Abstract
What is known and objective:
Pharmacogenomic biomarkers are now used in many clinical care settings and represent one of the successes of precision medicine. Genetic variants are associated with pharmacokinetic and pharmacodynamic changes leading to medication adverse effects and changes in clinical response. Actionable pharmacogenomic variants are common in transplant recipients and have implications for medications used in transplant, but yet are not broadly incorporated into practice.
Methods:
From the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group guidelines, and PharmGKB databases, 12 pharmacogenomic genes with 30 variants were selected and used to create diplotypes and actionable pharmacogenomic phenotypes. A total of 853 kidney allograft recipients who had genomic information available from a genome-wide association study were included.
Results:
Each recipient had at least one actionable pharmacogenomic diplotype/phenotype, whereas the majority (58%) had three or four actionable diplotypes/phenotypes and 17.4% had five or more among the 12 genes. The participants carried actionable diplotypes/phenotypes for multiple medications, including tacrolimus, azathioprine, clopidogrel, warfarin, simvastatin, voriconazole, antidepressants and proton-pump inhibitors.
What is new and conclusion:
Pharmacogenomic variants are common in transplant recipients, and transplant recipients receive medications that have actionable variants.
Clinical trial:
Genomics of Transplantation, clinicaltrials.gov (NCT01714440).
Keywords: clinical pharmacy, kidney transplantation, pharmacogenetics, pharmacogenomics
1. WHAT IS KNOWN AND OBJECTIVE
Pharmacogenomic biomarkers are emerging into clinical practice to guide selection of medications and doses. Clinical implementation of pharmacogenomics has been hastened by the several important initiatives. The Clinical Pharmacogenetic Implementation Consortium (CPIC), sponsored by the National Institute of Health (NIH), evaluates the pharmacogenomic literature and creates peer-reviewed, evidence-based actionable drug-gene practice guidelines.1 The Dutch Pharmacist Working Group (DPWG) is a European organization that peer reviews and develops pharmacogenomic-based therapeutic recommendations in the form of guidelines. The PharmGKB is a NIH-supported pharmacogenomic knowledge database that collects curates and disseminates knowledge about the impact of variation on drug response. There are also over 250 Food and Drug Administration (FDA)-approved drugs with genetic biomarker information provided in the drug labelling,2 and the FDA recently released a list of drug-gene pairs that they believe there is sufficient evidence that certain genotypes are likely to have altered drug metabolism and differential therapeutic and adverse effects.3
Application of pharmacogenomics is used most commonly to inform clinical therapeutic decisions in cancer, psychiatry, cardiovascular disease, and to prevent serious drug-related adverse reactions. Pharmacogenomic testing has not been routinely adopted in transplantation despite the availability of genomic biomarkers for medications that transplant recipients receive. In kidney transplantation, there is strong evidence for the effect of genetic variants in the cytochrome P450 (CYP) 3A5 gene on the pharmacokinetics of tacrolimus, especially in African American recipients.4-7 African Americans are much more likely to be carriers (>70%) of the CYP3A5*1 allele than Caucasians (5%-10%) which confers enhanced tacrolimus clearance and higher dose requirements relative to most Caucasians who generally carry two loss-of-function alleles (CYP3A5*3/*3).5 Although azathioprine is no longer routinely used as primary maintenance immunosuppression, it is used in recipients as an alternative for those developing unacceptable toxicities of mycophenolate. Thiopurine methyltransferase (TPMT) is involved in the biotransformation pathway of azathioprine and is well studied. Individuals who carry low function and non-functional TPMT variants are at higher risk of azathioprine-related myelosuppression.8 Transplant recipients often require cardiovascular therapies such as clopidogrel, warfarin and statins which have clinically relevant pharmacogenomic markers.9-11 Other medications such as selective serotonin reuptake inhibitors,12 tricyclic antidepressants,13 proton-pump inhibitors, voriconazole14 and oestrogen-containing products15,16 also have genomic biomarkers. In this study, we describe the frequency of pharmacogenomic diplotypes and the associated actionable phenotypes, as recommended by guidelines, in kidney transplant recipients.
2. METHODS
2.1. Study participants and data collection
We studied 853 kidney allograft recipients of European and African ancestry as determined through principal components analysis enrolled in the multicentre, prospective, observational Genomics of Kidney Transplantation (GEN03) study (NCT01714440) between 2012 and 2016.17 This study was approved by the Institutional Review Boards of the respective enrolling centres. Participants were eligible if they had end-stage renal dysfunction and were undergoing kidney or simultaneous kidney-pancreas transplant. These participants were enrolled in a genome-wide association study (GWAS) to identify biomarkers for immunosuppressants and kidney allograft outcomes. Induction and maintenance immunosuppression were centre-specific. All participants provided written informed consent at the time of transplant. Participants were selected for the current analysis if they were enrolled in the GEN03 study and had GWAS data available. Demographic and clinical outcome information was collected from the electronic health record and maintained in a central database (Rho, Inc).
2.2. DNA collection and genotyping
Recipient DNA was obtained at time of transplant from peripheral blood lymphocytes. DNA was isolated from lymphocytes after red blood cell lysis and centrifugation. Genotyping was conducted on the Affymetrix Transplant Array chip (Affymetrix) enriched with content for transplant outcomes and genes.18 The chip produced ~782 000 high-quality genomic markers. Extensive quality control was performed on the variants using previously described community standards.17 Imputation of unmeasured variants was conducted by using 1000 Genomes Phase 3 and Genome of the Netherlands v5 as reference panels, and after quality control, ~40M genotyped and imputed variants remained.18,19 INFO (information) scores assessing the quality of imputation were high and in general >0.8. Seven imputed variants were used in the analysis.
2.3. Identification and evaluation of actionable diplotypes/phenotypes
Actionable diplotypes and corresponding phenotypes for this study were identified by searching the CPIC guidelines,20 DPWG guidelines21 and PharmGKB,15 and a comprehensive list of actionable variants (known alleles which have implications, backed by scientific evidence, for clinical management) was created. Actionable drug-gene pairs were selected for the analysis if they had CPIC A or B, or PharmGKB 1A, 1B, 2A or 2B scientific levels of evidence, or DPWG guidelines. The GWAS panel contained 30 of the identified variants within 12 genes (Supplement table S1). Some variants within CPIC guidelines were not available on the GWAS panel including CYP2B6 and allelic repeats and/or copy number variants for the UGT1A1 and CYP2D6 genes and are excluded from the analysis.
Phenotypes were assigned using the GWAS variants as recommended by the CPIC guidelines for each drug-gene pair. Since phenotype terminology varies between CPIC guidelines, this analysis uses the terminology used by the respective drug-gene guideline. The phenotype for F5 was assigned based on published literature and data reported on the CPIC website and PharmGKB.15,16 The phenotypes for dihydropyrimidine dehydrogenase (DPYD) were assigned an activity score using an allele functionality score.22 DPYD HapB3 variant is made up of 3 single nucleotide polymorphisms, rs75017182, rs56038477 and rs56276561, but the GWAS panel contained only rs75017182 which is in strong linkage disequilibrium with the other two variants; the functionality score was determined using rs75017182. The activity scores were calculated as recommended, which was the sum of the two DPYD alleles of the lowest functionality scores.
3. RESULTS
3.1. Demographics and characteristics
A total of 853 kidney transplant recipients (678 European and 175 African ancestries) were studied (Table 1). The mean (±SD) age at the time of transplant was 49.76 (±14.42) years, and the majority of participants (62.6%) received a living donor transplant. Diabetes (20.9%) and glomerular disease (28.5%) were the most common primary causes of kidney disease. The presence of cardiovascular disease was common; 88.5% had hypertension, and 50.3% had hyperlipidaemia that required treatment at transplant. Other cardiovascular disorders were present in lower frequencies (<11%). Tacrolimus was the primary maintenance immunosuppressant used in 91% of recipients. Acute rejection developed in 10.1% by 6 months post-transplant.
TABLE 1.
Demographics and characteristics of kidney transplant recipient population (n = 853)
| Characteristics | Number (%), mean ± SD |
|---|---|
| Age at transplant (mean ± SD) | 49.76 ± 14.42 |
| Living donor | 534 (62.6) |
| Simultaneous pancreas-kidney | 35 (4.1) |
| Weight (kg) at transplant (mean ± SD) | 81.75 ± 20.56 |
| Male recipient | 521 (61.1) |
| Male donor | 419 (49.1) |
| Ancestry | |
| European | 678 (79.5) |
| African | 175 (20.5) |
| Cause of kidney diseasea | |
| Diabetes | 178 (20.9) |
| Glomerular disease | 243 (28.5) |
| Other | 156 (18.3) |
| Hypertension | 100 (11.7) |
| Polycystic kidney disease | 126 (14.8) |
| Unknown | 50 (5.9) |
| Cardiovascular disease prior to transplanta | |
| Myocardial infarction | 45 (5.3) |
| Stroke | 39 (4.6) |
| Hypertension | 755 (88.5) |
| Congestive heart failure | 47 (5.5) |
| CAD requiring drug treatment | 99 (11.6) |
| CAD requiring invasive or surgical procedures | 107 (12.5) |
| Hyperlipidaemia requiring drug treatment at transplant | 429 (50.3) |
| Diabetes at time of transplant | 250 (29.3) |
| Induction immunosuppression | |
| Monoclonal antibody | 339 (39.7) |
| Polyclonal antibody | 496 (58.1) |
| Combination | 18 (2.1) |
| Calcineurin inhibitor at baseline | |
| Tacrolimus | 776 (91.0) |
| Cyclosporine | 62 (7.3) |
| None | 15 (1.8) |
Abbreviation: CAD, coronary artery disease.
The number of cardiovascular diseases exceeds 100% since some individuals had multiple conditions.
3.2. Genotypes and phenotypes
All variants were in Hardy-Weinberg equilibrium. The GWAS panel did not contain the TPMT*4 and NUDT15*2 variants and were not included in the respective phenotype determination. TPMT*4 and NUDT15*2 are rare variants, and the number of actionable phenotypes for TPMT and NUDT15 may be slightly underestimated. The ancestry distribution and frequencies of the actionable phenotypes in the selected 12 genes are shown in Table 2. Our observed frequencies were similar to that observed by others.23
TABLE 2.
Phenotype and diplotype frequencies of 12 pharmacogenes in the study population by ancestry
| Gene | Phenotype | Diplotype | European American n (%)a |
African American n (%)a |
|---|---|---|---|---|
| CYP2C9 | Extensive metabolizer | *1/*1 | 433 (64.2) | 162 (92.6) |
| Intermediate metabolizer | *1/*2, or *1*3 | 213 (31.6) | 13 (7.4) | |
| Poor metabolizer | *2/*2, *3/*3, *2/*3 | 28 (4.2) | 0 (0.0) | |
| CYP2C19 | Ultra-rapid metabolizer | *17/*17 | 28 (4.1) | 7 (4) |
| Rapid metabolizer | *1/*17 | 189 (27.9) | 48 (27.6) | |
| Extensive metabolizer | *1/*1 | 257 (37.9) | 62 (35.6) | |
| Intermediate metabolizer | *1/*2, *1/*3, or *2/*17 | 186 (27.4) | 52 (29.9) | |
| Poor metabolizer | *2/*3, *2/*2, or *3/*3 | 17 (2.5) | 5 (2.9) | |
| CYP3A5 | Extensive metabolizer | *1/*1 | 4 (0.6) | 35 (20.1) |
| Intermediate metabolizer | *1/*3, *1/*6, or *1*7 | 74 (11.0) | 91 (52.0) | |
| Poor metabolizer | *3/*3, *6/*6, *7/*7, *3/*7, *3/*6, or *6/*7 | 597 (88.4) | 48 (27.6) | |
| CYP4F2 | Normal function | *1/*1 | 339 (55.2) | 132 (75.4) |
| Intermediate function | *1/*3 | 275 (4.8) | 38 (21.7) | |
| Decreased function | *3/*3 | 64 (10.4) | 5 (2.9) | |
| DPYD | Normal metabolizer | Activity Score: 2 | 627 (92.5) | 173 (98.7) |
| Intermediate metabolizer | Activity Score: 1-1.5 | 40 (5.9) | 2 (1.1) | |
| Poor metabolizer | Activity Score: 0-0.5 | 1 (0.1) | 0 (0.0) | |
| F5 | Normal risk | C/C | 637 (94.0) | 175 (100) |
| High risk for thromboembolism | C/T | 40 (6.0) | 0 (0.0) | |
| Higher risk for thromboembolism | T/T | 1 (0.1) | 0 (0.0) | |
| HLA-B*57:01 | Very low risk for hypersensitivity | *X/*X | 633 (93.4) | 171 (97.7) |
| High risk for hypersensitivity | *57:01/*X, or *57:01/*57:01 | 45 (6.6) | 4 (2.3) | |
| IFNL3 | Increased response | CC | 301 (44.4) | 28 (16.0) |
| Decreased response | CT or TT | 377 (55.6) | 147 (84.0) | |
| NUDT15b | Normal metabolizer | *1/*1 | 675 (99.7) | 172 (99.4) |
| Intermediate metabolizer | *1/*3 | 2 (0.3) | 1 (0.6) | |
| Poor metabolizer | *3/*3 | 0 (0.0) | 0 (0.0) | |
| SLCO1B1 | Normal function | *1a/*1b, *1a/*1a, or *1b/*1b | 478 (70.5) | 162 (92.6) |
| Intermediate function | *1a/*15, *1a/*17, *1b/*15, *1b/*17, *1a/*5, or *1b/*5 |
190 (28.0) | 13 (7.4) | |
| Low function | *5/*5, *5/*15, *5/*17, *15/*15, *15/*17, or *17/*17 | 10 (1.5) | 0 (0.0) | |
| TPMTc | Normal, high activity | *1/*1 | 600 (88.8) | 160 (91.4) |
| Intermediate activity | *1/*2, *1/*3A, *1/*3B, or *1/*3C | 73 (10.8) | 14 (8.0) | |
| Low activity | *3A/*3A, *2/*3A, *3C/*3A, or *3C/*2 | 2 (0.3) | 1 (0.6) | |
| VKORC1 | Low warfarin sensitivity | GG | 237 (35.0) | 139 (79.9) |
| Intermediate warfarin sensitivity | AG | 337 (49.7) | 32 (18.3) | |
| Warfarin sensitive | AA | 104 (15.3) | 3 (1.7) |
Abbreviations: CYP, cytochrome P450; DYPD, dihydropyrimidine dehydrogenase; F5, Factor V Leiden; HLA, major histocompatibility complex;IFN, interferon; NUDT, nudix hydrolase; SLCO, solute carrier organic anion transporter; TPMT, thiopurine S-methyltransferase; VKORC1, vitamin K epoxide reductase complex 1.
Phenotypes may not add up to 100% due to inconclusive alleles or missing phenotypes in some individuals.
NUDT15*2 is a rare variant and not included due to unavailability on the GWAS panel.
TPMT *4 is a rare variant and not included due to unavailability on the GWAS panel.
Every individual had at least 1 actionable phenotype, whereas the majority (58%) of patients had three or four actionable phenotypes among the 12 genes (Figure 1). About 4% of recipients carried 6, 7 or 8 actionable phenotypes. The recommended clinical actions for these phenotypes are shown in Supplemental table S2. An actionable CYP3A5 phenotype (extensive or intermediate metabolizer) for tacrolimus was present in 11.6% of European and 72.1% of African ancestry. An actionable TPMT phenotype for azathioprine was present in ~10% of recipients. An actionable CYP2C19 phenotype (intermediate or poor metabolizer) for clopidogrel was present in 30.5% of recipients, whereas an actionable CYP2C19 phenotype for voriconazole was present in 34.5% of individuals. A CYP2C19 actionable phenotype for omeprazole and pantoprazole was present in 4.1% of the population. A CYP2C9 actionable phenotype relevant to phenytoin and warfarin was present in 29.9% of individuals. HLA-B*57:01, an actionable variant for abacavir, was present in 5.7% of the recipients.
FIGURE 1.
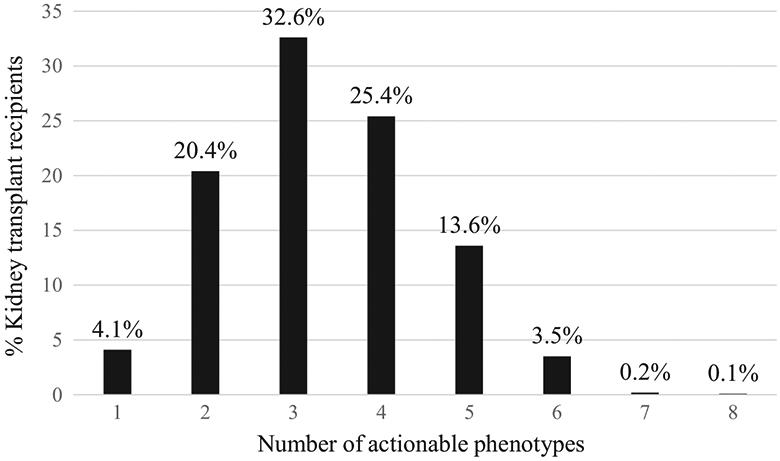
Transplant recipients with actionable pharmacogenomic phenotypes among 12 genes
4. DISCUSSION
Adverse drug reactions and poor drug efficacy are common problems encountered in transplantation. Pharmacogenomic markers are emerging as potential clinical tools to improve safety and efficacy by providing guidance for optimal dosing and/or drug selection. Every participant in our study had one or more actionable pharmacogenomic phenotypes which is consistent with observations in other studies.23 The majority (58%) carried three or four actionable pharmacogenomic phenotypes which may affect pharmacokinetics, pharmacodynamics or toxicity. When properly applied in practice, pharmacogenomic biomarkers reduce the frequency of toxicity due to supratherapeutic concentrations or avoidance in those at risk for hypersensitivity, and drug failure due to subtherapeutic concentrations of the active molecule.
The potential impact of pharmacogenetics has been evaluated in several studies. A recent Veteran Affairs (VA) study estimated the number of actionable variants in their population and analysed that along with known prescription use.24 They concluded that 54% of VA pharmacy users received at least one pharmacogenomic scientific level A evidence drug, 15.3% received two drugs, and 11.7% received 3 or more drugs. They concluded that testing would be most impactful for simvastatin, tramadol and warfarin in the veteran population. Vanderbilt University evaluated 56 medications in >50 000 medical home patients.25 They estimated that 64.8% of individuals would be exposed to at least one medication with an actionable variant within a 5-year period and that 398 severe adverse events could have been prevented by preemptive testing. A longitudinal evaluation of >73 million patients over a 4-year period with private insurance, Medicare Supplemental or Medicaid was conducted.26 At least one actionable pharmacogenomic drug was prescribed in approximately 50% of patients and 25%-33% received two or more actionable drugs. These drugs were mostly within the oncology, analgesic and cardiology therapeutic areas.
Multiple medications have sufficient scientific evidence to support evaluating pharmacogenomic testing to guide selection of drug and/or dose in transplantation. Supplement table S2 is a proposed list of variants that could be studied in transplantation, and the clinical implications and recommended action per CPIC guidelines. Tacrolimus is used in almost all kidney transplant immunosuppressive regimens, and CYP3A5*1 is the most actionable allele for this drug. It is associated with rapid tacrolimus metabolism, low trough concentrations and higher dose requirements, whereas CYP3A5*3, CYP3A5*6 and CYP3A5*7 are loss-of-function or low function variants associated with reduced metabolism and lower dose requirements.4 In those of African ancestry, the CYP3A5*1 allele is important and creates an intermediate metabolism phenotype in 52% (one *1 allele), extensive metabolism phenotype in 20.1% (two *1 alleles), and 27.6% (no *1 alleles) will carry the same genotype as most Caucasians. In Caucasians, only about 12% will carry one or more CYP3A5*1 and will require higher than expected tacrolimus doses which may be incorrectly attributed to non-adherence in the absence of pharmacogenomic knowledge. Therefore, race is an imprecise marker of tacrolimus metabolism and should not be used to select dose. Genotype-directed dosing for tacrolimus will be most useful in individuals who are intermediate or extensive CYP3A5 metabolizers, and in our study, this occurred in 11.6% and 72.1% of recipients of European and African ancestry, respectively. The results from the randomized-controlled trials evaluating genotype-directed dosing of tacrolimus in kidney transplantation have showed a large effect of variants on pharmacokinetics but the results for a reduction in acute rejection and long-term graft function have been mixed.27-29 In totality, the data in transplantation suggest that the use of genotype-directed dosing may reduce time to therapeutic range, the number of measured troughs and dose modifications, and unscheduled follow-up visits. Testing has been shown to be useful in the management of cardiovascular diseases, depression and anxiety and given the high prevalence of these conditions these variants are worthy of investigation in transplantation.
Implementation of pharmacogenomics in practice has been slow in transplantation. This is due to a number of factors including lack of clinical utility studies, knowledge of pharmacists and other health professionals on the scientific evidence and appropriate use of genotype information, lack of a clinical decision support systems for pharmacogenomics, misperceptions on the cost of testing, reimbursement concerns, for some lack of inhouse pharmacogenomic assays, lack of knowledge on how to select the best commercial testing platform and cost of institutional implementation of a programme. Predictive pharmacogenomic risk models for drug safety and efficacy are being developed and implemented in multiple therapeutic areas.30 Transplantation is an ideal setting for studying these strategies given the high medication burden and the use of multiple low therapeutic index drugs.
Pharmacogenomic testing can be performed preemptively or reactively. Preemptive testing is where results are placed in the medical record and an alert fires when an actionable drug-gene pair is detected at time of prescribing whereas reactive testing occurs ‘as needed’ usually after a patient develops an adverse drug effect or fails therapy. Many health systems implementing pharmacogenomics have employed preemptive, multigene testing since most patients are likely to be prescribed a drug with an actionable variant sometime in the future and the low marginal cost between a single gene test and multigene panel. In the case of transplantation, a preemptive, multigene panel prior to transplantation would have multiple advantages and future studies should test this approach. In the pretransplant period, a panel could studied as a tool to determine tacrolimus dose or select antimetabolite therapy. In the post-transplant period, the same panel could be studied for its utility in guiding statin therapy in those with non-responsive hyperlipidaemia or developing myalgias, antiplatelet therapy selection (clopidogrel vs other) for those with cardiovascular disease, select and/or determine proper dose of a selective serotonin reuptake inhibitors, tricyclic antidepressants, proton-pump inhibitors, 5-HT antagonists, voriconazole and warfarin, and identify patients at risk for azathioprine myelosuppression. CPIC guidelines are under development or consideration for opioids, antipsychotics, serotonin and norepinephrine reuptake inhibitors which are also used in transplant patients. In the future, pharmacogenomic markers will likely be measured using next-generation sequencing (NGS) platforms but at this time are not cost-effective. NGS has the advantage of measuring most variants allowing for the capture of all rare variants. This would be particularly useful for genes such as RYR1 where multiple rare risk variants (minor allele frequency <0.01) are known.
There are >250 drugs with pharmacogenomic biomarkers described under warnings and precautions, dose or adverse effect sections of drug labelling and others where testing is required prior to medication use by the FDA.2 Abacavir is an example of the later where testing for an HLA-B variant is required and clopidogrel labelling contains a boxed warning of diminished antiplatelet effectiveness in CYP2C19 poor metabolizers. In 2018, the FDA approved, for the first time, a direct to consumer pharmacogenomic assay by 23andMe. The assay is not approved for clinical use, and patients with potential actionable variants are advised to see their healthcare provider for confirmation and discussion of the findings. Given this and the number of health systems now conducting pharmacogenomic testing, it is likely that patients will present to transplantation with pharmacogenomic results in hand. Healthcare providers will be responsible for having sufficient knowledge to make prescription changes in response to actionable variants even though they did not order them.
There are a number of limitations in our study. Several actionable variants were not on our GWAS panel, and the number of actionable phenotypes in transplant recipients is higher than what we report here. Specifically, CYP2D6 is an important gene which metabolizes many drugs, has over 100 variants and copy number variants, and is included in several CPIC guidelines. Our GWAS panel also does not include the CYP2C9*8 and CYP2C9*11 variants which occur in African Americans and are associated with warfarin metabolism. We estimate that this would increase the number of intermediate or poor CYP2C9 metabolizers by ~15%. We also do not have a sufficient number of the relevant CYP2B6 variants on the panel to create the phenotypes for efavirenz as recommended by the CPIC guidelines.31 Another limitation is that we did not have sufficient number of patients of other ancestries, and these groups have allele frequencies that vary substantially from European and African. Therefore, our findings cannot be directly inferred to transplant centres with a significant number of Asians and Hispanic patients. We have used phenotype definitions used by CPIC; however, as the scientific data mature and new variants are discovered in these other populations, the phenotype definitions will evolve and become even more robust.
5. WHAT IS NEW AND CONCLUSION
This study highlights the potential utility of incorporating pharmacogenomic testing into drug decision-making in kidney transplantation, the potential to improve medication management and safety, and proposes a panel of variants that could be studied in transplant practice. We found that the majority of transplant recipients carried three or four actionable diplotypes/phenotypes and 17.4% had five or more actionable diplotypes/phenotypes among the 12 genes that would require dose modifications or medication changes if they were prescribed these medications. Pharmacogenomic testing is becoming more common due to the availability of a greater number of Clinical Laboratory Improvement Amendments (CLIA)-certified commercial platforms and low cost. In the future, pharmacogenomic variants will be likely assayed through whole genome sequencing. This provides the advantage that as new actionable variants are identified, they will be readily available from the sequence and eliminate the need for additional DNA collection and add on testing for new variants. Future research to define the clinical utility of testing, what patients should be tested, cost-effectiveness and finally the development of strategies to return results to patients such that the information will be available for future drug prescribing is needed.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the contributions of our generous patients. We also acknowledge the dedication and hard work of our coordinators at University of Alberta, Nicoleta Bobocea, Tina Wong, Adrian Geambasu and Alyssa Sader; University of Minnesota, Mandi DeGrote and Monica Meyers; Hennepin County Medical Center, Lisa Berndt; Mayo Clinic, Tom DeLeeuw; University of Alabama, Jacquelin Vaughn, Valencia Stephens and Tena Hilario. We also acknowledge the dedicated work of our research scientists Marcia Brott and Amutha Muthusamy.
Funding information
This project was supported by grants (U19-AI070119 and U01-AI058013) from the National Institute of Allergy and Infectious Disease (AM, PJ, AI, WO) and by Melendy Student Research Scholarships, University of Minnesota, College of Pharmacy (RP, TN). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or National Institute of Allergy and Infectious Disease.
APPENDIX A
GEN03 GENOMICS OF TRANSPLANTATION INVESTIGATORS
Arthur Matas, MD, Department of Surgery, University of Minnesota, Minneapolis, MN, matas001@umn.edu; J. Michael Cecka, MD, UCLA Immunogenetics Center, Los Angeles, CA, mcecka@ucla.edu; John Connett, PhD, Division of Biostatistics, University of Minnesota, Minneapolis, MN, john-c@biostat.umn.edu; Fernando G. Cosio, MD, Division of Nephrology, Mayo Clinic, Rochester, MN, Cosio.Fernando@mayo.edu; Robert Gaston, MD, Division of Nephrology, University of Alabama, Division of Nephrology, Birmingham, AL, rgaston@uab.edu; Rosalyn Mannon, MD, Division of Nephrology, University of Alabama, Division of Nephrology, Birmingham, AL, rmannon@uabmc.edu; Joseph P. Grande, MD, PhD, Mayo Clinic College of Medicine, Rochester, MN, Grande.Joseph@mayo.edu; Lawrence Hunsicker, MD, Nephrology Division, Iowa City, IA, lawrencehunsicker@uiowa.edu; Bertram Kasiske, MD, Division of Nephrology, Hennepin Healthcare, Minneapolis, MN, kasis001@umn.edu; and David Rush, MD, Health Sciences Center, Winnipeg MB, Canada, drush@exchange.hsc.mb.ca.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
GEN03 Investigators members presented in Appendix A.
DATA AVAILABILITY STATEMENT
The data with participant consent that support the findings of this study are openly available in dbGaP at https://www.ncbi.nlm.nih.gov/gap.
REFERENCES
- 1.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 2011;89(3):464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Table of Pharmacogenomic Biomarkers in Drug Labeling. https://www.fda.gov/media/124784/download. Accessed June 21, 2019. [Google Scholar]
- 3.Table of Pharmacogenetic Associations∣FDA. https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations. Accessed May 14, 2020. [Google Scholar]
- 4.Birdwell KA, Decker B, Barbarino JM.et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther. 2015;98(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson PA, Oetting WS, Brearley AM, et al. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation. 2011;91(3):300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oetting WS, Schladt DP, Guan W, et al. Genomewide association study of tacrolimus concentrations in African American kidney transplant recipients identifies multiple CYP3A5 alleles. Am J Transplant. 2016;16(2):574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oetting WS, Wu B, Schladt DP, et al. Genome-wide association study identifies the common variants in CYP3A4 and CYP3A5 responsible for variation in tacrolimus trough concentration in Caucasian kidney transplant recipients. Pharmacogenomics J. 2018;18(3):501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Relling MV, Schwab M, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin Pharmacol Ther. 2019;105:1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsey LB, Johnson SG, Caudle KE, et al. The Clinical Pharmacogenetics Implementation Consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96(4):423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin Pharmacol Ther. 2017;102(3):397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriyama B, Obeng AO, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin Pharmacol Ther. 2017;102(1):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.PharmGKB. https://www.pharmgkb.org. Accessed February 24, 2019.
- 16.Genes-Drugs - CPIC. http://cpicpgx.org/genes-drugs/. Accessed December 21, 2018.
- 17.Oetting WS, Wu B, Schladt DP, et al. Genetic variants associated with immunosuppressant pharmacokinetics and adverse effects in the DeKAF Genomics Genome-wide Association Studies. Transplantation. 2019;103(6):1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li YR, van Setten J, Verma SS, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med. 2015;7(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46(8):818–825. [DOI] [PubMed] [Google Scholar]
- 20.Guidelines - CPIC. https://cpicpgx.org/guidelines/. Accessed January 6, 2019.
- 21.Pharmacogenetics — KNMP.nl. https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica/pharmacogenetics-1/pharmacogenetics. Accessed June 21, 2019.
- 22.Amstutz U, Henricks LM, Offer SM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin Pharmacol Ther. 2018;103(2):210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush WS, Crosslin DR, Owusu-Obeng A, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100(2):160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chanfreau-Coffinier C, Hull LE, Lynch JA, et al. Projected prevalence of actionable pharmacogenetic variants and level A drugs prescribed among US veterans health administration pharmacy users. JAMA Netw Open. 2019;2(6):e195345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schildcrout JS, Denny JC, Bowton E, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92(2):235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samwald M, Xu H, Blagec K, et al. Incidence of exposure of patients in the United States to multiple drugs for which pharmacogenomic guidelines are available. PLoS One. 2016;11(10):e0164972.27764192 [Google Scholar]
- 27.Thervet E, Loriot MA, Barbier S, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther. 2010;87(6):721–726. [DOI] [PubMed] [Google Scholar]
- 28.Pallet N, Etienne I, Buchler M, et al. Long-term clinical impact of adaptation of initial tacrolimus dosing to CYP3A5 genotype. Am J Transplant. 2016;16(9):2670–2675. [DOI] [PubMed] [Google Scholar]
- 29.Shuker N, Bouamar R, van Schaik RHN, et al. A randomized controlled trial comparing the efficacy of Cyp3a5 genotype-based with body-weight-based tacrolimus dosing after living donor kidney transplantation. Am J Transplant. 2016;16(7):2085–2096. [DOI] [PubMed] [Google Scholar]
- 30.Lewis JP, Backman JD, Reny J, et al. Pharmacogenomic polygenic response score predicts ischemic events and cardiovascular mortality in clopidogrel-treated patients. Eur Hear J Cardiovasc Pharmacother. 2019. Epub ahead of print. 10.1093/ehjcvp/pvz045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desta Z, Gammal RS, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2B6 and efavirenz-containing antiretroviral therapy. Clin Pharmacol Ther. 2019;106(4):726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data with participant consent that support the findings of this study are openly available in dbGaP at https://www.ncbi.nlm.nih.gov/gap.


