Abstract
Receptor adenylate cyclases (RACs) on the surface of trypanosomatids are important players in the host–parasite interface. They detect still unidentified environmental signals that affect the parasites’ responses to host immune challenge, coordination of social motility, and regulation of cell division. A lesser known class of oxygen-sensing adenylate cyclases (OACs) related to RACs has been lost in trypanosomes and expanded mostly in Leishmania species and related insect-dwelling trypanosomatids. In this work, we have undertaken a large-scale phylogenetic analysis of both classes of adenylate cyclases (ACs) in trypanosomatids and the free-living Bodo saltans. We observe that the expanded RAC repertoire in trypanosomatids with a two-host life cycle is not only associated with an extracellular lifestyle within the vertebrate host, but also with a complex path through the insect vector involving several life cycle stages. In Trypanosoma brucei, RACs are split into two major clades, which significantly differ in their expression profiles in the mammalian host and the insect vector. RACs of the closely related Trypanosoma congolense are intermingled within these two clades, supporting early RAC diversification. Subspecies of T. brucei that have lost the capacity to infect insects exhibit high numbers of pseudogenized RACs, suggesting many of these proteins have become redundant upon the acquisition of a single-host life cycle. OACs appear to be an innovation occurring after the expansion of RACs in trypanosomatids. Endosymbiont-harboring trypanosomatids exhibit a diversification of OACs, whereas these proteins are pseudogenized in Leishmania subgenus Viannia. This analysis sheds light on how ACs have evolved to allow diverse trypanosomatids to occupy multifarious niches and assume various lifestyles.
Keywords: trypanosomatid, Kinetoplastida, adenylate cyclase, receptor, oxygen, phylogenetics
Significance
Adenylate cyclases (ACs) are a protein family that mediates cellular responses to external stimuli. In kinetoplastids, a group of organisms with well-known parasites among its ranks, genes encoding ACs have undergone considerable expansion and diversification. However, the evolutionary forces shaping this phenomenon are poorly understood. Here, we have studied all identifiable kinetoplastid ACs and show that there is a correlation with AC gene family expansion and lifestyle. Specifically, these proteins appear to mediate immune evasion, insect tissue navigation, or even interaction with endosymbionts. Thus, this work sheds light on how gene expansion can allow diverse kinetoplastids to occupy diverse ecological niches.
Introduction
Trypanosomatids are obligatory parasites of the class Kinetoplastea. These monoflagellated protists evolved into a highly diverse and speciose group, members of which are capable of colonizing various invertebrate and vertebrate hosts, as well as numerous plants. Several lines of evidence suggest that trypanosomatids were primarily parasites of insects that only later infected vertebrates and plants (Maslov et al. 2013; Lukeš et al. 2014). Consequently, only a relatively small fraction of trypanosomatids alternate between the invertebrate and vertebrate or plant hosts (Lukeš et al. 2018). Due to the clinical and economic relevance of life-threatening diseases, such as leishmaniases, African trypanosomiases, and Chagas disease, significant attention has been given to the study of their etiological agents belonging to the genera Leishmania and Trypanosoma (Büscher et al. 2017; Telleria and Tibayrenc 2017; Bruschi and Gradoni 2018). Not surprisingly, their distinct lifestyles are reflected in their respective gene pools. Indeed, genetic redundancy, generated by the expansions or contractions of certain protein families is one of the key factors behind the switch from the free-living to the parasitic life strategy (Jackson et al. 2016; Opperdoes et al. 2016).
One of the outcomes of the inaugural sequencing of the “TriTryp” genomes (Leishmania major, Trypanosoma brucei, and T. cruzi) was the detection of remarkable expansions of surface protein families involved in immune evasion, infection, and host–parasite interplay. These include gp85-transialidases, mucin-associated surface proteins, and variant and invariant surface glycoproteins (VSGs/ISGs). In all cases, these genes were found to be very variable in sequence, with a high degree of pseudogenization, and often located in the non-syntenic sub-telomeric regions (El-Sayed 2005).
Because dixenous Trypanosoma and Leishmania spp. alternate between the vertebrate and invertebrate hosts and undergo extensive differentiation throughout their life cycles, they are subject to a range of extracellular stimuli. However, little is known about the signaling pathways involved in such transitions (Shaw et al. 2019). Nevertheless, cyclic AMP (cAMP), a major universal second messenger that is generated by adenylate cyclases (ACs), has been demonstrated to play a role in diverse processes ranging from cell differentiation to quorum sensing (Gancedo 2013). Moreover, other cAMP signaling-related molecules, such as phosphodiesterases (PDEs), have been linked to the host–parasite interplay in trypanosomatids (Salmon et al. 2012a; Makin and Gluenz 2015; Saldivia et al. 2016; Musikant et al. 2017; Shaw et al. 2019). A prominent group in this respect is the family of ACs that is represented by receptor adenylate cyclases (RACs) and oxygen-sensing adenylate cyclases (OACs) in trypanosomatids. All these proteins contain an AC domain (Sen Santara et al. 2013; Salmon 2018). But in mammalian cells, the G protein-coupled receptors are physically separated from the effector AC domains, in the trypanosomatids RACs, both the ectodomain (ED) and the AC domain are on the same polypeptide chain, separated by the central transmembrane domain (TMD) that allows the protein to be properly embedded in the plasma membrane (Salmon 2018). A highly variable ED may facilitate the binding of yet unknown host-derived compounds (Saada et al. 2014; Makin and Gluenz 2015).
Interestingly, the T. brucei genome encodes ∼80 RACs (Salmon 2018), a prominent expansion when compared with other trypanosomatids, for which complete genomes are available (Lukeš et al. 2018). However, the reasons of this expansion remain elusive. The initially characterized RAC-encoding genes of T. brucei were found in the VSG “expression sites” (ES) and elsewhere in the genome. To distinguish between these two types, the ES-associated RACs were called ESAG4, whereas RACs outside the ES were termed GRESAG4 (genes related to ESAG4) and subdivided into subfamilies 4.1 to 4.4. Moreover, phylogenetic analysis detected also two ESAG4-like genes (ESAG4L) located outside the ES (Salmon et al. 2012b). ESAG4 is co-expressed with VSG genes in the flagellum of bloodstream form (BSF) of T. brucei that infects mammals (Paindavoine et al. 1992), but both GRESAG4 and ESAG4L genes were found to be constitutively expressed throughout its life cycle (Salmon et al. 2012b). The knock-out of ESAG4 did not result in a specific phenotype, RNAi-mediated knock-down of GRESAG4 and ESAG4L produced dramatic cytokinesis defects, suggesting their role in this process (Salmon et al. 2012b). Furthermore, both ablation or inhibition of phosphodiesterases PDEB1 and/or PDEB2 presented the same phenotype as ESAG4 RNAi, lending further support for the involvement of cAMP in BSF cytokinesis (Oberholzer et al. 2007; De Koning et al. 2012).
Functional analyses of a handful of T. brucei RAC proteins revealed their diverse roles in PCF and BSF. Some RACs localize to distinct subdomains of the flagellar membrane, including some located only in the flagellar tip (Saada et al. 2014). In PCF, RAC-produced cAMP functions as a second messenger involved in social motility (SoMo). Of all the RACs characterized so far, only those localized to the flagellar tip were associated with SoMo (Saada et al. 2014; Lopez et al. 2015). PDEs act as negative regulators of cAMP signaling by converting cAMP into AMP (Salmon 2018). In T. brucei, RNAi-mediated depletion of PDE led to the defect in social behavior directly correlated with an increase in the intracellular cAMP levels (Oberholzer et al. 2015). This defect, however, could be complemented by adding the wild-type T. brucei cells apparently providing yet unknown extracellular factor(s) required for SoMo, thus suggesting that regulation of SoMo in trypanosomatids might be even more complex than previously thought (Oberholzer et al. 2015). Recently, flagellar cAMP-mediated signaling associated with SoMo was found to be required for the passage of PCFs through the peritrophic matrix of the tsetse fly (Shaw et al. 2019). In BSF, the drop of RAC-generated cAMP impairs TNF-α production by macrophages, compromising the infection and, thus, increasing survival of the host. Based on this, it has been proposed that the expansion of RACs in the extracellular stage might have been triggered by a prolonged exposure of parasites to the immune system, and that the extensive diversification of their EDs may help to evade humoral responses (Salmon et al. 2012a).
Very little is known about proteins interacting with the extracellular domain of ACs, or about the downstream targets of cAMP. The first indirect evidence of a possible ligand of an unidentified T. cruzi AC was the α-d globin subunit from the kissing bug Triatoma sp., which activated and promoted the differentiation of epimastigotes into metacyclic trypomastigotes (Fraidenraich et al. 1993). Similarly, in salivarian trypanosomes, an extract from the tsetse fly hindgut induced the AC activity of both PCF and BSF. This implies the presence of a vector-derived ligand that is recognized by Trypanosoma RACs (Bachmaier et al. 2019). There is also evidence that progression of BSF into the non-dividing stumpy form is mediated, at least in part, by a cAMP-dependent mechanism (Van Den Abbeele et al. 1995; Vassella et al. 1997). However, other factors involved in this differentiation process include oligopeptides (Rojas et al. 2019), target of rapamycin (Barquilla et al. 2012), and AMP-activated kinase (Saldivia et al. 2016) pathways. Regarding the downstream effectors of cAMP signaling, it is well-established that protein-kinase A (PKA) in the mammalian cells becomes activated upon binding of cAMP and, in turn, phosphorylates other kinases, initiating a signaling cascade (Taylor et al. 2013). Interestingly, in T. cruzi, PKA interacts with trans-sialidases and the PI3 kinase VPS34, coordinating cAMP production (Bao, Weiss, et al. 2008, 2010). However, in T. brucei, PKA appears to be activated by cGMP rather than cAMP (Shalaby et al. 2001; Bachmaier et al. 2019). Nonetheless, T. brucei flagellar cAMP responsive proteins (CARPs 1–4) have been identified and validated as interesting drug targets by means of a PDE inhibition screening (Gould et al. 2013).Yet another variation has been described in L. major, where the PKA activation correlates with cAMP production and is linked to the parasite’s survival under hypoxic conditions via OACs, another significantly expanded group of ACs (Sen Santara et al. 2013). Both RACs and OACs appear to play critical roles in parasite survival and niche adaptation, yet the molecular mechanisms, governing their function, remain to be elucidated. Although ACs and cAMP signaling in trypanosomatids have been extensively studied, many important questions concerning extracellular ligands and downstream interactors remain unanswered. Finally, the phylogenetic distribution of RACs and OACs in trypanosomatids, especially their monoxenous representatives, remains understudied. To the best of our knowledge, the most comprehensive phylogenetic study of ESAG4 homologs encompassed just five species of the genus Trypanosoma (Jackson et al. 2013).
In the present work, we provide extensive large-scale phylogenetic analyses of ACs, identified in the genomes of parasitic trypanosomatids and a free-living eubodonid B. saltans. Our analysis sheds light on sequence diversity and domain architecture of RACs and OACs, their evolutionary origin, and the roles that pseudogenization and protein family expansion played in the evolutionary history of trypanosomatid ACs. Moreover, molecular analysis of several previously uncharacterized ACs in T. brucei and L. mexicana provides additional evidence for the functional role of the flagellum as an environmental sensor.
Materials and Methods
Phylogenetic and Clustering Analyses
A well-characterized T. brucei RAC sequence (Tb927.7.6050) (Paindavoine et al. 1992) was used as a query to perform a BLAST search with an E-value cut-off 1 × 10−20 against a dataset containing annotated proteins and genomes of 57 trypanosomatid species in addition to the eubodonid B. saltans (supplementary table S1, Supplementary Material online). Sequences were aligned using MUSCLE (Edgar 2004) and those, containing more than 90% of gaps, were excluded from further analyses. The resulting alignment was trimmed using TrimAl v.1.4.rev22 with the “strict” option (Capella-Gutierrez et al. 2009). A maximum-likelihood phylogenetic tree that included 631 sequences and 802 aligned amino acid positions was constructed using IQTree v. 1.5.3 with 1,000 standard bootstrap replicates and VT + F + G4 model (Minh et al. 2013). Bayesian tree was inferred in MrBayes v3.2.6 (Huelsenbeck and Ronquist 2001) with a mixed amino acid prior (WAG model was selected), four discrete γ categories and allowing for a proportion of invariable sites. The analysis was run for 1,300,000 generations, 25% of iterations were discarded as a burn-in and every 100th tree was sampled. Although it was not possible to obtain a tree with high support values for all nodes, because the dataset incorporates a high number of divergent sequences, sometimes partial and different in their domain architecture, its full topology can be visualized in Newick format (supplementary file S1, Supplementary Material online). The clustering analysis of salivarian trypanosome RACs (Trypanosoma clades I and II depicted in fig. 1) was performed using Clustal Omega multiple sequence alignment server with default parameters and visualized using Phylo.io (Robinson et al. 2016; Madeira et al. 2019). Within each group, sequences were aligned and compared regarding length, presence of putative domains (periplasmic-binding-, AC-, nucleotide-binding, globin, etc.) retrieved from Pfam and InterPro databases, and signal peptides/glycosyl phosphatidyl-inositol (GPI) anchor motifs were predicted using SignalP 5.0 and PredGPI with the default settings (Pierleoni et al. 2008; Almagro Armenteros et al. 2019). TMDs were predicted by TMHMM 2.0 (Krogh et al. 2001). Additionally, T. brucei RACs were further screened for preferential expression in PCF and BSF life cycle stages by using ribosome profiling data (Vasquez et al. 2014).
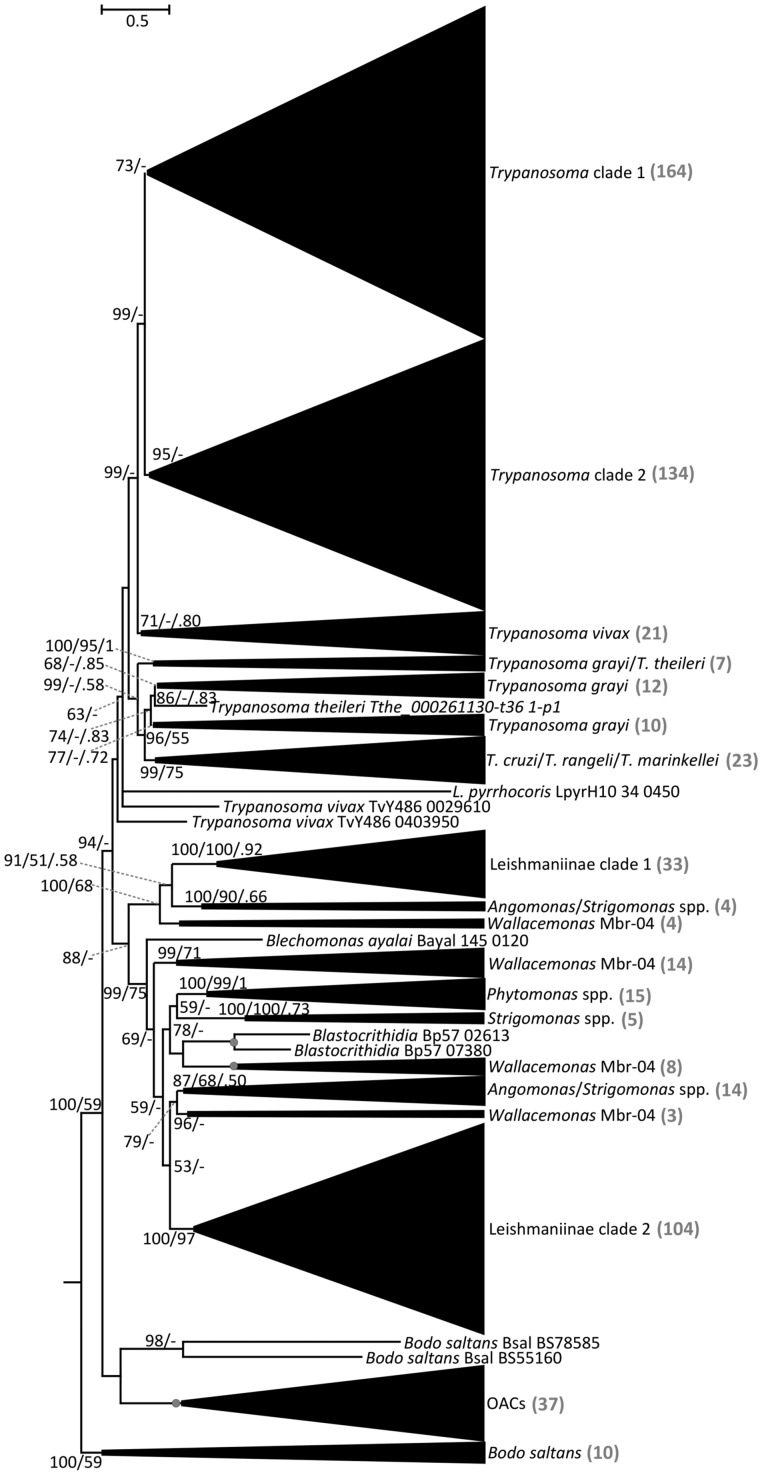
Fig. 1.
Phylogenetic tree of kinetoplastid ACs. Maximum-likelihood phylogenetic tree of kinetoplastid ACs based on a trimmed alignment of 631 sequences. Scale bar on top left represents 0.5 substitutions per site. The support values are in the following format: SH-aLRT support (%)/bootstrap support (%)/posterior probability. Only bootstrap support values ≥50% and posterior probabilities ≥0.50 are shown; nodes displaying maximal supports are marked with gray circles. “Trypanosoma clade 1” and “Trypanosoma clade 2” contain the genes of salivarian clades (T. congolense, T. brucei brucei, T. brucei gambiense, T. evansi, and T. equiperdum) upregulated in bloodstream and procyclic forms, respectively (see supplementary file S1, Supplementary Material online for details). OACs, oxygen-sensing adenylate cyclases from Leishmaniinae and other species (complete tree in fig 4). “Leishmaniinae clade 1” and “Leishmaniinae clade 2” contain homologs of leishmanial RAC-A and RAC-B sequences. The number of sequences within each collapsed clade is shown in gray color in the brackets.
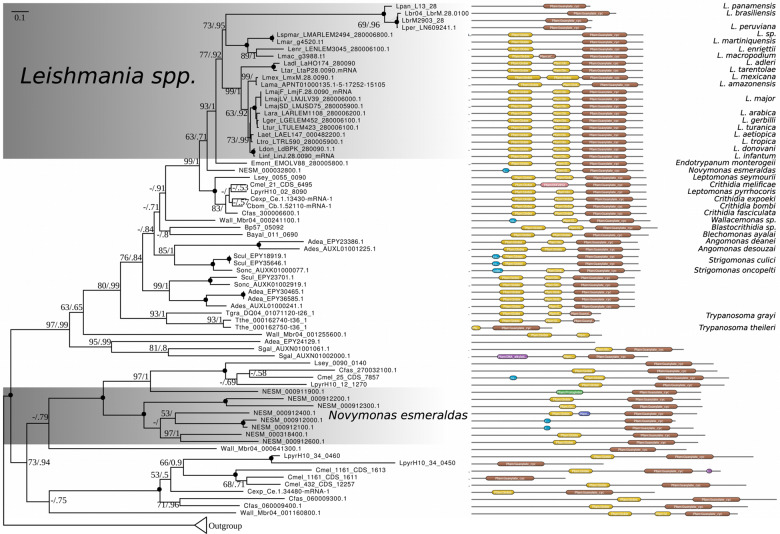
For the analysis of OACs, the protein sequence of L. major OAC (LmjF.28.0090) (Sen Santara et al. 2013) was used as a query for a BLAST search against the reference dataset (supplementary table S1, Supplementary Material online) with an E-value cut-off of 10−15. As described above, 206 OAC homologs were identified and aligned, with maximum likelihood and Bayesian trees inferred using the final alignment of 1,032 amino acids in the IQ-Tree v. 1.5.3 (VT + F + I + G4 model; 1,000 standard bootstrap replicates) and PhyloBayes MPI v.1.7b (GTR + CAT model; four discrete γ categories; 120,000 iterations), respectively. In Bayesian analysis, four chains were run and converged on the topology shown in figure 4. The initial 20% of cycles were discarded as burn-in, and every fifth tree was sampled.
Fig. 4.
Phylogenetic analysis of OACs in trypanosomatids. Maximum-likelihood phylogenetic tree of trypanosomatid OACs with nodes exhibiting maximal bootstrap support and posterior probability (inferred using PhyloBayes MPI) marked by black circles. Numbers at the nodes represent bootstrap supports/posterior probabilities. Scale bar on top left depicts number of substitutions per site. Domain structures for the protein sequences are depicted on the right. The length of the black line in each case is proportional to protein length; globin and AC domains are shown in yellow and brown, respectively. Other less significant domains (DUF and DNA alkylation) are also shown. Outgroup contains AC sequences closely related to OACs and, thus, retrieved by our searches, and yet lacking globin domain.
T. brucei Culture, Endogenous Tagging, and Transfection
T. b. brucei SMOX cells (Poon et al. 2012) were grown in SDM79 supplemented with 10% fetal bovine serum (Sigma-Aldrich). To assess the expression and subcellular localization of selected RACs, they were endogenously tagged on the C-termini with V5 or V5-mNeonGreen, as described elsewhere (supplementary table S2, Supplementary Material online) (Dean et al. 2015). For transfection, ∼107 exponentially growing cells were washed and re-suspended in Human T-Cell Nucleofector transfection solution (Lonza) according to the manufacturer’s instructions and transfected using the AMAXA program X-014. Twelve to 16 h post transfection, cells were diluted to the density of 0.2 cells/well in 96-well plates and incubated for 2 weeks in the presence of hygromycin or blasticidin (Merck) at 50 µg/ml. Plates were evaluated for the expression of V5-tagged proteins by Western blotting (WB) and immunofluorescence assay (IFA) analyses.
Immunofluorescence Microscopy and Immunoblotting Assays
Both IFA and WB analyses were performed as described previously (Durante et al. 2015) and the tagged proteins were visualized using primary monoclonal α-V5 (Thermo Fischer Scientific; 1:500 for IFA and 1:2,000 for WB), α-cMyc, and α-HA (both from Sigma-Aldrich; 1:2,000 for WB) antibodies, followed by secondary donkey α-mouse Alexa 488 antibody (Molecular Probes) for IFA and peroxidase-coupled rabbit α-mouse antibody (Sigma-Aldrich) for WB. Micrographs were taken with a Zeiss Axioplan-2 fluorescence microscope (Zeiss) and the acquired images were processed with ImageJ software (Schindelin et al. 2012).
Cultivation and Generation of L. mexicana Recombinant Cell Lines
L. mexicana promastigotes (MNYC/BZ/62/M379) were cultivated in complete M199 medium and transfected as described previously (Kraeva et al. 2019). All genetic manipulations followed the published LeishGEdit protocols (Beneke et al. 2017) to produce either mNeonGreen-Myc- or HA-V5-tagged OAC (encoded by LmxM.28.0090). For generation of the add-back strain, the LmxM.28.0090 gene was PCR amplified and cloned into the pLEXSY-sat2.1 vector (Jena Bioscience) with a C-terminal HA tag. The ablated strain was transfected with a linearized plasmid, selected in the presence of nourseothricin, and validated by WB.
RT-PCR and Growth Curve Analyses of L. mexicana Lines
For the RT-PCR validation of LmxM.28.0090 in Cas9, knock-out and add-back cell lines, total RNA from L. mexicana promastigotes of each strain was extracted using the TRI Reagent (Sigma-Aldrich). First-strand cDNA synthesis was done using RT-PCR QuantiTect Reverse Transcription kit (Qiagen). A Spliced Leader PCR (SL-PCR) was performed as described elsewhere (Hashimi et al. 2013) with SL for and RT rev primers (supplementary table S2, Supplementary Material online). Cell proliferation was analyzed as described previously (Durante et al. 2015) in three biological replicates (each in three technical replicates).
Results and Discussion
Adenylate Cyclase Family Is Expanded in Some Trypanosomatid Lineages
In order to obtain a list of putative ACs from fully annotated trypanosomatid genomes (supplementary table S1, Supplementary Material online), we conducted a BLAST search using the RAC gene Tb927.7.6050 as a query and retrieved 631 AC sequences in total. Interestingly, we could not identify any AC in the well-assembled genome of Paratrypanosoma confusum, a basally branching monoxenous parasite of mosquitoes (Flegontov et al. 2013). Understudied trypanosomatid OACs emerged as a separate well-supported clade in the ACs tree, which prompted us to investigate their phylogenetic distribution in more detail.
The BLAST output showed a remarkable expansion of the RAC family in T. b. brucei, retrieving a total of 88 sequences, which is in agreement with previous results (Salmon 2018). In the livestock-dwelling Trypanosoma congolense and T. evansi, RACs showed a similar degree of expansion, with 69 members each, whereas the human-dwelling T. b. gambiense carries 51 identified RACs (supplementary table S1, Supplementary Material online). In other trypanosomatids, the number of ACs per genome is significantly lower. On the higher end of AC repertoire spectrum, Wallacemonas sp. contains 30 genes. As this is an understudied group of monoxenous trypanosomatids (Kostygov et al. 2014), the reasons for this relatively high number of RACs remains mysterious. Members of the endosymbiont-harboring genus Angomonas display a highly variable number of RACs, ranging from 20 in A. ambiguus to only 3 in A. deanei, with a similar situation in the sister clade Strigomonas, for which a range between 4 and 11 RACs was documented in S. culicis and S. galati, respectively. Variable numbers of RACs were identified in the representatives of the dixenous genus Phytomonas, which are transmitted by hematophagous insects to plants (Jaskowska et al. 2015). The genome of P. serpens harbors 17 RACs, whereas the closely related strains Phytomonas sp. HART1 and Phytomonas sp. EM1 retain only one detectable RAC. Interestingly, two distantly related trypanosomatid lineages with intracellular stage in their life cycles (Leishmania spp. and Trypanosoma cruzi) possess a markedly lower number of RACs, ranging from 1 to 9, than the blood-dwelling salivarian trypanosomes. A recent study suggested that RACs repertoire in L. donovani may have been shaped by gene conversion (Rogozin et al. 2020). Finally, in B. saltans, the only publicly available bodonid genome, 13 RACs were identified (supplementary table S1, Supplementary Material online and fig. 1). But not on the scale of the RAC repertoire of salivarian trypanosomes, this number may represent an adaptation of B. saltans to detect various stimuli present in its aqueous environment. Certainly, the presence of RACs in a free-living kinetoplastid argues for their potential to recognize a wide spectrum of ligands, which are not limited to those encountered at the host–parasite interface.
Taken together, these results show that the prominent expansion of RAC repertoire observed in T. brucei is recapitulated in other dixenous salivarian trypanosomes that have an extracellular lifestyle and infect mammals, whereas in the dixenous intracellular parasites of the genera Leishmania and Trypanosoma their number is lower, yet still considerably higher than in the monoxenous trypanosomatids and free-living B. saltans (fig. 1). The unique pattern of distribution of RACs with remarkable differences between trypanosomatid lineages can be explained by two hypotheses. The first, more parsimonious scenario supported by the phylogenetic analysis and, thus, favored by us, suggests that the ACs family expansion observed in trypanosomatid parasites is a result of both multiple gene duplication events, which occurred in the common ancestor of the respective trypanosomatid groups and species-specific expansions. Gene duplication events were accompanied by sequence diversification of the ED triggered by extracellular environmental conditions, such as those encountered within the mammalian host. Indeed, sequence diversification in what were initially nonfunctional regions might have been turned into an immune evasion mechanism, as previously proposed (Salmon et al. 2012a; Salmon 2018). However, we cannot exclude the second remote possibility that the common ancestor of trypanosomatids might had been already endowed with a highly diverse RAC family, reduced or lost altogether in the descendant lineages.
Clustering of Adenylate Cyclases Correlates with Differential Expansions and Domain Architecture
The phylogenetic tree of all AC-bearing proteins in kinetoplastids retrieved by BLAST search with T. brucei Tb927.7.6050 as a query revealed the expansion of this protein family in the salivarian trypanosomatids T. congolense, T. b. brucei, T. b. gambiense, and the dyskinetoplastic strains of T. b. evansi and T. b. equiperdum (fig. 1 and supplementary table S1, Supplementary Material online) (Lai et al. 2008; Carnes et al. 2015). The salivarian ACs clustered together in two well-defined groups termed Trypanosoma clades 1 and 2. Interestingly, according to the available riboprofiling data (Vasquez et al. 2014), clades 1 and 2 are translationally upregulated in either BSF and PCF forms, respectively.
Within the salivarian trypanosomes, T. vivax forms a distinct clade that contains a lower RAC number. This reduction could be related to the fact that the parasite develops only in the cibarium and proboscis of the tsetse fly (Ooi et al. 2016). This is a much simpler developmental cycle within the vector as compared with T. brucei and T. congolense, which have to navigate a complex set of tissues, including the alimentary tract and salivary glands (Ooi et al. 2016; Gibson et al. 2017; Shaw et al. 2019). Thus, the reduced RAC repertoire in T. vivax implies that a complex developmental cycle within the insect vector may represent yet another driving force behind RAC expansion, along with the exposure of these extracellular parasites to the host immune system.
A remarkable AC family expansion was also documented in the Trypanosoma grayi clade, which belongs to the stercorarian trypanosomes (e.g., T. cruzi, T. cruzi marinkellei, and Trypanosoma rangeli) (Hoare 1929; Kelly et al. 2014; Lukeš et al. 2018). This observation lends further credence to host immunity being a driving force for the RAC expansion, given that T. grayi is a hemoflagellate, in contrast to the intracellular T. cruzi (Kelly et al. 2014). However, it should be noted that T. grayi is transmitted by tsetse flies, which may contribute to the RAC family expansion. Furthermore, but not definitely established, T. rangeli may also be extracellular, albeit nonpathogenic to mammals (Stoco et al. 2014). Within Leishmaniinae, ACs grouped into two main clusters (fig. 1, Leishmaniinae clades 1 and 2). These clades incorporate RAC-A and RAC-B sequences, as originally defined in Leishmania donovani (Sanchez et al. 1995). Lastly, one clade was exclusively composed of OACs, with representatives in several species, bearing a N-terminal globin domain together with the C-terminal AC domain (fig. 1, OACs). These results show that in addition to the above-mentioned variability in number of functional AC-encoding genes, trypanosomatid AC family displays a high heterogeneity regarding domain architecture.
Salivarian Trypanosome RACs Diversity Is Related to Gene Duplication
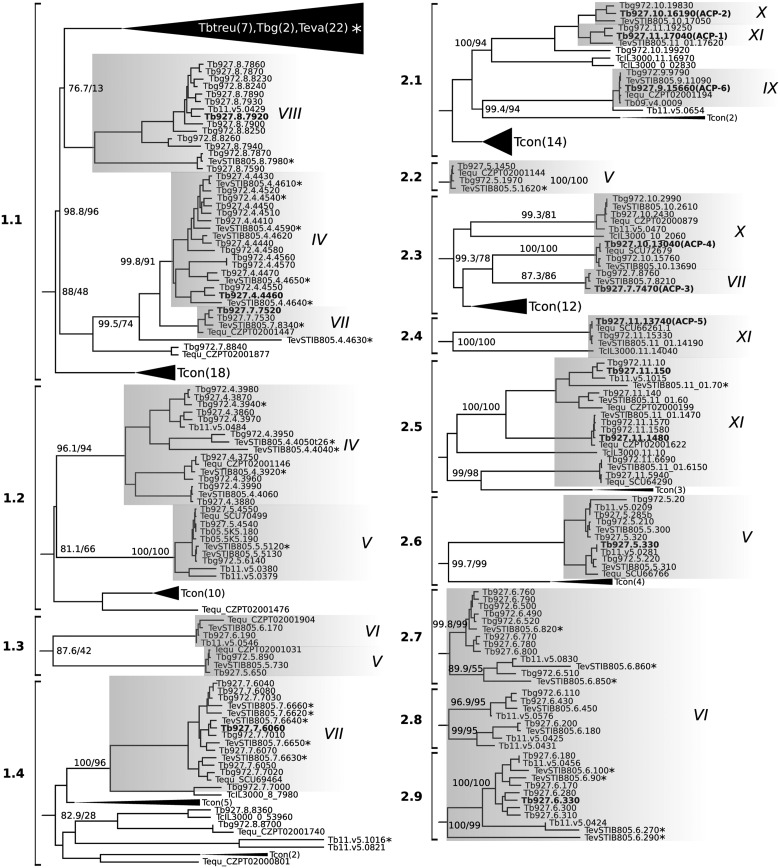
To highlight their diversification, we have focused on the phylogeny of all RACs encoded by the subspecies of T. brucei and the closely related salivarian trypanosomes. These genes grouped into 13 clusters (supplementary table S3, Supplementary Material online), which were further subdivided in 4 subclusters corresponding to Trypanosoma clade 1 in the complete phylogenetic tree (fig. 1) and therefore named 1.1 thru1.4, and 9 subclusters (2.1 to 2.9) composed of RACs belonging to Trypanosoma clade 2 (fig. 2).
Fig. 2.
Clustal-W analysis of the expanded salivarian RAC clades. Some clades containing sequences of T. evansi and T. congolense are collapsed. RACs with translational efficiency values retrieved from Vasquez et al. and selected for endogenous tagging of T. brucei procyclics are indicated in bold. The support values are in the following format: SH-aLRT support (%)/bootstrap support (%). Only support values ≥50% are shown. Flagellum-localized RACs (ACP1 thru 6) described by Saada et al. (2014) are in brackets. Shaded boxes represent distinct chromosomes indicated with roman numbers. Asterisk denote pseudogenes and no. of genes in collapsed clades are indicated in brackets.
Previously, some of T. brucei RACs termed ACP1–ACP6 were shown to localize into the flagellar tip and/or flagellar body subdomains (Saada et al. 2014). Furthermore, ACP1, 2, and 6 were linked to SoMo (Lopez et al. 2015). Interestingly, the SoMo RACs are clustered in 2.1 (fig. 2), which likely reflects their functional relatedness. Another interesting feature of the salivarian RACs clustering is the presence of several clusters of expanded T. congolense sequences that are mingled with most of the defined clusters. For example, cluster 2.1 also contains a subset of T. congolense RACs that one would predict could have an analogous role in putative SoMo in this species. This reinforces the concept of these two species being more closely related than generally appreciated (Gibson et al. 2017) and that gene duplication and diversification of RACs occurred early.
All RACs are known to contain the AC and TMD domains, whereas the signal peptide (SP) and ligand-binding extracellular domain are not always present (supplementary fig. S1A, Supplementary Material online) (Lukeš et al. 2018). All RACs evaluated herein lacked C-terminal GPI anchor sequences, whereas 34% of T. b. brucei and 42% of T. congolense RACs were predicted to carry the SP domain. All T. b. brucei full-length RACs displayed at least one TMD located between the AC and ED domains. In some RACs, an additional TMD identified in the N-terminal region, indicated the presence of a cleavable signal peptide not predicted by the SignalP program. Although most of T. b. brucei RACs displayed the conserved “canonical” receptor structure, 21% of them were truncated. In T. congolense, where most RACs cannot be assigned to a chromosome, this fragmentation was even more pronounced, accounting for 46% of all annotated RACs.
Taken together, the diversity of the expanded RACs family in T. b. brucei and T. congolense and the rest of salivarian trypanosomes can be best explained by extensive gene duplication. Although pseudogene assignment constitutes a nontrivial issue, and the genome assembly in T. congolense might not be impeccable, the fact that the fragments of RAC genes are prevalent in the chromosome-level assembly of T. b. brucei genome suggests that these sequences are under rapid evolution and rearrangement.
ESAG4 RACs Clustering Correlates with Stage-Specific and Upregulated Expression
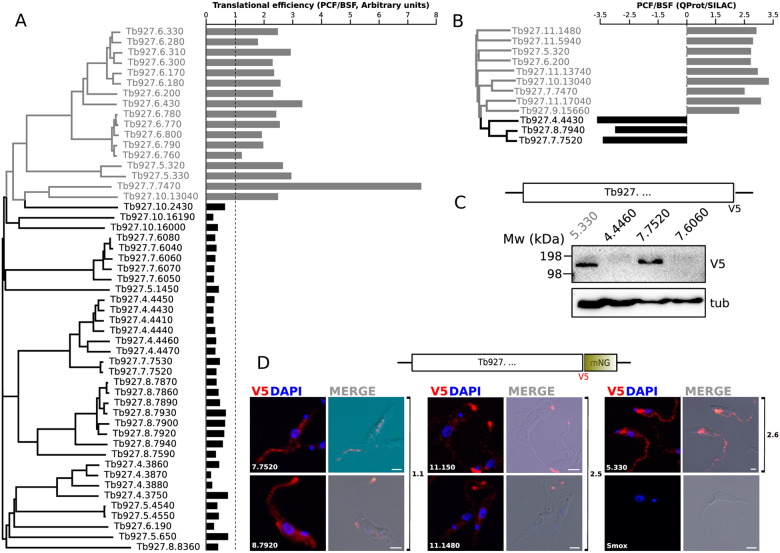
Taking advantage of the ribosome profiling data of PCF and BSFs (Vasquez et al. 2014), we determined whether the abundance of transcripts with enhanced translation efficiency in one of these life stages correlates with RAC protein sequence-based clustering. Remarkably, we found that RACs were split into two clusters that correlate well with the relative translational efficiency values, calculated as the translational efficiency in PCF divided by the translational efficiency in BSF (fig. 3A). RACs with relative translational efficiency values ≥ 1 (i.e., upregulated in PCF) accounted for 33% of the entire dataset and were preferentially located on chromosome 6, with some found on chromosomes 5, 7, and 10. Collectively, they formed clusters 2.3 and 2.6–2.9 in the clustering analyses and fell within Trypanosoma clade 2 on the phylogenetic tree (figs. 1 and 2). Interestingly, among these PCF-upregulated RACs were ACP3 (Tb927.7.7470) and ACP4 (Tb927.10.13040), in agreement with previous report (Saada et al. 2014). The remaining 67% were the RACs with relative translational efficiency ratios <1, hence the majority of RACs is upregulated in BSF (fig. 3A). These genes were primarily located on chromosomes 4, 5, 7, and 8, although some were found on chromosomes 6 and 10 (fig. 3A). The BSF-upregulated RACs that fell within Trypanosoma clade 1 on the phylogenetic tree formed clusters 1.1 to 1.4 (figs. 1 and 2). As opposed to PCF-upregulated RACs, this group exhibited subclustering which may indicate higher diversification that in turn could be attributed to the immune system-driven selective pressure in the vertebrate host.
Fig. 3.
In silico and in vivo analysis of selected RACs expression data. (A) Paired comparison between the clustering of RACs and riboprofiling data reported in Vasquez et al. (2014). The relative translational efficiency expressed as PCF/BSF ratio is plotted for each RAC. The base line of value 1 (equally translated RNAs) is indicated as a dotted line. Gray: PCF upregulated, black: BSF upregulated. (B) Relative abundance ratio (PCF/BSF) from quantitative proteomic data in SILAC experiments reported in Urbaniak et al. (2012). Gray: PCF upregulated, black: BSF upregulated. (C) WB determination of V5 C-terminally tagged RAC expression. Total lysates of T. brucei procyclic cells were separated by SDS-PAGE electrophoresis and transferred to PVDF membranes probed with mouse α-V5 antibody and monoclonal mouse α-tubulin as loading control. Molecular weights (kDa) are indicated at the left of the membrane. (D) Tagging and endogenous expression analysis of selected RACs. IFA analysis of the subcellular localization of endogenously V5::mNeonGreen tagged RACs. The data suggest the localization of RACs 11.150, 11.1480, and 8.7920 to the flagellar tip, but RACs 5.330, 7.7520 appear to be localized to the flagellar body and the cytoplasm. Parental SmOx cells are shown as a negative control. White bars: 2 µm.
There was a strong correlation between our translational efficiency analysis and BSF versus PCF stable isotope labeling by amino-acids in cell culture (SILAC) quantitative proteomic data (Urbaniak et al. 2012). Thus, RACs with higher translational efficiency in BSFs, also showed relative expression enriched in SILAC experiments (fig. 3B).
Many T. congolense RACs are intermingled with those of T. brucei in both clades 1 and 2 (fig. 2). This observation gives rise to the hypothesis that these corresponding RACs may be regulated in the life cycle stages occupying the mammalian host and insect vector as the cognate RACs in T. b. brucei. This observation also implies a duplication of RAC gene(s) in the common ancestor of T. congolense and T. brucei, which eventually evolved into functionally distinct proteins. This event was likely followed by further duplication of RACs in each species. More data are required to validate this correlation for other salivarian trypanosomes. Combined, our analyses imply the existence of sequence-derived constraints that may differentially affect translational efficiency in individual life stages of T. b. brucei.
Interestingly, except for a few interspersed long branches throughout the tree, most of the pseudogenes of T. b. evansi formed a monophyletic group that grouped together with cluster 1.1. This cluster, in turn, is composed of some T. brucei RACs that are upregulated in BSF according to the ribosome profiling analysis (Vasquez et al. 2014). As previously mentioned, many of the genes encoding RACs in the dyskinetoplastic (= with still visible kinetoplast that contains defective DNA) or akinetoplastic (= completely lacking the kinetoplast [Lai et al. 2008]) subspecies T. b. evansi and T. b. equiperdum are prone to pseudogenization. Because of a partial to complete loss of their mitochondrial genomes, dyskinetoplastic trypanosomes cannot differentiate into PCF, and, thus, do not thrive in the alimentary tract or reach the salivary glands of tsetse flies (Schnaufer et al. 2002; Lai et al. 2008). We expected to see that RACs upregulated in PCF would be preferentially pseudogenized. While some of these RACs were, we were surprised to observe that many of the RACs upregulated in BSF were also pseudogenized (figs. 2 and 3A). Three non-contradictory hypotheses could explain this phenomenon: 1) RACs are under extreme diversifying selection pressure, giving rise to its varied repertoire, in contrast to most genes in the T. brucei subspecies genome (Carnes et al. 2015); 2) RACs upregulated in BSF also play a role in PCF; 3) BSF-upregulated RACs may facilitate the colonization of mammalian tissues that are inaccessible to dyskinetoplastic trypanosomes. Consistent with the second hypothesis, the adipose tissue form of T. brucei undergoes β-oxidation of fatty acids presumably found in its milieu (Trindade et al. 2016), a process that could require at least some of the proteins encoded in the mitochondrial genome (Smith et al. 2017). This would imply that these RACs may facilitate invasion of the adipose tissue. More work is needed to verify this hypothesis.
For further analysis of their expression, RACs Tb927.4.4460 (henceforth 4.4460), 7.6060 and 7.7520 from clusters 1.1 and 1.4 (as representatives of PCF-upregulated Trypanosoma clade 1) and 5.330 (as representative of cluster 2.6 and Trypanosoma clade 2) were endogenously tagged with a C-terminal V5 epitope in T. b. brucei PCFs. WB analysis showed variable expression of V5 signal for these RACs (fig. 3C). A strong expression occurred in the case of 5.330, as it was expected for this RAC being translationally upregulated in PCFs according to the riboprofiling data (Vasquez et al. 2014). On the other hand, only very weak signal was detected for 4.4460- and 7.6060-tagged cell lines, again in concordance with their upregulated translational efficiency in BSFs. Although 7.7520 also represents a gene upregulated in BSF that was nonetheless detected by WB in PCF (fig. 3C), this result is not contradictory, given that this particular RAC showed BSF upregulation also in the SILAC experiments (Urbaniak et al. 2012) (fig. 3B), but was also detected in the surface proteomes of both stages (Shimogawa et al. 2015). We therefore conclude that WB analysis for 4.4460, 7.6060, and 5.330 seems to be correlated with both the proteomic and riboprofiling data. This points out the fact that the expression of certain RACs with presumably conserved roles both in PCF and BSF may occur regardless of low translational efficiency, thus supporting the first hypothesis proposed above. However, a caveat to this interpretation is that the C-terminal tagging also replaces the endogenous 3′ UTR (Dean et al. 2015), an element controlling gene expression in trypanosomatids (Maslov et al. 2019).
Endogenous Expression Reveals Conserved Location of ESAG4 RACs in Discrete Flagellar Domains
To investigate the expression and subcellular localization of selected members of the T. brucei RAC family, five PCF cell lines endogenously tagged with a C-terminal V5::mNeonGreen (V5NG) epitope were generated. The tagged RACs were selected according to the following criteria: 1) belonging to the clades correlated with enhanced stage-specific translational efficiency; 2) have not been studied previously; and 3) represent RACs with sequence divergence at the level of independently segregating genes, that is, unlikely to be products of duplication, and/or mapping to different chromosomes. Hence, the following genes were selected: Tb927.8.7920 (8.7920) and 7.7520 from clade 1.1; 11.150 and 11.1480 from clade 2.5; and 5.330 from clade 2.6 (fig. 3D).
IFA of the expressed V5NG fusions showed flagellar tip localization of three tagged RACs belonging to clades 1.1 (8.7920) and 2.5 (11.150, 11.1480). The same localization was reported for ACP1, ACP4, and ACP6, which were implicated in SoMo (Saada et al. 2014; Lopez et al. 2015) and belong to the clade 2.1 (fig. 2). Additionally, RACs 5.330 and 7.7520 were localized throughout the flagellar body, a localization also previously reported for ACP2 and ACP5, belonging to clades 2.1 and 2.4, respectively (Saada et al. 2014). No IFA signal was detected in the parental T. brucei cell line from which the tagged cell lines were derived, which thus served as a negative control lacking the V5NG epitopes. No correlation was observed between the tip or flagellar body localization and translational upregulation. Taken together, these results show that the majority of RACs from highly divergent clades display a conserved localization to flagellar subdomains, which is in agreement with previous reports for other members of the RACs family (Saada et al. 2014).
Distribution of OACs Is Associated with Complex Life Cycles and Endosymbionts
As mentioned earlier, another important subgroup of ACs is comprised of OACs (fig. 1). The tree topology, albeit with low support, suggests that OACs have emerged early in the evolution of trypanosomatids, before the diversification of RACs. They likely represent a trypanosomatid innovation, because we did not identify OACs in B. saltans. Deeper analysis led to the identification of OAC homologs in most species within the reference dataset. Exceptions that lack identifiable OACs are the basal trypanosomatid P. confusum (Flegontov et al. 2013), representatives of the genus Phytomonas with streamlined genomes (Jaskowska et al. 2015), and, importantly, members of the genus Trypanosoma (except for T. grayi and T. theileri) (fig. 4). Due to the low quality of genome assemblies of the two latter species, it remains to be confirmed, whether the identified partial OAC-encoding genes are indeed functional.
Most members of the genera Leishmania, Endotrypanum, Blechomonas, and Blastocrithidia possess a single OAC homolog in their genomes. However, monoxenous representatives of the subfamily Leishmaniinae (Kostygov and Yurchenko 2017) and members of the genus Wallacemonas (Kostygov et al. 2014) encode several OAC genes (fig. 4). Interestingly, Novymonas esmeraldas, possessing multiple endosymbionts in each cell (Kostygov et al. 2016), carries nine OAC-related genes, which is the highest documented number for any trypanosomatid. This prominent expansion of the OAC-related genes implies their possible functional association with the presence of the endosymbiont. Moreover, OACs from N. esmeraldas are longer and bear the N-terminal extensions, which have no identifiable functional domains and, thus, may carry yet undescribed targeting signals that direct them into the endosymbiont.
Finally, the genomes of Leishmania from subgenus Viannia, namely L. braziliensis, L. panamensis, and L. peruviana, contain easily identifiable pseudogenized OACs. These pseudogenes invariably retain the AC domains, but the globin domains were apparently degraded (fig. 4). Interestingly, members of subgenus Viannia also lack a gene encoding the Seltryp protein (Bonilla et al. 2016). This kinetoplastid-specific protein, found in all other trypanosomatids and B. saltans, has been hypothesized to be mitochondrion-targeted and involved in sulfide detoxification (Lobanov et al. 2006; Bonilla et al. 2016). Perhaps, the lack of functional OACs and Seltryp in Vianna is not a coincidence. Given that molecular oxygen is a reactant in sulfide detoxification, and the still uncharacterized Seltryp protein may catalyze other redox reactions to for example, mitigate oxidative stress (Labunskyy et al. 2014), there may be a link between the functions of OAC and Seltryp.
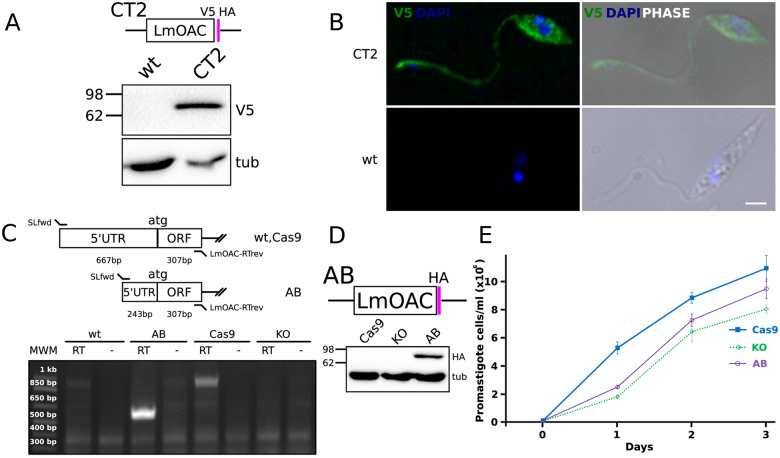
The OAC proteins were scarcely studied, with only one report assessing their function in L. major (Sen Santara et al. 2013). Thus, we decided to explore the single-copy OAC in L. mexicana (LmxM.28.0090-encoded LmOAC) (fig. 5). To this end, we generated several recombinant cell lines, in which LmOAC was tagged, knocked out, and added back. First, to assess the subcellular localization of LmOAC, we generated stable transfectants expressing a C-terminal V5-HA-tagged endogenous version of LmOAC, which was detected in WB as a specific ∼77 kDa band (fig. 5A). The IFA of the corresponding cell line of the L. mexicana promastigotes detected a cytoplasmic signal that also spread to the flagellum (fig. 5B). This localization was expected given lack of the SP N-terminal targeting signals for OACs (supplementary fig. S1B, Supplementary Material online). Considering that the L. major homolog of LmOAC was reported to be associated with growth defects under normoxic conditions (Sen Santara et al. 2013), we wondered whether a similar phenotype would be triggered in promastigotes following the disruption of LmOAC. The expression of LmOAC in wild-type and add-back L. mexicana promastigotes and the absence of the corresponding transcript in the knock-out strain was evaluated by RT-PCR analysis (fig. 5C). Finally, proper expression of LmOAC in the HA-tagged add-back cells was assessed by WB analysis and compared with the parental Cas9 and knock-out strains (fig. 5D). Comparison of the growth curves of three cell lines under normal conditions revealed a mild growth defect in cells lacking LmOAC (fig. 5E). This growth phenotype was only partially complemented by adding the gene back. Taken together, these results show that LmOAC displays a previously undocumented cytoplasmic localization in the promastigote stage. Additionally, ablation of this gene produced a growth defect that mimics the previously reported phenotype for L. major OAC (Sen Santara et al. 2013), confirming that these proteins are functional orthologs.
Fig. 5.
Characterization of OAC in L. mexicana. (A) WB analysis of the expression of C-terminally tagged V5-HA LmOAC CT2 cell line. Molecular weights are indicated at the left of membranes. Tubulin signal is shown as loading control. (B) Cytoplasmic and flagellar localization of V5-tagged LmOAC expression assessed by IFA analysis of CT2 L. mexicana promastigotes. White bar: 2 µm. (C) RT-PCR validation of Cas9, KO, and AB strains. PCR products obtained over RT+ and RT− (negative control) reactions performed over total RNA polyA+ of L. mexicana Cas9, KO, and AB strains are shown. An analysis figure is included in the upper panel. Product sizes (bp) are indicated over the molecular weight marker (MWM) on the left. (D) WB assessment of the expression of HA-tagged LmOAC in the AB cell line. Total lysates of Cas9, KO, and AB were probed with mouse α-HA monoclonal antibody. Tubulin expression is shown as loading control. (E) Standard growth curve of Cas9, KO, and AB promastigote cell lines. The total number of parasites (106 cells/ml) versus day of culture in normal conditions is shown.
Conclusion
To the best of our knowledge, the present study represents the most comprehensive analysis of AC-bearing proteins in kinetoplastids. Our analyses show that RACs greatly expanded in all the salivarian trypanosomes: T. congolense, T. b. brucei, T. b. evansi and T. b. gambiense and, to a lesser extent, in T. vivax. The reduced number of RACs in T. vivax may be correlated with its truncated life cycle in the insect. It appears that the RACs number differs in the salivarian (exclusively extracellular in the mammalian host and transmitted by insect inoculation) and stercorarian trypanosomes (intracellular and spread via fecal transmission) (Hoare 1972; Grisard 2002). Interestingly, T. grayi does have a greater RAC repertoire than related stercorarian trypanosomatids, perhaps due to its extracellular lifestyle and, subsequently, exposure to host immunity. This may indeed be associated with their failure to traverse the entire alimentary tract of their invertebrate host (Ooi et al. 2016). Another significant drop of the number of RACs is characteristic for the plant-infecting genus Phytomonas, whose genomes are extremely streamlined (Porcel et al. 2014). Taken together, our data are compatible with the scenario that RACs are massively expanded in salivarian trypanosomes in response to exposure to mammalian host immunity, as well as their complex life cycle within the insect host (Shaw et al. 2019).
Given their remarkable expansion, we performed a thorough analysis of the RACs family, centered at T. b. brucei. The topology of the phylogenetic tree where clades incorporating intermingled sequences of several species, as well as species-specific clades can be identified, suggests that there were multiple gene duplication events in the evolutionary history of the AC family. This situation also will allow researchers to make informed predictions about the function of T. congolense RACs based on their clustering with better studied T. brucei RAC clades. The remarkably high prevalence of pseudogenes, lacking either the PBP or canonical AC domains may be related to generation of the functional diversity, including the diversity in the binding affinities of PBP that functions as an extracellular binding domain in RACs.
Our phylogenetic reconstruction also revealed a remarkable correlation between sequence-based clustering and expression data from ribosome profiling (Vasquez et al. 2014) and stage-specific proteomics (Urbaniak et al. 2012), showing that RACs are divided into two major clades that are upregulated in BSF or PCF T. brucei. The selected RACs analyzed in this study localize to the flagellar tip or throughout the flagellar body, which is in good correlation with subcellular localization reported for other AC proteins (Saada et al. 2014). Therefore, our results further emphasize the notion of the flagellum acting as an antenna of extracellular signals.
Phylogenetic analysis of OACs, the other poorly characterized group of ACs, revealed their early emergence in the evolution of trypanosomatids, which likely happened prior to the diversification of RACs. OACs, absent in free-living B. saltans, appear to be a trypanosomatid innovation and are likely to be predominantly important for monoxenous species (potentially giving an additional advantage for the endosymbiont-bearing ones) with their life cycle confined to invertebrates. We found the OAC family to be expanded in monoxenous representatives of the subfamily Leishmaniinae, with an endosymbiont-harboring N. esmeraldas carrying the highest number of OAC-encoding genes among trypanosomatids, analyzed herein. Overall, the distribution of OACs implies their yet unknown function(s) in the complex life cycle of trypanosomatids, with the absence of globin-domain containing OACs in the genus Phytomonas possibly explained by lack of any hemoproteins in these trypanosomatids (Kořený et al. 2012). Reflecting the dearth of in vivo functional data on OACs, we have experimentally shown the expression of LmOAC in the L. mexicana promastigotes, where the depletion of this cytoplasmic protein causes a mild growth defect, resembling the results reported for its counterpart in L. major (Sen Santara et al. 2013).
Supplementary Material
Acknowledgments
We thank Isabel Roditi (University of Bern) for useful advice and stimulating discussion.
Funding
This work was supported by the following funding sources: Czech Science Foundation grants 20-23513S to H.H., 20-07186S to J.L., and 18-15962S to J.L. and V.Y.; ERC CZ grant LL1601 to J.L.; Czech Ministry of Education grant OP3V16_019/0000759 to J.L., V.Y., and H.H. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
Literature Cited
- Almagro Armenteros JJ, et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. [DOI] [PubMed] [Google Scholar]
- Bachmaier S, et al. 2019. Nucleoside analogue activators of cyclic AMP-independent protein kinase A of trypanosoma. Nat Commun. 10(1):1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Weiss LM, Braunstein VL, Huang H. 2008. Role of protein kinase A in Trypanosoma cruzi. IAI 76(10):4757–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Weiss LM, Ma YF, Kahn S, Huang H. 2010. Protein kinase A catalytic subunit interacts and phosphorylates members of trans-sialidase super-family in Trypanosoma cruzi. Microbes Infect. 12(10):716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A, et al. 2012. Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proc Natl Acad Sci U S A. 109(36):14399–14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneke T, et al. 2017. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R Soc Open Sci. 4(5):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla M, Krull E, Irigoín F, Salinas G, Comini MA. 2016. Selenoproteins of African trypanosomes are dispensable for parasite survival in a mammalian host. Mol Biochem Parasitol. 206(1–2):13–19. [DOI] [PubMed] [Google Scholar]
- Bruschi F, Gradoni L, editors. 2018. The leishmaniases: old neglected tropical diseases. Cham, Switzerland: Springer. [Google Scholar]
- Büscher P, Cecchi G, Jamonneau V, Priotto G. 2017. Human African trypanosomiasis. Lancet 390(10110):2397–2409. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, et al. 2015. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis. 9(1):e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning HP, et al. 2012. Pharmacological validation of Trypanosoma brucei phosphodiesterases as novel drug targets. J Infect Dis. 206(2):229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S, et al. 2015. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biol. 5(1):140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante IM, Cámara MDLM, Buscaglia CA. 2015. A novel Trypanosoma cruzi protein associated to the flagellar pocket of replicative stages and involved in parasite growth. PLoS One 10(6):e0130099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science. 309(5733):404–409. [DOI] [PubMed] [Google Scholar]
- Flegontov P, et al. 2013. Paratrypanosoma is a novel early-branching trypanosomatid. Curr Biol. 23(18):1787–1793. [DOI] [PubMed] [Google Scholar]
- Fraidenraich D, et al. 1993. Stimulation of Trypanosoma cruzi adenylyl cyclase by an α D-globin fragment from Triatoma hindgut: effect on differentiation of epimastigote to trypomastigote forms. Proc Natl Acad Sci U S A. 90(21):10140–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. 2013. Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol Rev. 88(3):645–668. [DOI] [PubMed] [Google Scholar]
- Gibson W, Kay C, Peacock L. 2017. Trypanosoma congolense: molecular toolkit and resources for studying a major livestock pathogen and model trypanosome. Adv Parasitol. 98:283–309. [DOI] [PubMed] [Google Scholar]
- Gould MK, et al. 2013. Cyclic AMP effectors in African trypanosomes revealed by genome-scale RNA interference library screening for resistance to the phosphodiesterase inhibitor CpdA. Antimicrob Agents Chemother. 57(10):4882–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisard EC. 2002. Salivaria or Stercoraria? The Trypanosoma rangeli dilemma. Kinetoplastid Biol Dis. 1(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, McDonald L, Stříbrná E, Lukeš J. 2013. Trypanosome letm1 protein is essential for mitochondrial potassium homeostasis. J Biol Chem. 288(37):26914–26925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare CA. 1929. Studies on Trypanosoma grayi. Trans R Soc Trop Med Hyg. 23(1):39–56. [Google Scholar]
- Hoare CA. 1972. The trypanosomes of mammals. A zoological monograph. Science. 169(4068):60. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Jackson AP, et al. 2013. A cell-surface phylome for African trypanosomes. PLoS Negl Trop Dis. 7(3):e2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, et al. 2016. Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr Biol. 26(2):161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskowska E, Butler C, Preston G, Kelly S. 2015. Phytomonas: trypanosomatids adapted to plant environments. PLoS Pathog. 11(1):e1004484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Ivens A, Manna PT, Gibson W, Field MC. 2014. A draft genome for the African crocodilian trypanosome. Sci Data. 1(1):140024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostygov AY, et al. 2016. Novel trypanosomatid-bacterium association: evolution of endosymbiosis in action. mBio 7(2):e01985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostygov AY, Grybchuk-Ieremenko A, Malysheva MN, Frolov AO, Yurchenko V. 2014. Molecular revision of the genus Wallaceina. Protist 165(5):594–604. [DOI] [PubMed] [Google Scholar]
- Kostygov AY, Yurchenko V. 2017. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasit. 64:020. [DOI] [PubMed] [Google Scholar]
- Kraeva N, et al. 2019. LmxM.22.0250-encoded dual specificity protein/lipid phosphatase impairs Leishmania mexicana virulence in vitro. Pathogens 8(4):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Labunskyy VM, Hatfield DL, Gladyshev VN. 2014. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 94(3):739–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D-H, Hashimi H, Lun Z-R, Ayala FJ, Lukes J. 2008. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc Natl Acad Sci U S A. 105(6):1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, Gromer S, Salinas G, Gladyshev VN. 2006. Selenium metabolism in Trypanosoma: characterization of selenoproteomes and identification of a Kinetoplastida-specific selenoprotein. Nucleic Acids Res. 34(14):4012–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MA, Saada EA, Hill KL. 2015. Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes. Eukaryot Cell 14(1):104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J, et al. 2018. Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol. 34(6):466–480. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V. 2014. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol. 195(2):115–122. [DOI] [PubMed] [Google Scholar]
- Madeira F, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makin L, Gluenz E. 2015. cAMP signalling in trypanosomatids: role in pathogenesis and as a drug target. Trends Parasitol. 31(8):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov DA, et al. 2019. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology. 146(1):1–27. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Votýpka J, Yurchenko V, Lukeš J. 2013. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol. 29(1):43–52. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musikant D, et al. 2017. Host Epac1 is required for cAMP-mediated invasion by Trypanosoma cruzi. Mol Biochem Parasitol. 211:67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M, et al. 2007. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J. 21(3):720–731. [DOI] [PubMed] [Google Scholar]
- Oberholzer M, Saada EA, Hill KL. 2015. Cyclic AMP regulates social behavior in African trypanosomes. mBio. 6(3):e01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CP, et al. 2016. The cyclical development of Trypanosoma vivax in the tsetse fly involves an asymmetric division. Front Cell Infect Microbiol. 6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes FR, Butenko A, Flegontov P, Yurchenko V, Lukeš J. 2016. Comparative metabolism of free-living Bodo saltans and parasitic trypanosomatids. J Eukaryot Microbiol. 63(5):657–678. [DOI] [PubMed] [Google Scholar]
- Paindavoine P, et al. 1992. A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol Cell Biol. 12(3):1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A, Martelli P, Casadio R. 2008. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 9(1):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon SK, Peacock L, Gibson W, Gull K, Kelly S. 2012. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2(2):110037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcel BM, et al. 2014. The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLoS Genet. 10(2):e1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson O, Dylus D, Dessimoz C, Rosenberg M. 2016. Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web. Mol Biol Evol. 33(8):2163–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I, Charyyeva A, Sidorenko I, Babenko V, Yurchenko V. 2020. Frequent recombination events in Leishmania donovani: mining population data. Pathogens 9(7):572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas F, et al. 2019. Oligopeptide signaling through TbGPR89 drives trypanosome quorum sensing. Cell 176(1–2):306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saada EA, et al. 2014. Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei flagellar membrane. Eukaryot Cell 13(8):1064–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivia M, Ceballos-Pérez G, Bart JM, Navarro M. 2016. The AMPKα1 pathway positively regulates the developmental transition from proliferation to quiescence in Trypanosoma brucei. Cell Rep. 17(3):660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D. 2018. Adenylate cyclases of Trypanosoma brucei, environmental sensors and controllers of host innate immune response. Pathogens 7(2):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D, et al. 2012. a. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science 337(6093):463–466. [DOI] [PubMed] [Google Scholar]
- Salmon D, et al. 2012. b. Cytokinesis of Trypanosoma brucei bloodstream forms depends on expression of adenylyl cyclases of the ESAG4 or ESAG4-like subfamily. Mol Microbiol. 84(2):225–242. [DOI] [PubMed] [Google Scholar]
- Sanchez MA, Zeoli D, Klamo EM, Kavanaugh MP, Landfear SM. 1995. A family of putative receptor-adenylate cyclases from Leishmania donovani. J Biol Chem. 270(29):17551–17558. [DOI] [PubMed] [Google Scholar]
- Sen Santara S, et al. 2013. Globin-coupled heme containing oxygen sensor soluble adenylate cyclase in Leishmania prevents cell death during hypoxia. Proc Natl Acad Sci U S A. 110(42):16790–16795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Domingo GJ, Stuart K. 2002. Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol. 32(9):1071–1084. [DOI] [PubMed] [Google Scholar]
- Shalaby T, Liniger M, Seebeck T. 2001. The regulatory subunit of a cGMP-regulated protein kinase A of Trypanosoma brucei. Eur J Biochem. 268(23):6197–6206. [DOI] [PubMed] [Google Scholar]
- Shaw S, et al. 2019. Flagellar cAMP signaling controls trypanosome progression through host tissues. Nat Commun. 10(1):803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa MM, et al. 2015. Cell surface proteomics provides insight into stage-specific remodeling of the host-parasite interface in Trypanosoma brucei. Mol Cell Proteomics. 14(7):1977–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Bringaud F, Nolan DP, Figueiredo LM. 2017. Metabolic reprogramming during the Trypanosoma brucei life cycle. F1000Res 6:683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoco PH, et al. 2014. Genome of the avirulent human-infective trypanosome-Trypanosoma rangeli. PLoS Negl Trop Dis. 8(9):e3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Zhang P, Steichen JM, Keshwani MM, Kornev AP. 2013. PKA: lessons learned after twenty years. Biochim Biophys Acta 1834(7):1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telleria J, Tibayrenc M, editors. 2017. American trypanosomiasis Chagas disease: one hundred years of research. Cambridge: Elsevier. [Google Scholar]
- Trindade S, et al. 2016. Trypanosoma brucei parasites occupy and functionally adapt to the adipose tissue in mice. Cell Host Microbe 19(6):837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Guther MLS, Ferguson MAJ. 2012. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One 7(5):e36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Abbeele J, et al. 1995. Trypanosoma brucei: stimulation of adenylate cyclase by proventriculus and esophagus tissue of the tsetse fly, Glossina morsitans morsitans. Exp Parasitol. 81(4):618–620. [DOI] [PubMed] [Google Scholar]
- Vasquez J-J, Hon C-C, Vanselow JT, Schlosser A, Siegel TN. 2014. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 42(6):3623–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Reuner B, Yutzy B, Boshart M. 1997. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 110(21):2661–2671 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.