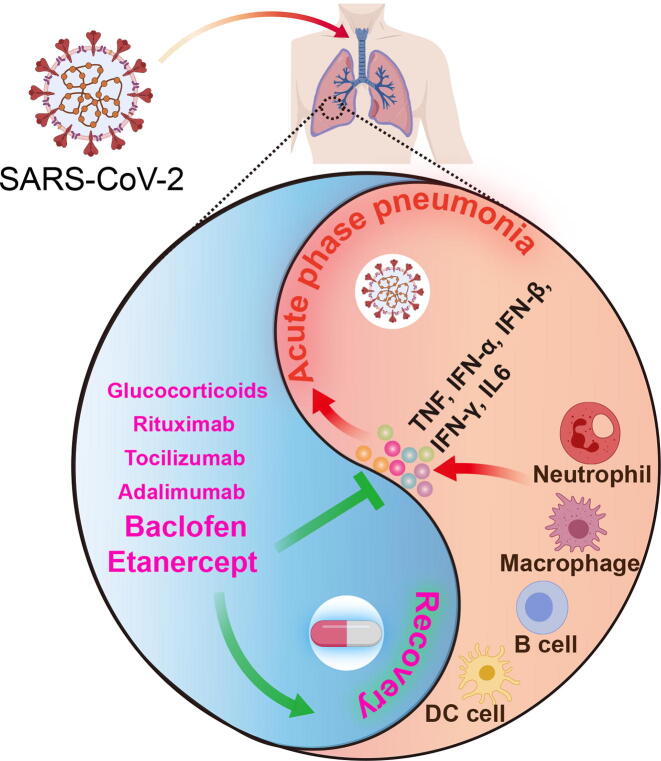
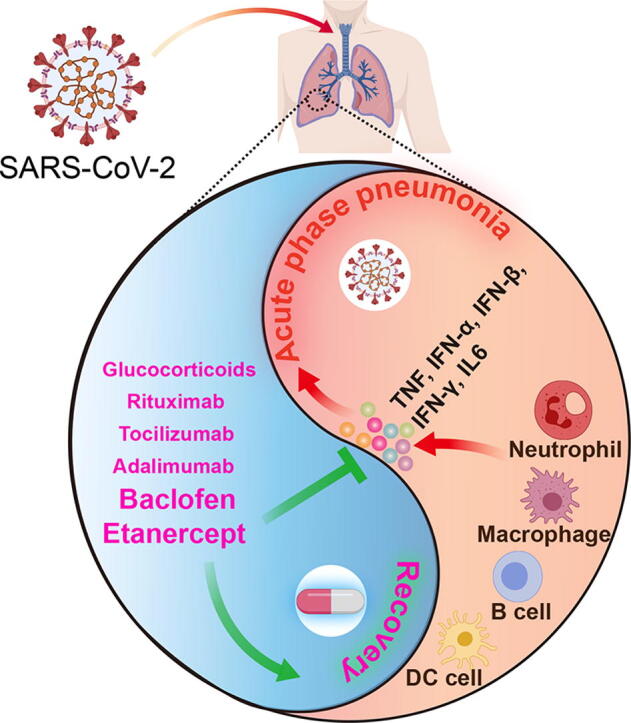
Graphical abstract
Abstract
The novel coronavirus SARS-CoV-2 is damaging the world’s social and economic fabrics seriously. Effective drugs are urgently needed to decrease the high mortality rate of COVID-19 patients. Unfortunately, effective antiviral drugs or vaccines are currently unavailable. Herein, we systematically evaluated the effect of SARS-CoV-2 on gene expression of both lung tissue and blood from COVID-19 patients using transcriptome profiling. Differential gene expression analysis revealed potential core mechanism of COVID-19-induced pneumonia in which IFN-α, IFN-β, IFN-γ, TNF and IL6 triggered cytokine storm mediated by neutrophil, macrophage, B and DC cells. Weighted gene correlation network analysis identified two gene modules that are highly correlated with clinical traits of COVID-19 patients, and confirmed that over-activation of immune system-mediated cytokine release syndrome is the underlying pathogenic mechanism for acute phase of COVID-19 infection. It suggested that anti-inflammatory therapies may be promising regimens for COVID-19 patients. Furthermore, drug repurposing analysis of thousands of drugs revealed that TNFα inhibitor etanercept and γ-aminobutyric acid-B receptor (GABABR) agonist baclofen showed most significant reversal power to COVID-19 gene signature, so we are highly optimistic about their clinical use for COVID-19 treatment. In addition, our results suggested that adalimumab, tocilizumab, rituximab and glucocorticoids may also have beneficial effects in restoring normal transcriptome, but not chloroquine, hydroxychloroquine or interferons. Controlled clinical trials of these candidate drugs are needed in search of effective COVID-19 treatment in current crisis.
1. Introduction
Based on the situation report of World Health Organization (WHO), as of December 8, 2020, the outbreak of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide with a total of 68,266,812 infections and 1,557,674 deaths [1]. The rapid propagation of COVID-19 makes it difficult to tackle. No effective drugs or vaccines also contributes to the uncontrollable situation. Researchers and scientists all over the world are racing against time to characterize the nature of this virus and find novel therapeutics for this virus [2], [3]. The most striking drug in the current COVID-19 crisis is remdesivir, a broad-spectrum antiviral medication with antiviral activity against several RNA viruses including SARS coronavirus and middle eastern respiratory syndrome virus (MERS) [4]. Significant outcome improvements were achieved for severe COVID-19 patients after treatment with compassionate-use of remdesivir [4]. However, another study from China with a randomized, double-blind, placebo-controlled and multicenter trial showed that remdesivir was not associated with statistically significant clinical benefits for severe COVID-19 patients [5]. Moreover, large scale trials of vaccines are still ongoing though the vaccine must be the line of defense to eventually eliminate this coronavirus. So how to reduce the mortality of infected patients before vaccines are approved, is now the most urgent task of researchers and physicians.
Fortunately, one of the most efficient ways to identify effective therapy is through repurposing existing clinic therapeutic drugs using public data since bioinformatic prediction [6] and machine learning [7] could provide novel biological insigths [8]. These drugs can be divided into two broad categories, those that can directly target the virus replication cycle, and those that can prevent the cytokine release syndrome (CRS) and severe inflammatory responses. In a recent study, researchers identified 11 possible covalent drugs targeting the main protease (3CLpro) of SARS-CoV-2 by a computer-aided drug discovery protocol [9]. In another study, researchers identified 69 compounds targeting 66 druggable human proteins or host factors from 332 high-confidence SARS-CoV-2-human protein–protein interactions (PPIs) assayed by affinity-purification mass spectrometry. According to the guidelines published by National Institutes of Health (NIH), remdesivir is the only drug recommended for the treatment of COVID-19 in hospitalized patients with severe disease [1]. Apart from the antiviral treatment strategy, targeting the inflammatory cascade should be considered, particularly in the most severe cases with acute respiratory distress syndrome (ARDS) and secondary hemophagocytic lymphohistiocytosis (sHLH) caused by CRS. CRS-induced ARDS and sHLH are commonly observed and associated with high mortality in patients with SARS-CoV-2 and SARS-CoV as well as in patients with MERS [10]. Several trials on evaluating the safety and effectiveness of immunosuppressants commonly used in rheumatic diseases are ongoing in patients with COVID-19 and CRS, some of which are achieving promising results [11]. For example, interleukin-6 inhibitors (e.g., sarilumab and tocilizumab) are potential immunosuppressive drugs for treatments of COVID-19 patients although there are insufficient data from clinical trials [1]. However, a systematic approach for drug repurposing based on big data analysis are lacking although the world’s largest clinical trial has launched to evaluate whether existing drugs work to treat people hospitalized with COVID-19 [12].
In the current study, we systematically evaluated the effect of SARS-CoV-2 on the transcriptome profiles of both blood and lung tissue from COVID-19 patients. Differential gene analysis revealed key pathways associated with SARS-CoV-2 infection. Moreover, we applied weighted correlation network analysis (WGCNA) to identify gene modules that are highly correlated with clinical traits of COVID-19 patients. Finally, we employed three state-of-the-art methods to predict the potential therapeutic drugs which possess rescue effect on COVID-19 gene signatures. Our results highlight the potential of TNFα inhibitor etanercept and GABABR agonist baclofen for COVID-19 treatment. We also suggest that adalimumab, tocilizumab, rituximab and glucocorticoids may have beneficial effects for COVID-19 treatment.
2. Methods
2.1. Data sources
RNA-seq or microarray raw data of GSE147507, GSE36177, GSE128101, GSE145918, GSE119939, GSE48027, GSE30351, GSE74235, GSE76492, GSE112101, GSE40165, GSE18948, GSE67368 and GSE54629 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). RNA-seq data of CRA002390 were downloaded from the National Genomics Data Center database (https://bigd.big.ac.cn/). NanoString nCounter transcriptomic profiling from E-MTAB-8871, and microarray data of E-TABM-116, E-TABM-642, E-GEOD-20272, and RNA-seq data of E-MTAB-5921 were downloaded from Array Express (https://www.ebi.ac.uk/arrayexpress/). Patients’ information was shown in Supplementary Table 1.
2.2. RNA-seq data analysis
According to our previous epigenetic study [13], the cutadapt (https://cutadapt.readthedocs.org/en/stable/) and FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) tools wrapped in Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) were used to trim low-quality bases (Q < 20) and adapter for raw sequences. Reads with length < 20 bp were removed after trimming. Cleaned RNA-seq reads were mapped to the hg19 genome for human and the mm10 genome for mouse using HISAT2 (v2.1.0) [14]. Duplicated reads for pair-end data were removed by SAMtools (v1.5) [15].
2.3. Identification of differential expressed genes (DEGs)
RNA-Seq data (GSE147507) for two uninfected human lung biopsies and two lung samples from COVID-19 deceased patient, and RNA-Seq data (CRA002390) for three PBMC samples from healthy donors and COVID-19 patients were analyzed by DESeq2 [16] to identify differentially expressed genes, respectively. Similar to our previous study [17], false discovery rate (FDR q-value) was calculated by adjusting P-values with the Benjamini-Hochberg method. Genes with FDR q-value < 0.05 and |Log2 (Fold change) | > 1 were considered as DEGs. The volcano plot was drawn using the ggplot2 package by R language.
2.4. Gene ontology (GO) pathway enrichment analysis of DEGs
Pathway enrichment analysis was performed by Metascape (https://metascape.org) [18]. Upregulated and downregulated DEGs were analyzed in this study, respectively. Pathway with P-value < 0.05 was considered significantly enriched pathway. Pathwaybubble plot was created by R package ggplot2.
2.5. Protein-protein interaction (PPI) network construction
STRING database version 11.0 (https://string-db.org/) was used to construct the PPI network with 0.9 highest confidence interaction score. Cytoscape (3.7.1) was used for visualizing the PPI network. The Molecular Complex Detection (MCODE) was applied to screen core modules in the PPI network with parameters as following: node score cutoff = 0.2, k-core = 2, and max. depth = 100, degree cutoff = 2.
2.6. Gene set enrichment analysis (GSEA)
To further identify the gene sets that were affected by COVID-19 infection, the whole gene expression profile of GSE147507, CRA002390 and E-MTAB-8871 were analyzed by GSEA with the curated gene sets (c2.all.v7.1.symbols.gmt) from Molecular Signatures Database (MSigDB, https://www.gsea-msigdb.org/) and cell markers of different cell types from CellMarker database (http://biocc.hrbmu.edu.cn/CellMarker/) [19]. For enrichment of the reversal effects of potential drugs, the gene signatures of GSE147507, CRA002390 and E-MTAB-8871 were used as the reference gene sets respectively. Gene sets with |NES| > 1 and Nom P-value < 0.05 were considered statistically significant. To directly evaluate the reversal effect of potential drugs, we developed a scoring system as showed below:
where and represent NES and P-value of downregulated genes from COVID-19 gene signature while and represent NES and P-value of upregulated genes from COVID-19 gene signature. Higher reversal score indicates a higher reversal capacity of potential drugs.
2.7. WGCNA analysis
Similar to our previous study [20], the weighted correlation network analysis (WGCNA) was used to construct the gene co-expression network. In this study, whole blood RNA NanoString nCounter transcriptomic profiling (E-MTAB-8871) from 10 healthy donors and a total of 9 days from illness onset to remission of a COVID-19 patient, with their clinic traits, were analyzed. We selected the power = 8 as the soft threshold to ensure a scale-free network. The topological overlap was calculated to measure network interconnectedness, and then average linkage hierarchical clustering was used to identify gene modules whose gene expression was highly correlated with the clinic traits from the gene co-expression network. The minimum number of genes in a module was set to 25. Throughout the analysis, we finally identified 6 modules. The expression of top 30 hub genes from brown and turquoise modules across healthy donors (shown as an average) and each day of illness progression of patients were shown in heatmap created by the R package pheatmap, and their PPI network was drawn by Cytoscape (v3.7.1).
2.8. Verify the utility of gene signature
The DEGs associated with the enriched pathway of GSE147507 and CRA002390, and genes from brown and turquoise modules of E-MTAB-8871 were used as processed gene signature for further analysis. Similar to our previous study [21], an over-representation method in WebGestalt webserver (http://www.webgestalt.org/) was used to enrich the gene signatures-related diseases.
2.9. Drug predicted by CREEDS and WebGestalt webserver
Three COVID-19 gene signatures in this study were put into WebGestalt webserver (http://www.webgestalt.org/), respectively [22]. A GSEA method in WebGestalt webserver with the minimum number of genes for a category = 4 was used to enrich potential COVD-19 therapeutic drugs from Drugbank (https://www.drugbank.ca/), and higher enrichment score indicates a higher similarity of the gene signature affected by the drug with our input COVID-19 gene signature. CREEDS webserver (http://amp.pharm.mssm.edu/creeds/) was further used for predicting the potential COVD-19 therapeutic drugs, whose affected gene signatures were opposite to our input COVID-19 gene signatures. Lower signed jaccard index indicates a better reversal effect [23].
3. Results
3.1. Gene signature of COVID-19 lung samples
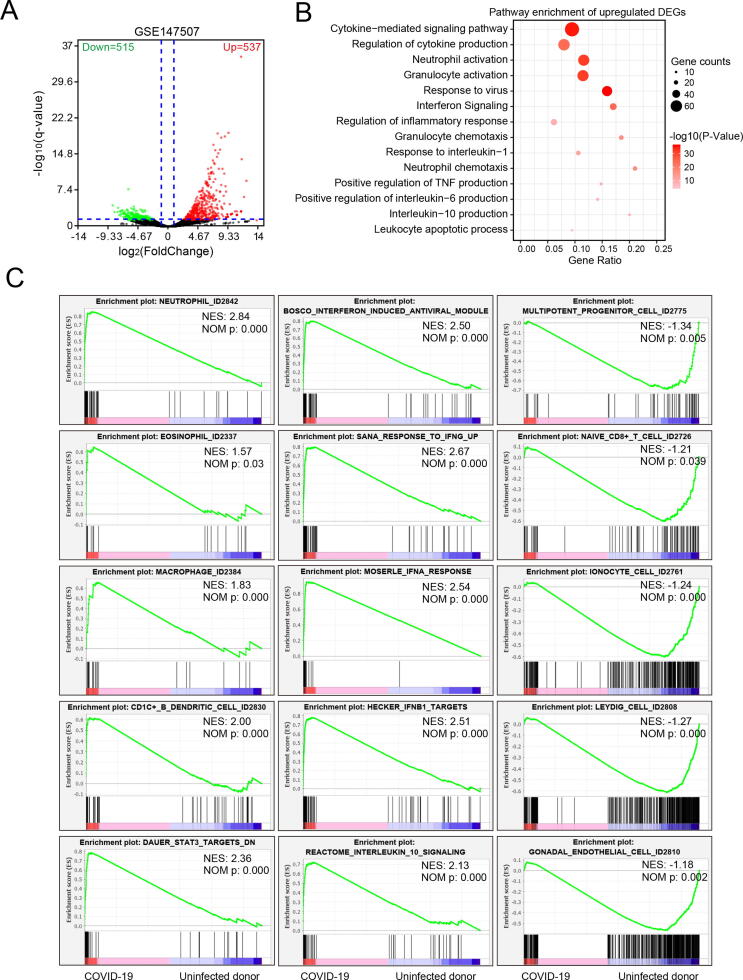
In order to get the gene signature of COVID-19, we first screened the differentially expressed genes (DEGs) of the RNA-seq transcriptome profiling from GSE147507 [24]. Two uninfected lung biopsies from two healthy male were used as the control group, and two lung biopsies derived from two male COVID19 deceased patient were used as the COVID-19 group. A total of 1052 genes were reserved with false discovery rate (FDR) q-value < 0.05 and |Log2 (Fold change) | > 1, including 537 upregulated DEGs and 515 downregulated DEGs (Fig. 1A and Supplementary Table 2). Pathway enrichment analyses showed that upregulated genes upon COVID-19 infection were mainly enriched in immune and inflammation-related pathways, such as cytokine-mediated signaling pathway, regulation of cytokine production, neutrophil activation, granulocyte activation, interferon signaling, and regulation of inflammatory response (Fig. 1B). This result was consistent with other reports that severe COVID-19 patients strongly associated with cytokine release syndrome (CRS), leading to acute respiratory distress syndrome (ARDS) and secondary haemophagocytic lymphohistiocytosis (sHLH), which are two of main causes of mortality [25], [26]. In contrast, downregulated genes were mainly associated with blood vessel development and nervous system development, suggesting that COVID-19 infection can cause damage to blood vessels and nervous systems in lung tissue (Supplementary Fig. 1A). Our gene set enrichment analysis (GSEA) further confirmed that COVID-19 infection was significantly enriched in cytokine-related gene sets, especially interferon-induced antiviral, including IFN-α, IFN-β and IFN-γ. Notably, COVID-19 infection was significantly positively enriched in neutrophil, eosinophil, macrophage and CDC1+ B dendritic cell, however, COVID-19 infection was negatively associated with multipotent progenitor cell, naïve CD8 + T cell, ionocyte cell, endothelial cell and leydig cell (Fig. 1C).
Fig. 1.
Transcriptome profiling of lung samples from uninfected donor and COVID-19 patients. RNA-Seq data (GSE147507) including two uninfected human lung biopsies and two lung samples from COVID-19 deceased patients were analyzed by DESeq2 differential expression analysis. A. Volcano plot of total 1052 differential expression genes with FDR q-value < 0.05. Red color represented upregulated genes in COVID-19 samples with Log2(fold change) > 1.0 and green represented downregulated genes in COVID-19 samples with Log2(fold change) < 1.0. B. Metascape pathway enrichment of 537 upregulated genes. C. Enrichment plots of GSEA correlation analyses for the whole gene expression profile of COVID-19 and donor lung samples by using curated gene sets (c2.all.v7.1.symbols.gmt) and cell marker gene set (CellMarker). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To increase the specificity of DEGs for following analysis, DEGs associated with pathways in Fig. 1B and Supplementary Fig. 1A was set as the gene signature of GSE147507 of which gene list was shown in Supplementary Table 3. To verify the utility of our processed gene signature, WebGestalt web tool was employed to enrich the gene signature-related diseases [22]. Results showed that our processed gene signature was highly related to influenza, hypersensitivity, chronic obstructive airway disease and pneumonia, indicating that this gene signature can fully represent whole DEGs of COVD-19 infection for further analysis (Supplementary Fig. 1B and Supplementary Table 4). Then, we constructed a protein–protein interaction (PPI) network of all genes of our processed gene signature (Supplementary Fig. 1C) and the molecular complex detections (MCODEs) clustering algorithm in Cytoscape was used to identify core cluster in PPI network. Finally, five most significant clusters were extracted, and pathway enrichment analysis of each cluster showed that cluster 1 was mainly associated with response to tumor necrosis factor (TNF), neutrophil degranulation, granulocyte activation, cytokine production, IL-1 and IL-10 signaling; cluster 2 was mainly associated with interferon signaling; cluster 3 was mainly associated with rRNA processing and ribosome biogenesis; cluster 4 and 5 were also mainly associated with neutrophil degranulation and granulocyte activation (Supplementary Fig. 2). Taken together, we identified a gene signature from two COVID-19 lung samples, and speculated that neutrophil, macrophage, B and DC cells mediated CRS, triggered specifically by IFN-α, IFN-β and IFN-γ, maybe the core mechanism of COVID-19-induced pneumonia.
3.2. Gene signature of COVID-19 peripheral blood mononuclear cells (PBMC) samples
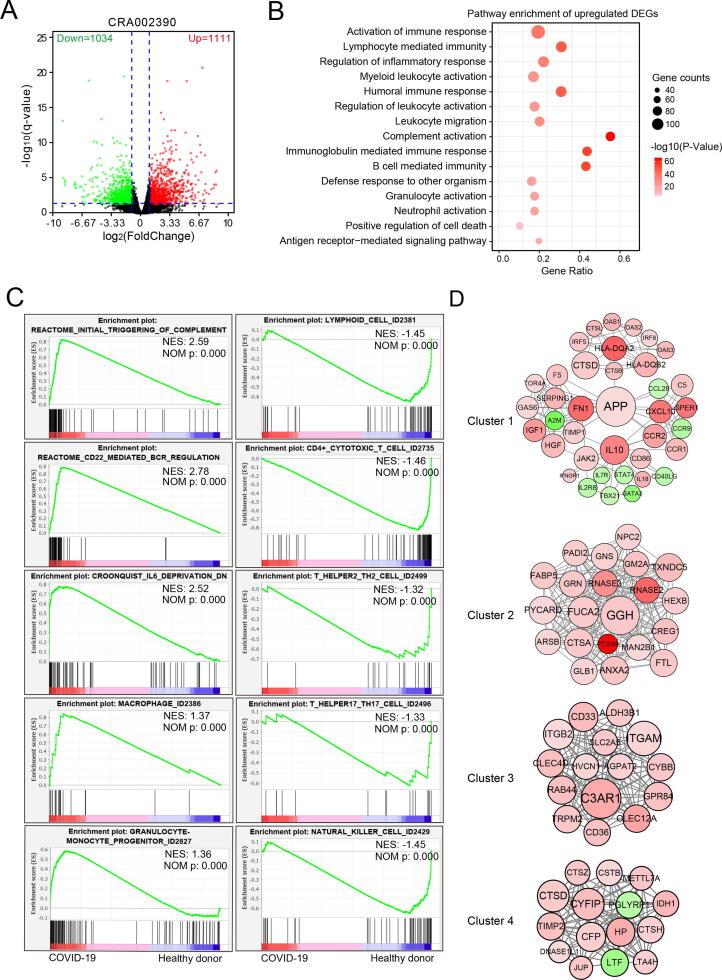
To avoid data bias from a single data source, we analyzed RNA-Seq data (CRA002390) containing three PBMC samples from healthy donors and three PBMC samples from COVID-19 patients [27]. We identified a total of 2145 DEGs, of which 1111 genes were upregulated and 1034 genes downregulated (Fig. 2A and Supplementary Table 5). Pathway enrichment analyses showed that upregulated DEGs induced by COVID-19 infection were mainly associated with following pathways: activation of immune response, lymphocyte-mediated immunity, humoral immune response, regulation of leukocyte activation, complement activation, B cell-mediated immunity, neutrophil activation, etc (Fig. 2B). However, downregulated DEGs mainly enriched in pathways including cell morphogenesis involved in neuron differentiation, axon development, T cell activation, Natural killer cell-mediated cytotoxicity, Th1 and Th2 cell differentiation, Th17 cell differentiation, etc (Supplementary Fig. 3A). Consistent with the results of pathway enrichment, our GSEA analysis further confirmed that COVID-19 infection was significantly enriched in the initial triggering of complement, CD22-mediated B cell receptor regulation, IL6 signaling, macrophage and granulocyte-monocyte progenitor. In contrast, COVID-19 infection was significantly negatively associated with the lymphoid cell, CD4 + cytotoxic T cell, Th17, Th2, and NK cell (Fig. 2C).
Fig. 2.
Transcriptome profiling of PBMC from healthy donors and COVID-19 patients. RNA-Seq data (CRA002390) including three PBMC samples from healthy donors and COVID-19 patients were analyzed by DESeq2 differential expression analysis. A. Volcano plot of total 2145 differential expression genes with FDR q-value < 0.05. Red color represented upregulated genes in COVID-19 samples with Log2(fold change) > 1.0, and green represented downregulated genes in COVID-19 samples Log2(fold change) < 1.0. B. Metascape pathway enrichment of 1111 upregulated genes. C. Enrichment plots of GSEA correlation analyses for whole gene expression profile using curated gene sets (c2.all.v7.1.symbols.gmt) and cell marker gene set (CellMarker). D. Four clusters of protein–protein interaction (PPI) network of gene signature. Node color density indicates the absolute value of the fold change of DEGs. Red indicates upregulated DEGs, and green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We then assumed DEGs associated with pathways in Fig. 2B and Supplementary Fig. 3A as the gene signature of CRA002390, of which gene list was shown in Supplementary Table 6. By using the WebGestalt webserver to verify its utility [22], we found that our processed gene signature was highly related to human T-lymphotropic virus 2 infections, bronchopneumonia, rheumatoid pneumoconiosis, lupus Erythematosus, arthritis, macrophage activation syndrome and anaphylaxis, indicating this gene signature has the transcriptome feature of virus-associated pneumonia (Supplementary Fig. 3B and Supplementary Table 4). We further constructed the PPI network of all genes in this gene signature (Supplementary Fig. 3C), and four most significant clusters were identified by the MCODEs clustering algorithm. Among them, cluster 1 was mainly associated with cytokine production, inflammatory response, platelet degranulation, IFN-γ signaling, TNF superfamily, IL-10; cluster 2, 3 and 4 were all significantly associated with neutrophil degranulation and activation (Fig. 2D and Supplementary Fig. 4). Taken together, the feature of the PBMC gene signature was similar with the gene signature from lung samples, and suggested that TNF, IFN-γ, IL-10 and IL6 secreted by neutrophil and macrophage, not T and NK cell, may play a critical role in the pathogenesis of COVID-19 infection.
3.3. WGCNA revealed gene-network modules associated with COVID-19
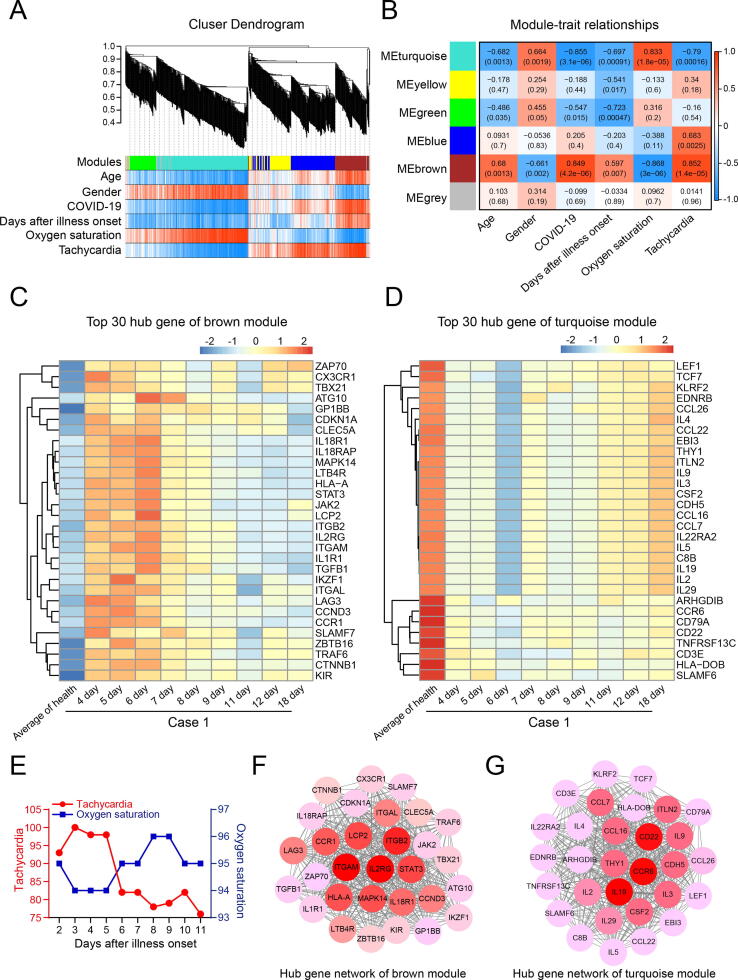
The weighted gene correlation network analysis (WGCNA) has been widely used to identify gene modules that highly correlated with clinical traits of patients [28], [29], [30]. A whole blood RNA NanoString nCounter transcriptomic profiling (E-MTAB-8871) from 10 healthy donors and total of 9 days from illness onset to remission of a COVID-19 patient (case 1) was downloaded and first analyzed by GSEA for quality evaluation [31]. Consistent with the GSEA data of COVID-19 lung samples of GSE147507 in Fig. 1C, the COVID-19 infection of this patient was also significantly enriched in IFN-α, IFN-β and IFN-γinduced antiviral pathway, inflammatory response lection/LPS, and apoptosis by serum deprivation (Supplementary Fig. 5A). Then, we applied WGCNA to analyze this transcriptomic profiling with patients’ clinical traits. Genes were assigned to five modules by hierarchical clustering of the weighting coefficient matrix, and unassigned genes were categorized into the grey module (Fig. 3A). Module-trait relationships were calculated by correlating the module’s eigengene to the clinical traits of the patient. And we found that the genes in brown module with the highest Pearson correlation with tachycardia (R = 0.852, P = 1.4e-5) and positive COVID-19 diagnosis, while brown module genes were significantly negatively correlated with oxygen saturation of patient (R = −0.868, P = 3.0e-6, Fig. 3B and Supplementary Table 7). In contrast, genes in the turquoise module were negatively correlated with tachycardia (R = −0.79, P = 1.6e-4) and positive COVID-19 diagnosis, and positively correlated with oxygen saturation of patient (R = 0.833, P = 1.8e-5, Fig. 3B and Supplementary Table 7). This result indicated that genes in the brown module were the driving genes of COVID-19 infection and mediated severe dyspnea, while genes in the turquoise module were the protective genes against COVID-19 infection.
Fig. 3.
Gene-network modules associated with COVID-19 revealed by WGCNA. Whole blood transcriptomic profiling (E-MTAB-8871) by RNA NanoString nCounter for 10 healthy donors and a total of 9 days from illness onset to remission of a COVID-19 patient, with their clinic traits were analyzed. A. Hierarchical cluster dendrogram showing different modules based on the unsupervised hierarchical clustering. B. Heatmap of the correlation between modules and clinical traits. Red indicates positive correlation, and blue indicates negative correlation. Person correlation value and P-value were shown in each heatmap. C-D. Expanded view of the expression of top 30 hub genes (weight analysis) in brown and turquoise modules across healthy donors (shown as an average) and each day of illness progression of patient case 1. E. Tachycardia and oxygen saturation traits of 2 to 11 day after illness onset of patient case 1. F-G. PPI network of top 30 hub genes in brown and turquoise modules. Node color intensity indicates the weight value in the weighted gene co-expression network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Hub genes in each module were analyzed by weight analysis (Supplementary Table 8), and the expression of the top 30 hub genes in brown and turquoise modules across healthy donors (shown as an average) and each day of illness progression of patient case 1 were shown in the heatmap (Fig. 3C, D). Interestingly, the expressions of all 30 hub genes in the brown module were increased with the dyspnea symptoms reached a nadir on day 4 and 5 post-illness onset, whereas these highly expressed genes gradually returned to normal level with the restoration of respiratory function from day 7 to day 18 (Fig. 3C, E and Supplementary Fig. 5B). Conversely, the expressions of all 30 hub genes in the turquoise module were markedly decreased with the dyspnea symptoms reached a nadir on day 4 and 5, and then gradually increased as the patient recovered from day 7 to day 18 (Fig. 3D, E and Supplementary Fig. 5B). Similar results were also found in another COVID-19 patient case 2 (Supplementary Fig. 5D-E), and his clinic traits of illness progression were shown in Supplementary Fig. 5C. Furthermore, the hub-gene-network analysis of gene signature revealed a hierarchical organization of highly connected genes in brown and turquoise modules. And we found ITGB2, ITGAM and IL2RG were the top 3 hub genes with the highest weight in the brown module network, which are highly expressed on neutrophils, and contribute to neutrophil activation and inflammation [32], [33], [34] (Fig. 3F). On the other hand, CD22, CCR6 and IL19 were found to be the top 3 hub genes in the turquoise module, and all these three genes are anti-inflammation genes and functionally important for preventing over-activation of the immune system and the development of autoimmune diseases [35], [36], [37] (Fig. 3G). We then assumed the genes in brown and turquoise modules as the gene signature of E-MTAB-8871 (Supplementary Table 7), and WebGestalt analysis also showed that this gene signature was highly related to juvenile rheumatoid arthritis, C-reactive protein increased, recurrent bronchitis, hypersensitivity, pneumonia, pulmonary fibrosis, fever, etc (Supplementary Fig. 6 and Supplementary Table 4). Therefore, our results extracted a powerful gene signature of E-MTAB-8871, and confirmed that over-activation of immune system-mediated CRS is the underlying pathogenic mechanism of the acute phase of COVID-19 patients, and suggest that anti-inflammatory therapies may benefit those patients.
3.4. A GSEA method to predict COVID-19 potential therapeutic drugs
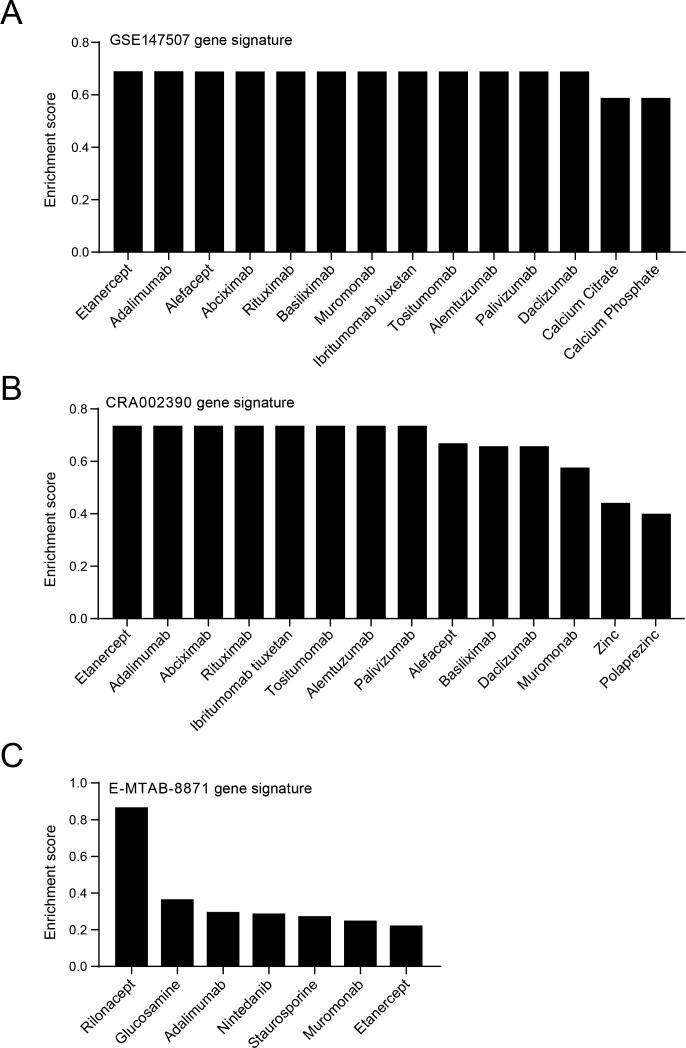
Currently, there are no effective drugs and vaccines for COVID-19 treatment or prevention. It is very urgent to find potential drugs to treat COVID-19 and save the patients. In this study, we utilized the targeting gene sets of 5825 drugs from drugBank and analyzed the similarity between drug targeting genes and COVID-19 gene signatures to predict potential therapeutic drugs. The drug targets were used as the reference gene sets, and three gene signatures, GSE147507, CRA002390 and E-MTAB-8871, were processed and analyzed by WebGestalt webserver using the GSEA enrichment method. Predicting results showed that targets affected by two TNFα inhibitors, etanercept and adalimumab, were positively correlated with gene signature of GSE147507, and several T cell or B cell inhibitors including alefacept, rituximab, basiliximab, muromonab, ibritumomab tiuxetan, tositumomab, alemtuzumab and daclizumab were also positively enriched (Fig. 4A and Supplementary Table 9). We also identified abciximab, a platelet aggregation inhibitor and palivizumab, a preventive drug for respiratory syncytial virus (RSV) infection, shared gene targets with COVID19 gene signature; Moreover, calcium citrate and calcium phosphate were also enriched (Fig. 4A and Supplementary Table 9). Analysis of the gene signature of CRA002390 revealed a very similar result with GSE147507, in which inhibitors of TNFα, T cell and B cell were enriched, and zinc, polaprezinc also shared similar COVID19 signature (Fig. 4B and Supplementary Table 9). Simultaneously, targets of TNFα inhibitors etanercept and adalimumab were also positively correlated with gene signature of E-MTAB-8871, and an IL1 inhibitor: rilonacept, an anti-inflammation drug for osteoarthritis treatment: glucosamine, idiopathic pulmonary fibrosis treatment medication: nintedanib and a T cell inhibitor: muromonab also shared similar COVID19 signature (Fig. 4C and Supplementary Table 9).
Fig. 4.
Predicting potential therapeutic drugs by a GSEA method with drugBank in WebGestalt webserver for COVID-19 gene signature. A. Enriched drugs for gene signature of GSE147507. Positively enriched drugs were shown. B. Enriched drugs for gene signature of CRA002390. C. Enriched drugs for gene signature of brown and turquoise modules of E-MTAB-8871. A high enrichment score indicates a higher similarity of the gene signature affected by the drug with our input COVID-19 gene signature.
3.5. The predicted drugs may reverse COVID-19 gene signature
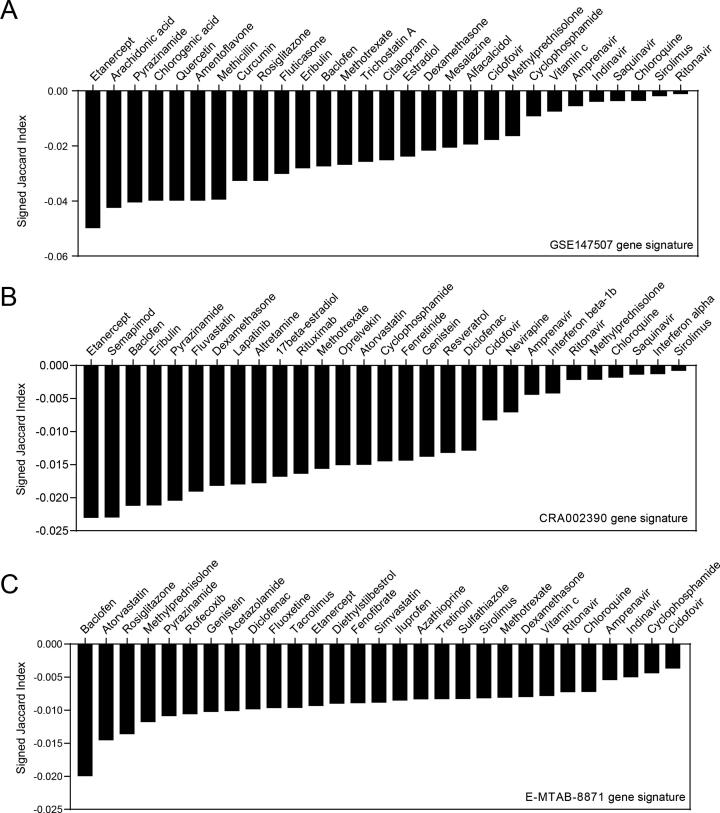
Since the WebGestalt webserver only analyzed the similarity between drug targets and COVID-19 gene signatures, whether these drugs can reverse COVID-19 gene signatures remains unknown. To solve this issue, we used CREEDS webserver to identify drugs that induce reciprocal changes to COVID-19 gene signature from a total of 6100 drugs. Drugs were ranked by the signed jaccard index, and a lower signed jaccard index indicates a potentially better reversal effect of COVID-19 gene signature [23]. We found that the expression of genes affected by TNFα inhibitor etanercept had the most opposite feature with the COVID-19 gene signatures, which ranked first in GSE147507 and CRA002390, and ranked middle in E-MTAB-8871 (Fig. 5). Notably, Baclofen, a γ-aminobutyric acid-B receptor (GABABR) agonist for muscle spasticity and multiple sclerosis treatment, was also found to be markedly opposite to three COVID-19 gene signatures [38], [39] (Fig. 5 and Supplementary Table 10). In contrast, drugs which are currently widely used by clinic or recommended by treatment guidelines, such as chloroquine, ritonavir, interferon and vitamin c, only showed modest counter effect to COVID-19 signatures (Fig. 5 and Supplementary Table 10). Glucocorticoids such as fluticasone, dexamethasone and methylprednisolone, also showed moderate counter effects (Fig. 5 and Supplementary Table 10).
Fig. 5.
Predicting potential therapeutic drugs by CREEDS webserver which measures opposite effects of drugs on gene expression of COVID-19. Drugs have potential reversal effects of COVID-19 gene signature of GSE147507, CRA002390 or E-MTAB-8871 were ranked by the signed jaccard index. Lower signed jaccard index indicates a better reversal effect.
3.6. Validate counter effects of potential drugs on COVID-19 gene signature
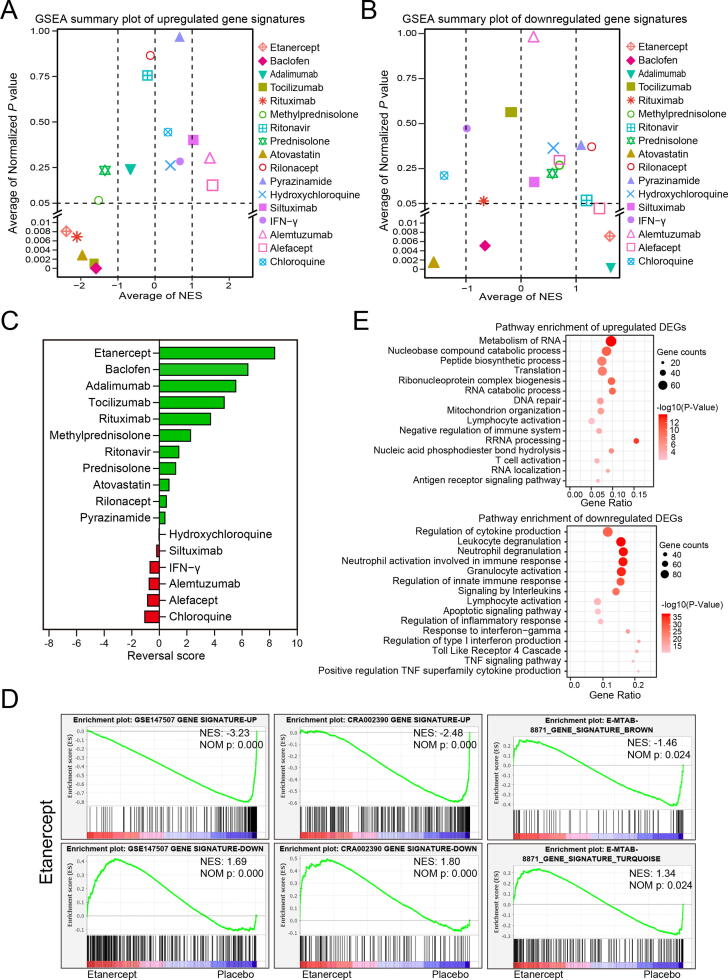
To further confirm the reversal effects of potential drugs, the transcriptome profiling of drugs was downloaded and then analyzed by GSEA. Enhancement or reduction of gene signatures of GSE147507, CRA002390 and E-MTAB-8871 were used as reference gene sets respectively. Two summary plots of GSEA results were shown the average NES and average normalized P value of each drug enriched with three COVID-19 gene signatures (Fig. 6A and B). Reversal score of each drug was calculated by a scoring system based on NES and normalized P value. Consistent with the results in Fig. 4 and Fig. 5, etanercept was the most effective drug that we screened out to reverse the COVID-19 gene signatures. Etanercept significantly decreased expression of genes that were upregulated in all three COVID-19 gene signatures, and significantly increased the expression of the downregulated genes in all three COID-19 gene signatures (Fig. 6A, B and D). Pathway enrichment analysis strongly confirmed that etanercept had a very desirable reversal effect on COVID-19-affected signaling pathway. The pathways enriched by etanercept downregulated genes were almost identical to the pathways enriched by COVID-19 upregulated genes (Fig. 6E, Fig. 1B and Fig. 2B). Etanercept upregulated genes mainly enriched in RNA metabolism, ribonucleoprotein complex biogenesis, mitochondrion organization, T cell activation, etc, which were also found to be enriched by COVID-19 downregulated genes (Fig. 6E, Supplementary Fig. 1A and Supplementary Fig. 3A). Baclofen was another potential drug which can significantly reverse the upregulated genes in three COVID-19 gene signatures, and increased the expression of the downregulated genes in gene signatures of GSE147507 (Supplementary Fig. 7A). These results suggest etanercept and baclofen are two promising drugs for COVID-19 treatment. In addition, another TNFα inhibitor, adalimumab was also found had significantly reversal effects on downregulated genes of all three COVID-19 gene signatures and on upregulated gene signature of GSE147507 (Supplementary Fig. 7B).
Fig. 6.
GSEA validation of the potential COVD-19 therapeutic drugs. A. GSEA summary plot of reversal effects of potential therapeutic drugs on upregulated gene signature of COVID-19. B. GSEA summary plot of reversal effects of potential therapeutic drugs on downregulated gene signature of COVID-19. NES > 1 means genes affected by drugs was positively correlated with COVID-19 gene signatures while NES < -1 means genes affected by drugs was negatively correlated with COVID-19 gene signatures. C. Reversal scores for predicting potential therapeutic drugs. Microarray profiling of whole blood leukocyte from15 healthy donors who pretreated with etanercept or placebo then received an intravenous injection of endotoxin challenge (GSE36177) was analyzed by GSEA, Over-expression (Up) or down-expression (Down) genes in gene signatures of GSE147507, CRA002390 and E-MTAB-8871 were used as reference gene sets. D. Reversal effect of etanercept on gene signatures of COVID-19. E. Metascape pathway enrichments of 860 upregulated and 730 downregulated DEGs (q-value < 0.01) induced by etanercept treatment, respectively.
Severe COVID-19 cases may benefit from IL6 inhibitors and its controlled clinical trials are still ongoing [1]. We found that tocilizumab had significantly reversal effects on upregulated genes of three COVID-19 gene signatures (Supplementary Fig. 7C), but no efficacy on downregulated genes of COVID-19 gene signatures. However, siltuximab was found no reversal effects on COVID-19 gene signatures (Supplementary Fig. 7D). Rituximab is an anti-CD20 antibody approved for treating lymphoma and rheumatoid arthritis. Rituximab has recently been reported to suppress the cytokine storm and is beneficial to COVID-19 patient [40]. We found rituximab had desirable reversal effect on upregulated genes of three COVID-19 gene signatures, while it would further decrease the expression of downregulated genes in gene signatures of CRA002390 and E-MTAB-8871 (Supplementary Fig. 8A). In addition, the application of glucocorticoids in COVID-19 treatment is also a controversial issue. Herein, we found that both methylprednisolone and prednisolone had desirable reversal effect on COVID-19 gene signatures, especially for reversing the upregulated genes of COVID-19 gene signatures (Supplementary Fig. 8B-C). Ritonavir has been reported for COVID-19 treatment by several articles [41], [42], [43] while we found ritonavir only had a very slight reversal effect on downregulated genes of E-MTAB-8871 gene signature, and there was no effect on the upregulated genes of COVID-19 gene signatures (Supplementary Fig. 8D). Moreover, Atorvastatin, a drug for treating high cholesterol, was also found to have the reversal effects for upregulated genes in three COVID-19 gene signatures (Supplementary Fig. 9A). However, atorvastatin further reduced the expression of COVID-19 downregulated genes (Supplementary Fig. 9A). Rilonacept (an IL-1 inhibitor) and pyrazinamide (an anti-tuberculous agent), were found only significantly reversed the expression of genes in the turquoise module of E-MTAB-8871 (Supplementary Fig. 9B-C).
However, chloroquine and hydroxychloroquine, the most controversial COVID-19 treatment drugs [44], [45], [46], [47] had no obvious effect to reverse COVID-19 gene signature. Actually, they may be harmful to COVID-19 treatment supported by evidence that chloroquine would exhaust the COVID-19 downregulated genes in GSE147507 gene signature, and hydroxychloroquine further increased the expression of COVID-19 upregulated genes in CRA002390 gene signature (Supplementary Fig. 9D and 10A). Consistently, our results strongly supported the COVID-19 treatment guidelines issued by USA NIH [1] that recommended against the use of interferons for the treatment of COVID-19. We found that IFN-γwas potentially harmful to COVID-19 treatment, evidenced by that it significantly exacerbated the expression of upregulated genes and decreased the expression of downregulated genes of GSE147507 gene signature (Supplementary Fig. 10B). Finally, alemtuzumab, an antibody binding to CD52 on mature lymphocytes and approved for chronic lymphocytic leukemia and multiple sclerosis treatment, and alefacept, a psoriasis treatment drug via interfering with CD2 to inhibit T cell activation, both of them have been found to further increase the upregulated genes of COVID-19 gene signature and may be harmful to COVID-19 treatment (Supplementary Fig. 10C-D). Collectively, through the GSEA analysis of drug’s transcriptome profiling, we are highly optimistic about the TNFα inhibitor etanercept and GABABR agonist baclofen for COVID-19 treatment, and suggest that adalimumab, tocilizumab, rituximab and glucocorticoids may also be potential drugs for COVID-19 treatment (Fig. 7). Controlled clinical trials of these candidate drugs for COVID-19 treatment are urgently needed. Nevertheless, our data suppose chloroquine, hydroxychloroquine and interferons should not be used for COVID-19 treatment.
Fig. 7.
Graph abstract of our findings.
4. Discussion
In this study, we identified three gene signatures from COVID-19 patients, and found the upregulated genes upon COVID-19 infection were mainly enriched in immune and inflammation-related pathway which related to IFN-α, IFN-β, IFN-γ, TNF and IL6 triggered cytokine storm mediated by neutrophil, macrophage, B and DC cells. Our findings are highly consistent with accumulating evidence suggesting that disease severity of COVID-19 was correlated with hypercytokinaemia which are characterized by increased IL-1β, IL-7, IL-10, IL6, TNFα, IFN-γ, granulocyte colony-stimulating factor (G-CSF), IP10/CXCL10, monocyte chemoattractant protein 1 (MCP-1) and macrophage inflammatory protein 1-α (MIP-1) [48], [49]. Neutrophil count in blood was reported remarkably higher in COVID-19 severe patients than that in mild cases [50]. An autopsy study also reported that the cell types of infiltrated immune cells in alveolar space and septa of COVID-19 patient were predominantly infiltrating CD68+ macrophages and CD20+ B cells [51]. CRS was found to be the major cause of severe pneumonia and morbidity in COVID-19 patients [52]. Thus, immunosuppressive and anti-inflammatory therapies were considered to be potentially effective COVID-19 treatment strategies [10]. In contrast, our study revealed that COVID-19 downregulated genes were mainly associated with blood vessel development, nervous system development, ionocyte cell, endothelial cell, leydig cell, T cell activation, NK cell-mediated cytotoxicity, Th1, Th2 and Th17 cell differentiation. COVID-19 causes pulmonary blood vessel damage which is usually manifested in severe endothelial injury and widespread thrombosis with microangiopathy [53]. The nervous system and the male fertility damage have also been reported in COVID-19 patients [54], [55], [56], [57]. In addition, our foundings also support the observations by previous reports that the CD8+ T cell and NK cell were decreased significantly in blood and CD4+ T were barely detectable in the pulmonary biopsy of COVID-19 patients, suggesting that the functional exhaustion of cytotoxic lymphocytes is another critical underlying mechanism of COVID-19 infection [50], [51], [58]. Furthermore, CD4+ T cells were activated to turn into pathogenic Th1 cells and generate granulocyte–macrophage colony-stimulating factor, augmenting expression of IL6 in CD14+ CD16+ monocytes on the SARS-CoV-2 infection [48]. Hence, SARS-CoV-2 infection may break down antiviral immunity at an early stage.
Aside from uncovering the pathogenic mechanism of COVID-19, another goal of this study is finding the potential drugs to decrease the high mortality rate of COVID-19 patients. However, the development of a novel drug will take over 10 years from target identification, target validation, hit discovery, lead optimization to preclinical and clinical trials [9]. Therefore, it seems impossible to develop novel drugs in the time frame needed to rescue the current SARS-CoV-2 crisis. Hence, our study tried to find potential therapies from existing clinical drugs by drug repurposing. Based on our bioinformatic analysis, we are highly confident that TNFα inhibitor etanercept and GABABR agonist baclofen would significantly benefit COVID-19 patients. Anti-tumor necrosis factor (TNF) antibodies have been approved for more than 20 years in severe cases of autoimmune inflammatory disease and anti-TNF therapies have previously been shown protective effects in lethal SARS-CoV infection [59], [60]. A recent case report also suggested that the use of etanercept before the viral infection was disassociated with a severe evolution of the COVID-19 [61]. TNF acts upstream of IL6, both of them are produced in most types of inflammation, especially in the acute phase, and anti-TNF therapy was postulated that the earlier the better after hospital admission of COVID-19 patients [60]. On the other hand, baclofen, a drug for muscle spasticity and multiple sclerosis treatment, has also been reported in recent years to attenuate basal and IFN-γ/LPS-induced release of the pro-inflammatory cytokines IL6 and TNFα in both in vitro and in vivo studies, suggesting that baclofen is a promising anti-inflammatory and immunomodulatory treatment [38], [39]. However, there are currently no sufficient clinical trial tests for TNFα inhibitors and baclofen so that we propose associated clinical trials should be initiated as early as it is practicable.
Our study also suggests that IL6 inhibitors, especially tocilizumab may be a potential drug for COVID-19 treatment. Consistent with our observations, tocilizumab was shown to immediately improve the clinical outcome of severe COVID-19 patients [62], and several clinical trials on the use of tocilizumab in patients with COVID-19 are ongoing [63]. Our results also found rituximab had significantly reversal effects on upregulated genes of three COVID-19 gene signatures, but it would further decrease the expression of downregulated genes of COVID-19 gene signatures. Recently, a case report speculated that rituximab may limit the cytokine storm and delay the worsening and need for mechanical ventilation [40], but another case report concluded that rituximab is responsible for long-lasting B-cell depletion and associated with high-risk fatal outcome of two COVID-19 patients [64]. Thus, more evidence is needed to settle the debate. In addition, our data supported the latest clinical trial results that no benefit was observed with lopinavir-ritonavir treatment for COVID-19 patients [65]. Pyrazinamide and atorvastatin were also high-ranked in CREEDS webserver results. Their anti-inflammatory and immune modulation activities have been discovered for many years [66], [67]. Notably, statins have been reported to reduce hospitalization rate and mortality caused by flu, and they have also been found to decrease the fatality rate of MERS [68], [69] infection. Most recently, molecular docking analysis revealed statins can directly bind to the SARS-CoV-2 main protease (Mpro), and suggested that statins could be efficient SARS-CoV-2 Mpro inhibitors although further experimental and clinical validations are needed [70], [71].
On the other hand, glucocorticoids are recommended to treat COVID-19 patients in the acute phase in China FDA guideline [72] while are against the routine use for critically ill patients with COVID-19 by USA FDA guideline [1]. According to our findings, methylprednisolone and prednisolone showed moderate reversal effect on COVID-19 gene signatures, especially for reversing the upregulated genes of COVID-19 gene signatures. These immunosuppressive agents could likely reduce hyper-inflammation in concurrent COVID-19 patients with rheumatic diseases by inhibiting gene expression of the cytokine factors [73]. Moreover, a recent study showed that an early short course of methylprednisolone in patients with moderate to severe COVID-19 reduced escalation of care and improved clinical outcomes [74]. The above facts are all in support of the China FDA guideline. However, the use of glucocorticoids therapy is still debated, and a common side effect of glucocorticoids is the increasing risk of infections, especially facilitating the worsening of a SARS-CoV-2 infection into severe acute respiratory distress syndrome [75]. Moreover, it has also been reported that glucocorticoids used in SARS and MERS patients did not improve mortality and resulted in delayed viral clearance [10]. Taken together, glucocorticoid regimens in COVID-19 treatments require more evidence-based validations.
Several herbal compounds were also enriched out by CREEDS webserver analysis, such as chlorogenic acid, quercetin, curcumin, genistein and resveratrol. Chlorogenic acid is one of the most bioactive compounds rich in the honeysuckle which is an antiviral herb widely used in Chinese medicine [76]. Molecular docking showed that chlorogenic could stably combine with Gln42/Asp38 in ACE2, which hindered the combination between SARS-CoV-2 S-protein and ACE2 [77]. Quercetin, a strong antioxidant and anti-inflammatory agent, has also been reported to inhibit the proteases of SARS and MERS [78], and a clinical trial (NCT04377789) to evaluate the preventive and therapeutic effect of quercetin on COVID-19 is also found on ClinicalTrials.gov. Moreover, curcumin was found to have a dual binding affinity of both SARS-CoV-2 S-protein and ACE2 [79], and can inhibit the release of numerous cytokines including IL1β, IL8, TNFα and MCP1 in a mouse model of virus-induced acute respiratory distress syndrome [80]. Similarly, genistein and resveratrol are both anti-inflammatory drugs and have also been shown significant interaction with the SARS-CoV-2 proteins by forming hydrogen bonds [81], [82], [83].
Aside from alleviating damage induced by the CRS, another effective treatment is to boost innate antiviral immune responses to SARS-CoV-2. Calcium, zinc and vitamin C were found in WebGestalt or CREEDS analysis. Undoubtedly, they are all widely reported to play key roles in the regulation of immunity and inflammation [84], [85], [86]. According to a new report, dietary supplements containing micronutrients (zinc and omega-3 fatty acid) and vitamins C and D are a safe, low-cost, and effective way of helping the immune system fight off COVID-19 and other acute respiratory tract diseases [87]. In contrast, our results suggest chloroquine, hydroxychloroquine and interferons should not be used for COVID-19 treatment, which is supported by the COVID-19 treatment guidelines from USA NIH who recommends against the use of chloroquine/hydroxychloroquine and interferon [1].
In conclusion, our study reveals that CRS mediated by neutrophil, macrophage, B and DC cells is the underlying pathogenic mechanism of the SARS-CoV-2 infection, especially when it is highly related to the symptoms of the acute phase, and suggests that the anti-inflammatory therapies may benefit COVID-19 patients. Our study also highlights the power of TNFα inhibitor etanercept and GABABR agonist baclofen in the reversal of COVID-19 genes signature, so we are highly optimistic about their clinical use for COVID-19 treatment and achieving a good clinical outcome. In addition, our results suggest that adalimumab, tocilizumab, rituximab and glucocorticoids may also be potential drugs, but our data suggests that chloroquine, hydroxychloroquine and interferons may not be beneficial to COVID-19 patient. We provide the therapeutic prospects from an integrated transcriptional analysis of COVID-19 patients and we believe validations of these candidates are urgently needed in the near future.
5. Key points
-
•
Neutrophil, macrophage, B and DC cells mediated IFN-α, IFN-β, IFN-γ, TNF and IL6 release is the underlying pathogenic mechanism of the SARS-CoV-2 infection.
-
•
Cytokine release syndrome is highly related to the acute phase pneumonia and suggests that the anti-inflammatory therapies may benefit COVID-19 patients.
-
•
Drug repurposing analysis revealed that etanercept and baclofen may be two promising drugs for the treatments of severe COVID-19 patients.
Adalimumab, tocilizumab, rituximab and glucocorticoids may also have beneficial for COVID-19 patients, but our results suggest thatchloroquine, hydroxychloroquine and interferons may not be beneficial to COVID-19 patient.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We are grateful to Dr. Benjamin R. tenOever (Icahn School of Medicine at Mount Sinai), Dr. Yu Chen (Wuhan University) and Dr. Jenny Guek Hong Low (SingHealth Duke-NUS Academic Medical Center) for their uploaded COVID-19-related RNA-seq raw data and NanoString nCounter transcriptomic profiling.
Contributors
FM, GL, YD designed the study. GL, FM collected transcriptome data. FM, GL, QL analyzed the data. GL, XZ, YD, FM wrote the paper. SR provided clinical interpretation and revised the paper. FM, YD, SR supervised the study.
Funding
Thanks the support from National Natural Science Foundation of China [H16-82003137], China.
Patient and public involvement
Patients and/or the public were not involved in the design, or conduct, or reporting or dissemination plans of this research. Patient consent for publication not required.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.056.
Contributor Information
Yali Dou, Email: yalid@med.umich.edu.
Fengbiao Mao, Email: maofengbiao@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Metascape pathway enrichment of downregulated genes, enriched diseases and PPI network of GSE147507 gene signature. A. Metascape pathway enrichment of 515 downregulated genes of GSE147507. B. Enrichment of related diseases of gene signature of GSE147507 using WebGestalt webserver. C. Protein-protein interaction (PPI) network of GSE147507 gene signature. Node color density indicates the absolute value of Log2(fold change) of DEGs. Red indicates upregulated DEGs, and green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity.
Five clusters and their enriched pathways of the GSE147507 PPI network. Node color density in clusters indicates the absolute value of Log2(fold change) of DEGs. Red indicates upregulated DEGs while green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity. Bubble plots of pathway enrichment in each cluster were shown.
Metascape pathway enrichment of downregulated genes, enriched diseases and PPI network of CRA002390 gene signature. A. Metascape pathway enrichment of 1034 downregulated genes of CRA002390. B. Enrichment of related diseases of gene signature of CRA002390 using WebGestalt webserver. C. PPI network of CRA002390 gene signature. Node color density indicates the absolute value of Log2(fold change) of DEGs. Red indicates upregulated DEGs and green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity.
Metascape pathway enrichment of the four clusters of CRA002390 PPI network.
GSEA analysis, patients’ clinic traits, expression heatmaps of brown and turquoise modules hub genes of E-MATB-8871. A. Enrichment plots of GSEA correlation analysis for 10 healthy donors and day 4, 5, 6 of case 1 of E-MTAB-8871 using curated gene sets (c2.all.v7.1.symbols.gmt). B-C. Clinic traits of case 1 and case 2 COVID-19 patients. D-E. Heatmaps of top 30 hub genes (weight ranked) in brown and turquoise modules across healthy donors (shown as an average) and each day of illness progression of patient case 2.
Enriched diseases of E-MTAB-8871 gene signature. Disease enrichment was analyzed by an over-representation method in WebGestalt webserver for the genes of brown and turquoise modules.
GSEA validation of the reversal effects of baclofen, adalimumab, tocilizumab and siltuximab. A. The RNA-seq of aortic tissues from mice treated with baclofen or vehicle (GSE128101) was analyzed by GSEA. B. RNA-seq of maternal blood neutrophils from the Rhesus intrauterine infection model treated without or with adalimumab (GSE145918) was analyzed by GSEA. C. Microarray of PBMC fromm juvenile idiopathic arthritis patients taken before and after tocilizumab treatment (GSE76492) was analyzed by GSEA. D. Microarray of ovarian cancer SKOV-3 and Caov-3 cells pre-treated without or with siltuximab, then treated with IL6 (E-GEOD-20272) was analyzed by GSEA.
GSEA validation of the reversal effects of rituximab, methylprednisolone, prednisolone and ritonavir. A. Whole-blood transcriptomic profiling for patients with rheumatoid arthritis who are non-responders or responders to rituximab treatments (GSE54629) was analyzed by GSEA. B. RNA-seq of circulating B cells from healthy human donors treated with methylprednisolone or vehicle in vitro (GSE112101) was analyzed by GSEA. C. Microarray of whole blood from dengue patients received prednisolone or placebo therapy (GSE40165) was analyzed by GSEA. D. Microarray of PBMC treated with placebo or ritonavir from healthy volunteers (E-TABM-642) was analyzed by GSEA.
GSEA validation of the reversal effects of atorvastatin, rilonacept, pyrazinamide and chloroquine. A. Transcription profiling of human quadriceps femoris muscle from patients received placebo or atorvastatin (E-TABM-116) was analyzed by GSEA. B. Microarray profiling of skin from diffuse cutaneous systemic sclerosis patients who pre- or post-treated with rilonacept (GSE119939) was analyzed by GSEA. C. Transcriptional array of lung tissue of tuberculosis-infected mice treated without or with pyrazinamide (GSE48027) was analyzed by GSEA. D. Microarray of human liver Huh7 cells treated with or without chloroquine (GSE30351) was analyzed by GSEA.
GSEA validation of the reversal effects of hydroxychloroquine, IFN-γ, alemtuzumab and alefacept. A. RNA-seq of PBMC from healthy participants treated without or with hydroxychloroquine (GSE74235) was analyzed by GSEA. B. RNA-seq of murine lung endothelial cells treated without or with IFN-γ (E-MTAB-5921) was analyzed by GSEA. C. Microarray profiling of T-cell prolymphocytic leukemia (T-PLL) from pre- or post-treated with alemtuzumab (GSE67368) was analyzed by GSEA. D. Microarray data from PBMCs of psoriasis patients who are non-responders or responders to alefacept treatment (GSE18948) was analyzed by GSEA.
References
- 1.Health. C-TGPNIo. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines; 2020. [PubMed]
- 2.Diao B, Wang C, Tan Y, Chen X, Liu Y, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol. 2020;11:827. [DOI] [PMC free article] [PubMed]
- 3.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;1–13 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y.i., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. The Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teng H, Wei W, Li Q, Xue M, Shi X, et al. Prevalence and architecture of posttranscriptionally impaired synonymous mutations in 8,320 genomes across 22 cancer types. Nucleic Acids Res. 2020;48(3):1192–205. [DOI] [PMC free article] [PubMed]
- 7.Wang H, Wang T, Zhao X, Wu H, You M, et al. AI-Driver: an ensemble method for identifying driver mutations in personal cancer genomes. NAR Genomics Bioinformatics. 2020;2(4). [DOI] [PMC free article] [PubMed]
- 8.Mao F., Xiao L., Li X., Liang J., Teng H., Cai W., Sun Z.S. RBP-Var: a database of functional variants involved in regulation mediated by RNA-binding proteins. Nucleic Acids Res. 2016;44(D1):D154–D163. doi: 10.1093/nar/gkv1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Zheng Q, Wang Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics; 2020. [DOI] [PMC free article] [PubMed]
- 10.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science:abb8925. [DOI] [PubMed] [Google Scholar]
- 11.Picchianti Diamanti A., Rosado M.M., Pioli C., Sesti G., Lagana B. Cytokine Release Syndrome in COVID-19 Patients, A New Scenario for an Old Concern: The Fragile Balance between Infections and Autoimmunity. Int J Mol Sci. 2020;21(9) doi: 10.3390/ijms21093330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biggest COVID-19 trial tests repurposed drugs first. Nat Biotechnol. 2020;38(5):510. [DOI] [PubMed]
- 13.Mao F., Liu Q., Zhao X., Yang H., Guo S. EpiDenovo: a platform for linking regulatory de novo mutations to developmental epigenetics and diseases. Nucleic Acids Res. 2018;46(D1):D92–D99. doi: 10.1093/nar/gkx918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T., Ruan S., Zhao X., Shi X., Teng H. OncoVar: an integrated database and analysis platform for oncogenic driver variants in cancers. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Lan Y., Xu J., Quan F., Zhao E. Cell Marker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Shi L., Wang Y., Zhong J., Zhao X. OncoBase: a platform for decoding regulatory somatic mutations in human cancers. Nucleic Acids Res. 2019;47(D1):D1044–D1055. doi: 10.1093/nar/gky1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y., Zhou B., Mao F., Xu J., Miao H. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell. 2018;34(4):643–58 e5. doi: 10.1016/j.ccell.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Monteiro C.D., Jagodnik K.M., Fernandez N.F., Gundersen G.W. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat Commun. 2016;7(1):1–11. doi: 10.1038/ncomms12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181(5):1036–45 e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C., Li S., Liu Y. Role of immunosuppressive therapy in rheumatic diseases concurrent with COVID-19. Ann Rheum Dis. 2020;79(6):737–739. doi: 10.1136/annrheumdis-2020-217460. [DOI] [PubMed] [Google Scholar]
- 26.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong Y., Liu Y., Cao L., Wang D., Guo M. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y., Coskun V., Liang A., Yu J., Cheng L. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161(5):1175–1186. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radulescu E., Jaffe A.E., Straub R.E., Chen Q., Shin J.H. Identification and prioritization of gene sets associated with schizophrenia risk by co-expression network analysis in human brain. Mol Psychiatry. 2020;25(4):791–804. doi: 10.1038/s41380-018-0304-1. [DOI] [PubMed] [Google Scholar]
- 30.Tang J., Kong D., Cui Q., Wang K., Zhang D. Prognostic Genes of Breast Cancer Identified by Gene Co-expression Network Analysis. Front Oncol. 2018;8:374. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jerke U., Rolle S., Dittmar G., Bayat B., Santoso S. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J Biol Chem. 2011;286(9):7070–7081. doi: 10.1074/jbc.M110.171256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiScipio R.G., Daffern P.J., Schraufstatter I.U., Sriramarao P. Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves alphaMbeta2 (CD11b/CD18) J Immunol. 1998;160(8):4057–4066. [PubMed] [Google Scholar]
- 34.Losse J., Zipfel P.F., Jozsi M. Factor H and factor H-related protein 1 bind to human neutrophils via complement receptor 3, mediate attachment to Candida albicans, and enhance neutrophil antimicrobial activity. J Immunol. 2010;184(2):912–921. doi: 10.4049/jimmunol.0901702. [DOI] [PubMed] [Google Scholar]
- 35.Rivino L., Gruarin P., Häringer B., Steinfelder S., Lozza L. CCR6 is expressed on an IL-10–producing, autoreactive memory T cell population with context-dependent regulatory function. J Exp Med. 2010;207(3):565–577. doi: 10.1084/jem.20091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang C., Magracheva E., Kozlov S., Fong S., Tobin G. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J Biol Chem. 2003;278(5):3308–3313. doi: 10.1074/jbc.M208602200. [DOI] [PubMed] [Google Scholar]
- 37.Clark E.A., Giltiay N.V. CD22: a regulator of innate and adaptive B cell responses and autoimmunity. Front Immunol. 2018;9:2235. doi: 10.3389/fimmu.2018.02235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crowley T., Cryan J.F., Downer E.J., O’Leary O.F. Inhibiting neuroinflammation: the role and therapeutic potential of GABA in neuro-immune interactions. Brain Behav Immun. 2016;54:260–277. doi: 10.1016/j.bbi.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Crowley T., Fitzpatrick J.-M., Kuijper T., Cryan J.F., O’Toole O. Modulation of TLR3/TLR4 inflammatory signaling by the GABAB receptor agonist baclofen in glia and immune cells: relevance to therapeutic effects in multiple sclerosis. Front Cell Neurosci. 2015;9:284. doi: 10.3389/fncel.2015.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilpain P., Le Bihan C., Foulongne V., Taourel P., Pansu N. Rituximab for granulomatosis with polyangiitis in the pandemic of covid-19: lessons from a case with severe pneumonia. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-217549. [DOI] [PubMed] [Google Scholar]
- 41.Hung I.-F.-N., Lung K.-C., Tso E.-Y.-K., Liu R., Chung T.-W.-H. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. The Lancet. 2020 doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia T., Wang Y. Coronavirus disease 2019 and transplantation: The combination of lopinavir/ritonavir and hydroxychloroquine is responsible for excessive tacrolimus trough level and unfavorable outcome. Am J Transplant. 2020 doi: 10.1111/ajt.15992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao B., Wang Y., Wen D., Liu W., Wang J. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yazdany J, Kim AH. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: what every clinician should know; 2020. [DOI] [PMC free article] [PubMed]
- 45.Ferner RE, Aronson JK. Chloroquine and hydroxychloroquine in covid-19; 2020. [DOI] [PubMed]
- 46.Guzik T.J., Mohiddin S.A., Dimarco A., Patel V., Savvatis K. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res. 2020 doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magagnoli J., Narendran S., Pereira F., Cummings T., Hardin J.W. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. medrxiv. 2020 doi: 10.1016/j.medj.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodriguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson B.S., Laloraya M. A cytokine super cyclone in COVID-19 patients with risk factors: the therapeutic potential of BCG immunization. Cytokine Growth Factor Rev. 2020;54:32–42. doi: 10.1016/j.cytogfr.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng M., Gao Y., Wang G., Song G., Liu S. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020:1–3. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao X.-H., He Z.-C., Li T.-Y., Zhang H.-R., Wang Y. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 2020;1–3 doi: 10.1038/s41422-020-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schett G., Sticherling M., Neurath M.F. COVID-19: risk for cytokine targeting in chronic inflammatory diseases? Nat Rev Immunol. 2020;20(5):271–272. doi: 10.1038/s41577-020-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y., Xu X., Chen Z., Duan J., Hashimoto K. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun. 2020 doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao L, Jin H, Wang M, Hu Y, Chen S, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol; 2020. [DOI] [PMC free article] [PubMed]
- 56.Abobaker A., Raba A.A. Does COVID-19 affect male fertility? World J Urol. 2020;1–2 doi: 10.1007/s00345-020-03208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma L., Xie W., Li D., Shi L., Mao Y. Effect of SARS-CoV-2 infection upon male gonadal function: A single center-based study. medRxiv. 2020 [Google Scholar]
- 58.Diao B., Wang C., Tan Y., Chen X., Liu Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feldmann M., Maini R.N., Woody J.N., Holgate S.T., Winter G. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. The Lancet. 2020;395(10234):1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duret P.M., Sebbag E., Mallick A., Gravier S., Spielmann L. Recovery from COVID-19 in a patient with spondyloarthritis treated with TNF-alpha inhibitor etanercept. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-217362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X., Han M., Li T., Sun W., Wang D. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020 doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Picchianti Diamanti A., Rosado M.M., Pioli C., Sesti G., Laganà B. Cytokine Release Syndrome in COVID-19 Patients, A New Scenario for an Old Concern: The Fragile Balance between Infections and Autoimmunity. Int J Mol Sci. 2020;21(9):3330. doi: 10.3390/ijms21093330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tepasse P-R, Hafezi W, Lutz M, Kühn J, Wilms C, et al. Persisting SARS-CoV-2 viremia after rituximab therapy: Two cases with fatal outcome and a review of literature. British Journal of Haematology. n/a(n/a). [DOI] [PMC free article] [PubMed]
- 65.Cao B., Wang Y., Wen D., Liu W., Wang J. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manca C., Koo M.-S., Peixoto B., Fallows D., Kaplan G. Host targeted activity of pyrazinamide in Mycobacterium tuberculosis infection. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0074082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeiser R. Immune modulatory effects of statins. Immunology. 2018;154(1):69–75. doi: 10.1111/imm.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehrbod P., Omar A.R., Hair-Bejo M., Haghani A., Ideris A. Mechanisms of action and efficacy of statins against influenza. Biomed Res Int. 2014;2014 doi: 10.1155/2014/872370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan S. Statins May Decrease the Fatality Rate of Middle East Respiratory Syndrome Infection. mBio. 2015;6(4):e01120. [DOI] [PMC free article] [PubMed]
- 70.Bifulco M., Gazzerro P. Statins in coronavirus outbreak: It's time for experimental and clinical studies. Pharmacol Res. 2020;156 doi: 10.1016/j.phrs.2020.104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reiner Z., Hatamipour M., Banach M., Pirro M., Al-Rasadi K. Statins and the COVID-19 main protease: in silico evidence on direct interaction. Arch Med Sci. 2020;16(3):490–496. doi: 10.5114/aoms.2020.94655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.China NHCotPsRo. National Health Committee of the People’s Republic of China (2020) Guideline for Diagnosis and Treatment of Novel Coronavirus Pneumonia (Trial Version Fourth); 2020.
- 73.Lu C., Li S., Liu Y. Role of immunosuppressive therapy in rheumatic diseases concurrent with covid-19. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-217460. [DOI] [PubMed] [Google Scholar]
- 74.Fadel R., Morrison A.R., Vahia A., Smith Z.R., Chaudhry Z. Early Short Course Corticosteroids in Hospitalized Patients with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Isidori A.M., Pofi R., Hasenmajer V., Lenzi A., Pivonello R. Use of glucocorticoids in patients with adrenal insufficiency and COVID-19 infection. Lancet Diabetes Endocrinol. 2020 doi: 10.1016/S2213-8587(20)30149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang B., Yang R., Zhao Y., Liu C.Z. Separation of chlorogenic acid from honeysuckle crude extracts by macroporous resins. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;867(2):253–258. doi: 10.1016/j.jchromb.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Yu J.-w., Wang L. Bao L-d. Exploring the Active Compounds of Traditional Mongolian Medicine in Intervention of Novel Coronavirus (COVID-19) Based on Molecular Docking Method. J Funct Foods. 2020;104016 doi: 10.1016/j.jff.2020.104016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Polansky H, Lori G. Coronavirus disease 2019 (COVID-19): first indication of efficacy of Gene-Eden-VIR/Novirin in SARS-CoV-2 infection. Int J Antimicrob Agents. 2020:105971. [DOI] [PMC free article] [PubMed]
- 79.Dandapat J, Jena A, Kanungo N, Nayak V, Chainy G. Catechin and Curcumin interact with corona (2019-nCoV/SARS-CoV2) viral S protein and ACE2 of human cell membrane: insights from Computational study and implication for intervention; 2020.
- 80.Avasarala S., Zhang F., Liu G., Wang R., London S.D. Curcumin modulates the inflammatory response and inhibits subsequent fibrosis in a mouse model of viral-induced acute respiratory distress syndrome. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0057285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salman S., Shah F.H., Idrees J., Idrees F., Velagala S. Virtual screening of immunomodulatory medicinal compounds as promising anti-SARS-COV-2 inhibitors. Future Virol. 2020 [Google Scholar]
- 82.de Sá Coutinho D, Pacheco M.T., Frozza R.L., Bernardi A. Anti-inflammatory effects of resveratrol: Mechanistic insights. Int J Mol Sci. 2018;19(6):1812. doi: 10.3390/ijms19061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Verdrengh M., Jonsson I., Holmdahl R., Tarkowski A. Genistein as an anti-inflammatory agent. Inflamm Res. 2003;52(8):341–346. doi: 10.1007/s00011-003-1182-8. [DOI] [PubMed] [Google Scholar]
- 84.Vaeth M., Eckstein M., Shaw P.J., Kozhaya L., Yang J. Store-operated Ca2+ entry in follicular T cells controls humoral immune responses and autoimmunity. Immunity. 2016;44(6):1350–1364. doi: 10.1016/j.immuni.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonaventura P., Benedetti G., Albarède F., Miossec P. Zinc and its role in immunity and inflammation. Autoimmun Rev. 2015;14(4):277–285. doi: 10.1016/j.autrev.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 86.Ang A., Pullar J.M., Currie M.J., Vissers M.C. Vitamin C and immune cell function in inflammation and cancer. Biochem Soc Trans. 2018;46(5):1147–1159. doi: 10.1042/BST20180169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calder P.C., Carr A.C., Gombart A.F., Eggersdorfer M. Optimal Nutritional Status for a Well-Functioning Immune System Is an Important Factor to Protect against Viral Infections. Nutrients. 2020;12(4) doi: 10.3390/nu12041181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metascape pathway enrichment of downregulated genes, enriched diseases and PPI network of GSE147507 gene signature. A. Metascape pathway enrichment of 515 downregulated genes of GSE147507. B. Enrichment of related diseases of gene signature of GSE147507 using WebGestalt webserver. C. Protein-protein interaction (PPI) network of GSE147507 gene signature. Node color density indicates the absolute value of Log2(fold change) of DEGs. Red indicates upregulated DEGs, and green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity.
Five clusters and their enriched pathways of the GSE147507 PPI network. Node color density in clusters indicates the absolute value of Log2(fold change) of DEGs. Red indicates upregulated DEGs while green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity. Bubble plots of pathway enrichment in each cluster were shown.
Metascape pathway enrichment of downregulated genes, enriched diseases and PPI network of CRA002390 gene signature. A. Metascape pathway enrichment of 1034 downregulated genes of CRA002390. B. Enrichment of related diseases of gene signature of CRA002390 using WebGestalt webserver. C. PPI network of CRA002390 gene signature. Node color density indicates the absolute value of Log2(fold change) of DEGs. Red indicates upregulated DEGs and green indicates downregulated DEGs. The node diameter and label font size indicate the degree of connectivity.
Metascape pathway enrichment of the four clusters of CRA002390 PPI network.
GSEA analysis, patients’ clinic traits, expression heatmaps of brown and turquoise modules hub genes of E-MATB-8871. A. Enrichment plots of GSEA correlation analysis for 10 healthy donors and day 4, 5, 6 of case 1 of E-MTAB-8871 using curated gene sets (c2.all.v7.1.symbols.gmt). B-C. Clinic traits of case 1 and case 2 COVID-19 patients. D-E. Heatmaps of top 30 hub genes (weight ranked) in brown and turquoise modules across healthy donors (shown as an average) and each day of illness progression of patient case 2.
Enriched diseases of E-MTAB-8871 gene signature. Disease enrichment was analyzed by an over-representation method in WebGestalt webserver for the genes of brown and turquoise modules.
GSEA validation of the reversal effects of baclofen, adalimumab, tocilizumab and siltuximab. A. The RNA-seq of aortic tissues from mice treated with baclofen or vehicle (GSE128101) was analyzed by GSEA. B. RNA-seq of maternal blood neutrophils from the Rhesus intrauterine infection model treated without or with adalimumab (GSE145918) was analyzed by GSEA. C. Microarray of PBMC fromm juvenile idiopathic arthritis patients taken before and after tocilizumab treatment (GSE76492) was analyzed by GSEA. D. Microarray of ovarian cancer SKOV-3 and Caov-3 cells pre-treated without or with siltuximab, then treated with IL6 (E-GEOD-20272) was analyzed by GSEA.
GSEA validation of the reversal effects of rituximab, methylprednisolone, prednisolone and ritonavir. A. Whole-blood transcriptomic profiling for patients with rheumatoid arthritis who are non-responders or responders to rituximab treatments (GSE54629) was analyzed by GSEA. B. RNA-seq of circulating B cells from healthy human donors treated with methylprednisolone or vehicle in vitro (GSE112101) was analyzed by GSEA. C. Microarray of whole blood from dengue patients received prednisolone or placebo therapy (GSE40165) was analyzed by GSEA. D. Microarray of PBMC treated with placebo or ritonavir from healthy volunteers (E-TABM-642) was analyzed by GSEA.
GSEA validation of the reversal effects of atorvastatin, rilonacept, pyrazinamide and chloroquine. A. Transcription profiling of human quadriceps femoris muscle from patients received placebo or atorvastatin (E-TABM-116) was analyzed by GSEA. B. Microarray profiling of skin from diffuse cutaneous systemic sclerosis patients who pre- or post-treated with rilonacept (GSE119939) was analyzed by GSEA. C. Transcriptional array of lung tissue of tuberculosis-infected mice treated without or with pyrazinamide (GSE48027) was analyzed by GSEA. D. Microarray of human liver Huh7 cells treated with or without chloroquine (GSE30351) was analyzed by GSEA.
GSEA validation of the reversal effects of hydroxychloroquine, IFN-γ, alemtuzumab and alefacept. A. RNA-seq of PBMC from healthy participants treated without or with hydroxychloroquine (GSE74235) was analyzed by GSEA. B. RNA-seq of murine lung endothelial cells treated without or with IFN-γ (E-MTAB-5921) was analyzed by GSEA. C. Microarray profiling of T-cell prolymphocytic leukemia (T-PLL) from pre- or post-treated with alemtuzumab (GSE67368) was analyzed by GSEA. D. Microarray data from PBMCs of psoriasis patients who are non-responders or responders to alefacept treatment (GSE18948) was analyzed by GSEA.
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.