Graphical abstract
Keywords: Gram-positive bacteria, Electroactive organisms, Extracellular electron transfer, Bioelectrochemical systems
Abstract
The growing interest on sustainable biotechnological processes for the production of energy and industrial relevant organic compounds have increased the discovery of electroactive organisms (i.e. organisms that are able to exchange electrons with an electrode) and the characterization of their extracellular electron transfer mechanisms. While most of the knowledge on extracellular electron transfer processes came from studies on Gram-negative bacteria, less is known about the processes performed by Gram-positive bacteria. In contrast to Gram-negative bacteria, Gram-positive bacteria lack an outer-membrane and contain a thick cell wall, which were thought to prevent extracellular electron transfer. However, in the last decade, an increased number of Gram-positive bacteria have been found to perform extracellular electron transfer, and exchange electrons with an electrode. In this mini-review the current knowledge on the extracellular electron transfer processes performed by Gram-positive bacteria is introduced, emphasising their electroactive role in bioelectrochemical systems. Also, the existent information of the molecular processes by which these bacteria exchange electrons with an electrode is highlighted. This understanding is fundamental to advance the implementation of these organisms in sustainable biotechnological processes, either through modification of the systems or through genetic engineering, where the organisms can be optimized to become better catalysts.
1. Introduction
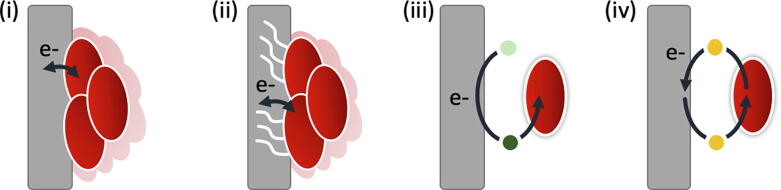
Electron transfer reactions are at the core of numerous biological processes, in particular in respiration. During respiration most microorganisms are able to convert biochemical energy into ATP. This usually involves a cascade of reactions where electrons are transferred, via several redox proteins, from an electron donor to an electron acceptor. Most forms of respiration involve a soluble compound as an electron acceptor (e.g. nitrate, oxygen, and sulfate), however there are others where solid compounds (e.g. metal oxides, electrodes) act as the electron acceptor [1]. In this case, the terminal electron acceptor is insoluble and cannot enter the cell, and the microorganisms must perform extracellular electron transfer (EET) to connect their electron transport chain to the solid electron acceptor [2], [3]. Today, it is well recognized that the reduction of solid electron acceptors occurs through two different mechanisms: directly, (i) through cell-surface proteins that interact with the solid electron acceptor (ii) or through cellular appendages, including electrically conductive pilus that form a bridge between the cell and the electron acceptor; or indirectly, (iii) through the use of chelators or siderophores that solubilize the solid electron acceptor and introduce them into the cell, or (iv) using soluble shuttles, such as organic compounds with quinones moieties, that interact with the electron acceptor outside of the cell [3], [4], [5] (Fig. 1).
Fig. 1.
Mechanisms of extracellular electron transfer (EET) processes. EET may occur through direct contact using cell-surface proteins, including multiheme c-type cytochrome (process i) or electrically conductive pilus (process ii), or through indirect electron transfer where chelators or siderophores solubilize the solid electron acceptor and transfer the electrons to the bacteria (process iii), or with soluble electron shuttles that mediate electron transfer between the cell and the solid electron acceptor (process iv). In this figure, the solid electron acceptor is represented in grey and bacteria are represented in red. Chelators/siderophores and electron shuttles are represented in green and in yellow, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Microorganisms that possess EET capabilities play a fundamental role in the geochemical cycle of several elements, including carbon and iron, and are also potential targets for numerous biotechnological applications, such as for the bioremediation of metal contaminated environments, production of energy and added-value compounds, or for biosensing [6], [7], [8], [9], [10]. Some of these organisms are also termed electroactive given their ability to exchange electrons with an electrode, in the so called bioelectrochemical systems (BES) [11], [12]. Microbial fuel cells (MFC) are one of the most studied BES [13]. A typical MFC is an electrochemical cell arranged in two chambers separated by a proton exchange membrane, containing an anode and a cathode. In the anode compartment, the microorganisms oxidize organic matter and use the electrode as the terminal electron acceptor [6]. Typically, the electrons collected at the electrode are then transferred to the cathode through an external wire, and are combined with oxygen to generate water. The electron flow between the anode and cathode enables the electrical power harvesting [6], [13]. The increased interest of this type of technology has boosted the application of BES, being currently explored for the production of electrical power, treatment of wastewaters, for electrosynthesis of added-value compounds and biofuels and for water desalination [14], [15].
Electroactive organisms can be found in all three domains of life, being ubiquitous in distinct environments, including lakes, soils as well as in deep-sea hydrothermal vents [11], [12]. Recently, is has been demonstrated that these organisms are also present in the human digestive system [16], [17], in the mouse gut microbiome [18], [19], [20] and oral plaque [21], with some of them associated with infectious diseases [22], [23].
Gram-negative mesophilic bacteria are one of the most studied class of electroactive organisms, with most of the knowledge being confined to the model organisms Geobacter sulfurreducens and Shewanella oneidensis MR-1 [2], [3], [24]. Nonetheless, Gram-positive bacteria have recently attracted the scientific attention, given their capacity in producing high levels of current in MFC [25], [26], and by being associated with infectious diseases in humans [16], [17], [22], [23]. Given the importance of these organisms in BES, research has been dedicated in exploring their use as catalysts in BES and in the understanding of their EET processes [27], [28], [29]. Only by understanding how electroactive organisms perform EET and exchange electrons to electrodes, it is possible to use them in biotechnological processes and start implementing BES in real-world applications. Indeed, knowledge on the molecular mechanisms of the EET processes performed by Gram-negative bacteria allowed to modify these organisms and improve their performance in BES [30], [31], [32], [33], [34], [35], [36], showing that synthetic biology field has the potential to advance the implementation of BES in the real-world [37].
As several reviews have been published on Gram-negative organisms [2], [3], [24], this review will focus on Gram-positive electroactive bacteria, in particular on the developments on the understanding of their EET mechanisms and on the cellular components involved in these processes.
2. Gram-positive electroactive bacteria
For several years Gram-positive bacteria were considered electrochemically inactive and EET was considered incompatible with this class of organisms [38], [39]. In contrast to Gram-negative bacteria, Gram-positive bacteria lack an outer-membrane, contain a thick cell wall (20–80 nm) composed by peptidoglycan, teichoic acids, and sometimes are covered by a glycoprotein S layer [40], [41], which were thought to prevent electron transfer to solid electron acceptors.
Nonetheless, when growing in co-cultures, some species of Gram-positive bacteria were showed to transfer electrons to an electrode in a MFC, being able to perform EET [39], [42], [43]. The first pure cultures of Gram-positive bacteria able to exhibit electrochemical activity were Clostridium butyricum [43], Desulfitobacterium hafniense [44], and Lactococcus lactis [45]. Studies on these organisms have demonstrated that Gram-positive organisms can perform indirect electron transfer, using electron shuttles excreted by them [45] or by other organisms [16], [39], [46]. But later, it was also shown that Gram-positive bacteria are able to perform direct electron transfer to electrodes, being this type of EET mechanism associated with a biofilm that is formed on the surface of the electrode [28], [47], [48]. Gram-positive bacteria can also perform EET by receiving electrons from a solid electron donor, including an electrode in microbial electrosynthesis (MES) [49], [50]. These devices have recently attracted the interest of the scientific and industrial community, given the ability to couple microbial metabolism to the production of valuable chemicals and fuels, with the reduction of CO2 [9].
Several electroactive Gram-positive bacteria are present as commensals in the intestines of numerous animals, whereas others are opportunistic pathogens [19], [21], [51], [52]. Examples of these are Lactococcus monocytogenes [51], Enterococcus faecalis [52], Enterococcus avium [21], Clostridium cochlearium [18] and Faecalibacterium prausnitzii [53], [54]. Indeed, in several of them the presence of an EET system was shown to be important for the colonization of pathogenic bacteria [21], [51].
Up to date more than 10 species of Gram-positive bacteria were identified as electroactive (Table 1).
Table 1.
List of Gram-positive bacteria described to be electroactive.
| Year | Microorganism | Source | Reference |
|---|---|---|---|
| 2001 | Clostridium butyricum EG3 | MFC containing starch processing wastewater | [43] |
| 2006 | Brevibacillus sp. PTH1 | MFC containing sludge and domestic and industrial wastewater | [55] |
| 2007 | Desulfitobacterium hafniense strain DCB2 | Deutsche Sammlung von Mikroorganismen und Zelkulturen (DSMZ) | [44] |
| 2008 | Thermincola potens strain JR | MFC containing sludge from thermophilic methanogenic anaerobic digester | [26] |
| 2008 | Thermincola carboxydophila | Sediment MFC containing marine marsh sediment | [56] |
| 2009 | Bacillus subtilis | Laboratory culture | [57] |
| 2009 | Lactococcus lactis | Meiji Milk Products Co., Ltd. | [45] |
| 2009 | Thermincola ferriacetica | DSMZ | [48] |
| 2012 | Faecalibacterium prausnitzii | DSMZ and human feces | [53] |
| 2014 | Enterococcus faecalis | MFC containing sludge | [58] |
| 2014 | Clostridium pasterianum DSM 525 | DSMZ | [49] |
| 2014 | Corynebacterium glutamicum | American Type Culture Collection (ATCC) | [50] |
| 2015 | Thermoanaerobacter pseudethanolicus | ATCC | [25] |
| 2016 | Clostridium beijerinckii IB4 | Mutant formed by ion implantation | [59] |
| 2017 | Bacillus thuringiensis | Laboratory culture | [60] |
| 2018 | Listeria monocytogenes | Unité des Interactions Bactéries Cellules laboratory's Listeria strain collection | [16] |
| 2018 | Bacillus megaterium strain LLD-1 | MFC containing sludge from JiMei wastewater treatment plant | [61] |
| 2019 | Bacillus cereus DIF1 | China Center for Type Culture Collection (CCTCC) | [62] |
| 2019 | Rhodococcus ruber DIF2 | CCTCC | [62] |
| 2019 | Clostridium cochlearium | DSMZ | [18] |
| 2020 | Paenibacillus dendritiformis MA-72 | Sediment MFC containing sediment from river Strum | [28] |
When compared with Gram-negative bacteria, the EET mechanisms of Gram-positive bacteria have been less explored. This is mainly associated with the difficulties encountered during growth as a pure culture in the laboratory, the lack of their genetic information, and, in most cases, the impossibility of genetically manipulate these organisms. Up to now, the EET mechanism of Gram-positive bacteria were only explored for a few organisms, showing that Gram-positive bacteria can perform direct and indirect electron transfer to solid electron acceptors. Understanding the processes by which Gram-positive bacteria perform EET is of significant relevance to enhance electroactivity and optimize BES, either by modifying electrode materials and bioreactor set-up, or through genetic modification where electroactive organisms can be genetically engineered to become better than their counterpart wild-type strains in terms of electroactivity and/or metabolic functionalities [36]. The main mechanisms performed by Gram-positive are described below, focusing on the molecular processes by which these organisms perform EET.
3. Indirect electron transfer in Gram-positive bacteria
Indirect electron transfer to solid electron acceptors takes place in the presence of small redox active compounds, that mediate electron transfer between the microorganism and the solid electron acceptor, or vice-versa. These redox compounds can be a metabolite produced by the microorganism (e.g. flavins, phenazines, quinones) [62], [63], [64] or an artificial electron shuttle that can be added to the system (e.g. poly(MPC-co-VF), macrocyclic cobalt hexamines, osmium redox polymers) [57], [65], [66].
The electroactivity of Gram-positive bacteria was first associated with their ability in using electron shuttles produced by Gram-negative bacteria belonging to Pseudomonas sp. [46]. Indeed, metabolites produced by Pseudomonas sp. were responsible for the Gram-positive bacterium Brevibacillus sp. PTH1 to achieve EET [39]. But later on, it was demonstrated that Gram-positive bacteria can also produce redox mediators, such as humic acids, quinones and flavins [16], [44], [61], [62], [67]. For example, Lactococcus lactis cells can catalyse EET to an electrode by the excretion of soluble quinones as redox mediators [45], while spore-forming bacteria belonging to Bacillus genus have the ability to excrete flavins [41], [61], [62], [68], [69], [70]. These flavins enable Bacillus sp. to mediate electron transfer to electrodes in MFC [62], [68], and provide a boost to electricity generation in microbial consortia with Gram-negative bacteria or yeasts [60], [69], [70]. Recently, it was also shown that flavins are secreted by Rhodococcus ruber DIF2, and that flavin mononucleotide (FMN) plays an important role in the EET of this bacterium to electrodes [62]. Though humic acids were shown to support the generation of electricity of Desulfitobacterium hafniense strain DCB2, the specific electron shuttle responsible to mediate electron transfer to the electrode was not identified [44].
The cofactor nicotinamide adenine dinucleotide (NAD) was also shown to play a significant role in the viability and electroactivity of B. subtilis when submitted to long-term exposure to harsh environments [71]. In these conditions B. subtilis can use EET as an electron communication pathway, where NAD is an essential participant to maintain its viability [71].
The incorporation of Bacillus cells in an anaerobic sludge also had a significant effect in the power generation in a MFC [72]. By promoting the formation of an electroactive biofilm and by supressing methanogenesis, Bacillus cereus enhanced current production in MFC [72]. Furthermore, a genetically modified strain of B. subtilis RH33 that produces high levels of riboflavin were also shown to boost electricity generation in a microbial consortium with S. oneidensis MR-1 [70], emphasising the importance of indirect electron transfer in EET of Gram-positive bacteria.
The electroactivity of Listeria monocytogenes has been observed almost three decades ago [73], but only recently it was demonstrated that this Gram-positive bacterium can use environmental flavins to shuttle electrons to solid electron acceptors [16]. An eight-gene locus was found to be responsible for the EET capacity, with a NADH dehydrogenase channelling electrons to a membrane-localized quinone pool, and an extracellular flavoprotein that, in conjunction with flavins, mediate electron transfer to extracellular acceptors [16]. This locus is present in numerous organisms within the Firmicutes class of organisms, which suggest that the flavin-based transfer mechanisms is highly conserved in Gram-positive bacteria [16], [51]. Indeed, this gene cluster was found to be important for EET to ferric iron in E. faecalis [22], indicating that the proteins involved in this process are similar in Gram-positive bacteria. It was proposed that an atypical NADH hydrogenase (NDH-3 in E. faecalis and NDH-2 in L. monocytogenes) couple the oxidation of NADH in the cytoplasm to reduction of demethylmenaquinone (DMK) (Fig. 2A). Given that DMK is the only quinone available in the membrane of E. faecalis, it can either be oxidised by cytochrome bd under aerobic conditions and in the presence of heme, or under anaerobic environments by EET processes [22], [52]. The lipoprotein PplA present at the surface of L. monocytogenes, was shown to bind FMN, being proposed to be an important component in the EET pathway of this Gram-positive bacterium, facilitating electron transfer between FMN and extracellular electron acceptors [16], [22]. Two small proteins, EetA and EetB were also found to be important for EET [16], [22]. EetA was predicted to be a membrane protein anchored to the outer-side of the cytoplasmic membrane, while EetB was demonstrated to be an integral membrane protein that contains four transmembrane segments and a large periplasmic loop (Fig. 2A). Although it was proposed that these proteins form a complex in the membrane their role remains to be elucidated [22]. Hederstedt and co-workers proposed that when osmium complex-modified redox polymers (OsRP) are used as mediators, E. faecalis uses a different EET pathway. This pathway does not depend on the NADH dehydrogenase and on EatA, suggesting that OsRP receives electrons directly from the DMK [22].
Fig. 2.
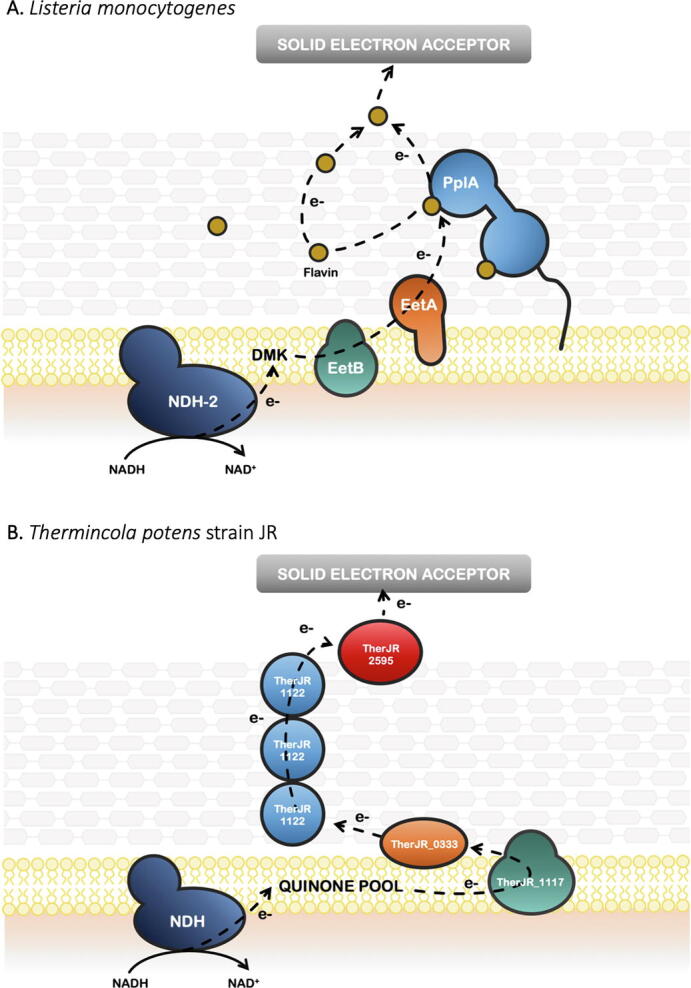
Model for the proposed EET processes pathway of L. monocytogenes (A) (adapted from [16]) and T. potens (B) (adapted from [29]).
The use of siderophores has also been observed in several electroactive Gram-positive bacteria [74], [75], [76]. In C. ferrireducens, a supplementary strategy for the utilization of siderophores in the reduction of iron oxides has been proposed [74]. This suggests that, as in Gram-negative bacteria [77], [78], [79], different EET pathways exist in Gram-positive bacteria, and that these may depend on the environment and growth conditions.
4. Direct electron transfer in Gram-positive bacteria
Most electroactive organisms use both types of EET processes to exchange electrons with an electrode, which make it difficult to identify solely the direct electron transfer process. Evidence of this type of mechanism is usually assessed by the formation of a biofilm, and by the lack of impact in current production after removing the medium or after adding redox mediators [47], [48].
Direct electron transfer to an electrode by Gram-positive bacteria was first identified in Thermincola sp. [48]. T. ferriacetica was capable of generating an electric current in an air–cathode MFC without the addition of soluble redox mediators [48]. The lack of electron shuttles, the formation of a biofilm on the electrode and the rapid current recovery by this strain after exchanging the media were shown to be consistent with direct EET. Furthermore, the Coulombic efficiency observed for this organism was higher than 95%, indicating that nearly all electrons were used for electrode reduction and not for the production of reduced organic compounds [48]. This behaviour was also observed for T. potens strain JR, suggesting that direct EET is characteristic of this genus [47]. In contrast to T. ferriacetica, T. potens strain JR could use soluble electron shuttles to transfer electrons to solid electron acceptors, although no soluble redox mediators have been identified in the MFC spent medium [47].
Genomic analysis of T. potens strain JR [80] have led to the proposal that multiheme c-type cytochromes (MHC) could be involved in EET in Gram-positive bacteria, as observed for Gram-negative electroactive bacteria. Indeed, trypsin-shaving LC-MS/MS experiments and surface-enhanced Raman spectroscopy allowed the identification of several MHC that could be involved in EET in this organism [29]. Although the identification of c-type cytochromes as key proteins for the reduction of insoluble electron acceptors has been previously observed for Gram-positive bacteria [74], this was the first time that an EET pathway was proposed for the transfer of electrons outside of the cell [29]. The proposed EET pathway is composed by four MHC, that were shown to be conserved among Thermincola sp. [81], suggesting that a general strategy for electron transfer may occur within this genus (Fig. 2B). As observed for several Gram-negative bacteria, the EET pathway in Thermincola sp. is composed by an inner-membrane MHC that receive electrons from the menaquinone pool, that then transfers the electrons to a periplasmic cytochrome. The transfer of electrons outside of the cell depends on a hexaheme cytochrome that was proposed to be embedded in the peptidoglycan at the cell wall and on the nine-heme cytochrome OcwA present at the cell-surface of these bacteria, responsible to reduce solid electron acceptors, electron shuttles and oxyanions [27], [29] (Fig. 2B). This arrangement is different from what is typical observed in electroactive Gram-negative bacteria, where porin-cytochrome complexes composed by a β-barrel porin and one or two MHC are responsible for electron transfer across the outer membrane [82].
Direct electron transfer was also observed for other Gram-positive bacteria including Chlostridium pasteurianum [49], P. dendritiformis MA-72 [28] and Carboxydothermus ferrireducens [74], although the molecular processes and the proteins involved are still unknown.
In Gram-negative bacteria, direct electron transfer was also observed to occur through electrically conductive pili [83] or outer membrane extensions [84]. These structures allow the microorganism to make an electrical connection between periplasmic carriers and the insoluble electron acceptors, that can be located as far as 15 μm away from the cell [85].
Filaments of the type IV pili were also reported for some Gram-positive bacteria [86]. Although most of the functions of type IV pili in Gram-positive bacteria have been associated with mobility and adherence to host cells [86], [87], pili-like appendages have been observed in C. ferrireducens grown on iron oxides [74], in Paenibacillus dendritiformis when growing on an electrode [28] and in E. faecalis pili revealed to contribute for EET [88]. It was demonstrated that E. faecalis biofilm matrix harboring iron sinks promotes EET and augments biofilm growth, increasing current generation in MFC [89]. The biofilm associated pilus (Ebp) was demonstrated to be a key player in this process by sequestering iron in close proximity to the cells, either as surface attached pili or within the biofilm matrix, enabling E. faecalis to use it as terminal electron acceptor [88].
Extracellular polymeric substances (EPS) surrounding electroactive organisms help cells to attach to solid minerals or surfaces, assisting biofilm formation and protecting them from unfavourable environments [90]. EPS were also demonstrated to have electroactive properties, due to the presence of nucleic acids, humic substances, flavins and even proteins that are redox-active or semiconductive [91], [92]. EPS from Bacillus sp. WS-XY1 was shown to be electroactive and of significant importance for EET in this bacterium [92].
5. Summary and outlook
EET between microbes and solid electron acceptors/donors, such as iron minerals or electrodes, is a widespread process that affect biogeochemical cycles, microbial ecology and that can be explored for the generation of electricity and chemicals in BES. For this reason, the elucidation of the mechanisms by which microorganisms perform electron transfer between intracellular and extracellular environments has been subject to widespread attention. Although most of electroactive organisms discovered to date are Gram-negative bacteria [11], [12], Gram-positive bacteria were also demonstrated to perform EET, being able to transfer electrons to solid compounds directly and indirectly. Like Gram-negative bacteria, some Gram-positive bacteria rely on MHC to transfer electrons to solid electron acceptors outside of the cell. Up to date the only cell-surface cytochrome from Gram-positive bacteria that have been characterized in detailed was the OcwA from T. potens, showing that it can reduce solid electron acceptors, soluble electron shuttles and oxyanions [27]. Nevertheless, the way this protein is anchored and arranged at the cell surface and how electrons are transferred across the cell wall are still to be determined. In the fermentative Gram-positive bacteria L. monocytogenes and E. faecalis flavin-interacting proteins represent the extracellular components of their EET machinery, facilitating electron transfer, via FMN, to extracellular electron acceptors [16]. Although a cluster of proteins has been proposed to be widespread within the Firmicutes phylum, and to be important for EET, the mechanisms by which these organisms exchange electrons with solid electron acceptors remains to be elucidated.
The recent studies on Gram-positive bacteria revealed that, as in Gram-negative bacteria, a diverse way to perform EET exists in this class of microorganisms, including direct and indirect electron transfer. This demonstrates that rather than being a specialized process, EET is a fundamental process of microbial metabolism that occurs in numerous organisms and across diverse environments. The characterization of these processes is crucial not only to unravel the involvement of microbial metabolism in the biogeochemical cycle of elements, but also to enhance the development of novel sustainable biotechnological processes, where these organisms can be employed. Indeed, although Gram-positive bacteria can be more sensitive to several compounds (e.g. antibiotics, cleaning agents) given the high permeability of the peptidoglycan layer, their thick cell wall in addition to their ability in forming spores, enables their use in more extreme conditions [18], [21], [26].
Although knowledge of the EET performed by Gram-positive bacteria have increased in the last years, much more studies are required to fully understand their molecular processes in exchanging electrons with solid electron acceptors or donors. As future work, studies on the proteins involved in these processes, as well as the understanding of their electron transfer mechanisms should be a priority. Only with this knowledge it will be possible to improve these organisms and boost their performance beyond their natural metabolic capabilities, allowing to advance the implementation of BES and expand their biotechnological application.
CRediT authorship contribution statement
Catarina M. Paquete: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by national funds through FCT– Fundação para a Ciência e a Tecnologia, I.P., Project UIDB/04612/2020, UIDP/04612/2020 and PTDC/BIA-BQM/30176/2017, and by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 810856.
References
- 1.Richardson DJ. Bacterial respiration: A flexible process for a changing environment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 2.Shi L., Dong H., Reguera G., Beyenal H., Lu A., Liu J. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 3.White G.F., Edwards M.J., Gomez-Perez L., Richardson D.J., Butt J.N., Clarke T.A. Mechanisms of Bacterial Extracellular Electron Exchange. Adv Microb Physiol. 2016;68:87–138. doi: 10.1016/bs.ampbs.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Gralnick J.A., Newman D.K. Extracellular respiration. Mol Microbiol. 2007;65:1–11. doi: 10.1111/j.1365-2958.2007.05778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa N.L., Clarke T.A., Philipp L.-A., Gescher J., Louro R.O., Paquete C.M. Electron transfer process in microbial electrochemical technologies: the role of cell-surface exposed conductive proteins. Bioresour Technol. 2018;255:308–317. doi: 10.1016/j.biortech.2018.01.133. [DOI] [PubMed] [Google Scholar]
- 6.Logan B.E., Hamelers B., Rozendal R., Schröder U., Keller J., Freguia S. Microbial fuel cells: Methodology and technology. Environ Sci Technol. 2006;40:5181–5192. doi: 10.1021/es0605016. [DOI] [PubMed] [Google Scholar]
- 7.Harnisch F., Schröder U. From MFC to MXC: chemical and biological cathodes and their potential for microbial bioelectrochemical systems. Chem Soc Rev. 2010;39:4433–4448. doi: 10.1039/c003068f. [DOI] [PubMed] [Google Scholar]
- 8.Pant D., Singh A., Van Bogaert G., Irving Olsen S., Singh Nigam P., Diels L. Bioelectrochemical systems (BES) for sustainable energy production and product recovery from organic wastes and industrial wastewaters. RSC Adv. 2012;2:1248–1263. doi: 10.1039/C1RA00839K. [DOI] [Google Scholar]
- 9.Prévoteau A., Carvajal-Arroyo J.M., Ganigué R., Rabaey K. Microbial electrosynthesis from CO2: forever a promise? Curr Opin Biotechnol. 2020;62:48–57. doi: 10.1016/j.copbio.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Prévoteau A., Rabaey K. Electroactive Biofilms for Sensing: Reflections and Perspectives. ACS Sensors. 2017;2:1072–1085. doi: 10.1021/acssensors.7b00418. [DOI] [PubMed] [Google Scholar]
- 11.Logan B.E., Rossi R., Ragab A., Saikaly P.E. Electroactive microorganisms in bioelectrochemical systems. Nat Rev Microbiol. 2019;17:307–319. doi: 10.1038/s41579-019-0173-x. [DOI] [PubMed] [Google Scholar]
- 12.Koch C., Harnisch F. Is there a Specific Ecological Niche for Electroactive Microorganisms? ChemElectroChem. 2016;3:1282–1295. doi: 10.1002/celc.201600079. [DOI] [Google Scholar]
- 13.Slate A.J., Whitehead K.A., Brownson D.A.C., Banks C.E. Microbial fuel cells: An overview of current technology. Renew Sustain Energy Rev. 2019;101:60–81. doi: 10.1016/j.rser.2018.09.044. [DOI] [Google Scholar]
- 14.Hernandez C.A., Osma J.F. Microbial electrochemical systems: Deriving future trends from historical perspectives and characterization strategies. Front Environ Sci. 2020;8:1–19. doi: 10.3389/fenvs.2020.00044. [DOI] [Google Scholar]
- 15.Harnisch F., Urban C. Electrobiorefineries : Unlocking the Synergy of Electrochemical and Microbial Conversions Angewandte. 2018:10016–10023. doi: 10.1002/anie.201711727. [DOI] [PubMed] [Google Scholar]
- 16.Light S.H., Su L., Rivera-Lugo R., Cornejo J.A., Louie A., Iavarone A.T. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature. 2018;562:140–144. doi: 10.1038/s41586-018-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinken A., Khan M.T., Paglia G., Rodionov D.A., Harmsen H.J.M., Thiele I. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol. 2014;196:3289–3302. doi: 10.1128/JB.01780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab L., Rago L., Koch C., Harnisch F. Identification of Clostridium cochlearium as an electroactive microorganism from the mouse gut microbiome. Bioelectrochemistry. 2019;130:107334. doi: 10.1016/j.bioelechem.2019.107334. [DOI] [PubMed] [Google Scholar]
- 19.Wang W., Du Y., Yang S., Du X., Li M., Lin B. Bacterial Extracellular Electron Transfer Occurs in Mammalian Gut. Anal Chem. 2019;91:12138–12141. doi: 10.1021/acs.analchem.9b03176. [DOI] [PubMed] [Google Scholar]
- 20.Rago L., Popp D., Heiker J.T., Harnisch F. Electroactive microorganisms in mouse feces. Electrochim Acta. 2021;365 doi: 10.1016/j.electacta.2020.137326. [DOI] [Google Scholar]
- 21.Naradasu D., Miran W., Sakamoto M., Okamoto A. Isolation and characterization of human gut bacteria capable of extracellular electron transport by electrochemical techniques. Front Microbiol. 2019;10:1–9. doi: 10.3389/fmicb.2018.03267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hederstedt L., Gorton L., Pankratova G. Two Routes for Extracellular Electron Transfer in Enterococcus faecalis. J Bacteriol. 2020;202:1–9. doi: 10.1128/JB.00725-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naradasu D., Guionet A., Okinaga T., Nishihara T., Okamoto A. Electrochemical Characterization of Current-Producing Human Oral Pathogens by Whole-Cell Electrochemistry. ChemElectroChem. 2020;7:2012–2019. doi: 10.1002/celc.202000117. [DOI] [Google Scholar]
- 24.Kumar A., Hsu L.H.H., Kavanagh P., Barrière F., Lens P.N.L., Lapinsonnière L. The ins and outs of microorganism-electrode electron transfer reactions. Nat Rev Chem. 2017;1:1–13. doi: 10.1038/s41570-017-0024. [DOI] [Google Scholar]
- 25.Lusk B.G., Khan Q.F., Parameswaran P., Hameed A., Ali N., Rittmann B.E. Characterization of Electrical Current-Generation Capabilities from Thermophilic Bacterium Thermoanaerobacter pseudethanolicus Using Xylose, Glucose, Cellobiose, or Acetate with Fixed Anode Potentials. Environ Sci Technol. 2015;49:14725–14731. doi: 10.1021/acs.est.5b04036. [DOI] [PubMed] [Google Scholar]
- 26.Wrighton K.C., Agbo P., Warnecke F., Weber K.A., Brodie E.L., DeSantis T.Z. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2:1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- 27.Costa N.L., Hermann B., Fourmond V., Faustino M.M., Teixeira M., Einsle O. How thermophilic Gram-positive organisms perform extracellular electron transfer: Characterization of the cell surface terminal reductase OcwA. mBio. 2019;10:1–13. doi: 10.1128/mBio.01210-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubenova Y., Hubenova E., Mitov M. Electroactivity of the Gram-positive bacterium Paenibacillus dendritiformis MA-72. Bioelectrochemistry. 2020;136 doi: 10.1016/j.bioelechem.2020.107632. [DOI] [PubMed] [Google Scholar]
- 29.Carlson H.K., Iavarone A.T., Gorur A., Yeo B.S., Tran R., Melnyk R.A. Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc Natl Acad Sci. 2012;109:1702–1707. doi: 10.1073/pnas.1112905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T., Yu Y.Y., Deng X.P., Ng C.K.K., Cao B., Wang J.Y. Enhanced Shewanella Biofilm Promotes Bioelectricity Generation. Biotechnol Bioeng. 2015;9999:1–9. doi: 10.1002/bit.25624. [DOI] [PubMed] [Google Scholar]
- 31.Silva A.V., Edel M., Gescher J., Paquete C.M. Exploring the Effects of bolA in Biofilm Formation and Current Generation by Shewanella oneidensis MR-1. Front Microbiol. 2020;11. doi: 10.3389/fmicb.2020.00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F., Li Y.Y., Cao Y.Y., Wang L., Liu C.C., Shi L. Modular engineering to increase intracellular NAD(H/+) promotes rate of extracellular electron transfer of Shewanella oneidensis. Nat Commun. 2018;9:3637. doi: 10.1038/s41467-018-05995-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min D., Cheng L., Zhang F., Huang X.-N., Li D., Liu D. Enhancing Extracellular Electron Transfer of Shewanella oneidensis MR-1 through Coupling Improved Flavin Synthesis and Metal-Reducing Conduit for Pollutant Degradation. Environ Sci Technol. 2017;51:5082–5089. doi: 10.1021/acs.est.6b04640. [DOI] [PubMed] [Google Scholar]
- 34.Li F., Wang L., Liu C., Wu D., Song H. Engineering exoelectrogens by synthetic biology strategies. Curr Opin Electrochem. 2018;10:37–45. doi: 10.1016/j.coelec.2018.03.030. [DOI] [Google Scholar]
- 35.TerAvest M., Tefft N.M. Reversing an extracellular electron transfer pathway for electrode-driven acetoin reduction. ACS Synth Biol. 2019;8(7):1590–1600. doi: 10.1021/acssynbio.8b00498. [DOI] [PubMed] [Google Scholar]
- 36.Chiranjeevi P., Patil S.A. Strategies for improving the electroactivity and specific metabolic functionality of microorganisms for various microbial electrochemical technologies. Biotechnol Adv. 2020;39 doi: 10.1016/j.biotechadv.2019.107468. [DOI] [PubMed] [Google Scholar]
- 37.Glaven S.M. Bioelectrochemical systems and synthetic biology: more power, more products. Microb Biotechnol. 2019;12:819–823. doi: 10.1111/1751-7915.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doyle L.E., Marsili E. Weak electricigens: A new avenue for bioelectrochemical research. Bioresour Technol. 2018;258:354–364. doi: 10.1016/j.biortech.2018.02.073. [DOI] [PubMed] [Google Scholar]
- 39.Pham T.H., Boon N., Aelterman P., Clauwaert P., De Schamphelaire L., Vanhaecke L. Metabolites produced by Pseudomonas sp. enable a Gram-positive bacterium to achieve extracellular electron transfer. Appl Microbiol Biotechnol. 2008;77:1119–29. doi: 10.1007/s00253-007-1248-6. [DOI] [PubMed] [Google Scholar]
- 40.Navarre W.W., Schneewind O., Wiley W., Schneewind O. Surface Proteins of Gram-Positive Bacteria and Mechanisms of Their Targeting to the Cell Wall Envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/MMBR.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrlich H.L. Are Gram-positive bacteria capable of electron transfer across their cell wall without an externally available electron shuttle? Geobiology. 2008;6:220–224. doi: 10.1111/j.1472-4669.2007.00135.x. [DOI] [PubMed] [Google Scholar]
- 42.Rabaey K., Verstraete W. Microbial fuel cells: Novel biotechnology for energy generation. Trends Biotechnol. 2005;23:291–298. doi: 10.1016/j.tibtech.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 43.Park H.S., Kim B.H., Kim H.S., Kim H.J., Kim G.T., Kim M. A novel electrochemically active and Fe(III)-reducing bacterium phylogenetically related to Clostridium butyricum isolated from a microbial fuel cell. Anaerobe. 2001;7:297–306. doi: 10.1006/anae.2001.0399. [DOI] [Google Scholar]
- 44.Milliken C.E., May H.D. Sustained generation of electricity by the spore-forming, Gram-positive, Desulfitobacterium hafniense strain DCB2. Appl Microbiol Biotechnol. 2007;73:1180–1189. doi: 10.1007/s00253-006-0564-6. [DOI] [PubMed] [Google Scholar]
- 45.Freguia S., Masuda M., Tsujimura S., Kano K. Lactococcus lactis catalyses electricity generation at microbial fuel cell anodes via excretion of a soluble quinone. Bioelectrochemistry. 2009;76:14–18. doi: 10.1016/j.bioelechem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Rabaey K., Boon N., Siciliano S.D., Verhaege M., Verstraete W. Biofuel Cells Select for Microbial Consortia That Self-Mediate Electron Transfer. Appl Environ Microbiol. 2004;70:5373–5382. doi: 10.1128/AEM.70.9.5373-5382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrighton K.C., Thrash J.C., Melnyk R.A., Bigi J.P., Byrne-Bailey K.G., Remis J.P. Evidence for direct electron transfer by a gram-positive bacterium isolated from a microbial fuel cell. Appl Environ Microbiol. 2011;77:7633–7639. doi: 10.1128/AEM.05365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshall C.W., May H.D. Electrochemical evidence of direct electrode reduction by a thermophilic Gram-positive bacterium Thermincola ferriacetica. Energy Environ Sci. 2009;2:699–705. doi: 10.1039/b823237g. [DOI] [Google Scholar]
- 49.Choi O., Kim T., Woo H.M., Um Y. Electricity-driven metabolic shift through direct electron uptake by electroactive heterotroph Clostridium pasteurianum. Sci Rep. 2014;4. doi: 10.1038/srep06961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki K., Tsuge Y., Sasaki D., Kondo A. Increase in lactate yield by growing Corynebacterium glutamicum in a bioelectrochemical reactor. J Biosci Bioeng. 2014;117:598–601. doi: 10.1016/j.jbiosc.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 51.Light S.H., Méheust R., Ferrell J.L., Cho J., Deng D., Agostoni M. Extracellular electron transfer powers flavinylated extracellular reductases in Gram-positive bacteria. Proc Natl Acad Sci U S A. 2019;116:26892–26899. doi: 10.1073/pnas.1915678116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pankratova G., Leech D., Gorton L., Hederstedt L. Extracellular Electron Transfer by the Gram-Positive Bacterium Enterococcus faecalis. Biochemistry. 2018;57:4597–4603. doi: 10.1021/acs.biochem.8b00600. [DOI] [PubMed] [Google Scholar]
- 53.Khan M.T., Duncan S.H., Stams A.J.M., Van Dijl J.M., Flint H.J., Harmsen H.J.M. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prévoteau A., Geirnaert A., Arends J.B.A., Lannebère S., Van De Wiele T., Rabaey K. Hydrodynamic chronoamperometry for probing kinetics of anaerobic microbial metabolism - Case study of Faecalibacterium prausnitzii. Sci Rep. 2015;5:1–13. doi: 10.1038/srep11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aelterman P., Rabaey K., The Pham H., Boon N., Verstraete W., Pham H.T. Continuous electricity generation at high voltages and currents using stacked microbial fuel cells. Commun Agric Appl Biol Sci. 2006;71:63–66. doi: 10.1021/es0525511. [DOI] [PubMed] [Google Scholar]
- 56.Mathis B.J., Marshall C.W., Milliken C.E., Makkar R.S., Creager S.E., May H.D. Electricity generation by thermophilic microorganisms from marine sediment. Appl Microbiol Biotechnol. 2008;78:147–155. doi: 10.1007/s00253-007-1266-4. [DOI] [PubMed] [Google Scholar]
- 57.Coman V., Gustavsson T., Finkelsteinas A., Von Wachenfeldt C., Hägerhäll C., Gorton L. Electrical wiring of live, metabolically enhanced Bacillus subtilis cells with flexible osmium-redox polymers. J Am Chem Soc. 2009;131:16171–16176. doi: 10.1021/ja905442a. [DOI] [PubMed] [Google Scholar]
- 58.Zhang F., Liu J., Ivanov I., Hatzell M.C., Yang W., Ahn Y. Reference and counter electrode positions affect electrochemical characterization of bioanodes in different bioelectrochemical systems. Biotechnol Bioeng. 2014;111:1931–1939. doi: 10.1002/bit.25253. [DOI] [PubMed] [Google Scholar]
- 59.He A.-Y., Yin C.-Y., Xu H., Kong X.-P., Xue J.-W., Zhu J. Enhanced butanol production in a microbial electrolysis cell by Clostridium beijerinckii IB4. Bioprocess Biosyst Eng. 2016;39:245–254. doi: 10.1007/s00449-015-1508-2. [DOI] [PubMed] [Google Scholar]
- 60.Jothinathan D., Wilson R.T. Comparative analysis of power production of pure, coculture, and mixed culture in a microbial fuel cell. Energy Sources, Part A Recover Util Environ Eff. 2017;39:520–527. doi: 10.1080/15567036.2016.1233306. [DOI] [Google Scholar]
- 61.You L.X., Liu L.D., Xiao Y., Dai Y.F., Chen B.L., Jiang Y.X. Flavins mediate extracellular electron transfer in Gram-positive Bacillus megaterium strain LLD-1. Bioelectrochemistry. 2018;119:196–202. doi: 10.1016/j.bioelechem.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Tian T., Fan X., Feng M., Su L., Zhang W., Chi H. Flavin-mediated extracellular electron transfer in Gram-positive bacteria: Bacillus cereus DIF1 and Rhodococcus ruber DIF2. RSC Adv. 2019;9:40903–40909. doi: 10.1039/c9ra08045g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marsili E., Baron D.B., Shikhare I.D., Coursolle D., Gralnick J.A., Bond D.R. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang B., Gao S., Xu Z., He H., Pan X. The Functional Mechanisms and Application of Electron Shuttles in Extracellular Electron Transfer. Curr Microbiol. 2018;75:99–106. doi: 10.1007/s00284-017-1386-8. [DOI] [PubMed] [Google Scholar]
- 65.Nishio K., Nakamura R., Lin X., Konno T., Ishihara K., Nakanishi S. Extracellular electron transfer across bacterial cell membranes via a cytocompatible redox-active polymer. ChemPhysChem. 2013;14:2159–2163. doi: 10.1002/cphc.201300117. [DOI] [PubMed] [Google Scholar]
- 66.Kracke F., Virdis B., Bernhardt P.V., Rabaey K., Krömer J.O. Redox dependent metabolic shift in Clostridium autoethanogenum by extracellular electron supply. Biotechnol Biofuels. 2016;9:1–12. doi: 10.1186/s13068-016-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pankratova G., Hederstedt L., Gorton L. Extracellular electron transfer features of Gram-positive bacteria. Anal Chim Acta. 2019;1076:32–47. doi: 10.1016/j.aca.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Nimje V.R., Chen C.Y., Chen C.C., Jean J.S., Reddy A.S., Fan C.W. Stable and high energy generation by a strain of Bacillus subtilis in a microbial fuel cell. J Power Sources. 2009;190:258–263. doi: 10.1016/j.jpowsour.2009.01.019. [DOI] [Google Scholar]
- 69.Wu S., Xiao Y., Wang L., Zheng Y., Chang K., Zheng Z. Extracellular electron transfer mediated by flavins in Gram-positive Bacillus sp. WS-XY1 and yeast Pichia stipitis. Electrochim Acta. 2014;146:564–7. doi: 10.1016/j.electacta.2014.09.096. [DOI] [Google Scholar]
- 70.Liu T., Yu Y.-Y., Chen T., Chen W.N. A synthetic microbial consortium of Shewanella and Bacillus for enhanced generation of bioelectricity. Biotechnol Bioeng. 2017;114:526–532. doi: 10.1002/bit.26094. [DOI] [PubMed] [Google Scholar]
- 71.Chen L., Cao C., Wang S., Varcoe J.R., Slade R.C.T., Avignone-Rossa C. Electron Communication of Bacillus subtilis in Harsh Environments. IScience. 2019;12:260–269. doi: 10.1016/j.isci.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Islam M.A., Ethiraj B., Cheng C.K., Yousuf A., Khan M.M.R. Electrogenic and Antimethanogenic Properties of Bacillus cereus for Enhanced Power Generation in Anaerobic Sludge-Driven Microbial Fuel Cells. Energy Fuels. 2017;31:6132–6139. doi: 10.1021/acs.energyfuels.7b00434. [DOI] [Google Scholar]
- 73.Deneer H.G., Boychuk I. Reduction of ferric iron by Listeria monocytogenes and other species of Listeria. Can J Microbiol. 1993;39:480–485. doi: 10.1139/m93-068. [DOI] [PubMed] [Google Scholar]
- 74.Gavrilov S.N., Lloyd J.R., Kostrikina N.A., Slobodkin A.I. Fe(III) Oxide Reduction by a Gram-positive Thermophile: Physiological Mechanisms for Dissimilatory Reduction of Poorly Crystalline Fe(III) Oxide by a Thermophilic Gram-positive Bacterium Carboxydothermus ferrireducens. Geomicrobiol J. 2012;29:804–819. doi: 10.1080/01490451.2011.635755. [DOI] [Google Scholar]
- 75.Fukushima T., Allred B.E., Raymond K.N. Direct evidence of iron uptake by the Gram-positive siderophore-shuttle mechanism without iron reduction. ACS Chem Biol. 2014;9:2092–2100. doi: 10.1021/cb500319n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukushima T., Allred B.E., Sia A.K., Nichiporuk R., Andersen U.N., Raymond K.N. Gram-positive siderophore-shuttle with iron-exchange from Fe-siderophore to apo-siderophore by Bacillus cereus YxeB. Proc Natl Acad Sci. 2013;110:13821–13826. doi: 10.1073/pnas.1304235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fonseca B.M.M., Paquete C.M.M., Neto S.E.E., Pacheco I., Soares C.M.M., Louro R.O.O. Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem J. 2013;449:101–108. doi: 10.1042/BJ20121467. [DOI] [PubMed] [Google Scholar]
- 78.Coursolle D., Gralnick J.A. Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol Microbiol. 2010:1–14. doi: 10.1111/j.1365-2958.2010.07266.x. [DOI] [PubMed] [Google Scholar]
- 79.Otero F.J., Chan C.H., Bond D.R. Identification of Different Putative Outer Membrane Electron Conduits Necessary for Fe(III) Citrate, Fe(III) Oxide, Mn(IV) Oxide, or Electrode Reduction by Geobacter sulfurreducens. J Bacteriol. 2018;200:1–20. doi: 10.1128/JB.00347-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Byrne-Bailey K.G., Wrighton K.C., Melnyk R.A., Agbo P., Hazen T.C., Coates J.D. Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J Bacteriol. 2010;192:4078–4079. doi: 10.1128/JB.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lusk B.G. Thermophiles; or, the Modern Prometheus: The Importance of Extreme Microorganisms for Understanding and Applying Extracellular Electron Transfer. Front Microbiol. 2019;10:1–10. doi: 10.3389/fmicb.2019.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edwards M.J., Richardson D.J., Paquete C.M., Clarke T.A. Role of multiheme cytochromes involved in extracellular anaerobic respiration in bacteria. Protein Sci. 2020;29:830–842. doi: 10.1002/pro.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reguera G., McCarthy K.D., Mehta T., Nicoll J.S., Tuominen M.T., Lovley D.R. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 84.Pirbadian S., Barchinger S.E., Leung K.M., Byun H.S., Jangir Y., Bouhenni R.A. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci. 2014;111:12883–12888. doi: 10.1073/pnas.1410551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reguera G., Kashefi K. The electrifying physiology of Geobacter bacteria, 30 years on. Adv Microb Physiol. 2019;74:1–96. doi: 10.1016/bs.ampbs.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 86.Melville S., Craig L. Type IV Pili in Gram-Positive Bacteria. Microbiol Mol Biol Rev. 2013;77:323–341. doi: 10.1128/mmbr.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mandlik A., Swierczynski A., Das A., Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lam L.N., Wong J.J., Matysik A., Paxman J., Chong K.K.L., Low P.M. Sortase-assembled pili promote extracellular electron transfer and iron acquisition in Enterococcus faecalis biofilm. BioRxiv. 2019:601666. doi: 10.1101/601666. [DOI] [Google Scholar]
- 89.Keogh D., Lam L.N., Doyle L.E., Matysik A., Pavagadhi S., Umashankar S. Extracellular electron transfer powers Enterococcus faecalis biofilm metabolism. mBio. 2018;9:1–16. doi: 10.1128/mBio.00626-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 91.Li S.W., Sheng G.P., Cheng Y.Y., Yu H.Q. Redox properties of extracellular polymeric substances (EPS) from electroactive bacteria. Sci Rep. 2016;6:1–7. doi: 10.1038/srep39098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao Y., Zhang E., Zhang J., Dai Y., Yang Z., Christensen H.E.M.M. Extracellular polymeric substances are transient media for microbial extracellular electron transfer. Sci Adv. 2017;3:1–9. doi: 10.1126/sciadv.1700623. [DOI] [PMC free article] [PubMed] [Google Scholar]