Abstract
Peptide nanofibers are useful for many biological applications, including immunotherapy, tissue engineering, and drug delivery. The robust lengthwise assembly of these peptides into nanofibers is typically difficult to control, resulting in polydisperse fiber lengths and an incomplete understanding of how nanofiber length affects biological responses. Here, rationally designed capping peptides control the length of helical peptide nanofibers with unique precision. These designed peptides bind the tips of elongated nanofibers to shorten and narrow their length distributions. Demonstrating their use as immunotherapies, capped nanofibers are preferentially cross-presented by dendritic cells compared to uncapped nanofibers. Due to increased cross-presentation, these capped nanofibers trigger stronger CD8+ T cell responses in mice than uncapped nanofibers without. This strategy illustrates a means for controlling the length of supramolecular peptide nanofibers to modulate their immunogenicity in the context of immunotherapies.
Keywords: self-assembling peptides, nanostructures, coiled coils, immunotherapies
Graphical Abstract
Body
Peptide biomaterials with fibrillar morphologies such as β-sheet peptides, worm-like micelles, and peptide amphiphiles have been explored towards numerous biomedical applications including scaffolds for tissue repair,[1–3] immunotherapies for infectious diseases,[4–7] cancer,[8] or inflammatory conditions,[9] and depots for the sustained delivery of drugs.[10] With considerable surface area, fibrillar peptide biomaterials are advantageous for generating highly multivalent displays of biomolecules. These materials can also be used to form hydrogels, which have been utilized for inducing anti-tumor and CD8+ T cell responses [11–13] Additionally, self-assembling materials can be engineered to be modular, where different co-assembling components can be integrated, adjusted, and optimized towards applications as diverse as vaccines or scaffolds for tissue engineering.[14,15]
Although almost all reported fibrillar peptide materials have exquisitely controlled widths, control of their lengths has remained challenging. The length of some self-assembling materials can be adjusted by extrusion,[16] ultrasound,[17] manipulating the concentration of self-assembling precursors,[18] or by stabilizing supramolecular assemblies with covalent linkages.[19,20] However, peptide nanofibers are highly dynamic, and self-assembled structures have a propensity to reform after physical disruption, leading to considerable polydispersity and difficulty in reliably adjusting nanofiber length.[21]
Accordingly, there are few systematic studies of how peptide nanofiber length affects the phenotype of immune responses against these materials, and differing results have been noted depending on immunization route and nanofiber composition. For example, when used for intranasal immunization, shortening of nanofibers via extrusion decreased their ability to raise CD8+ T cell responses,[7] whereas extrusion of nanofibers had a minimal effect on antibody responses elicited by sublingual immunization.[22] Interactions of size-controlled fibrillar materials with immune cell types have largely focused on evasion of antigen presenting cell (APC) uptake to prolong circulation time of nanomaterials for drug delivery, [16,23] rather than targeting specific APC engagement for immunotherapies. Most studies which examine the effects of nanomaterial size and shape on immune responses have employed spherical or low aspect ratio particles, rather than fibrillar structures. [24–26] Across different materials types, such studies have shown that nanomaterial size critically influences distribution through the lymphatic system, with additional factors such as shape, surface chemistry, and administration route also affecting the transport and ultimate immunogenicity of nanomaterials.[27,28] These physical parameters also influence cellular uptake and processing, thus modulating the phenotype of any subsequent immune responses.[16]
Fibrillar peptide biomaterials have been shown to be uniquely immunogenic, raising useful B cell and T cell responses towards a range of applications, including infectious diseases, [7,29] cancer, [8,30] drug addiction,[31] chronic inflammatory conditions,[9] and others. In each disease application context, success or failure hinges on raising not just a strong enough immune response, but one with the appropriate phenotype, and this phenotype varies considerably from one disease to the next. Given the significant influence of particle size and dimensions observed for other non-fibrillar materials platforms, we hypothesized that control over nanofiber length could be used to influence the trafficking, internalization, processing, and presentation of nanofiber-delivered peptide epitopes.
Although a few previous reports have indicated that aspect ratio can affect the cellular uptake and biodistribution of fibrillar materials, [16,32] there has been a lack of tools for finely adjusting nanofiber length to ascertain how this dimension influences the type of immune responses raised by nanofibers. Strategies for controlling the length of fibrillar materials have been described, but the disruption of fiber-forming peptides and proteins has been studied mainly in the context of pathogenic amyloids. β-sheet fibrillization in amyloids has been interrupted by molecules which bind the propagating fiber but do not allow stacking to continue, effectively forming a “cap” on the ends of fibers. [33–35] Restriction of lengthwise assembly has been achieved in other systems to study protein assembly or to design vehicles for drug delivery or other therapies. Notably, Shen, Baker and coworkers recently designed protein filaments which assemble directionally via monomers with two assembling interfaces.[36] Filaments of these proteins can be grown from surfaces and disassembled with capping agents containing only one assembling interface.[36] There are other self-assembling systems in which length control has been achieved, including peptide amphiphiles capped by lipids,[37] designed amphiphiles with differential stacking affinities,[38] self-assembling peptides with chemically protected N-termini, [39] and filamentous viruses with genetically encoded lengths.[32] Additional examples have demonstrated fine control over the diameter,[36] luminal structure,[40] and lateral assembly[19] of fiber- and filament-forming peptides.
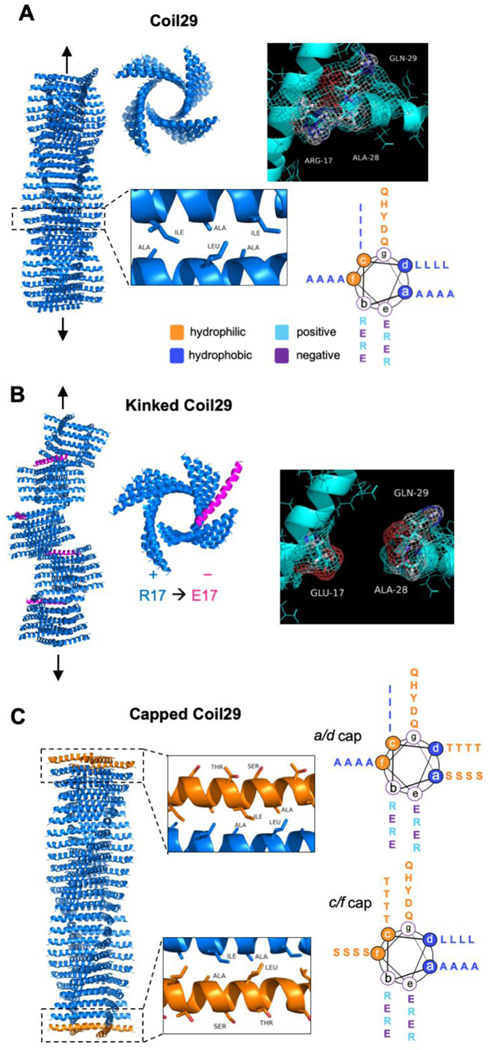
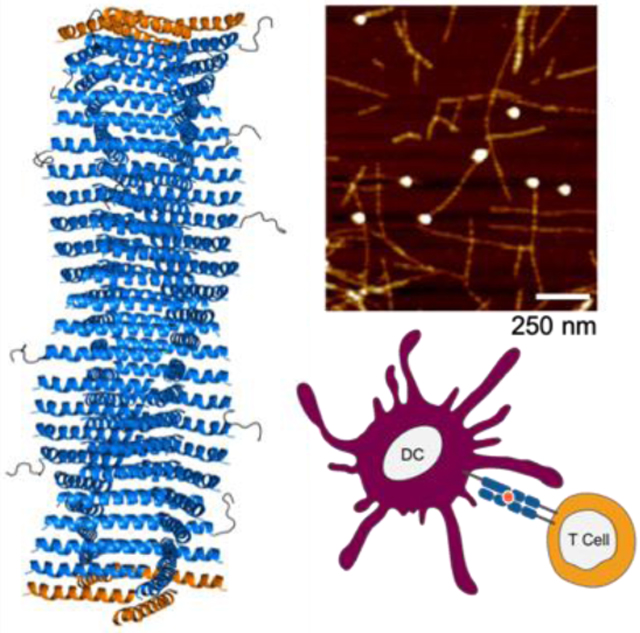
In this study, we designed a series of peptides that when mixed form long α-helical nanofibers with control over their lengths, then capitalized upon this system to study how nanofiber length influences the materials’ immunogenicity, focusing both on antibody and CD8+ T cell responses in mice. The strategy was based on the recently described Coil29 system, which forms high-aspect ratio nanofibers composed almost entirely of α-helical structure.[8,41] Coil29 has sequence-dependent structural flexibility and exhibits changes in assembly forms with single amino acid substitutions.[41] This structural specificity and control over assembly behavior, in addition to Coil29’s immunogenic properties, [8] made it a useful starting point for studying the impact of length on immunogenicity. Initially designed by Egelman, Conticello, and coworkers, Coil29 forms nanofibers with the structure shown in Scheme 1A, originally determined using cryo-electron microscopy.[41] In this structure, hydrophobic residues in the a/d and c/f positions of the α-helical heptad drive the stacking of the helices into long nanofibers, with the helices running perpendicular to the main axis of the nanofiber. Residues in the a and d positions form hydrophobic contacts with residues in the c and f positions of the subsequent peptide in a sterically complementary fashion (Scheme 1A), inducing directional elongation of nanofibers. Affinity of the C-terminus for the central Arg-Ala-Tyr-Ala-Arg (RAYAR) sequence causes the C-terminus to form a T-shaped junction with the central portion of the peptide, and the angle of this interaction results in a 4-armed, square-shaped structure in each layer of the fiber. Stacking of these squares produces the nanofiber structure shown in Scheme 1A, with the N-termini projecting radially from the fiber axis. We previously investigated Coil29 as a platform for developing vaccines and other immunotherapies, finding that when the peptide’s N-terminus is appended with competent B- and T-cell epitopes, it still forms regular nanofibers and is capable of raising humoral and cellular immune responses in the absence of adjuvant,[8] similar to other fiber-forming peptides.[9,42] In a recent publication by Wu et al., Coil29 nanofibers were additionally shown to be minimally inflammatory as they triggered significantly lower levels of inflammatory cytokines and cell types compared to the traditional adjuvants Alum and SAS.[43]
Scheme 1.
Structure of helical peptides and their assemblies. A) 3D structure of fiber-forming peptide Coil29 (PDB 3j89) and helical wheel projection of peptide sequence. From left to right: side view of nanofiber, top-down view of nanofiber, C-terminal interaction responsible for square formation. Lower cutout shows hydrophobic contacts that cause lengthwise elongation of nanofibers, and helical wheel shows peptide sequence. B) Design schematic for nanofibers incorporating kinking peptides and predicted changes caused by disruption of C-terminal interaction. C) Schematic of nanofibers terminated with capping peptides. Insets show predicted structure, with ends of fibers terminated with caps, leaving only hydrophilic residues on solvent-exposed helical faces. Helical wheel projections show mutations to hydrophobic residues in the Coil29 sequence.
We investigated two strategies for disrupting Coil29 assembly and shortening its nanofibers. First, to interrupt the interaction between the C-terminus and the central RAYAR sequence, we made the R17E mutation with the expectation that it would repel the C terminus from the inner face of the nanofiber, thus inducing “kinks” along the length of the nanofiber when intermixed with Coil29. We termed this sequence Coil29Kink (Scheme 1B). The R17E mutation was intended to flip the charge of R17 to a negative residue, and E was chosen over D because of its slightly larger size, which would more closely resemble the steric properties of the original R17 residue. By changing the charge of R17 but maintaining its relative size in the helical sequence, we hoped to preserve the helical structure of Coil29 while causing repulsion between E17 and the C-termini of other peptides with Coil29Kink.
As an alternative strategy, we also designed sequences in which residues at the a and d or c and f positions were mutated to Ser and Thr, in effect removing one of the hydrophobic faces of the helix from the fiber-forming sequence. We expected this modification would effectively “cap” the ends of Coil29 nanofibers (Scheme 1C) by interrupting the linear assembly of Coil29 and terminating the fiber ends with hydrophilic residues. We designed two separate peptides complementary to each end of Coil29 nanofibers, termed Coil29Caps. A full list of peptide sequences is given in Table S1.
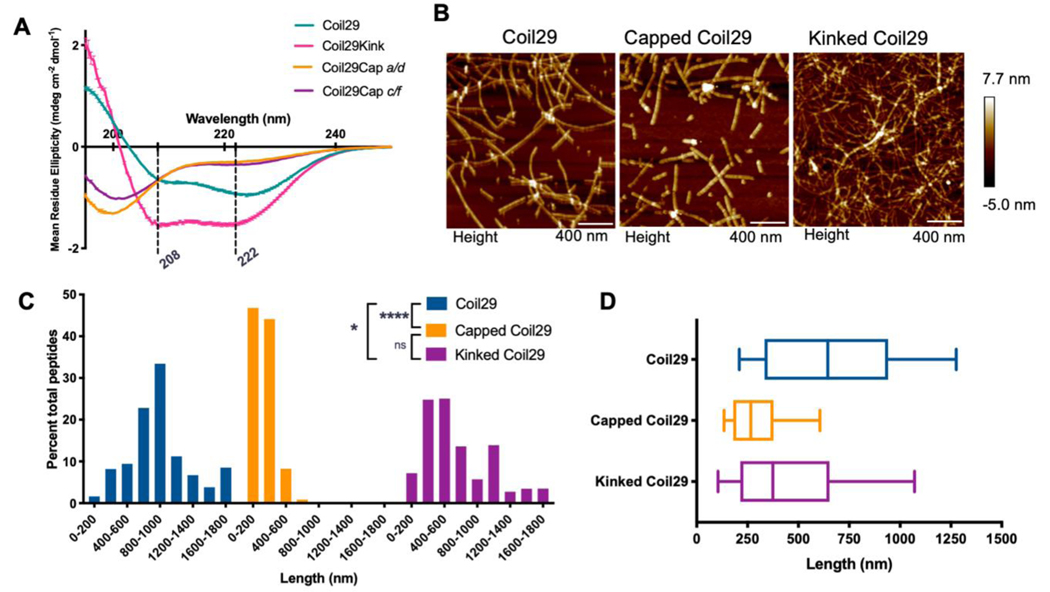
The described mutations altered the secondary structure of Coil29, as measured by circular dichroism (CD) (Figure 1A). The CD spectrum of Coil29 indicated an α-helical structure characteristic of coiled-coils, with minima at 208 nm and 222 nm having similar intensity. The R17E mutation in Coil29Kink altered the intensity of these minima, suggesting an α-helical structure not associated with coiled coil pairing. Coil29Caps did not show a specific secondary structure alone in solution, indicating that the series of mutations made along the hydrophobic helix faces removed the propensity of these peptides to form α-helices.
Figure 1. Morphological impacts of Coil29Caps and Coil29Kinks.
A) Circular Dichroism of individual peptides in PBS, diluted to 0.1 mM in water immediately prior to scanning. Reported spectra are averaged from 3 scans and error bars indicate standard deviation. B) AFM images of nanofibers formed from Coil29 alone (left), incorporating a 1:10 ratio of Coil29Caps:Coil29 (center) or incorporating a 1:10 ratio of Coil29Kink:Coil29 (right). C) Length distributions of nanofibers formed with capping and kinking peptides, calculated as percent of total peptides in fibers of a given length (n=3 images per condition). D) Box and whisker plots showing 25th percentile, 50th, and 75th percentile with error bars at 10 and 90 percentile range. Nanofiber lengths were compared using the Kruskal-Wallis test with Dunn’s multiple comparisons (*p<0.05, ****p<0.0001).
To test whether capping and kinking peptides could significantly alter the lengthwise assembly of Coil29 nanofibers, we first co-assembled Coil29Kink and Coil29Caps with Coil29 in a 1:10 molar ratio. These formulations are herein referred to as “Kinked Coil29” and “Capped Coil29”, respectively. The nanofibers formed from these mixtures and from Coil29 alone were imaged using atomic force microscopy (AFM) (Figure 1B). These fibers were then measured, and the lengths were analyzed by calculating the percent of the total material present in fibers of a given length. These length distributions indicated that the incorporation of Coil29Caps significantly shortened Coil29 nanofibers, while Coil29Kink had a more modest effect (Figure 1C, D). Because Coil29Caps induced a larger shift in nanofiber length distributions and reproducibly formed soluble nanofibers incorporating several different epitopes (Figure S1), we chose to characterize this system further and analyzed how shortened nanofibers impacted vaccine efficacy. Furthermore, when attempting to test Coil29Kink in formulations incorporating immunogenic epitopes, Coil29Kink often caused the formation of aggregates and precipitates, suggesting that it disrupted Coil29 nanofibers to an extent that they were no longer stable in solution.
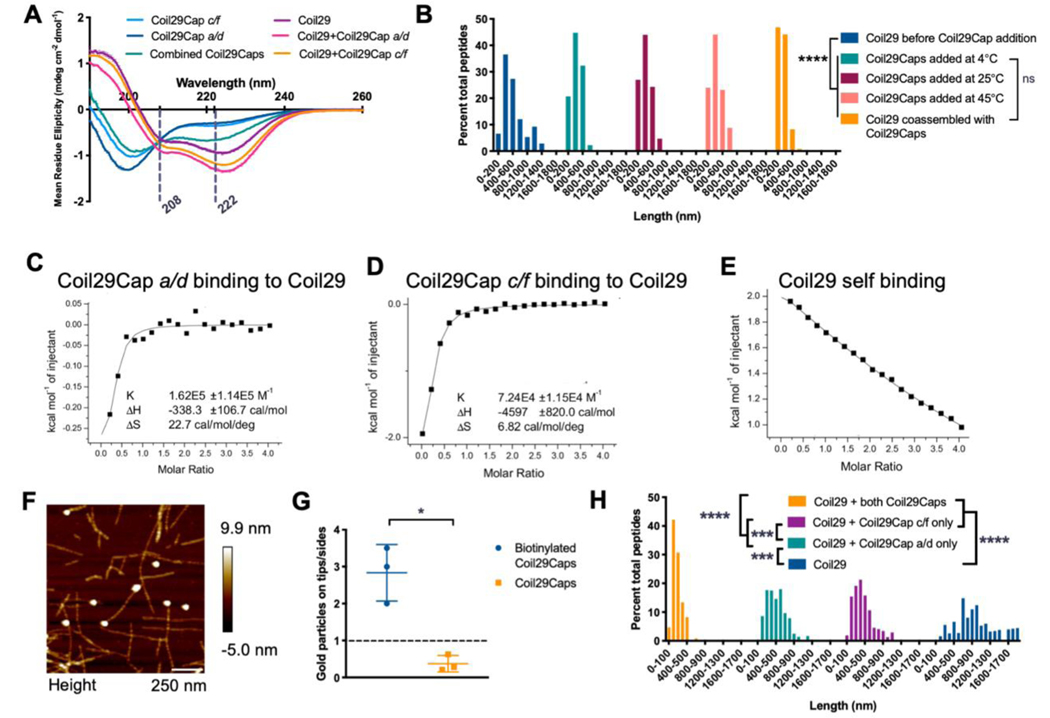
The significant shortening of nanofiber assemblies observed upon co-assembling Coil29Caps with Coil29 prompted further analysis of how Coil29Caps were influencing the assembly of Coil29. First, we examined the secondary structure of Coil29Caps in the presence of other helical peptides. Neither Coil29Cap a/d or Coil29Cap c/f showed a specific secondary structure alone in solution (Figure 2A). However, when these two peptides were mixed together, the CD spectrum exhibited a shift towards an α-helical conformation, indicating that the two capping peptides may interact with one another to induce a transition toward a helical secondary structure. This behavior indicated a model in which the hydrophobic faces of the two caps pair with each other, similar to the pairing observed in Coil29 assembly. Upon mixing the caps with Coil29, the CD spectrum of the mixture shows nearly identical structure to nanofibers formed from Coil29 alone and a loss of the minimum at 200 nm, indicating that nearly all peptides in the mixture adopted a helical conformation (Figure 2A). This induced helicity suggested helix-helix pairing of Coil29Caps with Coil29 in accordance with the designs proposed in Scheme 1C. The affinity of both of the individual Coil29Caps for Coil29 were next measured by isothermal titration calorimetry (ITC), where energetic changes consistent with saturation of a specific binding site was observed for both capping peptides (Figure 2 C,D). In comparison to Coil29Cap a/d, Coil29Cap c/f exhibited a higher affinity for Coil29, suggesting that it may have played a more significant role in fiber shortening than Coil29Cap a/d (Figure 2 C,D). Addition of Coil29Caps reached binding saturation, whereas Coil29 exhibited continuous binding when added to a solution of Coil29 peptides (Figure 2E). The stoichiometry of Coil29Cap binding to Coil29 was estimated from ITC data (Table S2) and suggested binding of one cap per eight Coil29 molecules for Coil29Cap c/f and one cap per four Coil29 molecules for Coil29Cap a/d. These values would fit with a model wherein Coil29 exists in tetrameric squares (see Scheme 1a) and Coil29Cap c/f could bind to either face of the Coil29 helix, whereas Coil29Cap a/d could bind to only one face of the Coil29 helix. However, this calculation depends on assuming the tetrameric structure and thus requires additional structural analysis for conclusive assignment of these stoichiometries. These results indicated that Coil29Caps spontaneously bound in a site-specific manner to a limited number of sites on Coil29, acting in accordance with the designs shown in Scheme 1.
Figure 2. Binding characterization of Coil29Caps and Coil29.
A) CD spectra of Coil29 and Coil29Caps, with dashed lines marking 208 nm and 222 nm. Peptides were prepared at 2 mM in PBS, diluted to 0.1 mM in water immediately prior to scanning. Reported spectra are averaged from 3 scans and error bars indicate standard deviation. (The CD spectra for Coil29Cap a/d and Coil29Cap c/f from Figure 1A are also displayed in Figure 2A for comparison to spectra from a mixture of both Coil29Caps.) B) Length distributions of Coil29 alone, after the addition of equimolar concentration of Coil29Caps, and co-assembled in a 1:10 ratio with Coil29Caps (n=3 images per condition). Nanofiber lengths were compared using the Kruskal-Wallis test with Dunn’s multiple comparisons (****p<0.0001). C-E) Wiseman plots showing changes in sample energy when Coil29Caps or Coil29 was titrated into Coil29, with data from fit equations using a 1 site binding model. F) Representative AFM image showing Coil29Caps labeled with gold nanoparticles. G) Quantification of Coil29Cap location on nanofiber structures compared to background labeling of Capped Coil29 nanofibers without biotin tag (n=3 images per condition). (*p<0.05, unpaired two-tail t test with Welch’s correction). H) Length distributions of nanofibers formed with one or both of Coil29Caps (n=3 images per condition). Nanofiber lengths were compared using the Kruskal-Wallis test with Dunn’s multiple comparisons (***p<0.001, ****p<0.0001).
Because we observed this exothermic, site-specific binding of Coil29Caps to Coil29, we investigated whether this interaction was energetically favorable enough to interrupt Coil29 fibers after they had already formed. To test this, we first imaged Coil29 nanofibers and then added Coil29Caps and re-imaged the fibers to determine if this addition induced structural changes. The addition of Coil29Caps shortened the average length of nanofibers after mixing them at multiple temperatures (Figure 2B). Coil29Caps begin to significantly shorten fibers within 1 minute of addition and the median length of nanofibers is minimized within 15 minutes of Coil29Caps addition and remains stable between 15 minutes and 24 hours (Figure S2). To visualize the location of Coil29Caps on Coil29 nanofibers, Coil29Caps were biotinylated, and assemblies were stained with 10 nm streptavidin gold particles. Addition of biotin to the N-terminal of Coil29Caps did not affect their ability to shorten Coil29 nanofibers (Figure S3). Strikingly, peptides labeled in this way indicated that Coil29Caps resided primarily on the tips of Coil29 nanofibers (Figure 2F,G). Because Coil29Cap c/f showed a higher affinity for Coil29 than Coil29Cap a/d when measured by ITC, we hypothesized that the shortening effects were dominated by Coil29Cap c/f and formulated nanofibers with Coil29 and only one of the capping peptides. Addition of only one Coil29Cap shortened nanofibers compared to those made from Coil29 alone but induced more modest changes compared to fibers formed with both Coil29Caps (Figure 2H). Furthermore, Coil29Cap affinity appears to dictate the extent of fiber shortening, as nanofibers formed with only Coil29Cap c/f had shorter and less disperse lengths than nanofibers formed with only Coil29Cap a/d. Taken together, these data indicated that the capping peptides acted in accordance with the designs mentioned above, where both Coil29Caps were required to stabilize the distinct upper and lower faces of Coil29 nanofibers.
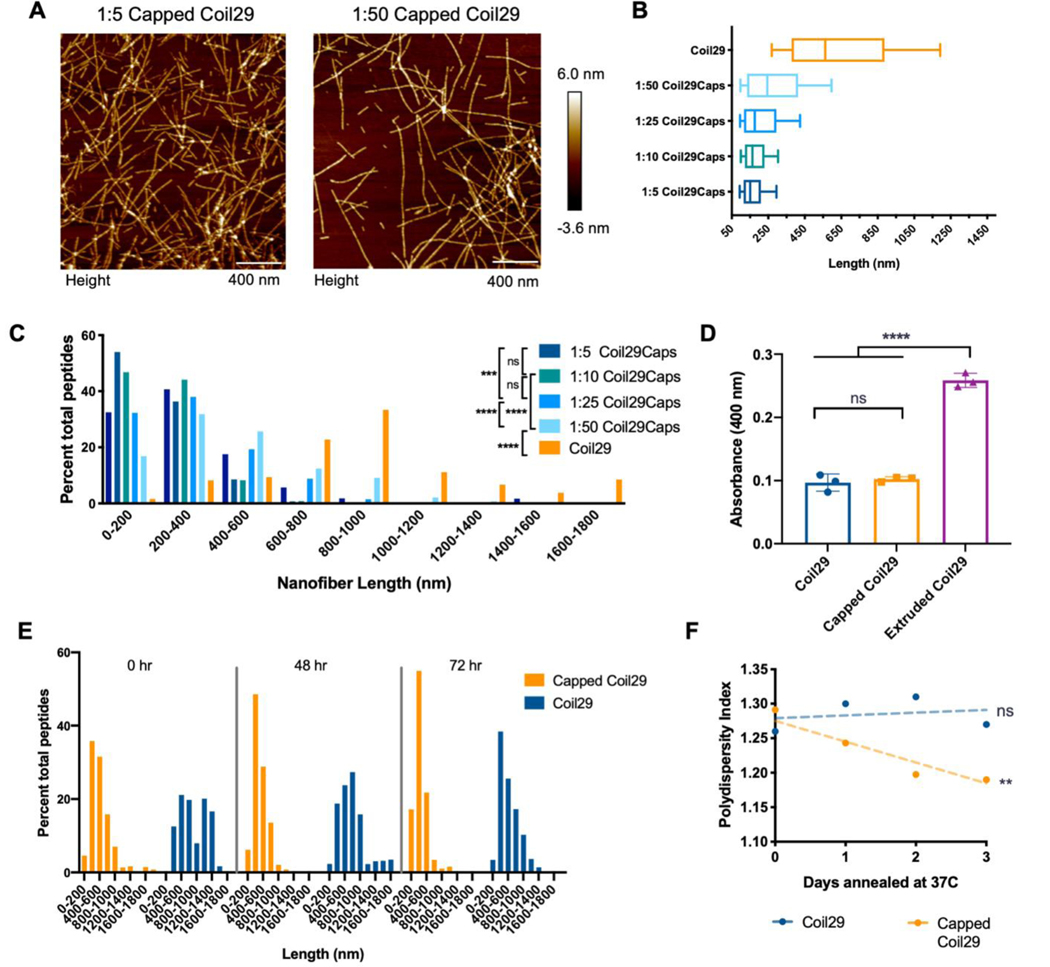
Owing to the robust shortening observed upon co-assembling Coil29Caps with Coil29 in a 1:10 ratio, we sought to tune the average length of the nanofibers by adjusting the ratio of Coil29 to Coil29Caps. Based on our observation of site-specific and saturating binding of Coil29Caps to Coil29 from ITC, we hypothesized that by altering the amount of Coil29Caps in solution, we could manipulate the frequency of capping events and tested the concentration of Coil29Caps necessary to saturate Coil29 nanofiber tips during assembly. By adjusting the rate of capping during assembly, we predicted that the formation of different lengths of Coil29 nanofibers would be dependent on Coil29Cap stoichiometry. In keeping with this hypothesis, ratios of Coil29Caps to Coil29 from 1:5 to 1:50 yielded nanofibers with distinct length distributions (Figure 3). Coil29 nanofibers co-assembled with varying ratios of Coil29Caps were annealed at 37°C for 1 hour prior to imaging to allow nanofiber lengths to equilibrate. Within this range, the mean and polydispersity of nanofiber lengths were inversely related to the content of Coil29Caps in assemblies, demonstrating that Coil29Caps shortened the nanofibers and decreased the variability of their lengths in a stoichiometric fashion (Figure 3A–C). Beyond this range, for example at ratios of 1:2, median length and polydispersity increased (10th percentile = 80 nm, median = 178 nm, 90th percentile = 492 nm), which we interpreted as indicating that cap pairing competed with Coil29 binding owing to the high concentration of the cap peptides. Given the well-behaved nature of the system at stoichiometries of 1:5 and greater, we focused on this range for subsequent experiments. Within the range of 1:5 to 1:50 Coil29Caps to Coil29, increasing ratios of Coil29Caps create nanofibers with decreasing polydispersity index (PDI) which eventually plateaus at PDI values of 1.2–1.3 (Figure S4), acting to create a controlled polymerization when Coil29 is formulated with Coil29Caps.
Figure 3. Capping produced stably shortened nanofibers.
A) Images of Coil29 nanofibers in PBS assembled with different molar ratios of Coil29Caps, with Coil29 concentration kept constant at 2mM and annealed at 37°C for 1 hour prior to imaging. B) Box and whisker plots showing 25th percentile, 50th, and 75th percentile with error bars at 10 and 90 percentile range and C) Length distributions of nanofibers formed with varying ratios of Coil29Caps (n=3 images per condition). Nanofiber lengths were compared using the Kruskal-Wallis test with Dunn’s multiple comparisons (***p<0.001, ****p<0.0001). D) Turbidity of 2mM solutions of nanofibers 3 hours after the addition of PBS, and immediately after extrusion. (n=3 independent samples, compared by one-way ANOVA with Tukey’s multiple comparisons.) E) Length distributions and F) polydispersity index of Coil29 nanofibers formed with and without Coil29Caps after annealing for up to three days, calculated from AFM measurements. (non-zero slope was non-significant for Coil29 fibers without Coil29Caps, **p<0.01 for Capped Coil29 fibers)
To determine if shortened nanofibers were stable at physiological temperatures, we annealed samples of Coil29, Capped Coil29, and Coil29 extruded through a 0.4 μm filter at 37°C for several days. Extrusion is commonly used for controlling the size of spherical liposomes or micelles, so we sought to compare the use of Coil29Caps to conventional size-control techniques. TEM imaging of these nanofibers indicated differences in the lateral association and aggregation of nanofibers that had been extruded or formulated with Coil29Caps (Figure S5). Observations from TEM imaging were consistent with turbidity measurements taken of these solutions, indicating that extrusion of Coil29 yielded aggregates, whereas capping did not (Figure 3D). We interpreted this data to indicate that extrusion did not produce nanofibers capable of remaining shortened at physiological temperatures, as exposed hydrophobic faces of the fibers were unstable in solution and likely to associate with neighboring fibers. When Capped Coil29 nanofibers were annealed over the course of several days, we observed decreasing polydispersity of nanofiber lengths, while Coil29 nanofibers without Coil29Caps did not display this property (Figure 3E). The PDI of Capped Coil29 nanofibers decreased linearly over 3 days (non-zero slope, p = 0.0069), while the PDI of Coil29 nanofibers showed no such linear trend (non-zero slope, p = 0.7831) (Figure 3F). The site-specific binding of Coil29Caps to Coil29, stoichiometric shortening of Coil29 nanofibers by Coil29Caps, and requirement of Coil29Cap a/d and Coil29Cap c/f indicate that Coil29Caps shorten nanofibers by binding to the ends of Coil29 nanofibers in a sterically complimentary and dose-dependent fashion. In accordance with the design described above, we observed behavior which matched the hypothesis that binding of Coil29Caps to Coil29 nanofibers created a non-assembling, hydrophilic surface at the growing edge of nanofibers, effectively “capping off” nanofiber tips and terminating assembly.
Given the control over Coil29 fiber formation provided by Coil29Caps, we sought to explore how Capped Coil29 nanofibers interacted with immune cells in comparison to Coil29 nanofibers without Coil29Caps. Uptake of nanofibers by dendritic cells was measured in vitro, and the ability of nanofibers to induce presentation of CD8+ T Cell epitopes by bone marrow-derived dendritic cells (BMDCs) was measured using a reporter CD8+ T cell line. The results from these in vitro studies then prompted investigation of Capped Coil29 and Coil29 nanofibers without Coil29Caps as vaccines for B cell and T cell responses in vivo. Nanofibers formed from Coil29 with fluorescent tags, B- and T-Cell epitopes were shortened by Coil29Caps in a similar fashion to those formed by unmodified Coil29 (Figure S1).
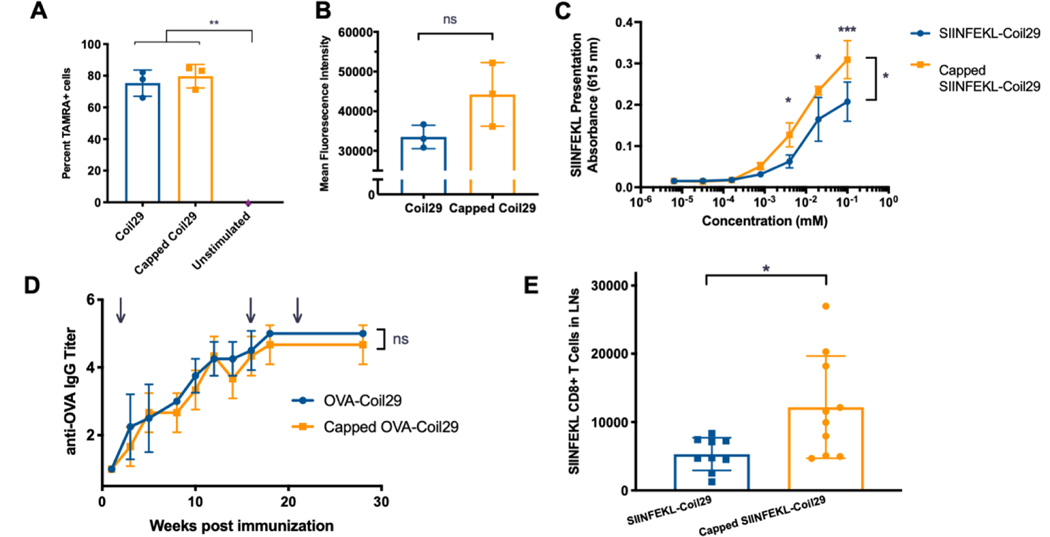
To probe how capping and nanofiber length change the cellular uptake of these materials under static conditions, we quantified the uptake of fluorescently labeled nanofibers by dendritic cells (DCs). Nanofibers were made from co-assemblies of Coil29 tagged with 5-carboxytetramethyltrhodamine (TAMRA), unmodified Coil29, and Coil29Caps. These nanofibers were then used to stimulate DC2.4 dendritic cells in culture, and uptake was measured by detecting cells positive for TAMRA by flow cytometry. DCs treated with Capped Coil29 and Coil29 nanofibers without Coil29Caps exhibited significantly more TAMRA+ cells than unstimulated cells, but no difference was detected between nanofiber-treated groups (Figure 4A). The amount of material taken up by cells was measured using the mean fluorescence intensity (MFI) of TAMRA+ cells and compared between DCs stimulated with Capped Coil29 and Coil29 fibers without Coil29Caps (Figure 4B). Although no statistically significant difference was measured, DCs which were exposed to Capped Coil29 fibers showed higher MFI, suggesting that formulation of nanofibers with Coil29Caps may have increased the total material acquired by antigen presenting cells. Coil29 and Capped Coil29 were both non-toxic to DC2.4 dendritic cells in culture and did not alter their ability to proliferate (Figure S7).
Figure 4. Nanofiber capping improved cross-presentation in vitro and CD8+ T cell responses in vivo.
A) Uptake of TAMRA-labeled Coil29 fibers (0.2 mM) by DC2.4 cells in vitro. (n=3 wells, compared by one-way ANOVA with Sidak’s multiple comparisons, **p<0.01). B) Mean Fluorescence Intensity of TAMRA+ cells from A (n=3 wells, compared by one-way ANOVA). C) Presentation of SIINFEKL peptide by mature BMDCs treated with Coil29 nanofibers bearing SIINFEKL peptides, measured by reporter B3Z CD8+ T cells. (n=3, compared by repeated measures ANOVA with Sidak’s multiple comparisons, *p<0.05, ***p<0.001) D) Endpoint IgG titer for mice immunized with Coil29 nanofibers bearing the OTII epitope OVA. Immunizations were given subcutaneously at time points indicated by arrows (n=5 mice, compared by repeated measures ANOVA). E) Numbers of CD8+ T cells specific to SIINFEKL epitope after 2 immunizations with SIINFEKL-Coil29 nanofibers. (n=10, pooled from 2 independent experiments, *p<0.05 compared by unpaired t-test with Welch’s correction).
To assess differences in intracellular processing and presentation, BMDCs were harvested from mice and matured with Flt-3 ligand before being incubated with nanofibers bearing the CD8+ T cell epitope SIINFEKL. Reporter B3Z T cells were then co-cultured with BMDCs and produced an optical signal to quantify the amount of SIINFEKL presented by BMDCs. In comparison to nanofibers formed from SIINFEKL-Coil29 without Coil29Caps, Capped SIINFEKL-Coil29 nanofibers were more efficiently cross-presented by BMDCs to present SIINFEKL on MHC I molecules (Figure 4C). Previous work found that zeta potential had a considerable influence on the uptake and presentation of self-assembled peptide nanofibers,[14] but in this case Capped SIINFEKL-Coil29 and SIINFEKL-Coil29 without Coil29Caps had very similar zeta potentials (Figure S6), ruling out surface charge as an explanation for their differential presentation. Together, the findings from this series of in vitro experiments indicated that Capped Coil29 nanofibers were internalized at similar levels to Coil29 nanofibers without Coil29Caps but underwent differential processing within the antigen presenting cells, leading to different levels of presented CD8+ epitopes on the surface of DCs.
To test the effects of nanofiber capping on humoral responses, Coil29 appended to the OTII epitope of ovalbumin (OVA-Coil29) was synthesized and assembled into nanofibers. OVA-Coil29 and Capped OVA-Coil29 nanofibers were administered subcutaneously, and antibody titers were monitored over 30 weeks (Figure 4D). Both OVA-Coil29 and Capped OVA-Coil29 nanofibers raised durable titers of IgG antibodies against the OTII epitope of similar magnitudes, indicating that capping had a minimal influence on humoral responses. Because we observed higher levels of SIINFEKL peptide cross-presentation by BMDCs in vitro, we investigated whether this produced a stronger CD8+ T cell response in vivo. Mice received two immunizations of SIINFEKL-Coil29 or Capped SIINFEKL-Coil29 nanofibers and were sacrificed 7 days after the second immunization. Lymph nodes were harvested and analyzed by staining for SIINFEKL-specific CD8+ T cells (Figure S8). The total number of SIINFEKL specific CD8+ T cells were significantly higher for mice immunized with Capped SIINFEKL-Coil29 nanofibers compared to mice immunized with SIINFEKL-Coil29 nanofibers without Coil29Caps (Figure 4E). These results indicated that Capped OVA-Coil29 nanofibers were capable of producing antibody responses equally as robust as OVA-Coil29 nanofibers without Coil29Caps but were more efficient at inducing CD8+ immune responses.
Utilizing short, rationally designed peptide sequences we have described the use of complementary and non-assembling interfaces to control the self-assembly of peptide nanofibers. The behavior observed in this system indicates that stabilization of assembling hydrophobic interfaces yields robust architectures which are stable over long periods of time at physiological temperatures. Furthermore, capping peptides enabled tuning of nanofiber length in much finer increments than existing systems.
We observed slight variations in the binding energy of Coil29Cap a/d and Coil29Cap c/f to Coil29 when compared by ITC. While it is unclear exactly what features dictated this difference, CD data indicates that Coil29Cap c/f may have had a slightly more helical structure that Coil29Cap a/d. This increased helicity could improve its ability to pair with Coil29 nanofibers leading to more exothermic binding and shorter nanofibers (Figure 2). Additionally, it is possible that the exposed, unpaired a/d and c/f faces of Coil29 had different stabilities, making them more likely to bind Coil29Caps. In the future, the ability to tune the affinity and conformation of capping agents with rationally or computationally designed sequences could be utilized for stabilization of self-assembling nanofibers and filaments at specific lengths or under particular conditions.
The design and application of capping peptides to control peptide nanofiber assembly has allowed for an investigation of how nanofiber aspect ratio influences immune responses. Whereas nanofiber capping did not significantly influence acquisition by dendritic cells or the overall titer of antibody responses, it did influence processing of the constituent peptides and loading on class-I MHC molecules, and it augmented subsequent CD8+ T cell responses. The ability to increase the magnitude of CD8+ T cell responses while maintaining high antibody titers may be advantageous for applications such as vaccines against cancer or infectious diseases, where engagement of multiple arms of the immune system is beneficial.[44,45]
At present, it remains to be investigated how the shorter assemblies came to promote CD8+ T cell responses, but multiple factors may have been at play. For example, shortened nanofibers may have been more capable of escaping endosomes into the cytosol to access cross-presentation pathways. Additionally, Capped Coil29 nanofibers were not only smaller, but for a given dose of antigen produced a greater number of particles, which may also have favored endosomal escape and afforded greater opportunities to load peptide into class-I MHC. Capped Coil29 nanofibers also likely had different surface chemistries at fiber tips and potentially had different levels of curvature, again potentially influencing cross-presentation. At present it is not known to what extent such factors may have contributed to the differential immune responses we observed, but independent testing of these factors may be of interest in the future.
In fibrillar peptide biomaterials, it is often difficult to achieve length control. Here, a capping peptide strategy was employed in an α-helical fibrillar system, enabling the tuning of peptide nanofiber length. The level of control afforded was capable of improving cross-presentation and subsequent CD8+ T cell responses against these materials in mice. As this class of materials is being investigated for a range of vaccines and immunotherapies, the present work informs future application towards conditions where maximization of a CD8+ T cell response is desired.
Experimental Section
Peptide Synthesis and Purification:
Peptides were synthesized by microwave-assisted solid phase synthesis on a CEM Liberty Blue synthesizer. After synthesis, peptides were cleaved from resin using a 95% trifluoracetic acid, 2.5% water, 2.5% triisopropylsilane cleavage cocktail and precipitated in diethyl ether. Purification was completed using reverse phase HPLC over a C12 Waters XBridge column. After separating fractions, peptide mass was confirmed using MALDI-ToF mass spectrometry. Purified peptides were frozen, lyophilized, and stored as dry powders at −20°C.
Nanofiber formation:
Nanofibers were prepared by dissolving peptides at a total concentration of 8mM fibrillizing sequences in acetate buffer (pH 4). After dissolving, peptides were stored at 4°C overnight, and the following day diluted to 2mM and brought to a salt concentration equal to 1x PBS. Fibers were then allowed to form at RT for 3 hours before use in assays or imaging. For Capped or Kinked Coil29 nanofibers, Coil29Caps or Coil29Kink were prepared as stock solutions in acetate buffer, and these solutions were used to dissolve Coil29, OVA-Coil29, TAMRA-Coil29 or SIINFEKL-Coil29. After solubilizing all components, nanofibers were formed as described above.
Nanofiber imaging:
For imaging, nanofibers were prepared as described above. For AFM imaging, mica substrates (Electron Microscopy Sciences) were cleaved immediately prior to sample preparation and nanofibers were diluted to a 0.2mM peptide concentration in water. 20 μL of diluted nanofibers were dropped on to the mica surface and allowed to incubate for 30 seconds before they were rinsed with ultrapure water and then dried under a stream of nitrogen. For gold-labeled nanofibers, a 1:100 dilution of streptavidin-conjugated 10 nm gold nanoparticles (Novus Biologicals, cat# 252–0200) were applied for 30 seconds before they were rinsed with ultrapure water and then dried under a stream of nitrogen. Imaging was completed using tapping mode on a Bruker AFM using Bruker RTESPA-300 silicon tips. For TEM imaging, nanofibers were diluted to a 0.2mM peptide concentration in PBS. 5 μL of diluted nanofibers was then dropped on to a formvar-coated copper grid (Electron Microscopy Sciences) and allowed to incubate for 2 minutes before being rinsed with water and stained with 5 μL of a 0.1% solution of uranyl acetate (Electron Microscopy Sciences). After 2 minutes, grids were rinsed with ultrapure water and wicked dry. Imaging was completed on a FEI Tecnai TEM microscope at 160 kV of power and 10,000 – 100,000x magnifications.
Nanofiber measurement and length distributions:
Nanofibers were measured from AFM images by a researcher blinded to the composition of each sample using ImageJ software. Histograms of length distributions were calculated using a script which added the total nanofiber lengths of a given image and then calculated the percentage of material presented in the image belonging to a give size range of nanofibers. The full analysis code can be found in Code S1. Conversion of length to peptide content was completed using measurements of the Coil29 crystal structure (PDB 3j89).
Circular Dichroism:
Secondary structures of peptides were determined by forming nanofibers as described above and then diluting peptides in water immediately prior to scanning. Scans were also taken with peptides in PBS and no changes were detected except for increased noise at low wavelengths. CD spectra were taken using a Chirascan Plus at the Macromolecular Interactions Facility at the University of North Carolina. CD spectra were taken with peptide solutions held at 25°C, from 185 to 260 nm, with a step size of 0.5 nm. Reported spectra are averaged from three separate scans, with error bars showing ±1 standard deviation.
Isothermal Titration Calorimetry:
ITC experiments were performed on a Malvern MicroCal Auto-iTC200 at the Macromolecular Interactions Facility at the University of North Carolina. To study Coil29 binding to Coil29Caps, Coil29Caps were first prepared at 8mM in acetate buffer (pH 4) as they would be prior to nanofiber formation and then diluted to 0.8 mM in acetate buffer (pH 4) before being placed in the reference cell. In two separate experiments, Coil29Caps were prepared separately at 1.6 mM in acetate buffer (pH 4) and injected into the reference cell every 3 minutes for 60 minutes. Thermal fluctuations were converted to kcal mol−1 injectant per molar ratio of injectant to reference material and then fitted to one site binding in Malvern software. A similar procedure was followed to assess Coil29 self-binding. A solution of 8 mM Coil29 in acetate buffer (pH 4) was diluted to 0.8 mM before performing the assay and placed in the reference cell and 1.6 mM Coil29 was injected every 3 minutes for 60 minutes. Data was not fit to any equation because it did not exhibit typical protein-ligand behavior as binding events never reached saturation.
Annealing and Turbidity Measurements:
Annealing of Coil29, Capped Coil29, and extruded Coil29 nanofibers was completed by first forming nanofibers as described above. Extruded nanofibers were passed through a 0.4 μm filter in a syringe extruder 25 times before use or analysis. For turbidity measurements, absorbance was read at 400 nm from 3 separate 2 μL samples on a Nanodrop 2000 (Thermo-Fisher) blanked with PBS. To study the structure of nanofibers under annealing conditions, they were incubated at 37°C for 5 days. Each day, samples were removed from the incubator and 10 μL was aliquoted for analysis by TEM imaging as described above. Samples were then returned to the incubator for the remainder of the experiment.
Cellular Uptake and Presentation:
DC uptake was measured using DC2.4 cells cultured in RPMI media. For uptake experiments, 250,000 cells per well were seeded in a 24-well plate overnight. TAMRA-OVA-Coil29 nanofibers were formed either from TAMRA-OVA-Coil29 (0.2 mM) and Coil29 (1.8 mM) or Coil29Caps (0.1 mM each) and TAMRA-OVA-Coil29 (0.2 mM) and Coil29 (1.8 mM) according to the process described above. TAMRA-OVA-Coil29 nanofibers were then diluted in fresh media and added on to DC cultures for 90 minutes. After incubation, cells were washed, trypsinized, and transferred to flow tubes for analysis by flow cytometry.
Cross-presentation was measured using a BMDC-B3Z assay. First, BMDCs were harvested from the femur of female C57BL/6 mice and cultured with Flt-3 ligand for 8 days to induce DC maturation. Non-adherent cells were then collected, counted, and transferred to a 96-well plate for use at a density of 1,500 cells per well. SIINFEKL-Coil29 nanofibers were formed either from SIINFEKL-Coil29 (0.5 mM) and Coil29 (1.5 mM) or Coil29Caps (0.1 mM each) and SIINFEKL-Coil29 (0.5 mM) and Coil29 (1.5 mM) according to the process described above. After seeding, BMDCs were treated with serial dilutions of SIINFEKL-Coil29 nanofibers, incubated for 2 hours, and then washed with fresh media. B3Z T cells were cultured separately for 1 passage, and then 100,000 cells added to each well containing BMDCs overnight. The amount of SIINFEKL presented by BMDCs was detected via induced activation of lacZ by B3Z T cells when their TCR is bound by a SIINFEKL-MHC I complex on BMDCs. A buffer containing IGEPAL detergent, β-mercaptoethanol, and chlorophenol red-β-D-galactopyranoside (CPRG) was then added to cocultures to allow detection of cleaved CPRG at 615 nm.
Cytotoxicity Assay:
DC2.4 dendritic cells were seeded in a 24-well plate at a density of 200,000 cells/well and allowed to adhere overnight. The following day, Coil29 and Capped Coil29 nanofibers and DMSO were diluted in fresh media at varying concentrations from 200 μM to 1 μM. Media was then added to triplicate wells of dendritic cells and allowed to incubate overnight. The following day, 1 μL of alamarBlue® was added to wells and the fluorescence intensity was read at 585 nm to determine the level of cell proliferation.
Animal immunizations:
Female C57BL/6 mice were immunized via subcutaneous injection with 100 μL of nanofiber solutions. For SIINFEKL-Coil29 immunizations, SIINFEKL-Coil29 (0.5 mM) was mixed with Coil29 (1.5 mM) as a dry powder prior to dissolving in PBS buffer as described above. Capped SIINFEKL-Coil29 nanofibers contained Coil29Caps (0.1 mM each), SIINFEKL-Coil29 (0.5 mM), and Coil29 (1.5 mM). For SIINFEKL-Coil29 immunizations, 8-week-old mice were given a priming immunization, boosted 2 weeks later, and sacrificed at 3 weeks post prime. A similar preparation was followed for OVA-Coil29 immunizations except that the fibers were composed of Coil29Caps (0.1 mM each), OVA-Coil29 (0.5 mM), and Coil29 (1.5 mM). For OVA-Coil29 immunizations, mice were immunized according to arrows on Figure 4. All experiments were performed under Duke University Institutional Animal Care and Use Committee protocol A264–18-11.
Measurement of Antibody Responses:
Blood was collected from mice immunized with OVA-Coil29 and Capped OVA-Coil29 nanofibers every 2 weeks over the course of the immunization regimen shown in Figure 4. After blood collection, serum was isolated via centrifugation and used for serum ELISAs against the OVA epitope. ELISAs were performed by coating high binding 96-well plates (Corning) with streptavidin overnight and treating with biotin-OVA the following day. After blocking plates with Superblock (Thermo Fisher Scientific), either serum or PBS was added in 100-fold dilutions and incubated for 2 hours before applying an HRP conjugated anti-IgG secondary antibody (Jackson, Cat #115–035-071). TMB substrate was added for 5 minutes and stopped with a phosphoric acid solution before absorbance is read at 450 nm. Endpoint titer is calculated by the number of 100-fold dilutions at which the signal from serum treated wells is greater than PBS treated wells by an absorbance greater than 0.2.
Measurement of T Cell Responses:
The induction of SIINFEKL specific CD8 T Cells after immunization with SIINFEKL-Coil29 or Capped SIINFEKL-Coil29 nanofibers was measured by isolating cells from lymph nodes and performing flow cytometry. Mice were immunized with SIINFEKL-Coil29 nanofibers at 3 weeks and 1 week prior to sacrifice and inguinal, axillary, and brachial lymph nodes were isolated, mechanically disrupted, passed through a 70 μm cell strainer, and counted with a Muse Cell Analyzer (Millipore Sigma). Cells were then washed with cold PBS with 2% Fetal Bovine serum, blocked with 2G4.2 antibody for 20 minutes, and stained with APC-tagged SIINFEKL tetramer for 1 hour at 37°C (NIH Tetramer Core Facility). After washing, surface molecules CD4 (FITC-tagged, clone GK1.5), CD3 (PE-tagged, clone UCHT1), CD19(PerCP/Cy5.5-tagged, clone 1D3), F4/80 (PerCP/Cy5.5-tagged, clone BM8), and CD8α (PE-Cy7-tagged, clone SK1) were stained with fluorescent antibodies (Biolegend) for 30 minutes and washed twice. Prior to flow cytometry, cells were stained with DAPI to detect live and dead populations. Flow cytometry was preformed using a BD Canto FACS instrument and signals were compensated using single color controls. Counts of antigen specific T cells were taken by gating SIINFEKL specific T cells (CD4−CD8+B220−F4/80−Tetramer+) and multiplying the percentage of the total population by the total cell count after harvesting lymph nodes.
Supplementary Material
Summary:
Rationally designed peptides control the assembly of α-helical peptide nanofibers, allowing the adjustment of nanofiber length within the range of >1μm to 100 nm. Shortened nanofibers, while raising equivalent B cell responses compared to full-length fibers, induce heightened CD8+ T Cell responses in mice.
Acknowledgements
We thank Vincent Conticello for helpful discussions. This research was supported by the National Institutes of Health (NIBIB 5R01EB009701) and the Biomedical Engineering Department of Duke University. MALDI was performed on an instrument supported by the North Carolina Biotechnology Center, grant 2017-IDG-1018. Circular dichroism was performed at the University of North Carolina at Chapel Hill using instruments supported by NIH grant P30CA016086. CNF and SHK were supported by National Science Foundation Graduate Research Fellowships DGE-1644868 and DGE-1644868, respectively.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
JHC and YW are inventors on United States and PCT patent applications describing the Coil29 system.
Contributor Information
Chelsea N Fries, Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
Yaoying Wu, Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
Sean H Kelly, Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
Michelle Wolf, Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
Nicole L Votaw, Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
Stefan Zauscher, Department of Mechanical Engineering and Materials Science, 3385 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA; Department of Chemistry, 3385 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA; Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
Joel H Collier, Department of Immunology, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA; Department of Biomedical Engineering, 1316 FCIEMAS, 101 Science Dr, Durham, NC 27708, USA.
References
- [1].Kumar VA, Shi S, Wang BK, Li I-C, Jalan AA, Sarkar B, Wickremasinghe NC, Hartgerink JD, J. Am. Chem. Soc 2015, 137, 4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI, Proc Natl Acad Sci USA 2010, 107, 3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hartgerink JD, Benaish E, Stupp SI, Science 2001, 294, 1684. [DOI] [PubMed] [Google Scholar]
- [4].Zhang H, Park J, Jiang Y, Woodrow KA, Acta Biomaterialia 2017, 55, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Black M, Trent A, Kostenko Y, Lee JS, Olive C, Tirrell M, Adv. Mater 2012, 24, 3845. [DOI] [PubMed] [Google Scholar]
- [6].Morris C, Glennie SJ, Lam HS, Baum HE, Kandage D, Williams NA, Morgan DJ, Woolfson DN, Davidson AD, Adv. Funct. Mater 2019, 29, 1807357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Si Y, Wen Y, Kelly SH, Chong AS, Collier JH, Journal of Controlled Release 2018, 282, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu Y, Norberg PK, Reap EA, Congdon KL, Fries CN, Kelly SH, Sampson JH, Conticello VP, Collier JH, ACS Biomater. Sci. Eng 2017, 3, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mora-Solano C, Wen Y, Han H, Chen J, Chong AS, Miller ML, Pompano RR, Collier JH, Biomaterials 2017, 149, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Karabin NB, Allen S, Kwon H-K, Bobbala S, Firlar E, Shokuhfar T, Shull KR, Scott EA, Nature Communications 2018, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Luo Z, Wu Q, Yang C, Wang H, He T, Wang Y, Wang Z, Chen H, Li X, Gong C, Yang Z, Adv. Mater 2017, 29, 1601776. [DOI] [PubMed] [Google Scholar]
- [12].Song H, Huang P, Niu J, Shi G, Zhang C, Kong D, Wang W, Biomaterials 2018, 159, 119. [DOI] [PubMed] [Google Scholar]
- [13].Xing R, Li S, Zhang N, Shen G, Möhwald H, Yan X, Biomacromolecules 2017, 18, 3514. [DOI] [PubMed] [Google Scholar]
- [14].Wen Y, Waltman A, Han H, Collier JH, ACS Nano 2016, 10, 9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hudalla GA, Sun T, Gasiorowski JZ, Han H, Tian YF, Chong AS, Collier JH, Nat Mater 2014, 13, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE, Nature Nanotech 2007, 2, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ke W, Lu N, Japir AA-WMM, Zhou Q, Xi L, Wang Y, Dutta D, Zhou M, Pan Y, Ge Z, Journal of Controlled Release 2020, 318, 67. [DOI] [PubMed] [Google Scholar]
- [18].Dong H, Paramonov SE, Hartgerink JD, J. Am. Chem. Soc 2008, 130, 13691. [DOI] [PubMed] [Google Scholar]
- [19].Thomas F, Burgess NC, Thomson AR, Woolfson DN, Angew. Chem 2015, 128, 999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sato K, Ji W, Palmer LC, Weber B, Barz M, Stupp SI, J. Am. Chem. Soc 2017, 139, 8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yokoi H, Kinoshita T, Zhang S, Proceedings of the National Academy of the Sciences 2005, 102, 8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kelly SH, Wu Y, Varadhan AK, Curvino EJ, Chong AS, Collier JH, Biomaterials 2020, 241, 119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Christian DA, Cai S, Garbuzenko OB, Harada T, Zajac AL, Minko T, Discher DE, Mol. Pharmaceutics 2009, 6, 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rincon-Restrepo M, Mayer A, Hauert S, Bonner DK, Phelps EA, Hubbell JA, Swartz MA, Hirosue S, Biomaterials 2017, 132, 48. [DOI] [PubMed] [Google Scholar]
- [25].Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF, Eur. J. Immunol 2008, 38, 1404. [DOI] [PubMed] [Google Scholar]
- [26].Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neil CP, Lee LK, Swartz MA, Hubbell JA, Nat Biotechnol 2007, 25, 1159. [DOI] [PubMed] [Google Scholar]
- [27].Benne N, van Duijn J, Kuiper J, Jiskoot W, Slütter B, Journal of Controlled Release 2016, 234, 124. [DOI] [PubMed] [Google Scholar]
- [28].Thomas SN, van der Vlies AJ, O’Neil CP, Reddy ST, Yu SS, Giorgio TD, Swartz MA, Hubbell JA, Biomaterials 2011, 32, 2194. [DOI] [PubMed] [Google Scholar]
- [29].Rudra JS, Mishra S, Chong AS, Mitchell RA, Nardin EH, Nussenzweig V, Collier JH, Biomaterials 2012, 33, 6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huang Z-H, Shi L, Ma J-W, Sun Z-Y, Cai H, Chen Y-X, Zhao Y-F, Li Y-M, J. Am. Chem. Soc 2012, 8730. [DOI] [PubMed] [Google Scholar]
- [31].Rudra JS, Ding Y, Neelakantan H, Ding C, Appavu R, Stutz S, Snook JD, Chen H, Cunningham KA, Zhou J, ACS Chem. Neurosci 2016, 7, 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shukla S, Eber F, Nagarajan A, DiFranco N, Schmidt N, Wen A, Eiben S, Twyman R, Wege C, Steinmetz N, Advanced Healthcare Materials 2015, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Smith T, Stains C, Meyer S, Ghosh I, J. Am. Chem. Soc 2006, 14456. [DOI] [PubMed] [Google Scholar]
- [34].Zheng J, Liu C, Sawaya MR, Vadla B, Khan S, Woods RJ, Eisenberg D, Goux WJ, Nowick JS, J. Am. Chem. Soc 2011, 133, 3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shaykhalishahi H, Mirecka EA, Gauhar A, Grüning CSR, Willbold D, Härd T, Stoldt M, Hoyer W, ChemBioChem 2014, 16, 411. [DOI] [PubMed] [Google Scholar]
- [36].Shen H, Fallas JA, Lynch E, Sheffler W, Parry B, Jannetty N, Decarreau J, Wagenbach M, Vicente JJ, Chen J, Wang L, Dowling Q, Oberdorfer G, Stewart L, Wordeman L, De Yoreo J, Jacobs-Wagner C, Kollman J, Baker D, Science 2018, 362, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Aluri S, Pastuszka MK, Moses AS, Mackay JA, Biomacromolecules 2012, 13, 2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee D-W, Kim T, Park I-S, Huang Z, Lee M, J. Am. Chem. Soc 2012, 134, 14722. [DOI] [PubMed] [Google Scholar]
- [39].Adler-Abramovich L, Marco P, Arnon ZA, Creasey RCG, Michaels TCT, Levin A, Scurr DJ, Roberts CJ, Knowles TPJ, Tendler SJB, Gazit E, ACS Nano 2016, 10, 7436. [DOI] [PubMed] [Google Scholar]
- [40].Thomas F, Dawson WM, Lang EJM, Burton AJ, Bartlett GJ, Rhys GG, Mulholland AJ, Woolfson DN, ACS Synth. Biol 2018, 7, 1808. [DOI] [PubMed] [Google Scholar]
- [41].Egelman EH, Xu C, Dimaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP, Structure 2015, 23, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rudra JS, Tian YF, Jung JP, Collier JH, Proc Natl Acad Sci USA 2010, 107, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wu Y, Kelly SH, Sanchez-Perez L, Sampson JH, Collier JH, Biomater. Sci 2020, 8, 3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Morera Y, Sánchez J, Bequet-Romero M, Selman-Housein K-H, de la Torre A, Hernández-Bernal F, Martín Y, Garabito A, Piñero J, Bermúdez C, de la Torre J, Ayala M, Gavilondo JV, Vaccine 2017, 35, 3582. [DOI] [PubMed] [Google Scholar]
- [45].Kazmin D, Nakaya HI, Lee EK, Johnson MJ, van der Most R, van den Berg RA, Ballou WR, Jongert E, Wille-Reece U, Ockenhouse C, Aderem A, Zak DE, Sadoff J, Hendriks J, Wrammert J, Ahmed R, Pulendran B, Proc Natl Acad Sci USA 2017, 114, 2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.