1381–1388. doi: 10.1128/mcb.9.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidary G, Fortini ME. Identification and characterization of the Drosophila tau homolog. Mech Dev. 2001;108:171–178. doi: 10.1016/s0925-4773(01)00487-7. [DOI] [PubMed] [Google Scholar]
- 34.Adams SJ, Crook RJ, Deture M, Randle SJ, Innes AE, Yu XZ, et al. Overexpression of wild-type murine tau results in progressive tauopathy and neurodegeneration. Am J Pathol. 2009;175:1598–1609. doi: 10.2353/ajpath.2009.090462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu M, Zhang S, Tian X, Wu C. Mask mitigates MAPT- and FUS-induced degeneration by enhancing autophagy through lysosomal acidification. Autophagy. 2017;13:1924–1938. doi: 10.1080/15548627.2017.1362524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberson ED, Halabisky B, Yoo JW, Yao J, Chin J, Yan F, et al. Amyloid-beta/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer’s disease. J Neurosci. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso AC, Grundke-Iqbal I, Iqbal K. Alzheimer’s disease hyperphosphorylated tau sequesters normal tau into tangles of filaments and disassembles microtubules. Nat Med. 1996;2:783–787. doi: 10.1038/nm0796-783. [DOI] [PubMed] [Google Scholar]
- 38.Lasagna-Reeves CA, de Haro M, Hao S, Park J, Rousseaux MW, Al-Ramahi I, et al. Reduction of Nuak1 decreases Tau and reverses phenotypes in a tauopathy mouse model. Neuron. 2016;92:407–418. doi: 10.1016/j.neuron.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iijima-Ando K, Sekiya M, Maruko-Otake A, Ohtake Y, Suzuki E, Lu B, et al. Loss of axonal mitochondria promotes tau-mediated neurodegeneration and Alzheimer’s disease-related tau phosphorylation via PAR-1. PLoS Genet. 2012;8:e1002918. doi: 10.1371/journal.pgen.1002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapoport M, Dawson HN, Binder LI, Vitek MP, Ferreira A. Tau is essential to beta -amyloid-induced neurotoxicity. Proc Natl Acad Sci U S A. 2002;99:6364–6369. doi: 10.1073/pnas.092136199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 42.Vossel KA, Zhang K, Brodbeck J, Daub AC, Sharma P, Finkbeiner S, et al. Tau reduction prevents Abeta-induced defects in axonal transport. Science. 2010;330:198. doi: 10.1126/science.1194653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, Wang JW, Yu W, Lu B. Phospho-dependent ubiquitination and degradation of PAR-1 regulates synaptic morphology and tau-mediated Abeta toxicity in Drosophila. Nat Commun. 2012;3:1312. doi: 10.1038/ncomms2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu H, Possenti A, Freer R, Nakano Y, Villegas NCH, Tang M, et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat Neurosci. 2019;22:47–56. doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson GR, Wiedau-Pazos M, Sang TK, Wagle N, Brown CA, Massachi S, et al. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34:509–519. doi: 10.1016/s0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Mandelkow E. Tau in physiology and pathology. Nat Rev Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 47.Guthrie CR, Kraemer BC. Proteasome inhibition drives HDAC6-dependent recruitment of tau to aggresomes. J Mol Neurosci. 2011;45:32–41. doi: 10.1007/s12031-011-9502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feuillette S, Miguel L, Frebourg T, Campion D, Lecourtois M. Drosophila models of human tauopathies indicate that Tau protein toxicity in vivo is mediated by soluble cytosolic phosphorylated forms of the protein. J Neurochem. 2010;113:895–903. doi: 10.1111/j.1471-4159.2010.06663.x. [DOI] [PubMed] [Google Scholar]
- 49.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 50.Li Z, Wang C, Wang Z, Zhu C, Li J, Sha T, et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575:203–209. doi: 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- 51.Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 52.DeVorkin L, Gorski SM. Monitoring autophagic flux using Ref(2)P, the Drosophila p62 ortholog. Cold Spring Harb Protoc. 2014;2014:959–966. doi: 10.1101/pdb.prot080333. [DOI] [PubMed] [Google Scholar]
- 53.Chen CS, Chen WN, Zhou M, Arttamangkul S, Haugland RP. Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. J Biochem Biophys Methods. 2000;42:137–151. doi: 10.1016/s0165-022x(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 54.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kathage B, Gehlert S, Ulbricht A, Ludecke L, Tapia VE, Orfanos Z, et al. The cochaperone BAG3 coordinates protein synthesis and autophagy under mechanical strain through spatial regulation of mTORC1. Biochim Biophys Acta Mol Cell Res. 2017;1864:62–75. doi: 10.1016/j.bbamcr.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Ji C, Tang M, Zeidler C, Hohfeld J, Johnson GV. BAG3 and SYNPO (synaptopodin) facilitate phospho-MAPT/Tau degradation via autophagy in neuronal processes. Autophagy. 2019;15:1199–1213. doi: 10.1080/15548627.2019.1580096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei Z, Brizzee C, Johnson GV. BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol Aging. 2015;36:241–248. doi: 10.1016/j.neurobiolaging.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 59.Bieniek KF, Murray ME, Rutherford NJ, Castanedes-Casey M, DeJesus-Hernandez M, Liesinger AM, et al. Tau pathology in frontotemporal lobar degeneration with C9ORF72 hexanucleotide repeat expansion. Acta Neuropathol. 2013;125:289–302. doi: 10.1007/s00401-012-1048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He H, Huang W, Wang R, Lin Y, Guo Y, Deng J, et al. Amyotrophic Lateral Sclerosis-associated GGGGCC repeat expansion promotes Tau phosphorylation and toxicity. Neurobiol Dis. 2019;130:104493. doi: 10.1016/j.nbd.2019.104493. [DOI] [PubMed] [Google Scholar]
- 61.Bolkan BJ, Kretzschmar D. Loss of Tau results in defects in photoreceptor development and progressive neuronal degeneration in Drosophila. Dev Neurobiol. 2014;74:1210–1225. doi: 10.1002/dneu.22199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berger Z, Roder H, Hanna A, Carlson A, Rangachari V, Yue M, et al. Accumulation of pathological tau species and memory loss in a conditional model of tauopathy. J Neurosci. 2007;27:3650–3662. doi: 10.1523/JNEUROSCI.0587-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamada K, Patel TK, Hochgrafe K, Mahan TE, Jiang H, Stewart FR, et al. Analysis of in vivo turnover of tau in a mouse model of tauopathy. Mol Neurodegener. 2015;10:55. doi: 10.1186/s13024-015-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khlistunova I, Biernat J, Wang Y, Pickhardt M, von Bergen M, Gazova Z, et al. Inducible expression of Tau repeat domain in cell models of tauopathy: aggregation is toxic to cells but can be reversed by inhibitor drugs. J Biol Chem. 2006;281:1205–1214. doi: 10.1074/jbc.M507753200. [DOI] [PubMed] [Google Scholar]
- 65.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 66.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 68.Lee MJ, Lee JH, Rubinsztein DC. Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol. 2013;105:49–59. doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Behl C. Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol Sci. 2016;37:672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.