Abstract
Sperm velocity is a key trait that predicts the outcome of sperm competition. By promoting or impeding sperm velocity, females can control fertilization via postcopulatory cryptic female choice. In Chinook salmon, ovarian fluid (OF), which surrounds the ova, mediates sperm velocity according to male and female identity, biasing the outcome of sperm competition towards males with faster sperm. Past investigations have revealed proteome variation in OF, but the specific components of OF that differentially mediate sperm velocity have yet to be characterized. Here we use quantitative proteomics to investigate whether OF protein composition explains variation in sperm velocity and fertilization success. We found that OF proteomes from six females robustly clustered into two groups and that these groups are distinguished by the abundance of a restricted set of proteins significantly associated with sperm velocity. Exposure of sperm to OF from females in group I had faster sperm compared to sperm exposed to the OF of group II females. Overall, OF proteins that distinguished between these groups were enriched for vitellogenin and calcium ion interactions. Our findings suggest that these proteins may form the functional basis for cryptic female choice via the biochemical and physiological mediation of sperm velocity.
Keywords: cryptic female choice, fertility, oocyte, sexual selection, sperm velocity
1 |. INTRODUCTION
In many species, females are capable of biasing sperm use and hence fertilization towards a preferred male via behavioural, morphological, physiological and biochemical mechanisms—a process called cryptic female choice (CFC) (Eberhard, 1996; Firman et al., 2017; Thornhill, 1983). This female-derived mechanism can function to increase the genetic quality of offspring (Jennions & Petrie, 2000; Kempenaers, 2007; Neff & Pitcher, 2005; Rosengrave et al., 2016; Simmons, 2005; Tregenza & Wedell, 2000). One potential mechanism of CFC through which sperm performance can be manipulated is via interactions with female reproductive fluids. These maternally derived fluids may be produced and released by the reproductive tract, or by fluids that surround the unfertilized ova. Recent research in both vertebrates and invertebrates indicates that sperm velocity, an important predictor of fertilization success across a range of different species (Gage et al., 2004; Gasparini et al., 2010), can be modified by female reproductive fluids (Yeates et al., 2013; Holt & Fazeli, 2015; Alonzo et al., 2016; Rosengrave et al., 2016; Lymbery et al., 2017; but see Kleppe et al., 2018)). For example, in broadcast spawners, such as the mussel (Mytilus galloprovincialis), sperm chemotaxis via exposure to ‘egg water’ differentially affects sperm velocity and patterns of motility depending on the combinatorial identities of the male and females contributing to the interaction (Lymbery et al., 2017).
In externally fertilizing fishes, where ova and sperm are simultaneously released into the aquatic environment, the maternally derived fluid surrounding the ova, known as ovarian fluid (OF), plays an important role in modulating sperm velocity across a range of fish species and therefore has been implicated as a mechanism of cryptic female choice (Alonzo et al., 2016; Gasparini & Pilastro, 2011; Poli et al., 2019; Rosengrave et al., 2008, 2016). For example, in Chinook salmon (Oncorhynchus tshawytscha), sperm velocity is determined via the interacting effect of male × female identity, and under sperm competition conditions, females bias fertilization towards the male with the fastest swimming sperm (Rosengrave et al., 2008, 2016). This female × male interaction also predicts embryos survival, highlighting the adaptive significance of this CFC mechanism (Rosengrave et al., 2016).
The mechanistic basis for postmating female × male interactions remains poorly understood, despite the mounting evidence that females play a role in moderating the outcome of sperm competition. Sperm attraction and sperm-activating proteins have been identified in broadcast spawning marine invertebrates (Kamei & Glabe, 2003; Stapper et al., 2015), and in the Pacific herring (Clupea pallasii; Oda et al., 1995). Likewise, egg surface proteins are known to mediate fertilization across disparate taxonomic groups, including rodents, fruit flies, sea urchins and seastars (Evans et al., 2012; Palmer & Swanson, 2012; Swanson & Vacquier, 2002; Vacquier & Swanson, 2011). As such, proteins are good candidates for mediating sperm function and, ultimately, fertilization success. Previous research from members of our team has qualitatively identified a marked variation between Chinook salmon females in the number and concentration of proteins present in OF (Johnson et al., 2014). Thus, a study that integrates OF, sperm velocity and fertilization outcomes was needed to advance our understanding of this phenomenon. Here, we have used a quantitative proteomic approach to investigate whether Chinook salmon OF proteome composition explains variation in sperm velocity and fertilization success.
2 |. MATERIALS AND METHODS
2.1 |. Study species and maintenance
As detailed in Rosengrave et al., 2016, chinook salmon were caught during their annual spawning run (April–May) in a trap located on the Kaiapoi River, a tributary of the Waimakariri River system, Canterbury, New Zealand. We studied sexually mature, 3-year-old, ‘hooknose’ males and females captured in 2011 (n = 17 males, 6 females). Fish were individually marked and maintained in a natural river-water raceway (12.5–13.8°C) at a hatchery (Salmon Smolt NZ, Canterbury, New Zealand) using standard husbandry procedures. Milt from males and OF from females was obtained as described previously (Rosengrave et al., 2016). A sample of OF from each female was immediately frozen in liquid nitrogen and stored at −80°C for protein analyses. All unfertilized ova, OF and milt samples were held at 4° C for up to five hours maximum. All animals were collected and maintained according to the standards of the Animal Ethics Committee for the University of Otago, New Zealand (protocol no. 13/10).
2.2 |. Measuring sperm velocity
The methods used to measure sperm velocity have previously been described in a publication by members of our team (Rosengrave et al., 2016). Sperm swimming speed for each male was measured at 10 s post-activation using a CEROS sperm tracker (v. 12, Hamilton-Thorne Research, Beverly, MA, USA). The sperm were assayed in a 100% concentration of OF. In total, an average of eight males was tested in the OF of each of the six females used in the study, resulting in the analysis of 54 male × female combinations. A comprehensive analysis of all male × female combinations was not possible due to practical sampling limitations (e.g. insufficient sperm or OF); however, we note that our experimental design allowed for the direct comparison of sperm swimming speed from males tested against both informative groups of females based on our proteomic analyses (see below). In line with previous studies from members of our team (Evans et al., 2013; Rosengrave et al., 2008, 2016; Rosengrave, Montgomerie et al., 2009; Rosengrave, Taylor et al., 2009), we use average path velocity (VAP, µm/s), which estimates the average velocity of sperm cells over a smoothed cell path, as our measure of sperm velocity. VAP ranged from 34.2 to 122 µm/s, with the maximum difference in VAP ranging from 27–88 µm/s within a given female’s OF. We note that VAP is strongly correlated with straight line velocity (VSL), the average velocity on a straight line between the start and the end point of the track, and curvilinear velocity (VCL µm/s), the actual velocity along the sperm’s trajectory, in this system (Evans et al., 2013).
2.3 |. Fertilization trials
We conducted in vitro fertilization (IVF) trials using the milt of each male combined with the unfertilized ova from each female—see Rosengrave et al. (2016) for IVF details. In total, six males were crossed with each of the six females used in the study, resulting in the analysis of 36 male × female combinations.
2.4 |. Ovarian fluid Sample preparation, iTRAQ labelling and peptide prefractionation
All OF samples were processed by a filter-aided sample preparation (FASP) following the procedure described by Wiśniewski et al., (2009). In brief, a sample volume of 100 µl of each OF (females 1–6, corresponding to tags 113–118 in the iTRAQ data) containing 200 µg of protein was supplemented with 100 µl denaturing solution containing 0.2% SDS, 40 mM Hepes pH 7.5, 2 mM EDTA, 2 mM EGTA, 10 mM tris(2-carboxyethyl)phosphine (TCEP), and one-fold protease inhibitor cocktail (Complete EDTA-free, Roche). The samples were denatured for 20 min at 45°C and then supplemented with 200 µl of detergent depletion solution containing 8 M urea and 100 mM triethylammonium bicarbonate (TEAB) in water. Each sample was loaded onto 0.5 ml Microcon centrifugal ultrafiltration units (10 kDa cut-off, Millipore) and washed three times with detergent depletion solution. Samples were then reduced, alkylated and buffer-exchanged into 200 mM TEAB by three consecutive washing steps. An aliquot of each sample was taken for protein measurement using the Bradford method. An aliquot of each sample containing 80 µg of protein was normalized to a volume of 40 µl with 100 mM TEAB and supplemented with 4 µg of trypsin in 5 µl of water. Proteins were digested over-night at 37°C and boosted with additional 4 µg of trypsin in the morning. After an incubation of 4 hr, peptides were labelled with the iTRAQ reagents according to the manufacturer’s protocol. The samples were then pooled, dried using a centrifugal vacuum concentrator and purified by solid phase extraction (SPE) on sep-pak C18 cartridges (Waters). Peptides of the pooled iTRAQ labelled sample were subjected to off-gel isoelectric focusing (IEF) using a 3,000 OFFGEL Fractionator (Agilent) and fractionated into 24 fractions along a pH gradient of 4 to 10. Each fraction was purified by SPE on sep-pak C18 cartridges, dried to completeness and reconstituted in 20 µl of 5% acetonitrile (ACN), 0.2% formic acid (FA) in water.
2.5 |. Liquid chromatography-coupled tandem mass spectrometry
Fractions were analysed in duplicate by nanoflow liquid chromatography-coupled LTQ Orbitrap tandem mass spectrometry. 5 µl aliquots were loaded onto an in-house packed emitter tip column (75 µm ID PicoTip fused silica tubing (New Objectives, Woburn, MA) packed with Luna C-18 material of 3µm bead size (Phenomenex) on a length of 15 cm) and peptides separated by a gradient of ACN in 0.2% aqueous FA. The gradient was developed in three steps, from (a) 5%–35% ACN (b) 35%–55% ACN and (c) 55%–95% ACN. The total gradient length including loading, washing and re-equilibration steps was 80 min for the first replicate and 100 min for the second replicate. The Orbitrap analyser was operated in full MS mode at a resolution of 60,000 at m/z 400 followed by eight data-dependent MS/MS scans. The eight data-dependent scans were performed as four collision-induced dissociation (CID) scans on the four strongest precursor ions for peptide identification followed by four high energy collision-induced dissociation (HCD) scans on the same four precursor ions for reporter ion quantification. CID fragment ions were detected in the LTQ ion trap and HCD fragment ions in the Orbitrap analyser at a resolution of 15,000 at m/z 400. Dynamic exclusion was enabled allowing two and three repeated fragmentation analyses on the same precursor ions during a 60 s time window for the first and second replicate measurement respectively.
2.6 |. Protein identification and iTRAQ quantitation
Raw spectra were processed and analysed using PEAKS Studio (Bioinformatics Solutions Inc.). Spectra were filtered to retain charges 2–8 and searched against the Oncorhynchus tshawytscha (assembly Otsh_v1.0) proteome using the PeaksDB search algorithm (Zhang et al., 2012). Parent mass error tolerance was set to 10.0 ppm and fragment mass error tolerance to 0.2 Da. Monoisotopic precursor mass search was employed with the cleavage enzyme set to Trypsin, allowing for up to 3 missed cleavages. Carbamidomethylation and iTRAQ 8plex modification for lysine and N-terminal were set as fixed modifications. Up to 3 variable post-translational modifications (PTMs) per peptide were allowed, which included oxidation (M), deamidation (NQ) and iTRAQ 8plex (Y). PeaksPTM (Han et al., 2011) and SPIDER (Han et al., 2005) searches were then used to identify additional PTMs and peptide spectral matches (PSMs) with single amino acid substitutions. Filtering criteria included −10LogP>= 23.7 (1% FDR) for peptides and −10LogP>= 20.0 (1.1% FDR) for proteins. This yielded a total of 28,904 PSMs across 4,629 peptides and 604 protein groups. Absolute normalized iTRAQ peak intensities were obtained from PEAKS Q, with a quantification mass tolerance of 0.1 Da, using −10LogP>= 23.7 (1% FDR) as the PSM filtering criteria. Protein abundance estimates were highly correlated between samples (pairwise average R = 0.86). The RAW data and peptide-spectrum match information are available via ProteomeXchange (PXD014551).
2.7 |. Proteome variation, statistical and gene ontology (GO) analyses
All statistical analyses was performed using R v 3.6.0 and 3.6.2 (R Core Development Team, 2019). Hierarchical clustering analyses were conducted using the Pvclust package (Suzuki & Shimodaira, 2006), including the Euclidean distance function with complete clustering and bootstrap analysis (10,000 iterations). Principle component analyses (PCA) of normalized protein intensities obtained from PEAKS Q were conducted using the FactoMineR package (Lê et al., 2008). A linear model was performed to assess correlations between protein abundance and sperm velocity data, using the lm function in R. Linear discriminant analysis (LDA) was performed using the lda function (Chang, 2015), with the protein abundance data centred and scaled. Significance of LDA correlations was assessed by MANOVA. The sperm velocity (VAP) analysis was restricted to proteins comprising at least 0.1% of the protein abundance to avoid issues associated with variance heterogeneity amongst low abundance proteins. GO terms were assigned to Chinook salmon genes using OrthoDB release 10 (Kriventseva et al., 2018) using functional annotations for the Actinopterygii taxonomic level. Chi-square with Yates’ correction was used to assess GO term enrichment. A two sample Mann–Whitney U test was used to compare sperm velocity and fertilization success between female cluster groups.
3 |. RESULTS
3.1 |. Ovarian fluid proteome composition
Tandem mass spectrometry analyses were conducted on Chinook salmon OF from six females collected during the 2011 spawning season. Analysis of two replicates (24 fractions each) resulted in 28,904 peptide-spectrum matches and the statistically robust identification of 604 protein groups (Table S1). To assess the quality of our proteome characterization, we compared our results to a previous Chinook salmon OF proteome (Johnson et al., 2014) and determined that 96.6% (168 of the 174) of previously identified proteins were in our expanded OF proteome. Our current OF proteome therefore represents an approximately 350% increase in the number of characterized OF proteins. To further assess our current dataset, we conducted a semi-quantitative comparison of the 168 proteins overlapping between studies and observed a highly significant correlation in abundance estimates (R = 0.95; p < .0001). Thus, our analysis achieved high proteome coverage and abundance estimates that were highly consistent with previous proteomic investigation.
Gene ontology (GO) revealed a proteome with high functional coherence, including 22 GO categories with > 20-fold enrichment relative to the remainder of the salmon genome (Table 1), and enrichment patterns that were highly concordant with previous analyses of salmon and trout OF (Johnson et al., 2014; Nynca et al., 2014). We identified 20 OF proteins with molecular weight below 15 kDa in this study. We have previously noted the importance of low-molecular-weight OF proteins on sperm VAP. VAP was comparable in sperm exposed to whole OF (mean 67.66 ± 17.86 SD, n = 5) and low-molecular-weight fractions (mean 66.60 ± SD 16.61; t4 = 0.37, p = .71), whereas VAP was lower when sperm were only exposed to high molecular weight OF fractions (mean 52.97 ± SD 12.78; t4 = −3.04, p = .0037; Johnson, unpublished data). The 20 OF proteins with molecular weight below 15 kDA included six with calcium ion binding annotations. Amongst them were five members of the S100 protein family, including S100-B-like, S100-A1-like, and ictacalcin-like proteins. Additionally, a suite of enriched immunity functional categories were observed, including leucocyte and lymphocyte mediated immune response, complement activation, adaptive immune response, regulation of response to wounding and acute inflammatory response. Enrichments were also observed in proteolysis regulation, a functional category commonly enriched in reproductive systems (Prokupek et al., 2008; Rettie & Dorus, 2012; Swanson et al., 2001, 2004), and categories related to blood plasma, including regulation of coagulation and haemoglobin complex.
TABLE 1.
Gene Ontology (GO) analysis of ovarian fluid proteome
| GO ID no. | Category name | Genome Count | Proteome Count | Enrichment | p-value |
|---|---|---|---|---|---|
| 0,004,869 | Cysteine-type endopeptidase inhibitor | 15 | 8 | 37.5 | 6.85E−57 |
| 0,072,376 | Protein activation cascade | 49 | 23 | 33.0 | 9.83E−153 |
| 0,002,443 | Leucocyte mediated immunity | 11 | 5 | 32.0 | 1.88E−28 |
| 0,002,449 | Lymphocyte mediated immunity | 11 | 5 | 32.0 | 1.88E−28 |
| 0,006,956 | Complement activation | 43 | 19 | 31.1 | 1.32E−117 |
| 0,002,460 | Adaptive immune response | 12 | 5 | 29.3 | 4.65E−26 |
| 0,050,818 | Regulation of coagulation | 10 | 4 | 28.1 | 2.96E−19 |
| 0,005,579 | Membrane attack complex | 10 | 4 | 28.1 | 2.96E−19 |
| 0,005,833 | Haemoglobin complex | 29 | 11 | 26.7 | 1.98E−56 |
| 0,006,959 | Humoral immune response | 57 | 21 | 25.9 | 1.09E−107 |
| 0,051,181 | Cofactor transport | 14 | 5 | 25.1 | 2.70E−22 |
| 1,903,034 | Regulation of response to wounding | 12 | 4 | 23.4 | 4.68E−16 |
| 0,004,859 | Phospholipase inhibitor activity | 10 | 3 | 21.1 | 3.00E−10 |
| 0,051,920 | Peroxiredoxin activity | 10 | 3 | 21.1 | 3.00E−10 |
| 0,002,250 | Adaptive immune response | 17 | 5 | 20.7 | 2.65E−18 |
| 0,004,866 | Endopeptidase inhibitor activity | 123 | 36 | 20.6 | 3.82E−146 |
| 0,010,951 | Negative endopeptidase regulation | 125 | 36 | 20.3 | 1.32E−143 |
| 0,002,526 | Acute inflammatory response | 14 | 4 | 20.1 | 9.11E−14 |
| 0,046,930 | Pore complex | 14 | 4 | 20.1 | 9.11E−14 |
Protein abundance analysis revealed that OF composition was dominated by a small number of highly abundant proteins. This included seven proteins that account for >50% and 101 proteins for >90% of total protein, respectively. The most abundant protein, vitellogenin-like, comprises 20.7% of OF protein. Additionally, 26 of the most abundant 34 proteins are blood plasma proteins and account for 45.4% of OF proteins. The majority of the remaining abundant proteins belong to two protein families: vitellogenins (3 proteins; 24.2% of total protein) and zona pellucida sperm-binding proteins (2 proteins; 3.9% of total protein).
3.2 |. Ovarian fluid proteome clustering and sperm velocity
As in Rosengrave et al. (2008), we observed evidence of male × female interactions (Figure S1), suggesting that VAP varies within each female’s OF according to individual male identity, though we do not have the necessary sample size and statistical power to analyse male × female interactions in relation to VAP. Instead, our study focuses on the more pronounced effects of how OF variation affects VAP. Hierarchical clustering analysis of OF proteome variation revealed two statistically robust clades (group I: females 1, 2 and 5; group II: females 3, 4 and 6; Figure 1a; confidence > 0.95 for each clade). Statistical analysis of VAP between groups revealed a significantly higher VAP in OF from females in group I (W = 234.5, p = .023; n = 23 male × female tests using group I females and 32 male × female tests using group II females) (Figure 1b). This relationship strengthened when the analysis was restricted to males that were tested in females from both groups (W = 367, p = .007; n = 20 male × female tests using group I females and 25 male × female tests using group II females). Lastly, a direct comparison of the 5 males that were tested against at least 2 of the 3 females within each group revealed a consistently higher average VAP in group I female OF (Figure 1c; W = 123, p = .05; n = 13 male × female tests using group I females and 13 male × female tests using group II females). In contrast, no significant difference was observed between groups for the percent of eggs fertilized (group I mean = 44.0 ± 5.12, n = 18; group II mean = 49.5 ± 5.92, n = 18; W = 136, p = .42), and this does not change when we restrict the dataset to only males that were crossed with both group I and group II females (group I mean = 46.0 ± 3.75, n = 155, group II mean = 53 ± 7.05, n = 13; W = 66, p = .23).
FIGURE 1.
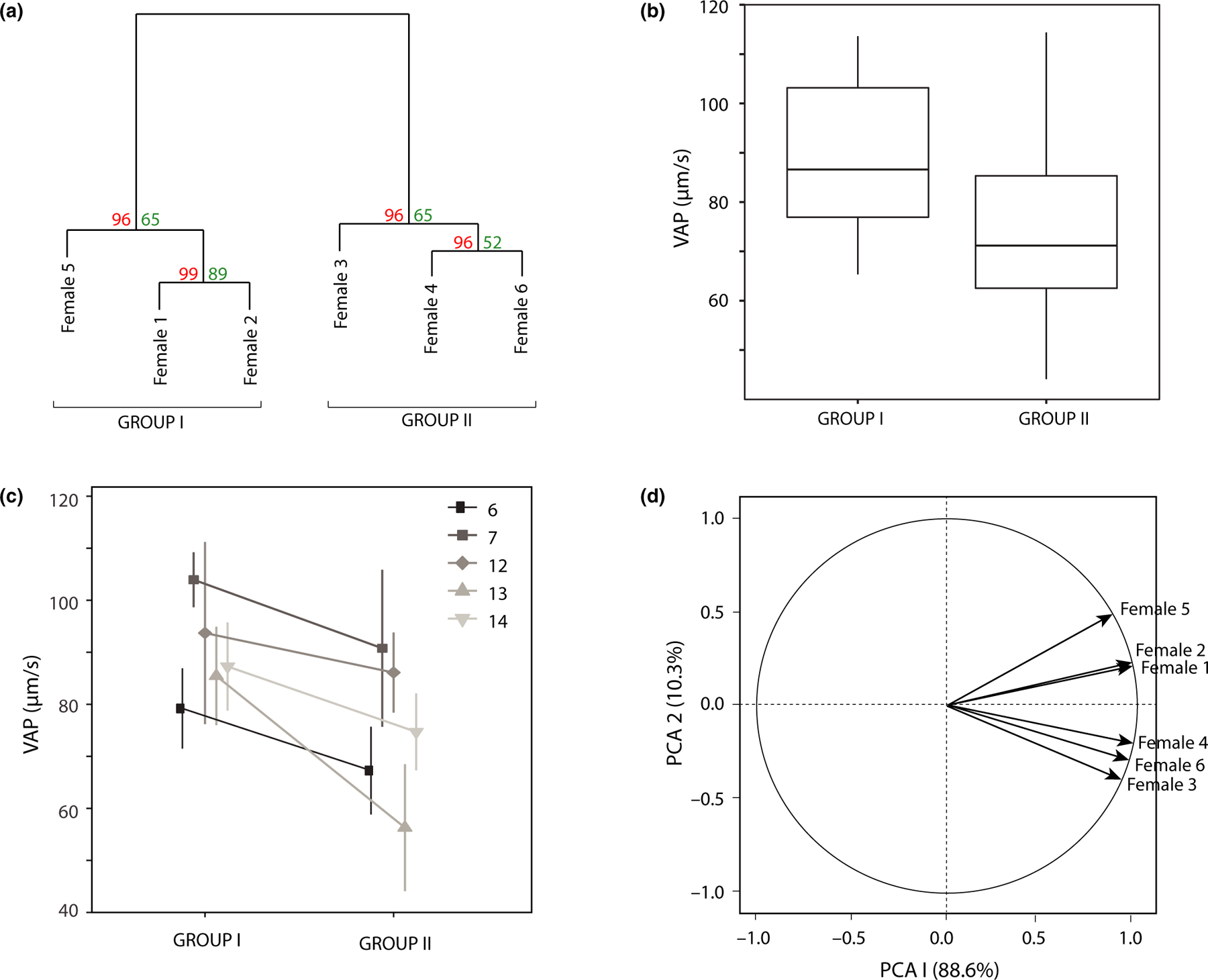
Ovarian fluid proteome variation analyses. (a) Hierarchical clustering analysis of OF proteomes using the Euclidean distance approach. Unbiased p-values (red; left) and bootstrap confidence (green; right) are indicated. (b) Whisker plot comparison of average path velocity (VAP) distributions of sperm exposed to OF fluid from the groups of females identified by hierarchical clustering above. (c) Mean ± standard error of VAP of the 5 males that were tested in at least 2 females in each OF cluster group. (d) Principal component (PC) analysis of OF proteomes. Protein abundances from all OF proteome were significantly positively correlated with PC1. Group I OF proteomes were significantly positively correlated, and group II OF proteomes were significantly negatively correlated with PC2, respectively
Next, a linear discriminant analysis (LDA) was used to identify proteins which best predicted OF group membership and several trends were observed (Table 2). First, LDA coefficients were negatively skewed overall, including 67 of the 84 proteins with significantly different abundances between groups based on MANOVA analysis. Proteins with negative LDA coefficients were more abundant in group II and, therefore, associated with decreased VAP. Amongst those with the greatest predictive power were alpha actinin-2 and alpha actinin-4 and filamin-B. These protein families are both involved in the remodelling of the actin cytoskeleton in mammalian spermatozoa (Roa-Espitia et al., 2016). Second, vitellogenin-like, the most abundant OF protein by far, was amongst the most predictive proteins that were more abundant in group I. OF proteomes in group I were therefore characterized by large quantitative increases in vitellogenin-like protein (and a small subset of other proteins), whereas OF proteomes in group II were characterized by a relative increase in abundance of a broad suite of proteins. As such, OF proteomes robustly cluster into two groups associated with differential sperm VAP and can be distinguished by the abundance of a restricted set of OF proteins.
TABLE 2.
Discriminant analysis of ovarian fluid proteins
| ID | Protein | LDA Coefficienta | p-value | Abundance rank |
|---|---|---|---|---|
| 112,265,290 | Alpha actinin−4-like | −0.039 | .0001 | 324 |
| 112,255,105 | Filamin-b | −0.017 | .0014 | 266 |
| 112,240,811 | Alpha actinin−1-like | −0.015 | .0048 | 334 |
| 112,232,253 | Prothrombin-like | −0.012 | .0080 | 69 |
| 112,264,761 | Ectonucleotide pyrophosphatase family 2-like | −0.012 | .0069 | 222 |
| 112,247,086 | Renin receptor-like | −0.011 | .0029 | 280 |
| 112,256,565 | Cartilage acidic protein 1 | −0.010 | .0232 | 307 |
| 112,242,209 | Coagulation factor vii-like | −0.009 | .0194 | 50 |
| 112,253,690 | Hemopexin-like | −0.009 | .0335 | 11 |
| 112,261,737 | Protein z-dependent protease inhibitor-like | −0.009 | .0057 | 271 |
| 112,217,447 | Proteasome subunit alpha type−6 | −0.009 | .0392 | 347 |
| 112,228,943 | Complement factor b-like | −0.008 | .0206 | 189 |
| 112,225,673 | Tubulin alpha chain-like | −0.008 | .0374 | 291 |
| 112,227,016 | Serotransferrin isoform x2 | −0.008 | .0471 | 4 |
| 112,217,872 | Alpha-1-antitrypsin homolog | −0.007 | .0871 | 7 |
| 112,233,757 | Wd repeat-containing protein 1-like | −0.007 | .0316 | 352 |
| 112,246,891 | Clathrin heavy chain 1 isoform x5 | 0.007 | .0075 | 346 |
| 112,229,908 | Palmitoyl-protein thioesterase 1-like | 0.008 | .0394 | 299 |
| 112,243,248 | Vitellogenin-like partial | 0.009 | .0276 | 9 |
| 112,214,915 | Papilin-like | 0.009 | .0118 | 393 |
| 112,251,203 | Vitellogenin-like | 0.009 | .0364 | 1 |
| 112,228,378 | Ribosomal oxygenase 2 | 0.010 | .0250 | 471 |
| 112,223,111 | Triosephosphate isomerase b-like | 0.011 | .0034 | 442 |
Proteins amongst the top 5% of LDA coefficients that also exhibited significant abundance differences between groups based on MANOVA.
3.3 |. Ovarian fluid proteome variation
To complement the hierarchical clustering and discriminant analysis, we analysed overall proteome variation using a principal component (PC) analysis. This analysis revealed that nearly all variation (98.85%) was captured by PC1 and 2 (Figure 1d). All OF proteomes were highly correlated with PC1, and the most abundant proteins had the highest squared cosines scores, whereas the least abundant have the lowest coordinates. We therefore concluded that PC1 captured shared variation in relative protein abundance across all samples. PC2, however, revealed loading relationships across samples that recapitulated the results from the hierarchical clustering analysis. Specifically, Females 1, 2 and 5 have positive loadings that are significantly correlated with OF protein abundances (p = 7.04E-7, 3.66E-6 and 9.57E-31, respectively) and Females 3, 4 and 6 have negative loadings that are inversely correlated with OF protein abundance (p = 7.60E-21, 4.16E-6 and 2.30E-11, respectively). This complementary analysis therefore recapitulated the groups identified by our hierarchical cluster analysis and revealed inter-proteome variation that was captured almost entirely by PC2.
To further investigate the variation captured by PC2, we performed a correlation analysis between PC2 loadings and sperm velocity across the six females in the study. PC2 was positively correlated, although not significantly, with sperm VAP (R = 0.59, p = .22). Next, correlations were conducted between PC2 loadings and protein abundance on a protein-by-protein basis, to delineate proteins that contribute most to variation in PC2. Not unexpectedly, these protein-by-protein relationships were significantly correlated with the results of our discriminant analysis (R = 0.81; p < .0001). This analysis identified seven proteins that positively correlated with PC2 loadings and, therefore, were more abundant in group I. Five of these were also identified in the LDA analysis as significantly more abundant in group I. These include all three highly abundant vitellogenin proteins. 119 proteins were negatively correlated with PC2 loadings and were therefore more abundant in group II. 60 of these were also identified in the LDA analysis. These proteins were enriched for peptidase activity (6.45X, p < .0001), calcium ion binding (2.92X, p = .0017), complement activation (66.98X, p < .0001), haemostasis (26.09X, p < .0001), a response to blood vessel injury and bleeding (19.12X, p < .0001) and actin filament organization (6.86X, p < .0001). Also notable amongst this protein set was sex hormone-binding globulin-like protein, which has been shown to decline prior to the sea bass reproductive season and may be correlated with seasonality or female quality (Miguel-Queralt et al., 2007). In summary, OF proteome variation captured by PC2 differentiates the OF proteome groups previously identified, was generally associated with a reproductive phenotype, and identifies additional proteins of potential relevance to OF fluid interactions with sperm.
3.4 |. Protein abundance associations with sperm VAP
To directly assess the association of individual protein abundance to sperm VAP, we performed a general linear model regression on a protein-by-protein basis. This revealed 12 proteins that achieved nominal significance at 0.05 without multiple testing corrections (Table 3). Consistent with previous analyses, all of these were negatively associated with VAP (i.e. more abundant where VAP is low) and found at higher abundances in group II. Interestingly, a homolog of pigment epithelium-derived factor-like protein (PEDF), the second most abundant of the significant proteins, has been suggested to play a role in the protection of spermatozoa in storage in rats (Conte et al., 2018). Additionally, a homolog of lipocalin-like protein mediates motility in mice (Lee et al., 2003). Although not significant, the top two positively correlated proteins with VAP were vitellogenin-like proteins, consistent with our complementary statistical approaches.
TABLE 3.
Linear model analysis of protein abundance and sperm VAP
| ID | Protein | p-value | Abundance rank |
|---|---|---|---|
| 112,255,040 | Phospholipid transfer protein-like | .0056 | 89 |
| 112,249,831 | Slit homolog 1 protein-like | .0056 | 49 |
| 112,264,335 | Serine-aspartate repeat protein F-like | .0083 | 6 |
| 112,231,811 | Fibrinogen gamma chain-like isoform X1 | .0103 | 47 |
| 112,228,606 | Pigment epithelium-derived factor-like | .0110 | 8 |
| 112,246,615 | Complement C3-like | .0145 | 22 |
| 112,215,456 | Lipocalin-like | .022 | 20 |
| 112,249,904 | Complement c1q-like protein 2 | .0253 | 17 |
| 112,257,419 | Complement component C9 | .0257 | 36 |
| 112,222,572 | Complement C3-like | .0274 | 91 |
| 112,240,139 | Fibrinogen beta chain | .0293 | 74 |
| 112,225,536 | Plastin-2 | .0227 | 104 |
4 |. DISCUSSION
In external fertilizers, OF has been demonstrated to mediate sperm velocity according to male and female identity and bias fertilization outcomes. Despite evidence of OF proteomic variation (this study and Johnson et al., 2014), there has been no previous investigation of the specific OF proteins that govern these specific male × female interactions. Using quantitative proteomics, we found that six OF proteomes robustly cluster into two groups using multiple, independent analyses (Figure 1; Tables 1–3). These groups can be distinguished by the abundance of a restricted set of OF proteins (Table 2) that associate with differential sperm velocity (VAP), but not fertilization success. Specifically, we found that exposure of sperm to OF from females that clustered in group I resulted in higher VAPs compared to sperm from the same males exposed to the OF from group II females. In order for OF proteins to be involved in CFC, they need to have a different effect on individual males. Although we do not show this directly, the OF proteins that distinguished between the two female cluster groups were enriched for vitellogenin and calcium ion interactions, and these proteins may potentially be involved in CFC. It is noteworthy that we did not observe associations with fertilization success in light of the previously demonstrated relationship between both of these phenotypes and sperm VAP (Rosengrave et al., 2016). This pattern could be due to the proximate relationship between OF and sperm phenotype via a direct physical interaction, although we cannot rule out that the absence of an effect could be to the limited number of wild-run males and females returning. Future studies will be needed to further explore potential associations between OF proteomic variation and fertilization outcomes.
Amongst the most abundant proteins we identified were two vitellogenin-like proteins, which were more abundant in the OF from group I females, that tended to increase sperm VAP. We described numerous vitellogenin family members in Chinook salmon OF, as we did in our previous study (Johnson et al., 2014), and these proteins are a common component of the OF of many species, including rainbow trout (Nynca et al., 2014; Rime et al., 2004) and Persian sturgeon (Keyvanshokooh & Vaziri, 2008). Vitellogenins are the main egg yolk precursor proteins in oviparous species and contribute a substantial portion of both the lipo- and phosphoproteins to the yolk. It is noteworthy that one vitellogenin subcomponent, phosvitin, is a potent chelator of metals such as Ca2+ (Mecham & Olcott, 1949) and mediates oxidative stress (Moon et al., 2014). Although we are unaware of any evidence that these vitellogenin characteristics influence sperm (but see below for a discussion on Ca2+ and sperm motility), vitellogenin has been demonstrated to be a binding partner for proteases in ascidian sperm (Akasaka et al., 2014). Therefore, OF vitellogenin protein variation should be investigated further to determine if it has a role in mediating sperm motility and cryptic female choice.
Amongst group II females, whose OF tended to reduce sperm VAP, alpha actinin and the actin binding protein filamin had the greatest predictive power. Filamins organize the actin cytoskeleton and maintain extracellular matrix connections by anchoring actin filaments to transmembrane receptors (Razinia et al., 2012). In addition to binding actin, filamins have more than 90 other binding partners including intracellular signalling molecules, receptors, ion channels, transcription factors, and cytoskeletal and adhesion proteins and are implicated in the regulation of a diverse array of cellular functions including motility, maintenance of cell shape and differentiation. There is some evidence that filamins might be important for sperm maturation and function from studies disrupting genes with which filamins interact (Jiang et al., 2008). However, a more compelling role exists; both alpha actinin and filamin are involved in the remodelling of the actin cytoskeleton in mammalian spermatozoa (Roa-Espitia et al., 2016). This remodelling is necessary for the acrosome reaction to function normally and for sperm to achieve adequate motility (Roa-Espitia et al., 2016). This process requires the assistance of numerous accessory proteins, but recent work suggests alpha actinin is important for the formation of a focal adhesion complex necessary for actin polymerization and remodelling during sperm capacitation (Roa-Espitia et al., 2016). The direction of abundance is however unexpected, with both proteins being more abundant in OF associated with reduced sperm VAP. In mammalian sperm, capacitation results in hypermotility, thus assuming this process is similar in fish we might have anticipated these proteins would exhibit increased abundance in group I. Further work is needed to explore this association.
Our data, and that from earlier studies (Rosengrave, Taylor et al., 2009), indicate that the regulation of calcium ions (Ca2+) is likely a key driver of the differential sperm function we observe amongst OFs. Our prior work demonstrated a significant negative relationship between duration of sperm movement and Ca2+ concentrations in OF (Rosengrave, Taylor et al., 2009), but was unable to explain the strong male by female interaction we observed in VAP (Rosengrave et al., 2008, 2016). Now, through our proteomic work, we have identified numerous proteins amongst our most significant hits that bind calcium (e.g. vitellogenins, B-cadherin-like protein and several others) or whose interactions are altered by the concentration of Ca2+ present (e.g. filamin-actin interactions). These new data provide a potential window into how variation in the abundance and perhaps diversity of these binding proteins might regulate free Ca2+ concentrations, mediating the difference in male sperm performance we observe amongst OF from different females.
In many species, sperm flagellar movement is regulated by calcium with changes in the concentration of intracellular Ca2+ reducing or blocking sperm function (Correia et al., 2015; Parodi, 2014; Sun et al., 2017). In mammals, Ca2+-independent flagellar dynein and ATP orchestrate the low-amplitude sinusoidal-activated motility of the tail, but hyperactivated motility requires Ca2+ to enter the sperm through specific channels, such as Catsper, as part of the capacitation process (Qi et al., 2007). Activation of sperm motility in teleost fish also requires Ca2+ influx (Cosson, 2019), but because teleost fish lack Catsper channels, Ca2+ signalling must involve a different process to those of marine invertebrates and mammals (Fechner et al., 2015). In fish, sperm flagella are able to respond to regulation by free calcium concentration by altering their beating pattern (Cosson, 2019). However, another potential mechanism may lie in the regulation of the focal adhesion complex, that both alpha actinin and filamin contribute to, which is important for capacitation and thus hyperactivation of mammalian sperm (Roa-Espitia et al., 2016). The interaction between filamin and actin is reduced in the presence of high Ca2+ and calmodulin (Nakamura et al., 2005); thus in the presence of high concentrations of Ca2+, we might observe reductions in the formation of the focal adhesion complex and thus hyperactivation in salmon sperm. Intraspecific variation in these complexes (and other Ca2+ proteins) in both sperm, OF and eggs might in part explain the pattern of male by female interactions we have described (Gessner, Johnson et al., 2017; Gessner, Nakagawa et al., 2017; Rosengrave et al., 2008, 2016).
We also identified significant functional enrichments amongst a broad set of immunity proteins, including several proteins with homology to the innate complement system (C1, C3 and C9), which are maternally transferred in rainbow trout (Løvoll et al., 2006). Complement components are important to nonimmunological processes as well, with complement protein C3 thought to function in the clearance of dead or dysfunctional sperm in the human female reproductive tract and other events leading up to fertilization (Anderson et al., 1993). The extent to which immunity proteins in OF contribute to ‘standard’ immunological processes or mechanisms solely related to reproduction (e.g. CFC), as has been suggested of reproductive proteins in sperm and seminal fluid (Dorus et al., 2012; Rowe et al., 2020), warrants further exploration.
It is worth noting that highly abundant proteins like vitellogenin (and albumin) could influence the viscosity of ovarian fluid as they do in blood (e.g. Joles et al., 1997; Saunders et al., 2000), which in turn could influence sperm swimming speed. Ovarian fluid viscosity is known to vary across females (Rosengrave, Taylor et al., 2009) and to influence sperm motility (Brokow, 1966; Butts et al., 2017; Hirai et al., 1997). Although we did not control for protein concentration during our CASA measurements, our proteomic analysis was done on equivalent amounts of protein per sample.
The actual mechanism(s) through which sperm function is impaired and/or enhanced to influence the sperm selection that we have observed in Chinook salmon remains unknown. Using quantitative proteomic data, we have identified a relatively small number of proteins in OF that explain the observed variation in sperm velocity, establishing a subset of proteins that warrant further examination. Many of these have direct or indirect interactions with Ca2+, and it may be that interaction between these two components of ovarian fluid mediates the male by female interactions that members of our team have previously described (Gessner, Johnson et al., 2017; Gessner, Nakagawa et al., 2017; Rosengrave et al., 2008, 2016). In addition to providing an inroad into the mechanistic basis of CFC, these OF proteins and knowledge about their interactions with other molecules (e.g. Ca2+) could be of utility in aquaculture and conservation of endangered fish species where sperm number or quality is limited.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marsha Villarroel for assistance with laboratory work. We thank the hatchery staff at Salmon Smolt, NZ, in particular Ben Divett, Karl French, Errol White, Tom Gough and Luke Price, for providing facilities, fish husbandry, gamete handling assistance and advice. We also thank Janine Wing, Tanya Blakely, Katherine McBride and Sara Ferreira for assistance in the field. This work was supported in part by grants from the Royal Society of New Zealand Marsden Fund (UOO913 to NJ), the Royal Society of New Zealand Fast-Start Grant (UOO1209 to PR), the Eunice Shriver National Institute for Child Health and Human Development (R21-HD088910 to SDX) and the National Science Foundation (DEB-1655840 to SD).
Funding information
National Science Foundation, Grant/Award Number: DEB-1655840 ; Royal Society of New Zealand, Grant/Award Number: UOO913, UOO1209
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/jeb.13717.
DATA AVAILABILITY STATEMENT
The 2011 sperm velocity and fertilization success data analysed in this study are available on dryad (http://dx.doi.org/10/5061/rspb.2016.0001). The RAW MS data and peptide-spectrum match information is available via ProteomeXchange (PXD014551). All other data are provided within the manuscript or supplemental material.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- Akasaka M, Kato KH, Kitajima K, & Sawada H (2014). Novel isoform of vitellogenin expressed in eggs is a binding partner of the sperm proteases, HrProacrosin and HrSpermosin, in the ascidian Halocynthia roretzi In Sawada H, Inoue N, & Iwano M (Eds.), Sexual Reproduction in Animals and Plants (pp. 131–139). Springer; Japan. [Google Scholar]
- Alonzo SH, Stiver KA, & Marsh-Rollo SE (2016). Ovarian fluid allows directional cryptic female choice despite external fertilization. Nature Communications, 7, 12452 10.1038/ncomms12452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Abbott AF, & Jack RM (1993). The role of complement component C3b and its receptors in sperm-oocyte interaction. Proceedings of the National Academy of Sciences, 90, 10051–10055. 10.1073/pnas.90.21.10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokow CJ (1966). Effects of increased viscosity on the movements of some invertebrate spermatozoa. Journal of Experimental Biology, 45, 113. [DOI] [PubMed] [Google Scholar]
- Butts IAE, Prokopchuk G, Kašpar V, Cosson J, & Pitcher TE (2017). Ovarian fluid impacts flagellar beating and biomechanical metrics of sperm between alternative reproductive tactics. The Journal of Experimental Biology, 220, 2210 10.1242/jeb.154195 [DOI] [PubMed] [Google Scholar]
- Chang J (2015). lda: Collapsed Gibbs Sampling Methods for Topic Models. R package version 1.4.2. Retrieved from https://rdrr.io/cran/lda/
- Conte MI, Cabrillana ME, Saez Lancellotti TE, Simon L, Funes AK, Cayado-Gutiérrez N, Tagle-Delgado MG, Vincenti AE, Lopez ME, Pietrobon EO, Fornes MW, & Monclus MA (2018). Pigment epithelium derived factor (PEDF) expression in the male tract of Wistar rats. Biochemical and Biophysical Research Communications, 504, 257–262. 10.1016/j.bbrc.2018.08.165 [DOI] [PubMed] [Google Scholar]
- Correia J, Michelangeli F, & Publicover S (2015). Regulation and roles of Ca2+ stores in human sperm. Reproduction (Cambridge, England), 150, R65–R76. 10.1530/REP-15-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson J (2019). Fish sperm physiology: structure, factors regulating motility, and motility evaluation In Morisawa M (Ed.), Sperm Cell Research in the 21st Century: Historial Discoveries to New Horizons (pp. 1–26). IntechOpen. [Google Scholar]
- Dorus S, Skerget S, & Karr TL (2012). Proteomic discovery of diverse immunity molecules in mammalian spermatozoa. Systems Biology in Reproductive Medicine, 58, 218–228. 10.3109/19396368.2012.700442 [DOI] [PubMed] [Google Scholar]
- Eberhard WG (1996). Female control: Sexual selection by cryptic female choice. Princeton University Press. [Google Scholar]
- Evans JP, Garcia-Gonzalez F, Almbro M, Robinson O, & Fitzpatrick JL (2012). Assessing the potential for egg chemoattractants to mediate sexual selection in a broadcast spawning marine invertebrate. Proceedings of the Royal Society B: Biological Sciences, 279, 2855–2861. 10.1098/rspb.2012.0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Rosengrave P, Gasparini C, & Gemmell NJ (2013). Delineating the roles of males and females in sperm competition. Proceedings of the Royal Society B: Biological Sciences, 280, 20132047 10.1098/rspb.2013.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechner S, Alvarez L, Bönigk W, Müller A, Berger TK, Pascal R, Trötschel C, Poetsch A, Stölting G, Siegfried KR, Kremmer E, Seifert R, & Kaupp UB (2015). A K(+)-selective CNG Channel Orchestrates Ca(2+) Signalling in Zebrafish Sperm, eLife, 4, e07624 10.7554/eLife.07624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman RC, Gasparini C, Manier MK, & Pizzari T (2017). Postmating female control: 20 years of cryptic female choice. Trends in Ecology & Evolution, 32, 368–382. 10.1016/j.tree.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage MJG, Macfarlane CP, Yeates S, Ward RG, Searle JB, & Parker GA (2004). Spermatozoal traits and sperm competition in Atlantic salmon: Relative sperm velocity is the primary determinant of fertilization success. Current Biology, 14, 44–47. 10.1016/j.cub.2003.12.028 [DOI] [PubMed] [Google Scholar]
- Gasparini C, Simmons LW, Beveridge M, & Evans JP (2010). Sperm swimming velocity predicts competitive fertilization success in the green swordtail Xiphophorus helleri. PLoS One, 5, e12146 10.1371/journal.pone.0012146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini G, & Pilastro A (2011). Cryptic female preference for genetically unrelated males is mediated by ovarian fluid in the guppy. Proceedings of the Royal Society B: Biological Sciences, 278, 2495–2501. 10.1098/rspb.2010.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner C, Johnson SL, Fisher P, Clarke S, Rutherford K, Symonds J, & Gemmell NJ (2017). Male–female relatedness at specific SNP-linkage groups influences cryptic female choice in Chinook salmon (Oncorhynchus tshawytscha). Proceedings of the Royal Society B: Biological Sciences, 284, 20170853 10.1098/rspb.2017.0853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner C, Nakagawa S, Zavodna M, & Gemmell NJ (2017). Sexual selection for genetic compatibility: The role of the major histocompatibility complex on cryptic female choice in Chinook salmon (Oncorhynchus tshawytscha). Heredity, 118, 442 10.1038/hdy.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, He L, Xin L, Shan B, & Ma B (2011). PeaksPTM: Mass spectrometry-based identification of peptides with unspecified modifications. Journal of Proteome Research, 10, 2930–2936. 10.1021/pr200153k [DOI] [PubMed] [Google Scholar]
- Han Y, Ma B, & Zhang K (2005). SPIDER: Software for protein identification from sequence tags with de novo sequencing error. Journal of Bioinformatics and Computational Biology, 3, 697–716. 10.1142/s0219720005001247 [DOI] [PubMed] [Google Scholar]
- Hirai M, Cerbito WA, Wijayagunawardane MPB, Braun J, Leidl W, Ohosaki K, Matsuzawa T, Miyazawa K, & Sato K (1997). The effect of viscosity of semen diluents on motility of bull spermatozoa. Theriogenology, 47, 1463–1478. 10.1016/S0093-691X(97)00136-2 [DOI] [PubMed] [Google Scholar]
- Holt WV, & Fazeli A (2015). Do sperm possess a molecular passport? Mechanistic insights into sperm selection in the female reproductive tract. Molecular Human Reproduction, 21, 491–501. 10.1093/molehr/gav012 [DOI] [PubMed] [Google Scholar]
- Jennions MD, & Petrie M (2000). Why do females mate multiply? A review of the genetic benefits. Bioliological Reviews of the Cambridge Philosophical Society, 75, 21–64. 10.1017/s0006323199005423 [DOI] [PubMed] [Google Scholar]
- Jiang S-T, Chiou Y-Y, Wang E, Lin H-K, Lee S-P, Lu H-Y, Wang C-K-L, Tang M-J, & Li H (2008). Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Human Molecular Genetics, 17, 3368–3379. 10.1093/hmg/ddn231 [DOI] [PubMed] [Google Scholar]
- Johnson SL, Villarroel M, Rosengrave P, Carne A, Kleffmann T, Lokman PM, & Gemmell NJ (2014). Proteomic analysis of chinook salmon (Oncorhynchus tshawytscha) ovarian fluid. PLoS One, 9, e104155 10.1371/journal.pone.0104155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joles JA, Willekes-Koolschijn N, & Koomans HA (1997). Hypoalbuminemia causes high blood viscosity by increasing red cell lysophosphatidylcholine. Kidney International, 52, 761–770. 10.1038/ki.1997.393 [DOI] [PubMed] [Google Scholar]
- Kamei N, & Glabe CG (2003). The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes & Development, 17, 2502–2507. 10.1101/gad.1133003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempenaers B (2007). Mate choice and genetic quality: A review of the heterozygosity theory. Advances in the Study of Behavior, 37, 189–278. 10.1016/S0065-3454(07)37005-8 [DOI] [Google Scholar]
- Keyvanshokooh S, & Vaziri B (2008). Proteome analysis of Persian sturgeon (Acipenser persicus) ova. Animal Reproduction Science, 109, 287–297. 10.1016/j.anireprosci.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Kleppe SA, Nordeide JT, Rudolfsen G, Figenschou L, Larsen B, Reiss K, & Folstad I (2018). No support for cryptic choice by ovarian fluid in an external fertilizer. Ecology and Evolution, 8, 11763–11774. 10.1002/ece3.4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, & Zdobnov EM (2018). OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Research, 47, D807–D811. 10.1093/nar/gky1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, & Husson F (2008). FactoMineR: An R package for multivariate analysis. Journal of Statistical Software, 25, 18 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- Lee Y-C, Liao C Jr, Li P-T, Tzeng W-F, & Chu S-T (2003). Mouse lipocalin as an enhancer of spermatozoa motility. Molecular Biology Reports, 30, 165–172. 10.1023/A:1024985024661 [DOI] [PubMed] [Google Scholar]
- Løvoll M, Kilvik T, Boshra H, Bøgwald J, Sunyer JO, & Dalmo RA (2006). Maternal transfer of complement components C3–1, C3–3, C3–4, C4, C5, C7, Bf, and Df to offspring in rainbow trout (Oncorhynchus mykiss). Immunogenetics, 58, 168–179. 10.1007/s00251-006-0096-3 [DOI] [PubMed] [Google Scholar]
- Lymbery RA, Kennington WJ, & Evans JP (2017). Egg chemoattractants moderate intraspecific sperm competition. Evolution Letters, 1, 317–327. 10.1002/evl3.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecham DK, & Olcott HS (1949). Phosvitin, the principal phosphoprotein of egg yolk. Journal of the American Chemical Society, 71, 3670–3679. 10.1021/ja01179a028 [DOI] [Google Scholar]
- Miguel-Queralt S, Blázquez M, Piferrer F, & Hammond GL (2007). Sex hormone-binding globulin expression in sea bass (Dicentrarchus labrax L.) throughout development and the reproductive season. Molecular and Cellular Endocrinology, 276, 55–62. 10.1016/j.mce.2007.06.009 [DOI] [PubMed] [Google Scholar]
- Moon SH, Lee JH, Lee M, Park E, Ahn DU, & Paik H-D (2014). Cytotoxic and antigenotoxic activities of phosvitin from egg yolk. Poultry Science, 93, 2103–2107. 10.3382/ps.2013-03784 [DOI] [PubMed] [Google Scholar]
- Nakamura F, Hartwig JH, Stossel TP, & Szymanski PT (2005). Ca2+ and calmodulin regulate the binding of filamin A to actin filaments. Journal of Biological Chemistry, 280, 32426–32433. 10.1074/jbc.M502203200 [DOI] [PubMed] [Google Scholar]
- Neff BD, & Pitcher TE (2005). Genetic quality and sexual selection: An integrated framework for good genes and compatible genes. Molecular Ecology, 14, 19–38. 10.1111/j.1365-294X.2004.02395.x [DOI] [PubMed] [Google Scholar]
- Nynca J, Arnold GJ, Fröhlich T, & Ciereszko A (2014). Shotgun proteomics of rainbow trout ovarian fluid. Reproduction, Fertility and Development, 27, 504–512. 10.1071/RD13224 [DOI] [PubMed] [Google Scholar]
- Oda S, Igarashi Y, Ohtake H, Sakai K, Shimizu N, & Morisawa M (1995). Sperm-activating proteins from unfertilized eggs of the Pacific herring, Clupea pallasii. Development, Growth & Differentiation, 37, 257–261. 10.1046/j.1440-169X.1995.t01-2-00003.x [DOI] [PubMed] [Google Scholar]
- Palmer MR, & Swanson WJ (2012). Evolution of sperm-egg interaction. Oxford University Press. [Google Scholar]
- Parodi J (2014). Motility, viability, and calcium in the sperm cells. Systems Biology in Reproductive Medicine, 60, 65–71. 10.3109/19396368.2013.869273 [DOI] [PubMed] [Google Scholar]
- Poli F, Immler S, & Gasparini C (2019). Effects of ovarian fluid on sperm traits and its implications for cryptic female choice in zebrafish. Behavioral Ecology, 30, 1298–1305. 10.1093/beheco/arz077 [DOI] [Google Scholar]
- Prokupek A, Hoffmann F, Eyun S-I, Moriyama E, Zhou M, & Harshman L (2008). An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution, 62, 2936–2947. 10.1111/j.1558-5646.2008.00493.x [DOI] [PubMed] [Google Scholar]
- Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, Kirichok Y, Ramsey IS, Quill TA, & Clapham DE (2007). All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proceedings of the National Academy of Sciences of the United States of America, 104, 1219–1223. 10.1073/pnas.0610286104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A language and environment for statistical computing. : R Foundation for Statistical Computing. [Google Scholar]
- Razinia Z, Mäkelä T, Ylänne J, & Calderwood DA (2012). Filamins in mechanosensing and signaling. Annual Review of Biophysics, 41, 227–246. 10.1146/annurev-biophys-050511-102252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettie EC, & Dorus S (2012). Drosophila sperm proteome evolution: Insights from comparative genomic approaches. Spermatogenesis, 2, 213–223. 10.4161/spmg.21748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rime H, Guitton N, Pineau C, Bonnet E, Bobe J, & Jalabert B (2004). Post-ovulatory ageing and egg quality: A proteomic analysis of rainbow trout coelomic fluid. Reprodive Biology and Endocrinolgy, 2, 26 10.1186/1477-7827-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa-Espitia AL, Hernández-Rendón ER, Baltiérrez-Hoyos R, Muñoz-Gotera RJ, Cote-Vélez A, Jiménez I, González-Márquez H, & Hernández-González EO (2016). Focal adhesion kinase is required for actin polymerization and remodeling of the cytoskeleton during sperm capacitation. Biology Open, 5, 1189 10.1242/bio.017558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengrave P, Gemmell NJ, Metcalf V, McBride K, & Montgomerie R (2008). A mechanism for cryptic female choice in chinook salmon. Behavioral Ecology, 19, 1179–1185. 10.1093/beheco/arn089 [DOI] [Google Scholar]
- Rosengrave P, Montgomerie R, & Gemmell NJ (2016). Cryptic female choice enhances fertilization success and embryo survival in chinook salmon. Proceedings of the Royal Society B: Biological Sciences, 283, 20160001 10.1098/rspb.2016.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengrave PR, Montgomerie R, Metcalf VJ, McBride K, & Gemmell NJ (2009). Sperm traits in Chinook salmon depend upon activation medium: Implications for studies of sperm competition in fishes. Canadian Journal of Zoology, 87, 920–927. 10.1139/Z09-081 [DOI] [Google Scholar]
- Rosengrave P, Taylor H, Montgomerie R, Metcalf V, McBride K, & Gemmell NJ (2009). Chemical composition of seminal and ovarian fluids of chinook salmon (Oncorhynchus tshawytscha) and their effects on sperm motility traits. Comparative Biochemistry and Physiology - Part A: Molecular & Integrative Physiology, 152, 123–129. 10.1016/j.cbpa.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Rowe M, Whittington E, Borziak K, Ravinet M, Eroukhmanoff F, Sætre G-P, & Dorus S (2020). Molecular diversification of the seminal fluid proteome in a recently diverged passerine species pair. Molecular Biology and Evolution, 37(2), 488–506. 10.1093/molbev/msz235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DK, Fowler O, & Smalley KN (2000). The effects of estradiol treatment on the blood viscosity of the bullfrog Rana catesbeiana. Transactions of the Kansas Academy of Science, 103, 38–45. 10.2307/3627934 [DOI] [Google Scholar]
- Simmons LW (2005). The evolution of polyandry: Sperm competition, sperm selection, and offspring viability. Annual Review of Ecology, Evolution, and Systematics, 36, 125–146. 10.1146/annurev.ecolsys.36.102403.112501 [DOI] [Google Scholar]
- Stapper AP, Beerli P, & Levitan DR (2015). Assortative mating drives linkage disequilibrium between sperm and egg recognition protein loci in the sea urchin Strongylocentrotus purpuratus. Molecular Biology and Evolution, 32, 859–870. 10.1093/molbev/msv010 [DOI] [PubMed] [Google Scholar]
- Sun X-H, Zhu Y-Y, Wang L, Liu H-L, Ling Y, Li Z-L, & Sun L-B (2017). The Catsper channel and its roles in male fertility: A systematic review. Reproductive Biology and Endocrinology, 15, 65 10.1186/s12958-017-0281-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, & Shimodaira H (2006). Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics, 22, 1540–1542. 10.1093/bioinformatics/btl117 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, & Aquadro CF (2001). Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 98, 7375–7379. 10.1073/pnas.131568198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, & Vacquier VD (2002). The rapid evolution of reproductive proteins. Nature Reviews Genetics, 3, 137–144. 10.1038/nrg733 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, & Aquadro CF (2004). Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics, 168, 1457–1465. 10.1534/genetics.104.030478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill R (1983). Cryptic female choice and its implications in the scorpionfly Harpobittacus nigriceps. The American Naturalist, 122, 765 10.1086/284170 [DOI] [Google Scholar]
- Tregenza T, & Wedell N (2000). Genetic compatibility, mate choice and patterns of parentage: Invited Review. Molecular Ecology, 9, 1013–1027. 10.1046/j.1365-294x.2000.00964.x [DOI] [PubMed] [Google Scholar]
- Vacquier VD, & Swanson WJ (2011). Selection in the rapid evolution of gamete recognition proteins in marine invertebrates. Cold Spring Harbor Perspectives in Biology, 3, a002931 10.1101/cshperspect.a002931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, & Mann M (2009). Universal sample preparation method for proteome analysis. Nature Methods, 6, 359 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Yeates SE, Diamond SE, Einum S, Emerson BC, Holt WV, & Gage MJG (2013). Cryptic choice of conspecific sperm controlled by the impact of ovarian fluid on sperm swimming behaviour. Evolution, 67, 3523–3536. 10.1111/evo.12208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, & Ma B (2012). PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Molecular & Cellular Proteomics, 11(M111), 10587 10.1074/mcp.M111.010587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


