Significance
The SARS-CoV-2 pandemic of 2020 has highlighted the great need for antiviral therapeutics. In this work, we identify a host factor, the SKI complex, is involved in replication of influenza and coronaviruses. Using computational modeling we were able to identify chemicals that interact with the host factor that display broad-spectrum antiviral activity against influenza, coronaviruses, and filoviruses, all of which cause significant morbidity and mortality. Broad-spectrum antiviral therapeutics are sorely needed to combat known and unknown viral pathogens and the work presented here is our first step to developing antivirals to target the SKI complex.
Keywords: broad-spectrum antiviral, coronavirus, influenza, filovirus, antiviral host factor
Abstract
The SARS-CoV-2 pandemic has made it clear that we have a desperate need for antivirals. We present work that the mammalian SKI complex is a broad-spectrum, host-directed, antiviral drug target. Yeast suppressor screening was utilized to find a functional genetic interaction between proteins from influenza A virus (IAV) and Middle East respiratory syndrome coronavirus (MERS-CoV) with eukaryotic proteins that may be potential host factors involved in replication. This screening identified the SKI complex as a potential host factor for both viruses. In mammalian systems siRNA-mediated knockdown of SKI genes inhibited replication of IAV and MERS-CoV. In silico modeling and database screening identified a binding pocket on the SKI complex and compounds predicted to bind. Experimental assays of those compounds identified three chemical structures that were antiviral against IAV and MERS-CoV along with the filoviruses Ebola and Marburg and two further coronaviruses, SARS-CoV and SARS-CoV-2. The mechanism of antiviral activity is through inhibition of viral RNA production. This work defines the mammalian SKI complex as a broad-spectrum antiviral drug target and identifies lead compounds for further development.
At the end of 2019 cases of pneumonia of unknown etiology were identified in China. In the first week of January, a novel coronavirus was identified as the cause and was found to be spreading between people. In the months since, the virus has spread around the world with over 40 million cases by October 2020. Among many things that the SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) outbreak has demonstrated is the immense need for both specific and broadly acting antiviral therapeutics to treat known viruses and those yet to emerge in the human population.
Here, we detail work identifying the SKI complex as a potential broad-spectrum, host-directed, antiviral target. The SKI complex is an RNA helicase and a cofactor for the cytosolic RNA exosome involved in various aspects of RNA metabolism (1–4). The SKI complex was originally identified in yeast as a result of the super killer (SKI) phenotype (5). In this phenotype, knockout of the yeast SKI genes allows production of a viral toxin that kills yeast cells; when present, the SKI complex degrades the viral RNA (6). The cytosolic RNA exosome has been suggested to have a role in replication of mammalian viruses such as influenza (7) and hepatitis B (8, 9). The TRAMP complex has a similar function to the SKI complex but acts as a nuclear cofactor of the RNA exosome. This protein complex has been linked to replication of multiple RNA viruses (10). Additionally, the enzymatic subunit of the SKI complex, SKIV2L, has been suggested to regulate the IFN response by modulating RIG-I (11), which could point toward a broader role in viral replication. These findings suggest that the SKI complex could impact mammalian virus replication, but the picture is far from clear.
Using yeast suppressor screening, we identified that influenza A virus (IAV) NS1 and Middle East respiratory syndrome coronavirus (MERS-CoV) ORF4a proteins have a genetic interaction with the SKI complex. Finding that both viral proteins have a genetic interaction with the yeast SKI complex we subsequently determined that siRNA knockdown of the human SKI complex resulted in significant reduction in replication of these two distinct viruses, suggesting the complex may be a broad-spectrum antiviral target.
Using the site identification by ligand competitive saturation (SILCS) in silico modeling approach (12) we identified a binding pocket on one of the subunits of the SKI complex and used this to screen for compounds with antiviral activity. From the selected compounds, experimental assays were used to identify three unique chemical compounds that were capable of inhibiting both IAV and MERS-CoV replication. Further experimental analyses show that the mechanism of antiviral action is through inhibition of viral RNA production. Moreover, our lead compound was found to inhibit replication of the filoviruses Ebola and Marburg, extending the broad-spectrum activity to a third viral family that causes significant human morbidity and mortality. Finally, we found that our lead compound has broad anticoronavirus activity, inhibiting severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2 replication.
Our work identifies a host factor involved in the replication of coronaviruses, influenza, and filoviruses. We have identified multiple chemical scaffolds that are modeled to interact with the SKI complex, and all show broad-spectrum antiviral activity. These compounds will act as the basis of structure–activity relationship studies in the search for more potent compounds with the potential to be developed into therapeutic agents. Developing broad-spectrum antivirals that target both the viruses themselves and the host they infect, that can be used in combination, may be the best approach to prepare for the next viral disease outbreak.
Results
The SKI Complex Has a Genetic Interaction with IAV NS1 and MERS-CoV ORF4a.
We have previously demonstrated that certain MERS-CoV proteins are capable of causing slow growth when expressed in the yeast Saccharomyces cerevisiae (13). In addition to MERS-CoV, proteins encoded by IAV are capable of causing a slow growth phenotype in S. cerevisiae (SI Appendix, Fig. S1A; 14). We focused our attention on the NS1 protein as we had previously validated its slow growth phenotype (14) and it has similarity to MERS-CoV ORF4a (the subject of our previous study) in being a double-stranded RNA binding protein that can inhibit the IFN response in mammalian cells. We performed suppressor screening in the yeast knockout library for IAV NS1 (see ref. 13 for details). We tested 101 yeast colonies that had a suppressor phenotype, which represented 69 unique genes. In follow-up validation experiments, 14 of these genes were determined to be bona fide suppressors (SI Appendix, Table S1).
The most frequent hit from the NS1 screening was the yeast gene YPR189W/SKI3. SKI3 along with SKI2, SKI7, and SKI8 form the yeast SKI complex. The gene YGL213C/SKI8 was also a validated suppressor for IAV NS1 (SI Appendix, Table S1) and SKI7 was a validated hit from our MERS-CoV ORF4a screen (13). We therefore directly investigated whether all of the yeast SKI genes would act as suppressors for each of the viral proteins. All of SKI2, SKI3, and SKI8 were potent suppressors for the NS1 slow growth phenotype, while SKI7 had only minimal effect (Fig. 1A). The suppressor phenotypes for ORF4a were milder than those seen for NS1; however, all of the SKI knockout strains gave an increase in growth rate compared to wild-type cells, with loss of SKI7 giving the largest increase (Fig. 1B). These alterations to growth rate were not the consequence of a loss of viral protein expression (SI Appendix, Fig. S1B). Overall, these data demonstrate that in S. cerevisiae, there is a functional genetic interaction between IAV NS1 and MERS-CoV ORF4a with the yeast SKI complex.
Fig. 1.
Yeast suppressor screening identifies the SKI complex as a suppressor of NS1 and ORF4a-mediated slow growth. (A) Yeast knockouts for each component of the SKI complex were transformed with the NS1 galactose-inducible expression plasmid or empty vector control (EV). Growth rate of these yeast was measured over a 48 h culture period. Mean OD600 between three independent colonies in two independent experiments is plotted with error bars being the SD. (B) As in A, but with an ORF4a expression plasmid. (C) HEK293T cells were cotransfected with plasmids to express HA-tagged human SKI genes (SKIV2L-HA, TTC37-HA, or WDR61-HA) with either IAV NS1-GFP or MERS-CoV ORF4a-GFP. After transfection, cells were lysed with RIPA lysis buffer and used to analyze whole cell lysate (WCL) or protein was used for HA immunoprecipitation, analyzing nonspecific bead binding (NS), flow through for unbound protein (FT) and the immunopreciptate (IP). In all cases, samples were separated by SDS/PAGE and Western blotted (WB) for GFP, HA, or tubulin (as loading and immunoprecipitation clearance control).
IAV NS1 and MERS-CoV ORF4a Bind to the Human SKI Complex.
The yeast SKI complex has a functional interaction with IAV NS1 and MERS-CoV ORF4a (Fig. 1 and SI Appendix, Table S1), suggesting that this protein complex may be involved with replication of these two viruses. The SKI complex is well conserved between yeast and mammalian cells. The mammalian homologs of SKI2, SKI3, and SKI8 are SKIV2L, TTC37, and WDR61, respectively (hereafter, the yeast genes and human genes will be identified by these different names). The mammalian homolog of SKI7 is poorly defined and we have excluded that from further study. To move from yeast to a mammalian system, we investigated whether the viral proteins bound to each of the components of the mammalian SKI complex by coimmunoprecipitation assays. Each of the SKI genes were HA-tagged and coexpressed with either NS1-GFP or ORF4a-GFP in HEK293T cells. Whole cell lysates were produced and used for Western blot or immunoprecipitation (IP) was performed for the HA-tag (Fig. 1C). As can be seen in the IP lanes, each of the SKI-HA genes were successfully immunoprecipitated. For all three of the SKI genes both NS1-GFP and ORF4a-GFP were also found in the IP fraction. Each viral protein was also found in the flow through fraction (FT), suggesting that while not all of the protein was bound to the SKI complex, a significant proportion of each viral protein interacted with the mammalian SKI complex.
The SKI Complex Is Required for IAV and MERS-CoV Replication.
The genetic evidence in yeast and the direct protein interaction in mammalian cells suggests the SKI complex may have a role in replication of IAV and MERS-CoV. To test this we analyzed two siRNA sequences targeting each of the three genes (SKIV2L, TTC37, and WDR61). A549 (IAV) or Huh7 (MERS-CoV) cells were transfected with these six individual siRNA sequences, a scrambled control, or were mock transfected for 3 d, prior to being infected with each virus for 24 h (multiplicity of infection [MOI] 0.01 and MOI 0.1 for IAV and MERS-CoV, respectively). After the infection, virus was collected and titered.
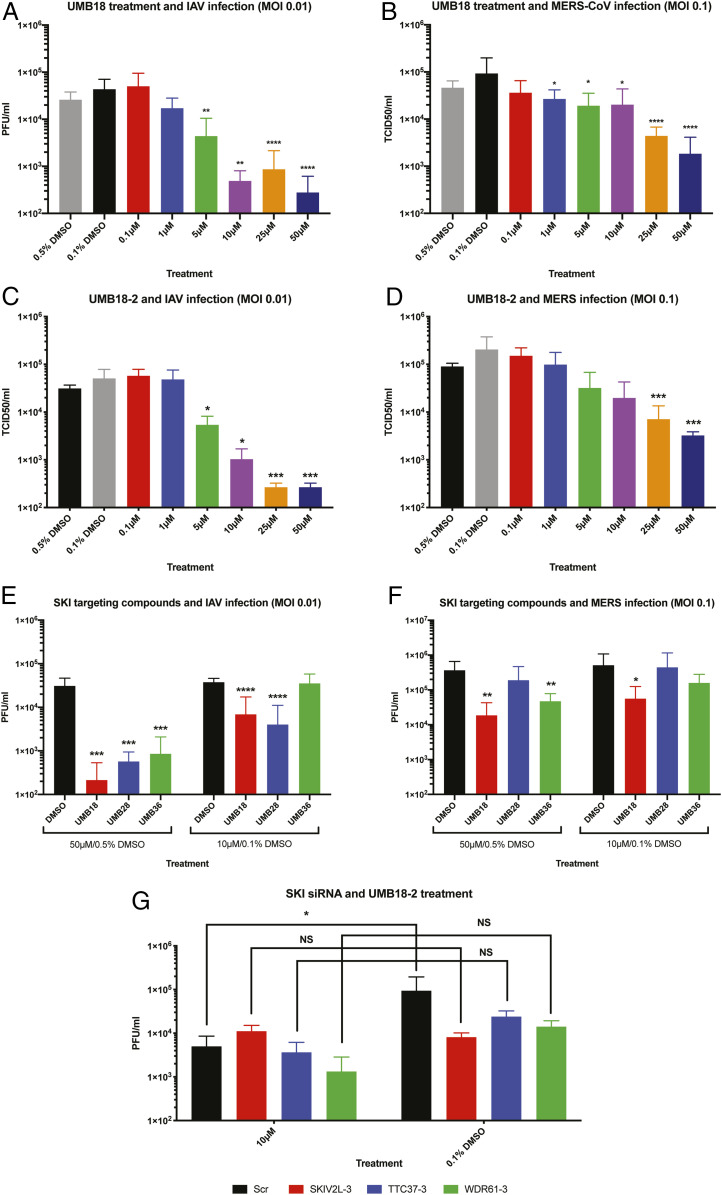
Both siRNA sequences against SKIV2L and TTC37 gave a significant reduction in IAV replication (Fig. 2A). The knockdown of WDR61 with one sequence also gave a significant reduction, while the other gave a significant enhancement in replication of IAV (Fig. 2A). For MERS-CoV, all but one of the siRNA sequences caused a significant reduction in replication (Fig. 2B). Owing to the discrepancy in the results of the two WDR61 sequences for IAV infection, we tested a third siRNA sequence for each of the SKI genes and found that all three inhibited IAV replication (Fig. 2C). These third sequences caused a statistically significant reduction in MERS-CoV replication, consistent with the other siRNA results (Fig. 2D). Knockdown of each of the SKI genes in A549 cells was confirmed by qRT-PCR (Fig. 2E) and at the protein level in both cell lines for SKIV2L (Fig. 2F) (we were unable to find usable antibodies for TTC37 and WDR61). Over the 3-d transfection time course, the two original siRNA sequences did not cause a significant reduction in cell viability as assessed by CellTiter-Glo (Fig. 2 G and H). Overall, we conclude that siRNA-mediated knockdown of each of the different components of the mammalian SKI complex negatively impact replication of IAV and MERS-CoV.
Fig. 2.
Knockdown of the SKI complex by siRNA inhibits replication of IAV and MERS-CoV. (A) A549 cells were transfected with siRNAs targeting the different components of the SKI complex using two unique sequences for each of the three target genes along with scrambled and mock controls. After 3 d of transfection, cells were infected with IAV at MOI 0.01. After 24 h, supernatant was collected, and viral titer assessed by plaque assay. Plotted is the mean plaque forming units (PFU)/mL from three independent experiments with error bars being SD. (B) As in A but with Huh7 cells and MERS-CoV infection at MOI 0.1. Virus titer was determined by TCID50 assay. Plotted is the mean TCID50/mL from three independent experiments with error bars being SD. (C) A third siRNA sequence for each of the three SKI genes was transfected into A549 cells for 3 d, at which point the cells were infected and assessed as in A. Plotted is a representative experiment of two showing the mean PFU/mL from triplicate wells of infection. (D) As in C but MERS-CoV infection of Huh7 cells. (E) A549 cells were transfected for 3 d as described and collected in TRIzol for qRT-PCR analysis of each of the SKI genes being targeted by siRNA (all three unique sequences). Data are a representative experiment of three performed in triplicate wells. PCR reads were normalized with GAPDH and fold change was set relative to scrambled siRNA transfected cells. (F) A549 and Huh7 cells were transfected with SKIV2L targeting siRNA (sequences 1 and 2) for 3 d prior to collection in RIPA lysis buffer. Samples were Western blotted for SKIV2L or tubulin as a loading control. Data are representative of two independent repeats. (G) A549 and (H) Huh7 cells were transfected with siRNAs targeting the SKI complex and cell viability was assessed over the 3-d period by CellTiter-Glo assay. Data are the mean relative luminescence set relative to scrambled control from a representative experiment performed in quadruplicate of three (A549) or two (Huh7) independent experiments. In all cases, t tests were performed for Scr control vs. siRNA transfected cells; ns, non-significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The SKI Complex Is a Potential Antiviral Target.
Our data suggest a genetic and physical interaction between viral proteins and the SKI complex and that the mammalian SKI complex may be important for replication of these two very different viruses. The SKI complex may therefore be a potential broad-spectrum antiviral target. No compounds targeting the SKI complex are available; as such we took an in silico modeling approach using the yeast SKI complex for which there are published three-dimensional (3D) structural data (15).
Ligand design efforts targeted the identification of compounds that would interact with the SKI complex, focusing on SKI8. The process involved the identification of regions on SKI8 in contact with other protein monomers in the complex (15) along with the identification of putative ligand binding sites using the SILCS approach (12). Based on SILCS hotspots and FragMap analysis (16) a putative binding site on the edge of the central region of the β-propeller of the SKI8 monomer was identified (Fig. 3 A and B). The region includes residues 20, 125, 188, 205, and 237 of SKI8. To initiate the screening of ligands targeting the binding pocket, the SILCS-Pharm approach (17) was applied to develop multiple pharmacophores for in silico screening of a database of ∼780,000 commercially available compounds. Upon visual inspection, two pharmacophores that each include four features (one of these shown in Fig. 3C) were selected for further in silico screening using the program Pharmer (18). For further detail on the computational modeling please see SI Appendix.
Fig. 3.
Modeling compounds to bind to the SKI complex and screening for antiviral activity. The 3D structure of SKI8 was subjected to SILCS simulations from which the (A) FragMaps (mesh representations for apolar [green, −0.9 kcal/mol], hydrogen-bond donor [blue, −0.6 kcal/mol], hydrogen bond acceptor [red, −0.6 kcal/mol], positive [cyan, −1.2 kcal/mol], and negative [orange −1.2 kcal/mol] functional groups) were calculated and the binding hotspots determined (all hotspots as blue spheres with those defining the identified binding site as larger spheres colored by ranking (low to high as blue to red). (B) Expanded view of putative binding site. (C) Putative binding site with the FragMaps, the pharmacophore features (spheres, aromatic [cyan] and hydrogen-bond donor [blue]), and the SILCS Monte Carlo conformational sampling docked orientation. Compounds predicted to bind were purchased and screened for antiviral activity. A549 cells were infected with IAV and treated with compounds as indicated. Virus was collected, and PFU/mL determined. (D) Compounds 1 to 20 were tested with single wells of infection. The 50 µM data are from one experiment, 10 µM data are from two independent experiments with error bars being SD. Dotted line to denote the DMSO control PFU/mL for ease of visualization. N.B. Compound 6 did not arrive with the order, it has not been deliberately omitted. (E) Compounds 21 to 40, data from two independent experiments with error bars being SD. UMB18 was included as a positive control. (F) Structural variants of UMB18 were investigated, data are from two independent experiments with error bars being SD. UMB18-2 was also blindly listed as UMB40. (G–J) Chemical structures of the four lead compounds.
Having mapped a potential compound docking site at the interface of SKI8 and SKI3, we purchased 39 compounds (in two sets) that were predicted to bind and were tested for antiviral activity in vitro. In the first batch of 20 compounds tested, one of these showed antiviral activity against IAV, hereafter referred to as UMB18 (Fig. 3D). Our second set had three further compounds that showed antiviral activity (UMB28, UMB36, and UMB40; Fig. 3E). Additionally, a set of chemical analogs to our initial hit of UMB18 were tested (Fig. 3F), none of which showed any greater antiviral activity. However, multiple compounds, for example UMB18-2, showed a similar level of inhibition. As part of the predictive process, structurally related compounds were not excluded; the compound coded UMB40 was also in the chemical analog set as UMB18-2, blindly revalidating this initial hit. We will refer to this compound as UMB18-2 for the remainder of the paper owing to the highly similar structures. Overall, we modeled a library of compounds to target the SKI complex and found four capable of inhibiting IAV infection. Of these chemicals, UMB18 and UMB18-2 are very structurally similar, differing by only a hydroxyl and fluoride group (Fig. 3 G and H) while the other two have distinct chemical structures (UMB28 and UMB36; Fig. 3 I and J), giving potential starting points for future development. UMB18 and UMB18-2 showed the greatest antiviral activity and will be approached as our lead compounds.
Investigation of Lead SKI Targeting Compounds for Antiviral Activity.
Having displayed inhibition of IAV infection at concentrations of 50 µM and 10 µM in our screening, we investigated the dose dependency of the compounds. Cells were treated with UMB18 across a broader range of concentrations and infected with IAV (Fig. 4A). These data demonstrate dose-dependent inhibition of IAV by UMB18 and an IC50 value of ∼5 µM. We similarly tested UMB18 against MERS-CoV and found it inhibited infection with a similar IC50 (Fig. 4B). These data suggest that UMB18 could potentially be a broad-spectrum antiviral compound. The antiviral activity is not a result of cell cytotoxicity; antiviral concentrations caused minimal toxicity as assessed by CellTiter-Glo in both cell lines used (SI Appendix, Fig. S2 A and B). UMB18-2 showed similar inhibitory profiles against both viruses (Fig. 4 C and D). Overall, UMB18 and UMB18-2 both appear to have antiviral activity against IAV and MERS-CoV.
Fig. 4.
SKI targeting compounds have antiviral activity against IAV and MERS-CoV. (A) A549 cells were infected with IAV at MOI 0.01 and treated with UMB18 at the indicated concentrations for 24 h. Based on the stock of compound, 0.5% DMSO acted as the vehicle control for 50 µM and 25 µM while 0.1% acted as the control for all other concentrations. Virus was collected after 24 h, and PFU/mL determined by plaque assay. Data are from three independent experiments performed in triplicate with mean PFU/mL displayed and error bars representing SD. (B) Huh7 cells were infected with MERS-CoV at MOI 0.1 and treated with UMB18 as in A. Virus was collected and titer determined by TCID50 assay. Data are from three independent experiments as in A. (C and D) Cells infected as in A and B but treated UMB18-2. Data are from a representative experiment of two, each performed in triplicate. (E) A549 cells were treated with UMB28 or UMB36 and compared with UMB18 at either 50 µM or 10 µM (with 0.5% or 0.1% DMSO being the appropriate negative controls) and infected with IAV at MOI 0.01. Virus was collected after 24 h, and PFU/mL determined by plaque assay. Data are from three independent experiments (one of a single well and two of triplicate wells) with the mean PFU/mL displayed and error bars being the SD. (F) As in E, but Huh7 cells were treated and infected with MERS-CoV at MOI 0.1 for 24 h. Virus titer was determined by TCID50 assay. Data are from three independent experiments all of triplicate wells with the mean TCID50/mL displayed and error bars being the SD. (G) A549 cells were siRNA transfected with sequence 3 of each of the SKI targeting siRNAs as used in Fig. 2C. Following the 3-d transfection, cells were infected with IAV (MOI 0.01) in the presence of 10 µM or 0.1% DMSO for 24 h. Virus was collected and PFU/mL determined by plaque assay. Data are from two independent experiments performed in triplicate wells. In all cases, except G, t tests were performed for vehicle control vs. drug-treated samples; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. For G, a two-way ANOVA Bonferroni multiple comparisons test was performed; NS, nonsignificant, *P < 0.05.
Assessment of Other Chemical Compounds Targeting the SKI Complex for Antiviral Activity.
In addition to UMB18 and UMB18-2, we also identified two further chemical structures that inhibited IAV infection (Fig. 3 I and J). While both had antiviral activity, neither appeared to be as potent as UMB18 in the initial screening (Fig. 3E). We further investigated this with more direct comparisons. At 50 µM, both UMB28 and UMB36 showed similar inhibition of IAV as UMB18, but both had reduced inhibition at 10 µM (Fig. 4E). Similar results were seen with MERS-CoV infection (Fig. 4F). Again, this inhibition was achieved at noncytotoxic concentrations (SI Appendix, Fig. S2 C and D). These data from Fig. 4 demonstrate UMB18 and UMB18-2 as our most potent antiviral compounds, but that different chemical structures also display broad-spectrum antiviral activity. Having multiple chemical structures may be useful for later development purposes.
The SKI Complex Is Required for Antiviral Activity of UMB18-2.
The compounds that have been screened for antiviral activity are predicted to target the SKI complex based on our in silico modeling data. However, we wanted to more directly assess this by testing whether antiviral activity was lost when the SKI complex was disrupted by siRNA. A549 cells were transfected with sequence 3 of each of the SKI targeting siRNAs that were found to inhibit IAV replication in Fig. 3C. After 3 d of siRNA transfection, cells were infected with IAV at MOI 0.01 and treated with 10 µM UMB18-2 or DMSO (dimethylsulfoxide) control, supernatant was then collected 24 h postinfection for titer. Importantly, when the SKI complex was disrupted by any of the siRNA sequences there was no significant difference between DMSO control and UMB18-2 treatment (Fig. 4G). However, there was still a significant reduction in viral replication in scrambled control cells between UMB18-2 treated and control (Fig. 4G). Most strikingly, the titer from SKIV2L (the enzymatic subunit) knockdown cells was nearly identical in UMB18-2-treated cells and DMSO control. These data demonstrate that the SKI complex is required for the antiviral activity of UMB18-2, suggesting the modeling has found compounds that do indeed target the complex.
UMB18 Inhibits Filovirus Infection.
We further investigated the breadth of antiviral activity of our lead compound UMB18. For this, we tested another family of viruses that cause severe human mortality, the filoviruses, specifically Ebolavirus (Makona strain, EBOV) and Marburg virus (Angola stain, MARV). Huh7 cells were treated with UMB18 across an 8-point dose curve and infected with EBOV (Fig. 5A) or MARV (Fig. 5B) at MOI 0.21 for 48 h. The percentage of inhibition was plotted along with the assessment of cytotoxicity at each concentration in the absence of infection. Toremifene citrate was used as a positive control for inhibition (19), and both were compared to DMSO as the negative control. Both viruses were found to be inhibited by UMB18 at noncytotoxic concentrations. EBOV appeared to be more sensitive to UMB18 with an IC50 calculated as ∼5 µM. MARV was comparably less sensitive with an IC50 ∼16 µM, but was still inhibited in the same concentration range. These data further extend our conclusion that UMB18 has broad-spectrum antiviral activity.
Fig. 5.
UMB18 inhibits filovirus infection. Huh7 cells were treated with UMB18 for test, with toremifene citrate (TOMF) as a positive control and DMSO as a negative control. Treatments were over an 8-point dose curve with threefold dilutions, each in triplicate. Cells were infected with (A) Ebola virus Makona strain (EBOV) or (B) Marburg virus Angola strain (MARV) for 48 h. Cells were fixed and labeled with antibodies to VP40 for each virus. Infected cells were detected by peroxidase secondary labeling to determine the percentage inhibition of infection by each treatment. Cytotoxicity is also displayed which was determined by CellTiter-Glo assay on uninfected samples. Data are from one representative experiment of two performed in triplicate. Dotted line is at 50% inhibition for determining IC50 values.
SKI Targeting Compounds Inhibit Production of Viral RNA.
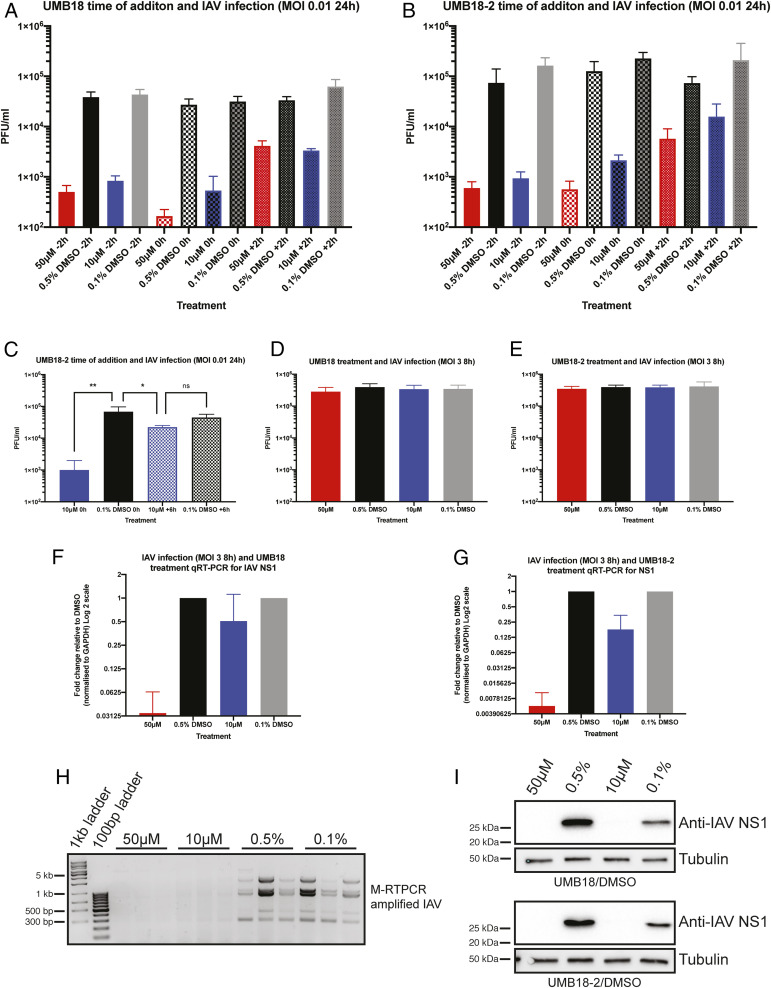
To better understand the mechanism of inhibition by our SKI targeting compounds we used a time-of-addition assay. In all experiments previously discussed, virus and compound were added to cells at the same time. In the time-of-addition assays, cells were either pretreated with compound for 2 h (−2 h), had the compound added with IAV as before (0 h), or compound was added 2 h after cells were infected (+2 h). Pretreatment of cells and addition of drug at the same time as virus led to a similar level of inhibition with both UMB18 and UMB18-2 (Fig. 6 A and B). Addition of compounds at 2 h after infection was started showed a marked reduction in the level of inhibition. However, the compounds were still able to inhibit infection compared to DMSO control. We therefore took a later time point of +6 h with UMB18-2 treatment at 10 µM. By 6 h, viral RNA production has reached a peak for IAV (20). As expected, there was a significant difference between UMB18-2 and DMSO control when both were added at 0 h (Fig. 6C). But importantly, there was no significant difference between DMSO control and UMB18-2-treated cells when drug was added at +6 h (Fig. 6C).
Fig. 6.
SKI targeting lead compounds inhibit viral RNA and protein production. Time-of-addition experiments were performed with IAV infection and UMB18 (A) and UMB18-2 (B). A549 cells were plated and treated with drug 2 h prior to infection (−2 h), at the time of infection (0 h), or 2 h after virus was added to cells (+2 h). After 24 h infection supernatant was collected and titer determined by plaque assay. Mean PFU/mL and SD are displayed from two independent experiments performed in triplicate with error bars being SD. (C) Experimental setup as described in B, but with UMB18-2 or DMSO control added at 0 h or +6 h. Data are from a representative experiment of two performed in triplicate. A one-way ANOVA was performed; ns, nonsignificant, *P < 0.05, **P < 0.01. (D) A549 cells were infected with IAV at MOI 3 for 8 h with UMB18 or (E) UMB18-2 treatment. Supernatant was collected and used to titer by plaque assay. Data are from three independent experiments performed on triplicate wells with mean PFU/mL displayed. (F and G) The same infected cells from D and E were collected in TRIzol and NS1 mRNA transcript analyzed by qRT-PCR. Input levels were normalized to GAPDH and fold change of transcript was determined relative to DMSO control. (H) Using the same extracted RNA as in G, an M-RTPCR protocol was used to amplify all IAV segments which were analysed on an agarose gel. Displayed are the amplifications from two independent wells for UMB18-2 at 50 μM and 10 μM and three wells for DMSO controls. (I) Infected and treated cells were also collected in RIPA lysis buffer and used for Western blotting of NS1. Displayed is a representative blot of the three independent repeats for each compound.
Since the SKI complex is involved in RNA metabolism, we directly investigated whether treatment with our lead compounds had an impact on viral RNA production, and by extension, viral protein production. Cells were treated with UMB18 or UMB18-2 and infected with IAV at MOI 3, to ensure all cells in the plate would be infected. After 8 h, supernatant was collected and cells were collected in TRIzol or RIPA lysis buffer for analysis of viral titer (Fig. 6 D and E) mRNA production (Fig. 6 F–H), and protein production (Fig. 6I). At an MOI 3 for an 8-h infection, neither concentration of either drug inhibited viral production (Fig. 6 D and E); however, there was dose-dependent inhibition of viral RNA production using NS1 as the reporter gene (Fig. 6 F and G). To further assess loss of IAV RNA we used a multisegment RT-PCR approach (M-RTPCR) (21) to amplify all segments of the IAV genome. We found that from cells treated with UMB18-2 there was a total loss of IAV RNA that could be amplified and detected by this protocol compared to DMSO controls (Fig. 6H), further suggesting that our compounds inhibit viral replication by inhibiting RNA production. In agreement with the lack of mRNA, a lack of NS1 protein was also observed when cells were treated with UMB18 or UMB18-2 (Fig. 6I). Overall, these data suggest that the mechanism of activity for our lead compounds is inhibition of viral RNA production.
SKI Complex Targeting Compounds Have Broad Anticoronavirus Activity.
Finally, we wanted to further extend our knowledge about the breath of antiviral activity by assessing the other highly pathogenic human coronaviruses, SARS-CoV and SARS-CoV-2. These two additional coronaviruses utilize a different cell surface receptor to MERS-CoV and there is therefore different cell line permissivity. Huh7 cells were used for the MERS-CoV work described in previous figures, but neither SARS-CoV nor SARS-CoV-2 infect these cells. The receptor for both of these additional coronaviruses is ACE2 (22–25). We utilized both A549 and Huh7 cells stably expressing ACE2 for SARS-CoV infections and treatment with UMB18-2. Virus was collected after 48 h from A549-ACE2 (where we found peak viral replication to be) or after 24 h from Huh7-ACE2 (to match the work in MERS-CoV infections) (Fig. 4) and titered by TCID50 assay. SARS-CoV was inhibited by UMB18-2, with 50 µM treatments showing large reduction in virus production and 10 µM showing more mild inhibition (Fig. 7 A–C).
Fig. 7.
UMB18-2 inhibits all highly pathogenic human coronaviruses in multiple cell lines. (A) A549-ACE2 cells were infected with SARS-CoV at MOI 0.1 and treated with UMB18-2 at 50 µM or 10 µM (with 0.5% or 0.1% DMSO being the appropriate negative controls) for 48 h. Supernatant was collected and used for TCID50 assay to determine viral titer. Mean TCID50/mL and SD are displayed from three independent experiments performed in triplicate. (B) As in A but infection at MOI 0.01. (C) Huh7-ACE2 cells were infected with SARS-CoV at MOI 0.1 and treated with UMB18-2. Viral production was analyzed as in A but after 24 h of infection. (D) Vero E6 cells were infected with SARS-CoV-2 at MOI 0.1 and treated with UMB18-2 as in A. Viral production was analyzed as in A, but after 24 h of infection. (E and F) As described in A and B, but with SARS-CoV-2 infection. (G and H) Cells that were infected in E and F were collected in TRIzol and used for qRT-PCR analysis. Primers targeting RdRp were used. Input levels were normalized to GAPDH RNA and fold change of transcript levels was determined relative to DMSO control for each concentration of compound. Data are from a representative experiment. (I and J) A549-DPP4 cells were infected with MERS-CoV in the same way as described for A and B. (K and L) MERS-CoV RNA analysis as described for G and H. In all cases t tests were performed for vehicle control vs. drug treated samples; *P < 0.05, **P < 0.01, ****P < 0.0001.
SARS-CoV-2 emerged during the preparation of this manuscript and there are still many unknowns regarding this virus. Interestingly, even though the virus utilizes the same cell surface receptor as SARS-CoV, we found that no infectious virus particles were released from the Huh7-ACE2 cells and therefore we used the same A549-ACE2 and Vero E6 cells to test in two different cell lines. When Vero cells were infected with SARS-CoV-2 at MOI 0.1 and treated with UMB18-2 for 24 h (Fig. 7D) there was significant inhibition of infectious virus production as measured by TCID50. In the A549-ACE2 cells there was a significant reduction in SARS-CoV-2 replication at either MOI 0.1 (Fig. 7E) or 0.01 (Fig. 7F) at both 50 µM and 10 µM. Similarly to IAV, treatment with UMB18-2 inhibited SARS-CoV-2 mRNA production (using RdRp as the target gene) (Fig. 7 G and H).
Finally, we wanted to further extend our MERS-CoV data and validate our results from Huh7 cells in an additional cell line. The MERS-CoV receptor is DPP4, so A549 cells stably expressing this protein were infected at MOI 0.1 or 0.01 for 48 h similarly to SARS-CoV and SARS-CoV-2 experiments. These data further confirm that MERS-CoV replication is inhibited by UMB18-2 treatment and that RNA production is inhibited (Fig. 7 I–L) as seen for IAV and SARS-CoV-2. Overall, these data suggest that SKI-targeting compounds may have broad antiviral activity against coronaviruses, targeting the three human pathogenic viruses of the family along with having antiviral activity against influenza and filoviruses, in various cell lines.
Discussion
The SKI complex is an RNA helicase complex that is a cofactor for the cytosolic RNA exosome and involved with various aspects of RNA metabolism (1–4). There are data demonstrating that the enzymatic subunit of the SKI complex is involved in regulation of the IFN response (11) and the RNA exosome has been linked with replication of various RNA viruses (7–10), but the role of the SKI complex in viral replication is poorly defined. Using a yeast suppressor screen, we identified a genetic interaction between proteins of IAV and MERS-CoV with the SKI complex, which developed into our identification of the SKI complex as a potential antiviral target. We have identified three chemical compounds that display broad-spectrum antiviral activity, with our lead compounds inhibiting influenza, all three pathogenic human coronaviruses and filoviruses, all of which cause significant human morbidity and mortality.
We investigated the role of the SKI complex in viral replication because of our work in yeast that suggested a genetic interaction between viral proteins and the yeast protein complex. We previously used suppressor screening to identify SIRT1 as a proviral factor for MERS-CoV replication (13). Here, we build on that work and added in screening data for IAV NS1. We found that both viral proteins coimmunoprecipitated with the component proteins of the mammalian SKI complex opening interesting avenues for future research, such as how this interaction may be beneficial for viral replication and how our identified compounds are disrupting this. We would predict other viruses have proteins that could similarly interact with the SKI complex since it appears to be involved in replication of multiple viral families.
Our data demonstrate that the mechanism of antiviral activity is an inhibition of viral RNA production. We arrive at this conclusion through multiple avenues as we directly demonstrate that there is a reduction in production of viral RNA for IAV, MERS-CoV, and SARS-CoV-2. Moreover, our time-of-addition experiments show reduced antiviral activity when compounds were added 2 and 6 h after infection with IAV. Data suggest that fusion of IAV with endosomes for entry occurs around 1.5 h postinfection (26), while replication of viral RNA reaches a peak around 6 h (20). Since addition of our chemicals at 2 h postinfection still caused a 1-log drop in virus production, we conclude that entry is not the main target for inhibition. However, there is a near complete loss of antiviral activity of UMB18-2 when added at 6 h postinfection, suggesting RNA production is targeted. The SKI complex has been suggested to play a role in the IFN response (11) and is an RNA helicase. We therefore hypothesize that either there is direct degradation of viral RNA when the SKI complex is disrupted or that the IFN response is activating antiviral pathways that inhibit viral RNA production. We can also not rule out roles of the SKI complex in protein translation since translation and transcription are intimately linked, especially for negative-sense RNA viruses such as influenza. These questions are the source of ongoing research.
At this stage, our compounds that display broad antiviral activity are only modeled to interreact with the SKI complex. We find that loss of the SKI complex through siRNA targeting removes the antiviral activity of UMB18-2 (Fig. 4G), suggesting a requirement for the complex to produce the observed antiviral activity. Future work is aimed at further analyzing whether the compounds bind directly to the SKI complex using structural biology approaches and investigating whether other roles of the SKI complex are impacted.
Viral infection can have a major burden on human health. Influenza has historically caused numerous large epidemics and pandemics such as 1918 Spanish ‘flu and 2009 swine ‘flu. Ebola has causes sporadic outbreaks since the 1970s, but in recent years these have been growing in scale. The 2014 West Africa Ebola outbreak saw over 28,000 people contract the disease causing over 11,000 deaths. Coronaviruses have always posed a threat of mass spread because of their respiratory transmission. In 2002 to 2003, the emergence of SARS-CoV infected over 8,000 people, killing around 10% in 9 mo, while MERS-CoV has sporadically spread since 2012, causing around 2,500 infections with a case fatality rate of around 35%.
The year 2020 has seen the rapid emergence of a novel human coronavirus, SARS-CoV-2, which rapidly spread after its identification in Wuhan, China, became a pandemic, and has infected over 40 million people and counting. The outbreaks mentioned above, and in particular this current pandemic, highlight the huge lack of broadly effective antiviral therapeutic options available for treatment. Currently, we are scrambling to identify drugs that may be able to inhibit SARS-CoV-2 and treat COVID-19 patients. As the global population increases and invades formerly uninhabited environments there will be more zoonotic viruses to emerge and spread in the human population; therefore developing broad-spectrum antiviral therapeutics is imperative. Two complimentary approaches for this are to find antivirals that target multiple viruses, such as nucleotide analogs (e.g., remdesivir) (27, 28), and antivirals that target the host. Combination therapy is a highly effective strategy to limit viral resistance as clearly demonstrated for HIV-1 with highly active anti-retroviral therapy and having multiple broad-spectrum approaches will be a powerful way to combat viral infection in the future.
Here, we demonstrate the SKI complex is a potential broad-spectrum, host-directed, antiviral target. Our lead compounds indicate that the SKI complex is druggable and compounds targeting this complex inhibit three distinct families of viruses that cause significant disease in humans. By developing broadly acting antiviral drugs now, we will be prepared to respond quickly and effectively to the next virus to emerge, no matter the origin.
Methods
Plasmids and Compounds.
Genes from influenza were synthesized by Biobasic Inc. using sequence information for the H1N1/CAL09 strain. Genes were cloned into a modified pRS413 plasmid containing a GAL1 promoter. See ref. 13 for further detail on the yeast plasmid and cloning. The ORF4a-GFP pCAGGS plasmid for mammalian cell expression was described in ref. 13. NS1-GFP was produced by cloning on the EcoR1/Xma1 sites between pRS413 and pCAGGS. For production of SKI-HA pCAGGS plasmids see SI Appendix. All SKI targeting compounds were purchased from the ChemBridge Hit2Lead library.
Yeast.
See ref. 13 for experimental details on yeast and SI Appendix.
Mammalian Cell Culture.
A549 and Huh7 cells were cultured in DMEM (Dulbecco's Modified Eagle Medium; Quality Biological), supplemented with 10% (vol/vol) fetal bovine serum (FBS; Sigma) and 1% (vol/vol) penicillin/streptomycin (pen/strep), 10,000 U/mL/10 mg/mL (Gemini Bio-Products). Vero E6 cells were cultured in DMEM supplemented with FBS and pen/strep, as A549 and Huh7, but additionally supplemented with 1% (vol/vol) l-glutamine (2 mM final concentration, Gibco). Cells were maintained at 37 °C and 5% CO2.
Viruses.
Details on stock production are provided in SI Appendix. Influenza A virus NL09 strain was a kind gift from Florian Kramer, Icahn School of Medicine at Mt. Sinai, New York, NY. MERS-CoV (Jordan strain: GenBank accession no. KC776174.1, strain MERS-CoV-Hu/Jordan-N3/2012). SARS-CoV MA15 has been described previously (29). Samples of SARS-CoV-2 were obtained from the CDC following isolation from a patient in Washington State (WA-1 strain: BEI #NR-52281). All coronavirus work was performed in a Biosafety level 3 laboratory and approved by our Institutional Biosafety Committee. Influenza work was performed at Biosafety level 2. The EBOV used in these studies was the Makona isolate, Ebola virus/H.sapiens-tc/GIN/14/WPG-C05 (EBOV/Mak, GenBank accession no. KP096420); this isolate was obtained from Gary Kobinger, Public Health Agency of Canada, Winnipeg, MB, Canada. The MARV used in these studies was the Angola isolate, Marburg virus/H.sapiens-tc/AGO/2005/Ang-1379v (MARV/Ang, BioSample accession no. SAMN05916381). EBOV and MARV stocks were generated as previously described (30). All procedures using live EBOV or MARV were performed under Biosafety level 4 conditions.
Viral Infections.
Details of viral infections are in SI Appendix.
siRNA Knockdown.
Cells were transfected with indicated siRNA purchased from Sigma using their Rosetta prediction algorithm and purchasing the top three ranked siRNA sequences. Scrambled siRNA was used as a control (MISSION siRNA Universal Negative Control #1 [Sigma]). Transfection protocol is detailed in SI Appendix.
RNA Extraction and qRT-PCR.
Cells were collected in TRIzol and RNA was extracted using Direct-zol RNA Miniprep Kit (Zymo Research) as per the manufacturer’s instructions. RNA was converted to cDNA using RevertAid RT Kit (Thermo Scientific), with 12 μL of extracted RNA per reaction. For qRT-PCR, 2 μL of cDNA reaction product was mixed with PowerUp SYBR Green Master Mix (Applied Biosystems) and gene-specific primers (SI Appendix, RT-PCR-Primers section). To normalize loading, GAPDH or 18S were used as housekeeping genes (18S was analyzed by TaqMan Gene Expression Assays (Applied Biosystems) and TaqMan Fast Advanced Master Mix). Fold change between drug treated and vehicle control was determined by calculating ΔΔCT after normalization to the housekeeper gene. Primer sequences are listed in SI Appendix.
M-RTPCR.
See ref. 21 for full detail of the M-RTPCR protocol. Briefly, A549 cells were infected with IAV at MOI 3 for 8 h and treated with UMB18-2 or DMSO control. Cells were collected in TRIzol and RNA was extracted and converted to cDNA as detailed above. From this reaction, 2 μL of cDNA was used in a PCR using Phusion Flash PCR Master Mix (Thermo Scientific) and M-RTPCR primers, MBTuni-12 (5′-ACGCGTGATCAGCAAAAGCAGG-3′) and MBTuni-13 (5′-ACGCGTGATCAGTAGAAACAAGG-3′). The reaction product was then separated on an agarose gel and imaged with a BioRad ChemiDoc system.
Western Blotting.
Western blots were performed as described in ref. 13. Primary antibodies used are as follows: rabbit anti-SKIV2L (61 μg/150 μL, Proteintech), rabbit anti-H1N1 NS1 (0.5 mg/mL, Genscript), and mouse anti-tubulin (clone DMA1A, Sigma). SKIV2L and tubulin-targeting antibodies were diluted 1:1,000 and NS1 targeting antibodies were diluted 1:300 for use. Secondary antibodies were used as follows: goat anti-rabbit horseradish peroxidase (HRP) (0.8 mg/mL, Thermo Scientific) and goat anti-mouse Alexa Fluor 546 (2 mg/mL, Life Technologies). HRP-conjugated secondary antibodies were diluted 1:10,000 and fluorescent secondary antibodies were diluted 1:2,000.
Coimmunoprecipitation.
Transfections and coimmunoprecipitation were performed as described in ref. 13.
CellTiter-Glo Assays.
Cells were plated in opaque 96-well plates 1 d prior to siRNA transfection. Plates were collected on days 1, 2, and 3 posttransfection and used for CellTiter-Glo Luminescent Cell Viability Assay (Promega) as per the manufacturer’s instruction. Luminescence was read using a Synergy HTX Multi-Mode plate reader. For assessing viability of cells treated with compounds, cells were plated 1 d prior to use and treated for 24 h prior to being used in CellTiter-Glo assays.
Computational Modeling.
Please see SI Appendix for full details on computational modeling.
Statistical Analysis.
All statistical analysis was performed with GraphPad Software and details are provided in the figure legends.
Supplementary Material
Acknowledgments
We kindly thank Emergent BioSolutions for financial support to perform these experiments. We also thank Dr. Natalie Thornburg (CDC) for the SARS-CoV-2 (WA1) strain. M.B.F. was supported in part by R21 AI153480-01. A.D.M. was supported by NIH GM131710 and the University of Maryland Baltimore Computer-Aided Drug Design Center. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services (DHHS) or of the institutions and companies affiliated with the authors. This project has been partially funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, to Laulima Government Solutions, LLC contract No. HHSN272201800013C and Battelle Memorial Institute former contract No. HHSN272200700016I. A.A., B.B., and C.C. performed this work as previous employees of Battelle Memorial Institute and current employees of Laulima Government Solutions, LLC. Subcontractors to Laulima Government Solutions, LLC who performed this work are: D.D., E.E., and F.F., all employees of Tunnell Government Services, Inc.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012939117/-/DCSupplemental.
Data and Materials Availability.
All study data are included in the article and supporting information.
References
- 1.Houseley J., Tollervey D., The many pathways of RNA degradation. Cell 136, 763–776 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Schneider C., Tollervey D., Looking into the barrel of the RNA exosome. Nat. Struct. Mol. Biol. 21, 17–18 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Johnson S. J., Jackson R. N., Ski2-like RNA helicase structures: Common themes and complex assemblies. RNA Biol. 10, 33–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Januszyk K., Lima C. D., The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 24, 132–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toh-E A., Guerry P., Wickner R. B., Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136, 1002–1007 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widner W. R., Wickner R. B., Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol. 13, 4331–4341 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rialdi A., et al. , The RNA exosome syncs IAV-RNAPII transcription to promote viral ribogenesis and infectivity. Cell 169, 679–692.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aly H. H., et al. , RNA exosome complex regulates stability of the hepatitis B virus X-mRNA transcript in a non-stop-mediated (NSD) RNA quality control mechanism. J. Biol. Chem. 291, 15958–15974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiromoto F., et al. , IL-1β/ATF3-mediated induction of Ski2 expression enhances hepatitis B virus x mRNA degradation. Biochem. Biophys. Res. Commun. 503, 1854–1860 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Molleston J. M., et al. , A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation. Genes Dev. 30, 1658–1670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckard S. C., et al. , The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat. Immunol. 15, 839–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guvench O., MacKerell A. D. Jr, Computational fragment-based binding site identification by ligand competitive saturation. PLoS Comput. Biol. 5, e1000435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weston S., et al. , A yeast suppressor screen used to identify mammalian SIRT1 as a proviral factor for Middle East respiratory syndrome coronavirus replication. J. Virol. 93, e00197-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basu D., et al. , Novel influenza virus NS1 antagonists block replication and restore innate immune function. J. Virol. 83, 1881–1891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halbach F., Reichelt P., Rode M., Conti E., The yeast ski complex: Crystal structure and RNA channeling to the exosome complex. Cell 154, 814–826 (2013). [DOI] [PubMed] [Google Scholar]
- 16.MacKerell A. D. Jr, Jo S., Lakkaraju S. K., Lind C., Yu W., Identification and characterization of fragment binding sites for allosteric ligand design using the site identification by ligand competitive saturation hotspots approach (SILCS-Hotspots). Biochim. Biophys. Acta Gen. Subj. 1864, 129519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu W., Lakkaraju S. K., Raman E. P., Fang L., MacKerell A. D. Jr, Pharmacophore modeling using site-identification by ligand competitive saturation (SILCS) with multiple probe molecules. J. Chem. Inf. Model. 55, 407–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koes D. R., Camacho C. J., Pharmer: Efficient and exact pharmacophore search. J. Chem. Inf. Model. 51, 1307–1314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen L. M., et al. , FDA-approved selective estrogen receptor modulators inhibit ebola virus infection. Sci. Transl. Med. 5, 190ra79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laske T., et al. , Model-based analysis of influenza A virus replication in genetically engineered cell lines elucidates the impact of host cell factors on key kinetic parameters of virus growth. PLoS Comput. Biol. 15, e1006944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B., Wentworth D. E., Influenza A virus molecular virology techniques. Methods Mol. Biol. 865, 175–192 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Li W., et al. , Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, e00127-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M., et al. , SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee I., Yamauchi Y., Helenius A., Horvath P., High-content analysis of sequential events during the early phase of influenza A virus infection. PLoS One 8, e68450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown A. J., et al. , Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 169, 104541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheahan T. P., et al. , Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, eaal3653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts A., et al. , A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3, e5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong Y., et al. , Evaluation of the activity of lamivudine and zidovudine against ebola virus. PLoS One 11, e0166318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.