Highlights
-
•
SncRNAs contribute to the progress of renal cell carcinoma.
-
•
SncRNAs are promising biomarkers for diagnosis and prognosis of renal cell carcinoma.
-
•
Despite the potential of sncRNA-based cancer therapy, some obstacles remain, including several severe adverse effect.
Keywords: Small noncoding RNA, Renal cell carcinoma, MicroRNA, Piwi-interacting RNA, TRNA-derived small RNA
Keywords: Abbreviations: 3′UTR, 3′ untranslated region; AGO, argonaute; ANG, angiogenin; ccRCC, clear cell renal cell carcinoma; DGCR8, diGeorge syndrome critical region 8; EMT, epithelial-mesenchymal transition; EXP5, exportin 5; LncRNA, long non-coding RNA; miRNA, microRNA; miRISC, miRNA-induced silencing complex; piRNA, piwi-interacting RNA; pRCC, papillary renal cell carcinoma; RCC, renal cell carcinoma; SncRNA, small non-coding RNA; tiRNA, tRNA halves; TKI, tyrosine kinase inhibitors; TRBP, transactivation-responsive RNA-binding protein; tRFs, tRNA-derived fragments; tsRNA, tRNA-derived small RNAs; Zuc, zucchini
Abstract
Noncoding RNAs are transcribed in the most regions of the human genome, divided into small noncoding RNAs (less than 200 nt) and long noncoding RNAs (more than 200 nt) according to their size. Compelling evidences suggest that small noncoding RNAs play critical roles in tumorigenesis and tumor progression, especially in renal cell carcinoma. MiRNA, the most famous small noncoding RNA, has been comprehensively explored for its fundamental role in cancer. And several miRNA-based therapeutic strategies have been applied to several ongoing clinical trials. However, piRNAs and tsRNAs, have not received as much research attention, because of several technological limitations. Nevertheless, some studies have revealed the presence of aberration of piRNAs and tsRNAs in renal cell carcinoma, highlighting a potentially novel mechanism for tumor onset and progression. In this review, we provide an overview of three classes of small noncoding RNA: miRNAs, piRNAs and tsRNAs, that have been reported dysregulation in renal cell carcinoma and have the potential for advancing diagnosis, prognosis and therapeutic applications of this disease.
Graphical abstract
Introduction
Renal cell carcinoma(RCC) is the sixth most frequently diagnosed cancer in men and the tenth in women worldwide [1]. RCC is particularly prevalent in Northern American, while the incidence rate is low in Africa. Those differences have been attributed to the differences in races, age structure of population and detection technology [2]. In addition, cigarette use, obesity, and hypertension, the most influential risk factors for RCC [1], also play a role in RCC prevalence. Annually, there are more than 175,000 RCC deaths worldwide [3]. Because of the high rate of metastasis and recurrence, it is conceivable that many patients are only diagnosed at the advanced stages of disease. Nearly one third of patients are diagnosed with metastatic RCC [4]; Moreover, RCC is highly resistant both to chemotherapy and radiotherapy. Although there has been extensive research on RCC, the specific mechanisms underlying carcinogenesis in the kidney are still poorly understood [5]. Hence, discovering the detailed molecular mechanism of RCC is of urgent importance for the development of effective treatments for RCC.
Initially, scientists focused their eyes on protein-coding genes to identify relevant proteins, when exploring the molecular mechanism of kidney carcinogenesis. However, there are only ∼2% of human DNA coding proteins and over 80% of the human genome is transcribed to various transcripts without protein-coding potential with the rapid development of high throughput sequencing. Currently, more and more research has been focused on non-coding RNAs (ncRNAs); once considered as “junk RNA”, it is now clear that many ncRNAs are participating in many critical physiological and pathological processes [6,7]. Aberrant expression of ncRNAs is frequently observed in different types of carcinomas, suggesting a key role for ncRNAs in carcinogenesis [8].
Non-coding RNAs can be grouped into several classes, grouped by size. Small ncRNAs (sncRNAs), which play a key role in carcinogenesis, include miRNAs, piRNAs and tsRNAs. Long ncRNAs (lncRNAs), comprising ncRNAs longer than 200 nucleotides(nt), include pseudogenes and circRNAs [9].
Although the current knowledge of ncRNAs, especially small ncRNAs, is still inadequate, the elevating evidences for their indispensable role in many biological processes suggest their considerable roles in carcinogenesis.This review summarizes the current knowledge of small ncRNAs, with emphasis on the roles of those molecules in renal cell carcinoma. We also highlight the possible utilization of sncRNAs as novel biomarkers and potential therapeutic targets in renal cell carcinoma [10].
Evidence acquisition
We accessed PubMed to search English-language articles up to June 2020, using a combination of the following terms: noncoding RNA, or small noncoding RNA, or microRNA, or miRNA, or piRNA, or piwi-interacting RNA, or TRF, or tRNA derived small fragments, and kidney cancer, or renal cell carcinoma or RCC.
MicroRNA
MiRNAs biogenesis and biological functions
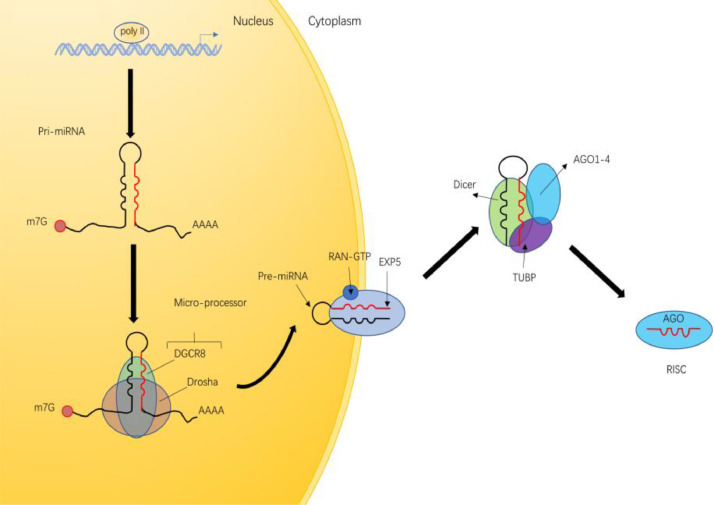
MiRNA is a type of sncRNAs about 22 nt in length. It guides Argonaute(AGO) proteins to target 3′ untranslated region (3′UTR) of mRNA, mediating posttranscriptional gene silencing. Since the first miRNA was reported in Caenorhabditis elegans in 1993 [11], thousands of miRNAs have been discovered in flies, plants and mammals. Canonical miRNAs are transcribed by RNA polymerase II (poly II) to generate the primary miRNAs (pri-miRNA) in the nucleus, where pri-miRNA is processed by 5′capping and 3′polyadenylation [12]. This initial event is followed by two processing steps: The first step is cleaved by a microprocessor, including DiGeorge syndrome critical region 8 (DGCR8) that recognizes pri-miRNAs and Drosha that cleaves the pri-miRNA to an approximately 70-nucleotide-long hairpin structure called pre-miRNA [13,14]. Next,Exportin-5 and RAN-GTP export pre-miRNA to the cytoplasm [15].In the second step, Dicer generates the mature 18∼25 nt miRNA duplex by binding and cleaving the dsRNA stem, during which the transactivation-responsive RNA-binding protein TRBP enhances the Dicer -mediated cleavage and alters miRNA guide-strand selection [16]. Next, Argonaute incorporates one strand of mature miRNA to form the miRNA-induced silencing complex (miRISC); the other strand is degraded [17,18] (Fig. 1).
Fig. 1.
Overview of the miRNA biogenesis pathway. Pri-miRNAs are transcribed by RNA polymerase II in the nucleus. The first step is mediated by a microprocessor, comprised of Drosha and DGCR8, to produce pre-miRNA. Next, pre-miRNAs are exported to the cytoplasm by EXP5. The second step is mediated by Dicer, TRBP and AGO1–4, during which pre-miRNA are processed. Finally, the remaining strand of miRNA is loaded on AGO to form RISC.
MiRNA is thought to exert its main function by binding to and silencing target mRNAs in a sequence-specific manner. MiRNA commonly targets 3′UTR sequences of mRNAs. The capability of miRNAs to combine with mRNAs depends largely on the level of complementarity between these molecules [13]. The miRNA-mRNA pair recruits RISC to regulate mRNA expression by hampering the translational process or mRNA decay. Thus, the miRNA expression can be related to clinical features and biological characteristics [19].
Although miRNAs exist and function mainly in the cytoplasm, there is still a group of miRNAs secreted extracellularly, that can be detected by fluid biopsy. Extracellular miRNAs have been extensively shown to participate in the regulation of multiple targets of recipient cells, and they trigger almost every pivotal process of tumor progression [20]. In addition, extracellular miRNAs can be detected in a convenient and non-invasive way, suggesting their potential as sources of important clinical information on the onset, progression, and recurrence of cancer and response to cancer treatment [19].
MiRNAs alteration in renal cell carcinoma
The function of miRNAs in the progression of RCC has been reported in extensive research. Here, we present a summary of some crucial miRNAs and their eventful roles in carcinogenesis (Table 1).
Table 1.
Altered microRNA (miRNA) expression in renal cell carcinoma.
| MiRNA | Expression | mRNA target | Reference |
|---|---|---|---|
| miR-200b | down | LAMA4 | [32] |
| miR-16 | up | SLC27A4, CD55 | [33] |
| miR-200c-3p | down | ZEB1 | [21] |
| miR-543 | up | KLF6 | [34] |
| miR-22 | down | PTEN | [25] |
| miR-19 | up | FRK | [26] |
| miR-124 | down | ZEB2 | [23] |
| miR-203 | down | ZEB2 | [23] |
| miR-182–5p | down | FLOT1 | [35] |
| miR-186 | down | SENP1 | [36] |
| miR-122 | up | FOXO3 | [37] |
| miR-30a-3p | down | WNT2 | [38] |
| miR-21 | up | PDCD4 | [103] |
| miR-195 | down | HMGA1 | [104] |
The transcriptional repressors ZEB1 and ZEB2, which control EMT (epithelial-mesenchymal transition) or MET (mesenchymal-epithelial transition), are inhibited by miR-200 family members [21]. Researchers found that the expression of ZEB1 could be downregulated by miR-200c to inhibit RCC progression [22]. Except from miR-200 family, Chen et al. also have shown that, in addition to the miR-200 family, both miR-124 and miR-206 can also suppress the EMT pathway via co-regulation of ZEB2 [23].
The key tumor suppressor gene PTEN is involved in many cell processes [24]. Fan et al. demonstrated that miR-22 is downregulated in RCC tumor tissues, and that miR-22 can exert its anti-tumor effect by targeting PTEN [25]. Jing et al. also found that miR-19 can promote RCC cells proliferation by targeting Fyn-related kinase (FRK), thus inhibiting FRK induced PTEN phosphorylation [26].
Several miRNAs have been used to characterize RCC through identifying different pathological characteristic, including clinical stage and pathological grade [19]. Analyzing 54 pairs of RCC tissues and adjacent normal tissues, Zaman et al. found an increase of miR-21 in malignant tissues compared to miR-21 levels in adjacent tissues. Expression levels of miR-21 were positively correlated with the stage of renal cancer [27]. In addition, miRNA can also be used to distinguish metastatic RCC from localized RCC. White et al. found elevated levels of MiR-149, miR-638 and miR-1915 in metastatic RCC, and downregulated miR-196a, miR-10b and miR-27b* [28].
MiRNA expression can also be used to distinguish different subtypes of RCC. Papillary renal cell carcinoma (pRCC), a heterogeneous group including various types of renal cell carcinoma, is the second most common type of RCC, accounting for nearly 10% of all RCC cases [29]. Powers et al. showed that miR-143, miR-126* and miR-126 are downregulated in pRCC compared with ccRCC tissues [30]. Xp11.2 translocation renal cell carcinoma (Xp11 tRCC) is a rare sporadic kidney cancer in children. Kurahashi et al. found that the expression level of miR-204–5p is markebly increased in the Xp11 tRCC mouse model compared to controls. In addition, miR-204–5p has also been detected in the urinary exosome in Xp11 tRCC mice, but not in controls, thus suggesting its potential as a diagnostic biomarker of Xp11 tRCC in humans [31].
MiRNAs as biomarkers
Accurate diagnosis is the foundation for precise, effective treatment of renal cell carcinoma. Since a growing number of studies have shown that almost all biomolecules (including small noncoding RNA such as miRNAs, piRNAs and tsRNAs) packed into extracellular vesicles which could be detected in the body fluid specimens [39], it is feasible and urgent to develop the miRNA-based non-invasive detection system [40]. The system would allows repeated sampling and real-time monitoring of disease progression and of response to treatment [41,42]. We present here some of miRNAs which might most effectively reflect the physiological and pathological characteristics of RCC (Table 2).
Table 2.
Clinical application of sncRNAs in renal cell carcinoma.
| RNA type | SncRNA | Expression | Potential clinical application | Biological Samples | Technique/Cohort | Reference |
|---|---|---|---|---|---|---|
| miRNA | miR-21 miR-142–5p miR-194 |
up | Prognostic markers | Tissue | qRT-PCR/54 ccRCC vs 54 N | [27] |
| miR-155–5p miR-210–3p |
up | Prognostic markers | Tissue | qRT-PCR/ 205 ccRCC vs 205 N |
[44] | |
| miR-122–5p miR-206 |
up | Prognostic markers | Serum | qRT-PCR/ 68 ccRCC,47 benign renal tumors and 28 N |
[45] | |
| miR-141 | down | Predictive markers | Tissue | qRT-PCR/9 poor responders to sunitinib vs 11 good responders | [46] | |
| miR-9–5p | up | Predictive markers | Tissue | TaqMan/41 responders to TKIs vs 19 non-responders | [47] | |
| miR-376b-5p | down | Predictive markers | Tissue | Microarray/25 responders to sunitinib vs 22 non-responders | [48] | |
| piRNA | piR-32,051 piR-39,894 piR-43,607 |
up | Prognostic markers | Tissue | Microarrays and qRT-PCR/Cohort 1: 18 ccRCC and 18 N Cohort 2: 68 cases of FFPE ccRCC |
[73] |
| piR-30,924 piR-57,125 piR-38,756 |
down | Prognostic markers | Tissue | Microarrays and qRT-PCR/13 metastatic ccRCC, 106 ccRCC and 77 N | [74] | |
| piR-51,810 piR-34,636 |
down | Prognostic markers | Serum | qRT-PCR/30 RCC vs 15 N | [75] | |
| piR-823 | down | Prognostic markers | Tissue | qRT-PCR/57 RCC vs 38 N | [71] | |
| tsRNA | 5′tRNA-Arg-CCT half 5′tRNA-Leu-CAG half 5′tRNA-Glu-CTC half 5′tRNA-Lys-TTT half |
down | Prognostic markers | Serum | qRT-PCR/ i)Tissue:95 RCC vs 50 N ii)Serum: 27 RCC vs 13 N |
[76] |
An increasing amount data supports the view that miRNAs could serve as novel prognostic biomakers. Using a combination miRNA strategy in a powerful study of 107 specimens, Lokeshwar et al. revealed that combining miR-21, miR-142–5p and miR-194 could significantly predict cancer metastasis and disease-specific mortality [43]. In a study of miR-210–3p and miR-155–5p expression in a cohort of 206 RCC patients, Zhang et al. reported that high expression of miR-210–3p and miR-155–5p is associated with elevated risk of relapse [44]. Another independent patient cohort study showed a strong correlation of miR-122–5p and miR-206 levels with progression-free periods and overall survival [45]. It has been recently reported that patients with high levels of miR-16 show a poor prognosis. In contrast, miR-484, miR-497, and miR-20 have been considered as early diagnosis indicators of RCC because they are differentially expressed only in stage I tumor tissues and blood specimens [33].
MiRNAs have potential not only for predicting the pathological characteristics and prognosis of RCC, but also for predicting responses to chemotherapy. Since tyrosine kinase inhibitors (TKI) such as sunitinib, pazopanib and sorafenib are frequently used in the first-line treatment of RCC patients, the prediction value of miRNAs in TKI response has attracted the attention of several researchers. Joost et al. found that miR-141 was markedly downregulated in tumor samples of poor responders compared to that of good responders showing at least 1-year progression-free survival. In vitro assays have shown that miR-141 can inhibit EMT by inhibiting ZEB transcriptional factors [46]. In another study analyzing nephrectomy specimens of 50 metastatic RCC patients, mRCC patients were divided into non-responders (progression < 3 months) and complete or partial responders (progression >6 months). Comparision of nonresponders and partial responders found that miR-9–5p levels significantly increased in non-responders compared to responders. Finally, the potential of miR-9–5p to predict patient response to TKIs was confirmed using Kaplan-Meier and Cox regression analyses [47]. Another interesting recent study analyzed miRNAs expression in two independent cohorts: an exploratory cohort containing 25 good responders (progression-free survival (PFS) >17 months) and 22 poor responders (PFS <7 months), and a validation cohort comprised of patients with primary resistance (PFS <5 months), patients with intermediate response levels (PFS >5 but <12 months), and patients with a long-term response (PFS >12 months). Finally, the potential of miR-376b-3p in predicting drug response has been demonstrated by its correlation with PFS in metastatic RCC treated with sunitinib [48].
MiRNA-based therapies
Several miRNAs have been reported to inhibit cancer cell growth and migration when overexpressed or inhibited, suggesting that proliferation and migration of cancer cell could be regulated by manipulating miRNA expression. Several methods for overexpressing or inhibiting miRNA expression are available. For example, artificial synthetic miRNA mimics or antagonists can be successfully transfected into cells to overexpress or inhibit the expression of target mRNAs. Strategies for therapeutic uses of miRNA are entering to a new era, with several ongoing phase I and phase II trials [49].
MRX34 (Mirna Therapeutics), a liposome miR-34a mimic, has completed the phase I trial in advanced solid tumors, including viral-related hepatoma, renal cell cancer, and melanoma. In a trial administering MRX34 to 47 adult patients twice a week, researchers found that one patient achieved partial remission and four patients maintained a stable disease lasting more than 4 cycles. Although researchers considered that MRX34 was acceptable safe, Dexamethasone premedication did not prevent occurrence of adverse events [50]. Despite the promise of this drug in treating some refractory solid tumor patients, even though 3 patients acquired partial remissions and the 16 patients remained stable for more than 4 weeks, the trial was terminated in 2016 because of severe immune-mediated adverse events that resulted in four deaths [51].
Another clinical trial, however, used miravirsen, an antisense oligonucleotide inhibitor targeting miR-122, to inhibit miR-122 in vivo that had been proved effective in reducing HCV (Hepatitis C) RNA levels of Hepatitis C patients in a dose-dependent reduction manner. HCV patients who received subcutaneous injection of miravirsen for give weeks showed a long-term decrease HCV RNA levels without any unmanageable adverse events [52].
However, there are still many obstacles to clinical application, with only a few ongoing clinical trials on miRNAs, despite many years of numerous preclinical studies on miRNA treatment. More attention should be focused on enhancing the stability of miRNA molecules in vivo and on designing optimal delivery systems with disease specificity and minimum toxicity [53].
Piwi-interacting RNA
Piwi-interacting RNAs biogenesis and functions
Piwi-interacting RNAs (piRNAs) is a class of sncRNAs with 21–35 nucleotides in length [54,55], characterized by 2′-O-methylation at the 3′ end, which can be used to distinguish it from other noncoding RNAs [56]. piRNAs are normally divided into three classes according to their origins: 1) transposon-derived piRNAs, 2) lncRNA-derived piRNAs and 3) mRNA-derived piRNAs [57]. The biogenesis and function of transposon-derived piRNAs have been studied more extensively than the other two classes of piRNAs. The biogenesis of piRNAs includes two separated steps: 1) primary biogenesis and 2) secondary biogenesis.
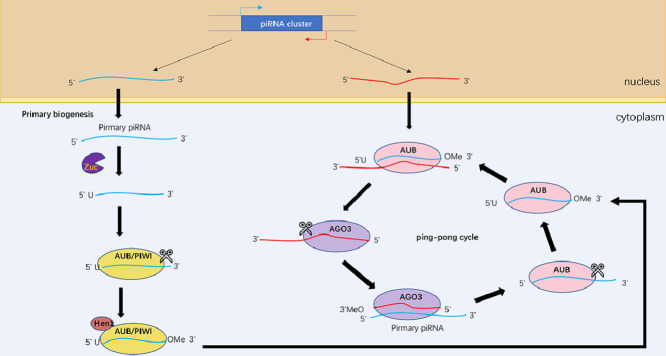
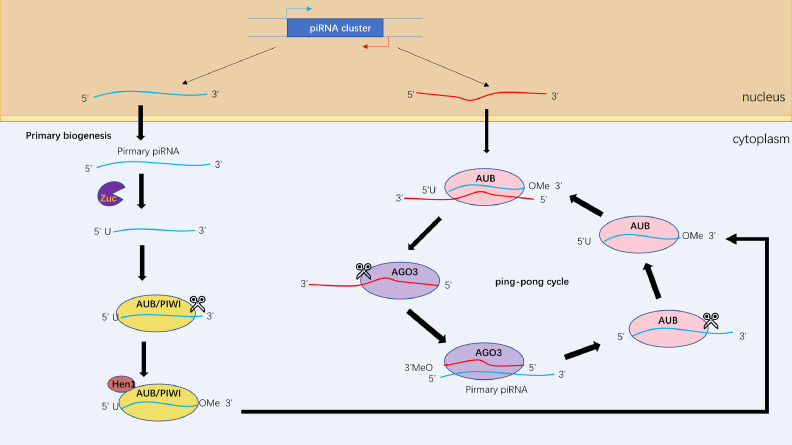
After transcription, the primary transcript (pri-piRNA) is cleaved by the ribo-endonuclease Zucchini (Zuc, also called PLD6 in mammals), generating piRNA intermediates with a 5′ uracil [58,59]. PiRNAs with uridine at the first nucleotides are preferentially loaded on PIWI proteins. Next, the 3′ fragment of immature piRNA is trimmed and 2′ O-methylated by the Hen1 methyltransferase. The final length of mature piRNA is considered to protect the piRNA from 3ʹ to 5ʹ exonucleases [60]. The secondary phase of biogenesis, known as the ping-pong cycle, begins with the mature piRNA. Aub/PIWI proteins bind to the mature piRNA, which targets the complementary transposon RNA sequence, and cleaves the 3′ fragment of transposon RNA in cytoplasm. The remaining transposon RNA is still bound to AGO3. The secondary piRNA is processed by trimming and 2′ O-methylating as in the primary process. There is a nearly 10-nucleotides overlap between the Aub-bound mature piRNA and the AGO3-bound secondary piRNA. Next, the piRNA, which is identical to the mature piRNA, is produced by piRNA-AGO3 complex induced antisense piRNA transcripts cleavage and they could continue to be amplified by repeating the second process [60]. In conclusion, this biogenetic process degrades the transposon RNA and amplifies the piRNA/PIWI complex [61] (Fig. 2).
Fig. 2.
The PiRNA biogenesis pathway. Pre-piRNAs are exported to cytosol after transcription, then cleaved by Zuc, resulting in the production of intermediate piRNAs intermediated with a 5′uracil. The 3′terminal of piRNA intermediates are trimmed and 2′-O-methylated by Hen1. In the secondary pathway, piRNAs bind to AUB or AGO3 proteins to form piRNA/AUB or piRNA/AGO3 complexes, which are complementary to each other, leading to transposon RNA cleavage and piRNA amplification.
PiRNAs accomplish their function through binding to PIWI proteins from Argonaute family. The piRNAs/PIWI complexes can silence the actively transcribed transposons, whose sequence is complementary to piRNAs, and alter the chromatin structure of target loci by recruiting histone methyltransferases [62]. In addition to chromatin silencing in the nucleus, piRNAs can also regulate mRNA expression in cytoplasm. For the nearly one third of mRNAs that carry transposon-derived sequence in their 3′-UTR, piRNA/PIWI complexes could recognize and eliminate these mRNAs in transcriptional level [63].
PiRNAs alteration in renal cell carcinoma
Since piRNAs were initially found to be important regulators in maintaining genomic stability of germ stem cells, it is plausible that similar self-renewal mechanisms are likely to occur in rapidly dividing cancer cells [64]. In support of this hypothesis, more and more studies have shown that piRNA expression is dysregulated in various types of cancer, including RCC, suggesting that piRNAs may exert tumor-suppressive or tumor-promoting effects in cancer cells. Moreover, other studies have revealed that piRNAs are strongly correlated with pathological and clinical tumor characteristics [65,66]. Next, we will discuss the vital role of specific piRNAs in development and progression of cancer, especially in renal cell carcinoma. piR-823 is an important piRNA that is dysregulated in various types of cancer. Yan et al. found that piR-823 expression is increased in multiple myeloma (MM) patients and that expression levels are correlated with clinical stage. Researchers have also found that inhibition of piR-823 can reduce expression of DNA methyltransferases DNMT3A and 3B and thus reduce the global DNA methylation level. Moreoever, piRNA-823 inhibition decreases proangiogenic activity in MM by reducing the secretion of vascular endothelial growth factor [67]. Unlike the case for MM, however, it has been reported that piR-823 is downregulated in kidney cancer, prostate cancer, and gastric cancer [68], [69], [70], [71]. In gastric cancer, the expression of piR-823 in tumor tissues was significantly lower than that in adjacent tissues. After increasing the level of piR-823, cell growth was inhibited. Also, in vivo mouse model confirmed piR-823 played a suppressive effect on gastric cancer [68]. Similarly, Lliev et al. analyzed piR-823 levels in 588 RCC and normal specimens, and found that, piR-823 was decreased in RCC tissues compared to adjacent normal tissues. Surpriginly, however, expression levels of piR-823 are also positively correlated with a poor prognosis, indicating the complex role of piR-823 in RCC pathogenesis [69].
Overall, the role of piRNAs in RCC has been little studied, and further research is needed to understand the mechanisms of piRNA in RCC progression and metastasis.
PiRNAs as biomarkers
Like miRNAs, piRNAs are expressed in a tissue-specific manner and have been shown to be differentially expressed in different types of tumors; their expression levels are also associated with histological grade and pathological characteristics of tumors, and with patient survival [72]. Here, we summarize several piRNAs with the potential to serve as biomarkers of RCC progression and metastasis (Table 2).
Li et al. reported that there were 19 piRNAs that are differentially expressed in benign and malignant renal tissues. Deep sequencning of 24 benign and ccRCC specimens revealed 46 piRNAs were associated with metastasis. In another 68-case ccRCC tissue validation cohort, researchers found that three piRNAs (piR-43,607, piR-39,894 and piR-32,051) were strongly associated with progression, metastasis, and prognosis of ccRCC [73]. Another similar study in RCC analyzed 106 RCC patient samples including 30 metastatic tissues, 76 localized tissues and normal samples. RT-qPCR assays revealed lower lexpression of piR-38,756, piR-57,125 and piR-30,924 in tumor samples compared with normal samples. In still another study, however, piR-57,125 showed lower expression in metastatic samples, but strong expression piR-30,924 and piR-38,756 in metastatic samples [74]. Recently, zhao et al. found that two mitochondrial piRNAs (piR-51,810 and piR-34,636) were downregulated in ccRCC, and that both piRNAs were predictors of cancer-specific and overall survival of ccRCC patients. However, the researchers found no significant differences in piRNA expression between ccRCC patients and non-malignant patients in serum samples [75].
TRNA-derived small RNAs
TRNA-derived small RNAs biogenesis and functions
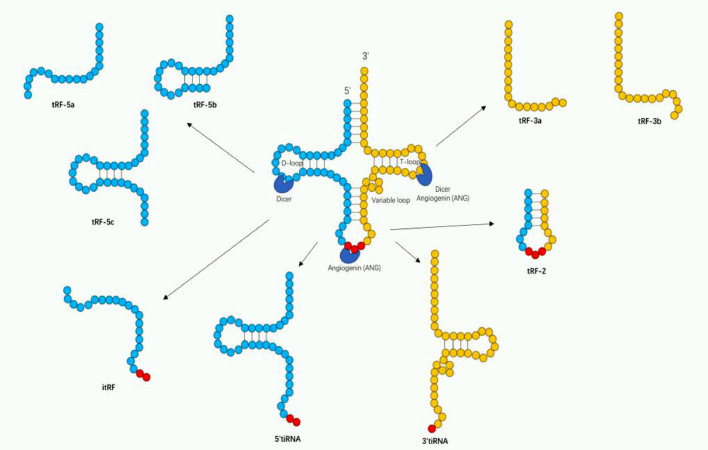
TRNAs are well characterized molecules that have been extensively studied for more than 50 years, and they have a well-defined role in protein translation. Besides their critical roles in basic protein synthesis, increasing evidence indicates that tRNAs also participate in several additional cellular processes by interacting with a range of proteins both in bacteria and eukaryotes [77]. With the recent development of high-throughput sequencing, it has been found that tRNAs are a critical source of small noncoding RNA [78]. tRNA-derived small RNAs (tsRNAs) are produced by tRNAs, and are cleaved by specifically nucleases under certain conditions such as stress and hypoxia [79]. There are two main groups of tsRNAs: 1) tRNA-derived fragments (tRFs) and 2) tRNA halves (tiRNAs), categorized according to different cleavage positions [80]. Based on their mapping positions on pre-tRNAs and mature tRNAs, tRFs and tiRNAs can be divided into several subclasses [81] (Fig. 3). tRFs are approximately 14–30 nt in length and can be grouped into 5 subclasses: tRF-1 s, tRF-3 s, tRF-5 s, tRF-2 s, itRFs [82]. tRF-1 s are derived from the 3′ end of mature tRNAs or precursor tRNAs cleaved by RNase Z in the nucleus and resulted in the presence of poly-U residues at the 3′ end of tRF-1 s [83]. tRF-2 s contain only anticodon stem loop sequences and are induced in hypoxic conditions [84]. Originating from the 3′ end of mature tRNAs, tRF-3 s can be produced in a Dicer-dependent or Dicer-independent manner [82]. Therefore, tRF-3 s can be subclassified as tRF-3a or tRF-3b due to different methods of cleavage. tRF-5 s, derived from 5′ end of mature tRNAs, are cleaved in the D-loop. Due to three specific lengths of tRF-5 s, they could be separated into three subclasses: tRF-5a, tRF-5b and tRF-5c [85]. i-tRFs come mainly from the internal region of mature tRNA [86].
Fig. 3.
Schematic presentation of tsRNAs biogenesis. tsRNAs include tRF-1 s, tRF-2 s, tRF-3 s (type a,b), tRF-5 s (type a, b and c) and i-tRFs. tRF-1 s are the maturation products of tRNAs (not shown in figure). The anticodon loop of mature tRNAs can be cleaved by Angiogenin (ANG) to produce 3′tiRNAs and 5′tiRNAs. tRF-2 s are generated from the anticodon stem loop of tRNAs. tRF-3 s, which are derived from the 3′end of tRNAs, are generated by Dicer or ANG -dependent cleavage in the T loop. Similarly, tRF-5 s are derived from the 5′end of tRNAs in Dicer-dependent cleavage in the D loop. i-tRFs come mainly from the internal region of tRNA.
As the earliest discovered tsRNAs, tiRNAs are produced by cleavage specifically in the anticodon loop of mature tRNAs [87]. Despite being known as stress-induced fragments, tiRNAs also exist in non-stressed conditions [88]. There are two subclasses of tiRNAs: 3′tiRNAs and 5′tiRNAs. As their name suggests, 5′tiRNAs replication start from the 5′ end of mature tRNAs and end at the anticodon loop. Similarly, 3′tiRNAs replication start from the anticodon loop and end at the 3′ end of mature tRNAs. Because of cleavage is conducted by angiogenin, tiRNAs have a 5′ hydroxyl group rather than a 5′ phosphate group, which is different from miRNAs and siRNAs [89,90].
Although the exact role of tsRNAs is still needs to be further elucidated, accumulating evidences suggest that they participate in various biological processes as we discussed in detail below. Firstly, tsRNAs could regulate the gene expression in an miRNA-like manner. Several studies have shown that both 5′tRFs and 3′tRFs can target the 3′UTR of specific mRNAs and repress their translation [79,91,92]. Wang et al. reported that 5′tiRNA which derived from 5′-tRNAGlu can suppress target genes in the cytoplasm and promote respiratory syncytial virus (RSV) proliferation [92]. Aside from the effets of tsRNAs on miRNA-like gene silencing, several studies suggest that tsRNAs can also repress the global translational levels [93]. Gebetsberger et al. found that 5′tRFVal binds to ribosomes directly and reduces protein synthesis under the stress conditions [84,94,95]. Moreover, tsRNAs can bind proteins to exert specific functions [84]. For example, YBX1 is an RNA-binding protein that can bind to some endogenous oncogene transcripts and enhance their stability. Several tsRNAs can bind to YBX1, resulting in the disassociation of the YBX1-oncogene transcripts complex, and in inhibition of tumor cell growth [96].
TsRNAs alteration in renal cell carcinoma
Although understanding of the role of tsRNAs in cancer is still in the early stage, increasing evidences suggest that tsRNAs play a vital role in various physiological processes, including differentiation, proliferation, division, RNA splicing, chromatin remodeling, transcription factor binding, among other process. It has been reported that tsRNAs have a functional role in many types of malignant cells. For example, some tsRNAs derived from tRNAGlu, tRNAAsp, tRNAGly, and tRNATyr are act as the tumor suppressors in breast cancer by binding to YBX1, as discussed above. However, there has been little research on tsRNA in renal cell carcinoma. With the development of new technologies such as high throughput sequencing, future research is likely to clarify the specific mechanisms of tsRNA in RCC.
TsRNAs as biomarkers
It is recognized that 5′tRNA halves circulate in the serum in a stable form and that their serum levels are fluctuate due to a variety of physiological changes [97]. What's more, numerous of tsRNAs have been shown to play a vital role in many types of malignancies [98], [99], [100]. Recent studies have revealed that tsRNAs can be used as diagnostic and prognostic biomarkers in breast cancer, oral squamous cell carcinoma and head and neck squamous cell carcinoma [101]. Here, we describe some potential tsRNA biomarkers in renal cell carcinoma (Table 2).
Analyzing a cohort containing 118 ccRCC and 78 normal tissues, Nientiedt et al. reported that 5′tRNA-Val-ACC is downregulated in renal cell carcinoma tissues [76]. A subsequent study was conducted by the same group indicated several potential diagnostic and prognostic biomarkers in ccRCC. By assessing the 5′tRNA expression levels in 95 ccRCC and 50 normal renal tissues, those investigators found that both 5′tRNA-Arg-CCT half, 5′tRNA-Leu-CAG half, 5′tRNA-Glu-CTC half, and 5′tRNA-Lys-TTT half were all downregulated in malignancies. Prognostic analysis also revealed that elevated levels of all of these tsRNA variants are associated with adverse clinical features [76].
Conclusion and future perspectives
As more researchers have focused on sncRNAs in the last two decades, growing evidence has revealed a close relationship between sncRNAs and cancer. MiRNAs, which have been extensively explored in the past decades, are considered as potential biomarkers or therapeutic targets in cancer therapy. There are several different views of miRNAs as therapeutic tools. At the moment, the greatest attention has been focused on those miRNAs that could inhibit cancer progression alone. However, another strategy that has not been extensively discussed is to utilize miRNAs to increase the sensitivity of cancer cells to chemotherapy [102]. Thus far, hundreds of preclinical experiments have indicated the vital role of miRNAs in cancer resistance. Thus miRNA formulations are promising materials for development as adjunctive therapy administered along with chemotherapy drugs.
Despite the promising future for miRNA therapy, there are still large obstacles to spanning the gap between basic research and clinical applications. Though many miRNAs are undergoing clinical trials, none of those miRNAs have yet been applied in routine clinical practice. Several reasons may explain this phenomenon. First, many miRNA biomarkers are selected in a group with a limited number of participants and without a validation cohort. Moreover, the patients included in clinical trials are usually selected in a way that is not representative of the broader patient population. Second, it is quite difficult to overcome the adverse effects of several miRNA-based therapeutic strategies: because miRNA can simultaneously regulate various targets, it is not easy to avoid off-target effects. For instance, MRX34, which was considered safe in animal models and early phase I clinical trials, nevertheless caused 4 patient deaths in later phase I clinical trials due to severe immune-mediated adverse effects. Last but not least, a more efficient delivery system should be developed to precisely deliver the therapeutic oligonucleotides to target sites and to maintain maximum stability of the therapeutic oligonucleotides in vivo. Apart from miRNAs, the roles of other types of sncRNAs in cancer have been less explored, especially in renal cell carcinoma. Since the expression levels of piRNAs and tsRNAs vary in different types of tumors and different stages of disease, more light should be shed on those sncRNAs to better understand the underlying mechanisms of renal cell carcinoma and to provide novel therapeutic targets for RCC.
Looking forward, it is conceivable that the evaluation of the expression levels of small noncoding RNA in patients’ tumor tissues and serum will been indispensable component of future approaches to cancer diagnosis and therapy. Through knowledge of specific sncRNA expression levels, we may someday be able to predict the pathological characteristics of tumors, and the overall patient prognosis and response to chemotherapy, and to apply targeted sncRNA treatment appropriate to the specific characteristics of the tumor and patient in question.
CRediT authorship contribution statement
Lifeng Ding: Writing - original draft. Minxiao Jiang: Writing - original draft. Ruyue Wang: Writing - original draft. Danyang Shen: Writing - review & editing. Huan Wang: Visualization, Validation. Zeyi Lu: Writing - original draft, Writing - review & editing. Qiming Zheng: Writing - original draft, Writing - review & editing. Liya Wang: Writing - review & editing. Liqun Xia: Conceptualization, Supervision. Gonghui Li: Conceptualization, Supervision.
Declaration of Competing Interest
The authors have no conflicts of interest to disclose.
Acknowledgments
This work was supported by Joint construction project of Zhejiang Province and Ministry (grant number: 2020388200); Key R & D plan of Zhejiang Province (grant number: 2019C03089) National Natural Science Foundation of China (Grant numbers: 81672520, 81870484, 81773789); Zhejiang Provincial Natural Science Foundation of China (grant number: LY17H160020); Zhejiang Medical and Health Plan Project (Grant number: 2019ZD007).
References
- 1.Capitanio U., Bensalah K., Bex A., Boorjian S.A., Bray F., Coleman J., Gore J.L., Sun M., Wood C., Russo P. Epidemiology of renal cell carcinoma. Eur. Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dy G.W., Gore J.L., Forouzanfar M.H., Naghavi M., Fitzmaurice C. Global burden of urologic cancers, 1990–2013. Eur. Urol. 2017;71:437–446. doi: 10.1016/j.eururo.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Patard J.-.J., Pignot G., Escudier B., Eisen T., Bex A., Sternberg C., Rini B., Roigas J., Choueiri T., Bukowski R., Motzer R., Kirkali Z., Mulders P., Bellmunt J. ICUD-EAU international consultation on kidney cancer 2010: treatment of metastatic disease. Eur. Urol. 2011;60:684–690. doi: 10.1016/j.eururo.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Clark D.J., Dhanasekaran S.M., Petralia F., Pan J., Song X., Hu Y., da Veiga Leprevost F., Reva B., Lih T.-S.M., Chang H.-.Y., Ma W., Huang C., Ricketts C.J., Chen L., Krek A., Li Y., Rykunov D., Li Q.K., Chen L.S., Ozbek U., Vasaikar S., Wu Y., Yoo S., Chowdhury S., Wyczalkowski M.A., Ji J., Schnaubelt M., Kong A., Sethuraman S., Avtonomov D.M., Ao M., Colaprico A., Cao S., Cho K.-.C., Kalayci S., Ma S., Liu W., Ruggles K., Calinawan A., Gümüş Z.H., Geiszler D., Kawaler E., Teo G.C., Wen B., Zhang Y., Keegan S., Li K., Chen F., Edwards N., Pierorazio P.M., Chen X.S., Pavlovich C.P., Hakimi A.A., Brominski G., Hsieh J.J., Antczak A., Omelchenko T., Lubinski J., Wiznerowicz M., Linehan W.M., Kinsinger C.R., Thiagarajan M., Boja E.S., Mesri M., Hiltke T., Robles A.I., Rodriguez H., Qian J., Fenyö D., Zhang B., Ding L., Schadt E., Chinnaiyan A.M., Zhang Z., Omenn G.S., Cieslik M., Chan D.W., Nesvizhskii A.I., Wang P., Zhang H., Hashimi A.S., Pico A.R., Karpova A., Charamut A., Paulovich A.G., Perou A.M., Malovannaya A., Marrero-Oliveras A., Agarwal A., Hindenach B., Pruetz B., Kim B.-.J., Druker B.J., Newton C.J., Birger C., Jones C.D., Tognon C., Mani D.R., Valley D.R., Rohrer D.C., Zhou D.C., Tansil D., Chesla D., Heiman D., Wheeler D., Tan D., Chan D., Demir E., Malc E., Modugno F., Getz G., Hostetter G., Wilson G.D., Hart G.W., Zhu H., Liu H., Culpepper H., Sun H., Zhou H., Day J., Suh J., Huang J., McDermott J., Whiteaker J.R., Tyner J.W., Eschbacher J., Chen J., McGee J., Zhu J., Ketchum K.A., Rodland K.D., Clauser K., Robinson K., Krug K., Hoadley K.A., Um K.S., Elburn K., Holloway K., Wang L.-.B., Blumenberg L., Hannick L., Qi L., Sokoll L.J., Cornwell M., Loriaux M., Domagalski M.J., Gritsenko M.A., Anderson M., Monroe M.E., Ellis M.J., Dyer M., Anurag M., Burke M.C., Borucki M., Gillette M.A., Birrer M.J., Lewis M., Ittmann M.M., Smith M., Vernon M., Chaikin M., Chheda M.G., Khan M., Roche N., Edwards N.J., Vatanian N., Tignor N., Beckmann N., Grady P., Castro P., Piehowski P., McGarvey P.B., Mieczkowski P., Hariharan P., Gao Q., Dhir R., Kothadia R.B., Thangudu R.R., Montgomery R., Jayasinghe R.G., Smith R.D., Edwards R., Zelt R., Bremner R., Liu R., Hong R., Mareedu S., Payne S.H., Cottingham S., Markey S.P., Jewell S.D., Patel S., Satpathy S., Richey S., Davies S.R., Cai S., Boca S.M., Patil S., Sengupta S., Carter S., Gabriel S., Thomas S.N., de Young S., Stein S.E., Carr S.A., Foltz S.M., Hilsenbeck S., Krubit T., Liu T., Skelly T., Westbrook T., Borate U., Velvulou U., Petyuk V.A., Bocik W.E., Chen X., Shi Y., Geffen Y., Lu Y., Wang Y., Maruvka Y., Li Z., Shi Z., Tu Z. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–983.e31. doi: 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G.K., Khatun J., Williams B.A., Zaleski C., Rozowsky J., Röder M., Kokocinski F., Abdelhamid R.F., Alioto T., Antoshechkin I., Baer M.T., Bar N.S., Batut P., Bell K., Bell I., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Falconnet E., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M.J., Gao H., Gonzalez D., Gordon A., Gunawardena H., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Luo O.J., Park E., Persaud K., Preall J.B., Ribeca P., Risk B., Robyr D., Sammeth M., Schaffer L., See L.-.H., Shahab A., Skancke J., Suzuki A.M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Ruan X., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T., Reymond A., Antonarakis S.E., Hannon G., Giddings M.C., Ruan Y., Wold B., Carninci P., Guigó R., Gingeras T.R. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong C.-.M., Tsang F.H.-C., Ng I.O.-L. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 9.Slack F.J., Chinnaiyan A.M. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens-Uzunova E.S., Böttcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y., Kim M., Han J., Yeom K.-.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beermann J., Piccoli M.-.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 14.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diederichs S., Haber D.A. Dual role for argonautes in MicroRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 18.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schubert M., Junker K., Heinzelmann J. Prognostic and predictive miRNA biomarkers in bladder, kidney and prostate cancer: where do we stand in biomarker development? J. Cancer Res. Clin. Oncol. 2016;142:1673–1695. doi: 10.1007/s00432-015-2089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grange C., Brossa A., Bussolati B. Extracellular vesicles and carried miRNAs in the progression of renal cell carcinoma. Int. J. Mol. Sci. 2019;20:1832. doi: 10.3390/ijms20081832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill L., Browne G., Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer. 2013;132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Chen X., Wang R., Xiao P., Xu Z., Chen L., Hang W., Ruan A., Yang H., Zhang X. MicroRNA-200c modulates the epithelial-to-mesenchymal transition in human renal cell carcinoma metastasis. Oncol. Rep. 2013;30:643–650. doi: 10.3892/or.2013.2530. [DOI] [PubMed] [Google Scholar]
- 23.Chen J., Zhong Y., Li L. MiR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC) J. Transl. Med. 2020;18:69. doi: 10.1186/s12967-020-02242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worby C.A., Dixon J.E. PTEN. Annu. Rev. Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 25.Fan W., Huang J., Xiao H., Liang Z. MicroRNA-22 is downregulated in clear cell renal cell carcinoma, and inhibits cell growth, migration and invasion by targeting PTEN. Mol. Med. Rep. 2016;13:4800–4806. doi: 10.3892/mmr.2016.5101. [DOI] [PubMed] [Google Scholar]
- 26.Jing Z.-.F., Bi J.-.B., Li Z.-.L., Liu X.-.K., Li J., Zhu Y.-.Y., Zhang X.-.T., Zhang Z., Li Z.-.H., Kong C.-.Z. MiR-19 promotes the proliferation of clear cell renal cell carcinoma by targeting the FRK-PTEN axis. Oncol. Target. Ther. 2019;12:2713–2727. doi: 10.2147/OTT.S199238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaman M.S., Shahryari V., Deng G., Thamminana S., Saini S., Majid S., Chang I., Hirata H., Ueno K., Yamamura S., Singh K., Tanaka Y., Tabatabai Z.L., Dahiya R. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0031060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White N.M.A., Khella H.W.Z., Grigull J., Adzovic S., Youssef Y.M., Honey R.J., Stewart R., Pace K.T., Bjarnason G.A., Jewett M.A.S., Evans A.J., Gabril M., Yousef G.M. MiRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br. J. Cancer. 2011;105:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barth D.A., Slaby O., Klec C., Juracek J., Drula R., Calin G.A., Pichler M. Current concepts of non-coding RNAs in the pathogenesis of non-clear cell renal cell carcinoma. Cancers Basel. 2019;11:1580. doi: 10.3390/cancers11101580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powers M.P., Alvarez K., Kim H.-.J., Monzon F.A. Molecular classification of adult renal epithelial neoplasms using microRNA expression and virtual karyotyping. Diagn. Mol. Pathol. 2011;20 doi: 10.1097/PDM.0b013e3181efe2a9. https://journals.lww.com/molecularpathology/Fulltext/2011/06000/Molecular_Classification_of_Adult_Renal_Epithelial.1.aspx [DOI] [PubMed] [Google Scholar]
- 31.Kurahashi R., Kadomatsu T., Baba M., Hara C., Itoh H., Miyata K., Endo M., Morinaga J., Terada K., Araki K., Eto M., Schmidt L.S., Kamba T., Linehan W.M., Oike Y. MicroRNA-204-5p: a novel candidate urinary biomarker of Xp11.2 translocation renal cell carcinoma. Cancer Sci. 2019;110:1897–1908. doi: 10.1111/cas.14026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Guan B., Liu J., Zhang Z., He S., Zhan Y., Su B., Han H., Zhang X., Wang B., Li X., Zhou L., Zhao W. MicroRNA-200b is downregulated and suppresses metastasis by targeting LAMA4 in renal cell carcinoma. EBioMedicine. 2019;44:439–451. doi: 10.1016/j.ebiom.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi Y., Wang L., Wang K., Peng Z., Ma Y., Zheng Z., Shang D., Xu W., Zheng J. New mechanistic insights of clear cell renal cell carcinoma from integrated miRNA and mRNA expression profiling studies. Biomed. Pharmacother. 2019;111:821–834. doi: 10.1016/j.biopha.2018.12.099. [DOI] [PubMed] [Google Scholar]
- 34.Yang F., Ma J., Tang Q., Zhang W., Fu Q., Sun J., Wang H., Song B. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Krüppel-like factor 6. Biomed. Pharmacother. 2018;97:616–623. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 35.Xu X., Wu J., Li S., Hu Z., Xu X., Zhu Y., Liang Z., Wang X., Lin Y., Mao Y., Chen H., Luo J., Liu B., Zheng X., Xie L. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol. Cancer. 2014;13:109. doi: 10.1186/1476-4598-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao D., Wu M., Ji L., Liu F., Liu Y. MicroRNA-186 suppresses cell proliferation and metastasis through targeting sentrin-specific protease 1 in renal cell carcinoma. Oncol. Res. 2018;26:249–259. doi: 10.3727/096504017X14953948675430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie W., Ni D., Ma X., Zhang Y., Gao Y., Peng C., Zhang X. MiR-122 promotes proliferation and invasion of clear cell renal cell carcinoma by suppressing Forkhead box O3. Int. J. Oncol. 2019;54:559–571. doi: 10.3892/ijo.2018.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L., Chen L., Wu T., Qian H., Yang S. MicroRNA-30a-3p functions as a tumor suppressor in renal cell carcinoma by targeting WNT2. Am. J. Transl. Res. 2019;11:4976–4983. https://pubmed.ncbi.nlm.nih.gov/31497214 [PMC free article] [PubMed] [Google Scholar]
- 39.Linxweiler J., Junker K. Extracellular vesicles in urological malignancies: an update. Nat. Rev. Urol. 2020;17:11–27. doi: 10.1038/s41585-019-0261-8. [DOI] [PubMed] [Google Scholar]
- 40.Tsiakanikas P., Giaginis C., Kontos C.K., Scorilas A. Clinical utility of microRNAs in renal cell carcinoma: current evidence and future perspectives. Expert Rev. Mol. Diagn. 2018;18:981–991. doi: 10.1080/14737159.2018.1539668. [DOI] [PubMed] [Google Scholar]
- 41.Sole C., Arnaiz E., Manterola L., Otaegui D., Lawrie C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019;58:100–108. doi: 10.1016/j.semcancer.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Oto J., Plana E., Sánchez-González J.V., García-Olaverri J., Fernández-Pardo Á., España F., Martínez-Sarmiento M., Vera-Donoso C.D., Navarro S., Medina P. Urinary microRNAs: looking for a new tool in diagnosis, prognosis, and monitoring of renal cancer. Curr. Urol. Rep. 2020;21:11. doi: 10.1007/s11934-020-0962-9. [DOI] [PubMed] [Google Scholar]
- 43.Lokeshwar S.D., Talukder A., Yates T.J., Hennig M.J.P., Garcia-Roig M., Lahorewala S.S., Mullani N.N., Klaassen Z., Kava B.R., Manoharan M., Soloway M.S., Lokeshwar V.B. Molecular characterization of renal cell carcinoma: a potential three-microRNA prognostic signature. Cancer Epidemiol. Prev. Biomark. 2018;27:464–472. doi: 10.1158/1055-9965.EPI-17-0700. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Ye Y., Chang D.W., Lin S.-.H., Huang M., Tannir N.M., Matin S., Karam J.A., Wood C.G., Chen Z.-.N., Wu X. Global and targeted miRNA expression profiling in clear cell renal cell carcinoma tissues potentially links miR-155-5p and miR-210-3p to both tumorigenesis and recurrence. Am. J. Pathol. 2018;188:2487–2496. doi: 10.1016/j.ajpath.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinemann F.G., Tolkach Y., Deng M., Schmidt D., Perner S., Kristiansen G., Müller S.C., Ellinger J. Serum miR-122-5p and miR-206 expression: non-invasive prognostic biomarkers for renal cell carcinoma. Clin. Epigenet. 2018;10:11. doi: 10.1186/s13148-018-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joost B., Olivier G., Pascal W., Benoit B., Patrick S., C. van K.L., Maarten A., den V., Joost O., Tania R., Johan S., Steven J., Hendrik V.P., Evelyne L. A possible role for microRNA-141 down-regulation in sunitinib resistant metastatic clear cell renal cell carcinoma through induction of epithelial-to-mesenchymal transition and hypoxia resistance. J. Urol. 2013;189:1930–1938. doi: 10.1016/j.juro.2012.11.133. [DOI] [PubMed] [Google Scholar]
- 47.Ralla B., Busch J., Flörcken A., Westermann J., Zhao Z., Kilic E., Weickmann S., Jung M., Fendler A., Jung K. MiR-9-5p in nephrectomy specimens is a potential predictor of primary resistance to first-line treatment with tyrosine kinase inhibitors in patients with metastatic renal cell carcinoma. Cancers Basel. 2018;10 doi: 10.3390/cancers10090321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovacova J., Juracek J., Poprach A., Kopecky J., Fiala O., Svoboda M., Fabian P., Radova L., Brabec P., Buchler T., Slaby O. MiR-376b-3p is associated with long-term response to sunitinib in metastatic renal cell carcinoma patients. Cancer Genom. Proteom. 2019;16:353–359. doi: 10.21873/cgp.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiler J., Hunziker J., Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 50.Beg M.S., Brenner A.J., Sachdev J., Borad M., Kang Y.-.K., Stoudemire J., Smith S., Bader A.G., Kim S., Hong D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong D.S., Kang Y.-.K., Borad M., Sachdev J., Ejadi S., Lim H.Y., Brenner A.J., Park K., Lee J.-.L., Kim T.-.Y., Shin S., Becerra C.R., Falchook G., Stoudemire J., Martin D., Kelnar K., Peltier H., Bonato V., Bader A.G., Smith S., Kim S., O'Neill V., Beg M.S. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer. 2020 doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 53.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 54.Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 55.Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ozata D.M., Gainetdinov I., Zoch A., O'Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 2019;20:89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- 57.Assumpção C.B., Calcagno D.Q., Araújo T.M.T., Batista dos Santos S.E., Ribeiro dos Santos Â.K.C., Riggins G.J., Burbano R.R., Assumpção P.P. The role of piRNA and its potential clinical implications in cancer. Epigenomics. 2015;7:975–984. doi: 10.2217/epi.15.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haase A.D., Fenoglio S., Muerdter F., Guzzardo P.M., Czech B., Pappin D.J., Chen C., Gordon A., Hannon G.J. Probing the initiation and effector phases of the somatic piRNA pathway in drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimasu H., Ishizu H., Saito K., Fukuhara S., Kamatani M.K., Bonnefond L., Matsumoto N., Nishizawa T., Nakanaga K., Aoki J., Ishitani R., Siomi H., Siomi M.C., Nureki O. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe T., Lin H. Posttranscriptional regulation of gene expression by piwi proteins and piRNAs. Mol. Cell. 2014;56:18–27. doi: 10.1016/j.molcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moyano M., Stefani G. PiRNA involvement in genome stability and human cancer. J. Hematol. Oncol. 2015;8:38. doi: 10.1186/s13045-015-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luteijn M.J., Ketting R.F. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 63.Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T., Waki K., Hornig N., Arakawa T., Takahashi H., Kawai J., Forrest A.R.R., Suzuki H., Hayashizaki Y., Hume D.A., Orlando V., Grimmond S.M., Carninci P. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 64.Weng W., Li H., Goel A. Piwi-interacting RNAs (piRNAs) and cancer: emerging biological concepts and potential clinical implications. Biochim. Biophys. Acta Rev. Cancer. 2019;1871:160–169. doi: 10.1016/j.bbcan.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo B., Li D., Du L., Zhu X. PiRNAs: biogenesis and their potential roles in cancer. Cancer Metas. Rev. 2020 doi: 10.1007/s10555-020-09863-0. [DOI] [PubMed] [Google Scholar]
- 66.Weng W., Liu N., Toiyama Y., Kusunoki M., Nagasaka T., Fujiwara T., Wei Q., Qin H., Lin H., Ma Y., Goel A. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer. 2018;17:16. doi: 10.1186/s12943-018-0767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan H., Wu Q.-.L., Sun C.-.Y., Ai L.-.S., Deng J., Zhang L., Chen L., Chu Z.-.B., Tang B., Wang K., Wu X.-.F., Xu J., Hu Y. PiRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- 68.Cheng J., Deng H., Xiao B., Zhou H., Zhou F., Shen Z., Guo J. PiR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Iliev R., Fedorko M., MacHackova T., Mlcochova H., Svoboda M., Pacik D., Dolezel J., Stanik M., Slaby O. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res. 2016;36:6419–6423. doi: 10.21873/anticanres.11239. [DOI] [PubMed] [Google Scholar]
- 70.Öner Ç., Coşan D.T., Çolak E. Estrogen and androgen hormone levels modulate the expression of PIWI interacting RNA in prostate and breast cancer. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0159044. e0159044–e0159044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iliev R., Stanik M., Fedorko M., Poprach A., Vychytilova-Faltejskova P., Slaba K., Svoboda M., Fabian P., Pacik D., Dolezel J., Slaby O. Decreased expression levels of PIWIL1, PIWIL2, and PIWIL4 are associated with worse survival in renal cell carcinoma patients. Oncol. Targets Ther. 2016;9:217–222. doi: 10.2147/OTT.S91295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H., Xu Z., Liu D. Small non-coding RNA and colorectal cancer. J. Cell. Mol. Med. 2019;23:3050–3057. doi: 10.1111/jcmm.14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y., Wu X., Gao H., Jin J.M., Li A.X., Kim Y.S., Pal S.K., Nelson R.A., Lau C.M., Guo C., Mu B., Wang J., Wang F., Wang J., Zhao Y., Chen W., Rossi J.J., Weiss L.M., Wu H. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol. Med. 2015;21:381–388. doi: 10.2119/molmed.2014.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Busch J., Ralla B., Jung M., Wotschofsky Z., Trujillo-Arribas E., Schwabe P., Kilic E., Fendler A., Jung K. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J. Exp. Clin. Cancer Res. 2015;34:61. doi: 10.1186/s13046-015-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao C., Tolkach Y., Schmidt D., Toma M., Muders M.H., Kristiansen G., Müller S.C., Ellinger J. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma. World J. Urol. 2019;37:1639–1647. doi: 10.1007/s00345-018-2575-1. [DOI] [PubMed] [Google Scholar]
- 76.Zhao C., Tolkach Y., Schmidt D., Kristiansen G., Müller S.C., Ellinger J. 5′-tRNA halves are dysregulated in clear cell renal cell carcinoma. J. Urol. 2018;199:378–383. doi: 10.1016/j.juro.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 77.Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., Yang X. TRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martens-Uzunova E.S., Olvedy M., Jenster G. Beyond microRNA – novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 79.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu L., Ge J., Li T., Shen Y., Guo J. TRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–37. doi: 10.1016/j.canlet.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 81.Shen Y., Yu X., Zhu L., Li T., Yan Z., Guo J. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J. Mol. Med. 2018;96:1167–1176. doi: 10.1007/s00109-018-1693-y. [DOI] [PubMed] [Google Scholar]
- 82.Zhu P., Yu J., Zhou P. Role of tRNA-derived fragments in cancer: novel diagnostic and therapeutic targets tRFs in cancer. Am. J. Cancer Res. 2020;10:393–402. https://pubmed.ncbi.nlm.nih.gov/32195016 [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang P., Yan F. TiRNAs & tRFs biogenesis and regulation of diseases: a review. Curr. Med. Chem. 2019;26:5849–5861. doi: 10.2174/0929867326666190124123831. [DOI] [PubMed] [Google Scholar]
- 84.Goodarzi H., Liu X., Nguyen H.C.B., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar P., Kuscu C., Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem. Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soares A.R., Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. WIREs RNA. 2017;8:e1423. doi: 10.1002/wrna.1423. [DOI] [PubMed] [Google Scholar]
- 87.Li S., Hu G.-.F. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J. Cell. Physiol. 2012;227:2822–2826. doi: 10.1002/jcp.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson D.M., Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 89.Couvillion M.T., Sachidanandam R., Collins K. A growth-essential tetrahymena piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 91.Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W.S., Green P.J., Barton G.J., Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q., Lee I., Ren J., Ajay S.S., Lee Y.S., Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. J. Am. Soc. Gene Ther. 2013;21:368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gebetsberger J., Zywicki M., Künzi A., Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012 doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Couvillion M.T., Bounova G., Purdom E., Speed T.P., Collins K. A tetrahymena piwi bound to mature tRNA 3’ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell. 2012;48:509–520. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dhahbi J.M., Spindler S.R., Atamna H., Yamakawa A., Boffelli D., Mote P., Martin D.I.K. 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu L., Li J., Gong Y., Wu Q., Tan S., Sun D., Xu X., Zuo Y., Zhao Y., Wei Y.-.Q., Wei X.-.W., Peng Y. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer. 2019;18:74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dhahbi J.M., Spindler S.R., Atamna H., Boffelli D., Martin D.I. Deep sequencing of serum small RNAs identifies patterns of 5’ tRNA Half and YRNA fragment expression associated with breast cancer. Biomark. Cancer. 2014;6:37–47. doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dhahbi J., Nunez Lopez Y.O., Schneider A., Victoria B., Saccon T., Bharat K., McClatchey T., Atamna H., Scierski W., Golusinski P., Golusinski W., Masternak M.M. Profiling of tRNA halves and YRNA fragments in serum and tissue from oral squamous cell carcinoma patients identify key role of 5’ tRNA-Val-CAC-2-1 half. Front. Oncol. 2019;9:959. doi: 10.3389/fonc.2019.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Victoria Martinez B., Dhahbi J.M., Nunez Lopez Y.O., Lamperska K., Golusinski P., Luczewski L., Kolenda T., Atamna H., Spindler S.R., Golusinski W., Masternak M.M. Circulating small non-coding RNA signature in head and neck squamous cell carcinoma. Oncotarget. 2015;6:19246–19263. doi: 10.18632/oncotarget.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nientiedt M., Deng M., Schmidt D., Perner S., Müller S.C., Ellinger J. Identification of aberrant tRNA-halves expression patterns in clear cell renal cell carcinoma. Sci. Rep. 2016;6:37158. doi: 10.1038/srep37158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rupaimoole R., Slack F. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 103.Fan B., Jin Y., Zhang H., Zhao R., Sun M., Sun M., Yuan X., Wang W., Wang X., Chen Z., Liu W., Yu N., Wang Q., Liu T., Li X. MicroRNA-21 contributes to renal cell carcinoma cell invasiveness and angiogenesis via the PDCD4/c-Jun (AP-1) signalling pathway. Int. J. Oncol. 2020;56:178–192. doi: 10.3892/ijo.2019.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong H., Sun S., Yan T., Liang C., Zhu J., Miao C., Qin C., Shao P., Wang Z., Li J., Li P. MicroRNA-195 inhibits proliferation and metastasis in renal cell carcinoma via regulating HMGA1. Am. J. Transl. Res. 2020;12:2781–2792. https://pubmed.ncbi.nlm.nih.gov/32655809 [PMC free article] [PubMed] [Google Scholar]