Abstract
Purpose
Resveratrol (Res) is a natural polyphenolic compound found in several plants and reported as a promising biological molecule with effective anti-fibrosis and anti-inflammatory activities. However, the underlying mechanism of Res on systemic sclerosis (SSc) remains unclear. In the study, we identified the key cellular signaling pathways involved in the Res regulatory process on SSc.
Methods
Res-targeted genes interaction network was constructed using the STITCH database, and the shared Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways involved in both SSc and Res-targeted genes were then identified. The top five enriched KEGG pathways were visualized by GOplot. KEGG pathways associated with Res-targeted genes were established by Pathway Builder Tool 2.0. Quantitative real-time PCR (qRT-PCR) was used to assess the expression of sirtuin 1 (SIRT1), mammalian targeted of rapamycin (mTOR), and cytokines.
Results
Enrichment analysis of Res-targeted genes showed 79 associated pathways, 27 of which were also involved in SSc. Particularly, SIRT1/mTOR signaling was found as one of the crucial regulatory pathways. In vitro results suggested that SIRT1-mediated mTOR degradation ameliorated bleomycin (BLM)-induced fibrosis and inflammation. Res was capable of elevating the SIRT1 level in fibroblasts and partially reversing mTOR-dependent induction of fibrosis and inflammation.
Conclusion
These results indicated that Res is a feasible and effective choice for SSc and therapeutic target of mTOR could be a potential alternative for treatment of SSc.
Keywords: resveratrol, systemic sclerosis, SIRT1, mTOR, signaling
Introduction
Systemic sclerosis (SSc) is an autoimmune disease characterized by microvascular damage, innate and adaptive immune dysfunction, and multiple organ fibrosis, seriously affecting the quality of a patient’s life and placing a heavy burden on society.1 A previous study reported that the quality of a patient’s life was significantly lower than the control group using a SF-36 health questionnaire.2 The functional damage of patients is gradually aggravated over the prolongation of time.3 However, the specific pathological mechanism of SSc and its etiological factors remain unclear. Thus, it is an urgent time to investigate the underlying mechanism of SSc and explore a feasible and effective drug to ameliorate the pathological progress of fibrosis and inflammation in SSc patients.
Resveratrol (Res, 3–4ʹ-5-trihydroxystilbene), a non-flavonoid polyphenolic compound, is an antitoxin produced when plants are stimulated. It can be synthesized in grape leaves and is a bioactive ingredient in grape wine and grape juice.4 Previous in vitro and in vivo experiments suggested that Res has antioxidant, anti-inflammatory, anti-cancer and cardiovascular protective effects.5 Currently, accumulative attention has been attracted to the therapeutic effect of Res on SSc due to its anti-fibrotic and anti-inflammatory activities.6 For example, it was recently reported that Res could alleviate SSc-related pulmonary fibrosis through suppression of pro-inflammatory and pro-fibrotic processes.7 Recent research also indicated that Res-induced SIRT1 inhibits transforming growth factor-β (TGF-β) induced cell proliferation and fibroblast differentiation.8 mTOR is a serine/threonine protein kinase which has been shown to be involved in the initiation of inflammatory cascades and fibroblast activation.9
Microarrays, a lab technique to detect thousands of gene expressions, have been widely applied to the investigation of the pathogenic processes governing the development of a variety of diseases. Integrating microarray and bioinformatic tools makes it an invaluable method for functional genomic studies.10 In the present study, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to both Res-targeted genes and SSc development were analyzed. The mammalian target of rapamycin (mTOR) signaling was identified as a crucial KEGG pathway related to Res-targeted genes. Therefore, we investigated the effects of Res on SSc while focusing on the mTOR signaling in the present study.
Materials and Methods
Reagent
Res was purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Bleomycin (BLM) was obtained from Enzo Life Sciences, Inc. (Farmingdale, NY, USA). qRT-PCR kit was purchased from Thermo Fisher Scientific Co. (Waltham, MA, USA). Small interfering RNA (siRNA) mTOR and the siRNA control were designed by Applied Biological Materials Inc. (Richmond, BC, Canada). Plasmid SIRT1 was purchased from the Public Protein/Plasmid Library (PPL, #BC012499). High-glucose Dulbecco’s modified eagle medium was purchased from Gibco BRL (Thermo Fisher).
Ethical Statement
Protocols of the present study were approved by the Ethics Committee of The Second Affiliated Hospital of Hainan Medical University (Haikou, China; 2019-019-23), and informed written consent was achieved from all participants. All experiments involved with human samples were conducted in compliance with the Declaration of Helsinki.
Skin Sample Collection
From August 2018 to May 2020, skin biopsy specimens of the affected skin were collected both from SSc patients, and the specimens fulfilled the American College of Rheumatology criteria for SSc. At the same time, skin biopsy specimens were obtained from normal controls who had no history of autoimmune and other dermal diseases.
Culture of Human Skin Fibroblasts (HSFs)
High-glucose Dulbecco’s modified eagle medium was used to culture HSFs (FuHeng Biology, Shanghai, China). HSFs were cultured at 37°C with 5% CO2 and 95% humidity. HSFs treated with BLM (100 mmol/mL) for 24 hours were selected for further research. To knockdown endogenous mTOR, siRNA mTOR transfection was performed by Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s protocol. qRT-PCR and ELISA analyses were performed 24 hours post-lipofection. The plasmid SIRT1 was purchased from the Public Protein/Plasmid Library (PPL, #BC012499). qRT-PCR and ELISA analyses were performed 24 hours after plasmid transfection.
qRT-PCR Analysis
The total RNA was isolated from cells and skin samples using TRIzol® reagent (Thermo Fisher Scientific). The reverse-transcription process was performed using ReverTra Ace® qPCR RT Master Mix (Toyobo Life Science, Osaka, Japan) to synthesize cDNA according to the manufacturer’s protocol. The relative mRNA levels were normalized to GAPDH levels (the internal control) and were calculated according to the 2−ΔΔCt approach. All experiments were conducted in triplicate, and the primer sequences of mRNAs are shown in Table 1.
Table 1.
microRNAs and mRNA Primer Sequences
| microRNA or Gene Names | Primer Sequence (5ʹ to 3ʹ) |
|---|---|
| hsa-mTOR-Forward | TCCAGGGCTTCTTCCGTT |
| hsa-mTOR-Reverse | GGGCTGTCGTGGTAGACTTAG |
| hsa-SIRT1-Forward | CGGAAACAATACCTCCACCTGA |
| hsa-SIRT1-Reverse | GAAGTCTACAGCAAGGCGAGCA |
| hsa-α-SMA-Forward | TGTATGTGGCTATCCAGGCG |
| hsa-α-SMA-Reverse | AGAGTCCAGCACGATGCCAG |
| hsa-Col-1a1-Forward | CCTCCAGGGCTCCAACGAG |
| hsa-Col-1a1-Reverse | TCTATCACTGTCTTGCCCCA |
| hsa-Col-1a2-Forward | GATGTTGAACTTGTTGCTGAGC |
| hsa-Col-1a2-Reverse | TCTTTCCCCATTCATTTGTCTT |
| hsa-IL-1β-Forward | AGCTACGAATCTCCGACCAC |
| hsaIL-1β-Reverse | CGTTATCCCATGTGTCGAAGAA |
| hsa-IL-6-Forward | ATGAACTCCTTCTCCACAAGC |
| hsa-IL-6-Reverse | AGGCGGTGCTTGTTCCTCA |
| hsa-GAPDH-Forward | ACCCACTCCTCCACCTTTGA |
| hsa-GAPDH-Reverse | CTGTTGCTGTAGCCAAATTCGT |
Enzyme-Linked Immunosorbent Assay (ELISA)
The concentrations of IL-1β and IL-6 in cell supernatants were measured with specific ELISA kits (Shanghai Biotechnology, Shanghai, China) in accordance with the manufacturer’s instructions.
Res-Targeted Genes Interaction Network Construction
Search Tool for Interacting Chemicals (STITCH) is a database that integrates over 430,000 chemicals into a single resource. Res-targeted genes were obtained in STITCH based on the following settings: first shell: 0 and up to 10; second shell: 0 and up to 10; third shell: 0 and up to 10. The first shell is directly associated with resveratrol. The second shell is secondarily associated with resveratrol through the first shell. The interaction network of Res and its targeted genes were constructed in STITCH online tool. To obtain the degree, betweenness, closeness of each gene in the network and better display the three layers of genes, the Res-targeted gene symbols were imported into Cytoscape 3.7.2 and calculated. Degree represented the degree of association of one node with all other nodes in the network. Closeness represented the closeness between a node and other nodes in the network. Betweenness was the number of times a node acted as the shortest bridge between two other nodes.
Shared KEGG Pathways Involved Both in SSc and Res-Targeted Genes
miRWalk2.0 is a comprehensive archive, supplying the largest available collection of predicted and experimentally verified genes and related information. miRWalk2.0 was used to retrieve human SSc KEGG pathways. Some functional associations such as gene class, protein class, KEGG, and WIKI pathways can be obtained from the database. The KEGG pathways information was downloaded. Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources consists of an integrated biological knowledgebase and analytic tools aimed at systematically extracting biological meaning from large gene/protein lists. DAVID was used to generate the Res-targeted genes enriched KEGG pathways. Shared KEGG pathways of SSc and Res-targeted genes were identified using Venn Diagram (http://www.ehbio.com/ImageGP/index.php/Home/Index/VennDiagram.html).
Identification of the Hub Genes
The enrichment information of the top KEGG pathways with p<0.01 was presented with GOplot, an R package for visually combining expression data with functional analysis. Genes involved in all the top three shared KEGG pathways were considered as hub genes.
Retrieval of the KEGG Pathways Related to Res-Targeted Genes
The top 3 shared KEGG pathways with smallest p values were selected. The KEGG pathways related to the Res-targeted genes were established with the ScienceSlides. Hub genes and their action mechanisms were shown in the schematic diagrams.
Statistical Analysis
GraphPad Prism 8.0 was used to perform the statistical analyses. Results were shown as the mean ± SD. Student’s t-test was applied to compare two groups, and one-way analysis of variance with Tukey’s post hoc test was used to compare three or more groups. P < 0.05 was considered as statistically significant.
Results
Res-Targeted Genes and the Interaction Network
In total, 30 Res-targeted genes were obtained in STITCH with the limitation of three shells. Then the interaction of Res-targeted genes was reconstructed in Cytoscape (Figure 1). TP53, SIRT1, PTGS1, SIRT3, ESR1, PPARG, NOS3, AKT1, SIRT5, PTGS2 were involved in the first shell. They were directly associated with resveratrol. ATM, BRCA1, FOXO1, MTOR, EP300, RICTOR, FOXO3, CDKN1A, KAT2B, MDM2 were involved in the second shell. HSP90AA1, HIPK2, NCOA3, CDKN2A, MAPK8, SRC, USP7, RCHY1, CREBBP, SP1 were involved in the third shell.
Figure 1.
The interaction network of the resveratrol-targeted genes. Red is first shell. Yellow is second shell. Green is third shell.
Identification of the Shared KEGG Pathways of SSc and Res-Targeted Genes
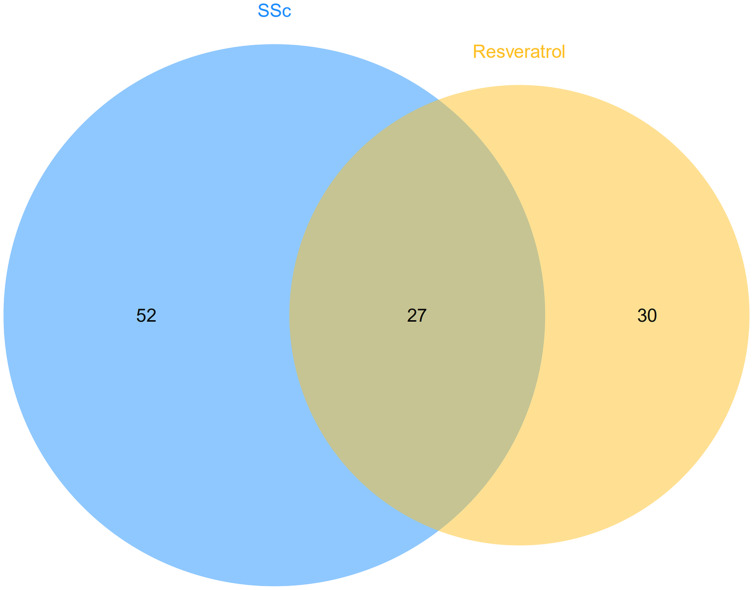
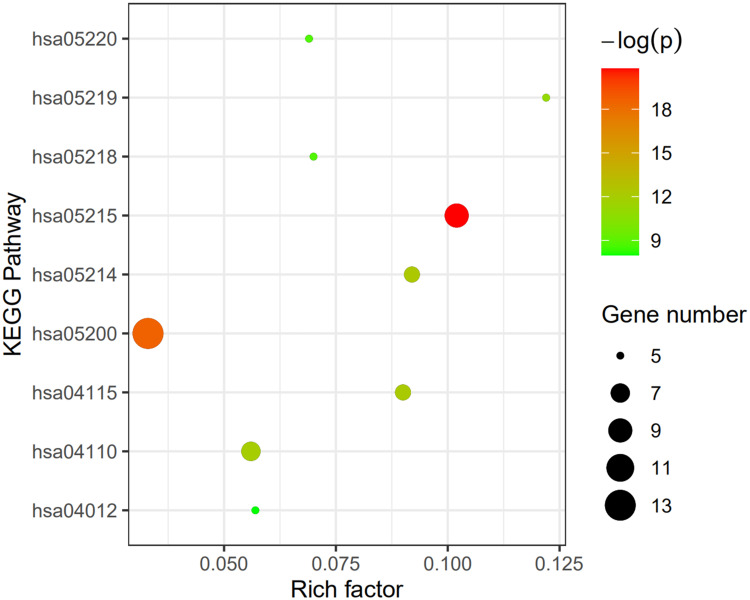
After the enrichment analysis of the Res-targeted genes in DAVID, 57 KEGG pathways were identified. 79 KEGG pathways involved in human SSc were retrieved using miRWalk2.0. 27 shared KEGG pathways of SSc and Res-targeted genes were identified (Figure 2 and Table S1). The top nine KEGG pathways with p <0.01 were Prostate cancer pathway, Pathway in cancer, Glioma pathway, p53 signaling pathway, Cell cycle pathway, Bladder cancer pathway, Melanoma pathway, Chronic myeloid leukemia pathway, and ErbB signaling pathway. The rich factor, -log(p), and gene number of the nine pathways are demonstrated in Figure 3. The detailed information of the top 3 KEGG pathways are shown in Table 2.
Figure 2.
Commen KEGG pathways of resveratrol-target genes and CCs. There are 57 resveratrol-target genes related KEGG pathways and 79 SSc related KEGG pathways. 27 shared KEGG pathways were obtained.
Figure 3.
Enrichment information of KEGG pathways with p<0.01. hsa05220: Chronic myeloid leukemia pathway, hsa05219: Bladder cancer pathway, hsa05218: Melanoma pathway, hsa05215: Prostate cancer pathway, hsa05214: Glioma pathway, hsa05200: Pathways in cancer, hsa04115: p53 signaling pathway, hsa04110: Cell cycle signaling pathway, hsa04012: ErbB signaling pathway.
Table 2.
The Information of Top 3 KEGG Pathways
| KEGG Pathway | Count | FDR | Genes |
|---|---|---|---|
| hsa05215: Prostate cancer | 9 | 2.1E-8 | CDKN1A, HSP90AA1, CREBBP, MDM2, EP300, AKT1, TP53, FOXO1, MTOR |
| hsa05200: Pathways in cancer | 13 | 1.4E-7 | CDKN1A, CREBBP, HSP90AA1, CDKN2A, PTGS2, FOXO1, MTOR, MAPK8, MDM2, AKT1, EP300, PPARG, TP53 |
| hsa05214: Glioma | 6 | 4.2E-5 | CDKN1A, CDKN2A, MDM2, AKT1, TP53, MTOR |
Identification of the Hub Genes
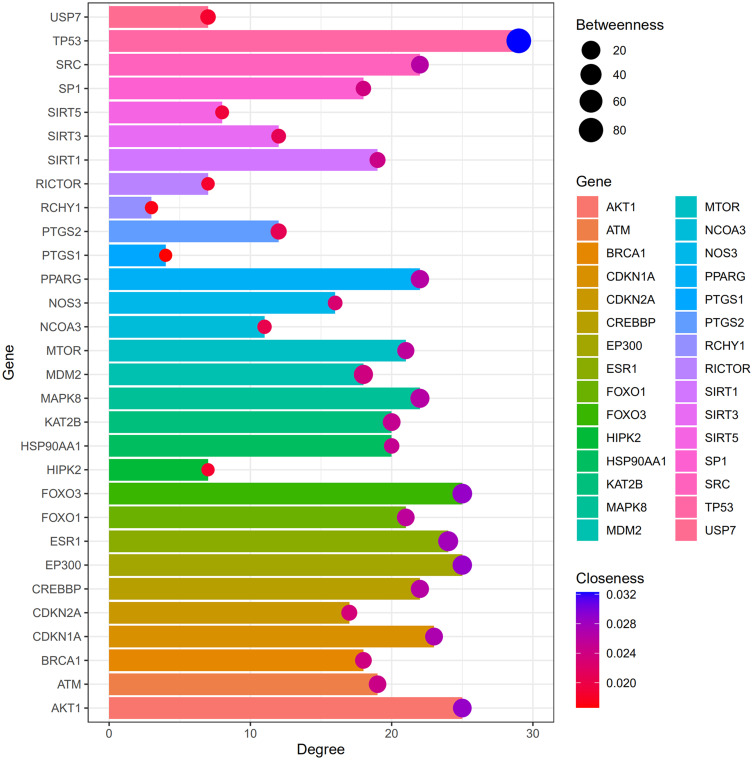
Among the 30 Res-targeted genes, AKT1, CDKN1A, TP53, MDM2, MTOR, HSP90AA1, EP300, CREBBP, FOXO1, PTGS2, PPARG, CDKN2A, MAPK8, RCHY1, and ATM were involved in the top three shared KEGG pathways. Degree, betweenness, and closeness information of all 30 Res-targeted genes are shown in Figure 4. The enrichment information of the top 9 KEGG pathways is shown in Figure 5. AKT1, CDKN1A, TP53, MDM2, MTOR are involved in all of the top three KEGG pathways and were identified as hub genes.
Figure 4.
Degree, betweenness, closeness of resveratrol-targeted genes.
Figure 5.
The resveratrol-targeted genes enrichment analysis information.
Retrieval of the KEGG Pathways Related to Res-Targeted Genes
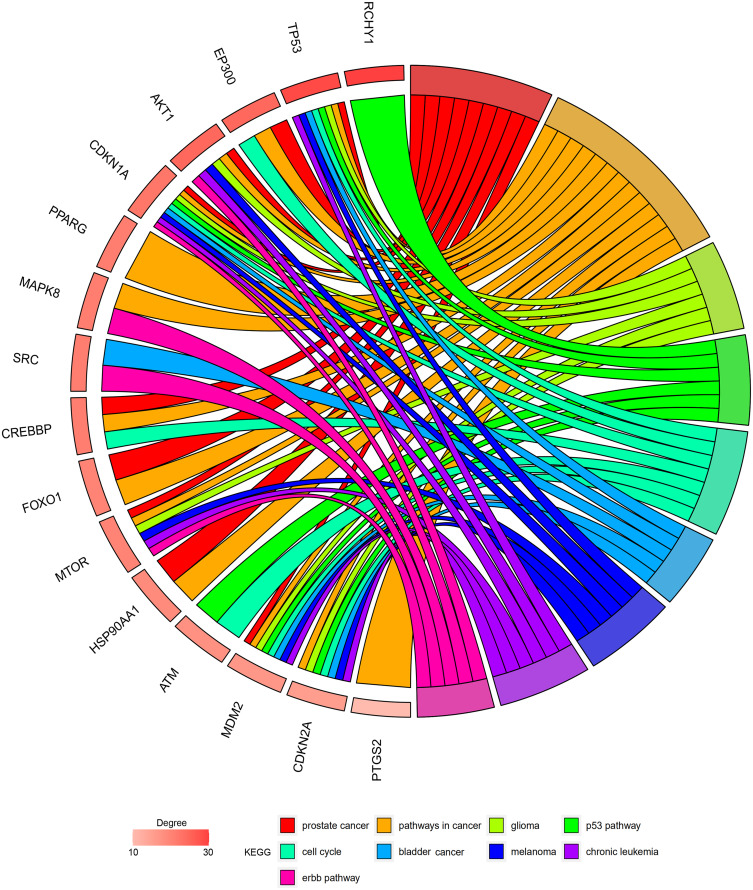
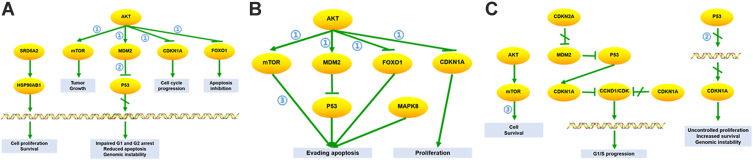
The Res-targeted genes related parts of the top three shared KEGG pathways are shown in Figure 6. They are associated with the cell proliferation, survival, tumor growth, impaired G1 and G2 arrest, reduced apoptosis, genomic instability, cell cycle progression, apoptosis inhibition, evading apoptosis, G1/S progression, uncontrolled proliferation. It can be found they may have biological action through PI3K-Akt signaling pathway, p53 signaling pathway, and mTOR signaling pathway.
Figure 6.
The resveratrol-targeted genes related part of the top three KEGG pathways. (A) Prostate cancer pathway: (1) PI3K-Akt signaling pathway and (2) p53 signaling pathway are involved in the process, resulting in impaired G1/G2 arrest, reduced apoptosis, genomic instability, cell proliferation, survival, tumor growth, cell cycle progression, apoptosis inhibitions. (B) Pathways in cancer: (1) PI3K-Akt signaling pathway and (3) mTOR signaling pathway are involved in the process, resulting in evading apoptosis and proliferation. (C) Glioma pathway: (2) p53 signaling pathway and (3) mTOR pathway are involved in the process, resulting in cell survival, G1/S progression, uncontrolled proliferation, increased survival, and genomic instability.
Res Inhibits Fibrosis and Inflammation Through Reduction of the Expression of mTOR
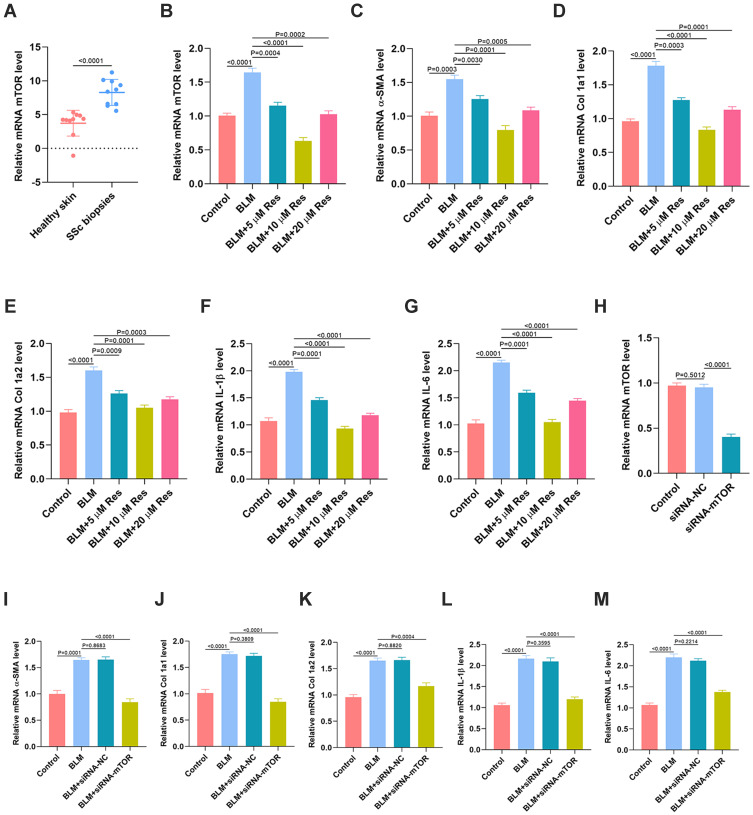
As seen in Figure 6, mTOR signaling was identified as a critical KEGG pathway related to Res-targeted genes. Thus, we next examined the association of Res and mTOR and their regulatory roles in fibrosis and inflammation. First, the gene expression of mTOR in skin tissues from both SSc patients and the healthy controls was measured by qRT-PCR. The result showed that mTOR level in the skin sample of SSc patients was significantly higher than the normal controls (Figure 7A). In vitro, BLM was used to treat HSFs to create a cellular model of fibroblast activation. The in vitro results indicated that Res was able to markedly reverse the high level of mTOR induced by BLM. 10 μM was shown as the optimum concentration of Res to exert its effective regulation (Figure 7B). Furthermore, the effect of Res on fibrosis was investigated in vitro and the results suggested that Res could significantly reverse the BLM-induced high expression of fibrotic genes, including α-SMA, Col 1a1, and Col 1a2 (Figure 7C–E). Similarly, the qRT-PCR results also indicated that Res could significantly reverse the BLM-induced expression of inflammatory cytokines including IL-1β and IL-6 (Figure 7F–G). Moreover, siRNA mTOR was used to knock down the expression of mTOR in HSFs (Figure 7H), and the results suggested that the high level of fibrotic and inflammatory markers in HSFs induced by BLM was significantly suppressed after the usage of siRNA of mTOR (Figure 7I–M).
Figure 7.
Res inhibits fibrosis and inflammation through reduction of the expression of mTOR. (A) The expression of mTOR in the skin samples from SSc patients and the normal controls was measured by qRT-PCR analysis, n = 10, per group; (B) The expression of mTOR in HSFs with different treatments was measured by qRT-PCR analysis; (C–E) The expression of fibrotic genes in the different groups was measured by qRT-PCR analysis; (F, G) The expression of inflammatory cytokines IL-1β and IL-6 in the different groups was measured by qRT-PCR analysis; (H) The expression of mTOR in HSFs with different treatments was measured by qRT-PCR analysis; (I–K) The expression of fibrotic genes in the different groups was measured by qRT-PCR analysis; (L, M) The expression of inflammatory cytokines IL-1β and IL-6 in the different groups was measured by qRT-PCR analysis. Data are the means ± SDs of three independent experiments.
Res Ameliorates SSc via Regulation of SIRT1/mTOR Signaling
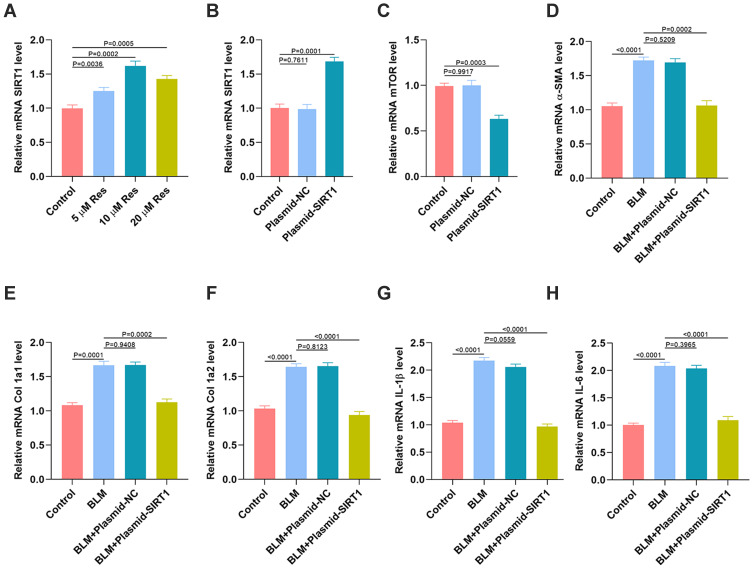
The previous studies demonstrated that SIRT1 is markedly elevated with the treatment of Res,11,12 and SIRT1 was identified as a key gene in the first shell of the Res-targeted genes. Thus, we then explored the relation between SIRT1 and mTOR and their regulatory effects on fibrosis and inflammation in vitro. As seen in Figure 8A, the qRT-PCR analysis indicated that the level of SIRT1 was significantly elevated with treatment of Res. 10 μM was the optimum concentration of Res to exert its effective regulation. Then, the SIRT1 plasmid was used to elevate the SIRT1 expression in HSFs (Figure 8B), and showed that the level of mTOR was significantly decreased after the SIRT1 plasmid interference (Figure 8C). Furthermore, the effect of SIRT1 on fibrosis was investigated in vitro. The results suggested that the BLM-induced high expression of fibrotic markers including α-SMA, Col 1a1, and Col 1a2, was significantly reversed after the interference of plasmid SIRT1 (Figure 8D–F). Similarly, the qRT-PCR results indicated that the BLM-induced expression of inflammatory markers including IL-1β and IL-6 was significantly reversed after the interference of plasmid SIRT1 (Figures 8G–H and S1).
Figure 8.
Res ameliorates SSc via regulation of SIRT1/mTOR signaling. (A) The expression of SIRT1 in HSFs with different treatments was measured by qRT-PCR analysis; (B) The expression of SIRT1 in HSFs after the SIRT1 plasmid inference was measured by qRT-PCR analysis; (C) The expression of mTOR in HSFs after the SIRT1 plasmid inference was measured by qRT-PCR analysis; (D–F) The expression of fibrotic genes in the different groups was measured by qRT-PCR analysis; (G, H) The expression of inflammatory cytokines IL-1β and IL-6 in the different groups was measured by qRT-PCR analysis. Data are the means ± SDs of three independent experiments.
Discussion
Currently, Res was widely demonstrated to play a significant role in the inhibition of development of SSc.13,14 However, little is known regarding the specific mechanism of its anti-fibrotic and anti-inflammation effect on SSc. In the present study, we identified mTOR signaling as a crucial KEGG pathway related to Res-targeted genes, and SIRT1 was found to be a key gene in the first shell of the Res-targeted genes. Furthermore, the elevated level of mTOR in the skin of SSc patients was validated via qRT-PCR measurement and its regulatory role in fibrosis and inflammation was demonstrated in vitro. In addition, the Res-dependent overexpression of SIRT1 was then verified to inhibit the expression of mTOR and ameliorate fibrosis and inflammation in vitro. Thus, our study indicates that Res may ameliorate SSc through activation of SIRT1 which reverses the enhancement of mTOR expression.
It is well known that mTOR is a crucial component of intracellular communication for many physiological and pathological processes.15 It was recently reported that mTOR phosphorylation was enhanced by the NF-κB activation while the mTOR inhibition suppressed NF-κB activation, which suggests a positive feedback loop of mTOR and NF-κB during inflammation.16 Consistent with this finding, we found a higher level of mTOR in the skin biopsies of SSc patients compared with the normal control. When treated with Res, the BLM-treated fibroblasts exhibited significantly decreased trend of the fibrotic and inflammatory cytokines. These findings suggested that mTOR activation is associated with the pathogenesis of SSc. Res may represent a potential therapeutic compound to prevent progressive fibrosis and inflammation in SSc patients.
Res is a non-flavonoid polyphenolic compound, which is widely demonstrated to have antioxidant, anti-inflammatory, anti-cancer and cardiovascular protection effects.17,18 Multiple signaling pathways are responsible for the generation and secretion of fibrotic and inflammatory cytokines, and play crucial roles in the pathogenesis of SSc.19,20 In the present study, we identified mTOR signaling as an important KEGG pathway regarding the Res-targeted genes, and many prior studies have demonstrated that mTOR is an important factor in SSc.21,22 Previous literature have also shown the mTOR signaling which may be regulated by the SIRT1 expression.23 For example, a recent study pointed out that SIRT1/mTOR signaling was the potential mechanism of the lycopene’s effect on the improved pain experience in the dorsal horn of burn injury rats.24 Our present study indicated that the Res-dependent overexpression of SIRT1 could markedly suppress the mTOR expression and exert a strong anti-fibrosis and anti-inflammation in vitro. Thus, we speculate that SIRT1/mTOR axis be the underlying mechanism of Res on SSc.
In this study, not only SIRT1 but also SIRT3 and SIRT5 were found directly targeted by Res. SIRT1 is localized to both the nucleus and cytoplasm, deacetylating intracellular signaling proteins, including histones, and regulate gene expression.25–27 SIRT3 and SIRT5 are exclusively localized to the mitochondria and regulate energy metabolism in response to mitochondrial stress.27–30 Whether SIRT3 and SIRT5 play a role in promoting SSc by Res needs to be investigated in future studies.
Conclusion
Our study determined that Res exhibited protective effects on SSc, as evidenced by suppressing the increase in fibrotic and inflammatory cytokines observed in vitro. The mainly involved mechanism was the anti-fibrosis and anti-inflammatory role of Res through SIRT1/mTOR axis. This study provided a potential candidate natural compound for the treatment of SSc.
Acknowledgments
This study was supported by the Research and Development fund of the Second Affiliated Hospital of Hainan Medical University (Hospital Fund Number: 2018–19).
Author Contributions
WC and BL designed the study. QY and QW collated the data, analyzed the data and produced the initial draft of the manuscript. QY, QW, XX, YX, JL, QL, MH, YC, BL and WC revised the figures and tables. QY, QW, BL and WC helped to draft the manuscript. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors declared no conflicts of interest.
References
- 1.Hoffmann-Vold AM, Molberg Ø. Detection, screening, and classification of interstitial lung disease in patients with systemic sclerosis. Curr Opin Rheumatol. 2020;32(6):497–504. doi: 10.1097/BOR.0000000000000741 [DOI] [PubMed] [Google Scholar]
- 2.Lo LMP, Taylor BV, Winzenberg T, et al. Estimating the relative contribution of comorbidities in predicting health-related quality of life of people with multiple sclerosis. J Neurol. 2020. doi: 10.1007/s00415-020-10195-w [DOI] [PubMed] [Google Scholar]
- 3.Geraldes R, Esiri MM, Perera R, et al. Vascular disease and multiple sclerosis: a post-mortem study exploring their relationships. Brain. 2020;143(10):2998–3012. doi: 10.1093/brain/awaa255 [DOI] [PubMed] [Google Scholar]
- 4.Rawat D, Chhonker SK, Naik RA, et al. Modulation of antioxidant enzymes, SIRT1 and NF-κB by resveratrol and nicotinamide in alcohol-aflatoxin B1-induced hepatocellular carcinoma. J Biochem Mol Toxicol. 2020:e22625. doi: 10.1002/jbt.22625 [DOI] [PubMed] [Google Scholar]
- 5.Fonseca-Santos B, Chorilli M. The uses of resveratrol for neurological diseases treatment and insights for nanotechnology based-drug delivery systems. Int J Pharm. 2020;589:119832. doi: 10.1016/j.ijpharm.2020.119832 [DOI] [PubMed] [Google Scholar]
- 6.Ahmad I, Hoda M. Attenuation of diabetic retinopathy and neuropathy by resveratrol: review on its molecular mechanisms of action. Life Sci. 2020;245:117350. doi: 10.1016/j.lfs.2020.117350 [DOI] [PubMed] [Google Scholar]
- 7.Chu H, Jiang S, Liu Q, et al. Sirtuin1 protects against systemic sclerosis-related pulmonary fibrosis by decreasing proinflammatory and profibrotic processes. Cell Mol Biol. 2018;58:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowosad A, Besson A. CDKN1B/p27 regulates autophagy via the control of ragulator and MTOR activity in amino acid-deprived cells. Autophagy. 2020;1–2. doi: 10.1080/15548627.2020.1831217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma D, Malik A, Balakrishnan A. et al. RIPK3 promotes mefv expression and pyrin inflammasome activation via modulation of mtor signaling. J Immunol.2020;ji2000244. doi: 10.4049/jimmunol.2000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Y, Cao F, Chen L, et al. Identification of key microRNAs and targeted genes for the diagnosis of bone nonunion. Mol Med Rep. 2020;21:1921–1933. doi: 10.3892/mmr.2020.10996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komorowska J, Wątroba M, Szukiewicz D. Review of beneficial effects of resveratrol in neurodegenerative diseases such as alzheimer’s disease. Adv Med Sci. 2020;65(2):415–423. doi: 10.1016/j.advms.2020.08.002 [DOI] [PubMed] [Google Scholar]
- 12.Viana-Mattioli S, Cinegaglia N, Bertozzi-Matheus M, et al. SIRT1-dependent effects of resveratrol and grape juice in an in vitro model of preeclampsia. Biomed Pharmacother. 2020;131:110659. doi: 10.1016/j.biopha.2020.110659 [DOI] [PubMed] [Google Scholar]
- 13.Gianchecchi E, Fierabracci A. Insights on the effects of resveratrol and some of its derivatives in cancer and autoimmunity: a molecule with a dual activity. Antioxidants (Basel). 2020;9. doi: 10.3390/antiox9020091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerr P, Palumbo-Zerr K, Huang J, et al. Sirt1 regulates canonical TGF-β signaling to control fibroblast activation and tissue fibrosis. Ann Rheum Dis. 2016;75(1):226–233. doi: 10.1136/annrheumdis-2014-205740 [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y, Hu F, Li Q, et al. Tanshinone IIA ameliorates the bleomycin-induced endothelial-to-mesenchymal transition via the Akt/mTOR/p70S6K pathway in a murine model of systemic sclerosis. Int Immunopharmacol. 2019;77:105968. doi: 10.1016/j.intimp.2019.105968 [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Akiyama K, Wang D, et al. mTOR inhibition rescues osteopenia in mice with systemic sclerosis. J Exp Med. 2015;212(1):73–91. doi: 10.1084/jem.20140643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mutlu E, Gibbs ST, South N, et al. Comparative toxicokinetics of trans-resveratrol and its major metabolites in harlan sprague dawley rats and B6C3F1/N mice following oral and intravenous administration. Toxicol Appl Pharmacol. 2020;394:114962. doi: 10.1016/j.taap.2020.114962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao X, Tian S, Fu M, et al. Resveratrol protects human bronchial epithelial cells against nickel-induced toxicity via suppressing p38 MAPK, NF-κB signaling, and NLRP3 inflammasome activation. Environ Toxicol. 2020;35(5):609–618. doi: 10.1002/tox.22896 [DOI] [PubMed] [Google Scholar]
- 19.Trapiella-Martínez L, Díaz-López JB, Caminal-Montero L, et al. Very early and early systemic sclerosis in the Spanish scleroderma registry (RESCLE) cohort. Autoimmun Rev. 2017;16(8):796–802. doi: 10.1016/j.autrev.2017.05.013 [DOI] [PubMed] [Google Scholar]
- 20.Gordon SM, Stitt RS, Nee R, et al. Risk factors for future scleroderma renal crisis at systemic sclerosis diagnosis. J Rheumatol. 2019;46(1):85–92. doi: 10.3899/jrheum.171186 [DOI] [PubMed] [Google Scholar]
- 21.Qi Q, Mao Y, Tian Y, et al. Geniposide inhibited endothelial-mesenchymal transition via the mTOR signaling pathway in a bleomycin-induced scleroderma mouse model. Am J Transl Res. 2017;9:1025–1036. [PMC free article] [PubMed] [Google Scholar]
- 22.Suto T, Karonitsch T. The immunobiology of mTOR in autoimmunity. J Autoimmun. 2020;110:102373. doi: 10.1016/j.jaut.2019.102373 [DOI] [PubMed] [Google Scholar]
- 23.Peng C, Fuchao C, Jiexin L, et al. Activation of the miR-34a-mediated SIRT1/mTOR signaling pathway by urolithin a attenuates d-galactose-induced brain aging in mice. Neurotherapeutics. 2019;16(4):1269–1282. doi: 10.1007/s13311-019-00753-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin Y, Jin-Feng W, Xiao-Hua X, et al. Effect of lycopene on pain facilitation and the SIRT1/mTOR pathway in the dorsal horn of burn injury rats. Eur J Pharmacol. 2020; 889:173365. [DOI] [PubMed] [Google Scholar]
- 25.Chang H-C, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138–145. doi: 10.1016/j.tem.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27(19):2072–2085. doi: 10.1101/gad.227439.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H-C, Chathuranga K, Lee J-S. Mechanisms and disease implications of sirtuin-mediated autophagic regulation. Exp Mol Med. 2019;51(12):1–11. doi: 10.1038/s12276-019-0299-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi J-E, Mostoslavsky R. Sirtuins, metabolism, and DNA repair. Curr Opin Genet Dev. 2014;26:24–32. doi: 10.1016/j.gde.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Nagasawa K, C. M, et al. Mitochondrial sirtuin network reveals dynamic SIRT3-dependent deacetylation in response to membrane depolarization. Cell. 2016;167(4):985–1000.e21. doi: 10.1016/j.cell.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Ven Robert AH, Santos D, Haigis Marcia C. Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol Med. 2017;23(4):320–331. doi: 10.1016/j.molmed.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]