Highlights
-
•
Integrative single-cell transcriptome analysis reveals the accumulation of fibroblasts in liver cancer tissues.
-
•
SPARCL1 expression distinguishes two subpopulations of the liver cancer-distinct fibroblasts.
-
•
SPARCL1 positive fibroblasts are related to favorable prognosis of liver cancer patients.
Keywords: Liver cancer, Single-cell transcriptome, Cancer-distinct fibroblast, SPARCL1
Abstract
Single-cell transcriptome analysis has provided detailed insights into the ecosystem of liver cancer. However, the changes of the cellular and molecular components of liver tumors in comparison with normal livers have not been described at single-cell level. Here, we performed an integrative single-cell analysis of both normal livers and liver cancers. Principal component analysis was firstly performed to delineate the cell lineages in liver tissues. Differential gene expression within major cell types were then analyzed between tumor and normal samples, thus resolved the cell type-specific molecular alterations in liver cancer development. Moreover, a comparison between liver cancer derived versus normal liver derived cell components revealed that two subpopulations of fibroblasts were exclusively expanded in liver cancer tissues. By further defining subpopulation-specific gene signatures, characterizing their spatial distribution in tumor tissues and investigating their clinical significance, we found that the SPARCL1 positive fibroblasts, representing a group of tumor vessel associated fibroblasts, were related to reduced vascular invasion and prolonged survival of liver cancer patients. Through establishing an in-vitro endothelial-to-mesenchymal transition model, we verified the conversion of the fetal liver sinusoidal endothelial cells into the fibroblast-like cells, demonstrating a possible endothelial cell origination of the SPARCL1 positive fibroblasts. Our study provides new insights into the cell atlas alteration, especially the expanded fibroblasts in liver cancers.
Introduction
Liver cancer, consisting mainly of hepatocellular carcinoma and intrahepatic cholangiocarcinoma, is the second most common cause of cancer death worldwide [1]. The therapeutic failure, drug resistance and high mortality of liver cancer have been proved to be highly related to the heterogeneity of malignant cells [2] and the intricate tumor microenvironment (TME) [3]. Thus, resolving the comprehensive cell atlas and molecular component of liver cancer is critical to understanding the tumor biology and developing new therapeutic strategies.
Through previous intensive single-cell studies, detailed insights into the ecosystem of liver cancer have been developed. The single-cell RNA sequencing (scRNA-seq) analysis of liver tumor tissues have identified that the biodiversity of malignant cells and the corresponding TME reprogramming were related to immune therapy responses and patient prognosis [4]. The landscape of liver infiltrating T cells was also uncovered by scRNA-seq [5], and then the global immune cells [6], which comprehensively delineated the composition, functional states, developmental trajectory and cellular interactions of immune cells in liver cancer. These studies have demonstrated that the interplays between malignant cells and their microenvironment played key roles in liver cancer development. However, the changes of liver resident cell components and their conditions, especially the stromal cells, during tumor development have not been described.
Here, with the availability of the scRNA-seq data from both normal liver tissues [7] and liver cancer tissues [4], we performed an integrative analysis and systematically compared the transcriptome within major cell types and cell components between tumor and normal liver samples. We identified two subpopulations of fibroblasts that exclusively expanded in liver cancers and described their transcriptomic differences. Through liver tumor tissue staining, we characterized their spatial distributions. The clinical significance of these cancer-distinct fibroblasts was further revealed by using their specific gene signatures. These results provided insights into the changes of stromal cells between tumor and normal liver tissues at single cell resolution.
Materials and methods
scRNA-seq data collection and processing
Our study investigated a total of 8439 cells from normal liver tissues (n = 5) and 9946 cells form liver cancer tissues (SET1: 5115 cells from 12 primary liver cancer patients; SET2: 4831 cells from 7 primary liver cancer patients). The raw digital gene expression matrix (UMI counts per gene per cell) data were downloaded from the Gene Expression Omnibus (GEO) public database at NCBI (GEO: GSE115469 and GSE125449) [4,7]. Samples of the two datasets were both prepared as outlined by the 10x Genomics Single Cell 3′ v2 Reagent Kit user guide and the sequencing reads have been previously processed using CellRanger (10x Genomics) analysis pipeline. In order to identify shared cell states that present across different datasets, we firstly normalized the different datasets by using SCTransform [8] which was designed to remove the influence of technical effects between datasets and cells. After normalization, we performed the integration method introduced by Seurat v3 [9] to obtain a new integrated matrix.
We then used the new integrated matrix for the downstream analysis. Principal component analysis (PCA) was used to reduce the number of dimensions representing each cell. The first 40 components were applied for shared nearest neighbor (SNN) clustering according to the elbow of the scree plot. Clusters were then visualized using Uniform Manifold Approximation and Projection (UMAP) as implemented in Seurat [10]. Differential gene expression between clusters was calculated using a likelihood-ratio test for single-cell gene expression implemented in Seurat at a family-wise error rate of 5%. Gene Set Enrichment Analysis (GSEA) was performed using clusterProfiler [11] package in R. To generate cellular trajectories (pseudotemporal dynamics), we used the Monocle 3 R package v0.2.2 [12]. All the rest analyses were performed using the Seurat package (version 3.1.2) in R (version 3.6.1).
Identification of parenchymal and non-parenchymal cell lineages
We annotated cell clusters based on the known cell lineage-specific marker genes: T cells (CD3E, CD8B, CD4, IL7R, TRDC), natural killer (NK) cells (NKG7, GNLY, CD7, KLRD1, KLRF1), monocytes (CD14, LYZ, MS4A7, FCER1A, FCGR3A), B/plasma cells (MS4A1, CD79A, FCRL5, IGHA1, IGHG1), fibroblasts (ACTA2, PDGFRA, PDGFRB, ASPN, DCN), endothelial cells (PECAM1, VWF, ENG, CDH5, RAMP3), and hepatocytes/malignant cells (ALB, EPCAM, KRT19, ALDH1A1, CD24).
Analysis of the cancer genome atlas (TCGA) liver tumor data
Gene expression and clinical data of TCGA liver hepatocellular carcinoma (LIHC, n = 368) and adjacent normal (n = 50) samples were downloaded from Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/) [13]. Paired Wilcoxon rank-sum test was used to compare the differences of gene expression between paired tumor and adjacent normal samples. Student's t-test was used to compare the differences in gene expression between vascular invasion groups (None, Micro and Macro). P-values were further adjusted with Benjamini and Hochberg (BH) method. Survival analyses were visualized using Kaplan-Meier curves and the statistical significance was estimated using Log-rank test. All the statistical tests and figures were implemented using R.
Immunohistochemistry (IHC) staining and immunofluorescent (IF) staining
Liver tumor tissue sections derived from 14 liver cancer patients were provided by Beijing Tongren Hospital of China in accordance with the ethical standards of the institutional research committee (project identification code TRECKY2019–077) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from the subjects. The tissue staining was performed following standard protocols from Servicebio Inc. (Wuhan Servicebio Technology Co., Ltd., Wuhan, Hubei, China). Briefly, the 4 µm paraffin sections were deparaffinized, antigen retrieved, blocked and then incubated with primary antibodies [anti-SPARCL1 (Abcam, cat. no. ab255597, 1:1000 for IHC, 1:100 for IF), anti-CD31 (Abcam, cat. no. ab28364, 1:100 for IF), anti-α-SMA (Sigma, cat. no. A5228, 1:100 for IF)]. IHC was detected with the HRP/DAB kit (Servicebio, cat. no. G1215); For Immunofluorescent staining, primary antibodies were followed by FITC, CY5 or CY3 labeled secondary antibodies (Servicebio) and mounted with mounting medium with DAPI counterstain (Servicebio). Bright field images were captured by TissueFAXS (TissueGnostics), fluorescent images were captured by Pannoramic digital slide scanner (3DHISTECH).
Cell culture
Primary human fetal liver sinusoidal endothelial cells were purchased from Pricells and cultured in EGM-2 medium (Lonza) at 37 °C with 5% CO2. Transforming growth factor β1 (TGF-β1) and interleukin-1β (IL-1β) were purchased from Peprotech and reconstituted following the manufacturer's protocol.
Quantitative RT-PCR (qRT-PCR)
RNA isolation, reverse transcription and qRT-PCR were performed as previously described [14]. All the real-time data were analyzed using the ΔΔCt approach with GAPDH as an endogenous control. The primers used are listed below: VECAD F/R: (5′-ACCGTGACGGAATCCTTCTCT-3′) (5′-GCTGGACTCCACTTTGCAC-3′); PECAM1 F/R: (5′-ACCGTGACGGAATCCTTCTCT-3′) (5′-GCTGGACTCCACTTTGCAC-3′); ACTA2 F/R: (5′-CAGGGCTGTTTTCCCATCCAT-3′) (5′-GCCATGTTCTATCGGGTACTTC-3′); MCAM F/R: (5′-GGTCGCTACCTGTGTAGGGA-3′) (5′-TGGACCCGGTTCTTCTCCT-3′); SNAIL F/R: (5′-TCGGAAGCCTAACTACAGCGA-3′) (5′-AGATGAGCATTGGCAGCGAG-3′); GAPDH F/R: (5′-GAGTCAACGGATTTGGTCGT-3′) (5′-TTGATTTTGGAGGGATCTCG-3′).
Results
Integration of single cells derived from normal liver tissues and liver cancer tissues
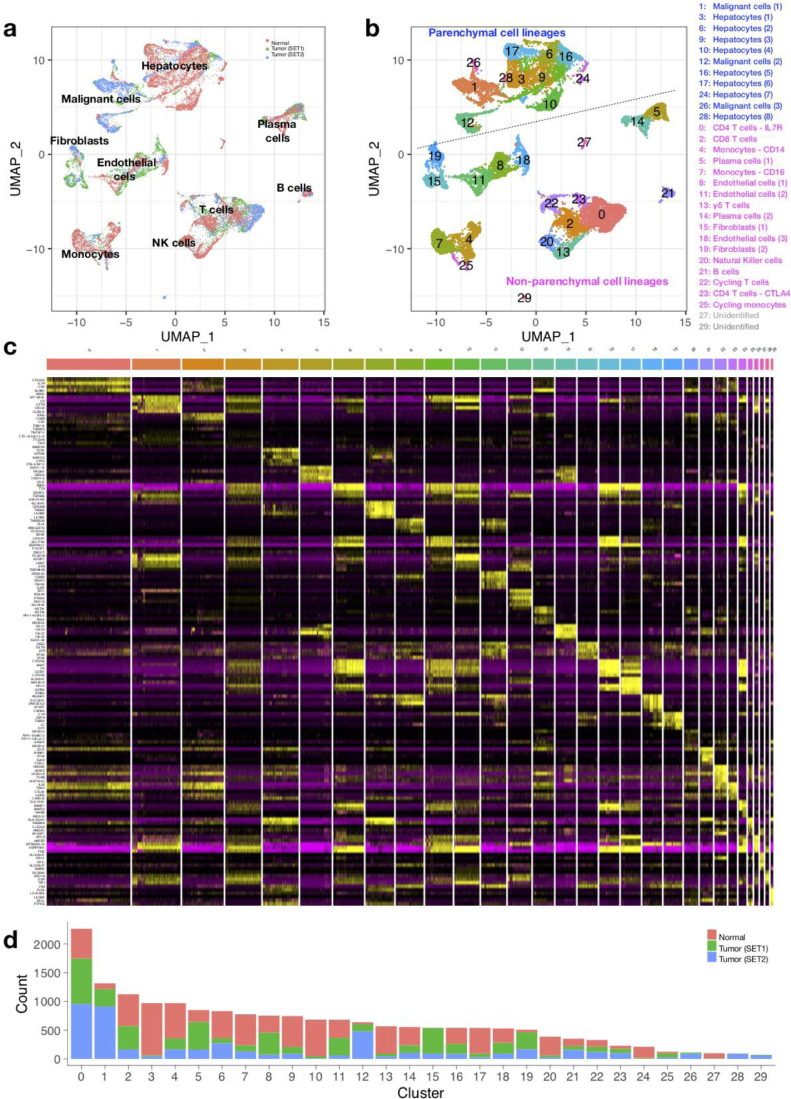
To investigate the difference in the landscape of cells between liver tumors and normal liver tissues, we first need to “anchor” normal and cancerous liver derived datasets together to enable the integrative single-cell measurements. By performing the Seurat alignment workflow, we assembled distinct scRNA-seq datasets into an integrated reference including 8439 cells from normal liver tissues (n = 5) and 9946 cells from liver tumor tissues (SET1: 5115 cells from 12 primary liver cancer patients; SET2: 4831 cells from 7 primary liver cancer patients). The UMAP analysis on the first 40 principle components of each cell in the integrated dataset revealed a total of 8 cell lineages (Fig. 1a), which were well annotated as T cells, B cells, plasma cells, monocytes, endothelial cells, fibroblasts, hepatocytes and malignant cells, according to the known cell lineage-specific marker genes (Fig. S1). To better delineate subpopulations of each cell type, all the cells were further clustered (Fig. 1b) and characterized by the top-ranking cluster-specific genes (Fig. 1c, Supplementary table). A total of 30 subclusters were revealed. Through mapping the cell sources to each cluster, we found that most of the clusters contained cells derived from both normal and cancerous liver tissues (Fig. 1d), thus allowing for our following comparative analysis to identify differentially expressed genes within certain cell type and cell component alterations during liver cancer development.
Fig. 1.
Overview of the single cells derived from normal liver tissues and liver cancer tissues. (a) UMAP plot of all the 18,385 cells from 5 normal liver tissues and 19 liver cancer tissues. Cells are colored by sample sources, normal (red), tumor set1 (green), tumor set2 (blue). (b) UMAP plot of all the single cells colored based on the 30 clusters defined by SNN clustering. (c) Heat map of the top-ranking cluster specific genes. Columns denote cells; rows denote genes. (d) Bar plot showing the number of cells in each cluster.
Comparative analysis of cell components and transcriptome changes in each cell cluster
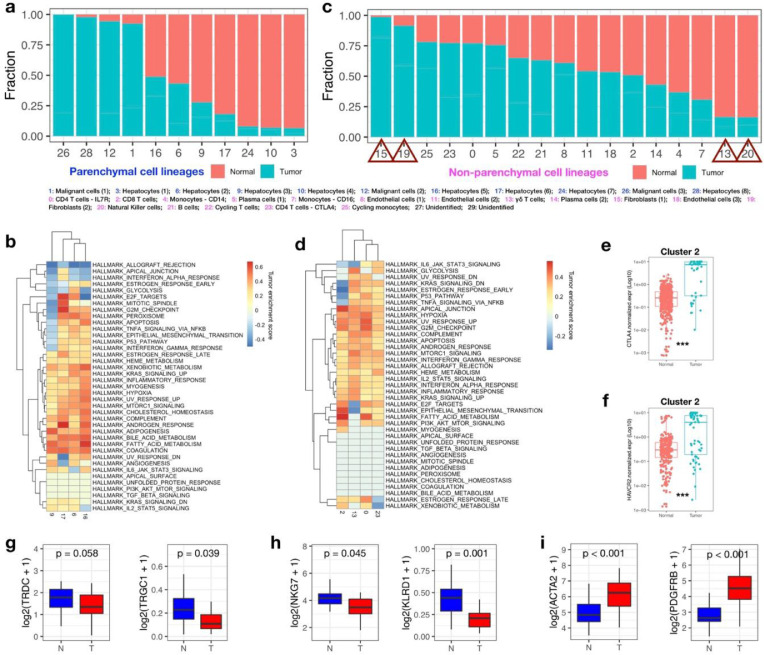
To delineate the cellular landscape alteration in the development of liver cancer, we analyzed the fraction of cells derived from different tissues in each cluster. In parenchymal cell lineages, the malignant cell clusters (cluster 1, 12, 26) were almost exclusively derived from tumor tissues; While some of the tumor-derived cells were clustered in the normal hepatocytes clusters (cluster 6, 9, 16, 17), suggesting that a portion of normal hepatocytes were reserved in the development of liver cancer (Fig. 2a). However, the transcriptome differences in cluster 6, 9, 16 and 17 between cells derived from tumor or normal tissues were still identified. The corresponding GSEA analysis on the differentially expressed genes between tumor and normal tissues suggested that tumor tissue derived hepatocytes are characterized by elevated fatty acid metabolism and bile acid metabolism, but show heterogeneity in proliferative potentials (pathways involving E2F targets, mitotic spindle and G2M checkpoint) (Fig. 2b).
Fig. 2.
Comparative analysis of single cells derived from liver cancers and normal livers. (a) Distribution of cells derived from normal or tumor tissues across parenchymal cell clusters. (b) GSEA analysis for genes differentially expressed between cancer and normal liver-derived hepatocytes within each cluster. Normalized enrichment score (NES) was used to indicate the enrichment of the related pathways. (c) Distribution of cells derived from normal or tumor tissues across non-parenchymal cell clusters. (d) GSEA analysis for genes differentially expressed between cancer and normal liver-derived T cells within each cluster. Normalized enrichment score (NES) was used to indicate the enrichment of the related pathways. (e, f) The expression of CTLA4 (e) and HACVR2 (f) in liver cancer or normal liver derived single cells within cluster 2 were shown. The Wilcoxon rank sum test were used for statistical analysis. *** p<0.001. (g-i) Boxplot showing the expression of marker genes of γδ-T cells (g), NK cells (h) and fibroblast cells (i), between paired tumor and adjacent normal samples in TCGA LIHC cohort. Paired Wilcoxon rank-sum test was used to compare the differences of gene expression between the two groups.
In non-parenchymal cell lineages, T cell clusters (cluster 0, 2, 13, 23), B cell clusters (cluster 5, 14, 21) and endothelial cell clusters (cluster 8, 11, 18) are all consist of cells derived from both tumor and normal tissues (Fig. 2c). In the T-cell lineage, the most prevalent non-parenchymal cell type in liver, we found that almost all of the highly expressed gene in tumor-derived T-cell sub-clusters were enriched in hypoxia, G2M checkpoint, allograft rejection and interferon-α (IFN-α)/ IFN-γ response pathways, indicating that T cells were activated under the influence of tumors (Fig. 2d). In consistent with previous findings that activated T cells in tumors express high level of immune checkpoint molecules, we also observed higher expression of CTLA4 (Fig. 2e) and HACVR2 (Fig. 2f) in tumor infiltrating CD8+ T cells (cluster 2). In addition to T cells, we also investigated the pathway alteration in B cell lineage. The enriched pathways (upregulated G2M checkpoint and E2F targets) in liver cancer-derived B cells within cluster 21 revealed the tumor-associated proliferation of B cells, while the differentially expressed genes within cluster 5 or 14 suggested inhibited cell proliferation and accumulated hypoxia pathways in tumoral plasma B cells (Fig. S2a). As to the endothelial cells (cluster 8, 11 and 18), we found the general upregulation of TGF-β signaling, G2M checkpoint and fatty acid metabolism in cancer-associated endothelial cells. While the cells in cluster 18, a cluster of ACKR positive cells responsible for fibrotic niche formation in cirrhotic livers [15], also showed distinct expression profile in liver cancers involving upregulated heme metabolism and inhibited bile acid metabolism (Fig. S2b).
Apart from the above clusters with mixtured cell originations, the top two clusters consisting mostly of normal liver-derived cells were annotated as NK cells (cluster 20) and γδ-T cells (cluster 13); In contrast, the top two clusters (cluster 15, 19) consisting mostly of cells from liver tumor tissues were both annotated as fibroblast cells. These results implied a possible decrease in the infiltration of γδ-T cells and NK cells and an accumulation of fibroblasts in liver tumors. Consistent with these observations, we also observed the downregulation of γδ-T cell markers (TRDC and TRGC1, Fig. 2g) and NK cell markers (NKG7 and KLRD1, Fig. 2h), and the upregulation of fibroblasts markers (ACTA2 and PDGFRB, Fig. 2i) in TCGA LIHC tumor samples compared with the paired adjacent normal samples.
Identifying sub-populations of the liver cancer-distinct fibroblasts
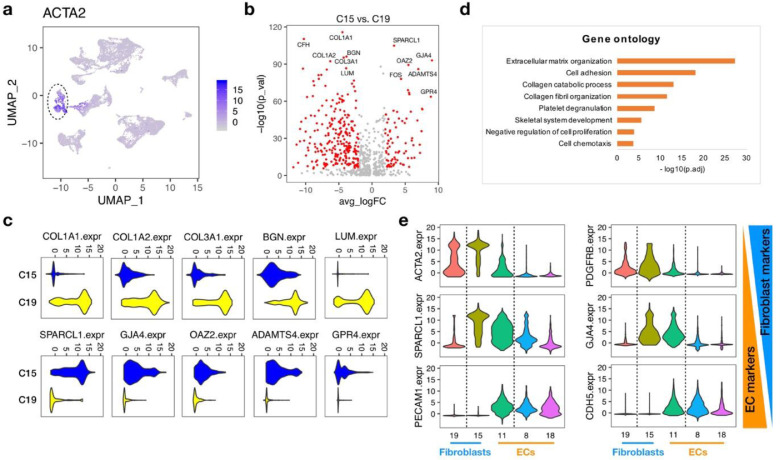
Fibroblasts are the major stromal cell type in the microenvironment of liver diseases, including liver cirrhosis and liver cancers. They have been reported to play important roles in the development of liver diseases and therefore the potential therapeutic targets [16]. Inspired by the observed accumulation of fibroblasts in liver tumors, we further investigated the heterogeneity of these expanded cancer-distinct fibroblasts and aimed to unravel their biological functions. Since the classical fibroblast marker ACTA2 did not discriminate between cluster 15 and cluster 19 (Fig. 3a), to fully annotate the two clusters of cancer-distinct fibroblasts, we analyzed differentially expressed genes between them. We found that fibrogenic genes (e.g., COL1A1, COL1A2, COL3A1) and proteoglycan encoding genes (e.g., LUM, BGN) were overexpressed in cluster 19 (Fig. 3b, c). Gene Ontology (GO) enrichment analysis further revealed that the upregulated genes in cluster 19 were significantly associated with biological processes including extracellular matrix organization and collagen fibril organization (Fig. 3d), indicating that cluster 19 could be functionally annotated as the fibrotic scar-associated fibroblast cells.
Fig. 3.
Subpopulations of cancer-distinct fibroblasts. (a) UMAP plot showing the expression of fibroblast cell marker, ACTA2, across all the cell lineages. (b) Volcano plot of differential expressed genes between the two clusters of cancer-distinct fibroblasts (Cluster 15 and Cluster 19). (c) Violin plots of the top-ranking differential expressed genes between the two clusters of cancer-distinct fibroblasts (Cluster 15 and Cluster 19). (d) Gene ontology analysis of genes highly expressed in Cluster 19. (e) Violin plots showing the expression of marker genes across the fibroblast cell clusters (C15, C19) and endothelial cell clusters (C8, C11, C18).
Compared with cluster 19, cluster 15 showed a quite different gene expression pattern (Fig. 3b, c). Although the upregulated genes in cluster 15 were not enriched in any GO terms, we found that many of which were simultaneously highly expressed in one subpopulation of the endothelial cells (cluster 11), especially the top-ranking genes, SPARCL1 and GJA4 (Fig. 3e). SPARCL1 has been reported to be expressed in confluent endothelial cells [17] and was one of the signature genes for tumor angiogenesis [18]. GJA4 was also demonstrated to play important roles in endothelial cells, including the formation of gap junctions [19], cell cycle regulation [20] and the production of nitric oxide [19,21]. Thus, different from the fibrotic scar-associated fibroblast cells (cluster 19), the cells in cluster 15 displayed a hybrid gene signature, with both features of fibroblast cells and endothelial cells, suggesting that they might be functionally associated with endothelial cells.
Spatial distribution of the cancer-distinct fibroblasts in liver tumor tissues
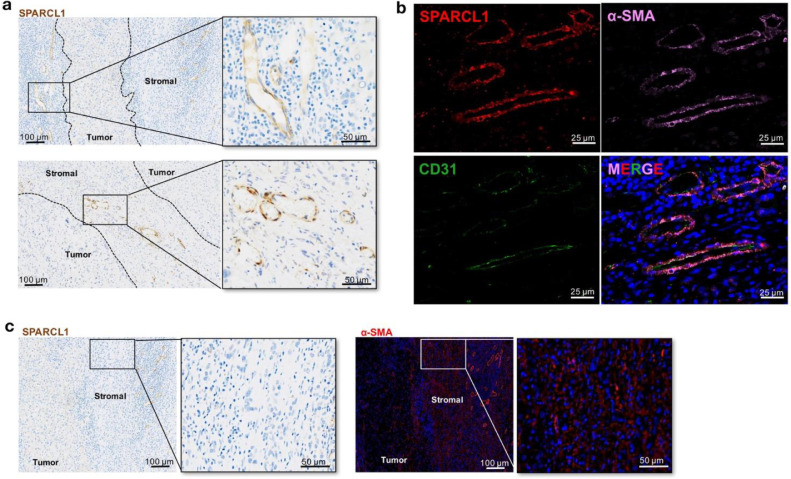
In order to further characterize the two clusters of cancer-distinct fibroblasts, we performed tissue stainings to investigate their spatial distributions in liver tumor tissues.
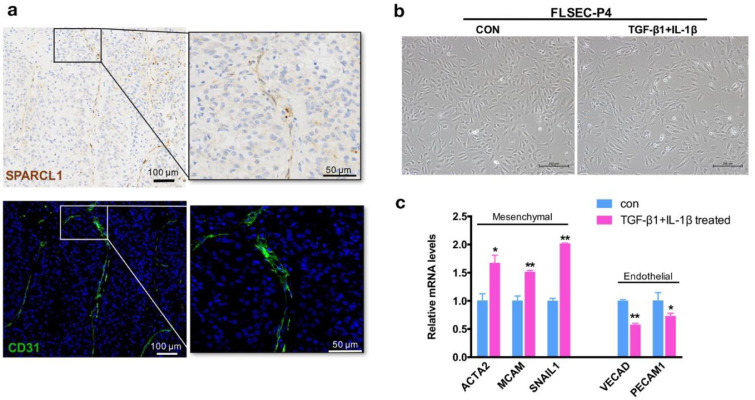
Firstly, the expression of SPARCL1, the top-ranking marker for cluster 15, was analysed by IHC staining. The staining result revealed that the specific SPARCL1 signals were located in the large blood vessels in the stromal niche of liver tumor, while the core area of the tumor tissues showed negative signals (Fig. 4a). To further confirm the fibroblast traits of these SPARCL1 positive cells, we co-stained the fibroblast cell specific marker α-SMA (ACTA2 gene) and endothelial cell specific marker CD31 (PECAM1 gene) (Fig. 4b). The SPARCL1 positive cells also expressed α-SMA but were lack of CD31 expression, which was consistent with the gene expression feature of cells in cluster 15. This observation suggested that cells in cluster 15 topographically localized in the stromal niche of liver tumor, representing a group of vessel associated fibroblasts.
Fig. 4.
Spatial distribution of the cancer-distinct fibroblasts in liver tumor tissues. (a) IHC staining for SPARCL1 expression in samples from liver cancer patients. Scale bar: 100 µm (left panel) and 50 µm (right panel). (b) Colocalization of SPARCL1, α-SMA and CD31 was determined by IF staining (SPARCL1: red; α-SMA: pink; CD31: green). Tissues were counterstained using DAPI (blue). Scale bar: 25 µm; (c) IHC staining for SPARCL1 and IF staining for α-SMA was performed on consecutive sections. Scale bar: 100 µm (left panel) and 50 µm (right panel).
Meanwhile, the tumor stromal areas were also filled with the α-SMA+SPARCL1−fibroblasts, i.e. cells belonging to cluster 19 (Fig. 4c). Previous study on fibrotic livers has demonstrated that the scar-associated mesenchymal (SAMes) cells, which expanded in cirrhotic livers, were mapped to the fibrotic niche [15]. The cells in cluster 19 showed great similarities with these SAMes cells, in both gene signatures and spatial distributions, suggesting that cluster 19 might primarily participate in the development of liver cirrhosis, and then the liver cancer.
Cluster 15 is related to favorable prognosis of liver cancer patients
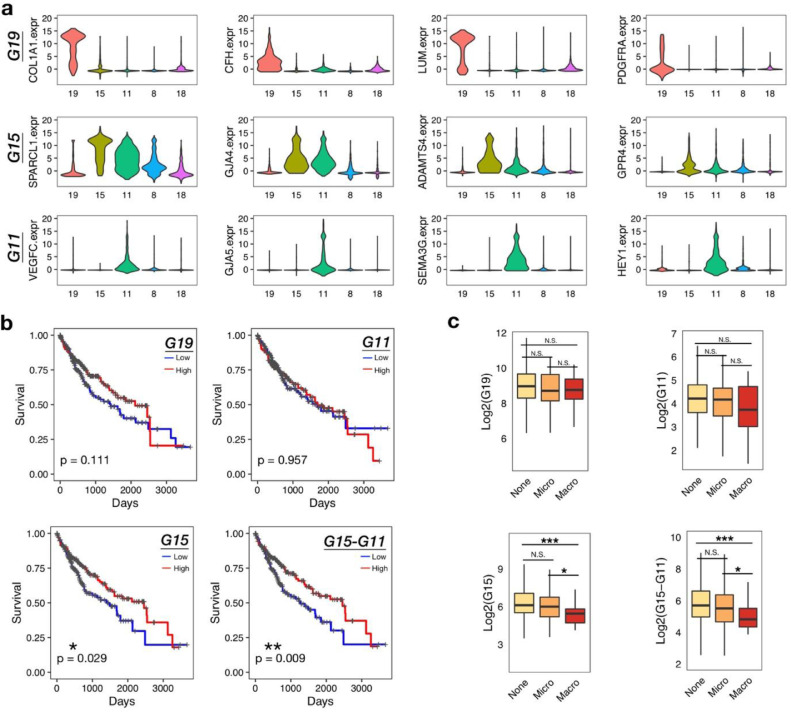
The abnormal expansion of fibroblasts in the liver tumor microenvironment prompted us to examine their association with patients’ clinical features. We firstly identified the cluster-specific signature genes to fully define the different clusters (Fig. 5a). Cluster 19 was well defined by the specific gene set G19 (COL1A, CFH, LUM and PDGFRA). Cluster 15 could be distinguished from cluster 19 by gene set G15 (SPARCL1, GJA4, ADAMTS4 and GPR4), however, G15 was also expressed in cluster 11. In order to further distinguish cluster 15 from cluster 11, we also identified cluster 11 specific gene set G11, including VEGFC, GJA5, SEMA3G and HEY1. Thus, the level of cluster 15 could be evaluated by G15 minus G11 (G15-G11). Then, the associations of each gene set with the survival time of patients were analyzed in the TCGA LIHC cohort (Fig. 5b). We found that the higher expression of gene set G15 in tumor tissues were correlated with prolonged survival time of patients, while G19 and G11 showed no correlations. Not only that, to exclude the impact of cluster 11 on the expression of G15, G15-G11 was also evaluated and showed a more significant correlation with good survival than G15.
Fig. 5.
Favorable clinical outcome of LIHC patients with high expression of C15 signature genes. (a) Violin plots showing the expression of cluster-specific genes across the two fibroblast cell clusters (C15, C19) and three endothelial cell clusters (C8, C11, C18). Genes in each row were grouped into the cluster-specific gene set, i.e. G19, G15 and G11. (b) Kaplan-Meier curves showing the association between the cluster-specific gene set scores and patient survival time in TCGA LIHC cohorts. The score was defined by the sum expression of the signature genes in each gene set. The high or low groups were determined by the median of each score. Statistical significance comparing different groups was evaluated using the Log-rank test. (c) Boxplots showing G19, G11, G15 and G15-G11 scores across vascular invasion groups in TCGA LIHC cohort (None, Micro and Macro). Student's t-test was used to compare the differences in gene expression between vascular invasion groups (None, Micro and Macro). P-values were further adjusted with Benjamini and Hochberg method. *adjusted p<0.05, ***adjusted p<0.001.
In addition, as Cluster 15 representing a group of vessel associated fibroblasts and tumor vascular invasion has been recognized as a crucial prognostic factor for liver cancer patients [22,23], we further analyzed the relationship between G15 expression and tumor vascular invasion. As shown in Fig. 5c, patients with higher G15 expression were more likely to be absent of vascular invasion, or only develop microvascular invasions; Lower G15 expression was related to macrovascular invasion, which involves main portal veins and predicts worse survival [23]. The similar trend was also observed in G15-G11, but not in G19 and G11, suggesting that the SPARCL1+GJA4+ADAMTS4+GPR4+ fibroblasts might play positive roles in protecting blood vessels from tumor cell invasions, which contribute to the favorable prognosis of liver cancer patients.
Investigating the possible origin of cells in cluster 15
Given that the SPARCL1 positive fibroblasts (cluster 15) shared similar gene signatures with one of the endothelial cell subpopulations (cluster 11), along with the vessel associated phenotype of cluster 15, there is a possibility that cells in cluster 15 originated from the endothelial cells in cluster 11. We thus performed a pseudotemporal analysis to investigate our hypothesis. The transcriptional profile of cells in cluster 11 and cluster 15 was mapped along a pseudotemporal trajectory, suggesting the possible transitions of cluster 11 towards cluster 15 (Fig. S3).
Then, we returned to the tissue staining to better define the identity of cluster 11. We found that the SPARCL1 and CD31 double stained cells were sporadically located in the sinusoidal structure in some of the liver tumors (Fig. 6a), suggesting that cluster 11 might be the liver sinusoidal endothelial cells (LSECs). With a process known as endothelial-to-mesenchymal transition (EndMT) [24], we cultured the human fetal LSECs (FLSECs) in vitro and treated FLSECs with TGF-β1 and IL-1β, a combination of potent EndMT inducers [25], to establish an in-vitro model of EndMT. After TGF-β1/IL-1β treatment for 4 days (FLSECs were treated in FBS-reduced medium to avoid overgrowth), the FLSECs changed to the more spindle-like shape (Fig. 6b). A significant increase of mesenchymal markers, ACTA2, MCAM and SNAIL1, along with the downregulation of endothelial markers VECAD and PECAM1 further pointed to the gain of fibroblastic properties in the treated endothelial cells (Fig. 6c). The specific expression profile of TGFBR2 (TGFBR1 and IL1R1 were not detectable in the datasets) in three clusters of endothelial cells also supported for the molecular basis of EndMT (Fig. S4).
Fig. 6.
TGF-β1 induces EndMT in FLSECs in vitro. (a) Spatial distribution of cells belonging to cluster 11 in liver tumor tissues. SPARCL1 expression was detected by IHC staining. Colocalization of CD31 (green) was determined by immunofluorescence staining on consecutive sections. Tissues were counterstained using DAPI (blue). Scale bar: 100 µm (left panel) and 50 µm (right panel). (b) Representative pictures of FLSECs maintained in EGM-2 medium with reduced fetal bovine serum (CON, day 4, picture in the left), or FBS reduced EGM-2 medium containing 10 ng/mL of TGF-β1 and 1 ng/mL of IL-1β (TGF-β1+IL-1β, treated for 4 days, in the right). Scale bar: 200 µm. (c) Comparison of the relative expression levels of mesenchymal markers and endothelial markers in TGF-β1/IL-1β treated FLSECs (4 days), with non-treated FLSECs as control. FLSECs were cultured and treated in the FBS reduced EGM-2 medium.
Based on the above evidences, we deduced that there might be a possibility of the transition of the sinusoidal endothelial cells to fibroblasts in liver cancer.
Discussion
Solid tumor tissue consists of complicated and heterogeneous groups of tumor cells, tumor associated stromal cells and reserved normal cells. Here, we conducted a novel integrative analysis of the single cells derived from both normal and cancerous livers, and found that in liver cancers, except for the malignant cells, the most significant alteration in cell atlas was the expansion of a population of ACTA2+ fibroblasts. The cancer associated fibroblasts, which were mainly studied through tissue staining or in vitro experiments, have been demonstrated to play tumor-promoting roles in liver cancer through multiple mechanisms, including direct interactions with tumor cells [26,27], or indirect effects on tumor cells by cross talking with endothelial cells [28] and immune cells [29,30]. However, in contrary to the previous conclusions, our single-cell analysis here revealed the SPARCL1 positive fibroblasts (cluster 15) were associated with favorable prognosis of liver cancer patients.
Functionally, SPARCL1 is a member of secreted protein, and has been identified as a tumor suppressor in prostate cancer [31], colorectal cancer [32] and gastric cancer [33], etc. In addition, a study in colorectal carcinoma demonstrated that the SPARCL1 expression was induced in endothelial cells by the pro-survival tumor microenvironment, the secreted SPARCL1 further maintained the vessel homeostasis and then contributed to the favorable prognosis of patients [17]. This is highly consistent with our observations on the positive roles of SPARCL1+ fibroblasts. Thus, here we speculate that the expanded SPARCL1+ fibroblasts in liver tumors may protect vessels from tumor cell invasions either through stabilizing the vessels or reducing the invasive ability of tumor cells.
Here, we also explored the possible origination of this group of protective fibroblasts by establishing an in-vitro EndMT model, which clarified the phenotypic conversion of the FLSECs into the fibroblast-like cells, as well as the induction of the fibroblast-specific marker. However, one limitation of this model is that the fetal liver derived sinusoidal endothelial cells might be different from adult livers. Besides, whether this phenomenon is also true in vivo and whether other possible origins of the expanded fibroblasts exist still ask for more intensive studies in the future.
In summary, with the availability of the scRNA-seq data from both normal liver tissues and liver cancer tissues, we performed an integrative analysis and systematically compared the cell components and transcriptome changes between tumor and normal liver samples. We identified two subpopulations of fibroblasts that exclusively expanded in liver cancers and further described their gene signature, spatial distribution and clinical significance. We demonstrated for the first time that SPARCL1 positive fibroblasts, representing a group of tumor vessel associated fibroblasts, were related to reduced vascular invasion and prolonged survival of liver cancer patients. Our study provides new insights into the fibroblasts that expanded in liver cancers at single cell resolution.
Declaration of Cmpeting Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
We thank Prof. Xin Wei Wang for the helpful discussions. This work was supported by grants from National Natural Science Foundation of China (81773137, 31801227), Guangzhou Health Care and Cooperative Innovation Major Project (201508020257, 201803040005), Beijing Hospitals Authority Youth Programme (QML20180203).
Author contributions
Conceptualization, H.Y.W., J.Z., W.Y. and X.T.P.; Methodology, H.Y.W., C.F., M.X.L., B.Z., Q.Z. and J.F.X.; Data curation, Z.J.; Formal analysis, J.Z.; H.Y.W.; Investigation, H.Y.W., J.Z., B.Z., Y.C.X., J.N.Z. and X.M.Y.; Writing—original draft preparation, H.Y.W., J.Z. and W.Y.; Writing—review and editing, all authors.; Supervision, W.Y. and X.T.P.; All authors have read and agreed to the published version of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100981.
Contributor Information
Jian Zhang, Email: jian_zhang@tju.edu.cn.
Wen Yue, Email: yuewen@bmi.ac.cn.
Xuetao Pei, Email: peixt@bmi.ac.cn.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019: cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Dang H., Wang X.W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 2018;50:e416. doi: 10.1038/emm.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amicone L., Marchetti A. Microenvironment and tumor cells: two targets for new molecular therapies of hepatocellular carcinoma. Transl. Gastroenterol. Hepatol. 2018;3 doi: 10.21037/tgh.2018.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L., Hernandez M.O., Zhao Y., Mehta M., Tran B., Kelly M. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019 doi: 10.1016/j.ccell.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng C., Zheng L., Yoo J.-.K., Guo H., Zhang Y., Guo X. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169 doi: 10.1016/j.cell.2017.05.035. 1342-1356.e16. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845. doi: 10.1016/j.cell.2019.10.003. e20. [DOI] [PubMed] [Google Scholar]
- 7.MacParland S.A., Liu J.C., Ma X.-.Z., Innes B.T., Bartczak A.M., Gage B.K. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu G., Wang L-G, Han Y., He Q.-.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ally A., Balasundaram M., Carlsen R., Chuah E., Clarke A., Dhalla N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H.-.Y., Zhang B., Zhou J.-N., Wang D-X, Xu Y.-C., Zeng Q. Arsenic trioxide inhibits liver cancer stem cells and metastasis by targeting SRF/MCM7 complex. Cell Death Dis. 2019;10 doi: 10.1038/s41419-019-1676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z., Dong C., Jiang K., Xu Z., Li R., Guo K. Heterogeneity of cancer-associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. J Hematol Oncol. 2019;12 doi: 10.1186/s13045-019-0782-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naschberger E., Liebl A., Schellerer V.S., Schütz M., Britzen-Laurent N., Kölbel P. Matricellular protein SPARCL1 regulates tumor microenvironment–dependent endothelial cell heterogeneity in colorectal carcinoma. J. Clin. Invest. 2016;126:4187–4204. doi: 10.1172/JCI78260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masiero M., Simões F.C., Han H.D., Snell C., Peterkin T., Bridges E. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell. 2013;24:229–241. doi: 10.1016/j.ccr.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pogoda K., Füller M., Pohl U., Kameritsch P. NO, via its target Cx37, modulates calcium signal propagation selectively at myoendothelial gap junctions. Cell Commun. Signal. 2014;12:33. doi: 10.1186/1478-811X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang J.S., Coon B.G., Gillis N., Chen Z., Qiu J., Chittenden T.W. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfenniger A., Derouette J.-P., Verma V., Lin X., Foglia B., Coombs W. Gap junction protein Cx37 interacts with endothelial nitric oxide synthase in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2010;30:827–834. doi: 10.1161/ATVBAHA.109.200816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonas S., Bechstein W.O., Steinmüller T., Herrmann M., Radke C., Berg T. Vascular invasion and histopathologic grading determine outcome after liver transplantation for hepatocellular carcinoma in cirrhosis. Hepatology. 2001;33:1080–1086. doi: 10.1053/jhep.2001.23561. [DOI] [PubMed] [Google Scholar]
- 23.Aa M., Ag S., Ja M., H Z., Ac Y. Vascular invasion and metastasis is predictive of outcome in barcelona clinic liver cancer stage c hepatocellular carcinoma. J. Natl. Compr. Canc. Netw. 2017;15:197–204. doi: 10.6004/jnccn.2017.0020. [DOI] [PubMed] [Google Scholar]
- 24.Zeisberg E.M., Potenta S., Xie L., Zeisberg M., Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 25.Xiong J., Kawagishi H., Yan Y., Liu J., Wells Q.S., Edmunds L.R. A metabolic basis for endothelial-to-mesenchymal transition. Mol. Cell. 2018;69:689–698. doi: 10.1016/j.molcel.2018.01.010. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C., Liu L., Chen X., Cheng J., Zhang H., Zhang C. LSD1 stimulates cancer-associated fibroblasts to drive notch3-dependent self-renewal of liver cancer stem–like cells. Cancer Res. 2018;78:938–949. doi: 10.1158/0008-5472.CAN-17-1236. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M., Yang H., Wan L., Wang Z., Wang H., Ge C. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020;73:1118–1130. doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 28.Gascard P., Tlsty T.D. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–1019. doi: 10.1101/gad.279737.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monteran L., Erez N. The dark side of fibroblasts: cancer-associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Y., Li H., Deng Y., Tai Y., Zeng K., Zhang Y. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis. 2018;9 doi: 10.1038/s41419-018-0458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley P.J., Marchionni L., Simons B.W., Ross A.E., Peskoe S.B., Miller R.M. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc. Nat. Acad. Sci. 2012;109:14977–14982. doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H., Zhang H., Ge W., Liu X., Loera S., Chu P. Secreted protein acidic and rich in cysteines-like 1 suppresses aggressiveness and predicts better survival in colorectal cancers. Clin. Can. Res. 2012;18:5438–5448. doi: 10.1158/1078-0432.CCR-12-0124. [DOI] [PubMed] [Google Scholar]
- 33.Hu H., Cai W., Zheng S., Ge W. SPARCL1, a novel prognostic predictive factor for gi malignancies: a meta-analysis. Cell. Physiol. Biochem. 2017;44:1485–1496. doi: 10.1159/000485584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.