Graphical abstract
Keywords: Gut microbiome, Metaproteomics, Functional network, Resistant starch
Abstract
Resistant starches (RS) are dietary compounds processed by the gut microbiota into metabolites, such as butyrate, that are beneficial to the host. The production of butyrate by the microbiome appears to be affected by the plant source and type of RS as well as the individual’s microbiota. In this study, we used in vitro culture and metaproteomic methods to explore individual microbiome's functional responses to RS2 (enzymatically-resistant starch), RS3 (retrograded starch) and RS4 (chemically-modified starch). Results showed that RS2 and RS3 significantly altered the protein expressions in the individual gut microbiomes, while RS4 did not result in significant protein changes. Significantly elevated protein groups were enriched in carbohydrate metabolism and transport functions of families Eubacteriaceae, Lachnospiraceae and Ruminococcaceae. In addition, Bifidobacteriaceae was significantly increased in response to RS3. We also observed taxon-specific enrichments of starch metabolism and pentose phosphate pathways corresponding to this family. Functions related to starch utilization, ABC transporters and pyruvate metabolism pathways were consistently increased in the individual microbiomes in response to RS2 and RS3. Given that these taxon-specific responses depended on the type of carbohydrate sources, we constructed a functional ecological network to gain a system-level insight of functional organization. Our results suggest that while some microbes tend to be functionally independent, there are subsets of microbes that are functionally co-regulated by environmental changes, potentially by alterations of trophic interactions.
1. Introduction
Prebiotics are functional compounds that modulate the gut microbiome, promoting the growth and activity of bacteria that are beneficial to human health [1]. This can enhance immune system function and protect from diseases [2]. Resistant starches (RS) are prebiotic polysaccharides that resist digestion by pancreatic amylase and are therefore not hydrolyzed to D-glucose in the small intestine. Current perspectives differ on the definitions and number of RS structural types [3], [4], [5], [6], [7]. Resistant starch occurs naturally in three different forms (RS1/RS2/RS3), and synthetically as fourth (RS4) [4], [5], [6], [7] and fifth (RS5) form [3], [5]. Among which, RS2-4 are more frequently studied for their health impacts, and there are fewer studies on RS1 and RS5 because they contain dietary fibre (RS1) or lipid component (RS5) that could be significant confounding factors on their potential effects [8]. Because they resist digestion in the small intestine, RS can reach the colon, where they can be fermented by the gut microbiome [9], [10]. RS have been linked to a number of host-beneficial effects when included in human diets [11]. Studies have shown that the effects of RS on the microbiome depend on the source and type of RS, and on individual variations in the human gut microbiomes. For example, an increase of the Bacteroidetes phylum relative to Firmicutes phylum was observed following an RS4-enriched diet and the opposite following an RS2-enriched diet in human individuals with potential metabolic syndrome [12], whereas in another study RS2 and RS4 in the diet resulted in vastly different effects on the composition of the gut microbiota in healthy human subjects [13]. Nevertheless, both RS types induced physiological changes that were highly similar [13]. In bacterial monoculture, RS3 from two different plants had different effects on short-chain fatty acid (SCFA) production levels in Clostridium butyricum and Eubacterium rectale [14]. Moreover, a study showed that fecal butyrate levels varied widely among 46 individuals given an RS dietary supplement [15].
There have been studies on microbiome responses to RS using metagenomics, metaproteomics and metabolomics approaches [10], [16], [17], [18], [19]. However, most studies remain at the stage of revealing taxonomic and functional responses. The ecological response of the gut microbiome under such nutrient fluctuations remains largely unexplored. The gut microbiome is a complicated ecosystem where interspecific interactions such as cross-feeding, competition, and mutualism occur. Although there have been some studies on building ecological network of gut microbiome species [20], [21], they are based on bacterial abundance information. The system-level insight of functional organization between microbiome members is a missing link in current studies. Metaproteomics technique based on liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) allows us to quantify functional protein expressions in different microbiome members, and thus can be an ideal tool to study the functional ecology of the gut microbiome [22].
We previously developed an in vitro culturing method to facilitate individualized evaluation of stimulus effects on the gut microbiome [23]. In this study, we used this culturing method to evaluate the effect of three commonly studied types of RS, namely RS2 (enzymatically-resistant starch), RS3 (retrograded starch) and RS4 (chemically-modified starch), on the functional profiles of in vitro gut microbiomes from seven healthy individuals. Through metaproteomics, protein-level variations between individual microbiomes responding to RS types were elucidated in this study. Based on these metaproteomics responses to the nutrient changes, we constructed an ecological network between microbiome members by computing their functional correlations. Through a network analysis approach [24], we revealed a mixed ecological network structure of the microbiome functional organization, which contains both modular sub-networks of individual genus and associated sub-networks between different microbial genera.
2. Results
2.1. Overview of RS effect on expressed metaproteome
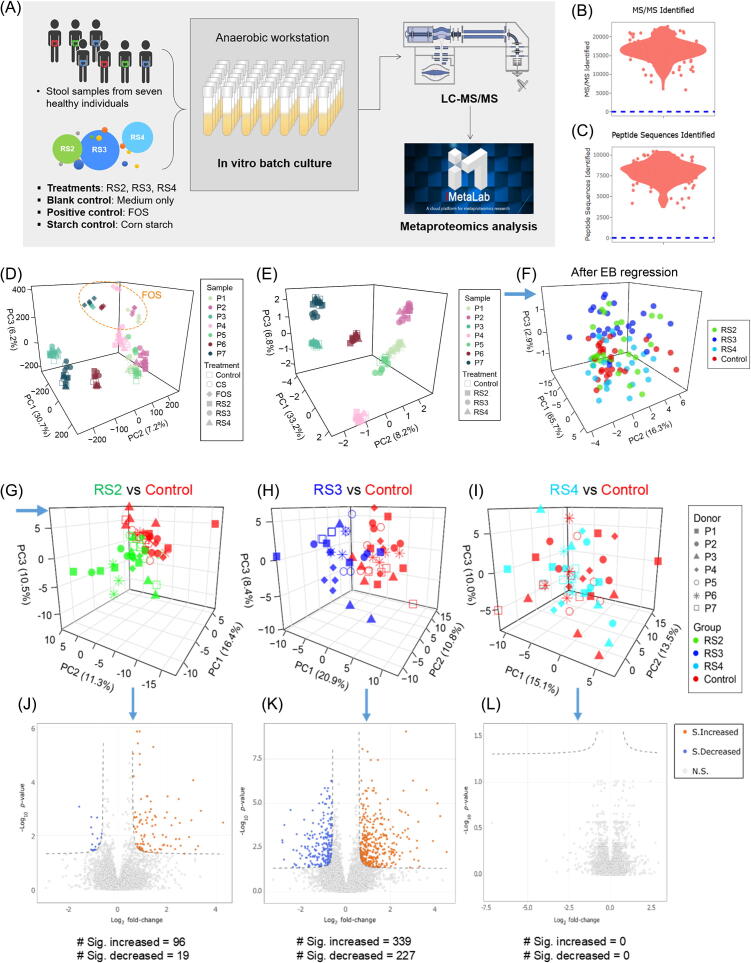
We first explored whether functional changes in individual microbiomes induced by RS are dependent on the type of starches and on individual microbiomes. Briefly, stool microbiomes from seven healthy individuals were cultured for 24 hr in the presence of RS or controls using our previously described method [23] (Fig. 1A). The microbiomes were treated with RS2(Hi Maize 260), RS3(Novelose 330), RS4(Fibersym RW), a non-resistant starch control (corn starch (CS)), a positive control (fructo-oligosaccharide (FOS) known to consistently and markedly shift the in vitro gut microbiome [25], [26], [27]) or a blank control containing no compounds. The samples were then subjected to metaproteomic analysis as previously described [23]. Altogether, the bioinformatic analysis of 157 LC-MS/MS raw files by MetaLab [28] identified 5,119,332 MS/MS spectra, 80,297 peptides and 21,240 protein groups using a false discovery rate (FDR) threshold of < 1% (Fig. 1B and C). There were 20,378 protein groups with at least one type of functional annotation; 93% had clusters of orthologous groups (COG) annotation, and 70% had Kyoto encyclopedia of genes and genomes (KEGG) annotation. 65,763 peptides were assigned to taxonomic lowest common ancestors (LCA) by MetaLab based on the pep2taxa database [28].
Fig. 1.
Experimental design and data overview. (A) Fecal microbiomes from seven individuals were cultured in medium (as the blank control) or culture medium containing one of the following materials: RS2 (Hi Maize 260), RS3(Novelose 330), RS4(Fibersym RW), FOS, or corn starch. Cultured microbiomes were analyzed using our metaproteomics workflow. (B) Number of MS/MS identified in the dataset. (C) Number of peptides identified in the dataset; (D) PCA generated using RS-treated, FOS-treated, CS-treated and blank control microbiomes; (E) PCA generated using RS-treated and blank control microbiomes; (F) PCA of RS-treated and blank control microbiomes after EB regression; (G)-(I) PCA plots of RS2-RS4 versus the blank control using EB-corrected data; (J)-(L) Volcano plots of RS2-RS4 versus the blank control generated using EB-corrected data. The p values were calculated using Wilcoxon test, and were adjusted using FDR method. Blue arrow means logical relationship in the data analysis workflow. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Principle component analysis (PCA) revealed that each individual’s RS treated samples clustered closely with the blank control and CS treatment, whereas FOS treatment, as expected, consistently shifted the microbiome metaproteomic profile for all individual microbiomes (Fig. 1D and E). As expected, the largest contributors to the differences observed by PCA were the individual microbiomes. We applied an empirical Bayesian (EB) regression approach [29] to remove the inter-individual variance as well as possible batch effects which overshadowed the responses to RS (Fig. 1F). This approach allows for the combination of multiple balanced experiments and is robust to small sample sizes [29]. By separating each individual’s microbiome cultures as a subgroup, the experimental design was balanced between subgroups. After EB correction, the inter-individual variance in our dataset was removed, and all subgroups were centered on the PCA (Fig. 1F). PCA using control and each RS types individually revealed that RS2 and RS3 altered the metaproteomic profiles compared to the blank control (Fig. 1G and H), whereas RS4-treated samples did not separate from the control (Fig. 1I). It was also clear from hierarchical clustering analysis (Supplementary Fig. S1) that samples treated by RS2 and RS3 were distinct from RS4 which clustered with the control. Analysis of the differentially expressed protein groups between each RS treatment and the control revealed that RS2 led to 96 protein groups with a significantly increased expression, and 19 with a significantly decreased expression (Fig. 1J). In contrast, RS3 resulted in 339 significantly increased and 227 significantly decreased protein groups (Fig. 1K). Since RS4 did not show any significant protein expression response (Fig. 1L), our subsequent analyses focused on RS2 and RS3.
2.2. Functional responses of individual microbiomes to RS
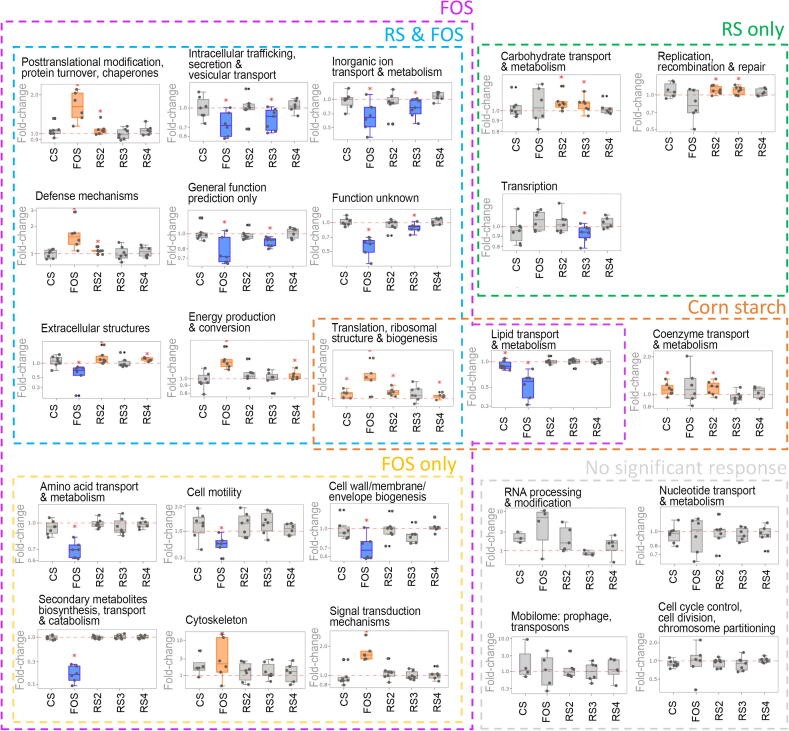
In our dataset, the protein groups were annotated to 24 COG categories (Fig. 2). The treatment with the positive control FOS showed that there were 16 significant changed COG categories (total abundance of their protein constituents compared to the blank control). The corn starch-treated microbiomes only had three significantly changed COG categories whereas microbiomes treated with the three RS showed a total of 12 significantly changed COG categories of which three COG categories were only changed in the presence of RS.
Fig. 2.
Responses of microbiome functional pathways. Response of COG categories different carbohydrates, in comparison to the blank control sample; the scale of y-axis was log10-transformed. Significant responses (Wilcoxon test, p < 0.05) were marked with “*”, significant increases are plotted in orange boxes and significant decreases are plotted in blue boxes. Dashed frames gathered significant changes that occurred under the treatment(s), labeled above each colored frame. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We next explored whether the functional changes in response to RS treatments were common across all individual microbiomes or specific to subgroups of microbiomes. We first looked at starch and sucrose metabolism, ABC transporters, and pyruvate metabolism pathways which were significantly increased by RS2 and RS3 treatment (Supplementary Fig. S2A–C). Most of the microbiomes showed increase in these three pathways in response to at least one of RS2 or RS3: 6/7 microbiomes showed increased starch sucrose metabolism (Supplementary Fig. S2D), 7/7 microbiomes showed increased ABC transporters (Supplementary Fig. S2E); and 6/7 microbiomes showed increased pyruvate metabolism pathways (Supplementary Fig. S2F). There were consistent responses of each downstream metabolic pathway between technical replicates (Supplementary Fig. S2D–F, H–M), supporting the reproducibility of the experiment. Finally, we investigated whether the downstream butyrate production pathway had similar profiles across the individual microbiomes. Interestingly, enzymes within the acetyl-CoA pathway of butyrate production showed different responses across individuals (Supplementary Fig. S2G–M) [30].
2.3. Taxon-specific functional enrichment of differential proteins
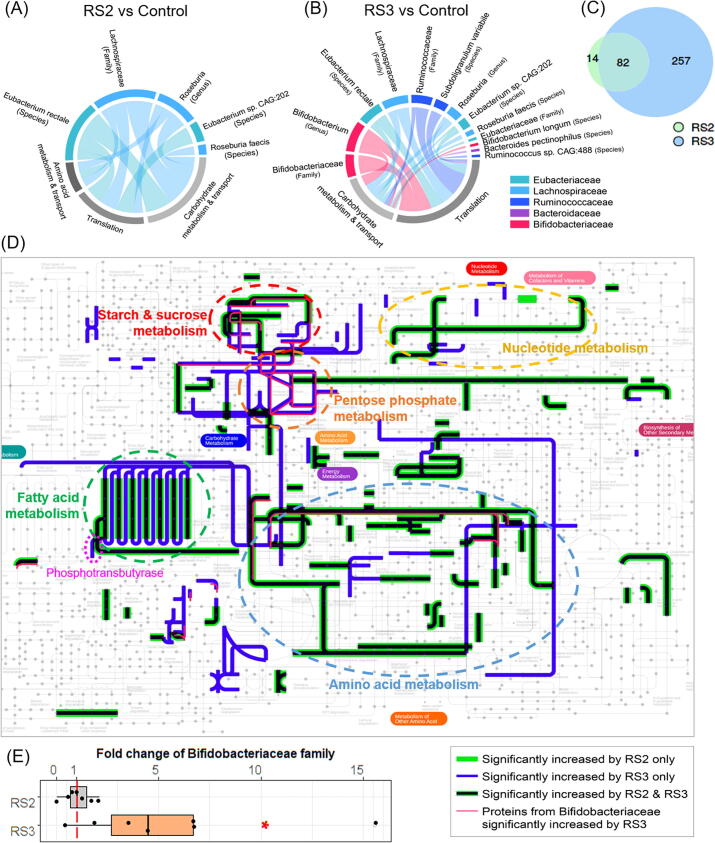
To find out the taxonomic origins of functional changes, taxon-specific functional enrichment (p < 0.05) of the proteins significantly increased by RS2 revealed an enrichment in carbohydrate metabolism & transport, amino acid metabolism & transport, as well as translation pathways of butyrate producers that belongs to families Eubacteriaceae and Lachnospiraceae (Figs. 3A and Supplementary Fig. S3). Enrichment of carbohydrate metabolism & transport, and translation pathways were also found with RS3. In addition to bacteria from families Eubacteriaceae and Lachnospiraceae, families Bifidobacteriaceae, bacteria from Ruminococcaceae and Bacteroidaceae also contributed to these functional enrichments in response to RS3(Fig. 3B and Supplementary Fig. S3).
Fig. 3.
Functional and taxonomic profiles of the significantly increased proteins. (A) Taxon-specific functional enrichment of the significantly increased protein groups in response to RS2; (B) Taxon-specific functional enrichment of the significantly increased protein groups in response to RS3; (C) Comparison of significantly increased protein groups between RS2- and RS3-treated groups; (D) pathways of the significantly increased protein groups; (E) fold change of the Bifidobacteriaceae family in RS2- and RS3-treated groups. Red asterisk indicates statistical significance using one-sided Wilcoxon test (p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, we found that most of the significantly upregulated proteins from RS2 (82 out of 96) were also up-regulated by RS3 (Fig. 3C). We mapped COG numbers corresponding to these significantly up-regulated protein groups to iPath 3 (https://pathways.embl.de/) [31]. We merged the result of RS2 and RS3 (Fig. 3D), the pathway changes caused by RS3 overlapped completely with those cause by RS2 (with only one exception in nucleotide metabolism). Commonly upregulated pathways in both RS groups included starch and sucrose metabolism, nucleotide metabolism, fatty acid metabolism and amino acid metabolism. Notably, proteins involved in pentose phosphate metabolism were only up-regulated by RS3, and more pathways of fatty acid metabolism were upregulated by RS3. Phosphotransbutyrase, a key enzyme in butyrate synthesis [32], [33], was significantly upregulated only by RS3.
The probiotic bacterial family Bifidobacteriaceae (as well as its genus Bifidobacterium and species Bifidobacterium longum) was only enriched in protein groups upregulated by RS3, and it was the most significantly enriched family in the list of RS3 responders (Supplementary Fig. S3). We compared the protein biomass between the control and RS groups based on summed peptide intensities unique to this family. RS3 showed an average fold-change of 5.6 compared to the control (Fig. 3E), which was significant (p < 0.05) by one-sided Wilcoxon test. We overlaid the protein groups corresponding only to Bifidobacteriaceae to iPath 3 (Fig. 3D, red line). This showed that Bifidobacteriaceae mainly contributed in starch and sucrose metabolism and pentose phosphate metabolism pathways, as well as part of amino acid metabolism pathway.
2.4. Ecological network of microbiome functional interaction
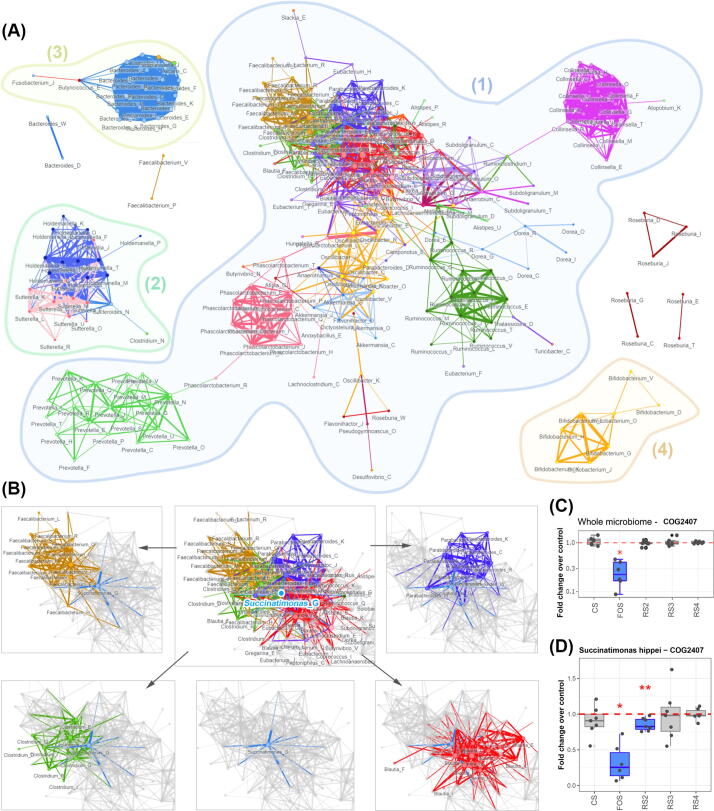
We constructed an ecological network of functional interaction in the gut microbiomes using the metaproteomics responses to nutrient changes. Using a correlation-based approach, a co-occurrence network of taxon-specific functions (microbial genera – COG categories) was built with the criteria that the Spearman's rank correlation coefficient rho > 0.7 and p < 0.05. Altogether, 97 microbial genera were included in this analysis. The analysis resulted in one larger network and a few smaller-scale networks (Fig. 4A). The larger network (network 1) composed of a few modular sub-networks that contained different COG categories in a same genus (e.g. Prevotella, Ruminococcus, Phascolarctobacterium, Collinsella), and sub-networks that are inter-connected by functions from different bacterial genera. One of the smaller network (network 2) was mainly composed of Sutterella and Holdemanella, these two bacteria were correlated by major functions such as energy production and conversion (COG category: [C]), cell wall/membrane/envelope biogenesis [M], amino acid transport and metabolism [E], intracellular trafficking, secretion, and vesicular transport [U]. Two other smaller networks were mainly composed of functions from Bacteroides and Bifidobacterium, respectively.
Fig. 4.
Ecological network of microbiome functional interaction. (A) Co-occurrence networks of taxon-specific functions. Labels of nodes are composed of a bacteria genus name followed by a COG category letter (genus_COG); (B) a detailed view of the sub-network centered by Succinatimonas_G; (C) response of enzyme COG2407 to different carbohydrates in the whole microbiome level, and (D) response of enzyme COG2407 specific to Succinatimonas hippei.
Interestingly, COG category [G], i.e. carbohydrate transport and metabolism from Succinatimonas was at the center of the multi-genera component of the network 1 (Fig. 4B), connecting modules of Faecalibacterium, Clostridum, Parabacteroides and Blautia. A major contributor to this Succinatimonas_G node was the significant change of COG2407, L-fucose isomerase of the L-fucose metabolism pathway. The expression of COG2407 in the whole microbiome was significantly reduced only by FOS treatment (Fig. 4C). Our taxon-specific functional analysis showed that COG2407 from Succinatimonas hippei was significantly decreased by both FOS and RS2 (Fig. 4D).
3. Discussion
In this study, we investigated the functional and taxonomic changes in individual microbiomes exposed to different type and sources of RS using in vitro culturing and metaproteomics. The effects of RS on the microbiomes were milder than the changes observed with FOS, which is known to induce taxonomic and functional changes in the human gut microbiome in vitro [25], [26], [27]. The microbiomes treated with RS maintained their individuality (Fig. 1D and E) whereas FOS treatment tended to regroup the microbiomes (Fig. 1D). Although the PCA showed only weak changes induced by RS treatments, we still observed many significantly shifted microbiome function in response to RS (RS2 and RS3), both by using paired comparisons for individual microbiomes (Fig. 2), and by using EB-transformed protein intensities (Fig. 3).
Interestingly, RS2 and RS3 are both derived from high amylose maize and had similar functional effects of increasing carbohydrate metabolism and transformation in butyrate producing bacteria from families Eubacteriaceae and Lachnospiraceae (Fig. 3A and B, and Supplementary Fig. S3). In addition, we observed highly overlapped functional pathway responses between RS2 and RS3 (Fig. 3D), which may also be due to their same origin of high amylose maize. Nevertheless, RS3 had higher number of enriched taxa corresponding to its functional response, and significantly increased proteins were found in Bifidobacteriaceae (Supplementary Fig. S3). Proteins from Ruminococcaceae and Bacteriodiaceae were also increased in response to RS3. In agreement with our findings, previous studies have reported increases of bacteria from families Bifidobacteriaceae [34], [35], [36], [37], [38], Ruminococcaceae [12], [19], [37], [38], Eubacteriaceae [12], [37], [39], Lachnospiraceae [40] and Bacteriodiaceae [12], [36] in response to different sources and forms of RS. Members of these bacterial families have been shown to be capable of metabolizing resistant starches [14], [41], [42], [43] due to their amylolytic activities [41], [43], and many are butyrate producers [44]. Our findings showed that the protein groups up-regulated by RS2 and RS3 were enriched in starch metabolism pathways. In response to RS3, Bifidobacteriaceae showed increases in the starch uptake and pentose phosphate pathways, which contributed significantly to the overall increase seen in the pentose phosphate pathway. The product of the pentose phosphate pathway, NADPH, is a major source of electrons for diverse anabolic biosynthetic processes, and therefore a primary reducing agent for biosynthesis pathways [45], including fatty acid metabolism and especially butyrate production. However, our results showed that the response of the pentose phosphate pathway was strongly related to the type of the RS, indicating that different types of RS may result in various effects on biosynthesis pathways including butyrate generation. Although RS2 and RS3 were both derived from high-amylose starch, they are not chemically identical (RS2 – hydrothermal treated, RS3 - retrogradation), which may contribute to the difference of functional and taxonomic responses observed in our study. RS4 is a type of chemically modified starch that is not naturally occurring. In this study, we did not observe significant protein changes induced by RS4, which is similar to a recent study reporting that an RS4 did not affect the microbiome [46].
In agreement with a single strain-based study on Eubacterium rectale [43], we found that microbiome functions of starch utilization and ABC transporters were increased in the presence of RS. The gut bacteria expressed enzymes for starch degradation, and the ABC glycan-binding proteins were increased to scavenge the liberated maltooligosaccharides and glucose [43]. Subsequently, the pyruvate metabolism pathways utilizing glucose were also significantly increased. These pathways were consistently increased across all individual microbiomes.
These results showed that we were able to observe significant, taxon-specific functional responses to the supplementation of these different carbohydrates using our metaproteomics approach. Revealing statistical significant changes in taxonomic and functional profiles are common approaches in current microbiome studies. However, an often-neglected fact is that microbiome members are functionally interconnected. Functional responses happen not only in individual taxa but also in their interactions. So here we used our metaproteomics dataset to perform a system-level analysis of the functional organization in the microbiomes. Functional co-occurrence network between different microbial genera showed a mixed network structure. The sub-network modules that primarily consisted of one bacterial genus suggested that nutrient alteration changed the abundance of this genus, and subsequently levels of different functions were altered proportionally. For example, Bifidobacterium had an independent network, suggesting that its functional responses were a direct result of the addition of different carbohydrates (significantly increased in RS3 and FOS). More interestingly, some genera are correlated by specific functions, suggesting that they may be co-regulated through certain forms of functional interactions. The multi-genera component of the network 1 suggested potential trophic interactions between Succinatimonas other genera. In our dataset, Succinatimonas hippei had a significant decrease in L-fucose isomerase. Studies have found a trophic interaction between bacterial species through degradation of L-fucose by a Bifidobacterium species and release constituents that can be utilized by other species [47], [48]. In our network, Eubacterium is also connected to Succinatimonas. To our best knowledge, there hasn’t been study on Succinatimonas utilization of L-fucose, but our result suggest that Succinatimonas may also be a cross-feeder. When the L-fucose pathway in Succinatimonas was decreased by RS2 and FOS, other genera may be affected due to the change in trophic level.
In summary, our study demonstrated that different types of RS have markedly variable functional effects on the human gut microbiome. We also demonstrated that inter-individual differences in microbiome pathway responses were considerable. Our taxon-specific functional analysis suggested carbohydrate metabolic pathway changes in certain groups of bacteria. Finally, we constructed a functional ecological network, which suggested that while some microbes showed genus-specific response to different carbohydrates, there are co-regulated sub-networks potentially due to trophic interaction between microbiome members.
4. Materials and methods
4.1. Resistant starches and controls
RS involved in this study were, RS2 [Hi Maize 260, Ingredion, Inc., Westchester, IL, USA], containing 60% of RS2; RS3 [Novelose 330, obtained from Ingredion, Inc., Westchester, IL, USA], containing 28–38% of RS3; and RS4 [Fibersym RW, obtained from MGP Ingredients, Atchison, KS, USA], containing 85% RS4. Due to the fact that the RS contain different proportion of non-resistant starch, we included corn starch as the negative control. Fructo-oligosaccharide (FOS) - Orafti P95 (BENEO, Inc., Parsippany, NJ, USA), known to consistently and markedly shift the in vitro gut microbiome [25], [26], was used as the positive control. Previous literature involving microbiome responses to prebiotics using fecal slurries inoculations with a 1% w/v prebiotic concentration [49], [50]; this concentration was used for this study’s culture samples. 0.04 g (1% w/v) of RS/corn starch/FOS was added to each corresponding culture tube. For a blank control, microbiome samples were cultured in the same medium but without any compounds added.
4.2. Stool sample collection
This study was following a human stool sample collection protocol (#2016-0585-01H) that is authorized through the University of Ottawa's Ottawa Health Science Network Research Ethics Board. Stool samples from seven healthy volunteers (22–39 years of age; males and females) were involved in this study. Exclusion criteria were: IBS, IBD, or diabetes diagnosis; antibiotic use or gastroenteritis episode in the last 3 months; use of pro-/pre-biotic, laxative, or anti-diarrheal drugs in the last month; or pregnancy. Each potential participant was assigned a number to blind researchers from sample identity. On the day of collection, the participant is given a kit containing a 50 mL BD Falcon™ tube containing 12.5 mL of pre-reduced PBS [311-010-CL; Fisher Scientific, Fair Lawn, NJ, USA] (had been placed in an anaerobic chamber 24 h before collection) and 0.0125 g of L-cysteine [C7352; MilliporeSigma, Oakville, ON, Canada] (0.1% w/v; added just before sample collection). Approximately 3 g of fresh stool sample was collected by each participant into buffer described above and returned to study coordinator within 30 min. Samples were then weighed and transferred into an anaerobic workstation (5% H2, 5% CO2, and 90% N2 at 37 °C), vortex mixed and filtered through sterile gauzes to remove solids.
4.3. In vitro culturing and prebiotic treatment
The microbiome inoculums were treated with the RS2/RS3/RS4/FOS samples through in vitro culturing using a bioanalytical assay testing, as previously reported culture medium [23]. Each participant's stool sample was cultured in 21 separate tubes: four technical replicates for each of the five different treatment conditions (RS2, RS3, RS4, FOS, and the blank), and one well for corn starch as a negative control, with an in-solution stool concentration of 2% w/v. An exception was individual sample P5, for which RS4 and FOS treatments were not performed. In each culture tube, 4 mL of a basic nutrient and salt medium was added prior to stool inoculation. This medium consisted of peptone water [70179; MilliporeSigma] (0.2 g; 0.2% w/v), yeast extract [212750; BD Biosciences, Sparks, MD, USA] (0.2 g; 0.2% w/v), monopotassium phosphate [P5655; MilliporeSigma] (0.045 g; 0.045% w/v), dipotassium phosphate [PX1570-1; EMD Millipore, Etobicoke, ON, Canada] (0.045 g; 0.045% w/v), sodium hydroxide [S8045; MilliporeSigma] (0.09 g; 0.09% w/v), sodium bicarbonate [SX0320-3; EMD Millipore] (0.4 g; 0.4% w/v), magnesium sulphate heptahydrate [230391; MilliporeSigma] (0.009 g; 0.009% w/v), calcium chloride [CX0156-1; EMD Millipore] (0.009 g; 0.009% w/v), bile salts [48305; MilliporeSigma] (0.05 g; 0.05% w/v), Tween 80 [P1754; MilliporeSigma] (200 µL; 0.2% v/v), and distilled water (~90 mL). Each 100 mL of medium was autoclaved, then, then supplemented with 0.4 g porcine gastric mucin [M1778; MilliporeSigma], 0.05 g L-cysteine, and 100 μl of 10 mg/ml Vitamin K1 [47773; MilliporeSigma] stock solution in 100% ethanol. Then the medium was corrected to pH ~ 7, and placed in an anaerobic chamber for 24 h. After inoculation, culture tubes were placed on a shaking platform (300 rpm) inside the anaerobic chamber at 37 °C for 24 h.
4.4. Cell washing, cell lysis, protein extraction and tryptic digestion
Cultured samples were centrifuged at 300 g/4 °C for 5 min to remove debris. The supernatant was collected and bacterial cells were pelleted by centrifuging for 20 min (14,000g/4°C). Bacterial pellets were then washed three times using cold PBS (centrifuged at 14,000g/4 °C). All samples were stored overnight in −80 °C, followed by a cell lysis procedure: 200 µL of a lysis buffer (10 mL recipe: urea [U5128; MilliporeSigma] (3.8 g, 38% w/v); 20% SDS [L3771; MilliporeSigma] (2 mL, 20% v/v); 1 M Tris-HCl [C4706; MilliporeSigma] (0.5 mL, 5% v/v); ddH2O (4 mL, 40% v/v); PhosSTOP™ tablet [4906837001; MilliporeSigma] (one tablet); cOmplete mini™ [4693124001; MilliporeSigma] (one tablet)) was added to each sample culture tube. Each sample was then ultrasonicated (25% amplitude) for 30 s, placed on ice for 30 s, then ultrasonicated again (same setting) for 30 s. Sample tubes were then centrifuged for 10 min (16,000g/8°C). Supernatants were transferred to new 2.0 mL Axygen® tubes, then 1 mL of a −20 °C pre-chilled precipitation solution was added (50% v/v acetone [A949-4; Fisher Scientific]; 50% v/v ethanol [1009; Commercial Alcohols, Tiverton, ON, Canada]; 0.1% v/v acetic acid [A38-212; Fisher Scientific]). Proteins in samples were then precipitated overnight at −20 °C.
Protein samples were washed for three rounds using cold (−20 °C) acetone. Then, samples were re-suspended in 100 µL of 6 M urea buffer (pH = 8.0), and a DC™ assay [Bio-Rad Laboratories, Mississauga, ON, Canada] was performed to determine protein content. 50 µg of protein from each sample was used for in-solution digestion. 2 µL of dithiothriotol (DTT) [43815; MilliporeSigma] was added to each tube, then tubes incubated with shaking for 30 min on an Eppendorf Thermomixer C (800 rpm/56 °C). 2 µL of iodoacetamide (IAA) [I1149; MilliporeSigma] was then added to each sample, and incubated in darkness for 40 min at rt. 1 µg trypsin [T1426; MilliporeSigma] was then added to digest the proteins with incubation at 37 °C for 24 h. Samples were then acidified to pH ~ 2–3 using 5% (v/v) formic acid and were desalted using C18 beads [ReproSil120 C18-AQ; Dr. Maisch GmbH, Ammerbuch-Entringen, DE]. Four samples were lost during sample processing, i.e. one replicate from P3-RS3, P3-FOS, P6-RS2 and P6-RS3.
4.5. LC-MS/MS analysis
All samples were run on Eksigent 425 nanoHPLC connected to Orbitrap Elite™ Hybrid Ion Trap Mass Spectrometer [Thermo-Fisher Scientific, Waltham, MA, USA] with a 240 min gradient. Peptides were separated with an in-house made column (75 μm i.d. × 15 cm) packed with reverse phase beads [1.9 μm/120 Å ReproSil-Pur C18 resin, Dr. Maisch GmbH]. The samples were loaded at 5% buffer A (0.1% formic acid in H2O) and analyzed by a gradient from 5 to 30% (v/v) buffer B (0.1% formic acid, 80% acetonitrile in H2O) at a flow rate of 300 nL/min. MS analysis was done with a full MS scan from 350 to 1750 m/z in Orbitrap, followed by data-dependent MS/MS scan of the 20 most intense ions in ion trap. Microbiome samples corresponding to each individual were run on LC-MS/MS in a randomized order. Spectral data were collected as *.RAW files.
4.6. Metaproteomic data processing and statistical analysis
Database search was performed automatically, following the MetaPro-IQ workflow using the MetaLab software (version 1.0) [28], with MaxQuant version 1.5.3.30 involved in the workflow. Carbamidomethyl (C) was set as fixed modifications, and Acetyl (Protein N-term) and Oxidation (M) modifications were included as variable modification in the database search space. The MetaLab database search results allows for protein identification and quantification and serves as a basis for identifying biochemical pathways that are differentially expressed. The output provides massive information on the dataset, including summary, peptides, protein groups, functional and taxonomic data tables, etc.
Data quality check was performed by submitting the summary.txt file to the MaxQuant Quick Summary app (https://shiny.imetalab.ca/MQ_summary/). LFQ intensities of protein groups were filtered with the criteria of having non-zero values in ≥ 4 samples in each individual subgroup, at least in four of the individual microbiomes. PCA of the protein groups data was then performed using R function prcomp() and visualized using R package “scatterplot3d”. To overcome inter-individual variance that were overriding RS responses, an Empirical Bayesian-based approach was performed with the online tool (https://shiny.imetalab.ca/batcheffect_explorer/). The corrected data was then scaled by sum sample wise, and then differentially expressed protein groups between two conditions were performed using our online iMetaShiny app - Differential Protein Analyzer (https://shiny.imetalab.ca/Volcano_plot/). Criteria and parameters used for statistical test and visualization were: |Fold change| > 1.5, non-parametric test (Wilcoxon test), FDR-adjusted p < 0.05, and curvature = 0.05. Here, we applied a smooth curve cut-off, which was first used in proteomics by Keilhauer et al [40]. The smooth curve is defined by the following equation: y = curvature/|x-“Log2FoldChangeCutOff”| + “-Log10pValueCutOff”.
Taxonomic and functional enrichment analysis were performed using the online tool Enrichment Analysis (https://shiny.imetalab.ca/metaproteomics_enrichment/), p value significances threshold for the enrichment analyses was set as < 0.05. Visualization of pathways was performed using COG accession numbers in iPath 3 (https://pathways.embl.de/)[31]. For functional.csv visualization, we made an R Shiny web page (https://leyuan.shinyapps.io/RS_data/) to compare COG and NOG categories, COG, NOG and KEGG accessions and names, as well as GO comparisons between each treatment and the blank control. Each data point represents the average fold-change of all technical replicates corresponding to one individual microbiome and one treatment. A Wilcoxon test was used to evaluate the statistical significance of the fold change in comparison to the blank control. To identify proteins that are unique to each family, we extracted the protein groups corresponding to this family by mapping its unique peptide IDs (in MetaLab.allPepTaxa.csv) to the protein IDs (in proteinGroups.txt). To summarize the contribution of Bifidobacteriaceae’s proteomic biomass relative to the whole microbiome, as a proxy of the relative total biomass, we summed the peptide intensities corresponding to the Bifidobacteriaceae and calculated its proportion of the total peptide biomass in each sample.
Network analysis was performed using the online tool Co-occurrence Analysis (https://shiny.imetalab.ca/matrix2network/). Taxon-specific intensities of COG functional categories were used as the input data, and genus matches were used as the meta data. Spearman method was selected to compute the correlations, with a threshold (absolute value) of rho > 0.7, and a p value threshold of 0.05.
5. Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017907. There were 21 additional samples in the same database search task that was not involved in this study (three other compounds tested with the same microbiomes). To ensure data analysis reproducibility, we uploaded all these 157 raw files to the Pride Database.
CRediT authorship contribution statement
Leyuan Li: Methodology, Investigation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. James Ryan: Conceptualization, Methodology, Investigation, Writing - original draf, Writing - review & editing, t. Zhibin Ning: Software, Resources, Data curation, Writing - review & editing. Xu Zhang: Methodology, Resources, Writing - review & editing. Janice Mayne: Resources, Project administration, Writing - review & editing. Mathieu Lavallée-Adam: Supervision, Resources, Funding acquisition, Writing - review & editing. Alain Stintzi: Conceptualization, Supervision, Funding acquisition, Writing - review & editing. Daniel Figeys: Conceptualization, Supervision, Writing - original draft, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: DF and AS have co-founded Biotagenics and MedBiome, clinical microbiomics companies. All other authors declare no potential conflicts of interest.
Acknowledgements
This work was supported by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-156 and OGI-149), the Natural Sciences and Engineering Research Council of Canada (NSERC, grant no. 210034), the Ontario Ministry of Economic Development and Innovation (ORF-DIG-14405 and project 13440), the W. Garfield Weston Foundation, and an NSERC Discovery Grant to M.L.A; L.L. and J.R. were funded by a stipend from the NSERC CREATE in Technologies for Microbiome Science and Engineering (TECHNOMISE) Program. The authors wish to thank Dr. Kendra Hodgkinson for editing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.042.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hutkins R.W. Prebiotics: why definitions matter. Curr Opin Biotechnol. 2016;37:1–7. doi: 10.1016/j.copbio.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schley P.D., Field C.J. The immune-enhancing effects of dietary fibres and prebiotics. Br J Nutr. 2002;87(S2):S221–S230. doi: 10.1079/BJNBJN/2002541. [DOI] [PubMed] [Google Scholar]
- 3.Fuentes-Zaragoza E. Resistant starch as prebiotic: a review. Starch – Stärke. 2011;63(7):406–415. [Google Scholar]
- 4.Yao N., Paez A.V., White P.J. Structure and Function of Starch and Resistant Starch from Corn with Different Doses of Mutant Amylose-Extender and Floury-1 Alleles. J Agric Food Chem. 2009;57(5):2040–2048. doi: 10.1021/jf8033682. [DOI] [PubMed] [Google Scholar]
- 5.Birt D.F. Resistant starch: promise for improving human health. Adv Nutr. 2013;4(6):587–601. doi: 10.3945/an.113.004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sajilata M.G., Singhal R.S., Kulkarni P.R. Resistant starch–A review. Compr Rev Food Sci Food Saf. 2006;5(1):1–17. doi: 10.1111/j.1541-4337.2006.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 7.Higgins J.A., Brown I.L. Resistant starch: a promising dietary agent for the prevention/treatment of inflammatory bowel disease and bowel cancer. Curr Opin Gastroenterol. 2013;29(2):190–194. doi: 10.1097/MOG.0b013e32835b9aa3. [DOI] [PubMed] [Google Scholar]
- 8.Guo J., Tan L., Kong L. Impact of dietary intake of resistant starch on obesity and associated metabolic profiles in human: a systematic review of the literature. Crit Rev Food Sci Nutr. 2020:1–17. doi: 10.1080/10408398.2020.1747391. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y. The Capacity of the Fecal Microbiota From Malawian Infants to Ferment Resistant Starch. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vital M. Metagenomic insights into the degradation of resistant starch by human gut microbiota. Appl Environ Microbiol. 2018;84(23):e01562–18. doi: 10.1128/AEM.01562-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashwar B.A. Preparation, health benefits and applications of resistant starch—a review. Starch – Stärke. 2016;68(3–4):287–301. [Google Scholar]
- 12.Upadhyaya B. Impact of dietary resistant starch type 4 on human gut microbiota and immunometabolic functions. Sci Rep. 2016;6(1):28797. doi: 10.1038/srep28797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.I. Martínez K.J., Duffy PR, Schlegel VL, Walter J., Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects PLoS One 5 2010 e15046. [DOI] [PMC free article] [PubMed]
- 14.Purwani E.Y., Purwadaria T., Suhartono M.T. Fermentation RS3 derived from sago and rice starch with Clostridium butyricum BCC B2571 or Eubacterium rectale DSM 17629. Anaerobe. 2012;18(1):55–61. doi: 10.1016/j.anaerobe.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 15.McOrist A.L. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. J Nutr. 2011;141(5):883–889. doi: 10.3945/jn.110.128504. [DOI] [PubMed] [Google Scholar]
- 16.Yu M. Microbiome-metabolomics analysis investigating the impacts of dietary starch types on the composition and metabolism of colonic microbiota in finishing pigs. Front Microbiol. 2019;10(1143) doi: 10.3389/fmicb.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bang S.-J. Effect of raw potato starch on the gut microbiome and metabolome in mice. Int J Biol Macromol. 2019;133:37–43. doi: 10.1016/j.ijbiomac.2019.04.085. [DOI] [PubMed] [Google Scholar]
- 18.Zybailov B.L. Metaproteomics reveals potential mechanisms by which dietary resistant starch supplementation attenuates chronic kidney disease progression in rats. PLoS ONE. 2019;14(1) doi: 10.1371/journal.pone.0199274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier T.V. Impact of dietary resistant starch on the human gut microbioCme metaproteome, and metabolome. mBio. 2017;8(5):e01343–17. doi: 10.1128/mBio.01343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venturelli O.S. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol. 2018;14(6) doi: 10.15252/msb.20178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Layeghifard M., Hwang D.M., Guttman D.S. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 2017;25(3):217–228. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L., Figeys D. Proteomics and metaproteomics add functional, taxonomic and biomass dimensions to modeling the ecosystem at the mucosal-luminal interface. Mol Cell Proteomics. 2020 doi: 10.1074/mcp.R120.002051. p. mcp.R120.002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L. Evaluating in vitro culture medium of gut microbiome with orthogonal experimental design and a metaproteomics approach. J Proteome Res. 2018;17(1):154–163. doi: 10.1021/acs.jproteome.7b00461. [DOI] [PubMed] [Google Scholar]
- 24.Ravasz E. Hierarchical organization of modularity in metabolic networks. Science. 2002;297(5586):1551. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 25.Liu F. Fructooligosaccharide (FOS) and galactooligosaccharide (GOS) increase bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci Rep. 2017;7(1):11789. doi: 10.1038/s41598-017-10722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivieri K. Prebiotic effect of fructooligosaccharide in the simulator of the human intestinal microbial ecosystem (SHIME® Model) J Med Food. 2014;17(8):894–901. doi: 10.1089/jmf.2013.0092. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X. In vitro metabolic labeling of intestinal microbiota for quantitative metaproteomics. Anal Chem. 2016;88(12):6120–6125. doi: 10.1021/acs.analchem.6b01412. [DOI] [PubMed] [Google Scholar]
- 28.Cheng K. MetaLab: an automated pipeline for metaproteomic data analysis. Microbiome. 2017;5(1):157. doi: 10.1186/s40168-017-0375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2006;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 30.Vital M., Howe A.C., Tiedje J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5(2):e00889–14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darzi Y. iPath3.0: interactive pathways explorer v3. Nucleic Acids Res. 2018;46(W1):W510–W513. doi: 10.1093/nar/gky299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter K.A. Sequence and arrangement of two genes of the butyrate-synthesis pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;134(1):107–111. doi: 10.1016/0378-1119(93)90182-3. [DOI] [PubMed] [Google Scholar]
- 33.RC Valentine, R.W., Purification and role of phosphotransbutyrylase. Journal of Biological Chemistry, 1960. 235(7): p. 1948-1952. [PubMed]
- 34.Alfa M.J. A randomized trial to determine the impact of a digestion resistant starch composition on the gut microbiome in older and mid-age adults. Clin Nutr. 2018;37(3):797–807. doi: 10.1016/j.clnu.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Metzler-Zebeli B.U. Resistant starch reduces large intestinal pH and promotes fecal lactobacilli and bifidobacteria in pigs. Animal. 2019;13(1):64–73. doi: 10.1017/S1751731118001003. [DOI] [PubMed] [Google Scholar]
- 36.Gopalsamy G. Resistant starch is actively fermented by infant faecal microbiota and increases microbial diversity. Nutrients. 2019;11(6):1345. doi: 10.3390/nu11061345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkataraman A. Variable responses of human microbiomes to dietary supplementation with resistant starch. Microbiome. 2016;4(1):33. doi: 10.1186/s40168-016-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kieffer D.A. Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am J Physiol Renal Physiol. 2016;310(9):F857–F871. doi: 10.1152/ajprenal.00513.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verberkmoes N.C. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 40.Keilhauer E.C., Hein M.Y., Mann M. Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS) Mol Cell Proteomics. 2015;14(1):120. doi: 10.1074/mcp.M114.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ze X. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6(8):1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y. The in vitro effects of retrograded starch (resistant starch type 3) from lotus seed starch on the proliferation of Bifidobacterium adolescentis. Food Funct. 2013;4(11):1609–1616. doi: 10.1039/c3fo60206k. [DOI] [PubMed] [Google Scholar]
- 43.Cockburn D.W. Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol Microbiol. 2015;95(2):209–230. doi: 10.1111/mmi.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe A.J. Glycolysis for microbiome generation. Microbiol Spect. 2015;3(3):p. doi: 10.1128/microbiolspec.MBP-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deehan E.C. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Schwab C. Trophic interactions of infant bifidobacteria and eubacterium hallii during l-fucose and fucosyllactose degradation. Front Microbiol. 2017;8:95. doi: 10.3389/fmicb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunesova V., Lacroix C., Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16(1):248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rycroft C.E. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J Appl Microbiol. 2001;91(5):878–887. doi: 10.1046/j.1365-2672.2001.01446.x. [DOI] [PubMed] [Google Scholar]
- 50.Olano-Martin E., Gibson G.R., Rastall R.A. Comparison of the in vitro bifidogenic properties of pectins and pectic-oligosaccharides. J Appl Microbiol. 2002;93(3):505–511. doi: 10.1046/j.1365-2672.2002.01719.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017907. There were 21 additional samples in the same database search task that was not involved in this study (three other compounds tested with the same microbiomes). To ensure data analysis reproducibility, we uploaded all these 157 raw files to the Pride Database.