Significance
Polycomb repressive complex 2 (PRC2) is a major epigenetic machinery that maintains transcriptionally silent heterochromatin in the nucleus and plays critical roles in embryonic development and oncogenesis. It is generally thought that PRC2 propagates repressive histone marks by modifying neighboring nucleosomes in strictly linear progression. However, the behavior of PRC2 on native-like chromatin substrates remains incompletely characterized, making the precise mechanism of PRC2-mediated heterochromatin maintenance elusive. Here we use single-molecule force spectroscopy and computational modeling to dissect the interactions between PRC2 and polynucleosome arrays. Our results provide direct evidence that PRC2 can simultaneously engage nonadjacent nucleosome pairs. The demonstration of PRC2's ability to bridge noncontiguous chromosomal segments furthers our understanding of how Polycomb complexes spread epigenetic modifications and compact chromatin.
Keywords: Polycomb-group protein, single-molecule force spectroscopy, molecular dynamics simulation, epigenetics, heterochromatin
Abstract
Polycomb repressive complex 2 (PRC2) installs and spreads repressive histone methylation marks on eukaryotic chromosomes. Because of the key roles that PRC2 plays in development and disease, how this epigenetic machinery interacts with DNA and nucleosomes is of major interest. Nonetheless, the mechanism by which PRC2 engages with native-like chromatin remains incompletely understood. In this work, we employ single-molecule force spectroscopy and molecular dynamics simulations to dissect the behavior of PRC2 on polynucleosome arrays. Our results reveal an unexpectedly diverse repertoire of PRC2 binding configurations on chromatin. Besides reproducing known binding modes in which PRC2 interacts with bare DNA, mononucleosomes, and adjacent nucleosome pairs, our data also provide direct evidence that PRC2 can bridge pairs of distal nucleosomes. In particular, the “1–3” bridging mode, in which PRC2 engages two nucleosomes separated by one spacer nucleosome, is a preferred low-energy configuration. Moreover, we show that the distribution and stability of different PRC2–chromatin interaction modes are modulated by accessory subunits, oncogenic histone mutations, and the methylation state of chromatin. Overall, these findings have implications for the mechanism by which PRC2 spreads histone modifications and compacts chromatin. The experimental and computational platforms developed here provide a framework for understanding the molecular basis of epigenetic maintenance mediated by Polycomb-group proteins.
Eukaryotic chromatin is modified by a plethora of epigenetic machineries that add or erase specific histone posttranslational modifications, thereby exerting transcriptional control (1). Many of these machineries are recruited to and stimulated by their own enzymatic products, leading to the modification of nearby nucleosomes and eventually the formation of large-scale chromatin domains. One preeminent example that employs such a positive feedback mechanism is the Polycomb repressive complex 2 (PRC2), which catalyzes methylation of the lysine 27 residue on histone H3 (2). The final catalytic product, trimethylated H3K27 (H3K27me3), is a hallmark of facultative heterochromatin. The PRC2 core complex comprises four subunits: EED, EZH2, SUZ12, and RBBP4. PRC2 is generally thought to spread H3K27me3 marks through a “read-and-write” mechanism: The EED subunit recognizes an existing H3K27me3 mark, which allosterically activates the methyltransferase EZH2 subunit of the same complex engaged with a neighboring nucleosome (3, 4). This proposed mechanism is supported by recent cryo-electron microscopy (EM) structures of PRC2:dinucleosome assemblies, in which EED interacts with an H3K27me3-containing nucleosome and EZH2 engages an adjacent unmodified nucleosome (5). PRC2 is also known to bind mononucleosomes and free DNA with nanomolar affinity (6–8). In addition, it was inferred from measuring the histone methyltransferase activities of PRC2 on designer chromatin substrates that PRC2 may be able to bridge distal nucleosomes and mediate chromatin looping (9, 10). However, this last binding mode has not yet been directly observed. In fact, the interaction between PRC2 and polynucleosome arrays has thus far been refractory to structural interrogation due to the prohibitive conformational heterogeneity associated with the assembled complexes. As a result, the occurrence of various possible binding modes of PRC2 on long chromatin substrates and their relative frequency remain unclear.
The behavior of the PRC2 core complex on chromatin is regulated by a multitude of factors including accessory subunits, histone modifications, DNA methylation, and RNA (11–13). The exact roles of these regulatory factors are often confounded by contradictory reports. For instance, AEBP2 and JARID2, two accessory proteins that interact with the core complex, define the PRC2.2 subtype (14, 15). It is still controversial whether these two subunits positively or negatively influence the H3K27me3 level of chromatin and whether they do so by changing the chromatin binding configuration or catalytic efficiency of PRC2 (7, 14, 16–25). It is plausible that chromatin contexts and cross-talk between multiple input signals dictate the specific effects exerted by these factors, the determination of which necessitates a quantitative assay with high sensitivity performed in a well-controlled biochemical system.
Single-molecule techniques have been instrumental in deciphering the mechanical properties of DNA, nucleosomes, and chromatin fibers (reviewed in refs. 26 and 27). In this work, we employed single-molecule force spectroscopy to probe the interactions between a major epigenetic machinery (PRC2) and various types of polynucleosome substrates. This approach circumvented the heterogeneity problem and yielded mechanical fingerprints of individual PRC2-bound chromatin complexes. We also performed large-scale computational simulations to corroborate the experimental results. The large size and complex protein–DNA contacts render chromatin a challenging system to study in silico. Although numerous coarse-grained models have been developed for chromatin structure characterization (28–32), they often lack sufficient molecular details necessary for describing the behavior of chromatin-binding complexes, especially those as intricate as PRC2. We applied a model with amino-acid and base-pair specificity to study protein–chromatin interaction at high chemical accuracy and structural resolution (33–38). A computational algorithm was further developed to efficiently probe the variation of PRC2 binding poses in response to changes in the chromatin conformation.
Our results revealed and quantitated a diverse repertoire of PRC2 binding modes on chromatin. Besides reproducing known PRC2 interaction with bare DNA, mononucleosomes, and neighboring nucleosome pairs, our data also provided direct evidence for frequent bridging of nonadjacent nucleosomes by PRC2, an activity associated with chromatin compaction. We showed that “1–3” nucleosome bridging is a preferred binding configuration for PRC2. Furthermore, we demonstrated that the distribution and stability of these binding modes are differentially modulated by AEBP2 and JARID2, as well as by the methylation and mutation levels of H3K27 in the nucleosome array. These findings offer fresh insight into the mechanism by which PRC2 mediates epigenetic spreading and heterochromatin maintenance.
Results
A Single-Molecule Platform to Dissect PRC2–Chromatin Interactions.
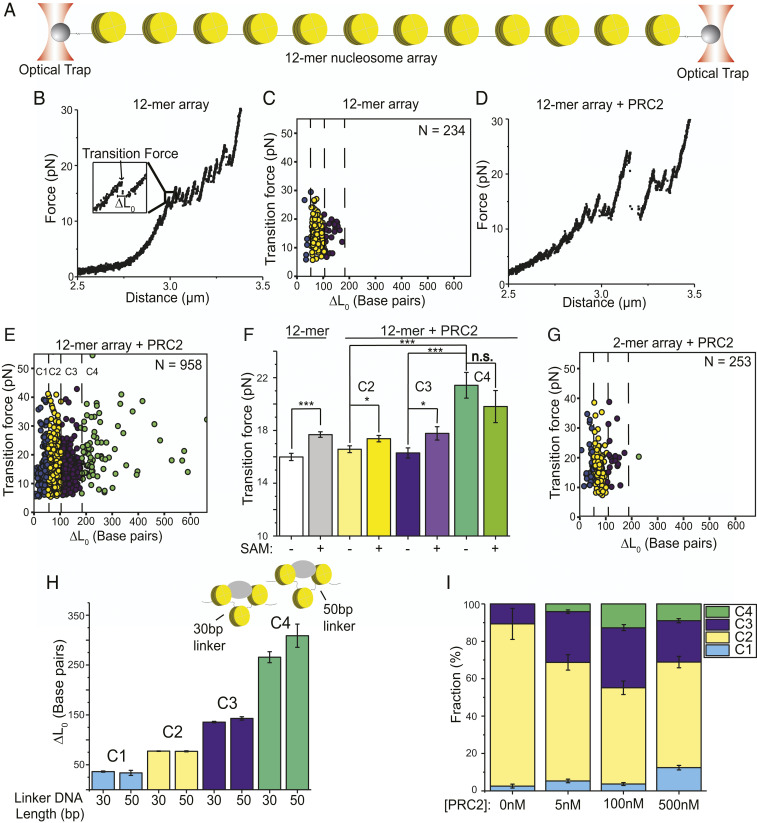
We constructed a DNA template harboring 12 repeats of the “601” nucleosome positioning sequence, each separated by 30 base pairs (bp) of linker DNA. After reconstitution with histone octamers, individual nucleosome arrays were tethered and mechanically stretched on a dual-trap optical tweezers instrument (Fig. 1A). Single-molecule force-extension curves displayed signature sawtooth patterns (Fig. 1B) consistent with previous results (39), with each abrupt transition signifying the stochastic unwrapping of a single nucleosome. The average force at which these transitions occurred is 16.0 ± 0.3 pN (mean ± SEM; SI Appendix, Table S1) and the average number of transitions observed on each array is 12 ± 1, suggesting that octamers were sufficiently loaded onto the DNA template. The contour length change (ΔL0) per transition, which reports the amount of DNA released upon force-induced disengagement, is 76 ± 1 bp (mean ± SEM), in good agreement with the unraveling of the inner DNA wrap around the histone octamer (Fig. 1C and SI Appendix, Fig. S1). The SD of ΔL0 is 11 bp, which likely reflects the structural pliability of nucleosomes (40). The unpeeling of the outer DNA wrap occurred in a gradual fashion at low forces (<5 pN) under our experimental condition, consistent with previous observations (39, 41).
Fig. 1.
Single-molecule force spectroscopy dissects PRC2 binding modes on nucleosome arrays. (A) Schematic of the dual-trap optical tweezers setup (not drawn to scale). A 12-mer nucleosome array is tethered to a streptavidin-coated bead via a 4-kbp-long biotinylated DNA handle on each side. (B) A representative force-extension curve for a 12-mer array. (Inset) Zoomed-in view of a force-induced transition. ∆L0 represents the contour length change associated with the transition. (C) Cluster analysis of individual transitions observed in the force-extension curves of 12-mer arrays. N denotes the number of transitions. (D) A representative force-extension curve for a 12-mer array incubated with 500 nM PRC2. (E) Cluster analysis of transitions for PRC2-bound 12-mer arrays. (F) Effect of the PRC2 cofactor SAM on the average transition force for 12-mer arrays and for different transition clusters of PRC2-bound 12-mer arrays. (G) Cluster analysis of transitions found in PRC2-bound dinucleosome substrates. (H) Average ∆L0 of each transition cluster for PRC2-bound 12-mer arrays with a 30-bp linker DNA versus those with a 50-bp linker DNA. (I) Cluster distribution for 12-mer arrays incubated with PRC2 at different concentrations. Data are presented as mean ± SEM. P values are determined by two-sample t tests (n.s., not significant, P ≥ 0.05; *P < 0.05; ***P < 0.001).
Next, we incubated the 12-mer arrays in a relaxed state (i.e., under zero force) with PRC2 core complexes at a concentration of 500 nM (SI Appendix, Fig. S2). At this concentration, the majority of nucleosomes in the array are expected to be PRC2-bound (7). In the meantime, some PRC2 complexes are expected to bind the free DNA as well (8). The tethered enzyme–substrate assemblies were then subjected to single-molecule pulling. Force-extension trajectories for these assemblies exhibited transitions of diverse sizes (Fig. 1D). Using a clustering algorithm based on the Bayesian information criterion (BIC) (SI Appendix, Fig. S3), we categorized all transitions into four distinct clusters (Fig. 1E and SI Appendix, Table S1). The first cluster (C1) includes transitions with ΔL0 smaller than 51 bp. We posited that PRC2 sequesters, perhaps bends, a stretch of free DNA located in the flanking or linker DNA regions of the nucleosome array and that the C1 transitions are due to force-induced dissociation of this stretch of DNA from PRC2. This interpretation is supported by the force-extension curves obtained with bare DNA substrates incubated with PRC2, which exhibited exclusively these small transitions (SI Appendix, Fig. S4). A recent atomic force microscopy (AFM) study also reported DNA looping by PRC2 (8), which likely represents the same phenomenon as shown here. Our data provide additional insight that this interaction is stable, withstanding an average force of 17 pN.
The second cluster (C2 in Fig. 1E) contains transitions between 52 and 104 bp in size with an average ΔL0 of 77 ± 1 bp, mimicking transitions observed in the absence of PRC2 that represent the unraveling of mononucleosomes (Fig. 1C). The average transition force for C2 was not significantly altered by the presence of PRC2 (16.2 ± 0.3 pN without PRC2 vs. 16.6 ± 0.3 pN with PRC2). Nonetheless, given the high PRC2 concentration used in this experiment, the C2 transitions likely contained disassembly events of both PRC2-bound mononucleosomes and unbound nucleosomes.
The third cluster (C3 in Fig. 1E) encompasses transitions between 105 and 179 bp in size. Besides the fraction that can be attributed to two mononucleosomes coincidentally unraveling at the same time (also seen in the absence of PRC2 but occurring with a lower frequency: 22% with PRC2 vs. 11% without PRC2), this cluster also corresponds to the binding mode in which PRC2 simultaneously engages two neighboring nucleosomes as seen in the PRC2:dinucleosome cryo-EM structure (5). The rationale for this interpretation is as follows: Disruption of the PRC2:dinucleosome complex would release ∼180 bp of DNA—two outer DNA wraps (∼150 bp) plus 30 bp of linker DNA; alternatively, it would release between 105 and 180 bp of DNA if all or part of the outer wrap from one of the PRC2-interacting nucleosomes is not subjected to PRC2 sequestration, hence already undone at a low force (SI Appendix, Fig. S5A).
Importantly, we also observed a significant population of transitions (86 out of 958) with ΔL0 greater than 180 bp (C4 in Fig. 1E). This is in stark contrast to the transition distribution for 12-mer without PRC2, where no C4 transitions were identified (0 out of 234). The average rupture force for C4 transitions is significantly higher than those for C2 and C3 (16.6 ± 0.3 pN for C2, 16.3 ± 0.4 pN for C3, and 21.4 ± 1.0 pN for C4; Fig. 1F and SI Appendix, Table S1), indicating that they represent a distinct PRC2 binding mode on chromatin. The large sizes of these transitions entail concurrent engagement of PRC2 with two distal sites. Specifically, if PRC2 bridges two nucleosomes that are separated by one spacer nucleosome (“Nuc1–3” mode), disruption of this linkage would release between 210 and 285 bp of DNA (SI Appendix, Fig. S5B). If PRC2 bridges a pair of nucleosomes that are separated by two or more spacer nucleosomes (Nuc1–4, Nuc1–5, etc.), transitions larger than 300 bp would be expected (SI Appendix, Fig. S5C). Because the majority of C4 transitions (78%) were between 180 and 300 bp in size, we speculated that Nuc1–3 is the preferred mode of PRC2–nucleosome interaction in this cluster.
Next, we examined the effect of S-adenosylmethionine (SAM)—the methyl donor for PRC2-catalyzed H3K27 methylation—on the transition forces. We found that SAM had a noticeable effect on the stability of nucleosome itself independent of PRC2, raising the average transition force from 16.0 ± 0.3 to 17.7 ± 0.2 pN (Fig. 1F). This difference is markedly larger than the uncertainty of our force measurements associated with the instrumental noise and analysis algorithm (<0.3 pN), hence representing a real change in the transition force. Therefore, the modest stabilization effect of the SAM cofactor on C2 and C3 transition forces in the presence of PRC2 most likely came directly from SAM (Fig. 1F). In contrast, the C4 transition force was not significantly affected by SAM, reinforcing the notion that C4 represents a distinct class of PRC2–nucleosome interactions.
Validation of the Ability of PRC2 to Bridge Nonadjacent Nucleosome Pairs.
Besides the model in which PRC2 makes specific contacts with a pair of distal nucleosomes, there is an alternative model that could also explain the observed large transitions in C4. In this model, PRC2 engages with two distal segments of the chromatin and slides on them until running into a pair of steric blocks (SI Appendix, Fig. S6A). To differentiate between these models, we placed two well-separated dCas9 roadblocks in the DNA template through single-guide RNA (sgRNA)-mediated targeting (SI Appendix, Fig. S6B). The nonspecific roadblock model predicts large force-induced transitions when the dCas9-bound DNA is incubated with PRC2, while the nucleosome-specific model does not. We did not observe such large transitions, even though there was clear evidence that dCas9 was stably bound to DNA (SI Appendix, Fig. S6 C–E). This result disfavors the nonspecific roadblock model and suggests that the C4 transitions are largely attributed to specific PRC2–nucleosome contacts rather than steric hindrance.
The interpretation that C4 transitions originated from PRC2 bridging two nonneighboring nucleosomes makes a few testable predictions. First, since this mode of PRC2 interaction requires a substrate harboring at least three nucleosomes, it should not be observed on arrays containing only two nucleosomes. We therefore constructed a dinucleosome substrate (SI Appendix, Fig. S1) and, satisfyingly, found that C4 was essentially absent from this substrate when incubated with PRC2 (Fig. 1G and SI Appendix, Table S1). The difference between the transition cluster distributions for PRC2-bound 12-mer and 2-mer arrays is very significant (Fig. 1E vs. 1G; P = 5.9 × 10−27, Kolmogorov–Smirnov test). This result rules out the possibility that the C4 cluster originates from PRC2 engaging purely free DNA, or engaging one nucleosome and one free DNA segment. It also argues against an altered pathway of DNA unwrapping from the nucleosome caused by PRC2 being the source of these large transitions.
Second, our interpretation of C4 transitions predicts that the amount of DNA released per transition should be dependent on the length of linker DNA between nucleosomes. To test this, we constructed a 12-mer nucleosome array with longer linkers (50 bp) and conducted single-molecule pulling experiments with this substrate (SI Appendix, Table S1). Our measurements showed that the linker DNA length selectively modulates the size of C4 transitions and that the average ΔL0 in C4 for the 50-bp-linker array is larger by ∼40 bp than that for the 30-bp-linker array (Fig. 1H). This magnitude of size change again suggests that the Nuc1–3 mode, which releases two linker DNA lengths upon disruption, is the predominant mode of PRC2 engagement in C4. We note that the difference in average ΔL0 for C3 transitions between 30-bp-linker and 50-bp-linker arrays is ∼7 bp, smaller than the predicted 20 bp (Fig. 1H). As mentioned above, some of the C3 transitions represent two mononucleosomes coincidentally unraveling together, independent of PRC2 engagement, which explains the relative insensitivity of the average ΔL0 in this cluster to the linker length.
Third, since the occurrence of C4 transitions strictly relied on the presence of PRC2 (Fig. 1C vs. Fig. 1E), we reasoned that the frequency of their occurrence should depend on the amount of chromatin-bound PRC2. We thus conducted pulling experiments with varying concentrations of PRC2 and indeed observed such dependence (Fig. 1I): The relative population of C4 transitions increased as the PRC2 concentration was raised from 5 nM to 100 nM and appeared to become saturated afterward. On the other hand, the relative population of C1 transitions, which represent free DNA sequestration by PRC2, continued to rise at higher PRC2 concentrations (Fig. 1I). This is in accordance with the higher dissociation constant value (∼900 nM) for PRC2-induced DNA looping obtained from the AFM study (8). Taking these pieces of experimental evidence together, we concluded that the most plausible interpretation for the C4 transitions is a nonadjacent nucleosome bridging ability possessed by PRC2.
In Silico Characterization of PRC2–Chromatin Interactions.
To further corroborate the experimental results, we developed a computational workflow that combined molecular dynamics simulations to sample polynucleosome configurations with rigid docking to explore the binding poses of individual PRC2 complexes (SI Appendix, Fig. S7A). In the following, we used this in silico platform to determine the set of low-energy poses that PRC2 adopts when in contact with tetranucleosome substrates.
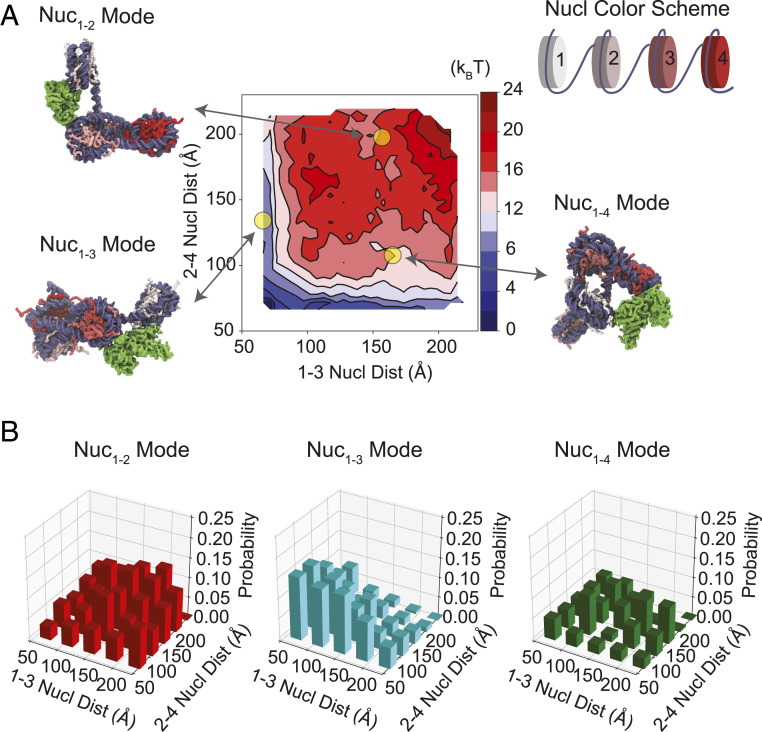
We first performed 25 independent molecular dynamics simulations to collect a total of 1,000 representative structures for the tetranucleosome with 30-bp linker DNA. To ensure that both extended and collapsed configurations of the tetranucleosome were included in this structural ensemble, we used harmonic restraints to bias each simulation toward different spatial distances between the 1–3 and 2–4 nucleosome pairs. These simulations were initialized with a tetranucleosome configuration obtained by sequentially extending the dinucleosome cryo-EM structure (5) with 30-bp linker DNA segments and mononucleosomes. A coarse-grained force field that models protein (42, 43) and DNA (44) molecules at a single-residue and single-bp resolution was used to provide an accurate description of protein–protein and protein–DNA interactions. Prior studies showed that this level of resolution is sufficient to accurately model the energetics of nucleosomal DNA unwrapping (36, 37) and internucleosome interaction (35). To further improve the computational efficiency, we modeled the core region of each nucleosome as rigid bodies while maintaining the flexibility of the outer layer DNA. As shown in Fig. 2A, these simulations covered a wide range of tetranucleosome configurations and allowed us to determine the free-energy surface for chromatin folding.
Fig. 2.
In silico analysis reveals the preferred mode of interaction between PRC2 and nucleosome pairs. (A) Thermodynamic stability (free energy) for the tetranucleosome with a 30-bp-long linker DNA as a function of the spatial distances between the 1–3 and 2–4 nucleosome pairs. The impact of PRC2 was not included when computing the free energy. Example chromatin configurations are shown on the side, with DNA in purple and the color of histone proteins varying from white to red as the nucleosome index increases. The PRC2 complex is colored in green and shown in its lowest-energy pose in the three examples. (B) Fraction of PRC2 engaging in different nucleosome-bridging modes (Nuc1–2, Nuc1–3, and Nuc1–4) at given tetranucleosome configurations grouped by its internucleosome distances.
Next, for each one of the tetranucleosome configurations, we applied a rigid docking procedure to determine the set of lowest-energy PRC2 binding poses. A structural model of the PRC2 core complex (SUZ12, EZH2, EED, and RBBP4) was constructed from several published crystal and cryo-EM structures (45–47) using homology modeling (48). The combination of several partially resolved structures ensured that full-length core subunits are represented in the model, including the N-terminal part of SUZ12 that is important for PRC2 recruitment (45, 49). Close examination of the docking results revealed that PRC2 can interact with adjacent nucleosomes using EED and EZH2 subunits (Nuc1–2 mode), consistent with the published cryo-EM structure (5). Repeating the docking procedure using the same PRC2:dinucleosome system as in the cryo-EM study led to similar structural configurations (SI Appendix, Fig. S7B). The computational model succeeded in reproducing the experimental structure as the top-ranking configuration. The agreement between simulated and experimental PRC2 binding poses lends support to the use of the computational protocol implemented here to study PRC2–chromatin interactions.
At many tetranucleosome configurations, we found that PRC2 can bridge nucleosome pairs with one (Nuc1–3 mode) or two (Nuc1–4 mode) spacers using EED and EZH2 subunits of the same complex. For small internucleosome distances, the Nuc1–3 mode is favored over Nuc1–2 and Nuc1–4 modes (Fig. 2B). We note that the results at different internucleosome distances cannot be compared directly against each other since they were obtained by simple counting of the docking results. With proper thermodynamic reweighting, however, the results can indeed be combined to provide a global estimation of the various PRC2 binding modes across the entire phase space (SI Appendix, In Silico Modeling and Simulations). We reweighted each PRC2–tetranucleosome assembly to account for both the binding energy between the two complexes and the free energy cost for the tetranucleosome to adopt the conformation in the assembly. The relative population of the Nuc1–3 mode was estimated to be 18%, in comparison with 4% for Nuc1–2 and 7% for Nuc1–4. The rest of the population displays a mononucleosome-binding or a free-DNA-binding pose of PRC2. These population estimations are for chromatin under zero tension. A better agreement with the experimental results could be obtained if only extended chromatin configurations, which are presumably more prevalent in optical trapping experiments, were included in the population estimation.
In addition, we applied the same modeling procedure to a tetranucleosome with 50-bp linker DNA. We again found ample examples in which PRC2 forms stable assemblies with nonadjacent nucleosome pairs. Extending the linker length to 50 bp generally increases the distances between nucleosomes and reduces the probability of PRC2 engaging with multiple nucleosomes. This reduction is more significant for the Nuc1–2 and Nuc1–4 modes than for Nuc1–3 (SI Appendix, Fig. S8), a trend that was observed in experimental results as well (C4 populates 9% of total transitions for the 30-bp-linker array vs. 15% for the 50-bp-linker array; SI Appendix, Table S1). Taken together, the computational modeling combined with the thermodynamic estimation corroborates the single-molecule data and suggests that bridging nonadjacent nucleosomes is an inherent property of the PRC2 core complex.
Differential Regulation of PRC2–Chromatin Interaction by AEBP2 and JARID2.
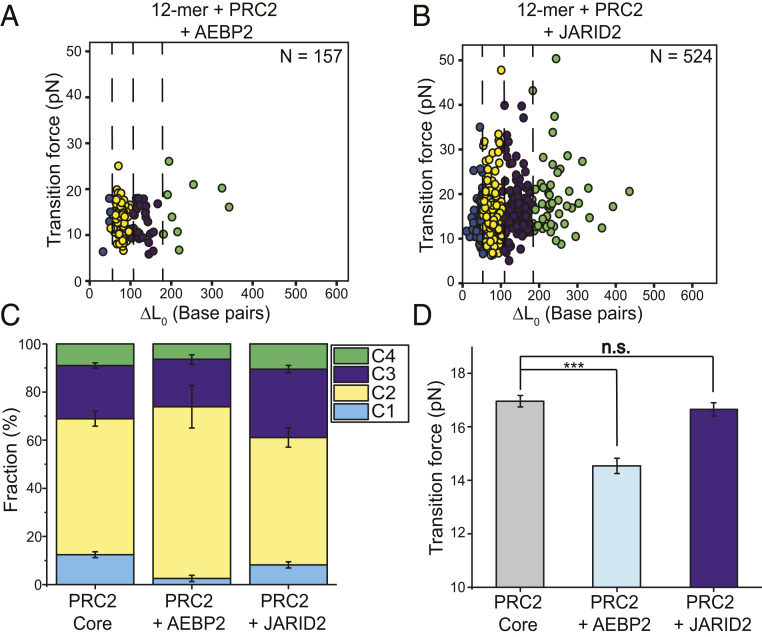
Next, we used the single-molecule experimental platform to assess the effects of two accessory subunits, AEBP2 and JARID2, on the binding of PRC2 to chromatin (Fig. 3 A and B and SI Appendix, Fig. S2). Interestingly, the addition of a stoichiometric amount of AEBP2 largely abolished C1 transitions, which correspond to free-DNA sequestration by PRC2 (Fig. 3C and SI Appendix, Table S1). There is a statistically significant difference between the transition cluster distributions for 12-mer arrays incubated with PRC2 core complexes and with PRC2–AEBP2 complexes (Fig. 1E vs. 3A; P = 0.00071, Kolmogorov–Smirnov test). Thus, AEBP2 either reduces DNA binding by PRC2 or alters its binding geometry such that it no longer sequesters DNA. The latter scenario is supported by the recent AFM study which showed that PRC2–AEBP2 mostly bends DNA without substantial wrapping (8). PRC2–AEBP2 can still bind nucleosomes and bridge nucleosome pairs (Fig. 3C). However, AEBP2 significantly lowered the average transition force (14.5 ± 0.3 pN for 12-mer + PRC2 + AEBP2 vs. 17.0 ± 0.3 pN for 12-mer + PRC2 core; Fig. 3D), indicating that AEBP2 destabilizes the engagement of PRC2 with chromatin.
Fig. 3.
Chromatin interaction of PRC2 core complex is differentially modulated by accessory subunits AEBP2 and JARID2. (A) Cluster analysis of transitions observed in the force-extension curves for 12-mer arrays incubated with PRC2–AEBP2 complexes. (B) Cluster analysis of transitions for 12-mer arrays incubated with PRC2–JARID2 complexes. N denotes the number of transitions. (C) Cluster distribution for 12-mer arrays incubated with PRC2 core, PRC2–AEBP2, and PRC2–JARID2 complexes. (D) Average transition force for 12-mer arrays bound to PRC2 core, PRC2–AEBP2, and PRC2–JARID2 complexes. Data are presented as mean ± SEM. n.s., P ≥ 0.05; ***P < 0.001.
On the contrary, the addition of JARID2 did not cause significant changes in the transition cluster distribution, nor did it significantly affect the average transition force (Fig. 3 B–D and SI Appendix, Table S1). These results are consistent with prior biochemical data suggesting that the effect of JARID2 on PRC2 function is mainly manifested in the allosteric regulation of the catalytic activity of EZH2 (7, 21, 22, 46). We do not, however, rule out that JARID2 may exert distinct effects under different chromatin contexts, synergizing with or antagonizing other regulatory signals (10, 50).
H3K27 Methylation and Mutation Levels Modulate PRC2 Interaction with Chromatin.
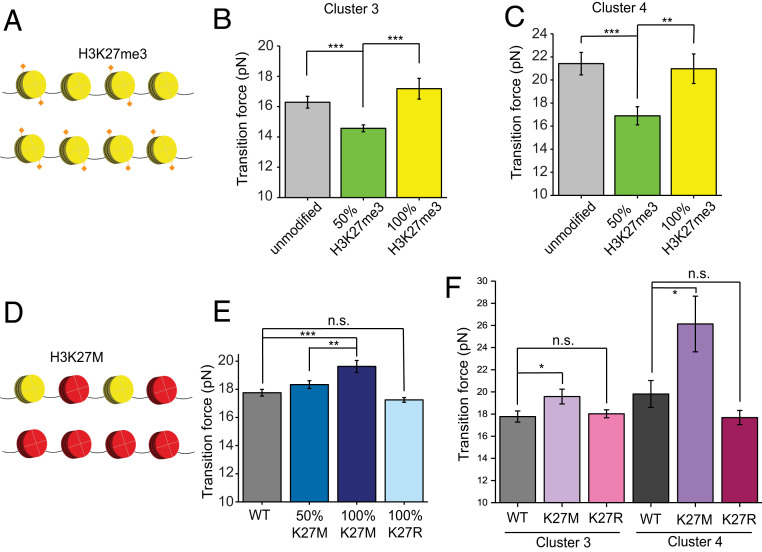
To examine how preexisting H3K27me3 marks affect the nucleosome-binding properties of PRC2, we constructed 12-mer arrays with purely H3K27me3-containing nucleosomes, as well as arrays with an equimolar mixture of unmodified and H3K27me3-containing nucleosomes that were randomly incorporated into the array (Fig. 4A). When pulling on the 50% H3K27me3 arrays incubated with PRC2, we found a significant reduction in the average rupture force compared to unmodified arrays for both C3 and C4 transitions (C3: 14.6 ± 0.2 pN for 50% H3K27me3 vs. 16.3 ± 0.4 pN for unmodified; C4: 16.9 ± 0.8 pN for 50% H3K27me3 vs. 21.4 ± 1.0 pN for unmodified; Fig. 4 B and C and SI Appendix, Table S1). Interestingly, 100% H3K27me3 arrays brought the transition force back to the level of unmodified arrays (Fig. 4 B and C and SI Appendix, Table S1).
Fig. 4.
Methylation and mutation levels of H3K27 affect the behavior of PRC2 on chromatin. (A) Cartoon of a nucleosome array containing 50% (Top) or 100% (Bottom) H3K27me3-modified nucleosomes. (B) Average C3 transition force for unmodified, 50% H3K27me3-modified, and 100% H3K27me3-modified 12-mer arrays in the presence of PRC2. (C) Average C4 transition force for unmodified, 50% H3K27me3-modified, and 100% H3K27me3-modified 12-mer arrays in the presence of PRC2. (D) Cartoon of a nucleosome array containing 50% (Top) or 100% (Bottom) H3K27M mutant nucleosomes. (E) Average overall transition force for wild-type, 50% H3K27M, 100% H3K27M, and 100% H3K27R 12-mer arrays incubated with PRC2 and SAM. (F) Average C3 and C4 transition forces for wild-type, 100% H3K27M, and 100% H3K27R 12-mer arrays in the presence of PRC2 and SAM. The methylated and mutant nucleosomes were randomly incorporated into the arrays at indicated molar ratios. Data are presented as mean ± SEM. n.s., P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
The lysine-to-methionine substitution of H3K27 (H3K27M) is prevalently found in pediatric brain cancers and associated with a global decrease of H3K27me3 via inhibition of PRC2 (51, 52). To evaluate how this oncogenic mutation impacts the PRC2–nucleosome interaction, we compared the force-extension trajectories for PRC2-bound 12-mer arrays that contain 0%, 50%, and 100% H3K27M mutant copies of H3 (Fig. 4D). We also included SAM in this experiment, as the inhibitory effect of H3K27M has been shown to rely on the presence of SAM (9, 52). We found that the mutation increased the overall average transition force in a dose-dependent manner (17.8 ± 0.3 pN for wild type, 18.3 ± 0.3 pN for 50% H3K27M, and 19.6 ± 0.5 pN for 100% H3K27M; Fig. 4E and SI Appendix, Table S1). An increased transition force by the K27M mutation was also observed when C3 and C4 transitions were selectively analyzed (C3: 17.8 ± 0.5 pN for wild type vs. 19.6 ± 0.7 pN for 100% H3K27M; C4: 19.8 ± 1.2 pN for wild type vs. 26.1 ± 2.6 pN for 100% H3K27M; Fig. 4F). These results suggest that H3K27M stabilizes the engagement of PRC2 with nucleosomes, lending support to the model in which H3K27M-containing nucleosomes trap PRC2 locally via strong contacts, thereby inhibiting the propagation of H3K27me3 marks (46, 51, 53). We also performed experiments with 12-mer arrays harboring 100% H3K27R nucleosomes as a nonsubstrate, noninhibitor control (9, 51). As expected, the lysine-to-arginine mutation did not elicit an appreciable effect on the transition forces, in contrast to the pathogenic K27M mutation (Fig. 4 E and F and SI Appendix, Table S1).
PRC2-Mediated Compaction of Nucleosome Arrays.
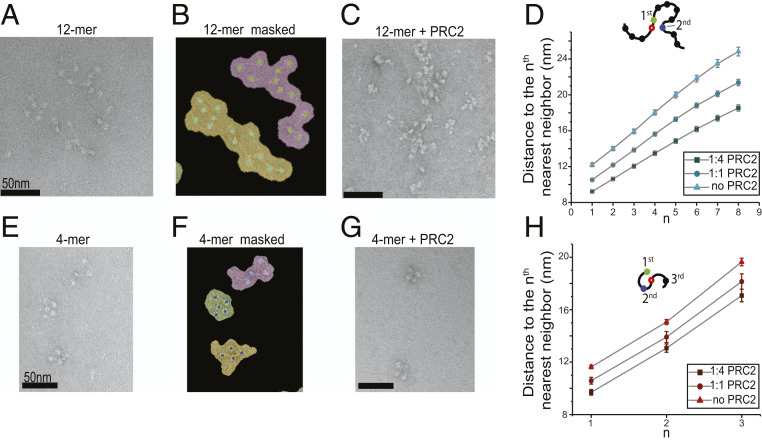
The ability of PRC2 to bridge pairs of nonadjacent nucleosomes entails a chromatin-compacting activity of the complex. To test this activity, we examined the architecture of wild-type 12-mer arrays using negative-stain EM (Fig. 5 A and B and SI Appendix, Fig. S9). We found that the arrays generally adopted more compact configurations when incubated with PRC2 (Fig. 5C). We quantified the level of compaction by measuring the average distance between nucleosomes within the same array. This analysis revealed that PRC2 condensed 12-mer arrays in a concentration-dependent manner (Fig. 5D). We then repeated the EM assay with tetranucleosome substrates and observed the same trend of PRC2-dependent compaction (Fig. 5 E–H). These results demonstrate that PRC2 is able to mediate chromatin compaction—a property conferred by bridging distal chromosomal segments. We note that, according to previous work, the chromatin-compacting capability of PRC2 can be further enhanced by replacing the EZH2 subunit with its homolog EZH1 (54).
Fig. 5.
PRC2 mediates the compaction of polynucleosomes. (A) A representative negative-stained electron micrograph of 12-mer nucleosome arrays. (B) The same micrograph as in A with binary masks depicting individualized nucleosome arrays. Nucleosomes inside their corresponding masks are shown as colored dots. (C) A representative micrograph of 12-mer arrays incubated with PRC2 at 1:4 molar ratio (150 nM array and 600 nM PRC2). (D) Average distance between a reference nucleosome (open circle) and its nth nearest neighbor within the same 12-mer array measured at different array:PRC2 mixing ratios. A total of 95 masks from 14 micrographs were analyzed for the 1:4 array:PRC2 ratio, 121 masks from 10 micrographs for the 1:1 array:PRC2 ratio, and 98 masks from 14 micrographs for the no-PRC2 condition. A higher PRC2 level correlates to a shorter distance, indicating a higher degree of array compaction. (E) A representative electron micrograph of tetranucleosome arrays. (F) The same micrograph as in E with binary masks depicting individualized arrays. (G) A representative micrograph of tetranucleosome arrays mixed with PRC2 at 1:4 molar ratio. (H) Average distance from a reference nucleosome (open circle) to its first, second, and third nearest neighbor within the same tetranucleosome array measured at different array:PRC2 mixing ratios. A total of 103 masks from 10 micrographs were analyzed for the 1:4 array:PRC2 ratio, 99 masks from 12 micrographs for the 1:1 array:PRC2 ratio, and 371 masks from 19 micrographs for the no-PRC2 condition. Data are presented as mean ± SEM.
Inferring Distal Nucleosome Contacts by PRC2 from Genome-Wide H3K27me3 Profiles.
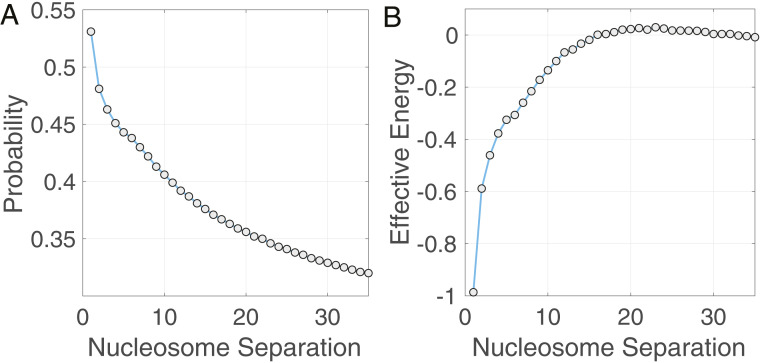
To evaluate the in vivo relevance of PRC2 engaged with distal nucleosomes, we analyzed the genome-wide profile of H3K27me3 from human cells (55). Specifically, we binarized the raw chromatin immunoprecipitation sequencing (ChIP-seq) data from the IMR90 lung fibroblast cell line with a Poisson background model at a resolution of 200 bp that approximates the nucleosome repeat length. From the processed data, we estimated the probability for the histone mark to appear simultaneously at two nucleosome sites of a given genomic separation by averaging the product of the ChIP-seq signal over the entire genome. As shown in Fig. 6A, H3K27me3 marks from nearest-neighbor nucleosomes are strongly correlated. Importantly, this correlation persists over much longer nucleosome separations, supporting the spreading of H3K27me3 over a long stretch of chromatin. The correlation between distant nucleosomes could in principle be explained by either a linear model or a loop-driven spreading model. These nucleosomes need not be in direct contact since correlation could arise if both nucleosomes are interacting with a common partner (such as PRC2). To distinguish a direct correlation from an indirect one and investigate which model best explains the ChIP-seq data, we used a long-range Ising model to fit the experimental data. Similar approaches have been used to analyze protein sequences and filter out indirect correlations to identify amino acid pairs that are in direct three-dimensional contact (56, 57). As detailed in SI Appendix, Information Theoretic Modeling of ChIP-seq Data, the model’s coupling parameters measure the direct contact strength between two nucleosomes. By explicitly considering coupling between distal nucleosomes, the energy function is general and can be used to describe both the linear and the looping model. For the linear model, the only nonzero coupling parameter would be the one that measures direct contacts between nearest-neighbor nucleosomes. On the other hand, for the looping model, nonzero interaction energies exist for long-range nucleosomes as well. The generic expression for the energy function is crucial and ensures that our analysis is not biased toward either model. We used the maximum entropy optimization algorithm introduced in ref. 58 to derive the set of least-biased parameters that can reproduce the correlation between histone marks at various nucleosome separations. Significant interaction energies were still found for genomic segments separated by more than 10 nucleosomes (Fig. 6B). Therefore, the looping model, but not the linear one, can fit the data well. The decrease in energy amplitude as the nucleosome separation increases could be due to contributions from the polymer configurational entropy. In sum, nucleosomes within facultative heterochromatin regions form distal contacts, potentially stabilized by long-range PRC2–chromatin interactions observed in this study, to facilitate histone mark spreading.
Fig. 6.
Analysis of H3K27me3 ChIP-seq data supports interaction between nonadjacent nucleosomes. (A) Probability for the H3K27me3 mark to simultaneously occur at two nucleosomes as a function of their separation length. (B) Effective interaction energy that measures the direct correlation between H3K27me3 occupancy at two nucleosomes as a function of their separation length. The energy profile supports the chromatin looping model. In contrast, for the linear model, only the energy between nearest-neighbor nucleosomes (a nucleosome separation of 1) has a nonzero value.
Discussion
PRC2 maintains transcriptional silencing of a large number of genes involved in development and cancer (59). Its function and regulation have been subjected to extensive investigations. However, conflicting results still exist regarding the mode of action of PRC2 on chromatin and the roles of diverse regulatory factors. In this work, we employ single-molecule force spectroscopy to extract the structural fingerprints of individual PRC2–polynucleosome assemblies. We observed a distinct cluster of large transitions (C4) that also feature higher rupture forces than transitions in other clusters for wild-type 12-mer arrays incubated with PRC2 core complexes, suggesting that these C4 transitions are associated with a low-energy PRC2–nucleosome interaction mode. This finding provides direct evidence for a stable PRC2–chromatin complex in which PRC2 mediates internucleosome contacts beyond nearest neighbors. Our data further suggest that this interaction geometry can accommodate different linker DNA lengths, even though its propensity is influenced by the linker DNA. Our experimental and computational analyses established the trinucleosome unit as a favored substrate for PRC2 engagement. The zigzag chromatin topology, which positions every other nucleosome into close proximity (60), may underlie this preferred mode of PRC2 binding.
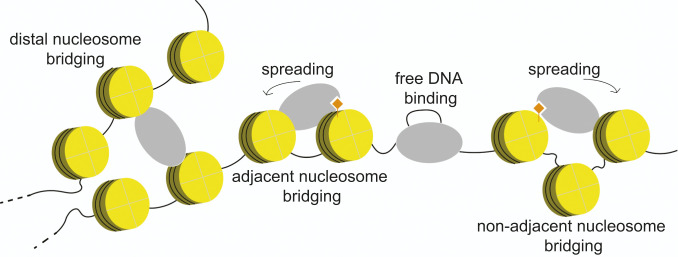
These long-range contacts have significant implications for the mechanism of H3K27me3 spreading (Fig. 7). It argues against models in which PRC2 propagates H3K27me3 along nucleosome arrays in strictly linear progression. Instead, PRC2 can bypass spacer nucleosomes and perhaps other roadblocks when carrying out the read-and-write function. As such, repressive marks can be spread to nucleosomes far away in sequence from the nucleation site. This feature could serve as the mechanistic basis for several epigenetic phenomena, such as memory and bistability (61, 62), and is necessary for establishing the H3K27me3 pattern in vivo (58). Another consequence of distal nucleosome bridging by PRC2, which is separable from its catalytic activity, is the physical compaction of nucleosome arrays (Fig. 7). This activity, combined with that of DNA compaction via bending and looping (8), likely constitutes another mechanism besides chemical modification that contributes to PRC2-dependent transcriptional repression. It is plausible that engaging noncontiguous chromosomal segments is a common characteristic of Polycomb-group proteins (63).
Fig. 7.
Working model for PRC2-mediated heterochromatin formation. PRC2 interacts with chromatin in multiple modes, carrying out enzymatic modification and nonenzymatic compaction of the chromatin, both of which contribute to transcriptional repression. In particular, the ability of PRC2 to simultaneously engage nonadjacent nucleosomes within the same domain, or perhaps distal nucleosomes from different domains, facilitates efficient spreading of repressive histone marks and mediates local or global chromatin condensation.
By subjecting PRC2–chromatin assemblies to mechanical perturbations, we obtained fresh insights into the strengths of different binding modalities for this important epigenetic machinery. Recent studies reported that PRC2–chromatin interactions are transient, allowing the enzyme to readily translocate on substrates for efficient spreading of repressive marks (52, 64). Our data here show that PRC2–nucleosome linkages can also withstand high tensions, exhibiting mechanical stability comparable to or higher than that of the nucleosome itself. It can be envisioned that PRC2 is performing a balancing act: Both stable and transient interactions are at work in order to accomplish de novo installation and self-propagation of H3K27me3. The strong interaction may be required for PRC2 recruitment to nucleation sites and for the deposition of initial H3K27me3 foci (49, 64), presumably with slow kinetics, whereas the weak contacts induced by, for example, the coexistence of activating (modified) and substrate (unmodified) nucleosomes, facilitate efficient spreading by making PRC2 more labile on chromatin. The latter scenario is directly supported by our single-molecule force measurements, which revealed that PRC2 binds less stably to partially H3K27me3-modified nucleosome arrays than to naïve and fully modified arrays, suggesting that PRC2 residing at the boundary between unmodified and modified nucleosome domains adopts a binding configuration that destabilizes its nucleosome engagement and allows its efficient spreading. In the future, it would be valuable to visualize the behaviors of PRC2 on chromatin substrates at different modification states using structural and single-molecule approaches.
In conclusion, our results unveil a diverse repertoire of PRC2 binding modes on chromatin. The in vivo concentration of PRC2 is estimated to range between 50 and 1,000 nM (52), similar to what we used in the experiments. Thus, these interaction modes are expected to contribute significantly to PRC2 function inside the nucleus, which is further regulated by the epigenetic and genetic states of the chromatin, as well as by the availability of various accessory factors. Future experiments are warranted to answer whether these interaction modalities are mediated by PRC2 monomers, dimers, or higher-order multimers (8, 65, 66). The experimental and computational platforms established in this work will facilitate continued exploration of the elaborate mechanisms that PRC2 and other epigenetic machineries utilize to achieve chromatin targeting and maintenance.
Materials and Methods
Bacterial Cell Culture.
Core histone expression vectors were transformed into Escherichia coli BL21 (DE3) cells and grown in lysogeny broth (Fisher Scientific) containing 100 µg/mL ampicillin and 34 µg/mL chloramphenicol at 37 °C, induced midlog phase with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and then harvested after 4 h (6,000 × g, 15 min, 4 °C). Vectors for DNA extraction (12 × 601 and mouse mammary tumor virus [MMTV]) were transformed into E. coli DH5α cells and grown in lysogeny broth containing 100 µg/mL ampicillin and 50 µg/mL kanamycin at 37 °C and harvested (6,000 × g, 15 min, 4 °C) after 16 to 18 h following the ZymoPURE protocol.
Insect Cell Culture.
All PRC2 proteins used in this study were prepared in Sf9 cells using a baculovirus system. Flag-tagged EZH2 and His-tagged EED, RBBP4, and SUZ12 were cloned into a pACEBac1 vector using the Multibac system (67). His-AEBP2 and Flag-JARID2 were prepared in their own individual pACEBac1 vectors. Plasmids were used to generate bacmids according to the manufacturer’s protocol (Multibac; Geneva Biotech); 2.5 µg of bacmid was transfected into 1.0 × 106 attached Sf9 cells in a six-well plate. Following transfection, cells were overlaid with 2 mL of fresh medium (Sf-900III SFM; Thermo Fisher Scientific) and incubated at 27 °C for 96 h in the dark. The supernatant was collected, filtered through 0.22 µm, and supplemented with 2% vol/vol fetal bovine serum (FBS) to produce the P1 virus. P2 virus was generated by infection of 10 mL of Sf9 cells (1.5 × 106 cells per mL) with 1 mL of P1 virus solution. Cells were grown at 27 °C in suspension culture until they reached <50% viability as monitored by trypan blue staining. Culture supernatant was collected, filtered, and supplemented with 2% vol/vol FBS. To generate the P3 virus, 300 µL of P2 virus solution was added to 50 mL of Sf9 cells (1.5 × 106 cells per mL). Cells were grown at 27 °C in suspension culture until they reached <50% viability. Culture supernatant was collected, filtered, and supplemented with 2% vol/vol FBS. For protein expression, a 1:100 dilution of P3 virus was added to Sf9 cells at 2.0 × 106 cells per mL density. After 48 h of incubation at 28 °C in the dark, cells were harvested by centrifugation.
Purification of PRC2 Complexes.
Sf9 cells were lysed by a Dounce homogenizer (Wheaton) in HEGN600 buffer (25 mM Hepes, pH 7.0, 600 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA; 10% vol/vol glycerol, and 0.02% Nonidet P-40). Soluble extracts were incubated with anti-Flag M2 affinity gel (100 µL resin per 100 mL cell culture) in HEGN350 (25 mM Hepes, pH 7.0, 350 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.02% Nonidet P-40) for 2 h at 4 °C. Bound proteins were eluted with HEGN350 containing 0.25 mg/mL Flag peptide (3 × 20 min at 4 °C). Eluted proteins were pooled, spin-concentrated in a Vivaspin centrifugal concentrator (molecular weight cutoff [MWCO] 30,000; Viva Products), and purified by size-exclusion chromatography on a Superose 6 column (GE Healthcare). Final products were eluted in 25 mM Hepes, pH 7.0, 350 mM NaCl, 2.5mM MgCl2, 10% glycerol, 0.02% Nonidet P-40, and 1 mM dithiothreitol (DTT). Fractions containing monomeric PRC2 (as analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis) were pooled, flash-frozen with liquid N2, and stored at −80 °C. Prior to use, the protein concentration was quantified by A280 and a bovine serum albumin standard curve.
Purification of Histone Proteins.
The human core histones (H2A, H2B, H3, and H4) were expressed in BL21 (DE3) cells. The cells were lysed and inclusion bodies were harvested through rounds of sonication and centrifugation (68). Histones were extracted from inclusion bodies with dimethyl sulfoxide and purified through ion-exchange chromatography. Octamers were reconstituted using salt dialysis and size-exclusion chromatography (Superdex 200 10/300) as described previously (68). H3K27me3-modified histone octamers were purchased from EpiCypher. K27M and K27R mutant H3 histones were cloned using site-directed mutagenesis.
Reconstitution of Nucleosome Arrays.
DNA templates were digested with BglI (New England Biolabs) from a plasmid containing 12 repeats of 601 nucleosome positioning sequences (69) in a pET28b backbone and then ligated with T4 DNA Ligase (New England Biolabs) to DNA handles containing two biotins on each end. Nucleosome arrays were formed on the DNA template described above through salt-gradient dialysis from 1.4 M KCl to 10 mM KCl in Slide-a-Lyzer MINI Dialysis units (7,000 MWCO) using a peristaltic pump set at a 1 mL/min flow rate for 6 h. MMTV DNA was added during dialysis to prevent octamer overloading. Nucleosome arrays were formed at a target final concentration of 1 µM.
Single-Molecule Experiments and Data Analysis.
Data acquisition.
Single-molecule experiments were performed at room temperature on a LUMICKS C-Trap instrument equipped with dual-trap optical tweezers (70). A computer-controlled stage enabled rapid movement of the optical traps within a five-channel flow cell; 3.23-µm streptavidin-coated polystyrene beads (Spherotech) were flowed into channel 1 and captured by optical traps. The traps were then moved to channel 2 that contained nucleosome array samples diluted in the imaging buffer (10 mM Tris⋅HCl, pH 8.0, 0.1 mM EDTA, 200 mM KCl, 0.5 mM MgCl2, 0.1% Tween, and 1 mM DTT). Nucleosome array tethers were formed between two beads under flow, which reduces the chance of sticking. The tether was then transferred to channel 3 (containing imaging buffer) or channel 4 (containing PRC2 samples) for data collection. The PRC2 core complex (or PRC2–AEBP2, PRC2–JARID2) was diluted to 500 nM (unless otherwise noted) in the same imaging buffer. For experiments with PRC2, there was free PRC2 in solution for the entire duration of the experiments. PRC2 was incubated with nucleosome arrays at zero force for 5 to 10 s prior to pulling. Where applicable, SAM was added at a final concentration of 1.4 µM. The tethers were subjected to mechanical pulling by moving one trap relative to the other at a constant velocity (0.1 µm/s), generating force-extension curves. The force measurement has a resolution (SD of the force over 10-s time windows) of 0.2 pN and a stability (peak-to-peak difference of the force over a 2-min interval) of <0.3 pN.
Force-extension analysis.
Single tethers were screened based on signatures in the force-extension curve (position of the first segment of the curve and a single-step tether breakage). At least 15 single tethers were analyzed for each condition. Only the force-extension data from the first pull of each tether were analyzed in order to eliminate ambiguities related to nucleosome dissociation. Force-extension traces were processed using a custom Force-Extension Analyzer software suite. Each trace was separated into segments of constant contour length (L0) based on disruption peaks. Disruption peaks were identified by applying a Butterworth low-pass filter to the trace and assigning transitions when the force decreased by a user-specified threshold, typically 0.2 pN. The unfiltered data for each segment were then fit with the extensible worm-like-chain (WLC) model: (71). In order to prevent overfitting and to allow for direct comparison of the array’s changes in L0, the persistence length (Lp) and elastic modulus (K0) were determined for each nucleosome array based on the first and last segments, respectively. Lp was determined by fitting the Marko–Siggia WLC model to the first segment in the low-force regime (72). Keeping this Lp fixed, the array’s K0 was determined by fitting the last segment, in the high-force regime, to the extensible WLC model. Each segment was subsequently fit to the extensible WLC model to determine its L0, maintaining Lp and K0 constant for a given trace. The average Lp value extracted from our data is 26 ± 4 nm. The average K0 value is 1,500 ± 200 pN. The somewhat low Lp values may be due to the specific imaging buffer used in the experiments or due to insufficient data points from the first segment to obtain an accurate value. Nonetheless, the contour length changes and transition forces extracted from the force-extension traces were insensitive to the choice of Lp values in the range between 25 and 50 nm. Relevant parameters (number of segments, transition force, and contour length change ΔL0) for each trace were exported. The code and tutorial of the Force-Extension Analyzer software is available at https://github.com/alushinlab/ForceExtensionAnalyzer.
Cluster assignment.
To identify the groups composing all observed transitions, the distribution of changes in contour length (ΔL0) was modeled using a Gaussian mixture model (GMM). Putative models were generated with varying numbers of components, and each model was assessed using the BIC score (SI Appendix, Fig. S3C). To evaluate the uncertainties of the BIC scores, bootstrapping was performed. Samples were drawn with replacement from the measured contour length changes, followed by clustering with an M-component GMM (M ranges from 1 to 10) and BIC scoring. This process was repeated 100 times to compute 95% confidence intervals for the BIC scores. Models with fewer than four components had significantly worse BIC scores than those with four or more components. Models with five or more components had BIC scores that were worse than or within the uncertainty of the four-component model. Furthermore, the models with four or five components had similar BIC scores and virtually the same cluster boundaries, with the only effective difference being that the cluster of large transitions would be further subdivided in the five-component model. Therefore, a four-component model was used in subsequent analyses. Each transition was then assigned to its cluster of maximum likelihood based on the GMM.
To further validate cluster robustness and to estimate uncertainties in the cluster boundaries, the following bootstrapping procedure was performed: Fifty percent of the transitions were randomly selected 100,000 times with replacement, and the GMM clustering analysis was performed on each subset. We found that 96.3% of the random subsets led to a nearly identical clustering pattern as the aggregated data; for the remaining 3.7% of subsets, clusters 1 and 2 collapsed into each other. This ambiguity was due to the relatively low frequency of transitions in cluster 1 in this small percent of randomly subsampled data and, thus, these models can be rejected in context of the bare DNA data shown in SI Appendix, Fig. S4. This bootstrapping procedure allowed us to estimate 95% confidence intervals for the cluster boundaries: 50.8 ± 1.3 bp, 104.4 ± 1.4 bp, and 179.0 ± 4.1 bp for the first, second, and third cutoffs, respectively. These results indicate that the four-cluster model is robust and the cluster boundaries are stable. Modeling was implemented in Python using the Scikit-learn library and default settings (73). Finally, kernel density estimation was performed to provide a smoothed depiction of the underlying distribution of contour length changes using the Scikit-learn library with a Gaussian kernel. A bandwidth of 6.6 bp was used.
Negative-Stain Electron Microscopy.
EM imaging.
The 12-mer and 4-mer nucleosome arrays were formed using salt dialysis as described above. Arrays were purified by MgCl2 precipitation (4 mM final MgCl2 concentration, centrifugation at 15,000 rpm for 10 min at 4 °C). PRC2 and nucleosome arrays were dialyzed into EM buffer (50 mM Hepes, pH 7.9, 50 mM KCl, and 1 mM TCEP). Samples were adsorbed to glow-discharged carbon-coated copper grids and stained with uranyl formate. Imaging was performed on a CM10 electron microscope at a nominal magnification of 52,000×.
Image analysis of electron micrographs.
The coordinates of nucleosomes in each micrograph were manually picked using the e2boxer tool in EMAN2 (74). These coordinates were assigned to nucleosome arrays by detecting whether they existed within masks encompassing the arrays. These masks were generated for each array as follows. Negative-stain micrographs were down-sampled after binning over 4 × 4 pixels to speed up computations. Uneven illumination was corrected by calculating an adaptive, local threshold and subtracting it from the down-sampled micrograph. These images were then entropy-filtered, followed by binarization at a fixed threshold per micrograph. Masks were slightly dilated and returned to the original scale. Picked coordinates within a mask were considered to be part of the same nucleosome array. For 12-mer arrays, only masks associated with five or more picked nucleosomes were selected for further analysis to evaluate the level of compaction.
Statistical Analysis.
Errors reported in this study represent the SEM. Unless noted otherwise, P values were determined from two-tailed two-sample t tests (n.s., not significant; P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001).
Descriptions of dCas9 roadblock experiments, in silico simulations, and ChIP-seq data modeling can be found in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank members of the S.L. laboratory for discussions, members of the T.W. laboratory for help with electron microscopy, S. Cai (Rockefeller University) for dCas9 purification, M. Newton (Imperial College London) for help with dCas9 sgRNA design, and J. Cabanas-Danés (LUMICKS) for technical assistance. S.L. acknowledges support from the Robertson Foundation, the Quadrivium Foundation, a Monique Weill-Caulier Career Award, a Sinsheimer Scholar Award, a Kimmel Scholar Award, and the NIH (DP2HG010510). B.Z. is supported by the NSF (MCB-1715859) and the NIH (R35GM133580).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003395117/-/DCSupplemental.
Data Availability.
All data are included in the paper and SI Appendix .
References
- 1.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Margueron R., Reinberg D., The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen K. H., et al. , A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Margueron R., et al. , Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poepsel S., Kasinath V., Nogales E., Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25, 154–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J., et al. , DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat. Struct. Mol. Biol. 24, 1039–1047 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Wang X., et al. , Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 24, 1028–1038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heenan P. R., Wang X., Gooding A. R., Cech T. R., Perkins T. T., Bending and looping of long DNA by Polycomb repressive complex 2 revealed by AFM imaging in liquid. Nucleic Acids Res. 48, 2969–2981 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diehl K. L., et al. , PRC2 engages a bivalent H3K27M-H3K27me3 dinucleosome inhibitor. Proc. Natl. Acad. Sci. U.S.A. 116, 22152–22157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge E. J., Jani K. S., Diehl K. L., Müller M. M., Muir T. W., Nucleation and propagation of heterochromatin by the histone methyltransferase PRC2: Geometric constraints and impact of the regulatory subunit JARID2. J. Am. Chem. Soc. 141, 15029–15039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laugesen A., Højfeldt J. W., Helin K., Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74, 8–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J. R., Lee C. H., Oksuz O., Stafford J. M., Reinberg D., PRC2 is high maintenance. Genes Dev. 33, 903–935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiles E. T., Selker E. U., H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43, 31–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hojfeldt J. W., et al. , Non-core subunits of the PRC2 complex are collectively required for its target-site specificity. Mol. Cell 76, 423–436.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healy E., et al. , PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol. Cell 76, 437–452.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Lee C. H., et al. , Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol. Cell 70, 435–448.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youmans D. T., Schmidt J. C., Cech T. R., Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 32, 794–805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H., Kang K., Kim J., AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 37, 2940–2950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grijzenhout A., et al. , Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development 143, 2716–2723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X., et al. , Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139, 1303–1314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanulli S., et al. , Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 57, 769–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Son J., Shen S. S., Margueron R., Reinberg D., Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 27, 2663–2677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasini D., et al. , JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Li G., et al. , Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24, 368–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng J. C., et al. , Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139, 1290–1302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chien F. T., van Noort J., 10 years of tension on chromatin: Results from single molecule force spectroscopy. Curr. Pharm. Biotechnol. 10, 474–485 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Ordu O., Lusser A., Dekker N. H., Recent insights from in vitro single-molecule studies into nucleosome structure and dynamics. Biophys. Rev. 8, 33–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlick T., Hayes J., Grigoryev S., Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 287, 5183–5191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clauvelin N., et al. , Nucleosome positioning and composition modulate in silico chromatin flexibility. J. Phys. Condens. Matter 27, 064112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norouzi D., Katebi A., Cui F., Zhurkin V. B., Topological diversity of chromatin fibers: Interplay between nucleosome repeat length, DNA linking number and the level of transcription. AIMS Biophys. 2, 613–629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong B. E., Brouwer T. B., Kaczmarczyk A., Visscher B., van Noort J., Rigid basepair Monte Carlo simulations of one-start and two-start chromatin fiber unfolding by force. Biophys. J. 115, 1848–1859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lequieu J., Córdoba A., Moller J., de Pablo J. J., 1CPN: A coarse-grained multi-scale model of chromatin. J. Chem. Phys. 150, 215102 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Chang L., Takada S., Histone acetylation dependent energy landscapes in tri-nucleosome revealed by residue-resolved molecular simulations. Sci. Rep. 6, 34441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenzaki H., Takada S., Partial unwrapping and histone tail dynamics in nucleosome revealed by coarse-grained molecular simulations. PLOS Comput. Biol. 11, e1004443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moller J., Lequieu J., de Pablo J. J., The free energy landscape of internucleosome interactions and its relation to chromatin fiber structure. ACS Cent. Sci. 5, 341–348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lequieu J., Córdoba A., Schwartz D. C., de Pablo J. J., Tension-dependent free energies of nucleosome unwrapping. ACS Cent. Sci. 2, 660–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons T., Zhang B., Critical role of histone tail entropy in nucleosome unwinding. J. Chem. Phys. 150, 185103 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Zhang B., Zheng W., Papoian G. A., Wolynes P. G., Exploring the free energy landscape of nucleosomes. J. Am. Chem. Soc. 138, 8126–8133 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Brower-Toland B. D., et al. , Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 1960–1965 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall M. A., et al. , High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16, 124–129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mihardja S., Spakowitz A. J., Zhang Y., Bustamante C., Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. U.S.A. 103, 15871–15876 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noel J. K., Whitford P. C., Sanbonmatsu K. Y., Onuchic J. N., SMOG@ctbp: Simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 38, W657–W661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noel J. K., et al. , SMOG 2: A versatile software package for generating structure-based models. PLOS Comput. Biol. 12, e1004794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freeman G. S., Hinckley D. M., Lequieu J. P., Whitmer J. K., de Pablo J. J., Coarse-grained modeling of DNA curvature. J. Chem. Phys. 141, 165103 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Chen S., Jiao L., Shubbar M., Yang X., Liu X., Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol. Cell 69, 840–852.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Justin N., et al. , Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasinath V., et al. , Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940–944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eswar N., et al. , Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 15, 5.6.1-5.6.30 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Højfeldt J. W., et al. , Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 25, 225–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper S., et al. , Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis P. W., et al. , Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stafford J. M., et al. , Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci. Adv. 4, eaau5935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatavosian R., et al. , Live-cell single-molecule dynamics of PcG proteins imposed by the DIPG H3.3K27M mutation. Nat. Commun. 9, 2080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margueron R., et al. , Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kundaje A. et al.; Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stein R. R., Marks D. S., Sander C., Inferring pairwise interactions from biological data using maximum-entropy probability models. PLOS Comput. Biol. 11, e1004182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie W. J., Zhang B., Learning the formation mechanism of domain-level chromatin states with epigenomics data. Biophys. J. 116, 2047–2056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comet I., Riising E. M., Leblanc B., Helin K., Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 16, 803–810 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Grigoryev S. A., et al. , Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. U.S.A. 113, 1238–1243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dodd I. B., Micheelsen M. A., Sneppen K., Thon G., Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 129, 813–822 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Erdel F., Greene E. C., Generalized nucleation and looping model for epigenetic memory of histone modifications. Proc. Natl. Acad. Sci. U.S.A. 113, E4180–E4189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon J. A., Kingston R. E., Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oksuz O., et al. , Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell 70, 1149–1162.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davidovich C., Goodrich K. J., Gooding A. R., Cech T. R., A dimeric state for PRC2. Nucleic Acids Res. 42, 9236–9248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S., Jiao L., Liu X., Yang X., Liu X., A dimeric structural scaffold for PRC2-PCL targeting to CpG island chromatin. Mol. Cell 77, 1265–1278.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berger I., et al. , The multiBac protein complex production platform at the EMBL. J. Vis. Exp. 77, e50159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vary J. C. Jr, Fazzio T. G., Tsukiyama T., Assembly of yeast chromatin using ISWI complexes. Methods Enzymol. 375, 88–102 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Hashemi Shabestari M., Meijering A. E. C., Roos W. H., Wuite G. J. L., Peterman E. J. G., Recent advances in biological single-molecule applications of optical tweezers and fluorescence microscopy. Methods Enzymol. 582, 85–119 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Wang M. D., Yin H., Landick R., Gelles J., Block S. M., Stretching DNA with optical tweezers. Biophys. J. 72, 1335–1346 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marko J. F., Siggia E. D., Stretching D. N. A.. Macromolecules 28, 8759–8770 (1995). [Google Scholar]
- 73.Pedregosa F., et al. , Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 74.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the paper and SI Appendix .