ABSTRACT
The SARS-CoV-2, a newly identified β-coronavirus, is the causative agent of the third large-scale pandemic from the last two decades. The outbreak started in December 2019 in Wuhan City, Hubei province in China. The patients presented clinical symptoms of dry cough, fever, dyspnea, and bilateral lung infiltrates on imaging. By February 2020, The World Health Organization (WHO) named the disease as Coronavirus Disease 2019 (COVID-19). The Coronavirus Study Group (CSG) of the International Committee on Taxonomy of Viruses (ICTV) recognized and designated this virus as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The SARS-CoV-2 uses the same host receptor, angiotensin-converting enzyme 2 (ACE2), used by SARS-CoV to infect humans. One hypothesis of SARSCoV-2 origin indicates that it is likely that bats serve as reservoir hosts for SARSCoV-2, being the intermediate host not yet determined. The predominant route of transmission of SARS-CoV-2 is from human to human. As of May 10th 2020, the number of worldwide confirmed COVID-19 cases is over 4 million, while the number of global deaths is around 279.000 people. The United States of America (USA) has the highest number of COVID-19 cases with over 1.3 million cases followed by Spain, Italy, United Kingdom, Russia, France and Germany with over 223.000, 218.000, 215.000, 209.000, 176.000, and 171.000 cases, respectively.
Keywords: Coronavirus; spike protein, SARS-CoV-2 [Supplementary Concept]; Diagnosis
INTRODUCTION
The coronaviruses (CoVs) are a family of enveloped positive-stranded RNA viruses broadly distributed among mammals and birds that cause respiratory and intestinal infections in animals and humans, and in some cases neurologic illness or hepatitis ( 1 , 2 ). The CoVs have been the causative agent of two large-scale pandemics in the past two decades: 1) Severe acute respiratory syndrome (SARS) in 2002 and 2003 in Guangdong province, China ( 3 , 4 ); and 2) Middle East respiratory syndrome (MERS) in 2012 in Middle Eastern countries ( 5 , 6 ). After these two pandemic events, several pieces of evidence suggested possible future disease outbreaks: 1) CoVs undergo genetic recombination ( 7 ), which may lead to new evolving genotypes; 2) the presence of a large reservoir of SARS-related coronaviruses (SARSr-CoVs) in horseshoe bats in China ( 8 , 9 ); and 3) previous studies determined that some bat SARSr-CoVs have the potential to infect humans ( 2 , 10 – 13 ).
The SARS-CoV-2, previously known as the 2019-novel coronavirus 2019-nCoV ( 14 ) ( Figure-1 ), is a newly identified β-coronavirus that caused an epidemic of acute respiratory syndrome in humans, which started in December 2019 in the context of a seafood market in Wuhan, China ( 14 ). Later, in February 2020, The World Health Organization (WHO) named the disease as corona-virus disease 2019 (COVID-19). The COVID-19 has now progressed to be transmitted by human-to--human ‘contact’ and spread within few months not only throughout China but also worldwide, affecting over 4 million people and killing more than 279.000 of them in 187 countries as of May 10th 2020 ( 15 ). Typical clinical symptoms of COVID-19 patients are fever, dry cough, breathing difficulties, headache and pneumonia and in some cases gastrointestinal infection symptoms.
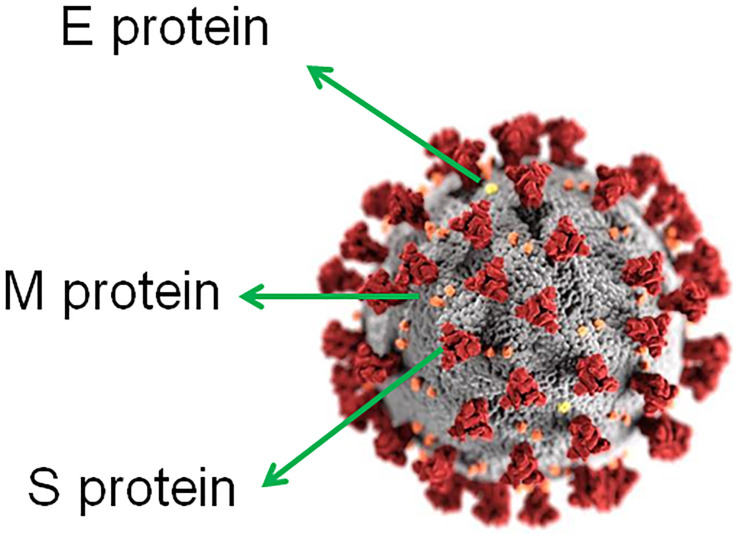
Figure 1. Ultrastructural morphology exhibited by coronaviruses.
E protein, small envelope (E) protein; M protein, matrix (M) protein; S protein, spike (S) glycoprotein (homotrimer). Adapted from “Centers for Disease Control and Prevention (CDC)/ Alissa Eckert, MS; Dan Higgins, MAMS”.
GENERAL CHARACTERISTICS OF SARS-CoV-2
1 - Classification
The CoVs were previously classified based on serologic (cross-) reactivity involving the structural protein spike (S) glycoprotein until the classification shifted to comparative sequence analysis of replicative proteins ( 16 , 17 ). The SARS-CoV-2 has been reported as the seventh corona-virus known to infect humans ( 14 , 18 ). SARS-CoV, MERS-CoV and SARS-CoV-2, all β-CoVs, can cause severe respiratory disease in humans. The other four human CoVs, two α-CoVs HCoV-NL63 and HCoV-229E, and two β-CoVs HCoV-OC43 and HCoV-HKU1, cause mild respiratory symptoms ( 2 , 13 , 14 ). The SARS-CoV-2 clusters with SARS-CoVs in trees of the species Severe acute respiratory syndrome-related coronavirus and genus Betacoronavirus ( 14 , 18 , 19 ) ( Figure-2 ). Based on phylogeny and taxonomy, the Coronavirus Study Group (CSG) of the ICTV recognized this virus as a sister to severe acute respiratory syndrome coronaviruses (SARS-CoVs) of the species Severe acute respiratory syndrome-related coronavirus and designated it as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ( 19 ).
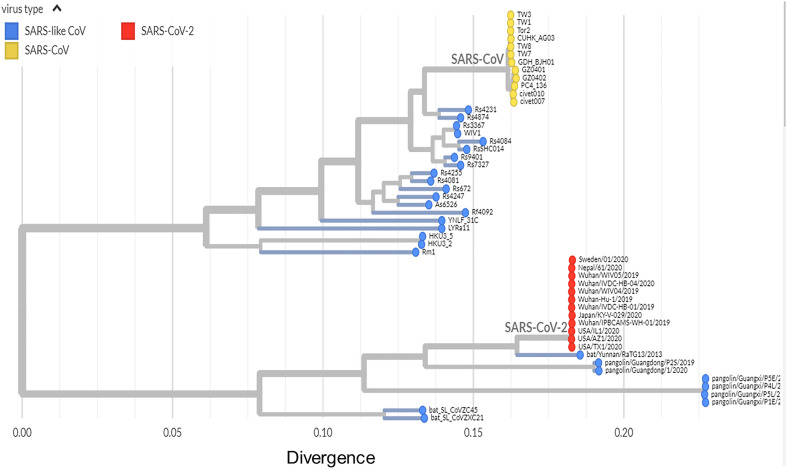
Figure 2. Phylogeny of SARS-like betacoronaviruses including novel coronavirus SARS-CoV-2.
Phylogenetic tree including 52 genomes. Red dots, SARS-CoV-2 coronaviruses from the COVID-19 epidemic; yellow dots, SARS-CoV coronaviruses from the 2002-03 SARS outbreak; and blue dots, SARS-like coronaviruses. Adapted from “github.com/blab/sars-like-cov”; “Built with blab/sars-like-cov and maintained by Trevor Bedford and Emma Hodcroft”.
2 - Structure and Mechanism
The first complete SARS-CoV-2 virus genome has been reported to be 29.9 kilobases (Gen-Bank accession number MN908947) ( 18 ), which consists of six major open-reading frames (ORFs) that are common to CoVs, and a number of other accessory genes ( 14 , 18 ). Four ORFs of SARS-CoV-2 genome encode four essential structural proteins: ( 1 ) spike (S) glycoprotein (S1 and S2 subunits) that attaches to the host receptor through the receptor binding domain (RBD) (S1 subunit), determines the virus host range (S1 subunit), and mediates virus-cell membrane fusion (S2 subunit); ( 2 ) matrix (M) protein that mediates nutrients transport across the transmembrane, bud release and envelope formation; ( 3 ) small envelope (E) protein; and ( 4 ) nucleocapsid (N) protein which interfere with the host innate immune response ( 20 ) ( Figure-1 ). The spike glycoprotein from CoVs forms homotrimers protruding from the viral surface and mediating the entry of the virus genome into the host cells ( 21 ). Therefore, it constitutes the main target of neutralizing antibodies after infection and hence, the focus of vaccine designing ( 22 ). Two structural acquisition of SARS-CoV-2 spike (S) glycoprotein have not been found in lineage B β-coronavirus: 1) a functional polybasic (furin) cleavage site at the junction between the S1/ S2 subunits which is cleaved during biogenesis; and 2) three adjacent predicted O-linked glycans ( 22 ). Curiously, the acquisition of polybasic cleavage sites in the hemagglutinin protein from low-pathogenic avian influenza viruses turns them into highly pathogenic forms ( 23 ). The introduction of the predicted O-linked glycans could build a ‘mucin-like domain’, like those found in Ebola and Marburg viruses, that shields select immunodominant epitopes on the SARS-CoV-2 spike protein ( 24 ). O-glycosylated ‘mucin-like domains’ may physically hinder the interaction between virus-infected cells and immune cells ( 25 ).
The SARS-CoV-2 uses the same host receptor, angiotensin-converting enzyme 2 (ACE2), used by SARS-CoV to infect humans ( 14 ). ACE2 is a metalloprotease expressed in the cells of the lung, intestine, liver, heart, vascular endothelium, testis and kidney ( 2 ). In addition, the SARS-CoV-2 seems to have an RBD that binds with high affinity to ACE2 from humans and other species with high receptor homology ( 26 ). Six amino acids present in the RBD of the spike protein are essential for binding to host ACE2 receptors, and for establishing the host range of SARS-CoV-like viruses. Interestingly, five of these six amino acids differ between SARS-CoV-2 and SARS-CoV ( 26 ).
3 - Theories of SARS-CoV-2 Origins
It has been reported that MERS-CoV originated from bats, being dromedary camels the reservoir host triggering the spillover to humans ( 27 ). However, palm civets and racoon dogs have been indicated as an intermediate host for zoonotic transmission of SARS-CoV bridging bats and humans ( 28 ). In this sense, the intermediate host of SARS-CoV-2 remains unknown. Nevertheless, Ge et al. ( 9 ) proposed that some bat SARS-like coronaviruses (SL-CoVs) may directly infect human cells without an intermediate host. They were able to isolate a live bat SL-CoV (bat SL-CoV-WIV1, Figure-2) from bat fecal samples, which shares 99.9% sequence identity to Rs3367 ( Figure-2 ), a bat coronavirus from Chinese horseshoe bats in Yunnan, China, and uses the ACE2 receptor from humans, civets and Chinese horseshoe bats for cell entry ( 9 ). Later in 2015, Menachery et al. ( 11 ) validated this hypothesis by synthetically producing an infectious recombinant virus from a bat coronavirus SHC014 (RsSHC014, Figure-2) that could efficiently replicate both in vitro in primary human airway cells, and in vivo in mouse lung. Thus, they emphasized the potential risk of SARS-CoV re-emergence from viruses currently circulating in bat populations without the necessity of an intermediate host ( 11 ).
Andersen et al. ( 29 ) have recently postulated two hypotheses that could explain the origin of SARS-CoV-2: 1) Natural selection in an animal host before zoonotic transfer; and 2) Natural selection in humans following zoonotic transfer. Regarding the first hypothesis, it is likely that bats serve as reservoir hosts for SARSCoV-2 ( 30 , 31 ), since the genome sequence of SARS-CoV-2 shares a 96.2% identity at the whole genome level with that of bat CoV RaTG13 ( 14 ) (phylogenetic proximity in the clade SARS-CoV-2//bat/Yunnan/RaTG13/2013, Figure-2). Nonetheless, Wu et al. ( 18 ) reported that bats were not available for selling in the seafood market, where the first COVID-19 cases appeared ( 18 ). They found that the SARS-CoV-2 virus strain designated as WHCV (GenBank accession number MN908947) shares a nucleotide identity of 89.1% with bat SARS--like CoV isolated from bat (Bat-SL-CoVZC45- Gen-Bank accession number MG772933) that had been previously collected from Zhoushan City, Zhejiang province, China, between 2015 and 2017 ( 32 ) (bat_SL_CoVZC45, Figure-2). Interestingly, the spike glycoprotein from some pangolin CoVs shows high similarity to SARS-CoV-2 in the RBD, which includes all six key RBD residues ( 33 ), supporting the existence of alternative intermediate host like pangolins, snakes and turtles ( 34 ), though not yet identified. This also indicates that all six key RBD residues may have been already present in the virus that jumped to humans ( 29 ). In terms of the second hypothesis, Andersen et al. ( 29 ) proposed that the genomic characteristics acquired from the progenitor of SARS-CoV-2, which would prime a pandemic outbreak, have taken place initially in humans during an undetected human-to--human transmission.
TRANSMISSION
The common transmission routes of SARS-CoV-2 include: 1) Direct exposure with cough, sneeze and droplet inhalation within a range of about 1.8 meters; and 2) Contact transmission through contact with oral, nasal, and eye mucous membranes ( 35 ). It has been also suggested that the SARS-CoV-2 transmission is not only limited to the respiratory tract ( 36 ), the eye mucosa may provide the virus with the portal to enter the body ( 32 ). Similarly, saliva may also directly or indirectly transmit SARS-CoV-2 ( 37 ). This is especially important during dental procedures, since aerosols and droplets mixed with patient's saliva and even contaminated blood with virus are generated ( 38 ). In similar way, Wax et al. ( 39 ) suggested that SARS-CoV-2 may be airborne through aerosols formed during medical procedures ( 39 ). In this sense, indirect contact via contaminated surfaces is another possible cause of infection. Interestingly, the presence of SARS-CoV-2 in fecal swabs (29%) and blood (1%) from infected individuals indicated the possibility of multiple transmission routes; however, no individuals contained detectable viral RNA in their urine ( 40 ). Additionally, it has been reported that contact with asymptomatic patients may represent another form of virus transmission ( 41 ). In this aspect, an epidemiological model published at the beginning of the outbreak in China suggested that subclinical infections may have been the source of a majority of infections ( 42 ).
International actions have been taken to reduce the social viral transmission by implementing “physical distancing” strategies, such as staying at least two meters apart from other people, not gathering in groups, considering delivery services, using cloth face cover to protect mouth and nose when around others or when going out in public, working from home when possible, avoiding the use of public transportation, implementing digital/distance learning. “Quarantine” has been employed to keep someone who might have been exposed to COVID-19 away from others, and “isolation” to separate sick people from healthy ones. Those actions have impacted on the viral transmission profile in those countries that followed the guidelines from the “Centers for Disease Control and Prevention” (CDC) ( Figures 3A and B ).
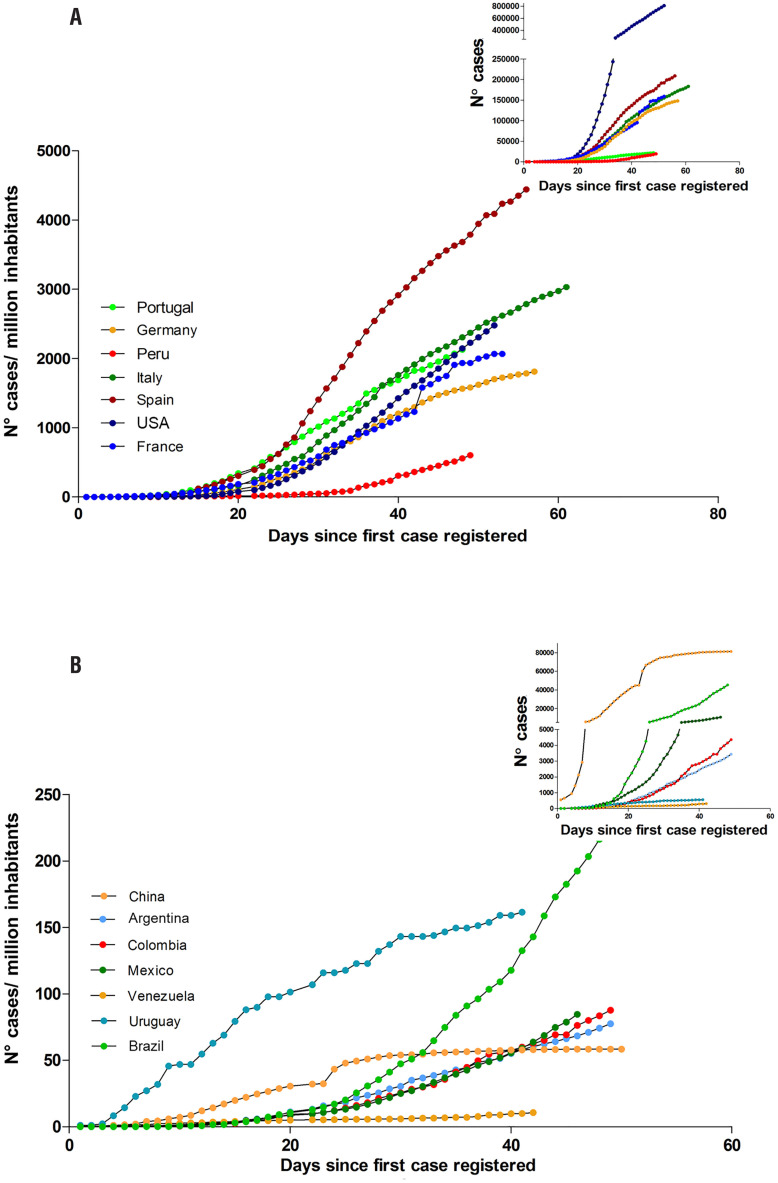
Figure 3. SARS-CoV-2 transmission.
Number of cumulative cases per million of inhabitants: Both graphs (A and B) show the daily evolution of the number of detected cumulative cases of COVID-19 until 50 days after the first reported case normalized to each country's population. In the upper right boxes the absolute number of cases during the same time period is shown. (A) Transmission of SARS-CoV-2 in countries (Portugal, Germany, Peru, Italy, Spain, USA, France) which number of cases/million inhabitants is greater than 500 (cumulative cases/ million inhabitants). (B) Transmission of SARS-CoV-2 in countries (China, Argentina, Colombia, Mexico, Venezuela, Uruguay, Brazil) which number of cases/million inhabitants is smaller than 500 (cumulative cases/ million inhabitants). The data used for the construction of the curves were obtained from the maps of John Hopkins University https://coronavirus.jhu.edu/map.html ( 15 ).
DISEASE PATHOPHYSIOLOGY AND THE IMMUNE RESPONSE
The clinical manifestations of COVID-19 range from mild to severe compromise, with few cases showing a fatal course. The most common reported symptoms are fever, cough, myalgia or fatigue, followed by pneumonia, and dyspnea, whereas less common reported symptoms include headache, diarrhea, and hemoptysis ( 43 ). Patients with mild symptoms were reported to recover after one week while severe cases experienced progressive respiratory failure due to alveolar damage, likely leading to death ( 43 ). Although the exact pathophysiological mechanisms underlying SARS-CoV-2 disease are not properly understood, genomic similarities to SARS-CoV may allow to infer the accompanying inflammatory response as being involved in the development of severe pneumonia ( 44 , 45 ) ( Table-1 ).
Table 1. Summary of the symptoms recorded in 191 COVID-19 confirmed patients hospitalized in Jinyintan Hospital or Wuhan Pulmonary Hospital before January 31st, 2020 (45). (n= 191).
| Fever (temperature >37.3°C) | Cough | Sputum | Myalgia | Fatigue | Diarrhoea | Nausea or vomiting |
|---|---|---|---|---|---|---|
| 180 (94%) | 151 (79%) | 44 (23%) | 29 (15%) | 44 (23%) | 9 (5%) | 7 (4%) |
Histopathological observations of pulmonary lesions from SARS cases not only show nonspecific inflammatory responses such as edema and inflammatory cell infiltration but also a severe exfoliation of alveolar epithelial cells, alveolar septal widening, damage to alveolar septa, as well as alveolar space infiltration in a distinctly organized manner. SARS-CoV infection can cause pathological changes, degeneration, infiltration, and hyperplasia. Damage to the pulmonary interstitial arteriolar walls indicates that the inflammatory response plays an important role throughout the course of disease despite (or beyond) the pathogenic effect of Coronaviruses ( 46 ).
Even though SARS-CoV-2 is less lethal than MERS-CoV, up to 10-20%, people over 60 years and those with underlying medical co-morbidities, are more likely to develop a severe disease characterized by interstitial pneumonia and acute respiratory distress syndrome (ARDS) or even septic shock. Likewise, it is common to observe high levels of acute-phase reactants and features from the macrophage activation syndrome such as hyperferritinaemia, hepatic dysfunction and diffuse intravascular coagulation ( 44 ). Case definition guidelines consider symptoms like fever, decrease in lymphocytes and white blood cells, new pulmonary infiltrates on chest radiography, and no improvement in symptoms after 3 days of antibiotics treatment ( 43 ).
During a viral infection, the host mounts an immune response (IR) addressed to contain the infection. Recent advances in the knowledge of the innate IR against viruses point out that this type of IR inhibits virus replication, promotes virus clearance, induces tissue repair, while promoting a prolonged adaptive IR against the viruses. In most cases, pulmonary and systemic inflammatory responses associated with coronavirus are mediated by innate immune mechanisms upon virus recognition. However, an exacerbated IR also plays an immunopathogenic role, accounting for pulmonary tissue damage, functional impairment, and reduced lung capacity ( 43 ). The damaged cells induce innate inflammation in the lungs, largely mediated by proinflammatory macrophages and granulocytes. Such lung inflammation further emerges as the main cause of life-threatening respiratory disorders in severely ill patients ( 47 ) ( Figure-4 ).
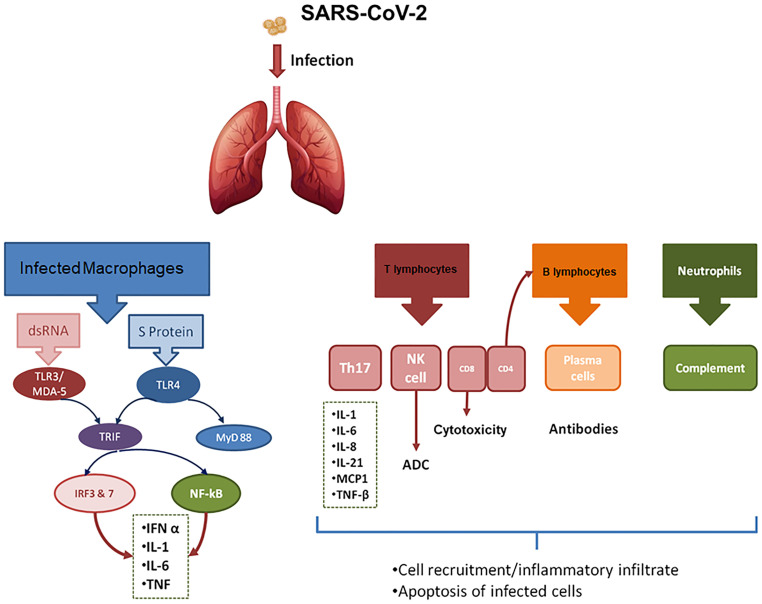
Figure 4. Diagram of the immune response to infection with SARS-CoV-2.
The intracellular response of the infected macrophage is shown on the left side of the figure. As shown, both protein S and viral RNA will induce the production of pro-inflammatory cytokines as well as type 1 interferons. The main activation pathways are those that lead to nuclear translocation of NF-kB and IRF (Interferon Response Factors). On the right side are shown the cells of the innate as well as the adaptive immune response that are involved in antiviral immunity, with the mechanisms of cytotoxicity as well as those of humoral immunity being the most relevant. TLR4, Toll Like receptor 4; MDA-5, Melanoma Differentiation Antigen 5; IFN, Type I Interferon; NK cells, Natural Killer Cells; IRF3 and 7, Interferon Response Factors; TNF-β, Tumor necrosis factor-β; MCP-1, Monocyte Chemoattractant Protein-1; Th17, T-helper 17 cells.
When a virus invades the host, the viral nucleic acid is initially recognized by Pattern Recognition Receptors, like Toll Like receptor 4 (TLR4) or Melanoma Differentiation Antigen 5 (MDA-5), that recognize S protein or nucleic acids, respectively. A signaling cascade is then activated to promote the synthesis of type I interferons (IFN-alpha and IFN--beta). Type I IFNs subsequently activate the downstream JAK-STAT signal pathway, promoting the expression of IFN-stimulated gene(s). As host's major antiviral molecules, IFNs limit virus spread, play a promoting role for macrophage phagocytosis of antigens, as well as Natural Killer (NK) cells restriction of infected target cells and T/B cells. It follows, that blocking the production of IFNs has a direct effect on the survival of the virus within the host ( 46 ) ( Figure-4 ).
Cytokine deregulation was also thought to underlie ARDS development. Apparently, SARS-CoV2, induces abnormally low levels of antiviral cytokines, particularly type I interferons, which form part of the very early IR to viral infections ( 44 ). Such lack of an antiviral innate IR may favor a poorly controlled viral replication with progressive increases in viral load and the accompanying pro inflammatory systemic response. This situation continues until the appearance of the adaptive IR, which brings viral replication under control. Concerning SARS-CoV2, its clinical severity is related to the high viral load and the intense inflammatory response as evidenced by serum cytokine profiles and histopathology ( 2 ).
Moving to antiviral adaptative IR, CD4+ T cells, and CD8+ T cells particularly play a significant antiviral role, with the former promoting the production of virus-specific antibodies by activating T-dependent B cells; and CD8+ T cytotoxic cells, killing viral infected cells. Of note, CD8+ T cells account for about 80% of total infiltrative inflammatory cells in the lung interstitium from SARS-CoV- infected patients, being involved in coronaviruses clearance of infected cells as well as immune injury ( 46 ). Additionally, T helper cells produce pro inflammatory cytokines via the NF-kB signaling pathway. Cytokine dysregulation is of particular interest in patients with COVID-19, who have higher levels of inflammatory cytokines. However, what is more interesting is that, as seen during the SARS outbreak, some cytokines seem to be up-regulated, especially in patients with more severe disease. IL-17 cytokines recruit monocytes and neutrophils to the site of infection which in turn activate other downstream cytokine and chemokine cascades, such as IL-1, IL-6, IL-8, IL-21, TNF-β, and MCP-1 ( 44 , 46 ). Some studies showed that the levels of inflammatory cytokines are high in the lungs of COVID-19 patients like TNF-α and IL-1. Besides disease severity correlated with TNF-α, IL-6 and IL-10 levels ( 44 ) ( Figure-4 ).
On the other hand, a worth considering question deals with the generation of immune memory to SARS-CoV-2. Considering the knowledge gathered from another coronaviruses, in SARS convalescents patients, memory T cell responses are directed at SARS-CoV structural proteins. These responses are found to last up to 11 years after infection. There is also evidence for an absence of cross-reactivity of these CD8+T cell responses against the MERS-CoV ( 46 ).
In summarizing, the IR induced by SARS-CoV-2 infection is two phased. During the incubation period and non-severe stages, a specific adaptive immune response is required to eliminate the virus and to preclude disease progression to advanced disease. Therefore, strategies to boost immune responses (anti-sera or pegylated IFNα) at this stage are certainly welcome. However, when a protective IR is impaired, the virus will propagate favoring a substantial destruction of affected tissues, especially in tissues that have high ACE2 expression ( 47 ). In turn, damaged cells will fuel innate-mediated inflammation in the lungs largely mediated by pro inflammatory macrophages and granulocytes. As stated, lung inflammation therefore emerges as a critical factor for life-threatening respiratory disorders at the severe stage ( 47 ).
THE DIAGNOSIS OF COVID-19 PATIENTS
Intensive testing of suspected cases to identify COVID-19 infected people is critical to avoid the further spread of infection. The in vitro diagnostic as-says based on viral nucleic acid detection using real--time reverse transcriptase polymerase chain reaction (RT-PCR) (~80% sensitivity) remain the standard of reference ( 48 – 51 ) ( Table-2 ). The assay duration has been shortened from 2-3 hours to 45 minutes; however, it is unable to detect the SARS-CoV-2 in early stages of viral infection, giving false negatives in people infected up to two weeks after symptom onset. Possible reasons for the low detection efficiency could be low patient viral load or improper clinical sampling. In this sense, chest radiography and computed tomography (CT) (~65% sensitivity) represent a complementary diagnostic tool that allows physicians to effectively make a diagnosis, reaching in many cases a higher sensitivity (~91%) by combining both tools ( 52 ).
Table 2. Summary of in vitro diagnostic assays based on SARS-CoV-2 viral nucleic detection.
| Assay type/ name | Target genes | Samples | Assay duration | Sensitivity and/or detection limit | Company/Authors |
|---|---|---|---|---|---|
| Real time RTPCR |
|
|
N.R. |
|
Tib-Molbiol, Berlin, Germany, published by Corman et al. (49) |
| One-step real time RTPCR |
|
|
> 1 hour | <10 RNA copies/ reaction | Published by Chu et al. (50) |
| Real time RTPCR (COVID-19-RdRp/Hel assay) |
|
|
N.R. |
|
Published by Chan et al. (51) |
| Real-time RT-PCR rapid test (Xpert® Xpress SARSCoV-2 test) | Two target genes (E and N2) |
|
45 min | 0.0100 plaque forming units (PFU)/mL | Cepheid, USA |
| Vivalytic COVID-19 | SARS-CoV-2 and nine other respiratory |
|
< 2.5 hour | N.R. | Bosch, Germany & Randox Laboratories, UK |
| Test (rapid test) |
Viruses
|
||||
| Abbott ID NowTM— COVID-19 test (rapid test) | Throat, nasal, nasopharyngeal and oropharyngeal swabs | 5 min | N.R. | ABBOTT, USA |
Real-time RT-PCR = real-time reverse transcriptase polymerase chain reaction; N.R = not reported; UK = United Kingdom
Other in vitro diagnostic assays, such as several serological immunoassays (rapid lateral flow immunoassay (LFIA) tests, automated chemiluminescence immunoassay (CLIA), and manual ELISA) detect SARS-CoV-2 viral proteins and antibodies like IgM and IgG, in the serum or plasma. The detection of IgM ranges from 10 to 30 days after SARS-CoV-2 infection; however, that of IgG from 20 days onwards ( 48 ) ( Table-3 ).
Table 3. Summary of rapid in vitro diagnostic serological immunoassays based on SARS-CoV-2 detection of viral proteins and antibodies.
| Assay type/ name | Target proteins | Samples | Assay duration | Sensitivity and/or detection limit | Company/Authors |
|---|---|---|---|---|---|
| DZ-Lite SARSCoV-2 CLIA * |
|
|
~ 50 tests/hour |
|
Diazyme, USA |
|
|
30 min | N.R. | Snibe Co, China | |
| COVID-19 IgM/IgG Rapid Test ** |
|
|
10-15 min |
|
BioMedomics, USA |
| SARS-CoV-2 Rapid Test ** |
|
|
20 min |
|
Pharmacyt AG, Germany |
| DPP COVID-19 Serological Point-of-Care Test ** |
|
|
15 min | N.R. | Chembio Diagnostics, USA |
= automated chemiluminescence immunoassay (CLIA);
= rapid lateral flow immunoassay (LFIA).
Additionally, other routine blood examinations are used to monitor the status of COVID-19 infection, such as liver and kidney function, myocardial markers, myoglobin, ferritin, erythrocyte sedimentation rate, C-reactive protein (CRP), procalcitonin (PCT), lactate, D-dimer, complete blood count, coagulation profile, urine routine test, creatine kinase, lactate dehydrogenase, electrolytes and inflammatory factors (interleukin (IL)-6, IL-10, TNF-α). Monitoring CRP and PCT levels help to distinguish whether there was bacterial infection in the lung. D-dimer infers the risk for blood clotting (thrombosis) and/or thrombotic embolism. It has been observed that in most severe COVID-19 patients, the D-dimer level is significantly increased showing frequent clotting disorders and microthrombotic formations. Quantifying inflammatory factors, especially IL-6, may help to preliminarily evaluate the immune status of patients in terms of the cytokine release syndrome ( 45 ).
REFERENCES
- 1.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; 1. Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193-292. [DOI] [PMC free article] [PubMed]
- 2.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; 2. Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660-94. [DOI] [PMC free article] [PubMed]
- 3.Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]; 3. Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967-76. [DOI] [PubMed]
- 4.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]; 4. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953-66. [DOI] [PubMed]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. Erratum in: N Engl J Med. 2013;369:394. [DOI] [PubMed] [Google Scholar]; 5. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814-20. Erratum in: N Engl J Med. 2013;369:394. [DOI] [PubMed]
- 6.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; 6. Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995-1007. [DOI] [PMC free article] [PubMed]
- 7.Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, Chan KH, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; 7. Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, Chan KH, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80:7136-45. [DOI] [PMC free article] [PubMed]
- 8.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]; 8. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676-9. [DOI] [PubMed]
- 9.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]; 9. Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535-8. [DOI] [PMC free article] [PubMed]
- 10.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]; 10. Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040-5. [DOI] [PMC free article] [PubMed]
- 11.Menachery VD, Yount BL, Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. Erratum in: Nat Med. 2016;22:446. [DOI] [PMC free article] [PubMed] [Google Scholar]; 11. Menachery VD, Yount BL Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508-13. Erratum in: Nat Med. 2016;22:446. [DOI] [PMC free article] [PubMed]
- 12.Wang N, Li SY, Yang XL, Huang HM, Zhang YJ, Guo H, et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; 12. Wang N, Li SY, Yang XL, Huang HM, Zhang YJ, Guo H, et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol Sin. 2018;33:104-107. [DOI] [PMC free article] [PubMed]
- 13.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; 13. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181-92. [DOI] [PMC free article] [PubMed]
- 14.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; 14. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-3. [DOI] [PMC free article] [PubMed]
- 15.The Johns Hopkins Center for Health Security (JHCHS) [[cited 2020 May 10]];Homepage [Internet] https://www.centerforhealthsecurity.org/ [Google Scholar]; 15. The Johns Hopkins Center for Health Security (JHCHS) | Homepage [Internet]. [cited 2020 May 10]. https://www.centerforhealthsecurity.org/
- 16.de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; 16. de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790-2. [DOI] [PMC free article] [PubMed]
- 17.Gorbalenya AE, Snijder EJ, Spaan WJ. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; 17. Gorbalenya AE, Snijder EJ, Spaan WJ. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004;78:7863-6. [DOI] [PMC free article] [PubMed]
- 18.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. Erratum in: Nature. 2020;580:E7. [DOI] [PMC free article] [PubMed] [Google Scholar]; 18. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-9. Erratum in: Nature. 2020;580:E7. [DOI] [PMC free article] [PubMed]
- 19.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Severe acute respiratory syndrome-related coronavirus : The species and its viruses – a statement of the Coronavirus Study Group. BioRxiv. 2020 Available at. < https://www.biorxiv.org/content/10.1101/2020.02.07.937862v1 >. [Google Scholar]; 19. Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Severe acute respiratory syndrome-related coronavirus : The species and its viruses – a statement of the Coronavirus Study Group. BioRxiv. 2020. Available at. <https://www.biorxiv.org/content/10.1101/2020.02.07.937862v1>
- 20.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; 20. Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325-328. [DOI] [PMC free article] [PubMed]
- 21.Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; 21. Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93-116. [DOI] [PMC free article] [PubMed]
- 22.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.:e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; 22. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281-92.e6. [DOI] [PMC free article] [PubMed]
- 23.Alexander DJ, Brown IH. History of highly pathogenic avian influenza. Rev Sci Tech. 2009;28:19–38. doi: 10.20506/rst.28.1.1856. [DOI] [PubMed] [Google Scholar]; 23. Alexander DJ, Brown IH. History of highly pathogenic avian influenza. Rev Sci Tech. 2009;28:19-38. [DOI] [PubMed]
- 24.Machiels B, Lété C, Guillaume A, Mast J, Stevenson PG, Vanderplasschen A, et al. Antibody evasion by a gammaherpesvirus O-glycan shield. PLoS Pathog. 2011;7:e1002387. doi: 10.1371/journal.ppat.1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]; 24. Machiels B, Lété C, Guillaume A, Mast J, Stevenson PG, Vanderplasschen A, et al. Antibody evasion by a gammaherpesvirus O-glycan shield. PLoS Pathog. 2011;7:e1002387. [DOI] [PMC free article] [PubMed]
- 25.Noyori O, Matsuno K, Kajihara M, Nakayama E, Igarashi M, Kuroda M, et al. Differential potential for envelope glycoprotein-mediated steric shielding of host cell surface proteins among filoviruses. Virology. 2013;446:152–161. doi: 10.1016/j.virol.2013.07.029. [DOI] [PubMed] [Google Scholar]; 25. Noyori O, Matsuno K, Kajihara M, Nakayama E, Igarashi M, Kuroda M, et al. Differential potential for envelope glycoprotein-mediated steric shielding of host cell surface proteins among filoviruses. Virology. 2013;446:152-61. [DOI] [PubMed]
- 26.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020:94–94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; 26. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94. [DOI] [PMC free article] [PubMed]
- 27.Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; 27. Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140-5. [DOI] [PMC free article] [PubMed]
- 28.Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. Erratum in: J Virol. 2006;80:7786. [DOI] [PMC free article] [PubMed] [Google Scholar]; 28. Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892-900. Erratum in: J Virol. 2006;80:7786. [DOI] [PMC free article] [PubMed]
- 29.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; 29. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450-2. [DOI] [PMC free article] [PubMed]
- 30.Giovanetti M, Benvenuto D, Angeletti S, Ciccozzi M. The first two cases of 2019-nCoV in Italy: Where they come from? J Med Virol. 2020;92:518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]; 30. Giovanetti M, Benvenuto D, Angeletti S, Ciccozzi M. The first two cases of 2019-nCoV in Italy: Where they come from? J Med Virol. 2020;92:518-21. [DOI] [PMC free article] [PubMed]
- 31.Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212–104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]; 31. Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. [DOI] [PMC free article] [PubMed]
- 32.Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7:154–154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; 32. Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7:154. [DOI] [PMC free article] [PubMed]
- 33.Zhang T, Wu Q, Zhang Z. Pangolin homology associated with 2019-nCoV. BioRxiv. 2020 Available at. < https://www.biorxiv.org/content/10.1101/2020.02.19.950253v1 >. [Google Scholar]; 33. Zhang T, Wu Q, Zhang Z. Pangolin homology associated with 2019-nCoV. BioRxiv. 2020. Available at. <https://www.biorxiv.org/content/10.1101/2020.02.19.950253v1>
- 34.Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARSCoV-2. J Med Virol. 2020 doi: 10.1002/jmv.25726. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]; 34. Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARSCoV-2. J Med Virol. 2020. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 35.Lu CW, Liu XF, Jia ZF. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; 35. Lu CW, Liu XF, Jia ZF. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. [DOI] [PMC free article] [PubMed]
- 36.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. Erratum in: Lancet. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; 36. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. Erratum in: Lancet. 2020. [DOI] [PMC free article] [PubMed]
- 37.To KK, Tsang OT, Chik-Yan Yip C, Chan KH, Wu TC, Chan JMC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]; 37. To KK, Tsang OT, Chik-Yan Yip C, Chan KH, Wu TC, Chan JMC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020. ahead of print. [DOI] [PMC free article] [PubMed]
- 38.Wei J, Li Y. Airborne spread of infectious agents in the indoor environment. Am J Infect Control. 2016;44(9) Suppl:S102–S108. doi: 10.1016/j.ajic.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; 38. Wei J, Li Y. Airborne spread of infectious agents in the indoor environment. Am J Infect Control. 2016;44(9 Suppl):S102-8. [DOI] [PMC free article] [PubMed]
- 39.Wax RS, Christian MD. Practical recommendations for critical care and anesthesiology teams caring for novel coronavirus (2019-nCoV) patients. Can J Anaesth. 2020;67:568–576. doi: 10.1007/s12630-020-01591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; 39. Wax RS, Christian MD. Practical recommendations for critical care and anesthesiology teams caring for novel coronavirus (2019-nCoV) patients. Can J Anaesth. 2020;67:568-76. [DOI] [PMC free article] [PubMed]
- 40.Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]; 40. Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386-389. [DOI] [PMC free article] [PubMed]
- 41.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]; 41. Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382:970-1. [DOI] [PMC free article] [PubMed]
- 42.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]; 42. Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science. 2020;368:489-493. [DOI] [PMC free article] [PubMed]
- 43.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9:29–29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; 43. Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9:29. [DOI] [PMC free article] [PubMed]
- 44.Sarzi-Puttini P, Giorgi V, Sirotti S, Marotto D, Ardizzone S, Rizzardini G, et al. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin Exp Rheumatol. 2020;38:337–342. [PubMed] [Google Scholar]; 44. Sarzi-Puttini P, Giorgi V, Sirotti S, Marotto D, Ardizzone S, Rizzardini G, et al. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin Exp Rheumatol. 2020;38:337-42. [PubMed]
- 45.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. Erratum in: Lancet. 2020;395:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]; 45. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054-62. Erratum in: Lancet. 2020;395:1038. [DOI] [PMC free article] [PubMed]
- 46.Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–32. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]; 46. Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424-32. [DOI] [PMC free article] [PubMed]
- 47.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; 47. Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451-4. [DOI] [PMC free article] [PubMed]
- 48.Vashist SK. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics (Basel) 2020;10:202–202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]; 48. Vashist SK. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics (Basel). 2020;10:202. [DOI] [PMC free article] [PubMed]
- 49.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045–2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]; 49. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. [DOI] [PMC free article] [PubMed]
- 50.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]; 50. Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66:549-555. [DOI] [PMC free article] [PubMed]
- 51.Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH, et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J Clin Microbiol. 2020:58–58. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; 51. Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH, et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J Clin Microbiol. 2020;58. [DOI] [PMC free article] [PubMed]
- 52.Guanmin Jiang, Xiaoshuai Ren, Yan Liu, Hongtao Chen, Wei Liu, Zhaowang Guo, et al. Application and optimization of RT-PCR in diagnosis of SARS-CoV-2 infection. medRxiv. 2020 Feb 25; [Google Scholar]; 52. Guanmin Jiang, Xiaoshuai Ren, Yan Liu, Hongtao Chen, Wei Liu, Zhaowang Guo, et al. Application and optimization of RT-PCR in diagnosis of SARS-CoV-2 infection. medRxiv 2020.02.25.