Abstract
Metabolomics is a promising technology for elucidating the mechanisms of metabolic syndrome (MetS). However, measurements in patients with MetS under different conditions vary. Metabolomics experiments in different populations and pathophysiological conditions are, therefore, indispensable. We performed a serum metabolomics investigation in untreated patients with MetS in the Chinese population. Untreated patients with MetS were recruited to this study. Metabolites were measured using a traditional 1H nuclear magnetic resonance (NMR) experiment followed by principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS‐DA). Key metabolic pathways were identified by searching the Kyoto Encyclopedia of Genes and Genomes Pathway Database. A total of 28 patients with MetS and 30 healthy subjects were enrolled. All patients were untreated because they were unaware of or neglected to treat their MetS. By 1H NMR, we identified 49 known substances. Following PCA and OPLS‐DA, 36 metabolites were confirmed to be closely associated with MetS compared with the control group; 33 metabolites were increased, whereas 3 metabolites were reduced. Importantly, 14 metabolites that changed in the serum of these untreated patients with MetS were previously unreported. Pathway analysis revealed the top 15 metabolic pathways associated with untreated MetS, which included 3 amino acid metabolic pathways. Our data suggest that untreated patients exhibit a worse pathophysiologic manifestation, which may result in more rapid progression of MetS. Thus, we propose that health education be reinforced to improve the public’s knowledge, attitude, and practice regarding MetS. The rates of “untreated” patients due to unawareness and neglect must be reduced immediately.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Metabolic syndrome (MetS) does not have any notable symptoms in the mild stage, therefore, many patients with MetS do not undergo any effective treatment (untreated) because of unaware or neglect of having MetS. Metabolomics have been a promising technology for elucidating the mechanisms of MetS.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ We aimed to identify changes in serum metabolites in these “untreated” patients with MetS compared with healthy subjects by using a simple 1H nuclear magnetic resonance experiment.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ We found that untreated patients with MetS have special characteristics, such as insufficient BCAA and vitamin B12 intake; tendency toward ketosis, kidney damage, and increased HbA1c; insulin resistance; and dysregulated lipid metabolism, oxidative signaling, and inflammatory response.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Our data indicated a worse pathophysiological state in these untreated patients, which may deteriorate MetS, or induce serious complications. Thus, we propose that health education be reinforced to improve the public’s knowledge, attitude, and practice regarding MetS.
Metabolic syndrome (MetS) is a common pathophysiological state characterized by obesity, insulin resistance, hypertension, hyperglycemia, and hyperlipidemia that is closely associated with type 2 diabetes (T2D) and cardiovascular disease. It has been a global public health concern with high incidence and prevalence. However, MetS does not have any notable symptoms in the mild stage, therefore, many patients with MetS do not undergo any effective treatment (untreated) because of unaware or neglect of having MetS. In the United States, ~ 12.2% of adults suffering from T2D, within 23.8% of people, did not know they had T2D. It has been estimated that prevalence of prediabetes MetS is about three times more than that of T2D. 1 Thus, ~ 25% of adults in the United States have MetS. 2 In China, the situation is far from optimism. The prevalence of MetS in China is about 15.5%. 3 However, it remains unknown how many patients with MetS in China leave the MetS untreated. Many patients do not seek medical service unless they are suffering from serious complications. Understanding the actual pathophysiological state of these “untreated” patients, and taking measures to stop the “untreated” situation has been a vital problem faced by the health administration in each country.
Currently, metabolomics has been a promising technology for elucidating the mechanisms of MetS. Because it can observe multiple metabolites simultaneously, it has been used for exploring novel biomarkers and therapeutic targets for MetS. A recent systematic review found that most of the studies reported higher branch chain amino acids (BCAAs), aromatic amino acids, in comparison with the normal subjects, however, results varied from different studies. 4 A study based on a cohort in Singapore found higher levels of BCAAs in the blood in subjects with impaired fasting blood glucose (FBG). 5 However, one study in Japan 6 and another study in Korea 7 got the contradictory results. They found the blood BCAAs were higher in the normal subjects. We have known that MetS is influenced by many complicated factors, such as different genetic factors, including genetics and environmental factors (diet, lifestyle, etc.), which likely greatly affect metabolomics results because they affect metabolic processes. 4 In this regard, metabolomic studies in different populations and groups with different lifestyles are important. Several studies have used metabolomics in Chinese populations. A 36‐patient study used a urinary metabolomics approach to compare metabolites in patients with MetS and healthy subjects and found that eight metabolites, including BCAAs, short‐chain acylcarnitine, tricarboxylic acid cycle intermediates, and glucuronidated products, were different in patients with MetS. 8 A later study on 976 adults in Shanghai, performed plasma metabolomics and found that 36 known and 10 unknown metabolites were associated with T2D. 9 These studies provided metabiological data for Chinese patients with MetS, but they were limited in that one study focused on urinary metabolomics and one study included patients with all stages of T2D.
We hypothesized that “untreated” patients with MetS have a worse pathophysiological state compared with healthy controls. In the present study, we enrolled patients with MetS from a physical examination center in China who had not received treatment for MetS, including medication or therapeutic lifestyle changes (exercise, diet, etc.). We aimed to identify changes in serum metabolites in these “untreated” patients compared with healthy subjects by using a simple 1H nuclear magnetic resonance (NMR) experiment. We believe that the data obtained will improve our understanding of the pathophysiological state of patients with MetS who do not undergo any effective intervention.
METHODS
Participants
Patients with MetS and healthy controls were recruited from November 2016 to July 2017 in a physical examination center of our institute. All patients were diagnosed with MetS per the Guidelines for the Prevention and Treatment of Type 2 Diabetes in China (the 2017 edition). 10 Briefly, (i) abdominal obesity (central obesity): male waist circumference ≥ 90 cm, women ≥ 85 cm; (ii) hyperglycemia: FBG ≥ 6.1 mM or 2 hours after glucose load ≥ 7.8 mM and/or diagnosed T2D; (iii) hypertension: blood pressure ≥ 130/85 mmHg and/or diagnosed hypertension; (iv) high triglycerides (TGs): TG ≥ 1.70 mM; and (v) high‐density lipoprotein cholesterol (HDL‐C) < 1.04 mM. The inclusion criteria were patients with MetS who had not undergone effective interventions against MetS, including medications (hypoglycemic agents or insulin), therapeutic lifestyle changes (exercise, diet, yoga, etc.), traditional Chinese medicines, or acupuncture. The exclusion criteria were (i) patients with MetS who had undergone effective interventions against MetS (listed above); (ii) type 1 diabetes, gestational diabetes, secondary hyperlipidemia obesity, or hypertension; (iii) severe heart, liver, kidney, or other organ diseases; and (iv) psychopathy. This study was designed and performed as per the Declaration of Helsinki of the World Medical Association (2000) and was approved by the Ethics Committee of the Fujian University of Traditional Chinese Medicine (approval number 2012‐04). All experimental protocols were formally explained to the participants and/or their relatives and informed consent was obtained.
Acquisition of the clinical data and serum sample
General clinical data associated with MetS including height, body weight, waistline circumference, blood pressure, and blood lipid levels and a fasting venous blood sample were acquired during the physical examination. All involved participants underwent 12‐hour abrosia before blood sample collections (from the previous 8:00 pm to 8:00 am on the sampling day), which were collected at 8:00 am. Blood samples (5 mL) were collected and centrifuged for 15 minutes at 3,500 rpm. Serum samples were preserved in a −81°C refrigerator for subsequent experiments.
1H NMR examination
A standard 1H NMR experiment was used for metabolomic analysis. 11 Briefly, samples were defrosted at room temperature. Each sample (200 µL) was mixed with 400 uL buffer solution composed of 50% heavy water (Cambridge Isotope Laboratories, Tewksbury, MA, USA), 45 mM Na2HPO4/K2HPO4, and 0.9% NaCl (pH 7.4) at room temperature, and vortex‐mixed followed by centrifugation (4°C; 16,099 g; 10 minutes). 12 Then, 550 uL supernatant was transferred into a 5‐mm NMR tube for 1H NMR (Varian 600 MHz spectrometer with 599.93 MHz resonance frequency of 1H). Transverse relaxation weighting experiments were performed as per the Carr–Purcell–Meiboom–Gill sequence with water peak suppression. Parameters were set as follows: relaxation delay 2.0 seconds, acquisition time 1.5 seconds, spectral width 12,000 Hz, temperature 25°C, total echo time 100 ms, and accumulation 64 times.
Raw data were processed according to previous studies. 13 , 14 , 15 Briefly, the raw data of free induction decays were first processed with TopSpin software (version 3.0; Bruker Biospin, Karlsruhe, Germany). All free induction decay data were treated with zero‐filling to 64‐K points. Fourier transformation was performed after being multiplied by an exponential function to a 1.0 Hz line‐broadening factor. Subsequently, we manually phased and baseline‐corrected the 1H NMR spectra, which were referenced to the doublet of 1H α‐glucose at δ 5.23. 16 Quality control was performed with rejection of the spectra with severely distorted baselines or poor water suppression. 17 Then the 1H‐NMR spectra were converted to ASCII files by using MestReNova software (version 9.0.1; Mestrelab Research, Spain), and were then imported into ‘‘R’’ (http://cran.rproject.org) for further analysis. We segmented the 1H NMR spectra between δ 0.5 and 9.0 into consecutive nonoverlapping regions with δ 0.002 chemical shift bins. We selected the bin size of δ 0.002 as the primary chemical shift bin width according to a previous study. 18 The regions of water resonance (δ 4.52–5.00) were excluded for eliminating the baseline effects. Then, we calculated the peak area of each bin. Normalization were performed with the TopSpin software by calculating the value of (area of each integrated segment/ total area of the spectrum) to compensate for the concentration differences among the samples. To achieve better clarification, data in the region of δ 6.0–9.0 were magnified 20 times compared with the corresponding region of δ 0.5–5.0.
Principal component analysis and orthogonal partial least squares discriminant analysis
Use of 1H NMR spectra alone cannot directly determine the differences in metabolites between the two groups. Therefore, we performed principal component analysis (PCA) analysis to identify differences and similarities in serum metabolism. Data were analyzed using SIMCA software (version 14.1; MKS Data Analytics Solutions, Umea, Sweden) for multivariate statistical analysis. Metabolite signals in the 1H NMR serum profiles were first examined by unsupervised PCA, which reduced the dimensionality of the data and summarized the similarities and differences between the two groups using score plots. The interpretability and predictability of the model (R2X; presents the interpretation rate of X variable and predictive Q2) were calculated and evaluated.
Subsequently, to explore the specific discriminant information between the two groups, orthogonal partial least squares discriminant analysis (OPLS‐DA) was performed to filter out orthogonal variables in metabolites that were not associated with categorical variables and to analyze nonorthogonal and orthogonal variables separately. 19 , 20 To evaluate the reliability of the OPLS‐DA model, we performed 200 random permutations. The statistically significant metabolites were then analyzed and summarized by calculating the corresponding correlation coefficients, as previously described. 21 Briefly, we multiplied the loading value with the square root of its SD, which was compared with the Corresponding Correlation Coefficient Critical Value Table. The metabolites that had significantly different levels between the groups were identified.
Identification of key metabolic pathways
To investigate the key metabolic pathways represented by the differentially measured metabolites determined by the above experiments, we searched in the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database to identify the key metabolic pathways closely associated with MetS. 22
Statistical analysis
Statistical analyses were performed using SPSS software (version 23.0.0; IBM, Chicago, IL).
Data were presented as means ± SD. A two‐sample t‐test was used to identify differences in data that were attributed to the normal distribution and homogeneity of variance. A rank sum test was used for those data in non‐normal distribution. A χ2 test was utilized for comparing genders. P < 0.05 was considered statistically significant.
RESULTS
A total of 58 participants were enrolled in this study: 28 patients with MetS (12 men and 16 women16) and 30 healthy controls (10 men and 20 women). The average patient’s age was 51.1 ± 14.6 years and the average healthy participant’s age was 44.0 ± 10.6 years. No significant differences were found in gender or age distribution between the groups. The clinical characteristics of enrolled participants are listed in Table 1 . All indices were significantly worse in the untreated MetS group, especially waistline circumference, body mass index, FBG, and TG. All patients were untreated because they were unaware of or neglected their MetS. After the study, patients were educated regarding the necessity of therapeutic interventions against MetS and all patients willingly initiated therapeutic interventions based on their actual conditions.
Table 1.
Clinical characteristics of enrolled participants
| Healthy control (N = 30) | MS patients (N = 28) | |
|---|---|---|
| Age, years | 44.0 ± 10.6 | 51.1 ± 14.6 |
| Gender, M/F | 10/20 | 12/16 |
| Ethnicity | Chinese | Chinese |
| Waistline, cm | 80.58 ± 8.96 | 95.14 ± 7.72** |
| BMI | 23.25 ± 3.01 | 35.96 ± 14.35** |
| SBP, mmHg | 114.47 ± 10.04 | 132.28 ± 21.43** |
| DBP, mmHg | 71.60 ± 10.70 | 84.80 ± 11.55** |
| BG, mM | 5.15 ± 0.70 | 10.38 ± 3.95** |
| Triglycerides, mM | 1.66 ± 1.97 | 2.31 ± 1.59** |
| HDL‐C, mM | 1.32 ± 0.28 | 1.27 ± 0.41** |
BG, blood glucose; BMI, body mass index; DBP, diastolic blood pressure; HDL‐C, high‐density lipoprotein cholesterol; SBP, systolic blood pressure.
Data are presented as means ± SD, ** means P < 0.01, patients with metabolic syndrome vs. healthy controls.
Figure 1 shows representative 1H NMR spectra of serum from patients with MetS and healthy controls. A total of 49 known substances were preliminarily identified according to their chemical shifts, including carbohydrate compounds (such as α and β glucose), amino acids (such as 1‐methylhistidine, glutamate, glycine, isoleucine, leucine, lysine, methionine, N,N‐dimethylglycine, phenylalanine, sarcosine, threonine, tyrosine, and valine), and lipid compounds (such as 3‐hydroxybuyarate, acetate, acetoacetate, acetone, alanine, choline, lipid, –CH2–C = O, lipid, –CH = CH–, lipid, –CH2–CH = CH–, lipid, = CH–CH2–CH–, very low‐density lipoprotein (VLDL), CH3‐(CH2)n‐VLDL, –CH2–CH2–C = O; Figure 1 ).
Figure 1.
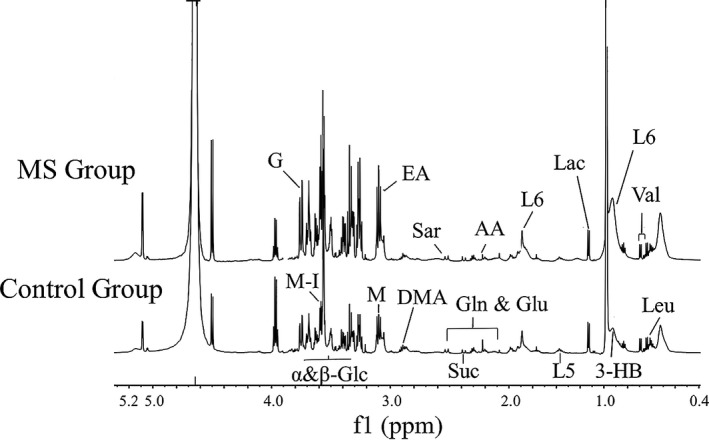
Representative 1H nuclear magnetic resonance (NMR) spectra of serum from patients with metabolic syndrome (MetS) and healthy controls (δ 0.5–5.0 and δ 6.0–9.0). 3‐HB, 3‐hydroxybutyrate; AA, acetoacetate; DMA, dimethylamine; EA, ethanolamine; G, glycerol; Glc, glucose; Gln, glutamine; Glu, glutamate; L5: very low‐density lipoprotein (VLDL), –CH2–CH2‐C = O; L6: lipid, –CH2–CH = CH–; Lac, lactate; Leu, leucine; m‐I, myo‐Inositol; Sar, sarcosine; Suc, succinate; Val, valine.
The cumulative interpretation rate of the PCA model was R2X = 0.926 and Q2 = 0.863. The distribution of the scatter points in the PCA score plot revealed great differences between the two groups (Figure 2a ). However, within the healthy control group, the distribution was more scattered, with a poorer clustering effect. We generated an OPLS‐DA model and the results were R2X = 0.683, R2Y = 0.919, and Q2 = 0.894, indicating that 68.3% of the X variables and 91.9% of the Y variables were applied to the construction of the model, and the accuracy of the grouping was 89.4% (Figure 2b ). The data of 200 permutation tests showed that when the abscissa was equal to 0, R2 = (0.0, 0.0928) and Q2 = (0.0, −0.303), demonstrating the good quality and reliability of the OPLS‐DA model (Figure 3 ). Therefore, we identified 36 serum metabolites with significant differences between the two groups (Table 2 ). We found that the most predominant differential metabolites were: (i) MetS > control for: 1‐methylhistidine (1‐MH), lactate, VLDL, CH3‐(CH2)n‐, low‐density lipoprotein (LDL), CH3‐(CH2)n‐, lipid, –CH2‐CH = CH–, 3‐hydroxybutyrate (3‐HB), tyrosine, lysine, lipid, –CH2–C = O, ethanolamine, and malonate; and (ii) MetS < control for: glycine, glycerophosphorylcholine, and leucine.
Figure 2.
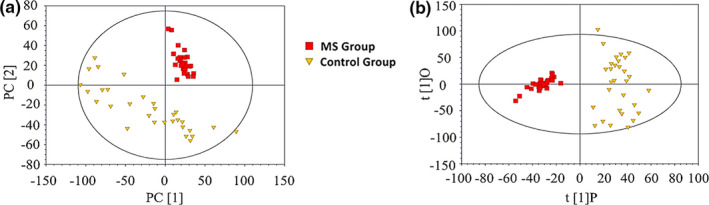
Score plots for patients with metabolic syndrome (MetS) and healthy controls. (a) The principal component analysis (PCA) score plot shows preliminary separation between patients with MetS and healthy controls. (b) The orthogonal partial least squares discriminant analysis (OPLS‐DA) score plot shows clear separation between patients with MetS and healthy controls. Red blocks represent data from patients with MetS; yellow triangles represent data from healthy controls.
Figure 3.
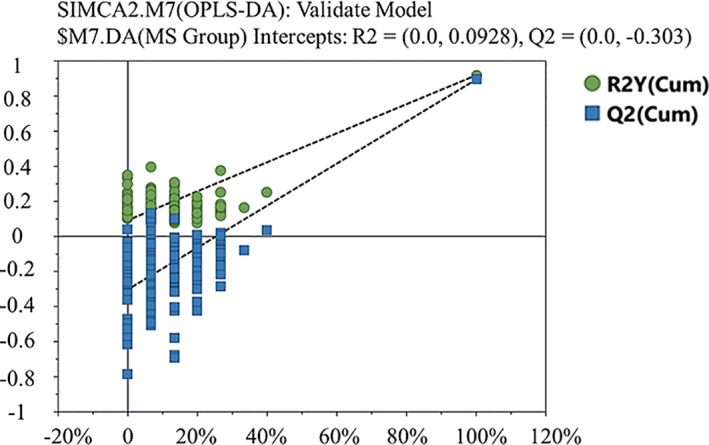
Statistical validation of the orthogonal partial least squares discriminant analysis (OPLS‐DA) model in 200 random permutation tests. Green dots are the R2 values and blue blocks are the Q2 values. When the abscissa equals 0, R2 = (0.0, 0.0928) and Q2 = (0.0, −0.303). The model was confirmed to be of good quality.
Table 2.
The OPLS‐DA correlation coefficient (concentration change) of significantly different metabolites between patients with MetS and control participants
| Metabolites | Chemical shift (ppm) | Correlation coefficients |
|---|---|---|
| 1‐Methylhistidine | 7.07(s),7.81(s) | 0.960 ↑ |
| 3‐Hydroxybuyarate | 1.20(d),2.31(dd),2.41(dd),4.16(m) | 0.904 ↑ |
| Acetate | 1.92(s) | 0.803 ↑ |
| Acetoacetate | 2.28(s) | 0.392 ↑ |
| Acetone | 2.23(s) | 0.642 ↑ |
| Alanine | 1.48(d) | 0.621 ↑ |
| Choline | 3.20(s) | — |
| Citrate | 2.53(d),2.68(d) | 0.798 ↑ |
| Creatine | 3.04(s),3.93(s) | — |
| Dimethylamine | 2.72(s) | — |
| Ethanol | 1.19(t) | 0.821 ↑ |
| Ethanolamine | 3.15(t) | 0.862 ↑ |
| Formate | 8.46(s), | — |
| Glutamate | 2.08(m),2.12(m),2.35(m) | 0.782 ↑ |
| Glutamate and glutamine | 3.78(t) | 0.791 ↑ |
| Glutamine | 2.14(m),2.45(m) | 0.587 ↑ |
| Glycerophosphorylcholine | 3.23(s) | ‐0.463 ↓ |
| Glycerol | 3.58(m),3.66(dd),3.79(m) | 0.843 ↑ |
| Glycine | 3.56(s) | ‐0.649 ↓ |
| Hypoxanthine | 8.19(s),8.21(s) | — |
| Isobutyrate | 1.09(d) | — |
| Isoleucine | 0.94(t),1.01(d) | 0.472 ↑ |
| LDL, CH3–(CH2)n‐ | 0.85(br),1.28(br) | 0.914 ↑ |
| Leucine | 0.96(t) | ‐0.410 ↓ |
| Lipid, –CH2‐C = O | 2.24(br) | 0.864 ↑ |
| Lipid, –CH = CH– | 5.31(br) | 0.563 ↑ |
| Lipid, –CH2‐CH = CH– | 2.02(br) | 0.904 ↑ |
| Lipid, c | 2.78(br) | — |
| Lysine | 1.73(m),1.91(m),3.03(t),3.76(t) | 0.889 ↑ |
| Lysine | 1.73(m),1.91(m),3.03(t),3.76(t) | 0.843 ↑ |
| Malonate | 3.11(s) | 0.852 ↑ |
| Methanol | 3.36(s) | 0.816 ↑ |
| Methionine | 2.14(s),2.65(t) | 0.635 ↑ |
| myo‐Inositol | 3.28(t),3.56(dd),3.61(m),4.06(t) | — |
| N,N‐Dimethylglycine | 2.93(s) | 0.467 ↑ |
| N‐Acetylglycoprotein | 2.04(s) | — |
| Phenylalanine | 7.33(d),7.37(t),7.42(m) | 0.820 ↑ |
| Phosphocholine | 3.21(s) | 0.576 ↑ |
| Pyruvate | 2.37(s) | 0.773 ↑ |
| Sarcosine | 2.73(s) | — |
| Threonine | 4.25(m) | — |
| Trimethylamine N‐oxide | 3.27(s) | 0.752 ↑ |
| Tyrosine | 6.90(d),7.19(d) | 0.896 ↑ |
| Valine | 0.99(d),1.04(d) | — |
| VLDL, CH3‐(CH2)n‐ | 0.88(br) | 0.916 ↑ |
| VLDL, ‐CH2‐CH2‐C = O | 1.58(br) | 0.761 ↑ |
| α‐Glucose | 3.42(t),3.54(dd), 3.72(t),3.84(m) | 0.796 ↑ |
| β‐Glucose | 3.41(t),3.46(dd),3.49(t),3.73(dd),3.90(dd) | 0.829 ↑ |
The cutoff value for each contrast was set at 0.361; the symbol “—” indicates a correlation coefficient < 0.361.
Multiplicity: br, broad resonance; d, doublet; dd, doublet of doublets; q, quartet; m, multiple; s, singlet; t, triplet.
LDL, low‐density lipoprotein; MetS, metabolic syndrome; OPLS‐DA, orthogonal partial least squares discriminant analysis; VLDL, very low‐density lipoprotein.
By KEGG pathway analysis, the top 15 metabolic pathways were: (1) aminoacyl − tRNA biosynthesis; (2) glycolysis or gluconeogenesis; (3) nitrogen metabolism; (4) alanine, aspartate, and glutamate metabolism; (5) synthesis and degradation of ketone bodies; (6) valine, leucine, and isoleucine biosynthesis; (7) methane metabolism; (8) propanoate metabolism; (9) D‐glutamine and D‐glutamate metabolism; (10) valine, leucine, and isoleucine degradation; (11) butanoate metabolism; (12) phenylalanine metabolism; (13) glycine, serine, and threonine metabolism; (14) taurine and hypotaurine metabolism; and (15) citrate (TCA) cycle (Figure 4 ).
Figure 4.
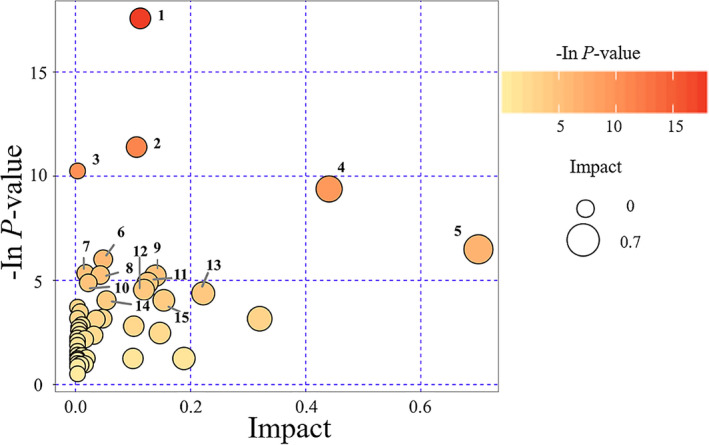
Pathway analysis of patients with metabolic syndrome (MetS) and healthy controls. The top 15 pathways with major changes (P < 0.05) were identified. Bubble color represents the P value: deeper colors represent smaller P values, indicating larger differences. The size of the bubble represents the impact of the pathway during topological analysis. Larger size represents higher impact.
DISCUSSION
In the present study, we used traditional 1H NMR followed by PCA and OPLS‐DA, and identified 36 significantly altered metabolites in the serum of Chinese patients with untreated MetS compared with healthy controls. The top 15 altered key metabolic pathways were identified by searching the KEGG Pathway Database. To the best of our knowledge, this is the first study of patients who were not undergoing any effective interventions for MetS. We believe that the findings of this study provide a deeper understanding of the pathophysiological state of untreated patients with MetS in the Chinese population.
Ethically, studying “untreated” patients is a challenge because all patients deserve appropriate treatments. In the present study, all patients lacked any effective treatments for their MetS because they were unaware of or neglected it. In advanced countries like the United States, approximately one fourth of the patients with T2D are not aware that they have it. 1 These data are not available in China. In our physical examination center, we enrolled 28 untreated patients with MetS during the 8‐month recruiting timeframe. These patients were untreated, not for economic reasons, but due to lack of knowledge or recent physical examination. Therefore, it is urgently necessary for global governments to strengthen health education to improve the knowledge, attitude, and practice (KAP) regarding MetS.
The untreated patients in this study presented with typical characteristics of MetS including larger waistlines and body mass index, hyperglycemia, and hyperlipidemia (Table 1 ). Levels of protective HDL‐C were significantly lower than in healthy subjects. We believe that data acquired from these patients were accurately representative of the pathophysiological state of patients with MetS, because the patients did not receive treatments that would have likely affected the sensitive metabolomic experiment.
The 1H NMR experiment identified 49 known substances and the subsequent PCA and OPLS‐DA analyses were indispensable to determine differences between patients with MetS and healthy subjects. The PCA indicated a high discriminability between the patient and control groups (Figure 2 ), and the OPLS‐DA along with the results of 200 permutation tests indicated the good quality of the model (Figure 3 ). Thus, the corresponding coefficients of serum metabolites in the two groups were determined.
Blood glucose and HDL‐C are the most important indices in MetS, such that higher blood glucose and HDL‐C were expected. Some metabolites were also identified in previous serum or plasma metabolomics studies in subjects with T2D 4 : in contrast to previous studies, 4 , 9 we found that glutamine was higher and leucine was lower in the MetS group. Interestingly, we found that some serum metabolites were unreported in previous studies: 3‐HB, ethanolamine, formate, LDL, methionine, N,N‐dimethylglycine, phosphocholine, trimethylamine N‐oxide, malonate, methanol, acetate, and ethanol were higher, whereas glycerophosphorylcholine was lower in the MetS group. Acetoacetate, acetone, citrate, and 1‐MH were higher in serum in our study, whereas previous studies reported these same changes but in urine.
Some amino acids (alanine, glutamate, isoleucine, phenylalanine, tyrosine, and glycine) exhibited a similar change observed in previous studies, whereas glutamine and leucine did not. Valine was reportedly higher in patients with T2D, 4 , 9 whereas, in the present study, no significant difference was found. A previous study 4 found that alanine, glycine, isoleucine, and tyrosine were the most important amino acids changed in patients with T2D because they were altered in plasma, serum, and urine. Our study is in accordance with this report. Together, the biggest differences in our study compared with prior findings are regarding leucine and valine, two BCAAs. In this study, leucine was significantly lower in patients with MetS, whereas valine did not differ with MetS. Although isoleucine showed a significant difference, the correlation coefficient was only 0.472. The metabolism of the BCAAs is quite complicated. The circulating levels of BCAAs are affected by many factors, such as the food intake 23 and BCAA catabolic enzymes. 24 It has been reported that the expression of the BCAA catabolic enzymes is downregulated in MetS subjects, 25 which might be a physiological basis of enhancements of circulating BCAAs. However, the cause of decrease of the leucine in this study is uncertain. One explanation is due to the different physiological effects of leucine and isoleucine. Zhang et al. reported that leucine, rather than isoleucine and valine, upregulates the expression of the amino acid transporters by activation of the PI3K/Akt/mTOR and ERK signaling pathways. 26 Another study found that isoleucine has a stronger effect (vs. leucine and valine) to decrease the circulating glucose in normal rats. 27 Thus, metabolisms of leucine, isoleucine, and valine in the patients with untreated MetS might be different. Another explanation is the low intake of BCAAs in these patients, but the effects of the intake of BCAAs on MetS remain controversial. 24 Additionally, the small sample size of the present study is likely to cause a bias. These issues require further investigation in our future study.
As for the lipid profile, we found that all the lipid compounds were significantly higher in the MetS group (Table 2 ). These data were in agreement with a previous study in young adults with obesity. 28 Along with the data of LDL and VLDL, impairment of lipid metabolism in these untreated patients with MetS has been verified.
Several serum metabolites were reported for the first time in this study, including 3‐HB, which was significantly higher in the MetS group. The 3‐HB is a major physiological ketone that serves as an alternative energy source during food deprivation. In a calorie restriction study in mice, retinal 3‐HB was elevated with increased body weight and calorie intake. 29 Several studies have found higher 3‐HB in the plasma and urine of patients with T2D, including data from a Chinese cohort study. 9 However, a Korean cohort study did not mention 3‐HB. 7 Factors that affect 3‐HB are unclear. For the untreated patients with MetS in this study, we suggest that the elevated 3‐HB is a result of a compensatory response because they have a ketogenic tendency.
Ethanolamine serum levels also increased with MetS in this study. Previous studies reported that ethanolamine may improve kidney injury in the mouse T2D model. 30 , 31 We speculate that enhancement of serum ethanolamine might indicate the high risk of kidney injury in these untreated patients. N,N‐dimethylglycine and trimethylamine N‐oxide are closely associated with elevated HbA1c. 32 Because these patients were untreated, increased N,N‐dimethylglycine and trimethylamine N‐oxide indicate the tendency of HbA1c to be elevated.
Methionine is associated with many metabolic processes, including lipid metabolism. It has been well‐documented that methionine restriction ameliorates lipid metabolism dysfunction, insulin resistance, and oxidative stress. 33 In these untreated patients, higher serum methionine levels may indicate a worsened physiological state of MetS. Phosphocholine is attributed to toxic lipid species, and was found to be higher in diabetic mice. 34 It is normal that phosphocholine serum levels are higher in these patients. Formate is associated with vitamin B12 metabolism. Elevated formate levels have been reported with vitamin B12 deficiency 35 ; thus, higher formate levels in these patients suggest vitamin B12 deficiency. LDL is characteristic of MetS, and it was significantly higher in these patients. Glycerophosphorylcholine is protective against oxidative stress, inflammation, etc., 36 , 37 and, thus, it is reasonable that it is reduced in the serum of these untreated patients. The increased serum levels of malonate, methanol, acetate, and ethanol require further investigation.
We identified the top 15 metabolic pathways altered with MetS by pathway analysis (Figure 4 ). Sun et al. identified 17 metabolic pathways altered in patients with T2D 4 and 8 pathways were altered in both our study and theirs (numbered based on their appearance in the top 15 list of this study): (1) aminoacyl − tRNA biosynthesis; (3) nitrogen metabolism; (4) alanine, aspartate, and glutamate metabolism; (6) valine, leucine, and isoleucine biosynthesis; (7) methane metabolism; (9) D‐glutamine and D‐glutamate metabolism; (10) valine, leucine, and isoleucine degradation; and (13) glycine, serine, and threonine metabolism. Importantly, three amino acid metabolic pathways were emphasized in both studies: (4) alanine, aspartate, and glutamate metabolism; (9) D‐glutamine and D‐glutamate metabolism; and (13) glycine, serine, and threonine metabolism. These pathways are, therefore, very likely associated with development of MetS, and, thus, should be considered as potential novel targets for treating MetS.
The present study focused on untreated patients with MetS. Compared with the previous studies in patients with MetS, we found that untreated patients with MetS have special characteristics, such as insufficient BCAA and vitamin B12 intake; tendency toward ketosis, kidney damage, and increased HbA1c; insulin resistance; and dysregulated lipid metabolism, oxidative signaling, and inflammatory response. These characteristics present a worse pathophysiological state in these patients that could worsen MetS or induce serious complications. Our results suggest that being unaware or neglectful of MetS, resulting in an “untreated” situation, is extremely dangerous. Effective interventions must be adopted to improve the KAP concerning MetS so that this “untreated” situation is stopped immediately.
CONCLUSIONS
We performed a metabolomics investigation in untreated patients with MetS in the Chinese population. Our 1H NMR data identified 36 serum metabolites closely associated with untreated MetS and the top 15 enriched metabolic pathways, including three amino acid metabolic pathways. Importantly, we identified 14 novel serum metabolites that were altered in these untreated patients and were not reported in previous studies. These data indicate that untreated patients with MetS have several characteristics that could exacerbate MetS or induce serious complications. Thus, we propose that health education must be reinforced to improve the public’s KAP regarding MetS. Now is the time to stop the “untreated” situation resulting from unawareness and neglection.
Funding
The study is supported by grants from the National Natural Science Foundation of China (Grants No. 81873234 and 81273666), grant from General Project of Natural Science Foundation of Fujian Province (2018J01875), grant from the Fujian Province 2011 Collaborative Innovation Center for Traditional Chinese Medicine Health Management (JG2017004‐Collaboration), and grants from the Japan Society for the Promotion of Science (Grant‐in‐Aid for Young Scientists, Type B, No. 20791025 and Grant‐in‐Aid for Scientific Research C, General, No. 24592157, 15k10358, and 18K08991).
Conflict of Interest
The authors declared no competing interests for this work.
Author Contributions
T.A. and Y.L. wrote the manuscript. T.A. and B.G. designed the research. Y.L., Y.W., Y.Z., P.Z., S.C., T.A., and BG performed the research. Y.L., Y.W., Y.Z., P.Z., S.C., and B.G. analyzed the data. T.A. and B.G. contributed new reagents/analytical tools.
Supporting information
Data S1.
Acknowledgment
The authors thank Enago (www.enago.jp) for the English language review.
Contributor Information
Tetsuya Asakawa, Email: asakawat1971@gmail.com.
Bizhen Gao, Email: gbz688@163.com.
References
- 1. Saklayen, M.G. The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 20, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Ferranti, S.D. et al Prevalence of the metabolic syndrome in American adolescents: findings from the Third National Health and Nutrition Examination Survey. Circulation 110, 2494–2497 (2004). [DOI] [PubMed] [Google Scholar]
- 3. Wang, Y. , Mi, J. , Shan, X.Y. , Wang, Q.J. & Ge, K.Y. Is China facing an obesity epidemic and the consequences? The trends in obesity and chronic disease in China. Int. J. Obes. (Lond). 31, 177–188 (2007). [DOI] [PubMed] [Google Scholar]
- 4. Sun, Y. , Gao, H.Y. , Fan, Z.Y. , He, Y. & Yan, Y.X. Metabolomics signatures in type 2 diabetes: a systematic review and integrative analysis. J. Clin. Endocrinol. Metab. 105, 1000–1008 (2020). [DOI] [PubMed] [Google Scholar]
- 5. Xu, F. et al Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry‐based metabolomics. J. Clin. Endocrinol. Metab. 98, E1060–E1065 (2013). [DOI] [PubMed] [Google Scholar]
- 6. Nagata, C. et al Branched‐chain amino acid intake and the risk of diabetes in a Japanese community: the Takayama study. Am. J. Epidemiol. 178, 1226–1232 (2013). [DOI] [PubMed] [Google Scholar]
- 7. Yun, J.H. et al Metabolomics profiles associated with HbA1c levels in patients with type 2 diabetes. PLoS One 14, e0224274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu, Z.R. , Ning, Y. , Yu, H. & Tang, N.J. A HPLC‐Q‐TOF‐MS‐based urinary metabolomic approach to identification of potential biomarkers of metabolic syndrome. J Huazhong Univ. Sci. Technolog. Med. Sci. 34, 276–283 (2014). [DOI] [PubMed] [Google Scholar]
- 9. Yu, D. et al Plasma metabolomic profiles in association with type 2 diabetes risk and prevalence in Chinese adults. Metabolomics 12, 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Society, C.D. Guidelines for the prevention and treatment of type 2 diabetes in China (2017 edition) [in Chinese]. Chinese J. Pract. Inter. Med. 38, 292–344 (2018). [Google Scholar]
- 11. Beckonert, O. et al Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2, 2692–2703 (2007). [DOI] [PubMed] [Google Scholar]
- 12. Zhang, X. et al Human serum metabonomic analysis reveals progression axes for glucose intolerance and insulin resistance statuses. J. Proteome Res. 8, 5188–5195 (2009). [DOI] [PubMed] [Google Scholar]
- 13. Sun, L.W. et al (1)H‐Nuclear magnetic resonance‐based plasma metabolic profiling of dairy cows with clinical and subclinical ketosis. J. Dairy Sci. 97, 1552–1562 (2014). [DOI] [PubMed] [Google Scholar]
- 14. Wang, P.R. , Wang, J.S. , Yang, M.H. & Kong, L.Y. Neuroprotective effects of Huang‐Lian‐Jie‐Du‐Decoction on ischemic stroke rats revealed by (1)H NMR metabolomics approach. J. Pharm. Biomed. Anal. 88, 106–116 (2014). [DOI] [PubMed] [Google Scholar]
- 15. Jin, B. et al Nuclear magnetic resonance‐assisted metabolic analysis of plasma for mild gestational diabetes mellitus patients. Metab. Syndr. Relat. Disord. 15, 439–449 (2017). [DOI] [PubMed] [Google Scholar]
- 16. Dona, A.C. et al Precision high‐throughput proton NMR spectroscopy of human urine, serum, and plasma for large‐scale metabolic phenotyping. Anal. Chem. 86, 9887–9894 (2014). [DOI] [PubMed] [Google Scholar]
- 17. McClay, J.L. et al (1)H nuclear magnetic resonance metabolomics analysis identifies novel urinary biomarkers for lung function. J. Proteome Res. 9, 3083–3090 (2010). [DOI] [PubMed] [Google Scholar]
- 18. Purohit, P.V. , Rocke, D.M. , Viant, M.R. & Woodruff, D.L. Discrimination models using variance‐stabilizing transformation of metabolomic NMR data. OMICS 8, 118–130 (2004). [DOI] [PubMed] [Google Scholar]
- 19. Verwaest, K.A. et al (1)H NMR based metabolomics of CSF and blood serum: a metabolic profile for a transgenic rat model of Huntington disease. Biochim. Biophys. Acta. 1812, 1371–1379 (2011). [DOI] [PubMed] [Google Scholar]
- 20. Rezaei‐Tavirani, M. et al Advantage of applying OSC to (1)H NMR‐based metabonomic data of celiac disease. Int. J. Endocrinol. Metab. 10, 548–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cai, A. et al A pilot metabolic profiling study of patients with neonatal jaundice and response to phototherapy. Clin. Transl. Sci. 9, 216–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nie, C. , He, T. , Zhang, W. , Zhang, G. & Ma, X. Branched chain amino acids: beyond nutrition metabolism. Int. J. Mol. Sci. 19, 954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weng, L. et al Association of branched and aromatic amino acids levels with metabolic syndrome and impaired fasting glucose in hypertensive patients. Metab Syndr. Relat. Disord. 13, 195–202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lackey, D.E. et al Regulation of adipose branched‐chain amino acid catabolism enzyme expression and cross‐adipose amino acid flux in human obesity. Am. J. Physiol. Endocrinol. Metab. 304, E1175–E1187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang, S. et al Leucine stimulates ASCT2 amino acid transporter expression in porcine jejunal epithelial cell line (IPEC‐J2) through PI3K/Akt/mTOR and ERK signaling pathways. Amino Acids 46, 2633–2642 (2014). [DOI] [PubMed] [Google Scholar]
- 27. Doi, M. , Yamaoka, I. , Fukunaga, T. & Nakayama, M. Isoleucine, a potent plasma glucose‐lowering amino acid, stimulates glucose uptake in C2C12 myotubes. Biochem. Biophys. Res. Commun. 312, 1111–1117 (2003). [DOI] [PubMed] [Google Scholar]
- 28. Pasanta, D. , Chancharunee, S. , Tungjai, M. , Kim, H.J. & Kothan, S. Effects of obesity on the lipid and metabolite profiles of young adults by serum (1)H‐NMR spectroscopy. PeerJ. 7, e7137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Izuta, Y. et al Ketone body 3‐hydroxybutyrate mimics calorie restriction via the Nrf2 activator, fumarate, in the retina. Aging Cell 17, e12699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han, P. et al Niclosamide ethanolamine improves kidney injury in db/db mice. Diabetes Res. Clin. Pract. 144, 25–33 (2018). [DOI] [PubMed] [Google Scholar]
- 31. Han, P. et al Niclosamide ethanolamine improves diabetes and diabetic kidney disease in mice. Am. J. Transl. Res. 10, 1071–1084 (2018). [PMC free article] [PubMed] [Google Scholar]
- 32. Friedrich, N. et al Identification of urine metabolites associated with 5‐year changes in biomarkers of glucose homoeostasis. Diabetes Metab. 44, 261–268 (2018). [DOI] [PubMed] [Google Scholar]
- 33. Yin, J. et al Metabolic regulation of methionine restriction in diabetes. Mol. Nutr. Food Res. 62, e1700951 (2018). [DOI] [PubMed] [Google Scholar]
- 34. Li, W. et al Profile of cardiac lipid metabolism in STZ‐induced diabetic mice. Lipids Health Dis. 17, 231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacMillan, L. et al Cobalamin deficiency results in increased production of formate secondary to decreased mitochondrial oxidation of one‐carbon units in rats. J. Nutr. 148, 358–363 (2018). [DOI] [PubMed] [Google Scholar]
- 36. Strifler, G. et al Targeting mitochondrial dysfunction with L‐alpha glycerylphosphorylcholine. PLoS One 11, e0166682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsubara, K. et al The delaying effect of alpha‐glycerophosphocholine on senescence, transthyretin deposition, and osteoarthritis in senescence‐accelerated mouse prone 8 mice. Biosci. Biotechnol. Biochem. 82, 647–653 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.


