Significance
Schaftoside and isoschaftoside are important plant defense chemicals and bioactive natural products widely present in higher plants. Although a number of plant CGTs have been reported, we know very little about the CGTs catalyzing the biosynthesis of flavonoid di-C-glycosides with different sugar residues. Here we reveal the biosynthesis of (iso)schaftosides in plants is sequentially catalyzed by a pair of homologous but functionally different enzymes (CGTa and CGTb). Crystal structure analysis and key amino acids mutagenesis could switch the function of SbCGTb to SbCGTa. We further reveal this pathway is general for higher plants. Our results provide a platform to efficiently synthesize (iso)schaftosides and to understand their plant defense mechanisms.
Keywords: flavonoid C-glycoside, C-glycosyltransferase, schaftoside, crystal structure, catalytic mechanism
Abstract
Schaftoside and isoschaftoside are bioactive natural products widely distributed in higher plants including cereal crops and medicinal herbs. Their biosynthesis may be related with plant defense. However, little is known on the glycosylation biosynthetic pathway of these flavonoid di-C-glycosides with different sugar residues. Herein, we report that the biosynthesis of (iso)schaftosides is sequentially catalyzed by two C-glycosyltransferases (CGTs), i.e., CGTa for C-glucosylation of the 2-hydroxyflavanone aglycone and CGTb for C-arabinosylation of the mono-C-glucoside. The two enzymes of the same plant exhibit high homology but remarkably different sugar acceptor and donor selectivities. A total of 14 CGTa and CGTb enzymes were cloned and characterized from seven dicot and monocot plants, including Scutellaria baicalensis, Glycyrrhiza uralensis, Oryza sativa ssp. japonica, and Zea mays, and the in vivo functions for three enzymes were verified by RNA interference and overexpression. Through transcriptome analysis, we found homologous genes in 119 other plants, indicating this pathway is general for the biosynthesis of (iso)schaftosides. Furthermore, we resolved the crystal structures of five CGTs and realized the functional switch of SbCGTb to SbCGTa by structural analysis and mutagenesis of key amino acids. The CGT enzymes discovered in this paper allow efficient synthesis of (iso)schaftosides, and the general glycosylation pathway presents a platform to study the chemical defense mechanisms of higher plants.
Flavonoid C-glycosides are an important class of bioactive natural products in plants (1–3). Schaftoside and isoschaftoside are a pair of flavonoid di-C-glycosides (Fig. 1A). They possess a variety of biological activities, including antirespiratory syncytial virus, antidiabetic, antihypertensive, hepatoprotective, anti-inflammatory, and antioxidant activities to mammals, indicating their potential applications as drugs or dietary supplements (4–6). While most flavonoid C-glycosides only occur in certain plant families, schaftoside and isoschaftoside are popularly present in higher plants. They have been reported in, at least, 184 plant species of 39 families (SI Appendix, Fig. S1A and Table S1). Particularly, they are present in cereal crops, such as rice, maize, wheat, sorghum, as well as in medicinal herbs, such as licorice root (G. uralensis) and baical skullcap root (S. baicalensis) (7, 8). Moreover, (iso)schaftosides are important plant defense compounds. Their production is related with drought tolerance of wheat leaves and ultraviolet resistance of rice seeds (9, 10). They could kill root-knot nematode (Meloidogyne incognita) and the brown planthopper (Nilaparvata lugens) (11, 12), and inhibit the parasitic witchweed Striga hermonthica (13). The amounts of (iso)schaftosides increased significantly in rice leaves after brown planthopper infestation (14). We also found the contents of (iso)schaftosides in rice shoots increased by around 40% during 6 and 24 h after penetration of the rice blast fungus Magnaporthe oryzae (SI Appendix, Fig. S2).
Fig. 1.
The biosynthetic pathway for (iso)schaftosides in S. baicalensis and functional characterization of SbCGTa and SbCGTb. (A) The structures of schaftoside (4a, apigenin 6-C-β-d-glucoside-8-C-α-l-arabinopyranoside) and isoschaftoside (4b, apigenin 6-C-α-l-arabinopyranoside-8-C-β-d-glucoside). (B) Pictures of the S. baicalensis plant and the Chinese herbal medicine Huang-Qin. (C) High-performance liquid chromatography (HPLC) chromatograms for functional characterization of SbCGTa and SbCGTb with 2 and 3 as substrates. (D) Mass spectra and tandem mass spectrometry (MS/MS) fragmentations of 3 and 4. (E) A proposed biosynthetic pathway for schaftoside and isoschaftoside. ∇, the products were purified in this study and identified by NMR; Δ, the products were identified by comparing with reference standards; *, the NMR spectral data were reported in this work. The 1, naringenin; 2, 2-hydroxynaringenin; 3, 2-hydroxynaringenin mono-C-glucoside; 3a, isovitexin; 3b, vitexin; 4, 2-hydroxynaringenin 6,8-di-C-glucoside-arabinoside. Glc, glucosyl residue; Ara, arabinosyl residue.
To dissect the biosynthetic pathway of (iso)schaftosides is critical to understand the chemical defense mechanisms of plants and may facilitate the medicinal use of (iso)schaftosides. Although biosynthesis of the aglycone apigenin has been extensively studied, little is known about the di-C-glycosylation pathway (15). The glycosylation reactions are usually catalyzed by CGTs. Due to low amino acid sequence similarity of CGTs, only 44 functional CGTs have been characterized from plants, thus far (16–27, SI Appendix, Table S2). These CGTs can be classified into two types (SI Appendix, Fig. S1B). The first type directly transfers one glucosyl residue to C-6 or C-8 of a flavone or isoflavone aglycone (20–23). At least, 38 CGTs belong to the second type, which could attach one glucosyl residue to a 2-hydroxyflavanone aglycone to form a flavone C-glycoside after dehydration. Among them, a few CGTs, including GgCGT we recently discovered from Glycyrrhiza glabra, could attach two glucosyl residues to form di-C-glucosides (19). However, no CGTs have been reported to catalyze the biosynthesis of (iso)schaftosides, which contain glucose and arabinose (Ara) residues at C-6 and C-8. It is unknown whether (iso)schaftosides are catalyzed by the first or second type CGTs, and whether the two different sugar residues are transferred by one or two CGTs. Moreover, almost all of the known plant CGTs prefer uridine 5′-diphosphate glucose (UDP-Glc) as the sugar donor, and very few CGTs have high catalytic efficiency with UDP-Ara.
In this paper, we report a two-step di-C-glycosylation pathway sequentially catalyzed by two 2-hydroxyflavanone-type CGTs, i.e., CGTa and CGTb, for the biosynthesis of (iso)schaftosides. The CGTa/b pairs were characterized in seven monocots and dicots, and homologous genes were discovered in 119 other higher plants. Furthermore, we resolved the crystal structures of five CGTa/b enzymes to elucidate mechanisms for their different catalytic features.
Results
Molecular Cloning and Characterization of SbCGTa and SbCGTb in S. baicalensis.
To discover the CGTs catalyzing the biosynthesis of (iso)schaftosides, we initially analyzed the transcriptome data of the medicinal plant S. baicalensis, the roots of which are used as the famous Chinese herbal medicine Huang-Qin (Fig. 1B). Ten typical CGT sequences were used as templates (SI Appendix, Table S2). The BLAST search yielded two candidate genes, SbCGTa and SbCGTb (SI Appendix, Table S3). They share low identity (∼22%) with Scutellaria O-glycosyltransferases (28, 29) but high identity (∼43%) with known 2-hydroxyflavanone CGTs (FcCGT and CuCGT) (27). The genes were cloned into the pET28a(+) vector and expressed in the BL21(DE3) Escherichia coli strain. The recombinant proteins were purified by Ni-NTA affinity chromatography for functional characterization. Interestingly, although SbCGTa and SbCGTb share high sequence identity (62%), they showed remarkably different catalytic features in sugar donor and acceptor selectivities. At the presence of UDP-Glc, SbCGTa efficiently catalyzed 2-hydroxynaringenin (2) into its mono-C-glucoside (3) but could not further convert 3 into 4 when UDP-Ara was added (Fig. 1 C and D and SI Appendix, Fig. S3). In contrast, SbCGTb preferred UDP-Ara to UDP-Glc as a sugar donor and efficiently converted 3 to 4 at the presence of UDP-Ara. With the addition of hydrochloric acid (150 mM) (17), 4 was completely converted into schaftoside (4a) and isoschaftoside (4b). Then we measured the kcat/Km values (catalytic efficiency) for SbCGTa and SbCGTb with different substrates. Consistent with the functional characterization results, the kcat/Km value (1.529 ± 0.160 μM−1 s−1) for SbCGTa with 2-hydroxynaringenin and UDP-Glc was 2,316 times higher than the value with 2-hydroxynaringenin mono-C-glucoside and UDP-Glc, and 339 times higher than 2-hydroxynaringenin and UDP-Ara. For SbCGTb, the kcat/Km value (0.128 ± 0.012 μM−1 s−1) with 2-hydroxynaringenin mono-C-glucoside and UDP-Ara was 213 times higher than with 2-hydroxynaringenin and UDP-Ara, and 118 times higher than 2-hydroxynaringenin mono-C-glucoside and UDP-Glc (SI Appendix, Fig. S4). The above results indicated the biosynthesis of (iso)schaftosides involved two steps of enzymatic C-glycosylation reactions, which were sequentially catalyzed by SbCGTa and SbCGTb. To our knowledge, this is a previously unidentified di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides.
The Biosynthetic Pathway of (iso)schaftosides in S. baicalensis.
Naringenin (1) is a key precursor for the biosynthesis of (iso)schaftosides. We cloned the flavanone 2-hydroxylase (SbF2H, previously reported as SbFNSII-2, GenBank accession No. KT963454, ref. 30) and coexpressed it with SbCGTa in yeast to produce 3a and 3b (SI Appendix, Fig. S5). Based on these results, we proposed the biosynthetic pathway of (iso)schaftosides in S. baicalensis as shown in Fig. 1E. The underground part of S. baicalensis mainly contained 4′-dehydroxylated derivatives of (iso)schaftosides, i.e., 4′a and 4′b, which may be biosynthesized by the same SbCGTa/b enzymes (SI Appendix, Fig. S6 A and B). The structures for most of the biosynthetic intermediates were fully identified by NMR spectroscopy and high-resolution mass spectrometry or by comparing with reference standards (SI Appendix, Figs. S7–S11).
CGTa/b Enzymes in Other Plants.
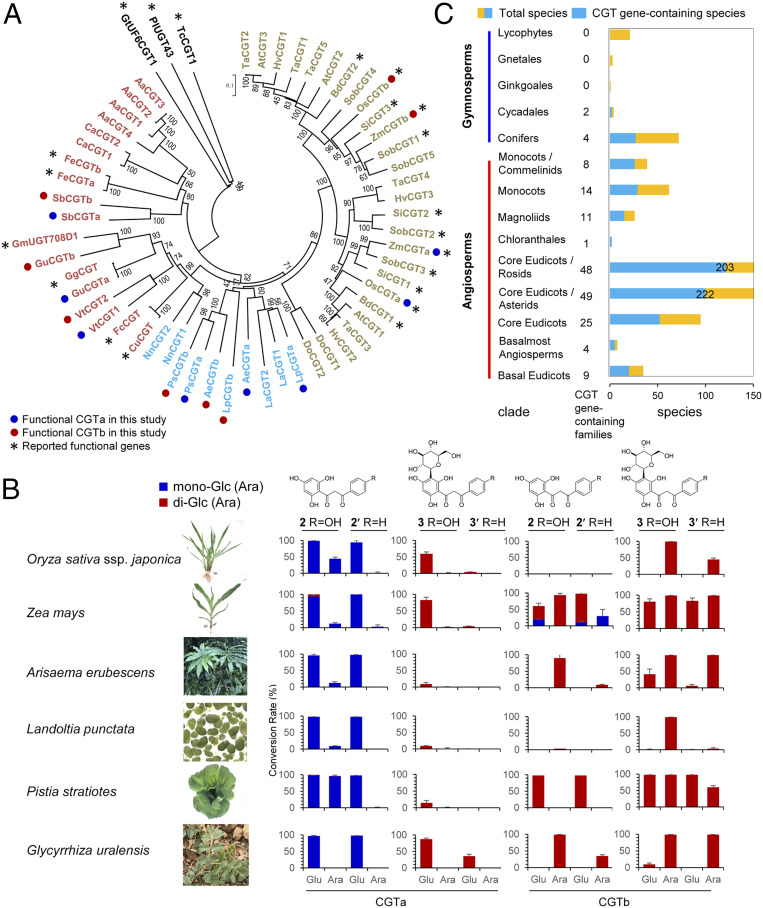
To explore the presence of similar CGTa/b enzymes in other plants, we analyzed the transcriptomes of 19 (iso)schaftosides-containing species. The gene sequences of ten known C-glycosyltransferases and SbCGTa/b were used as templates for the BLAST search. Together with SbCGTa/b, a total of 49 open reading frames (ORFs) were discovered (Fig. 2A and SI Appendix, Table S3). Then we extracted RNA from six plant species, including O. sativa ssp. japonica, Z. mays, Arisaema erubescens, Pistia stratiotes, Landoltia punctata, and G. uralensis. The 12 corresponding ORFs were cloned and expressed in E. coli, and the functions were characterized by in vitro enzyme activity assay. Similar to SbCGTa/b, the CGTa/b enzymes from the same plant share high sequence identity (49–68%) but exhibited remarkably different catalytic features. OsCGTa, ZmCGTa, AeCGTa, LpCGTa, PsCGTa, and GuCGTa showed similar functions to SbCGTa by converting 2 into the mono-C-glucoside 3. On the other hand, OsCGTb, ZmCGTb, AeCGTb, LpCGTb, PsCGTb, and GuCGTb showed similar catalytic features as SbCGTb and utilized UDP-Ara to produce 4 (Fig. 2B and SI Appendix, Fig. S12). All of the above CGT enzymes were reported in this work, except for OsCGTa, OsCGTb, ZmCGTa, and ZmCGTb, which had been previously reported as UGT708A3, UGT708A2, UGT708A6, and UGT708A11, respectively (18). These results indicate the biosynthetic pathway of (iso)schaftosides is common for the seven dicot and monocot plants.
Fig. 2.
CGTa and CGTb enzymes catalyze the biosynthesis of (iso)schaftosides in different plants. (A) Phylogenetic analysis of 49 CGT genes discovered from 19 plants (SI Appendix, Table S3) and previously reported functional CGT genes. Enzymes labeled in the tawny color represent 2-hydroxyflavanone CGTs from monocots (terrestrial); cyan, from aquatic plants; red, from dicots (terrestrial). The three non-2-hydroxyflavanone CGTs were labeled in black. (B) Functions of CGTs from six plant species, and their sugar donor and acceptor selectivities. The HPLC chromatograms are given in SI Appendix, Fig. S12. (C) Distribution of 2-hydroxyflavanone CGT genes in higher plants based on CNGB search.
The Two-Step di-C-glycosylation Pathway Is General for Higher Plants.
To further discover homologous genes of CGTa/b in an even broader spectrum of higher plants, we analyzed the transcriptome data of 1,163 species from 487 families in the CNGB 1KP database (31) using the conserved “DPF” motif for previously reported 2-hydroxyflavanone CGTs (16–19, 32). A total of 648 potential CGT genes were discovered in 399 species, including angiosperms (370/692 species, 53.5%) and gymnosperms (29/101 species, 28.7%) (Fig. 2C). Among them, 119 plants contained, at least, two candidate genes, which may correspond to CGTa/b enzymes (SI Appendix, Table S4). All of the 282 genes from these 119 plants were clustered with the UGT708 family in the phylogenetic tree and could be 2-hydroxyflavanone CGTs (SI Appendix, Fig. S13). These results suggest the two-step di-C-glycosylation pathway is general for the biosynthesis of (iso)schaftosides in higher plants.
CGTa/b Contribute to the Biosynthesis of (iso)schaftoside In Vivo.
To verify the functions of CGTa and CGTb enzymes in plants, first we studied the correlation between gene expression and (iso)schaftoside contents in S. baicalensis (Fig. 3A). During the period from day 50–75, the leaves of S. baicalensis contained relatively high amounts of (iso)schaftosides. This trend was consistent with the high expression of SbCGTa/b genes on day 75. On day 120, both the gene expressions and the chemical levels decreased remarkably (Fig. 3 B and C). The roots of S. baicalensis exhibited almost the same time-course pattern for pinocembrin C-glycosides (SI Appendix, Fig. S6C). These results indicate the expression of SbCGTa/b was correlated with the biosynthesis of (iso)schaftosides. Furthermore, we established RNA interference (RNAi) and overexpression hairy root systems to confirm the in vivo functions of CGTa/b in S. baicalensis and G. uralensis (SI Appendix, Fig. S14). In the hairy root tissue cultures of the SbCGTb-RNAi line, SbCGTb showed 55% lower expression than the empty vector (EV) control group. Because the hairy roots contained similar chemicals to the plant roots (rich in pinocembrin C-glycosides but not naringenin C-glycosides) (30, 33), we determined the contents of the di-C-glycosides 4′, 4′a, and 4′b. Their concentrations decreased markedly by 85–90% (Fig. 3D). In the meantime, levels of the mono-C-glucosides 3′, 3′a, and 3′b increased significantly. Although the SbCGTa-RNAi line was not established after many attempts, we obtained the RNAi lines for both GuCGTa and GuCGTb in G. uralensis. In the GuCGTa-RNAi line, GuCGTa showed 72% lower expression than the EV group. The contents of 3a and 3b decreased by 69%, and 4a and 4b decreased by 56%. The GuCGTb-RNAi line showed 99% lower expression of GuCGTb than the EV group, and the contents of 4, 4a, and 4b decreased by 77–86% (Fig. 3E). Contents of the substrates 3a and 3b increased by 3.7-fold, and the GuCGTa byproducts 5 and 5a increased by 2.9-fold. In the GuCGTa-overexpression line, GuCGTa showed 24.8-fold higher expression than the EV group, and the contents of 5 and 5a increased by 10.9-fold. In the GuCGTb-overexpression line, GuCGTb showed 20-fold higher expression than the EV group, and the contents of 4, 4a, and 4b increased significantly by 36–86% (Fig. 3F). These results confirmed that CGTa/b sequentially catalyzed the di-C-glycosylation in the biosynthesis of (iso)schaftosides and analogs in plants.
Fig. 3.
The in vivo functions of CGTa and CGTb enzymes in S. baicalensis and G. uralensis. (A) Pictures of S. baicalensis at different growth periods. (B) The contents of secondary metabolites in the biosynthetic pathway of (iso)schaftosides in the leaves of S. baicalensis on day 25, 50, 75, and 120 (n = 3). The solid and dotted lines represent quantification (calibrated with reference standards) and semiquantification (calculated with the regression equations of analogous compounds), respectively (SI Appendix, Table S6). (C) Gene expression levels of SbCGTa and SbCGTb in the leaves of S. baicalensis on day 50, 75, and 120 (n = 3). (D) Gene expressions and compounds concentrations in the SbCGTb-RNAi line and the EV group (n = 3). (E) Gene expressions and compounds concentrations in the GuCGTa/b-RNAi lines and the EV group (n = 5, SI Appendix, Table S5). (F) Gene expressions and compounds concentrations in the GuCGTa/b overexpression lines and the EV group (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the EV group by Student’s t test. The 5, 2-hydroxynaringenin 6,8-di-C-glucoside; 5a, vicenin-2. For the structures of 3′, 3′a, 3′b, 4′, 4′a, 4′b, see SI Appendix, Fig. S6.
Crystal Structures of Five CGTs and the Catalytic Mechanisms.
To explore the catalytic mechanisms of CGTa/b enzymes, we obtained five crystal structures, including SbCGTa•UDP (PDB ID: 6LG0), SbCGTb•UDP-Glc (6LFZ), LpCGTa•UDP (6LG1), LpCGTb (6LFN), and ZmCGTa•UDP (6LF6) with resolutions from 2.05 to 3.0 Å (Fig. 4A and SI Appendix, Fig. S15). All of the structures showed a typical GT-B fold with two Rossmann-like β/α/β domains (34–38, SI Appendix, Fig. S16). For previously reported plant glycosyltransferases, a histidine (His) residue of the 5′-end (between 10 and 30 bp) is critical to initiate the catalytic reaction through deprotonation of the substrate. This residue was mapped to His24 (H24) of SbCGTa and His23 of SbCGTb. The enzyme glycosylation activities could decrease remarkably when these histidine residues were mutated into alanine (A) (SI Appendix, Fig. S17). Analysis of the crystal structures further indicated the catalytic procedures for the di-C-glycosylation reaction. His24 of SbCGTa was close to 2-OH of 2-hydroxynaringenin (2) and could initiate the glycosylation reaction to produce 3. Next, His23 of SbCGTb was close to 6-OH of 3, and initiated the second-step glycosylation to produce 4, which could be further converted into (iso)schaftosides (4a and 4b) after dehydration. This mechanism was similar to that of GgCGT (19).
Fig. 4.
Structural basis for CGTa and CGTb enzymes. (A) Crystal structures of five CGTs. (B) The 11 key differential amino acids between SbCGTa (cyan) and SbCGTb (orange) around the active pocket. (C) The C-glycosylation conversion rates of wild-type (WT) SbCGTa (SbA), SbCGTb (SbB), and mutants. (D) Structural basis for different substrate selectivities between SbCGTb and its mutant (SbCGTb-R94M/I143M/V144T/T145S/H194D/G275T/P374Q). The methods for homology modeling and molecular docking are given in the SI Appendix, Supplementary Text.
Structural Basis for Different Catalytic Features of CGTa/b Enzymes.
To investigate the mechanisms for different catalytic features of CGTa/b enzymes, we superimposed the structures of SbCGTa and SbCGTb. Although the two structures are highly homologous with an rmsd of 0.38 Å, they showed significant differences in 11 amino acids around the active pocket (Fig. 4B). Similar differences were also observed for LpCGTa/b. Next we constructed mutants of SbCGTa and SbCGTb by reciprocally exchanging these 11 residues. While the activity was abolished for the SbCGTa-11aa-mutant, the SbCGTb-11aa mutant showed similar functions as SbCGTa by catalyzing the C-glucosylation of 2 to 3 (Fig. 4C). In the meantime, this mutant lost the native function to catalyze 3 to di-C-glycosides. To further identify the key amino acids of SbCGTb, we constructed a series of single- or multiple-site mutants (Fig. 4C and SI Appendix, Fig. S18). Among them, the SbCGTb-R94M/I143M/V144T/T145S/H194D/G275T/P374Q mutant had almost the same function and Km values as SbCGTa, although its catalytic efficiency (kcat/Km) was relatively low (SI Appendix, Fig. S4). Then we constructed the structure model of this mutant through MODELER and docked 2 with the mutant and SbCGTb, respectively. Structural analysis revealed the mutation could affect the shape of the binding pocket and, thus, affect the binding with the substrate (Fig. 4D). For native SbCGTb, C-3 and 2-OH of 2 were far away from the anomeric carbon of UDP-Glc (5.32 Å) and the key amino acid His23 (4.27 Å), respectively, indicating 2 was not a suitable substrate for SbCGTb. In the mutant model, however, 2 moved closer to the anomeric carbon (4.43 Å) and His23 (2.65 Å) and, thus, gained the function of CGTa.
TS/TA144 as a Sequence Marker for CGTa.
The TS144 or TA144 amino acids were highly conserved in all of the seven functional CGTa enzymes (SI Appendix, Fig. S19). When TS was mutated to SL residues of LpCGTb, the catalytic activities of SbCGTa, LpCGTa, GuCGTa, PsCGTa, and OsCGTa were almost abolished. Molecular docking revealed that T144 could form a hydrogen bond with 6-OH of UDP-Glc, and S145 may stabilize UDP-Glc through hydrophilic effects. The TS/TA rule also applied to the 49 CGT genes of this study and was verified by the functional characterization of VtCGT1 and VtCGT2 from Viola tricolor (SI Appendix, Table S3 and Fig. S20). Therefore, TS/TA144 may be used as a marker to identify CGTa enzymes.
Discussion
Although a number of plant C-glycosyltransferases have been reported, none of them could efficiently synthesize flavonoid di-C-glycosides with different sugar residues. In this study, we found the biosynthesis of schaftoside and isoschaftoside in plants was sequentially catalyzed by two homologous CGTs with different catalytic features, namely, CGTa and CGTb. The CGTa enzymes had the same function as previously reported 2-hydroxyflavanone CGTs by catalyzing mono-C-glucosylation of the aglycone. The CGTb enzymes could further catalyze C-arabinosylation of the mono-C-glucosides, with high selectivity with UDP-Ara. We also preliminarily interpreted mechanisms for the different catalytic selectivities of CGTa and CGTb by crystal structure analysis and mutagenesis of key amino acids.
Both the CGTa and the CGTb enzymes had broad substrate promiscuity and could be used to synthesize diverse di-C-glycosides of flavonoids and other 2ʹ,4ʹ,6ʹ-trihydroxyacetophenone units with glucosyl, arabinosyl, xylosyl, or galactosyl residues at C-6 and C-8 (SI Appendix, Fig. S21). The enzymatic C-glycosylation reactions were simple, green, and regiospecific and were more efficient than chemical methods (39–41). It was noteworthy that all of the seven CGTb enzymes could utilize UDP-Ara and UDP-Xyl as favorite sugar donors, and that most of them could also accept UDP-Glc and UDP-Gal. This sugar donor selectivity was remarkably different from previously reported CGTs. Our studies had shown that GgCGT and TcCGT1 preferred UDP-Glc and UDP-Xyl, and the stereoconfiguration of sugar 4-OH played a critical role for the catalytic efficiency (19, 20). The catalytic mechanisms of CGTb enzymes discovered in this paper warrant to be studied in the future.
It was particularly interestingly to find that the di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides may be general for higher plants. This result explains the widespread distribution of (iso)schaftosides, which was confirmed by qualitative and quantitative liquid chromatography/mass spectrometry (LC/MS) analyses of 29 plant species in this study (SI Appendix, Fig. S22 and Table S5). Given the widespread occurrence and conserved functions, CGTa/b genes may be derived from an ancient ancestor (42, 43). We further discovered that monocots and dicots showed different cluster patterns for CGTa/b genes in the phylogenetic tree (Fig. 2A). For monocots, OsCGTa, ZmCGTa, as well as OsCGTb and ZmCGTb were grouped together. In contrast, the CGTa/b genes from the same plant were respectively grouped together for the dicots. The aquatic plants were located between the monocots and the dicots. The CGTa/b enzymes could be an ideal research target to unveil the mechanisms for molecular evolution of plant secondary metabolism.
In conclusion, we report that the biosynthesis of (iso)schaftosides is sequentially catalyzed by two homologous but functionally different C-glycosyltransferases CGTa and CGTb. The transcriptome analysis of 1,163 species indicates this two-step di-C-glycosylation pathway is general for higher plants. A total of 14 CGTa/b enzymes from seven plants were cloned and characterized, and the in vivo functions of three enzymes were verified in S. baicalensis and G. uralensis by RNAi and overexpression experiments. Based on the crystal structures of five CGTs, we partly interpreted mechanisms for the different substrate selectivities of CGTa/b and established mutants for multiple key amino acids to switch the function of SbCGTb to SbCGTa. The general biosynthetic pathway established in this paper is valuable for plant evolution studies as well as for further understanding on the mechanisms of (iso)schaftosides as plant defense compounds. Moreover, given the noticeable amounts of (iso)schaftosides and their biosynthetic analogs in cereal crops and medicinal herbs, their effects on human health deserve to be further evaluated.
Materials and Methods
Plant Materials and Chemicals.
The seeds of S. baicalensis Georgi, G. uralensis Fisch., Z. mays L., and O. sativa L. ssp. japonica were sown in the laboratory under natural conditions. For RNAi experiments, leaf explants were collected from 3-mo-old S. baicalensis. Aseptic seedlings of G. uralensis were cultured on solid B5 medium (sugar free), and 3-wk-old seedlings were collected for RNAi experiments. Information on the other plants is given in SI Appendix, Table S5. Compounds 1, 1′, and 3a were purchased from Chengdu Must Biotechnology Co., Ltd. (China); 3b, 3′b, 4a, 4b, 4′a, 4′b, and 5a were previously purified and characterized in our laboratory (44); 2 and 2′ were prepared by alkaline hydration of apigenin and chrysin, respectively (24). Details for quantitative chemical analysis of S. baicalensis are given in SI Appendix, Table S6.
Molecular Cloning and Expression of CGTs.
The total RNA was extracted from 10-d-old seedlings using the TransZol method and was used to synthesize the first-stranded complementary DNA (cDNA) using TransScript one-step genomic DNA (gDNA) removal and cDNA synthesis SuperMix (Transgen Biotech, China). The candidate genes were amplified using TransScript KD Plus or Fast-Pfu DNA polymerases (Transgen Biotech, China) and were verified by sequencing. The candidate CGTs were cloned into pET28a(+) vectors (Invitrogen, US) by quick change and homologous recombination method at the BamHI site. The recombinant vectors were transformed into E. coli BL21(DE3) cells for heterologous expression. Single colonies harboring the desired expression construct were inoculated into the LB culture medium (50-μg/mL kanamycin) and were incubated at 37 °C. After optical density at 600 nm reached 0.6, the E. coli cells were incubated with 0.1-mM IPTG at 16 °C for 20 h. The cells were then harvested by centrifugation. The recombinant proteins were purified using a Ni-affinity column (TransGen Biotech, China, SI Appendix, Fig. S23) and were concentrated using Amicon Ultra-15 Ultracel-30K (Merck Millipore, Germany). The mutants of CGTs were obtained by a fast mutagenesis system (Transgen Biotech, China). The gene sequences of SbCGTa/b-11aa mutants and related primers were synthesized by Tsingke Biological Technology (China) (SI Appendix, Table S7).
Functional Characterization of CGTs and Scaled-Up Reactions.
The analytical reactions were carried out in 100-μL systems, containing 50-mM phosphate (pH 8.0), 0.1-mM substrate, 0.25-mM sugar donor, and 10 μg of purified enzyme (30 °C, 2 h). The reactions were terminated with 200-μL methanol. The mixtures were centrifuged at 21,130 × g for 15 min and then analyzed by ultraperformance LC/MS. A 2-μL aliquot of sample was injected for analysis. An Agilent 1290 series ultra-HPLC (UHPLC) system was used (Agilent Technologies, Germany). Samples were separated on an Agilent Zorbax Eclipse Plus C18 column (4.6 × 100 mm, 1.8 μm) protected with an Agilent 1290 Infinity in-line filter. The mobile phase consisted of methanol (A) and water containing 0.1% formic acid (v/v, B). A gradient elution program was used: 0 min, 20% A; 3 min, 45% A; 4 min, 45% A; 8.5 min, 100% A; 12.5 min, 100% A. The flow rate was 0.4 mL/min. The column temperature was 30 °C. The detection wavelength was 300 nm. MS analysis was performed on a Q-exactive hybrid quadrupole-orbitrap mass spectrometer equipped with a heated electrospray ionization (ESI) source (ThermoFisher Scientific, US). The parameters were as follows: sheath gas pressure, 45 arb; auxiliary gas pressure, 10 arb; discharge voltage, 4.5 kV; capillary temperature, 350 °C. The MS resolution was 70,000 full width at half maximum (FWHM), and the MS/MS resolution was 17,500 FWHM.
For scaled-up preparation of 3, 3′, and 4′, the system contained 2-mL buffer (50-mM phosphate, pH 8), 40-μL sugar acceptor (50 mM, dissolved in dimethyl sulfoxide), 50-μL sugar donor (50-mM, UDP-Glc or UDP-Ara), and 100-μg enzyme (SbCGTa or SbCGTb). The reactions were incubated at 30 °C for 2 h and were terminated by methanol. The mixtures were centrifuged at 21,130 g for 15 min and the supernatants were concentrated and dissolved in methanol. The glycosylated products were separated by reversed-phase semipreparative HPLC, and the structures were characterized by high resolution-ESI-MS and NMR.
Biochemical Properties of CGTs.
The biochemical properties of SbCGTa and SbCGTb were investigated using UDP-Glc as the sugar donor and 2-hydroxynaringenin (2) and 2-hydroxynaringenin mono-C-glucoside (3) as the sugar acceptors, respectively. UDP-Ara was not used here due to its high price. The effects of reaction time, pH, temperature, and divalent metal ions were studied (SI Appendix, Fig. S24). Kinetic parameters of the catalytic reactions were calculated using 2 and 3 as the sugar acceptors and UDP-Glc and UDP-Ara as the sugar donors, respectively (SI Appendix, Fig. S4). Assays were performed in a final volume of 50 μL, consisting of 50-mM Na2HPO4-NaH2PO4 buffer (pH 8), 1,000 μM of UDP-Glc/Ara, and different concentrations of sugar acceptor (2.5–200 μM). All of the reactions were conducted at 30 °C for 10 min and then terminated with 100-μL ice-cold methanol and centrifuged at 21,130 g for 15 min for further analysis. The experiments were performed in triplicate, and the mean values were used.
Transcriptome and Phylogenetic Analyses.
The transcriptome data of 19 plants were analyzed (SI Appendix, Table S8). Ten previously reported CGT genes and SbCGTa/b were used as query sequences for BLAST search (SI Appendix, Table S2). A total of 49 candidate CGT genes were screened by the e value (less than e−21) and the conserved domain (DPF) as shown in SI Appendix, Table S3. In addition, potential CGT genes were searched using the 1,000 Plant Transcriptomes Project of China National GeneBank (https://www.cngb.org). A total of 22 genes was selected as query sequences, including the 12 reported CGT genes and the 10 new genes identified in this study. A total of 2,596 potential genes was obtained through BLASTX using the default parameters. These sequences were aligned using MEGA6 software and were screened using the conserved DPF domain (Dataset S1). Molecular phylogenetic analysis was conducted using MEGA6 software with the maximum likelihood method. The bootstrap consensus tree inferred from 1,000 replicates was taken to represent the evolutionary history of the taxa analyzed.
RNAi and Overexpression Experiments in S. baicalensis and G. uralensis.
For RNAi experiments, nonhomologous DNA regions of SbCGTb, GuCGTa, and GuCGTb were amplified using primers listed in SI Appendix, Table S7. The PCR products were subcloned into pDONR207 vectors using the Gateway BP Clonase II Enzyme Mix (Invitrogen, US) and then cloned into the RNAi vector pK7WGIGW2R using the Gateway LR Clonase II Enzyme Mix according to the manufacturer’s instructions. The plasmids for RNAi were transformed into Agrobacterium rhizogenes A4 by electroporation. The reconstructed strains were used to induce hairy roots from leaf explants of S. baicalensis and aseptic seedlings of G. uralensis. For overexpression experiments, the full-length DNA regions of GuCGTa and GuCGTb were amplified using primers given in SI Appendix, Table S7. The PCR products were inserted into the overexpression vector pK7WG2R. The other procedures were the same as described above for RNAi experiments. The transformation and induction of hairy roots were carried out following previous reports, and positive hairy roots were screened by DsRed label (45, 46). For S. baicalensis, the positive hairy roots were cultured in liquid B5 medium containing 400-mg/L cefotaxime for 50 d. For G. uralensis, the positive hairy roots were cultured in solid B5 medium containing 500-mg/L cefotaxime for 30 d. The samples were then freeze dried and extracted in a sonicator with 50% methanol for 0.5 h. The extracts were filtered through 0.22-μm membranes before UHPLC/Orbitrap-MS analysis (SI Appendix, Table S5).
Real-Time qPCR Analysis.
Total RNA, extracted from the hairy roots or leaves using the RNAprep Pure Plant Kit (TianGen Biotech, China), was used to synthesize the first-stranded cDNA using TransScript one-step gDNA removal and cDNA synthesis SuperMix (Transgen Biotech, China) according to the manufacturer’s instructions. RT-qPCR was performed using TransStart Green qPCR SuperMix (Transgen Biotech, China) on an Agilent MX3005P real-time PCR system (Agilent Technologies, US). The relative amounts of the target genes were evaluated based on the relative expression index of messenger RNA using the 2−ΔΔC(T) method, and β-actin was used as the reference. The primers are given in SI Appendix, Table S7.
Crystallization of CGTs.
The full-length cDNA of SbCGTa, SbCGTb, LpCGTa, LpCGTb, and ZmCGTa was, respectively, cloned into a modified pET-28a(+) vector. A polyhistidine tag followed by a tobacco etch virus protease cleavage sequence was added in front of the N-terminus of the target protein to facilitate purification. The five proteins were, respectively, expressed in the E. coli BL21(DE3) strain and were purified by Ni-NTA affinity chromatography. Purified CGT proteins were incubated with 5-mM UDP-Glc or UDP-Ara at 4 °C for 1 h before crystallization (19, 20). The crystals were prepared by hanging or sitting drop vapor diffusion (−1 μL protein and 1-μL reservoir solution). Crystals were frozen in the reservoir solution with 30% glycerol or glycol (SI Appendix, Table S9).
Crystal Structures Determination and Refinement.
The diffraction data of ZmCGTa, LpCGTa•UDP, and SbCGTb•UDP-Glc crystals were collected at beamlines BL18U1 Shanghai Synchrotron Radiation Facility (SSRF). The data for SbCGTa•UDP and LpCGTb crystals were collected at beamlines BL19U1 SSRF. All of the data were processed with HKL-3000 (47). The structures were solved by molecular replacement with Phaser (48). ZmCGTa•UDP used the previously reported UGT72B1 structure (PDB ID: 2VCE) as the search model (34) (sequence identity, 27.27%). SbCGTb•UDP-Glc used the GgCGT structure (PDB ID:6L5P) as the model (19) (sequence identity, 42.11%). SbCGTa•UDP used the SbCGTb•UDP-Glc structure as the model (sequence identity, 61.72%). LpCGTb used ZmCGTa as the model (sequence identity, 43.15%). LpCGTa•UDP used LpCGTb as the model (sequence identity, 48.57%). Crystallographic refinement was performed repeatedly using Phenix and COOT (49, 50). The refined structures were validated by Phenix and the PDB validation server (https://validate-rcsb-1.wwpdb.org/). The final refined structures were deposited in the Protein Data Bank with the access PDB ID codes 6LF6 (ZmCGTa•UDP), 6LG1 (LpCGTa•UDP), 6LFN (LpCGTb), 6LG0 (SbCGTa•UDP), and 6LFZ (SbCGTb•UDP-Glc). The diffraction data and structure refinement statistics were summarized in SI Appendix, Table S10. The protein model of SbCGTb mutant was established using MODELER (51), and molecular docking of CGTs with UDP-sugar or substrate was conducted by Autodock (52).
Supplementary Material
Acknowledgments
We thank Professor Qing Zhao and Dr. Meng-Ying Cui at Shanghai Chenshan Plant Science Research Center of the Chinese Academy of Sciences for their technical help in the RNAi experiments; Dr. Fen Liu and Dr. Xu-Lin Sun at the State Key Laboratory of Natural and Biomimetic Drugs of Peking University for their assistance in NMR spectra acquisition; and staffs of BL18U1 and BL19U1 beamlines at National Center for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility, Shanghai, China, for assistance during data collection. This work was supported by National Natural Science Foundation of China (Grants 81725023, 81891010/81891011, 81973448, and 81921001) and the Beijing Natural Science Foundation (Grant JQ18027).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012745117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information; GenBank, https://www.ncbi.nlm.nih.gov (accession nos. MK894443, MK894444, MK894446–MK894455, Q5VMI0, NM_001139178, MT433767, and MT433768); the China National GeneBank, https://db.cngb.org/blast/blast/blastx/; and the Protein Data Bank, www.wwpdb.org (PDB ID codes 6LF6, 6LG1, 6LFN, 6LG0, and 6LFZ).
References
- 1.Talhi O., Silva A.-M.-S., Advances in C-glycosylflavonoid research. Curr. Org. Chem. 16, 859–896 (2012). [Google Scholar]
- 2.Xiao J., Capanoglu E., Jassbi A.-R., Miron A., Advance on the flavonoid C-glycosides and health benefits. Crit. Rev. Food Sci. Nutr. 56 (suppl. 1), S29–S45 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Yang B., Liu H.-L., Yang J.-L., Gupta V.-K., Jiang Y.-M., New insights on bioactivities and biosynthesis of flavonoid glycosides. Trends Food Sci. Technol. 79, 116–124 (2018). [Google Scholar]
- 4.Kant K., Lal U.-R., Ghosh M., Computational breakthrough of natural lead hits from the genus of Arisaema against human respiratory syncytial virus. Pharmacogn. Mag. 13 (suppl. 4), S780–S785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Melo G.-O., et al. , C-glycosylflavones from the aerial parts of Eleusine indica inhibit LPS-induced mouse lung inflammation. Planta Med. 71, 362–363 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Liu M., et al. , Schaftoside alleviates HFD-induced hepatic lipid accumulation in mice via upregulating farnesoid X receptor. J. Ethnopharmacol. 255, 112776 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Song W., et al. , Biosynthesis-based quantitative analysis of 151 secondary metabolites of licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal. Chem. 89, 3146–3153 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Qiao X., et al. , A targeted strategy to analyze untargeted mass spectral data: Rapid chemical profiling of Scutellaria baicalensis using ultra-high performance liquid chromatography coupled with hybrid quadrupole orbitrap mass spectrometry and key ion filtering. J. Chromatogr. A 1441, 83–95 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Ma D., Sun D., Wang C., Li Y., Guo T., Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol. Biochem. 80, 60–66 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Galland M., et al. , Compartmentation and dynamics of flavone metabolism in dry and germinated rice seeds. Plant Cell Physiol. 55, 1646–1659 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Du S.-S., et al. , Nematocidal flavone-C-glycosides against the root-knot nematode (Meloidogyne incognita) from Arisaema erubescens tubers. Molecules 16, 5079–5086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao P.-Y., et al. , Schaftoside interacts with NICDK1 protein: a mechanism of rice resistance to brown planthopper, Nilaparvata lugens. Front Plant Sci 9, 710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan Z.-R., Midega C.-A.-O., Bruce T.-J.-A., Hooper A.-M., Pickett J.-A., Exploiting phytochemicals for developing a ‘push-pull’ crop protection strategy for cereal farmers in Africa. J. Exp. Bot. 61, 4185–4196 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Uawisetwathana U., et al. , Global metabolite profiles of rice brown planthopper-resistant traits reveal potential secondary metabolites for both constitutive and inducible defenses. Metabolomics 15, 151 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Nabavi S.-M., et al. , Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 38, 107316 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Brazier-Hicks M., et al. , The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 284, 17926–17934 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirade Y., et al. , Identification and functional analysis of 2-hydroxyflavanone C-glucosyltransferase in soybean (Glycine max). FEBS Lett. 589, 1778–1786 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Sun Y., et al. , Pathway-specific enzymes from bamboo and crop leaves biosynthesize anti-nociceptive C-glycosylated flavones. Commun. Biol. 3, 110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M., et al. , Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J. Am. Chem. Soc. 142, 3506–3512 (2020). [DOI] [PubMed] [Google Scholar]
- 20.He J.-B., et al. , Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angew. Chem. Int. Ed. Engl. 58, 11513–11520 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Sasaki N., et al. , Identification of the glucosyltransferase that mediates direct flavone C-glucosylation in Gentiana triflora. FEBS Lett. 589, 182–187 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Li C., Zhou C., Li J., Zhang Y., Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata. Plant J. 90, 535–546 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Mashima K., Hatano M., Suzuki H., Shimosaka M., Taguchi G., Identification and characterization of apigenin 6-C-glucosyltransferase involved in biosynthesis of isosaponarin in wasabi (Eutrema japonicum). Plant Cell Physiol. 60, 2733–2743 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Nagatomo Y., et al. , Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (buckwheat) cotyledon. Plant J. 80, 437–448 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Chen D., et al. , Probing the catalytic promiscuity of a regio- and stereospecific C-glycosyltransferase from Mangifera indica. Angew. Chem. Int. Ed. Engl. 54, 12678–12682 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Chen D.-W., et al. , Probing and engineering key residues for bis-C-glycosylation and promiscuity of a C-glycosyltransferase. ACS Catal. 8, 4917–4927 (2018). [Google Scholar]
- 27.Ito T., Fujimoto S., Suito F., Shimosaka M., Taguchi G., C-Glycosyltransferases catalyzing the formation of di-C-glucosyl flavonoids in citrus plants. Plant J. 91, 187–198 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Noguchi A., et al. , Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21, 1556–1572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., et al. , Highly promiscuous flavonoid 3-O-glycosyltransferase from Scutellaria baicalensis. Org. Lett. 21, 2241–2245 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Zhao Q., et al. , A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci. Adv. 2, e1501780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.One Thousand Plant Transcriptomes Initiative , One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson A. E., Tian L., Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 100, 1273–1288 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Zhao Q., et al. , Two CYP82D enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4′-deoxyflavones in Scutellaria baicalensis. Mol. Plant 11, 135–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brazier-Hicks M., et al. , Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 20238–20243 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao H., et al. , Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17, 3141–3154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Offen W., et al. , Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S.-G., Salomon E., Yu O., Jez J.-M., Molecular basis for branched steviol glucoside biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 116, 13131–13136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zong G., et al. , Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-l-rhamnose and rhamnosylation mechanism. Plant J. 99, 257–269 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Yang Y., Yu B., Recent advances in the chemical synthesis of C-glycosides. Chem. Rev. 117, 12281–12356 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Shie J.-J., et al. , Regioselective synthesis of di-C-glycosylflavones possessing anti-inflammation activities. Org. Biomol. Chem. 8, 4451–4462 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Ho T.-C., Kamimura H., Ohmori K., Suzuki K., Total synthesis of (+)-vicenin-2. Org. Lett. 18, 4488–4490 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Davison A., Blaxter M., Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol. Biol. Evol. 22, 1273–1284 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Miettinen K., et al. , The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat. Commun. 8, 14153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji S., et al. , Anti-H1N1 virus, cytotoxic and Nrf2 activation activities of chemical constituents from Scutellaria baicalensis. J. Ethnopharmacol. 176, 475–484 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Zhao Q., et al. , The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol. Plant 12, 935–950 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Kereszt A., et al. , Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 48.McCoy A.-J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams P.-D., et al. , PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W.-G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trott O., Olson A.-J., AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information; GenBank, https://www.ncbi.nlm.nih.gov (accession nos. MK894443, MK894444, MK894446–MK894455, Q5VMI0, NM_001139178, MT433767, and MT433768); the China National GeneBank, https://db.cngb.org/blast/blast/blastx/; and the Protein Data Bank, www.wwpdb.org (PDB ID codes 6LF6, 6LG1, 6LFN, 6LG0, and 6LFZ).