Significance
We have developed a fluorescent protein, mGreenLantern, that features exceptionally high brightness in mouse, bacterial, and human cells (up to sixfold brighter than EGFP) and have demonstrated its superior ability to highlight neuronal morphology compared to EGFP and EYFP. Screening fluorescent protein mutants based on whole-cell brightness while evaluating expression kinetics in lysate enabled us to identify variants exhibiting striking divergences between their computed spectroscopic brightness and actual performance in cells. mGreenLantern additionally features unusually high chemical and thermodynamic stability and is compatible with existing GFP filter sets, excitation sources, commercial EGFP antibodies, expansion microscopy, and whole-brain tissue clearing. Our hypothesis-driven engineering strategy represents a generalizable method with great potential to enhance the performance of constitutive reporters and GFP-based biosensors.
Keywords: GFP, fluorescent protein, neurobiology, imaging, protein engineering
Abstract
Although ubiquitous in biological studies, the enhanced green and yellow fluorescent proteins (EGFP and EYFP) were not specifically optimized for neuroscience, and their underwhelming brightness and slow expression in brain tissue limits the fidelity of dendritic spine analysis and other indispensable techniques for studying neurodevelopment and plasticity. We hypothesized that EGFP’s low solubility in mammalian systems must limit the total fluorescence output of whole cells, and that improving folding efficiency could therefore translate into greater brightness of expressing neurons. By introducing rationally selected combinations of folding-enhancing mutations into GFP templates and screening for brightness and expression rate in human cells, we developed mGreenLantern, a fluorescent protein having up to sixfold greater brightness in cells than EGFP. mGreenLantern illuminates neurons in the mouse brain within 72 h, dramatically reducing lag time between viral transduction and imaging, while its high brightness improves detection of neuronal morphology using widefield, confocal, and two-photon microscopy. When virally expressed to projection neurons in vivo, mGreenLantern fluorescence developed four times faster than EYFP and highlighted long-range processes that were poorly detectable in EYFP-labeled cells. Additionally, mGreenLantern retains strong fluorescence after tissue clearing and expansion microscopy, thereby facilitating superresolution and whole-brain imaging without immunohistochemistry. mGreenLantern can directly replace EGFP/EYFP in diverse systems due to its compatibility with GFP filter sets, recognition by EGFP antibodies, and excellent performance in mouse, human, and bacterial cells. Our screening and rational engineering approach is broadly applicable and suggests that greater potential of fluorescent proteins, including biosensors, could be unlocked using a similar strategy.
Modern neuroscience research is placing increasing demands on fluorescent protein (FP) technology for robust performance in new applications, including high-resolution interrogation of intact biological systems through tissue clearing and expansion. With single-circuit studies and two-photon (2P) imaging in the living brain becoming increasingly routine, there is a strong need for brighter genetically encoded tags with efficient folding, broad dispersal, and fast expression in brain tissue to facilitate unambiguous discrimination of individual neurons. Limitations in FP brightness have led researchers to use various enhancement strategies, including intracellular dye injection into GFP-expressing neurons (1) and antibody labeling even in transgenic animals (2), which are strategies that consume additional time and resources and present their own set of limitations (3).
The brightest GFPs recently developed are Clover (4), mClover3 (5), and mNeonGreen (6), but they exhibit only minor differences in quantum yield (QY) and extinction coefficient (EC) values, suggesting that these properties may have reached a local maximum in FPs that contain a GYG-type chromophore (5). However, a biomolecule’s brightness in vitro hardly matters if it does not develop efficiently in a cell, and it has long been known that a considerable fraction of FP molecules misfold and/or fail to form a functional chromophore at 37 °C (7), resulting in aggregation-prone products incapable of fluorescence (8).
We therefore reasoned that enhancing the folding efficiency and solubility of FPs in mammalian systems should yield a larger fraction of functional emitting FP molecules per cell, increasing the cell’s total brightness and improving detection by fluorescence microscopy, independent of the FP’s molecular brightness (QY × EC). Aided by the wealth of structure–activity information available for Aequorea victoria FPs (avFPs), we chose to forego random mutagenesis and instead rationally introduced sets of known avFP “folding mutations” into GFP templates, beginning with Clover, and screened mutants from each round of selection for brightness in Escherichia coli, brightness in mammalian cells, and then for rapid chromophore maturation. The GFPs were compared in vitro and in vivo against six widely used FPs.
The best-performing mutant, mGreenLantern, is a bright, monomeric FP tailored for accelerated expression and high brightness in living cells, including neurons. The rational engineering of mGreenLantern, and its demonstrated superiority in neuronal highlighting experiments compared to enhanced green fluorescent protein (EGFP) and enhanced yellow fluorescent protein (EYFP), show that biological fluorophores can be optimized for robust expression in living cells with minimal impact on their intrinsic spectral properties. mGreenLantern expresses rapidly and brightly in mouse, human, and bacterial cells, indicating that it will be useful in a broad range of applications and organisms.
Results
Development and Screening of “Mini Libraries.”
To test our hypothesis that folding mutations can influence the brightness of FP-expressing cells, we introduced four substitutions that are absent in Clover but important for conferring solubility and brightness at 37 °C in Emerald GFP, particularly when used in combination: F64L/S72A/I167T/S175G (omitting N149K) (8, 9). When we screened transformants from the multisite mutagenesis reaction by eye using 470-nm transillumination, the brightest E. coli colonies sequenced as the quadruple mutant, which we will refer to as “FOLD1.” Interestingly, purified FOLD1 protein approximated Clover’s EC (∼100 mM−1⋅cm−1), but its QY had decreased from 0.75 to 0.70 (SI Appendix, Table S1), indicating that features apart from molecular brightness must be influencing the whole-cell fluorescence output.
We searched for patterns of mutations in the literature reported to contribute to brightness- and folding-related enhancements in diverse avFPs and applied them to the brightest variants uncovered from each round of mutagenesis, beginning with FOLD1. We used multisite mutagenesis to produce “mini libraries” of up to 3, 7, and 15 unique variants when using sets of two, three, and four primers, respectively, selecting the brightest E. coli colonies by eye for sequencing, downstream mutagenesis, and incorporation into a mammalian coexpression plasmid for quantitative comparison. Among the substitutions tested were the monomerizing mutations A206K, L221K, and F223R (10); E124V, K101E, T105Y, G232D, and D234N mutations, which are present in some fast-folding and well-expressed GFP variants (11, 12); S147P, discovered in an early avFP mutant with high thermostability (13); N149K, I167T, and S72A, which appear in Emerald (14) and are known to influence maturation (15); deletion of G4 (ΔG4), reported in a thermostable GFP (16); and N149Y, from mClover3 (5). Mutants are summarized in SI Appendix, Table S2.
Cellular Brightness.
To quantify and compare the brightness of chemically transfected mammalian cells (“cellular brightness”) under standardized conditions, we cloned FPs into a mammalian coexpression plasmid with the FP inserted N-terminal to a 2A linker followed by a C-terminal mCherry (Fig. 1 A, Inset). This system generates the two FPs in a roughly 1:1 stoichiometric ratio and thereby controls internally for transfection-related variability by allowing normalization to the mCherry signal (17).
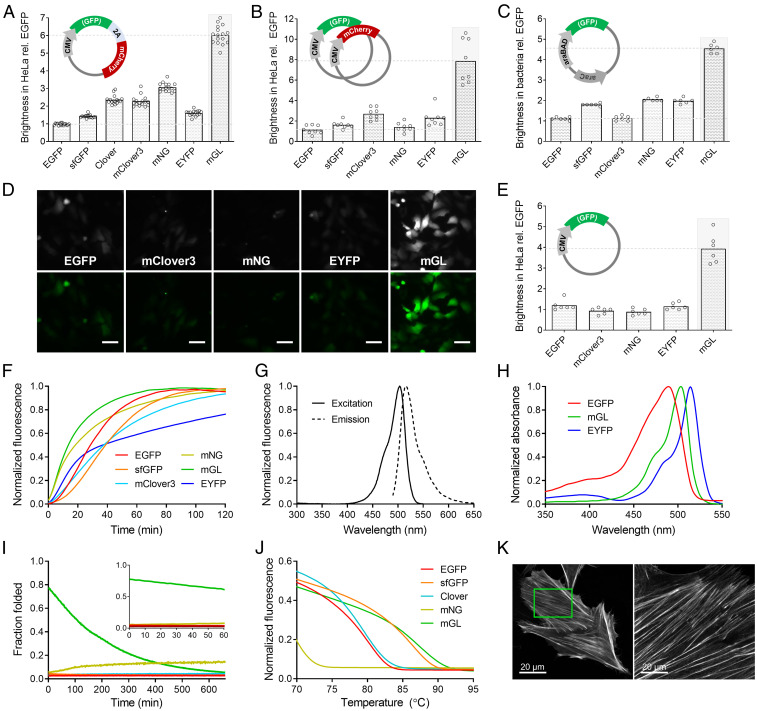
Fig. 1.
Biochemical properties and brightness of FPs in cultured cells. (A) Brightness of FPs in HeLa cells 48 h after transfection with a single coexpression plasmid that produces an FP and mCherry in a 1:1 stoichiometric ratio. (B) Brightness of FPs in HeLa cells cotransfected with an FP and mCherry from separate plasmids. (C) Brightness of FPs in E. coli after overnight expression from the pBAD vector. (D) Cytosolic FPs in HeLa cells imaged 48 h after chemical transfection using plasmids from B, excluding mCherry. All FPs were imaged using identical settings on a widefield microscope. The same image is shown in grayscale and standard green pseudocolor. Magnification, 10×. (Scale bars, 50 µm.) (E) Average brightness of HeLa cells expressing the FPs in D, n = 6 replicates, ≥300 cells analyzed per transfection per FP. (F) Maturation of FP chromophores from lysed bacteria as described in Methods. (G) Excitation and emission spectra of mGreenLantern. (H) Unlike EGFP and EYFP, mGreenLantern (mGL) does not absorb 405-nm near-UV light. (I) Kinetic unfolding of purified FPs in a denaturing solution of guanidinium HCl, 6.3 M, pH 7.5 (GdnHCl). (J) Melting of purified FPs in a thermal cycler. (K) HeLa cells expressing an actin-mGreenLantern fusion protein. (Left) Live cells before the expansion microscopy (proExM) process. (Right) A different representative cell after proExM, imaged in the expanded state in ddH2O. Both images were collected at 63× magnification. (Scale bars for both images: 20 µm.) The green box indicates approximate size of postexpansion field of view based on resolution.
Clover and mClover3 did not differ in cellular brightness, whereas mNeonGreen was 1.5-fold brighter than mClover3 despite similar molecular brightness. EYFP and superfolder GFP (sfGFP) were both dimmer than Clover and 1.8- and 1.5-fold brighter than EGFP, respectively (Fig. 1A). Cellular brightness trends were consistent across all three mammalian cell lines tested (SI Appendix, Fig. S1). HeLa cells expressing FOLD1 were 3.3-fold brighter than EGFP and 2.2-fold brighter than Clover, a difference that cannot be explained by molecular brightness alone (SI Appendix, Table S1).
Based on cellular brightness data and visual comparison of E. coli plates, we suspected that the fluorescence intensity had already peaked with mF1Y (FOLD1-A206K/N149Y) and was declining in subsequent mutants, as exemplified by our most extensively modified variant, mF3CPK (SI Appendix, Table S2). Initially, we had produced the double mutant, FOLD1-A206K/N149Y, before the single mutant, FOLD1-A206K, so we did not know whether individual or concerted effects accounted for the improvement in cellular brightness. While producing the FOLD1-V206K single mutant, we serendipitously introduced V206K into mF2BK for comparison, expecting that it would have little to no effect on brightness, since A206K has been reported previously as neutral to brightness (18). However, after overnight incubation, we observed green colonies that glowed brilliantly under ambient lighting in the way that other mutants typically appeared only after several days of refrigeration.
When expressed in HeLa cells, mF2BK (V206) was 3.0-fold brighter than EGFP, but the V206K substitution in this FP background (mGreenLantern) doubled that value to 6.3-fold greater brightness than EGFP. Encouraged by the results, we introduced D234N into mF2BKK, but the resulting mutant lost the benefit conferred by A206K, suggesting that a delicate balance of concerted effects contribute to cellular brightness in this background.
To confirm the reliability of the coexpression system used in Fig. 1A, we transfected HeLa cells with two plasmids, one coding for a GFP and another for mCherry (Fig. 1B). Separately in E. coli, we expressed the FPs using arabinose-inducible bacterial pBAD plasmids (Fig. 1C). In each case, we observed similar hierarchical trends in relative brightness between the GFPs. Likewise, when we transfected HeLa cultures and analyzed the mean fluorescence intensity of cells using matched imaging settings on a widefield microscope (Fig. 1D), mGreenLantern cells were at least 3.2-fold brighter than those expressing EGFP, mClover3, mNeonGreen, or EYFP (Fig. 1E). Together, the data demonstrate that mGreenLantern’s brightness in cells was superior regardless of expression system, imaging method, or host organism.
Chromophore Maturation.
Since efficient chromophore maturation largely defines the latency from folding to fluorescence in autocatalytic FPs (8, 19), we evaluated the chromophore maturation kinetics of select mutants and controls. Chromophore cyclization is an O2-dependent process (20) that can be inhibited by inducing protein expression under hypoxia and measured by kinetic sampling of emission intensity after reintroduction of O2 in the bacterial lysate.
Of the FPs tested, mF1Y (FOLD1-A206K/N149Y) had the fastest maturation half-time, at 7.2 min, followed by mF2BK (V206) and mF2BKK (K206) (mGreenLantern) at 8.5 and 13.5 min, respectively (SI Appendix, Table S1). EYFP matured in 33.0 min, but it showed atypical maturation kinetics consisting of an extended plateau phase that we did not observe in the other FPs (Fig. 1F). Based on the high cellular brightness (630% greater than EGFP) and rapid maturation (207% faster than EGFP) of mGreenLantern, we selected it for further characterization.
Biochemical Characterization.
Spectroscopy.
mGreenLantern is compatible with common EGFP filter sets and the popular 488-nm argon-ion laser line for widefield and confocal microscopy. Its excitation and emission peaks of 503/514 nm (Fig. 1G) additionally make it well matched to newer solid-state 491-nm lasers and 505-nm light-emitting diodes (LEDs). The QY and EC of mGreenLantern (ϕ = 0.72, ε = 101 mM−1⋅cm−1) compare favorably with those of other bright GFPs such as Clover (ϕ = 0.75, ε = 97 mM−1⋅cm−1), and its low pKa of 5.6 indicates greater acid resistance than EGFP (Table 1). We also noted that mGreenLantern completely lacks a 405-nm absorbance peak, unlike EGFP (Fig. 1H) and Clover (21), suggesting that it will be better suited for multicolor imaging with T-Sapphire (22) or other FPs with a large Stokes shift by eliminating the problematic cross-excitation artifacts observed during UV excitation of EGFP (23).
Table 1.
Spectroscopic characterization of FPs
| Protein | λex | λem | ϕ | ε, M−1⋅cm−1 | Molecular brightness* | Cellular brightness† | pK* | Maturation, min | Tm, °C | Oligomeric state‡ |
| EGFP | 488 | 507 | 0.71 | 54,000 | 38 | 1.0 | 6.0 | 28 | 80.3 | Weak dimer§ |
| EYFP | 513 | 527 | 0.62 | 94,000 | 58 | 1.8 | 6.8 | >37 | ND | ND |
| Superfolder GFP | 485 | 508 | 0.67 | 51,000 | 34 | 1.5 | 6.2 | 37 | 86.4 | Weak dimer§ |
| Clover | 505 | 515 | 0.75 | 97,000 | 72 | 2.2 | 6.5 | 15 | 79.5 | Weak dimer§ |
| mClover3 | 506 | 516 | 0.80 | 100,000 | 80 | 2.1 | 6.1 | 37 | 80.1 | ND |
| mNeonGreen | 506 | 516 | 0.78 | 115,000 | 90 | 3.3 | 5.6 | 18 | 68.0 | Monomer§ |
| mGreenLantern | 503 | 514 | 0.72 | 102,000 | 74 | 6.3 | 5.6 | 14 | 87.2 | Monomer |
Molecular brightness = (ϕ × ε)/103, where ϕ is quantum yield and ε is extinction coefficient at the absorbance peak (λex ± 2 nm).
Brightness of FP-expressing BE (2)-M17 human neuroblastoma cells, relative to EGFP, using the P2A quantitative coexpression system (Fig. 1A and Methods). These cellular brightness values are also listed in SI Appendix, Table S1.
Here, we define “monomer” as OSER assay score >80%; “weak dimer,” 50 to 79%. We are not aware of OSER scores for EYFP and mClover3.
Cited from Cranfill et al. (24) and interpreted as described in footnote ‡. All other data in the table were experimentally determined in our laboratory for this study.
Considering the striking differences in cellular brightness between the FPs, we chose to recharacterize all controls used in this study (EGFP, sfGFP, Clover, mClover3, mNeonGreen, and EYFP) spectroscopically alongside mGreenLantern and several mutants (SI Appendix, Table S1). Our experimentally determined values compared favorably with those published in a comprehensive recent study (24).
Chemical and thermal stability.
We next asked whether a reduced rate of degradation or improved thermodynamic stability might contribute to mGreenLantern’s high cellular brightness. As a proxy for cellular persistence, we measured the susceptibility of FPs to chemical and thermal denaturation. Kinetic unfolding experiments have been useful for identifying and characterizing robustly stable FPs (12, 25), so we dispensed purified FPs into a strongly denaturing solution of 6.3 M guanidinium HCl, pH 7.5 (GdnHCl), and measured the rate of fluorescence quenching. All FPs except mGreenLantern denatured instantly. Strikingly, mGreenLantern persisted for 100 s before reaching half the fluorescence of its native control wells, and its emission was still measurable after 10 min in the denaturing solution (Fig. 1I).
We assessed thermostability by melting purified FP in a real-time PCR machine programmed to increase temperature by 0.3 °C/s while collecting fluorescence measurements. EGFP’s melting temperature of 80.3 °C was consistent with independent data (16) and approximately equaled that of Clover and mClover3. mNeonGreen showed the poorest thermostability and melted at 68.0 °C, while mGreenLantern just surpassed sfGFP at 87.2 °C (Fig. 1J and Table 1).
Tissue expansion and whole-brain clearing.
mGreenLantern’s tolerance of chemical and thermal denaturation was superior to that of all other FPs tested, so we hypothesized that these properties, along with its high brightness, would make mGreenLantern especially useful in protein-retention expansion microscopy (proExM), a technique that enables diffraction-limited superresolution imaging through expansion of hydrogel-infused tissue or cells with ultrapure water (26). During proExM, FPs are exposed to harsh conditions known to diminish fluorescence, including aldehyde fixation, detergents, proteases, halide concentrations up to 1 M, and in some protocols, high temperature and GdnHCl treatment (27). mGreenLantern survived proExM and facilitated visualization of delicate actin fibers of cultured HeLa cells, without antibody enhancement steps (Fig. 1K).
To determine whether mGreenLantern could retain fluorescence after solvent-based clearing of an intact mouse brain, we used an established retrograde viral strategy to localize mGreenLantern to the nuclei of supraspinal neurons, which are cells that form descending projections to the spinal column and are of considerable importance in spinal injury (28, 29). The organic solvents and tissue dehydration processes used in many clearing techniques are known to quench FP emission partially or totally, and although anti-FP immunostaining can improve signal, antibodies require considerable incubation time (up to weeks) and special protocols to penetrate into deep brain structures (30, 31). Therefore, a tissue-clearing compatible FP would be highly desirable for saving time and money while minimizing background fluorescence and avoiding the problem of antibody penetration.
rAAV2-retro-CAG-H2B-mGreenLantern virus was injected into the spinal column of adult mice. Whole brains were cleared using the three-dimensional imaging of solvent-cleared organs (3DISCO) technique (30) as described in Methods, followed by light sheet microscopy and three-dimensional (3D) reconstruction, again without antibodies. We observed brightly fluorescent nuclei that were readily distinguishable from background and clustered in the expected brain regions including motor cortex, hypothalamus, brainstem, and red nucleus, among others (SI Appendix, Fig. S2 and Movie S1).
The data demonstrate that mGreenLantern will be useful for a variety of tissue-processing applications that are known to reduce the emission of FPs through spatial separation and/or chemical quenching, thereby allowing greater fluorescence retention and signal-to-noise ratio in superresolution imaging of neuronal processes and 3D visualization of the intact brain.
Antibody recognition.
mGreenLantern eliminates the time-consuming immunostaining steps of proExM and 3DISCO, but antibodies serve myriad purposes beyond signal enhancement, including immunoprecipitation, immunopanning, nanobody purification, and Western blotting. We tested three commercially available antibodies for compatibility by immunocytochemistry and found that all of them recognized mGreenLantern and did not recognize mNeonGreen, which is derived from Branchiostoma lanceloatum rather than Aequorea victoria (SI Appendix, Fig. S3). This result, and the 91% sequence identity shared with EGFP, strongly implies that mGreenLantern is readily compatible with existing technology for EGFP affinity-related applications.
Monomericity.
We continued characterizing mGreenLantern and found that it behaves as a monomer in vitro and in vivo, with pure protein still eluting as a monomer by gel filtration chromatography at 95 µM concentration (SI Appendix, Fig. S4A). Since chromatography performance does not always predict behavior in cells (24), we additionally performed the organized smooth endoplasmic reticulum (OSER) assay (32) and observed that mGreenLantern showed as little aggregation as mEGFP, the monomeric FP standard (SI Appendix, Fig. S4 B and C). As expected, Clover and tdTomato behaved as weak and strong dimers, respectively.
When fused with various subcellular localization sequences, mGreenLantern-tagged proteins localized properly to common targets including actin, tubulin, mitochondria, clathrin, endoplasmic reticulum, and nucleus (SI Appendix, Fig. S5 A–F). Together, the data demonstrate that mGreenLantern behaves as an inert monomeric tag with superior performance to EGFP and Clover, which are not strictly monomeric.
Photostability.
Bleaching of mGreenLantern during laser-scanning confocal microscopy follows a monoexponential decay pattern typical of GFPs (SI Appendix, Fig. S5G) after completion of a minor Emerald-like effect (33) during the first several seconds of high-intensity illumination (SI Appendix, Fig. S5H). When analyzed using normalized bleaching plots beginning at time 0 to include the Emerald-like behavior, mGreenLantern’s photostability was better than Venus (33, 34) and lower than Clover (4). We have not observed accelerated bleaching during routine confocal or widefield imaging, perhaps due to the very low laser power demands of mGreenLantern, but we suspect that it may be possible to exploit the photostability difference between EGFP and mGreenLantern using photostability contrast techniques to resolve discrete cellular structures by spectral multiplexing (35).
Soluble protein yield.
To confirm a link between cellular brightness and soluble protein production in cells, we induced FP expression in E. coli overnight and analyzed the soluble and insoluble fractions using sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis, with total protein mass loaded equally per lane. Upon quantifying the intensity of the ∼27-kDa band that represents the FP monomer, we noted that mGreenLantern total protein yield exceeded that of EGFP, mClover3, and mNeonGreen combined (SI Appendix, Fig. S6 A and B). We observed similar trends in HEK293T and BE(2)-M17 cells, with mGreenLantern soluble protein yield exceeding that of the other FPs by approximately threefold (SI Appendix, Fig. S6 C and D). Greater soluble yield in mGreenLantern-expressing cells relative to other FPs may be an important component of its cellular brightness.
mGreenLantern Improves Cellular Labeling in the Mouse Brain.
We next wondered to what extent the improved maturation kinetics, brightness, and soluble protein yield of mGreenLantern would translate to functional improvements in highlighting and filling of long-range neuronal projections relative to EYFP. EYFP is frequently used in circuit-tracing studies, but it has been shown to produce inhomogeneous fluorescence in dendrites, perhaps due to uneven cytoplasmic distribution (2).
Neurotropic AAV2/1-EF1α-DIO-mGreenLantern-WPRE-pA or EYFP virus was prepared and co-injected stereotaxically with AAV-Cre into visual cortex at a 10:1 particle ratio. mGreenLantern brightly highlighted discrete neurons without excessive interference or overlap at only 7 d postinjection (d.p.i.) (Fig. 2A), and regional morphology was clearly discernible (Fig. 2B).
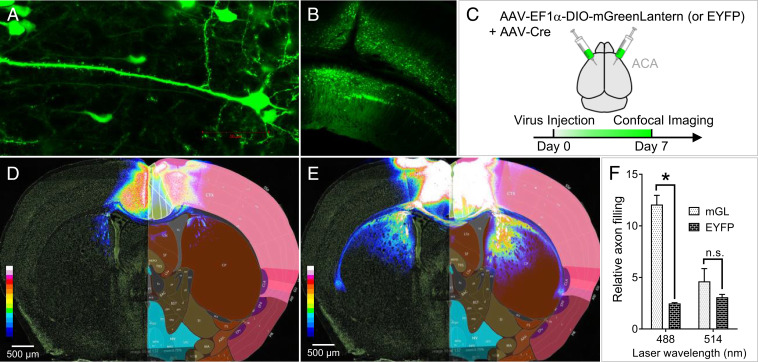
Fig. 2.
mGreenLantern expresses efficiently in the mouse brain and robustly illuminates long-range neuronal projections. (A) At 7 d post injection (d.p.i.), individual mGreenLantern-expressing neurons of the visual cortex were readily discernible using 63× magnification, (B) as well as the characteristic organization of visual cortex layer VI and hippocampus subfield CA1 at 10× magnification. (C) Injection strategy to label neurons using mGreenLantern and Cre virus mixture in a 10:1 particle ratio. (D) EYFP injected into the ACA as depicted in A did not effectively highlight neuronal projections from ACA to striatum at 14 d.p.i. Rather, fluorescence was primarily restricted to the ACA injection area. Grayscale confocal fluorescence microscopy images were converted to 16-color heat maps and overlaid with corresponding sections from the Allen Brain Atlas from an age-matched mouse for visual reference. (E) mGreenLantern fluorescence at 14 d.p.i. was clearly visible in neurons originating from ACA cell bodies with axonal projections radiating through striatum, corpus callosum, and claustrum. (F) Area of projections in corpus callosum and striatum relative to ACA expression is quantified; n = 3 mice, two-way ANOVA, *P < 0.05.
Having optimized our expression protocol, we injected mGreenLantern or EYFP virus alongside AAV-Cre into the anterior cingulate area (ACA) of individual mice (Fig. 2C), because this strategy would allow axons projecting to distant brain regions to be visualized in the striatum and corpus callosum in the same coronal plane. After 14-d expression, brains were coronally sectioned and imaged to examine the differences in extent of spread between EYFP and mGreenLantern. Remarkably, whereas the EYFP signal was generally limited to the ACA injection region (Fig. 2D), ACA cells expressing mGreenLantern showed bright projections radiating deep into the striatum and corpus callosum nearly 4 mm away (Fig. 2E).
We next sought to characterize whether the distal labeling observed was related to overall larger area of ACA labeling. We therefore normalized the area of FP signal in the striatum and corpus callosum to the area of FP in the ACA injection site. Furthermore, since mGreenLantern and EYFP are maximally excited by the 488- and 514-nm laser lines, respectively, we excited each FP with the two lasers individually under identical imaging conditions for comparison. Long-range projections of mGreenLantern-expressing neurons were clearly more visible than EYFP regardless of 488- or 514-nm excitation (SI Appendix, Fig. S7). Notably, we observed a 4.8-fold improvement in axon filling for mGreenLantern compared to EYFP in vivo (Fig. 2F), which is more than double the predicted in vitro molecular brightness from 488-nm excitation (ϕ × ε488). These results further suggest that molecular brightness is not enough to predict brightness of FP-expressing cells.
Rapid Expression of mGreenLantern In Vivo.
Next, we sought to determine whether the rapid chromophore maturation of mGreenLantern in vitro would translate to accelerated expression in vivo compared to EYFP, a popular fluorescent marker due to its long history, presence in numerous channelrhodopsin fusions, and its modestly improved brightness relative to EGFP (36). Rapid FP expression is particularly important in developmental biology experiments, when early time points for visualization are desired but often difficult to access due to long expression lag times.
We injected a mixture of AAV2/1-EF1α-DIO-mGreenLantern-WPRE-pA (or EYFP) plus AAV-Cre plus Alexa 594-coated latex beads (“tracer”) into the ACA of adult mice and collected coronal sections at 3, 7, and 14 d.p.i. (Fig. 3A). The tracer reaches maximal dispersal after 48 h and its signal and localization are stable for several weeks in the brain (37), allowing technical injection variability to be addressed by normalizing to the area of tracer fluorescence. In this experiment, we define “fluorescence” as the mean pixel intensity of the FP multiplied by its detectable fluorescence area (which increases over time), and divide the product by the area of detectable tracer area [which remains stable (37) or would be expected to decay similarly across conditions] (Fig. 3A).
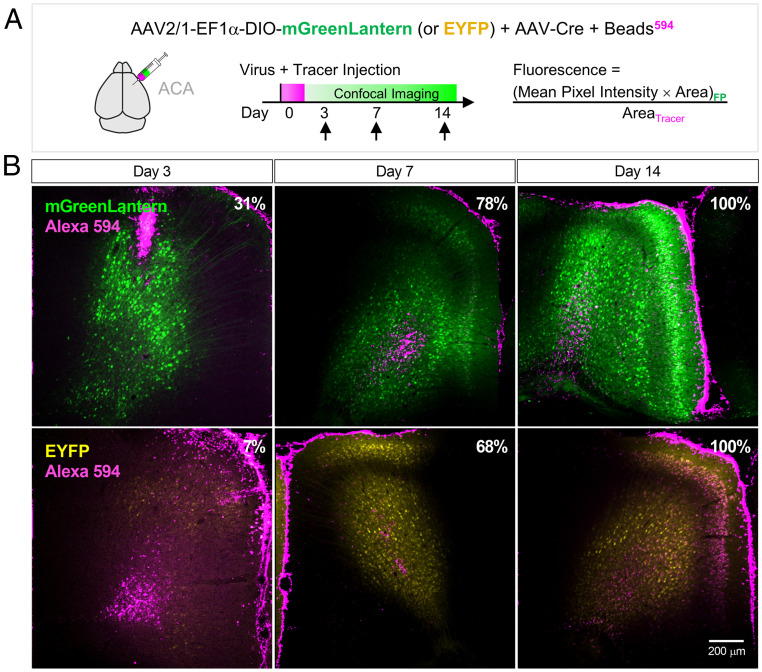
Fig. 3.
In vivo expression kinetics of virally transduced mGreenLantern and EYFP in mouse. (A) AAV2/1-EF1α-DIO-mGreenLantern-WPRE-pA or EYFP virus co-injected with AAV-Cre and Alexa 594-coated latex beads (tracer) into anterior cingulate area (ACA). (B) mGreenLantern and EYFP expression in ACA at the indicated time points. Gain was decreased equally for all samples at day 3 to avoid mGreenLantern oversaturation; settings were maintained for day 14. Percentages in Top Right represent the area filled relative to day 14 for each FP. Magnification, 10×. See SI Appendix, Fig. S8 for quantitation.
mGreenLantern displayed fourfold greater fluorescence area in the ACA than did EYFP at 3 d.p.i., with projections and cell bodies visible, whereas most EYFP fluorescence fell below the detection threshold and did not reveal cell morphology (Fig. 3B). At high magnification, we could observe neuronal substructures for mGreenLantern after only 7 d.p.i. (Fig. 4A). Using 488-nm excitation, the fluorescent area, mean pixel intensity, and fluorescence as defined in Fig. 3A, were all greater for mGreenLantern than EYFP (SI Appendix, Fig. S8). Values at 514 nm hardly differed in vivo, even though EYFP would have been predicted to show greater brightness than mGreenLantern based on spectroscopic data at 514 nm (ϕ × ε514 = 56 and 31, respectively). Collectively, the data suggest that faster maturation is a component of the greater in vivo expression of mGreenLantern.
Fig. 4.
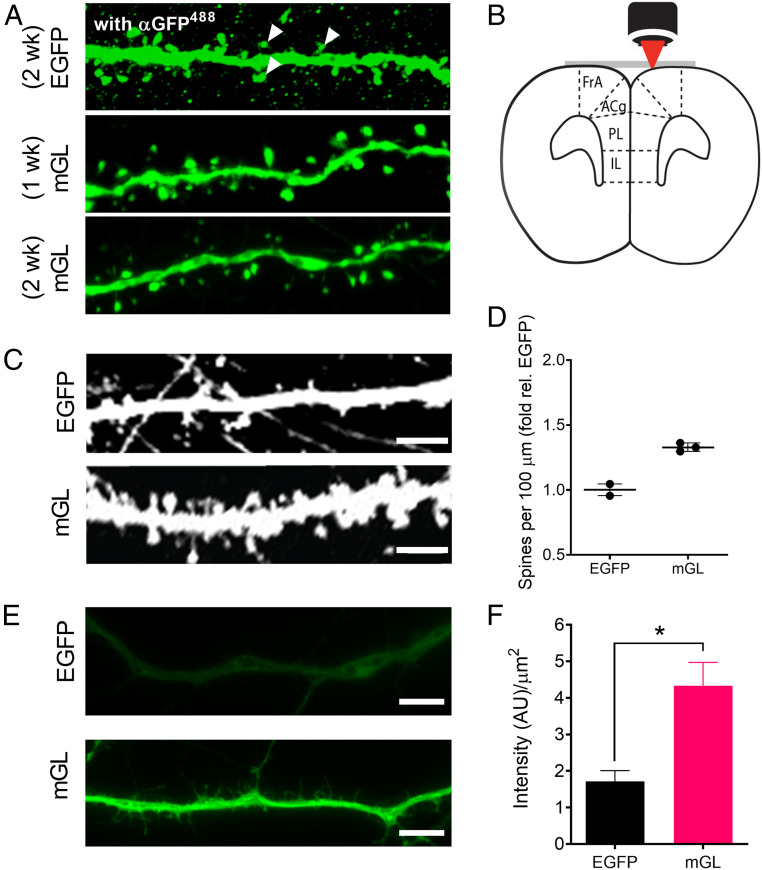
mGreenLantern (mGL) improves visualization of neuronal morphology in vivo and ex vivo. (A) Representative mGL-expressing neurons in ACA after 1- and 2-wk expression without antibody enhancement, compared to EGFP stained with α-GFP antibody at 2 wk. White wedges suggest the difficulty of distinguishing true neuronal substructures from background puncta after immunohistochemistry (IHC). Confocal microscopy, nonidentical gain settings. Magnification, 100×. (B) Schematic for two-photon (2P) imaging of the living mouse brain through a surgically implanted cranial window. (C) Representative images of neurons expressing EGFP and mGreenLantern in the cortex of live mice using 2P microscopy through a cranial window. Identical imaging settings. (Scale bar, 5 µm.) (D) Density of dendritic spines observed per 100 µm, normalized to EGFP. n = 95 and 101 neurons quantified from two and three mice for EGFP and mGreenLantern, respectively (mean ± SD). (E) Representative widefield images of neurons dissected from E17 mouse hippocampus and transduced with AAV2/1-CAG-EGFP or mGreenLantern virus after 10 d of culture. The neurons were fixed with PFA and imaged using identical excitation and acquisition settings with a standard EGFP filter set. Magnification, 63×. (Scale bar, 25 µm.) (F) Brightness of primary neurons from a minimum of three independent cultures, each with 20 random images quantified per condition. Mean ± SEM; Student’s t test, *P < 0.05.
We next asked whether the fast expression we observed after viral transduction was specific to that system or generalizable to transgenic animals with expression regulated by an endogenous promoter. We therefore introduced our Cre-dependent viral vector into the ACA of transgenic parvalbumin-Cre (PV) driver mice and perfused them at 24, 48, or 72 h. The earliest signs of mGreenLantern expression in PV neuron cell bodies were detectable within 48 h (SI Appendix, Fig. S9). We expect that the rapid expression of mGreenLantern in animals may facilitate developmental neuroscience experiments by allowing injections to occur at later postnatal stages, when the skull is further developed, thereby improving coordinate-based targeting.
Improved Detection of Neuronal Morphology In Vivo and Ex Vivo Using mGreenLantern.
Spine analysis is an important and common technique in neuroscience for measuring plastic changes at brain synapses, but spine resolution is often poor with EGFP and EYFP (38). Antibody enhancement can amplify FP signal, but such treatment increases background—often introducing puncta that could be mistaken for spines (Fig. 4A, white arrowheads)—while consuming additional time and resources. To determine whether mGreenLantern could offer improved spine detection relative to EGFP during 2P imaging of live animals, we used a dual viral approach to obtain sparse labeling of pyramidal cells.
Projection neurons from prefrontal cortex (PFC) to dorsal striatum (DS) were labeled by injecting FP virus into PFC and Cre virus into DS. A cranial window was implanted over the brain (Fig. 4B), and mice were allowed 2-wk recovery before imaging to allow the cranial window to clear completely. We captured representative images of FP-expressing neurons using a 2P microscope (Fig. 4C) and compared the number of spines visible per 100 µm for EGFP and mGreenLantern neurons (Fig. 4D), selecting dendrites of similar width and relative distance from the cortical surface for quantification (SI Appendix, Fig. S10). mGreenLantern neurons consistently appeared brighter and ∼40% more spines per unit length were visible in the mGreenLantern neurons compared to EGFP neurons, in accord with evidence presented in other figures that mGreenLantern is brighter and fills neuronal processes more rapidly. mGreenLantern is compatible with all optical equipment suited for EGFP and should be useful for longitudinal 2P studies in living mice.
We next compared the brightness of EGFP and mGreenLantern in cultured hippocampal neurons dissociated from embryonic day 17 (E17) mouse embryos. Primary neurons were transduced at 10 d in vitro (DIV) with AAV2/1-CAG-EGFP or mGreenLantern virus adjusted to equal titer. Cells were fixed 4 d after transduction, and images were collected with a widefield microscope. Neuronal processes were visible in the ex vivo mGreenLantern neurons but barely detectable in the EGFP neurons under the same imaging settings (Fig. 4E). In this experiment, mGreenLantern-expressing neurons were 2.5-fold brighter than those with EGFP (Fig. 4F). Altogether, mGreenLantern is brighter than EGFP in cells and animals and improves detection of neuronal morphology in vivo and in vitro, including dendritic spines, regardless of imaging modality or expression method.
Discussion
Summary.
Screening FPs using cellular brightness and maturation kinetics allowed us to produce a GFP, mGreenLantern, with neuron-highlighting capabilities that further surpassed those of EGFP and EYFP in brightness, spine detection, and expression rate in mice. mGreenLantern shows exceptional brightness in chemically transfected human cells, virally transduced mouse brain tissue, dissociated primary neurons, and bacteria, providing a link between FP engineering, characterization, and direct application.
mGreenLantern was 190% brighter than mNeonGreen, 260% brighter than Clover, and 620% brighter than EGFP when expressed in HeLa cells (Fig. 1A) and showed faster and complete maturation (Fig. 1F). The brightness of mGreenLantern was superior regardless of mammalian cell line chosen (SI Appendix, Fig. S1), the specific organism or induction system (Fig. 1 A–C), or the imaging and analysis method used (Fig. 1 D and E). In support of these observations and the underlying hypothesis, we have shown that HEK293T human embryonic kidney cells, BE(2)-M17 human neuroblastoma cells, and E. coli all produced at least three times more mGreenLantern soluble protein than EGFP, mClover3, or mNeonGreen (SI Appendix, Fig. S6).
Of course, many mechanisms converge on cellular brightness, which is a multifactorial phenotype. We did not directly assess degradation or cellular persistence of mGreenLantern but, given its exceptionally slow unfolding rate and high thermodynamic stability (Fig. 1 I and J), perhaps prolonged accumulation of the FP in cells and/or resistance to enzymatic degradation might also contribute to its greater soluble and insoluble total protein yield.
Without screening FPs in human cells quantitatively using “cellular brightness” and chromophore maturation measurements, it would have been difficult to recognize and differentiate mGreenLantern from other mutants with high “molecular brightness,” a number that we have shown greatly underestimates its brightness in practice. For example, the molecular brightness (QY × EC) of mClover3 is 8% greater than that of mGreenLantern, but cells expressing mGreenLantern were at least 300% brighter than mClover3 when quantified using any of three different expression systems. Likewise, using biochemical data alone, one would predict 20% lower brightness of mGreenLantern compared to mNeonGreen (ϕ = 0.72 vs. 0.78 and ε = 101 vs. 115 mM−1⋅cm−1, respectively) (Table 1), a prediction that we have shown diverges considerably from the reality of mGreenLantern’s superior brightness in actual biological systems including human, mouse, and bacteria.
With the ubiquity of FPs in biological studies and the stunning advances in FP technology during recent years (39, 40), scientists are confronted with an oftentimes bewildering array of fluorophore options that have important bearing on experimental design and outcomes. Complicating matters further, few FPs have been directly compared to multiple common controls for actual cellular brightness performance in the original publications, or for antibody compatibility. Our study directly demonstrates the effectiveness of mGreenLantern in neuronal tracing experiments in mouse tissue and the living brain, as well as in common human cell lines, ex vivo mouse embryonic neurons, and bacteria.
Of the few studies that have specifically compared the brightness of newly developed GFPs in mammalian cells, one showed that Clover, mClover3, and mNeonGreen generally did not differ significantly (5), similar to what we observed in three different human cell lines (SI Appendix, Fig. S1). Another study compared EGFP and mNeonGreen fluorescence in Caenorhabditis elegans embryos and adult worms using a variety of germline knockins and summarily found no significant differences between EGFP and mNeonGreen fluorescence intensity (41), consistent with our analysis of widefield images of transfected HeLa cells expressing cytosolic GFPs (Fig. 1 D and E). Additionally, our data reveal minimal differences in soluble protein yield between EGFP and mNeonGreen in E. coli and in two different human cell lines (SI Appendix, Fig. S6), in contrast to mGreenLantern, which produces over threefold more soluble protein. Our study underscores the value of whole-cell brightness measurements for predicting cellular imaging performance and in the evaluation of new fluorophores.
We compared mGreenLantern to six common FPs that we rigorously recharacterized spectroscopically to improve the accuracy of quantitative comparisons (SI Appendix, Table S1). This approach, while laborious, provides important validation of previous measurements and ultimately improves consensus in the field by decreasing reliance on individual cited literature values that may have been accepted and propagated for years without reevaluation by updated or unified methods. For example, we have noted that our experimentally determined value of the sfGFP EC in this study and in our previous work (21) was ∼32% lower than the reported value from the original sfGFP manuscript (42), but agrees with a recent study that comprehensively compared 16 different GFPs/YFPs under identical experimental conditions (24). Far from trivial, these measurements have important bearing on structure–activity interpretation, such as when comparing photophysical mechanisms underlying the behavior of EGFP, sfGFP, and Clover (21), and certainly in forward development of tags and biosensors.
During preparation of the manuscript, an extraordinarily bright RFP, mScarlet, was evolved from a synthetic consensus template and further evolved using a compelling application of the quantitative coexpression system in E. coli, essentially merging a live-cell maturation assay with cellular brightness in bacteria (43). Our study offers a screening strategy in mammalian cells and provides a broad view of interwoven features that converge on cellular brightness, but the semiautomated library screening approach of mScarlet may well be more efficient if sophisticated digital colony imaging equipment is available to the FP developer. We primarily screened bacteria by eye and cloned the brightest mutants into coexpression cassettes for brightness quantitation in live cells, a slower approach necessitating tightly focused mini libraries and a recursive strategy guided by sequencing data and structure–activity insights to be practical. Nonetheless, our effort demonstrates that fluorophores with dramatically improved brightness, chemical resistance, thermodynamic stability, and solubility, can be rationally engineered from templates such as EGFP that have already been exhaustively mined over two decades of research, without requiring semisaturation or random mutagenesis to do so.
Excitingly, numerous studies reporting well-folded FPs have independently uncovered similar or identical mutations (12, 36, 38)—even groups of mutations (12)—from diverse phenotypic screens, and such studies equipped us with structure–function data that we could operationalize into an ordered search for brighter mutants in this study (SI Appendix, Table S2). This convergence of folding mutations might indicate a peak in the fitness of the avFP folding landscape, but perhaps there is still room for growth, as we have shown here that specific combinations of “folding mutations” were more effective at improving cellular brightness than their total number alone. An exciting implication of our study is that existing biosensors such as GCaMP6 (44), for which structural (45) and mechanistic (46) studies are available, might be enhanced using a similar rational design approach to improve brightness and solubility when expressed in brain tissue while preserving sensor function, perhaps with concomitant decreases in cytotoxicity.
Mutational Analysis.
mGreenLantern differs from EGFP, sfGFP, and Clover by 21, 13, and 10 mutations, respectively (SI Appendix, Table S3). Relative to Clover, 7 of 10 mutations in mGreenLantern are surface exposed, and 5 of 7 of those are Lys, Arg, or Asp, which presumably improve hydrophilicity. To disrupt the avFP dimer interface aggressively and produce a true monomer, we introduced the three best-characterized monomerizing mutations, A206K/L221K/F223R (10), replacing hydrophobic residues with positively charged hydrophilic side chains. We also included N149K, suspected to contribute to the high brightness of Emerald (9, 14), with the supposition that additional positively charged residues would further repel the symmetry-related monomer. Indeed, mGreenLantern behaves as a monomer in vitro even at high protein concentration (SI Appendix, Fig. S4A), and in vivo (SI Appendix, Fig. S4 B and C), so its high cellular brightness and considerable thermodynamic stability are not due to dimer effects. Despite modifications to the protein surface, mGreenLantern still properly reacts with anti-EGFP antibodies (SI Appendix, Fig. S3).
mGreenLantern is based on Clover and it also features a GYG chromophore, unlike the TYG chromophore of EGFP. As we have previously shown for Clover, loss of the T65–E222 hydrogen bond found in EGFP (and stabilization of the protonated E222 by another side chain and/or by the chromophore imidazolinone moiety) appears to be most important for favoring the deprotonated Clover chromophore at the expense of the protonated form, resulting in lower 405-nm absorbance compared to EGFP (21). Based on studies of wtGFP point mutants (20) and our observation of the low pKa common to all FOLD1 variants and Emerald GFP (24), we suspect that the I167T mutation helps stabilize the deprotonated chromophore and contributes to loss of the 405-nm absorbance, although we cannot make definitive mechanistic conclusions without an mGreenLantern crystal structure.
We were unable to crystallize mGreenLantern using sparse matrix robotic screening or manually selected conditions from multiple protein preparations. Perhaps our monomerization effort disrupted the most efficient crystal packing orientations that would facilitate nucleation of crystal formation under those conditions. We have instead illustrated the structural locations of mGreenLantern mutations manually using the Clover structure (Protein Data Bank ID code 5WJ2) for spatial reference (SI Appendix, Fig. S11).
Benefits of Fast Maturation and Expression.
Fast expression after viral transduction must depend on at least the viral system, the FP itself, and the transduced cell’s endogenous machinery. It is possible that expression time may vary in other viral systems or when other brain regions are transduced. However, we suspect that mGreenLantern will perform similarly well in multiple brain regions and organisms, since the properties of maturation and brightness in cells appeared to be consistent in mice, humans, and bacteria. mGreenLantern may also be useful for early reporting of chemical transfection efficiency in cultured cells, since mGreenLantern expression is apparent in a matter of hours. Additionally, our viral transduction methods describe a simple approach for sparse highlighting of neurons using mGreenLantern that produces a labeling pattern reminiscent of a Golgi stain, improving visual discrimination of individual cells and potentially aiding neuronal tracing studies.
Conclusion
Altogether, mGreenLantern overcomes multiple limitations of FPs traditionally used in the neuroscience field and can directly replace nonmonomeric and slowly expressing genetically encoded neural highlighting constructs such as ChR2-EYFP, EGFP, and EYFP, having over threefold greater brightness than the latter, much faster and complete maturation, no visible oligomerization, greatly improved chemical and thermal stability, and superior highlighting of projection neurons. The high chemical and thermal stability of mGreenLantern make it useful for advanced tissue processing applications increasingly used in neuroscience and histology, including expansion microscopy and tissue clearing. mGreenLantern expresses rapidly in bacterial, mouse, and human cells, and can seamlessly replace EGFP and EYFP in biological systems that demand the greatest in vivo brightness and fastest expression.
Methods
Cellular Brightness.
Low-passage HeLa (ATCC CCL-2), BE(2)-M17 (ATCC CRL-2267), and HEK293T cells were routinely cultured in OptiMEM (Gibco) supplemented with 5% fetal bovine serum (VWR) and penicillin–streptomycin (Gibco).
Cells were passaged into 96-well black clear-bottom tissue culture treated assay plates (Corning) and grown for 24 h in media without phenol red before transfection with 0.2 µg of DNA per well using a single pcDNA3.1-[FP]-P2A-mCherry plasmid, or cotransfected with 0.2 µg each of pcDNA3.1-[FP] and pcDNA3.1-mCherry individual plasmids. Fluorescence was measured on a BioTek H1 Synergy microplate reader 48 h after transfection using endpoint scans (λex/λem = 495/525 nm for GFPs and 585/615 nm for mCherry). After subtracting blanks, GFP fluorescence was divided by mCherry signal, normalized to the EGFP ratio, and plotted in GraphPad Prism.
To determine cellular brightness using widefield microscopy, HeLa cells were transfected using 0.2 µg of pcDNA3.1-[FP] in 96-well format. After 48 h, cells were washed twice with PBS, pH 7.4 (Gibco), and allowed to incubate in fresh phenol red-free culture media for 30 min. Imaging was performed on a Keyence BZ-X700 All-in-One Fluorescence Microscope equipped with a 20× air objective and computer-controlled motorized stage. After briefly confirming the presence of cells using brightfield, focusing was manually adjusted for each well, and all wells were imaged under identical excitation and acquisition settings with a GFP filter cube (Keyence). Tile scans were collected for each well and analyzed in ImageJ by thresholding, quantifying mean fluorescence intensity of at least 300 cells per FP for each of six independent transfections, and normalizing the average values per transfection to an EGFP control.
Bacterial Brightness and Protein Solubility.
Bacteria were thawed from glycerol stocks and grown overnight in LB supplemented with 100 µg/mL ampicillin. The next day, all cultures were adjusted to the same optical density and used as starter cultures to inoculate at a 1:100 (vol/vol) ratio fresh media plus 0.2% arabinose for induction. Cultures were grown overnight at 37 °C with shaking at 275 rpm to ensure thorough oxygenation.
The following day, 100 µL of overnight culture was added in replicate to black 96-well clear-bottom assay plates (Corning), and fluorescence was measured using λex/λem = 495/525-nm detection settings on a BioTek Synergy H1 microplate reader. Fluorescence values were background subtracted, normalized to the EGFP control sample, and the bacterial brightness data were plotted in GraphPad Prism. The remaining cultures were pelleted and frozen at −80 °C.
To obtain lysate from bacterial cultures, cultures were thawed on ice and resuspended in ice-cold PBS. Resuspended cultures were sonicated identically on ice, centrifuged at 4 °C for 15 min at 20,000 × g, and the clarified lysate containing soluble protein was collected. To wash the insoluble pellet and remove residual soluble protein, the pellet was resuspended in PBS, centrifuged as described, and the supernatant was discarded. The washed insoluble pellet was resuspended in PBS and very briefly sonicated to homogenize it.
To obtain soluble lysate from mammalian cultures, HEK293T and BE(2)-M17 cells were passaged onto six-well plates and grown for at least 24 h before transfection using Turbofect (Thermo Fisher) according to manufacturer instructions. Cells were harvested by trypsinization 48 h later and washed three times with cold PBS before storage at −80 °C. Pellets were lysed by repeated freeze–thaw from liquid nitrogen to 37 °C water, resuspended in ice-cold RIPA buffer supplemented with phenylmethylsulfonyl fluoride protease inhibitor, and briefly sonicated on ice. Lysate was clarified by centrifugation as described.
Total protein concentration of clarified lysate was determined using the BCA Protein Assay (Pierce) with BSA standard curve. Aliquots of clarified lysate were adjusted using identical final concentration and volume using PBS, boiled at 100 °C for 5 min in SDS loading dye containing 0.2 M DTT, and allowed to cool passively to room temperature (RT). Samples were checked briefly using a 470-nm LED to confirm total loss of fluorescence, i.e., complete denaturation, before loading onto mini 4 to 15% or 12% Tris-Glycine gels (Bio-Rad) for bacterial or mammalian protein, respectively. Gels were stained with Coomassie reagent (Thermo Scientific) and imaged on a Bio-Rad ChemiDoc XRS+ scanner using identical exposure times. Band intensity was analyzed by densitometry in Fiji (ImageJ) using the Analyze → Gels function. Averages of three and two experimental replicates are presented for the bacterial and mammalian gels, respectively.
Mammalian Cell Culture and Imaging.
For localization, photobleaching, OSER assay, and proExM experiments, cells were passaged into 35-mm tissue culture plates containing a 22-mm glass bottom (MatTek) and grown for at least 24 h before transfection with Turbofect (Thermo Fisher) and 1-µg plasmid. Live cells were imaged 12 to 18 h after transfection using a Zeiss LSM 880 laser-scanning confocal microscope equipped with computer-controlled Zeiss Enhanced Navigation (ZEN) software, an argon-ion laser with 488- and 514-nm filters, 10×, 40×, or 63× differential interference contrast (DIC) objectives, and a high-sensitivity gallium arsenide phosphide (GaAsP) photodetector, or a lower-sensitivity photomultiplier tube (PMT) for the high-intensity photobleaching experiments.
OSER Assay.
The OSER assay was performed as described (32) under the mentioned transfection and culture conditions. Only healthy cultures of mycoplasma-free HeLa cells were imaged. Highly stressed or mitotic cells with sheeted ER, lobed nuclei, or indistinct nuclei, were excluded from analysis. In general, ∼100 cells were analyzed per FP for each of three experimental replicates and compared to mEGFP and Clover.
Photobleaching.
Photobleaching experiments were performed on a Zeiss LSM 880 confocal microscope with a Plan-Apochromat 40×/1.3 Oil DIC UVVIS-IR objective using live HeLa cells in phenol red-free media transfected as described. Pinhole was set to 1 Airy unit (“0.9-µm section”), scan time of 316 ms, pixel size of 0.83 µm, pixel dwell of 4.12 µs, 256 × 256-px frames, and 12-bit depth. The 488-nm argon-ion laser power was measured at the objective with a Thorlabs PMD100 power meter equipped with S130VC photodetector (Thorlabs). Laser power was initially set to the minimum necessary to identify suitable regions for bleaching before being increased to 77 µW at t = 0. Images from the H2B-[FP] transfected live HeLa cells were collected in the 490- to 650-nm emission range using ZEN acquisition software (Zeiss). Scaling of the initial emission rate to 1,000 photons/s per molecule at t = 0 s was performed as described (47).
Kinetic Unfolding.
Purified protein in TNG buffer (50 mM Tris⋅HCl, 150 mM NaCl, 10% glycerol, pH 7.4) was adjusted to 1 µM and diluted 10-fold directly in a black 96-well clear-bottom assay plate containing 7 M GdnHCl in TNG buffer to a final concentration of 6.3 M GdnHCl and 0.1 µM FP. At the same time, protein from the same stock was dispensed into TNG buffer without GdnHCl, and the plate was quickly sealed with clear optical adhesive to prevent evaporation. Fluorescence measurements began after an initial ∼15-s lag time, with λex/λem = 495/525 nm at 2-min interval for a total of 11 h. Curves were plotted as the fluorescence ratio of unfolded to native protein over time.
Thermostability.
FP was adjusted to 1 µM final concentration in PBS and 50 µL was dispensed into replicate wells of a clear 96-well qPCR plate. The plate was sealed with optically clear adhesive and heated in a Bio-Rad C1000 Touch thermal cycler equipped with a CFX96 Real-Time System and FAM filter. Temperature was held at 25 °C for 15 s to confirm consistent readings before the samples were heated at a rate of 0.3 °C/min to a final temperature of 100 °C. Melting temperatures were determined using the Bio-Rad CFX96 software and melting curve derivatives. Curves were plotted by normalizing each data point to the individual FP’s average initial value from the 25 °C baseline hold.
Expansion Microscopy (proExM).
HeLa cells were grown and transfected in 35-mm imaging plates as described, using pLifeAct-mGreenLantern. After live imaging, proExM was performed according to established protocols (26, 27), without immunostaining.
Antibody Compatibility.
HEK293T cells were transfected with pcDNA3.1-[FP] and cultured as described. After 48-h expression, cells were fixed with room temperature 4% paraformaldehyde (PFA) for 15 min, washed 3 × 5 min with PBS, and incubated in 0.1% Triton X-100 and 5% donkey serum in PBS (“blocking solution”) for 60 min at RT. Primary antibody (goat α-GFP polyclonal, Abcam, #ab6673; or goat α-GFP polyclonal, Novus, #NB1001770; or Ms α-GFP monoclonal, Life Technologies, #A-11122) was applied in fresh blocking solution at 1:1,000 dilution overnight at 4 °C. The next day, cells were washed 3 × 5 min in PBS and incubated with donkey α-goat IgG Alexa Fluor 555 secondary antibody (Invitrogen; #A-21432) for exactly 2 h at RT. Cells were washed, stained with DAPI, washed again with PBS, mounted using Vectashield antifade medium (Vector Laboratories), and sealed with nail polish that was allowed to dry fully before imaging on a Nikon Eclipse 80i microscope.
Visualization of Long-Range Projections and In Vivo Expression Rate.
Animals.
Male C57BL/6 mice (Charles River) and PV-Cre mice (The Jackson Laboratory; stock #017320) were group-housed under a standard 12-h light/dark cycle in a temperature and humidity-controlled vivarium and were provided with food and water ad libitum.
Stereotactic surgery.
Mice were anesthetized with 2% isoflurane and head-fixed over a heating pad in a mouse stereotactic apparatus (Narishige International). A volume of 350 nL of AAV2/1-EF1α-DIO-mGreenLantern-WPRE-pA (obtained from Boston Children’s Hospital Viral Core, Boston, MA) or AAV2/1-EF1α-DIO-EYFP-WPRE-pA (obtained from University of Pennsylvania Vector Core, Philadelphia, PA) was injected with AAV1-hSyn-Cre-WPRE-hGH in a 10:1 ratio of FP:Cre virus into visual cortex; or into the ACA to visualize long-range projections (n = 3 mice per FP); or into the ACA at a 15:1 (FP:Cre):Tracer ratio to visualize the rate of in vivo expression (n = 2 hemispheres per FP). Tracer was Alexa 594-coated latex beads (Lumafluor). ACA injection coordinates relative to bregma were as follows: anteroposterior, +0.4 mm; mediolateral, +0.2 mm; dorsoventral, −0.5 mm. Mice were isolated postsurgery until fully awake and immediately returned to their home cage.
For expression in transgenic animals, PV-Cre mice (The Jackson Laboratory; stock #017320) aged postnatal day 60 (P60) to P90 were injected with 500 nL of AAV2/1-EF1α-DIO-mGreenLantern-WPRE-hGH (obtained from Boston Children’s Hospital Viral Core, Boston, MA) with animal and surgical methods as previously described.
Tissue preparation and histology.
At the specified time points, mice underwent transcardial perfusion with PBS followed by 4% PFA in PBS. Brains were postfixed in 4% PFA overnight and coronally sectioned at 50 µm on a vibratome (Leica; VT1000 S).
Confocal Imaging and Analysis.
Imaging was performed on a Zeiss LSM 710 confocal microscope (Zeiss) using a calibrated 10× air objective (or 63× oil-immersion objective where specified). Images were collected using both 488- and 514-nm lasers separately while preserving identical settings for each FP (i.e., two sets of images) unless otherwise specified. The tracer was imaged using 561-nm laser excitation. Pinhole was 25 µm (1 Airy unit); resolution, 1,024 × 1,024 px; 0.6 scan zoom; averaging = 4 (line); scan speed = 8.
To visualize projection neurons, tile scans of whole coronal sections were acquired from tissue harvested 14 d (or 3, 7, and 14 d for the in vivo expression experiment) after injection, as described. Tiles were stitched in ImageJ, displayed using the ImageJ 16-color rainbow lookup table, and superimposed on an Allen Brain Atlas diagram at the coronal depth with corresponding anterior/posterior distance to bregma. “Axon filling” was quantified by measuring the area of the striatum and corpus callosum with positive fluorescence, defined as >2 SD below the mean pixel intensity for the given FP/laser line combination, and normalizing that value to the fluorescent ACA area defined in the same manner. The bar graph was generated in GraphPad Prism analyzed using two-way ANOVA with Sidak’s multiple-comparisons test, n = 3 mice per FP. Likewise, for the in vivo expression experiment, “fluorescence” was calculated as described in Fig. 3A and plotted for each FP/laser line combination.
Two-Photon Imaging and Spine Analysis.
Animals.
C57BL/6J mice at 6 to 8 wk of age on surgery day were used for spine imaging experiments. Mice had ad libitum access to food and water and were group housed (two to five mice per cage) under a 12-h light/dark cycle (lights on 06:00). All procedures were overseen by and adherent to the rules set forth by the Weill Cornell Medical College Institutional Animal Care and Use Committee.
Viral vectors.
A dual viral approach was used to obtain sparse labeling of pyramidal cells. rAAV8-CaMKIIa-mCherry-Cre was obtained from the University of North Carolina at Chapel Hill Vector Core (titer, 4.7 × 1012 particles/mL). Virus was diluted 10-fold using sterile PBS. This solution was then mixed with equal volume of either (undiluted) AAV2/1-EF1α-DIO-mGreenLantern-WPRE-pA (obtained from Boston Children’s Hospital Vector Core), AAV8-Syn-DIO-EGFP (obtained from the University of Pennsylvania Vector Core), or AAV2/1-EF1α-DIO-EYFP-WPRE-hGH (obtained from the University of Pennsylvania Vector Core). To label PFC neurons projecting to dorsal striatum, 500 nL of AAV2/1-EF1α-DIO-mGreenLantern-WPRE-pA virus was injected into PFC and 500 nL of AAV1-Cav2-Cre was injected into dorsomedial striatum (Cranial window surgery).
Cranial window surgery.
Anesthesia was induced using isoflurane (induction, 5%; maintenance, 1 to 2%). Dexamethasone (1 mg/kg, i.p.) and metacam (1 mg/kg, i.p.) were administered prophylactically to reduce brain swelling and pain, respectively. After animals were placed in a stereotactic frame (Kopf Instruments), sterile eye lubricant (Puralube; Fisher Scientific) was administered to prevent corneal drying, and a microwavable heating pad (Snugglesafe) was used to maintain body temperature.
Scalp fur was trimmed, and the skull surface was exposed with a midline scalp incision. Bupivicaine (0.05 mL, 5 mg/mL) was administered topically as a second prophylactic analgesic. A small circular section of skin (∼1-cm diameter) was excised using surgical scissors (Fine Science Tools). The periosteum was bluntly dissected away, and bupivacaine (0.05 mL, 5 mg/kg) was topically applied. The region to be imaged (dorsal PFC) was identified using stereotaxic coordinates (1.7 mm anterior to bregma; 0.35 mm lateral from the midline). A custom-made circular titanium headplate was attached to the skull using dental cement (C&B Metabond; Parkell). The titanium head plate was then screwed into a custom-built fork fixed to a solid metal base.
Under a continuous gentle flow of PBS (137 mM NaCl, 27 mM KCl, 10 mM phosphate buffer; VWR), a ∼4-mm circular section of the skull—slightly larger than the window—was removed using a 0.5-mm burr (Fine Science Tools) and a high-speed hand dental drill (Model EXL-M40; Osada), taking great care not to compress brain tissue or damage the sagittal venous sinus. The dura beneath the craniotomy was delicately removed using fine forceps (Fine Science Tools). Sugi swabs (John Weiss and Son, Ltd.) were used to absorb trace bleeding.
The AAV mixture (100 nL) was injected at a rate of 150 nL/min using a Nanofil syringe with a 33-G beveled needle and pump (World Precision Instruments). After the injection, the needle was kept in place for 2 min to allow time for diffusion of the viruses prior to removing the needle from the brain.
A 3-mm glass coverslip (Warner Instruments) was gently placed over the brain. Veterinary adhesive (Vetbond; Fisher Scientific) was used to form a seal between the coverslip and the skull. A layer of Metabond was then applied for added durability. Metacam (1 mg/kg, i.p.) was administered as an analgesic 24 h after surgery, and as needed thereafter.
Two-photon imaging and spine analysis.
Images were acquired using a commercial 2P laser-scanning microscope equipped with a scanning galvanometer and a Spectra-Physics Mai Tai DeepSee laser tuned to 920 nm. Images acquired through a cranial window were obtained using a 25×, 1.05 numerical aperture water-immersion objective with a 2-mm working distance (Olympus). Fluorescence was detected through GaAsP PMTs using the Fluoview acquisition software (Olympus), and images were collected in the green channel using a F30FGR bandpass filter (Semrock). Z-stacks were acquired (640 × 640 px, 4-µs pixel dwell time, 1-μm step size) with 3× digital zoom. Spine imaging experiments occurred under KX anesthesia (ketamine, 100 mg/mL, and xylazine, 10 mg/mL, at dosages of 0.1 mL/10 g body weight). Spine density of PFC neurons was determined by counting total spines per 100 µm from each of 95 and 101 dendritic segments and normalizing to the EGFP mean (n = 2 and 3 mice for EGFP and mGreenLantern, respectively).
Brightness of FP-Expressing Neurons Ex Vivo.
Cell culture.
Pregnant female mice were killed and hippocampus from E17 embryos were dissected. Following incubation in Trypsin-EDTA 0.25% for 10 min, hippocampal cells were mechanically dissociated with a fire-polished Pasteur pipette. Neurons were plated at 1.5 × 104 cells/cm2 in 24-well poly-l-lysine–coated plates in plating media (MEM containing 2 mM glutamine, supplemented with 10% FBS, 1 mM pyruvate, 0.37% glucose, and 25 U/mL penicillin/streptomycin). Medium was changed within 2 to 24 h of plating to a serum-free medium containing Neurobasal with B-27 supplement, 0.5 mM glutamine, 25 µM glutamate, and 25 U/mL penicillin/streptomycin and AraC (cytosine-1-B-d-arabinofuranoside). Cells were maintained at 37 °C and 5% CO2. Primary hippocampal neurons were infected at DIV10 with AAV2/1-CAG-EGFP or mGreenLantern virus diluted to 2.5 × 109 particles/mL (Duke Viral Vector Core) in the same media. After 4 d (DIV14) of viral infection, cells were fixed for immunocytochemistry. All reagents used to prepare primary neuronal cultured were purchased from Thermo Fisher, except glucose, which was from Sigma.
Immunocytochemistry.
After 4 d of viral infection, cells were briefly washed with Hanks’ balanced salt solution (HBSS) (Thermo Fisher) and fixed with prewarmed 4% PFA/4% sucrose solution for 15 min at room temperature, protected from light. After fixation, cells were washed several times with HBSS. Coverslips were mounted with ProLong Gold antifade reagent (Invitrogen), and imaging was performed within 48 h of fixation.
Neuronal imaging and analysis.
Imaging was performed within 48 h of fixation using a Nikon TE2000 inverted fluorescence microscope equipped with a 60× oil objective and ANDOR camera (Zyla Camera sCMOS). For fluorescence quantitation, 20 random images were acquired per coverslip under the same acquisition parameters for all samples. Background was subtracted using NIS-Elements AR (version 4.20) software before quantitation. Statistical analysis was performed using GraphPad Prism, version 8.2.0, software. Statistical significance was considered at *P < 0.05, **P < 0.01, and ***P < 0.001 between the means of a minimum of three cultures per conditions using Student’s t test. Results are expressed as mean ± SEM. All experiments were performed with at least three independent neuronal primary cultures.
Supplementary Material
Acknowledgments
We thank Ce Feng Liu and Melissa G. McKenzie for a critical reading of the manuscript. We thank Ryan W. Logan for early testing of mGreenLantern. Kiryl Piatkevich trained B.C.C. and E.M.N. in proExM. We are grateful to academic viral vector cores for producing high-quality virus, including Chen Wang and his group at Boston Children’s Hospital. P.T. acknowledges NIH Grant R01 NS083983-06 from M.G.B.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000942117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Harvey A. R., et al. , “Use of GFP to analyze morphology, connectivity, and function of cells in the central nervous system” in Viral Applications of Green Fluorescent Protein: Methods and Protocols, Hicks B. W., Ed. (Humana Press, 2009), pp. 63–95. [DOI] [PubMed] [Google Scholar]
- 2.Porrero C., Rubio-Garrido P., Avendaño C., Clascá F., Mapping of fluorescent protein-expressing neurons and axon pathways in adult and developing Thy1-eYFP-H transgenic mice. Brain Res. 1345, 59–72 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Lulevich V., Shih Y. P., Lo S. H., Liu G. Y., Cell tracing dyes significantly change single cell mechanics. J. Phys. Chem. B 113, 6511–6519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam A. J., et al. , Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajar B. T., et al. , Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner N. C., et al. , A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack B. P., Valdivia R. H., Falkow S., FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Stepanenko O. V., Stepanenko O. V., Kuznetsova I. M., Verkhusha V. V., Turoverov K. K., “Chapter Four - Beta-Barrel Scaffold of Fluorescent Proteins: Folding, Stability and Role in Chromophore Formation” in Beta-Barrel Scaffold of Fluorescent Proteins: Folding, Stability and Role in Chromophore Formation, Jeon K. W., Ed. (Academic Press, 2013), pp. 221–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teerawanichpan P., Hoffman T., Ashe P., Datla R., Selvaraj G., Investigations of combinations of mutations in the jellyfish green fluorescent protein (GFP) that afford brighter fluorescence, and use of a version (VisGreen) in plant, bacterial, and animal cells. Biochim. Biophys. Acta 1770, 1360–1368 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y., Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Duwé S., et al. , Expression-enhanced fluorescent proteins based on enhanced green fluorescent protein for super-resolution microscopy. ACS Nano 9, 9528–9541 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Fisher A. C., DeLisa M. P., Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS One 3, e2351 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimata Y., Iwaki M., Lim C. R., Kohno K., A novel mutation which enhances the fluorescence of green fluorescent protein at high temperatures. Biochem. Biophys. Res. Commun. 232, 69–73 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Cubitt A. B., Woollenweber L. A., Heim R., “Understanding structure-function relationships in the Aequorea victoria Green fluorescent protein” in Methods in Cell Biology, Sullivan K. F., Kay S. A., Eds. (Elsevier, 1999), Vol. 58, pp. 19–30. [DOI] [PubMed] [Google Scholar]
- 15.Iizuka R., Yamagishi-Shirasaki M., Funatsu T., Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal. Biochem. 414, 173–178 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Arpino J. A. J., Reddington S. C., Halliwell L. M., Rizkallah P. J., Jones D. D., Random single amino acid deletion sampling unveils structural tolerance and the benefits of helical registry shift on GFP folding and structure. Structure 22, 889–898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goedhart J., et al. , Quantitative co-expression of proteins at the single cell level–application to a multimeric FRET sensor. PLoS One 6, e27321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goedhart J., et al. , Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3, 751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craggs T. D., Green fluorescent protein: Structure, folding and chromophore maturation. Chem. Soc. Rev. 38, 2865–2875 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Heim R., Prasher D. C., Tsien R. Y., Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 91, 12501–12504 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell B. C., Petsko G. A., Liu C. F., Crystal structure of green fluorescent protein clover and design of clover-based redox sensors. Structure 26, 225–237.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Zapata-Hommer O., Griesbeck O., Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 3, 5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ai H. W., Olenych S. G., Wong P., Davidson M. W., Campbell R. E., Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: Identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 6, 13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cranfill P. J., et al. , Quantitative assessment of fluorescent proteins. Nat. Methods 13, 557–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepanenko O. V., Stepanenko O. V., Kuznetsova I. M., Verkhusha V. V., Turoverov K. K., Sensitivity of superfolder GFP to ionic agents. PLoS One 9, e110750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tillberg P. W., et al. , Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asano S. M., et al. , Expansion microscopy: Protocols for imaging proteins and RNA in cells and tissues. Curr. Protoc. Cell Biol. 80, e56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soderblom C., et al. , 3D imaging of axons in transparent spinal cords from rodents and nonhuman primates. eNeuro 2, ENEURO.0001-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Maunze B., Wang Y., Tsoulfas P., Blackmore M. G., Global connectivity and function of descending spinal input revealed by 3D microscopy and retrograde transduction. J. Neurosci. 38, 10566–10581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ertürk A., et al. , Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Richardson D. S., Lichtman J. W., SnapShot: Tissue clearing. Cell 171, 496–496.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Costantini L. M., Fossati M., Francolini M., Snapp E. L., Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic 13, 643–649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaner N. C., Steinbach P. A., Tsien R. Y., A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Nagai T., et al. , A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Orth A., et al. , Super-multiplexed fluorescence microscopy via photostability contrast. Biomed. Opt. Express 9, 2943–2954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liske H., Qian X., Anikeeva P., Deisseroth K., Delp S., Optical control of neuronal excitation and inhibition using a single opsin protein, ChR2. Sci. Rep. 3, 3110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katz L. C., Burkhalter A., Dreyer W. J., Fluorescent latex microspheres as a retrograde neuronal marker for in vivo and in vitro studies of visual cortex. Nature 310, 498–500 (1984). [DOI] [PubMed] [Google Scholar]
- 38.Verstraelen P., et al. , Image-based profiling of synaptic connectivity in primary neuronal cell culture. Front. Neurosci. 12, 389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean K. M., Palmer A. E., Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol. 10, 512–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez E. A., et al. , The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heppert J. K., et al. , Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol. Biol. Cell 27, 3385–3394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Bindels D. S., et al. , mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Chen T.-W., et al. , Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding J., Luo A. F., Hu L., Wang D., Shao F., Structural basis of the ultrasensitive calcium indicator GCaMP6. Sci. China Life Sci. 57, 269–274 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Barnett L. M., Hughes T. E., Drobizhev M., Deciphering the molecular mechanism responsible for GCaMP6m’s Ca2+-dependent change in fluorescence. PLoS One 12, e0170934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaner N. C., et al. , Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.