Significance
Aneuploidy, or an imbalanced chromosome content, is a hallmark of cancer. Paradoxically, aneuploidy is associated both with poor prognosis in cancer and with slow growth in primary cells. Here, we bridge these two seemingly contradictory observations: the cell cycle defects caused by aneuploidy protect cancer cells from chemotherapeutic damage. Our study may inform treatment decisions by impacting our understanding of how sensitive a particular tumor may be to certain chemotherapies.
Keywords: aneuploidy, cell cycle, chemotherapy resistance, cisplatin, paclitaxel
Abstract
Aneuploidy, defined as whole chromosome gains and losses, is associated with poor patient prognosis in many cancer types. However, the condition causes cellular stress and cell cycle delays, foremost in G1 and S phase. Here, we investigate how aneuploidy causes both slow proliferation and poor disease outcome. We test the hypothesis that aneuploidy brings about resistance to chemotherapies because of a general feature of the aneuploid condition—G1 delays. We show that single chromosome gains lead to increased resistance to the frontline chemotherapeutics cisplatin and paclitaxel. Furthermore, G1 cell cycle delays are sufficient to increase chemotherapeutic resistance in euploid cells. Mechanistically, G1 delays increase drug resistance to cisplatin and paclitaxel by reducing their ability to damage DNA and microtubules, respectively. Finally, we show that our findings are clinically relevant. Aneuploidy correlates with slowed proliferation and drug resistance in the Cancer Cell Line Encyclopedia (CCLE) dataset. We conclude that a general and seemingly detrimental effect of aneuploidy, slowed proliferation, provides a selective benefit to cancer cells during chemotherapy treatment.
Early observations of cancer noted that cancer cells often possess an abnormal number of chromosomes. About 90% of solid tumors are aneuploid, meaning they harbor a chromosome count that is not a multiple of the haploid complement. Thus, aneuploidy is more common than any individual gene mutations in cancer (1). This abnormal DNA content leads to changes in RNA and protein expression and many phenotypic changes (2–5). Notably, aneuploidy is significantly associated with poor patient prognoses, both in pan-cancer analyses and cancer-type-specific studies (6–13).
Despite its prevalence in cancer, recent studies have revealed that aneuploidy causes a wide variety of cellular stresses. As changes in gene copy number generally result in changes in gene expression, stoichiometric imbalances in protein complexes lead to increased protein misfolding and proteotoxic stress due to an increased demand for protein quality-control machinery (14–16). Aneuploidy also alters the metabolic landscape of cells and causes genome instability (5, 17–20). These aneuploidy-associated stresses lead to cell cycle changes and slow proliferation (4, 21). Specifically, aneuploidy causes G1 delays in the yeast, Saccharomyces cerevisiae, and lengthens G1 and S phase in mammalian cells under most circumstances (20, 22, 23).
How can a feature of cancer be associated with poor patient prognosis if it slows cancer’s growth? One possibility is that aneuploidy increases a tumor’s resistance to treatment. In cancer, drug resistance has, among other parameters, been associated with cells missegregating chromosomes at a higher rate, a phenomenon known as chromosomal instability (CIN). CIN and aneuploidy exhibit positive feedback, with each driving the other. Colon cancer cell lines with CIN have increased multidrug resistance, and patients with high CIN tumors have a worse prognosis than those with chromosomally stable cancers (24). The connection between CIN and drug resistance has been attributed to increased heterogeneity and, hence, subclones within the tumor that are drug resistant. However, other studies have found that extremely high levels of CIN increase patient survival (25–27). Extreme aneuploidy stress could synergize with the antiproliferative effects of cancer drugs and may prevent cells from “holding onto” a drug-resistant karyotype.
Specific aneuploidies also drive drug resistance in human cancer cells. In DLD1, a colon cancer cell line, two trisomic derivatives (trisomy 7 and trisomy 13) grew better than their euploid counterpart when treated with 5-fluorouracil (28). Whether aneuploidy-induced copy number changes of specific genes or some general feature of the aneuploid state drive chemotherapy resistance is currently unknown. Here we investigate this question, focusing on cis-diamminedichloroplatinum(II) (cisplatin) and paclitaxel, two front line chemotherapeutics that are used to treat highly aneuploid cancers such as bladder, ovarian, testicular, breast, and nonsmall cell lung cancer. We show that aneuploidy causes resistance to these chemotherapeutics through a general feature of the aneuploid condition: slowed proliferation. Equalizing growth of aneuploid and euploid control cells eliminates differences in drug resistance between them. Finally, we provide evidence that our findings are clinically relevant. Aneuploidy and chromosomal instability correlate with slowed proliferation and drug resistance in the Cancer Cell Line Encyclopedia (CCLE) dataset. We conclude that proliferative index is a major determinant of chemotherapy efficacy and that aneuploidy’s role in causing chemotherapy resistance is mediated at least in part by its adverse effects on cell proliferation.
Results
Trisomy Increases Resistance to Frontline Chemotherapeutics.
To test how aneuploidy affects chemotherapeutic response, we chose two commonly used chemotherapies that inhibit cell proliferation by very different mechanisms: cisplatin, a platinum-based drug that causes DNA damage by binding DNA in a non-cell-cycle-specific manner, and paclitaxel (Taxol), a microtubule stabilizing agent that arrests cells in mitosis. For cisplatin, we tested the response of multiple trisomic mouse embryonic fibroblast (MEF) cell lines and their euploid littermate controls (4). Because MEFs are not very responsive to paclitaxel, we tested the efficacy of paclitaxel against a pseudodiploid colon cancer cell line, HCT116, and four aneuploid derivatives of this cell line (22).
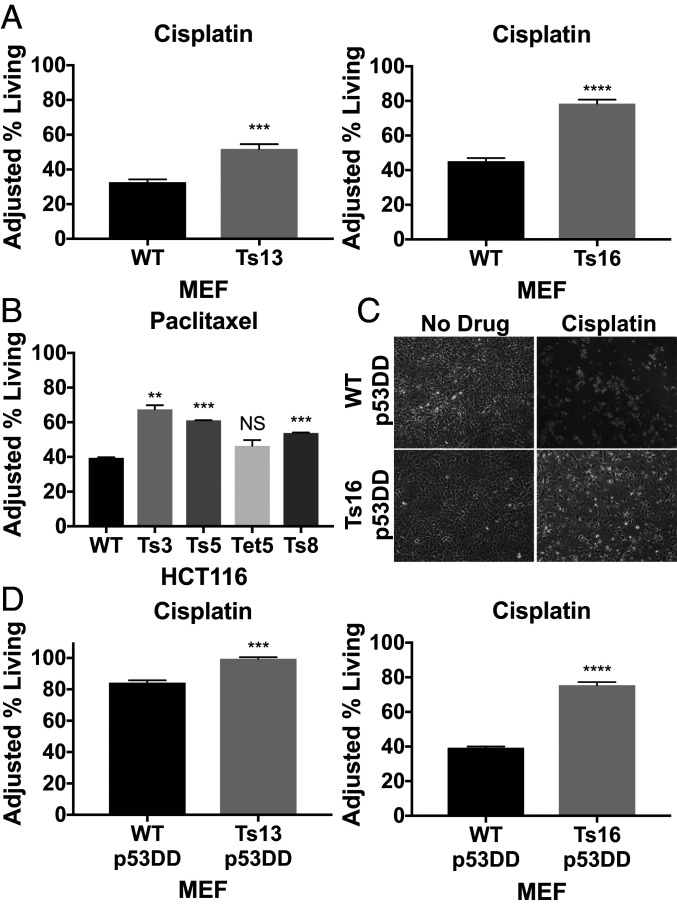
To determine how aneuploidy impacts cisplatin response, we compared the drug response of four trisomy 13 MEF lines (Ts13) and four trisomy 16 MEF lines (Ts16), all from different litters, to their euploid littermate controls. We treated cells with cisplatin (15 μM) for 48 h and then identified living, dead, and early apoptotic cell populations by flow cytometry using Annexin V and DAPI staining. We found that in three of the four trisomy 13 lines a greater percentage of cells were alive following drug treatment compared to their euploid controls (Fig. 1A and SI Appendix, Fig. S1A). Similarly, all four trisomy 16 cell lines exhibited increased survival following cisplatin treatment relative to euploid controls (Fig. 1A and SI Appendix, Fig. S1B). We conclude that trisomies 13 and 16 confer decreased cisplatin sensitivity, although we do note that not all lines exhibited this phenotype. Variability among MEF lines directly derived from mouse embryos is a well-documented phenomenon (29) and likely reflects variability in identity of outgrowing cells.
Fig. 1.
Trisomy 13 and 16 increases chemotherapeutic resistance in a p53-independent manner. (A) Litter-matched euploid and trisomic MEFs (trisomy 13 Left, trisomy 16 Right) were treated with 15 μM cisplatin or no drug for 48 h, collected, stained with Annexin V and DAPI, and analyzed by flow cytometry (n = 3). Adjusted % living = (100 – Avg. % living no drug) + % living drug. (B) Aneuploid HCT116 and HCT116 control cells were treated with 20 nM paclitaxel for 72 h, and the adjusted percentage of living cells was determined (n = 2). (C) Representative images of Ts16 p53DD and WT p53DD cells following treatment. (D) Litter-matched euploid and trisomic MEFs expressing p53DD were treated with 30 μM (Ts13) or 20 μM (Ts16) cisplatin or no drug for 48 h and the percentage of adjusted living cells was determined (n = 3). Unpaired t tests (NS, not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001).
To test whether trisomy affects the response to paclitaxel, we treated HCT116 and four trisomic/tetrasomic derivatives (trisomy 3, trisomy 5, tetrasomy 5, and trisomy 8) with 20 nM paclitaxel for 72 h. Three of the four aneuploid derivatives (trisomy 3, trisomy 5, and trisomy 8) exhibited significantly increased survival during paclitaxel treatment compared to their pseudodiploid parental line (Fig. 1B).
Previously isolated derivatives of trisomy 3 HCT116 lines that had lost the additional chromosome after growth as a xenograft (called 3-3 p.x. 1, 2, or 3) (21) enabled us to assess whether paclitaxel resistance was indeed due to trisomy 3. This control was important because, unlike the trisomic MEFs that were obtained from crosses (4), trisomic HCT116 lines were generated by microcell-mediated chromosome transfer, which involves drug selection and a single cell cloning step (22). These procedures could introduce genetic alterations that might confer the observed drug resistance. The validity of this concern is illustrated by the resistance of trisomy 3 HCT116 cells to cisplatin (SI Appendix, Fig. S2A). Derivatives of this cell line that had reverted to disomy retained this resistance (SI Appendix, Fig. S2B), indicating that the resistance was conferred by genomic alterations other than trisomy 3.
We tested the response of two trisomy 3 postxenograft cell lines, that were now disomic for chromosome 3, to paclitaxel. One cell line showed moderate resistance to paclitaxel relative to pseudodiploid cells but significantly less resistance than its trisomy 3 prexenograft counterpart (about 7.8% more living cells in the 3-3 p.x. 1 cell line than wild-type [WT] p.x. 1 compared to 27.9% more surviving cells in the true trisomy 3 cell line than in wild type; see SI Appendix, compare SI Appendix, Fig. S2C and Fig. 1B). The other disomy 3 HCT116 cell line (3-3 p.x. 2) showed no significant difference in survival compared to the pseudodiploid control (SI Appendix, Fig. S2D). We conclude that a variety of different trisomic karyotypes can cause resistance to chemotherapeutics that act by different mechanisms.
Trisomy-Mediated Resistance to Chemotherapeutics Is Largely Independent of p53.
The tumor suppressor p53 is critical for how cells respond to various stresses (reviewed in ref. 30). A recent study attributed aneuploidy-induced 5-hydroxyurea resistance to p53 activity, as p53 knockout eliminated the drug resistance of aneuploid cells (31). Cisplatin is known to induce both p53-dependent and p53-independent responses in cells (32). Thus, we tested whether cisplatin resistance of trisomic MEFs depended on p53 function.
To down-regulate p53 activity, we expressed a highly effective and well-studied dominant negative form of p53, p53DD, in our euploid and aneuploid MEFs (33). We then measured viability of these cells following 30 μM (trisomy 13 and control) or 20 μM (trisomy 16 and control) cisplatin treatment. Because p53DD expression has a significant impact on survival following drug treatment in both euploid and aneuploid cells (34), we used a higher concentration of cisplatin. This difference in drug concentrations makes a direct comparison of cell survival between wild-type and p53DD-expressing cells not possible. It was nevertheless clear that trisomy 13 and trisomy 16 cells expressing p53DD showed greater resistance to drug treatment than euploid control cells (Fig. 1C). We conclude that the increased survival of trisomic cells expressing p53DD indicates that this resistance is at least partially independent of p53. Whether expression of p53DD narrowed the survival gap between trisomy and euploid cells at all remains to be determined.
G1 Cell Cycle Delay Causes Chemotherapeutic Resistance.
Aneuploidy causes cell cycle delays, especially in G1 and S phase (20, 22, 23). Many frontline chemotherapeutics target specific phases of the cell cycle and were discovered based on their ability to kill rapidly growing cancers or cancer cell lines. These observations prompted us to ask whether cell cycle delays in G1, like those caused by aneuploidy, increase resistance to cisplatin and paclitaxel.
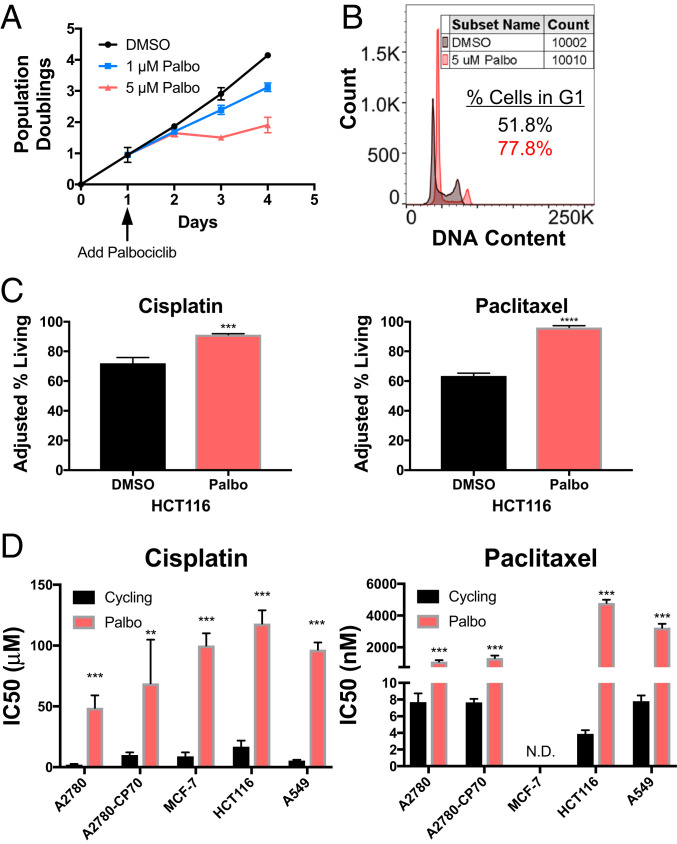
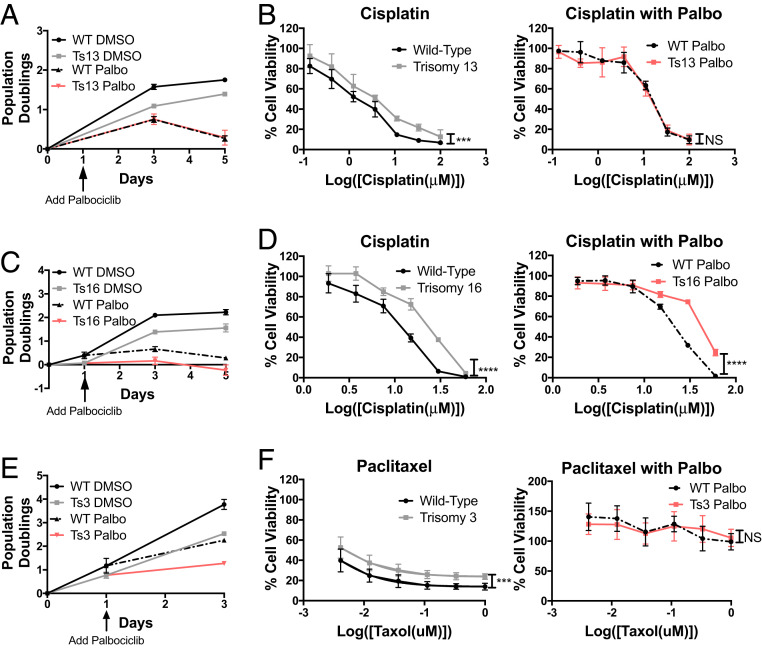
The CDK 4/6 inhibitor palbociclib (PD 0332991) causes cells to slow in G1 (35). This is also true in HCT116 cells; palbociclib delays HCT116 cells in G1 in a dose-dependent manner (Fig. 2 A and B). Previous studies had demonstrated that palbociclib reduces the efficacy of some chemotherapies, including anthracyclines and taxanes (36, 37). To determine whether palbociclib confers chemotherapy resistance to cisplatin and paclitaxel in our cell lines, we treated HCT116 cells with 5 μM palbociclib for 24 h followed by treatment with cisplatin or paclitaxel in the continuous presence of palbociclib for 48 h, and then measured cell viability. Treating cells with palbociclib was sufficient to increase cell survival in response to cisplatin and paclitaxel (Fig. 2C). This effect of the CDK 4/6 inhibitor was not specific to HCT116 cells. Palbociclib significantly increased the IC50 (drug concentration at which 50% of cells are viable) of cisplatin in five cancer cell lines and increased the IC50 of paclitaxel in four cancer cell lines (Fig. 2D). The IC50 of paclitaxel in MCF-7 cells was not determined, as it was greater than all tested concentrations.
Fig. 2.
Delaying cells in G1 increases their chemotherapeutic resistance. (A) Growth curve of HCT116 cells treated with 0, 1, or 5 µM palbociclib (n = 3). Arrow indicates palbociclib addition. (B) DNA content analysis of at least 10,000 HCT116 cells treated with 5 μM palbociclib or DMSO for 24 h. (C) HCT116 cells were treated with 5 μM palbociclib or DMSO for 24 h. Thereafter, cells were incubated with 5 μM palbociclib or DMSO together with 20 µM cisplatin or 25 nM paclitaxel for 48 or 72 h, respectively. The adjusted percentage of living cells = [(100 – Avg. % living no drug) + % living drug] was determined (n = 3). Unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (D) The indicated cell lines were treated with various concentrations of cisplatin (Left) or paclitaxel (Right) after being pretreated with DMSO or palbociclib. The drug concentration at which 50% of cells were alive (IC50) was determined by MTT assay (n ≥ 3). N.D., not determined due to the IC50 being above all tested concentrations. Unpaired t test (**P < 0.01, ***P < 0.001, ****P < 0.0001).
To ensure that palbociclib mediated drug resistance by delaying cells in G1, we performed two controls. First, we repeated the experiment in a cell line lacking the retinoblastoma gene (Rb; MDA-MB-468). Inactivation of RB eliminates the palbociclib-induced G1 delay (SI Appendix, Fig. S3A). If the drug-induced G1 delay was responsible for the observed resistance to chemotherapeutics, palbociclib should not cause resistance in the RB− cell line. This was indeed the case: MDA-MB-468 cells were either equally or more sensitive to cisplatin and paclitaxel following palbociclib treatment relative to dimethyl sulfoxide (DMSO) (SI Appendix, Fig. S3B).
In the second control experiment, we induced a G1 delay via a different mechanism: serum starvation. Serum starvation slows proliferation of A549 cells (SI Appendix, Fig. S3C). Growth of A549 cells in medium supplemented with 0.3% serum caused pronounced resistance to cisplatin and paclitaxel (SI Appendix, Fig. S3D). We conclude that G1 delays can decrease a cell’s sensitivity to cisplatin and paclitaxel.
Transient G1 Arrest in the Presence of Drug Allows for Long-Term Survival and Cell Cycle Reentry after Treatment.
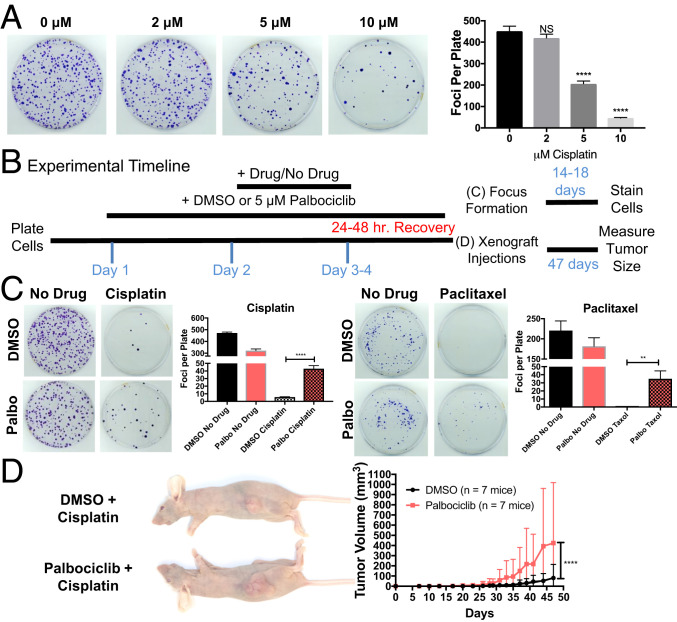
Transient G1 arrest during chemotherapy exposures could delay apoptosis or increase senescence, rather than cause true, long-term protection from the drug. To test this possibility, we followed cell proliferation long term by analyzing the ability of drug-treated cells to form foci. This assay reported long-term survival in a dose-dependent manner as judged by the decreased ability of HCT116 cells to form foci with increasing cisplatin concentrations (Fig. 3A). Thus, by counting focus number, we were able to assess the number of cells that survived drug treatment and then restarted proliferation.
Fig. 3.
G1 delay causes long-term survival following cisplatin and paclitaxel treatment. (A) Cells were treated with the indicated concentrations of cisplatin for 5 h, washed thoroughly with PBS, and 1,000 cells were plated on a 10-cm dish (n = 3). After 18 d, number of foci was determined. Representative images are shown on the Left, quantifications on the Right. Unpaired t tests (NS, not significant, ****P < 0.0001). (B) Scheme of focus formation assay and xenograft growth assay. (C) Cells were treated with DMSO or palbociclib as outlined in B followed by incubation with 7.5 μM cisplatin or 25 nM paclitaxel for 24 (cisplatin) or 48 (paclitaxel) hours. Cells were then allowed to recover for an equal length of time in DMSO or palbociclib. A total of 1,000 cells were plated on a 10-cm dish and number of foci was determined after 14 (paclitaxel) or 18 (cisplatin) days (n = 3). Representative images are shown on the Left, quantifications on the Right. Unpaired t tests (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (D) Cells were treated with DMSO or palbociclib as outlined in B followed by incubation with 7.5 μM cisplatin for 24 h. Cells were then allowed to recover for 24 h in medium with DMSO or palbociclib. A total of 500,000 cells were injected into the rear flanks of a Nu/J mouse (n = 7) and tumor volume was measured with calipers every 1 to 3 d. Representative images at the endpoint are shown on the Left, quantifications on the Right. Nonlinear regression curve fit and extra-sum-of-squares F test (****P < 0.0001).
We treated cells with DMSO or palbociclib for 24 h, added the chemotherapeutic for 24 to 48 h, washed out the chemotherapeutic, allowed cells to recover in fresh medium with palbociclib or DMSO for 24 h, and then plated 1,000 cells in the absence of all drugs (focus formation assay) or injected cells subcutaneously (s.c.) into a mouse to measure tumor formation in the animal (xenograft growth; Fig. 3B).
To form a focus in a low-density plating setting, cells must have a high proliferative capacity; thus, cells capable of forming a focus in this assay have likely repaired or avoided the chemotherapy’s damaging effects. Even with this significant challenge to cellular fitness, palbociclib-treated cells were significantly better at forming foci than untreated cells (Fig. 3C). To determine whether cells recovered/avoided chemotherapy damage sufficiently to form tumors in vivo, we repeated the experiment outlined in Fig. 3B but then injected cells s.c. into the flanks of seven mice and measured tumor growth over time (Fig. 3B). Cells treated with cisplatin alone, were able to form tumors in only three of the seven injected mice (Fig. 3D). In contrast, cells treated with cisplatin and palbociclib formed tumors in six mice. Furthermore, cells protected by palbociclib formed larger tumors (Fig. 3D). We conclude that the chemotherapeutic resistance of G1-delayed cells allows for full recovery and continued proliferation after removal of the drugs.
Delaying Cells in G1 Protects Them from Chemotherapy-Induced Damage.
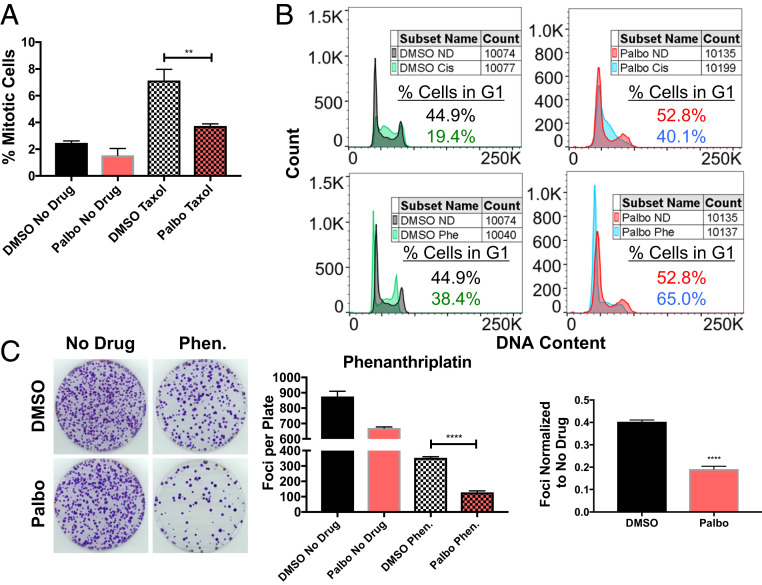
Palbociclib could confer chemotherapy resistance by preventing the drug from causing damage (e.g., reducing the drug’s intracellular concentration or preventing the cell from reaching the cell cycle stage where it causes damage) and/or by increasing repair of the damage. Paclitaxel kills cells during mitosis. By delaying cells in G1, palbociclib could prevent cells from reaching the cell cycle stage, mitosis, when paclitaxel is effective. Indeed, fewer cells arrested in mitosis during paclitaxel treatment when cells were also treated with palbociclib (Fig. 4A). Cisplatin causes much of its damage in S phase as cells try to undergo DNA replication with platinum cross-linked to their DNA (38, 39). Thus, a G1 delay could prevent DNA damage by preventing cells from replicating DNA in the presence of cisplatin. Indeed, cisplatin treatment of exponentially growing cells causes a G2 arrest. Palbociclib prevented this G2 arrest; the drug held cells in G1 throughout cisplatin treatment (Fig. 4 B, Top row). We conclude that delaying cells in G1 reduces paclitaxel’s and cisplatin’s ability to create cellular damage by preventing them from reaching the cell cycle stages where they are most effective.
Fig. 4.
Mechanisms of G1 delay induced chemotherapy resistance. (A) HCT116 cells were pretreated with 5 μM palbociclib or DMSO for 24 h, and then incubated with 20 nM paclitaxel for 24 h in the presence of DMSO or palbociclib. Cells were then stained with 10 μM Hoechst 33342 and at least 200 cells/well were counted to determine the percentage of cells with condensed, mitotic chromosomes (n = 3 wells). (B) HCT116 cells were treated with 5 μM palbociclib or DMSO for 24 h, followed by incubation with 20 μM cisplatin (cis), 3 μM phenanthriplatin (phe), or no drug (ND) in the presence of DMSO or palbociclib for 24 h. Cells were then washed with PBS, fixed in 70% ethanol, stained with DAPI, and at least 10,000 cells were analyzed by flow cytometry. No-drug sample plots are shown twice for comparison to each drug-treated plot. (C) HCT116 cells were treated with 5 μM palbociclib or DMSO for 24 h, and then incubated in medium containing 3 μM phenanthriplatin (Phen.) in the presence of DMSO or palbociclib for 24 h. Cells were allowed to recover for 24 h in DMSO or palbociclib containing medium, before plating to determine their ability to form foci (n = 3). Representative images on Left, quantifications Middle, and normalized quantifications Right. Unpaired t tests (**P < 0.01, ***P < 0.001, ****P < 0.0001).
Another platinum drug, phenanthriplatin, arrests and kills cells in G1 (39). We predicted that lengthening time in G1 would increase sensitivity to a drug that kills cells in G1. We first confirmed that phenanthriplatin treatment arrested many more cells in G1 than cisplatin treatment; palbociclib further increased the percentage of cells treated with these DNA damaging agents in G1 (Fig. 4 B, Bottom row). Consistent with phenanthriplatin damaging cells in G1, we found that phenanthriplatin was much more toxic when combined with palbociclib than with DMSO (Fig. 4C). We conclude that a delay in G1 impacts chemotherapy response by controlling whether a cancer cell reaches the cell cycle phase during which the drug is active; drugs that are active in G1 cells may be more effective against cells that spend longer in G1.
G1 Arrest Affects Intracellular Platinum Concentrations.
Because platinum can cross-link to DNA at any point in the cell cycle, the cells that successfully proliferate in our focus formation assay must eventually undergo DNA replication and deal with any platinum left in their DNA. Based on this consideration, we explored how G1-delayed cells recover from cisplatin to restart proliferation. We first tested how platinum concentrations change over time in the DNA of G1-arrested cells. We examined the quantity of DNA platination immediately following drug treatment and 24 h thereafter. We found less platinum in the DNA of palbociclib-treated cells immediately following a 5-h cisplatin treatment. Twenty-four hours later, DNA platination was reduced in both conditions, but importantly, platinum levels were still lower in palbociclib-treated cells (SI Appendix, Fig. S4A). Thus, by the time palbociclib-treated cells reenter S phase, they harbor less platinum cross-linked to their DNA; this likely reduces the amount of DNA damage that occurs when replicating platinum cross-linked DNA.
The observation that intra-DNA platinum concentrations were different even before the 24-h recovery was surprising; it suggested that G1 arrest reduces intracellular platinum concentrations or limits DNA-platinum binding (i.e., reduced platinum uptake vs. equal platinum uptake but the platinum binds to different macromolecules). To determine whether G1-delayed cells harbored less platinated DNA because of decreased platinum uptake, we measured total cellular platinum 3, 5, or 10 h after cisplatin treatment. G1-delayed cells harbored less platinum per biomass (SI Appendix, Fig. S4B). We conclude that delaying cells in G1 by palbociclib treatment either decreases cisplatin uptake or increases drug efflux. As a result, less platinum reacts with DNA and, presumably, creates less DNA damage. For comparison, phenanthriplatin, which is imported better and exported worse than cisplatin (40), reached equal or greater concentrations in palbociclib-treated cells compared to control cells (SI Appendix, Fig. S4C). While conclusions are complicated by the fact that phenanthriplatin affects cells in a different manner than cisplatin (39), it was interesting that when platinum uptake is equal between G1-delayed and control cells, the differential response to platinum-based chemotherapeutics was no longer evident. We conclude that palbociclib confers drug resistance by at least two mechanisms: 1) G1 delay shortens the relative window of the cell cycle where many chemotherapies act; and 2) G1 delay reduces intracellular drug concentrations (of at least cisplatin).
A third possible cisplatin resistance mechanism we examined was whether DNA repair was increased in G1-delayed cells. We inhibited the DNA damage checkpoint and repair using either a combination of Ku60019 + rabusertib (ATM and Chk1 inhibitors, respectively) or Ku55933 + BAY1895344 (ATM and ATR inhibitors, respectively). The drugs efficiently inhibited the checkpoint pathway as judged by loss of DNA damage-induced Chk1 phosphorylation (SI Appendix, Fig. S5 A and B). However, inhibiting the DNA damage checkpoint pathway did not affect cisplatin resistance of palbociclib-treated cells (SI Appendix, Fig. S5 C and D). This finding suggested that DNA repair is not necessary for G1-delayed cells to be more resistant to cisplatin treatment. The effects of ultraviolet (UV) radiation on palbociclib-treated cells further supported this conclusion. Like platinum adducts, UV-induced thymidine dimers are primarily repaired by nucleotide excision repair (NER). If improved NER was responsible for the resistance of palbociclib-treated cells to cisplatin, such cells should also be resistant to UV radiation. This was not the case (SI Appendix, Fig. S5 E and F). Thus, the sensitivity of G1 cells to UV demonstrates that increased DNA repair was not the reason for G1-induced cisplatin resistance. In summary, we conclude that decreased cisplatin exposure during S phase and reduced intracellular cisplatin concentration, rather than increased DNA damage repair, are the major differentiators between how actively dividing cells and G1-arrested cells respond to cisplatin.
Aneuploidy and G1 Delays Do Not Cause Changes to a Cell’s Apoptotic Threshold.
Each cell has a balance of pro- and antiapoptotic factors; shifts in this balance alter a cell’s sensitivity to stress by changing the threshold for when a cell will induce apoptosis. If aneuploidy or G1 delay raised this apoptotic threshold, this could confer resistance to many drugs. We used a well-established technique called BH3 profiling to assess the apoptotic threshold of trisomic and G1-delayed HCT116 cells (due to poor growth of MEFs, we did not analyze apoptotic threshold in these cells) (41). Cytochrome C release was not decreased significantly in the trisomic cell lines or cells treated with palbociclib (SI Appendix, Fig. S6 A and B). We conclude that, at least in HCT116 cells, neither trisomy nor palbociclib treatment alters the apoptotic threshold to confer paclitaxel resistance.
Slow Proliferation Is a Cause of Chemotherapy Resistance in Aneuploid Cells.
Our results show that slowing cell proliferation by delaying cells in G1 is sufficient to increase resistance to paclitaxel and cisplatin. Could slowed proliferation explain the increased resistance of trisomic cells to these chemotherapeutics? To address this question, we asked whether equalizing cell division length between euploid and trisomic cells by extending the time these cells spend in G1, would make trisomic and euploid cells equally resistant to the drugs.
We treated trisomy 13 and 16 and euploid control cells with 5 μm palbociclib. This slowed proliferation rate to similar levels (Fig. 5 A and C). Trisomy 13 MEFs displayed resistance to cisplatin relative to euploid littermate controls when grown in the absence of palbociclib, but when Ts13 and euploid MEFs proliferate at the same rate, they were equally sensitive to cisplatin (Fig. 5B). This suggests that their difference in proliferation rate was responsible for their differences in cisplatin response. The resistance of trisomy 16 cells to cisplatin was also due to slow growth, but only in part. When the proliferation rate of trisomy 16 and euploid cells was equalized by palbociclib treatment, aneuploid and euploid cells were equally resistant to cisplatin at low concentrations (Fig. 5D). At high cisplatin concentrations trisomy 16 cells remained less sensitive to cisplatin than slow-growing euploid cells (Fig. 5D). We conclude that slow growth contributes to the cisplatin resistance of trisomy 13 and 16 cells. However, in the case of trisomy 16 additional karyotype-specific characteristics contribute to resistance to high concentrations of the drug.
Fig. 5.
Aneuploidy-induced cell-division delays confer chemotherapy resistance. (A) Growth curve of euploid and trisomy 13 MEFs treated with DMSO or palbociclib (5 μM) (n = 3). (B) Euploid and trisomy 13 cell lines were grown in the presence or absence of palbociclib (5 μM) and then treated with varying concentrations of cisplatin in the presence or absence of palbociclib (n ≥ 3). (C) Growth curve of euploid and trisomy 16 MEFs treated with DMSO or palbociclib (5 μM) (n = 3). (D) Euploid and trisomy 16 cell lines were grown in the presence of DMSO or palbociclib (5 μM) and then treated with varying concentrations of cisplatin in the presence or absence of palbociclib (n ≥ 3). (E) Growth curve of trisomy 3 HCT116 and control cells treated with DMSO or palbociclib (5 μM) (n = 3). (F) Trisomy 3 and control HCT116 cells were grown in the presence of DMSO or palbociclib (5 μM) and then treated with varying concentrations of paclitaxel in the presence of DMSO or palbociclib (n ≥ 3). Nonlinear regression curve fit and extra-sum-of-squares F test (NS, not significant, ***P < 0.001, ****P < 0.0001).
To determine whether slow proliferation contributed to the paclitaxel resistance of trisomy 3 HCT116 cells, we equalized growth by palbociclib treatment (Fig. 5E). Under these growth conditions, both control and trisomy 3 cells exhibited similar degrees of resistance to paclitaxel. Thus, when grown at similar rates, euploid and aneuploid cells are equally unresponsive to paclitaxel treatment. We conclude that slow growth of aneuploid cells confers resistance to the chemotherapeutics cisplatin and paclitaxel. As illustrated by the resistance pattern of trisomy 16 MEFs, additional mechanisms, however, can also contribute.
High Aneuploidy Score and Slow Proliferation are Associated with Drug Resistance in Cancer Cell Lines.
Our results suggested that addition of a single additional chromosome confers resistance to the commonly used chemotherapeutics cisplatin and paclitaxel. Our data further indicated that this is, in part, due to aneuploidy slowing cell proliferation. These observations beg the question as to whether aneuploidy has similar effects in cancer cells that often have complex aneuploid karyotypes and that are more adapted to the aneuploid state. To address this possibility, we examined drug response data of almost 1,000 cell lines in the CCLE.
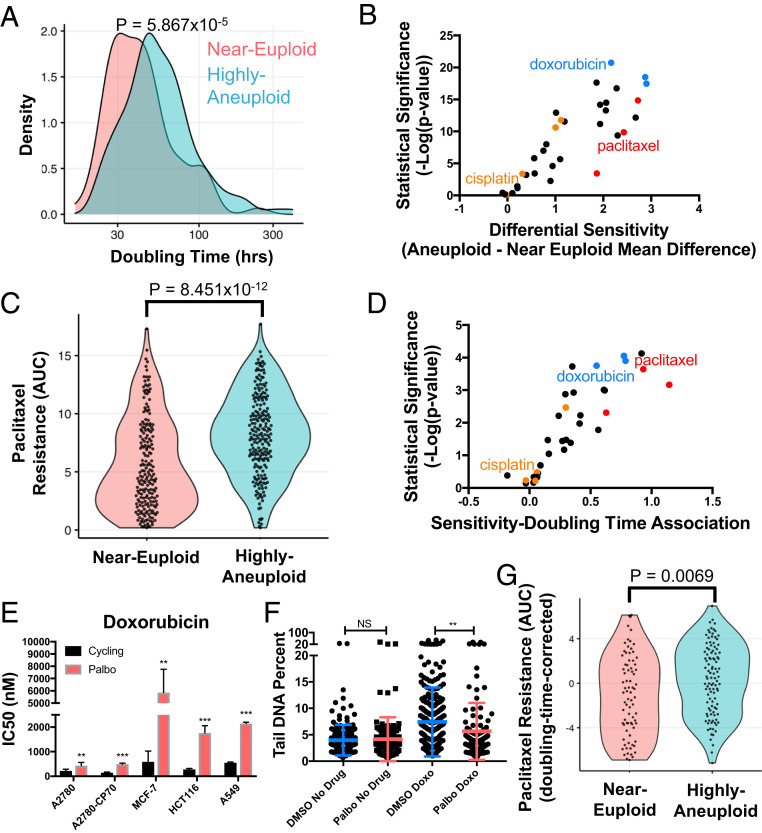
First, we quantified the degree of aneuploidy in each cell line by estimating the number of chromosome arm-level gains or losses relative to background ploidy, using published copy number profiles generated with the ABSOLUTE algorithm (CCLE phase 2). We then created two groups of cell lines, an aneuploid and a near-euploid group, defined as the top and bottom quartiles of the number of arm-level copy number variants (CNVs), respectively (quartiles and aneuploidy scores from ref. 42). We then compared the doubling times of these cell line groups to determine whether aneuploidy was associated with slow proliferation even in these highly adapted cancer cell lines. We found that cells with a high degree of aneuploidy proliferated more slowly than near-euploid cells (Fig. 6A). This finding suggested that even when given time to evolve, cancer cells, like primary cells, have difficulty overcoming the antiproliferative effects of aneuploidy. We note that the associations between aneuploidy scores, proliferation rates, and drug responses varied in significance when using doubling time measurements or drug response data generated by the Sanger Institute (43). Differences between supposedly identical cell lines could be a plausible explanation for why such associations do not hold across datasets generated at different institutions (44). We therefore focused our analyses on datasets that were generated from the same cell lines in the same institute (SI Appendix, Supplemental Note S1).
Fig. 6.
Degree of aneuploidy correlates with slow growth and chemotherapy resistance across cancer cell lines. (A) Histogram of doubling times of highly aneuploid (top quartile) and near-euploid cell lines (bottom quartile) from the Cancer Cell Line Encyclopedia obtained from the Broad Institute’s RNAi dependency screen. P value represents a two-sided Wilcoxon rank-sum test. (B) Association between drug response and degree of aneuploidy for cell lines from the CCLE was calculated for each drug (listed in SI Appendix, Table S2). Each data point shown is a different chemotherapy plotted by log(P value) and effect size (representing the shift in the mean of the AUC between the near-euploid and aneuploid groups). The farther to the right a drug is, the more resistant aneuploid cancer cell lines are to that drug compared to near-euploid lines. (C) For every drug and cell line in the CTD2 dataset, a CellTiterGlo assay was performed and the resulting curve from various drug concentrations was integrated to give an AUC that represents the cell line’s response to drug (45). Here, cell lines’ AUC values for paclitaxel are compared between the aneuploid and near-euploid cell line groups. P value represents a two-sided Wilcoxon rank-sum test. (D) Association between drug response and doubling time of cell lines from the CCLE was calculated for each drug (listed in SI Appendix, Table S3). Each data point shown is a different chemotherapy plotted by log(P value) and effect size of the association. (P < 0.0001, one sample t test). (E) The indicated cell lines were treated with various concentrations of doxorubicin in the presence of DMSO or palbociclib (5 μM) after being pretreated for 24 h with DMSO or palbociclib. IC50 was determined by MTT assay (n ≥ 3). Unpaired t test (**P < 0.01, ***P < 0.001). (F) HCT116 cells were grown in medium containing 5 μM palbociclib or DMSO for 24 h. Cells were then treated with 200 ng/mL doxorubicin for 24 h in the presence of DMSO or palbociclib. To measure degree of DNA damage, an alkaline comet assay was performed and the percent of DNA in the tail of each comet was counted (n = ≥100 cells/condition). Unpaired t test (NS, not significant, **P < 0.01). (G) Comparison of the response to paclitaxel between the aneuploid and near-euploid cell line groups after regressing out the relationship between proliferation rate (log[doubling time]) and paclitaxel response. P value represents a two-sided Wilcoxon rank-sum test.
Next, we sought to examine the association between aneuploidy and chemotherapy response in the CCLE cell lines. We measured the association between drug response and aneuploidy for chemotherapeutics that specifically inhibit cell division in the Broad Institute’s drug screen in the Cancer Target Discovery and Development (CTD2) database (45). Across a large number of antiproliferative chemotherapies, we observed a strong association between degree of aneuploidy and chemotherapy resistance (Fig. 6B). Specifically, of the 34 chemotherapies tested, 29 showed decreased efficacy in aneuploid compared to near-euploid cell lines (false discovery rate [FDR] < 0.1), and none of the 34 chemotherapies showed the opposite relationship. These drug associations remained similarly biased toward aneuploidy-induced drug resistance when we included non-cell-cycle-specific chemotherapies in the analysis (SI Appendix, Fig. S7A).
Examining the effects of individual drugs, we observed that aneuploid cancer cells were significantly more resistant to paclitaxel than near-euploid cell lines (P = 8.451 × 10−12, Wilcoxon rank-sum test; Fig. 6C). We, however, did not detect a significant association between aneuploidy levels and cisplatin resistance in this dataset (P = 0.3386). This was not surprising because all compounds in the CTD2 drug screen were dissolved in DMSO, which is known to inactivate cisplatin (note, we confirmed that dissolving cisplatin in DMSO reduces its efficacy and that the cisplatin used in the CTD2 drug screen had very poor efficacy compared to other drugs in the screen; see SI Appendix, Fig. S7 B and C) (45–47). Beyond paclitaxel, we identified many other chemotherapeutics that showed an association between aneuploidy and drug resistance. Other drugs that, like paclitaxel, affect microtubule assembly (red in Fig. 6B) such as vincristine and docetaxel, were highly biased in their effectiveness based on aneuploidy levels. We also identified inducers of DNA damage (cytarabine, clofarabine, and doxorubicin, in blue in Fig. 6B), DNA alkylating agents, and other platinum drugs (chlorambucil and oxaliplatin, in orange in Fig. 6B) to be less effective in highly aneuploid cancer cells than in near-euploid cancer cells.
Next, we examined how proliferation rate correlated with drug response in the CCLE dataset. For each drug, we calculated the strength and significance of the association between proliferation rate (log[doubling time]) and drug response. We observed a striking association between doubling time and chemotherapy resistance: slow proliferation associated with poor drug response for 21/34 chemotherapies (FDR < 0.1), and no drugs showed the opposite trend (Fig. 6D). Interestingly, slow proliferation was associated with resistance to many of the same drugs that were less effective in aneuploid cancer cells than in near diploid cancer cells. Drugs that impact microtubule assembly like paclitaxel, vincristine, and docetaxel (red in Fig. 6D) were some of the least effective drugs against cells with long doubling times. Similarly, drugs that induce DNA damage largely during S phase (doxorubicin, cytarabine, clofarabine in blue in Fig. 6D) were ineffective against aneuploid cells and against slowly dividing cells. We were able to confirm that slow growth indeed causes doxorubicin resistance, increasing the IC50 of doxorubicin in all five tested cell lines and increasing HCT116 survival in a viability assay (Fig. 6E and SI Appendix, Fig. S7D). Palbociclib treatment furthermore reduced the ability of doxorubicin to cause DNA damage as judged by comet assays (Fig. 6F). We conclude that doxorubicin, a drug identified in the CCLE dataset as responsive to degree of aneuploidy and cell division length, much like paclitaxel, causes less damage in slowly dividing cells.
As in our association analysis of degree of aneuploidy and chemotherapy resistance, we observed no association between doubling time and response to platinum drugs, including cisplatin. Inactive compound is likely the cause for this absence of association. We conclude that slowed proliferation and high levels of aneuploidy confer resistance to many chemotherapeutics that target cell division.
Slowed Proliferation Is Largely Responsible for Aneuploidy-Induced Chemotherapy Resistance in Cancer Cell Lines.
Is slowed cell proliferation the cause of aneuploidy-induced paclitaxel resistance in the CCLE cancer cell lines? To address this question, we controlled for proliferation rate (using linear regression) and reexamined the association between paclitaxel response and degree of aneuploidy. We found that these proliferation-rate-corrected paclitaxel responses were still significantly weaker in aneuploid cancer cell lines compared to near-euploid cancer cell lines (P = 0.0069; Wilcoxon rank-sum test); however, the robustness of this association was greatly reduced (compare Fig. 6 C and G). We observed similar results for other chemotherapies. After regression, 22/34 chemotherapies still exhibited more efficacy in near-euploid than in highly aneuploid cancer cell lines (FDR < 0.1), but the strength and significance of the observed correlations were greatly weakened (SI Appendix, Fig. S7E). We conclude that aneuploidy’s antiproliferative effects are a major contributing factor to aneuploidy-induced chemotherapy resistance. The remaining drug resistance, after regressing out proliferation rate effects, suggests that other aspects of the aneuploid condition likely contribute as well.
Discussion
Studies of cells that exhibit chromosomal instability or that harbor aneuploid karyotypes have shown that most aneuploid karyotypes are detrimental to cells, causing slow growth and cellular stress (reviewed in ref. 48). Although it has been proposed that cancers may find “optimal” karyotypes that increase oncogene expression and decrease tumor suppressor activity (49), an evolving tumor that frequently missegregates chromosomes will experience many “detrimental” karyotypes on its way to this optimal karyotype. Our analysis of almost 1,000 highly evolved cancer cell lines in the CCLE reflects this. Highly aneuploid cancer cell lines generally proliferate more slowly than near-euploid cell lines. We note that studies of The Cancer Genome Atlas and various tumors have suggested that aneuploidy in tumors is correlated with rapid proliferation (50–52); however, studies of tumors may be confounded by many other factors, like tumor grade. The association between aneuploidy and slowed proliferation that we observed in cancer cell lines, even when cancer cells had time to adapt to their aneuploid karyotypes, suggests that aneuploidy is still associated with poor proliferation relative to euploidy. It thus appears that aneuploid cancer cells cannot entirely escape the aneuploidy-associated stresses.
Mechanisms of Aneuploidy-Induced Drug Resistance.
Despite this antiproliferative effect of whole chromosome gains and losses even in highly evolved cancer cell lines, aneuploidy is nevertheless associated with poor prognosis (6–13). Our analysis of the effects of aneuploidy on chemotherapy response offers an explanation for this paradox. Based on the fact that many different trisomies, trisomy 13 and 16 in MEFs and trisomy 3, 5, and 8 in HCT116 cells, conferred resistance to chemotherapeutics we hypothesized that a general feature of the aneuploid state, slowed progression through G1 and S phase, causes this resistance. Our observation that euploid cells, slowed in G1 by treatment with palbociclib, are also resistant to these drugs, is consistent with this idea. The fact that leveling the proliferation rate between euploid and aneuploid cells largely equalizes their resistance to paclitaxel and cisplatin indicates that aneuploidy-induced slowed proliferation and drug resistance are causally related. Examination of drug responses of cell lines from the Cancer Cell Line Encyclopedia further supports this conclusion. In these cell lines, we saw surprisingly strong associations between aneuploidy, slow growth, and resistance to a large number of common chemotherapeutics. Many mechanisms have been described that can confer chemotherapy resistance, all of which could have masked this association, but slowed proliferation appears to be the dominant mechanism whereby aneuploidy confers drug resistance in cancer cell lines. Regressing out the effects of proliferation rate on the aneuploidy, associations significantly weakened but did not entirely eliminate them. Overall, our studies of chemotherapy response in five different engineered trisomies and almost 500 naturally aneuploid cancer cell lines (the top and bottom quartile of aneuploid cell lines in the CTD2 dataset) lead to a remarkable conclusion: the very fact that aneuploidy hampers cell proliferation bestows resistance to frontline chemotherapeutics, such as paclitaxel and cisplatin.
How do delays in G1 of the cell cycle, like those caused by aneuploidy, increase resistance to cell-cycle-specific drugs, like paclitaxel? Our data demonstrate that slowing cells in G1 causes resistance to chemotherapeutics by a variety of mechanisms that all reduce the drug’s ability to damage cells. The mechanism by which G1 delay causes paclitaxel resistance is intuitive. Paclitaxel causes cell death by inducing chromosome missegregation (53). Delaying cells in G1 prevents cells from reaching the cell cycle stage where the drug exerts its antiproliferative effects. We suspect that delaying cells in G1 confers resistance to many drugs that target a specific cell cycle stage (e.g., doxorubicin).
Resistance to cisplatin was more surprising because a recent study had shown that, in ovarian cancer cell lines, inactivation of CDK6 caused increased sensitivity to cisplatin treatment (54). In that study CDK6 was shown to play an important role in activating ATR during a cell’s DNA damage response to platinum compounds. When cells were treated in combination with, or after cisplatin treatment, arrest in G2/M and synergistic killing was observed (54). We found that when cells were treated with palbociclib prior to cisplatin treatment, to mimic the G1 delay caused by aneuploidy, cells were resistant to the chemotherapeutic. To understand why the sequence in which cells are treated with palbociclib and cisplatin affects treatment outcome, we must explore how slowing cells in G1 causes cisplatin resistance.
Cisplatin binds to DNA in a non-cell-cycle-specific manner but damages DNA during transcription and replication. Much like paclitaxel resistance, a delay in G1 is likely to confer resistance by limiting entry into S phase in the presence of cisplatin, which will greatly reduce cisplatin’s ability to damage DNA. However, G1 delay-induced resistance to cisplatin may go beyond whether or not a cell enters S phase in the presence of the drug; our data suggest that a second mechanism may be mitigating cisplatin’s ability to damage DNA: decreased intracellular platinum concentrations in G1 cells. Our results suggest that either decreased drug uptake or increased drug efflux lowers the amount of platinum-DNA adducts in G1-delayed cells. Cisplatin uptake is thought to occur mostly by passive diffusion (55), but some transporters of metal cations, like the copper transporter CTR1, have been implicated, both, in cisplatin uptake and export (56). We observed no changes in the expression of CTR1 in palbociclib-treated cells (SI Appendix, Fig. S8). While more mechanistic research is needed to understand why a cell in G1 is protected from cisplatin, we conclude that G1 delays shield cells from the damaging effects of the drug. We further note that this shielding of cells from DNA damage explains why G1 delay-induced resistance appears to be epistatic to the downstream role that CDK6 plays in repairing DNA damage.
Our work also indicates that the role of p53 in mediating cisplatin resistance also merits further investigation. A recent study found that p53 confers aneuploidy-induced drug resistance to 5-hydroxyurea in a chromosomally unstable HCT116 cell line (31). Our analysis revealed that in trisomic MEFs, p53 is certainly not the only factor that confers resistance to cisplatin. Overexpression of a dominant negative form of p53 did not significantly affect cisplatin resistance of trisomic MEFs. Many factors, including use of different cell lines, drugs, or manner in which p53 was inactivated, could explain why we did not observe a role for p53 in mediating cisplatin resistance. However, most notable is the fact that inactivation of p53 in chromosomally unstable HCT116 cells leads to improved proliferation, whereas inactivation of p53 has little effect on cell division length in trisomic MEFs (21). If, as our data indicate, cell division rate is the major determinant of drug resistance, any genetic alteration that improves cell proliferation should increase the sensitivity of cells to chemotherapeutics that target cell proliferation.
Aneuploidy-Induced Chemotherapy Resistance beyond Slowed Proliferation.
While we have found that proliferation rate is largely responsible for cisplatin and paclitaxel resistance, aneuploidy likely confers resistance to chemotherapies by additional mechanisms. Aneuploidy increases genomic instability, potentially giving cells more chances to acquire mutations that confer drug resistance. For example, chromosomal instability helps cancer cells evade oncogene addiction, allowing them to be more flexible in their response to drugs that target specific oncogenes (57). Furthermore, aneuploidy causes nongenetic heterogeneity (58). Recent studies have shown that nongenetic variability in cancer cells allows for drug-induced reprogramming of a cell into a stably drug-resistant cell state (59). Thus, aneuploidy could drive drug resistance by driving both genetic and epigenetic changes. Indeed, highly aneuploid cancer cell lines are more resistant to drugs in general, not just chemotherapies (42). Karyotype-specific mechanisms could also contribute. At high cisplatin concentrations, trisomy 16 cells showed drug resistance relative to euploid cells even when proliferation rates were equalized by palbociclib treatment. This suggests that gain of chromosome 16 confers cisplatin resistance through another mechanism beyond slowed proliferation. Notably, ERCC4 is located on mouse chromosome 16; working together with ERCC1, this gene plays a key role in nucleotide excision repair and has been implicated in cisplatin response (60). Beyond ERCC4, we identified 31 other genes on mouse chromosome 16 that fall under the gene ontology term “drug resistance” (SI Appendix, Table S1). Further studies are needed to determine which of the genes encoded on chromosome 16 confer increased resistance of trisomy 16 cells to cisplatin. Identifying karyotypes that cause specific drug resistances is also likely to increase our ability to predict how a tumor will respond to specific drugs.
Therapeutic Implications.
Our focus formation assay was designed to mimic a cycle of chemotherapy treatment, in which tumors are treated with drugs that will eventually wash out from the body and give the cancer cells an opportunity to reenter the cell cycle. Although patients typically go through multiple rounds of drug-treatment cycles, lengthening a tumor cell’s time in G1 will help it survive and recover from each of those cycles. We found that chemotherapies in use today, even those described as non-cell-cycle specific, are largely ineffective at damaging cells in G1, allowing them to recover and continue growing in the long term. These experiments strongly argue against combining general chemotherapeutics with palbociclib treatment. However, a survey of the ClinicalTrials.gov website revealed that currently at least four clinical trials have been registered that utilize these combinations. Notably, clinical trials have begun attempting to combine palbociclib with common chemotherapeutics such as paclitaxel, abraxane, 5-fluorouracil, oxaliplatin, cisplatin, or carboplatin (ClinicalTrials.gov identifiers: NCT02897375, NCT01522989, NCT01320592, and NCT02501902). Importantly, the timing and order or drug dose in these clinical trials does vary, which may play a critical role in patient outcome. Previous studies had already raised the possibility that palbociclib may antagonize the effects of doxorubicin (36), but we demonstrate here that CDK 4/6 inhibition administered before chemotherapy can antagonize the efficacy of even more chemotherapeutics that act by a variety of mechanisms. However, it is important to note that palbociclib can be used effectively in combination with chemotherapies when the drug is given after chemotherapy treatment. In fact, when given in this sequence, palbociclib significantly enhanced chemotherapy (61). Timing and order of drug treatment and Rb mutation status of the tumor may prove to be crucial in these drug trials in order to avoid the protective effects of palbociclib.
In summary, we show here that aneuploidy’s slowing of proliferation confers resistance to chemotherapeutics. Chemotherapeutics that were first identified for their ability to kill rapidly dividing cancer cells cause less cellular damage and stress in cells that proliferate slowly. Ultimately, this emphasizes the need for identifying compounds that can kill a cell in any stage of the cell cycle or that target the aneuploid state of cancer cells.
Methods
Cell Lines.
MEFs, HCT116, A549, and MCF7 cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen 11995) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine. A2780 and A2780 CP/70 cells were grown in RPMI (Invitrogen 11875) supplemented with 20% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine. P53DD protein expression was validated in relevant MEFs by Western blot analysis; its ability to interfere with the cellular p53 response was confirmed by examining p21 expression following doxorubicin treatment (SI Appendix, Fig. S9).
HPLC Analysis Showing Palbociclib Does Not React with Cisplatin.
The following drug solutions were prepared in Hepes buffer containing 20 mM Hepes and 150 mM NaCl: cisplatin (0.2 mM), palbociclib (0.1 mM), and cisplatin (0.2 mM) + palbociclib (0.1 mM). The solutions were incubated at 37 °C and analyzed on an Agilent 1200 Series High Performance Liquid Chromatography (HPLC) system at 0, 6, and 24 h (SI Appendix, Detailed Methods). We observed no changes in the spectra when palbociclib and cisplatin were incubated together, suggesting that cisplatin and palbociclib do not react under these conditions (SI Appendix, Fig. S10).
Cell Viability, Focus Formation, and Xenograft Analysis.
A total of 2 × 105 cells were plated on a six-well plate and incubated for 24 h. In experiments involving the use of palbociclib, medium was then replaced with medium containing DMSO or 5 μM palbociclib. The following day medium was replaced with medium containing the chemotherapeutic at the indicated concentrations. Following drug treatment (48 to 72 h), trypsinized cells, medium, and the phosphate buffered saline (PBS) wash were collected and stained with 1× Annexin V-allophycocyanin (APC) and 1 μg/mL DAPI in 1× Annexin binding buffer (Thermo Scientific A35110). The percentage of live (APC negative, DAPI negative), apoptotic (APC positive, DAPI negative), and dead (DAPI positive) cells was determined by flow cytometry.
In focus formation assays, a similar scheme was followed but with an additional drug washout and recovery for 24 to 48 h in medium with only DMSO or palbociclib (no other drugs) before plating 1,000 cells/10-cm plate. Fourteen to 18 days later, plates were stained with 0.5% crystal violet in 20% methanol, washed, and number of foci per plate was counted (SI Appendix, Detailed Methods). For xenograft studies, a similar scheme was followed except that 500,000 cells were injected into the rear flank of a Nu/J mouse (Jackson Research Laboratories, stock No. 002019) at the end of “recovery” and tumor growth was measured with a caliper (SI Appendix, Detailed Methods). These animal studies were approved by the Massachusetts Institute of Technology (MIT) Institutional Animal Care and Use Committee.
MTT Assays.
Cells were plated in 96-well plates. The following day, samples were pretreated with 5 μM palbociclib or 0.1% DMSO. For drug treatment, medium was replaced with fresh medium with or without palbociclib and with the indicated concentrations of chemotherapeutic for 48 to 72 h. Cells were then incubated with the tetrazolium dye MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 2 to 4 h. MTT was solubilized and absorbance at Optical Density (OD) = 590 nm was measured on a plate reader. Absorbance was normalized with “no drug” control wells and negative controls. IC50 values are defined as drug concentrations at which 50% of cells retained viability.
Platinum Uptake Measurements.
HCT116 cells were seeded on 10-cm plates and incubated for 24 h at 37 °C. Cells were then incubated in medium containing DMSO or 5 μM palbociclib for a 24-h pretreatment. Cells for whole-cell uptake studies were then treated with cisplatin (20 μM) or phenanthriplatin (3 μM) for 3, 5, or 10 h at 37 °C. Cells for DNA incorporation studies were treated with DMSO or 5 μM palbociclib and 20 μM cisplatin for 5 h. Medium was removed and cells were washed three times with PBS (in the 24-h sample for DNA incorporation, medium was then replaced with fresh medium with DMSO or palbociclib). For whole-cell platinum studies, cells were counted and an average cell volume was determined on a Beckman Coulter Counter (biomass = cell number × average cell volume); cells were then digested using 70% HNO3 (600 µL) for 5 h at room temperature. For DNA-platinum experiments, DNA was collected using a DNAzol solution (Thermo Fisher Scientific 10503027). The platinum content was analyzed by inductively coupled plasma mass spectrometry (ICP-MS). Platinum concentration was determined by comparison to a standard calibration curve made with 0.1, 1, 10, and 100 parts per billion of platinum. DNA concentration in each sample was measured on a nanodrop.
Cell Cycle Analysis.
HCT116 cells (1.5 × 106) were plated on a 15-cm plate and incubated for 24 h at 37 °C. Cells were pretreated with DMSO or 5 μM palbociclib for 24 h. The following day, medium was replaced and cisplatin (20 μM), phenanthriplatin (3 μM), or no drug was added. After 24 h, cells were collected, fixed with 70% ethanol, washed with PBS, stained with 1 μg/mL DAPI (Thermo Fisher Scientific D1306), and analyzed by flow cytometry. Percentage of cells in G1 was determined as the percentage of cells in the first (2 n) DNA content peak using FlowJo Analysis Software.
Computational Analyses and Data Sources.
Cell line doubling-time measurements were obtained from Tsherniak et al. (62). Drug response area under the curve (AUC) values were obtained from the Cancer Therapeutic Response Portal (CTRP) dataset (v2, 2015; https://ctd2.nci.nih.gov/dataPortal/). The list of chemotherapies was manually curated (SI Appendix, Tables S2–S4). Aneuploidy was quantified by estimating the total number of arm-level gains and losses for each cell line, based on the published ABSOLUTE copy number dataset (63). Specifically, we estimated the (segment length-weighted) median modal copy number across segments for each chromosome arm, and then called copy number gains/losses by comparing these arm-level copy number estimates to the cell lines’ background ploidy. Near-euploid and aneuploid cell line groups were defined by taking the bottom quartile based on arm-level CNVs (min = 0, max = 7) and the top aneuploidy quartile (min = 22, max =36), respectively. Cell line aneuploidy scores for the entire CCLE can be found in Cohen-Sharir et al. (42). The association between cell lines’ sensitivity to each drug and the aneuploidy groups (as well as measured log doubling times) was assessed by linear regression analysis, using the R package limma (64). Reported P values are given by empirical-Bayes moderated t statistics, and q values are estimated using the Benjamini–Hochberg method (65).
Statistics.
All statistical methods are described in the figure legends. All unpaired t tests were performed using Prism graphing software. Comparisons of drug response across drug concentrations in MTT assays were done using Prism graphing software. Focus formation assays were performed with technical replicates due to the large number of plates and for the sake of accurately plating 1,000 cells/plate; all other experiments were performed using biological replicates. A nonlinear (or linear for Fig. 5F) regression was performed for each line and an “extra sum of squares F test” was performed in order to ask whether one curve fit both lines being compared. Wilcoxon rank-sum tests were performed using R (see Computational Analyses and Data Sources for more details).
Supplementary Material
Acknowledgments
We thank Pei-Hsin Hsu for taking pictures of the mice in this paper. We thank Michael Hemann and members of the A.A. laboratory for their helpful comments and discussion regarding this project and manuscript. We thank the Swanson Biotechnology Center for equipment, resources, and technical support, specifically the Koch Institute Flow Cytometry core and High Throughput Sciences core. J.M.R. was supported by a David H. Koch predoctoral fellowship. This work was supported by NIH grant CA206157 to A.A., who is an Investigator of the Howard Hughes Medical Institute; the Paul F. Glenn Center for Biology of Aging Research at MIT; and the Ludwig Center at MIT. U.B.-D. is an Azrieli Faculty Fellow. This work was also supported by NIH grants CA034992 to O.Y. and S.J.L. and CA211184 to O.Y., who is a Pew Scholar. A.L. is a Ludwig Investigator.
Footnotes
Competing interest statement: A.A. and G.J.P.L.K. are coapplicants on a grant. They have not collaborated directly on this project.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009506117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Weaver B. A. A., Cleveland D. W., Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18, 658–667 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Dephoure N., et al. , Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3, e03023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres E. M., Springer M., Amon A., No current evidence for widespread dosage compensation in S. Cerevisiae. Elife 5, e10996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams B. R., et al. , Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passerini V., et al. , The presence of extra chromosomes leads to genomic instability. Nat. Commun. 7, 10754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duijf P. H. G., Schultz N., Benezra R., Cancer cells preferentially lose small chromosomes. Int. J. Cancer 132, 2316–2326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sciallero S., et al. , DNA aneuploidy is an independent factor of poor prognosis in pancreatic and peripancreatic cancer. Int. J. Pancreatol. 14, 21–28 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Dieterich B., et al. , The prognostic value of DNA ploidy and S-phase estimate in primary breast cancer: A prospective study. Int. J. Cancer 63, 49–54 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Alcaraz A., et al. , Aneuploidy and aneusomy of chromosome 7 detected by fluorescence in situ hybridization are markers of poor prognosis in prostate cancer. Cancer Res. 54, 3998–4002 (1994). [PubMed] [Google Scholar]
- 10.Smith J. C., Sheltzer J. M., Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife 7, e39217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasudevan A., et al. , Single-chromosomal gains can function as metastasis suppressors and promoters in colon cancer. Dev. Cell 52, 413–428.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stopsack K. H., et al. , Aneuploidy drives lethal progression in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11390–11395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-David U., Amon A., Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Oromendia A. B., Amon A., Aneuploidy: Implications for protein homeostasis and disease. Dis. Model. Mech. 7, 15–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnelly N., Passerini V., Dürrbaum M., Stingele S., Storchová Z., HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 33, 2374–2387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santaguida S., Vasile E., White E., Amon A., Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 29, 2010–2021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheltzer J. M., et al. , Aneuploidy drives genomic instability in yeast. Science 333, 1026–1030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheltzer J. M., A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 73, 6401–6412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J., Pavelka N., Bradford W. D., Rancati G., Li R., Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet. 8, e1002719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santaguida S., et al. , Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheltzer J. M., et al. , Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31, 240–255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stingele S., et al. , Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8, 608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorburn R. R., et al. , Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol. Biol. Cell 24, 1274–1289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee A. J. X., et al. , Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858–1870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birkbak N. J., et al. , Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamal-Hanjani M., et al. , Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann. Oncol. 26, 1340–1346 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Andor N., et al. , Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutledge S. D., et al. , Selective advantage of trisomic human cells cultured in non-standard conditions. Sci. Rep. 6, 22828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singhal P. K., et al. , Mouse embryonic fibroblasts exhibit extensive developmental and phenotypic diversity. Proc. Natl. Acad. Sci. U.S.A. 113, 122–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vousden K. H., Lu X., Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2, 594–604 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Fang X., et al. , p53 mediates hydroxyurea resistance in aneuploid cells of colon cancer. Exp. Cell Res. 376, 39–48 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Zamble D. B., Jacks T., Lippard S. J., p53-Dependent and -independent responses to cisplatin in mouse testicular teratocarcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 95, 6163–6168 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman T., et al. , Tissue-specific inactivation of p53 tumor suppression in the mouse. Genes Dev. 10, 826–835 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Chappell W. H., et al. , p53 expression controls prostate cancer sensitivity to chemotherapy and the MDM2 inhibitor Nutlin-3. Cell Cycle 11, 4579–4588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fry D. W., et al. , Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3, 1427–1438 (2004). [PubMed] [Google Scholar]
- 36.McClendon A. K., et al. , CDK4/6 inhibition antagonizes the cytotoxic response to anthracycline therapy. Cell Cycle 11, 2747–2755 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean J. L., McClendon A. K., Knudsen E. S., Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J. Biol. Chem. 287, 29075–29087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fichtinger-Schepman A. M., van der Veer J. L., den Hartog J. H., Lohman P. H., Reedijk J., Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: Formation, identification, and quantitation. Biochemistry 24, 707–713 (1985). [DOI] [PubMed] [Google Scholar]
- 39.Bruno P. M., et al. , A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 23, 461–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park G. Y., Wilson J. J., Song Y., Lippard S. J., Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc. Natl. Acad. Sci. U.S.A. 109, 11987–11992 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan J., Montero J., Rocco J., Letai A., iBH3: simple, fixable BH3 profiling to determine apoptotic priming in primary tissue by flow cytometry. Biol. Chem. 397, 671–678 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Cohen-Sharir Y., et al. , Selective vulnerability of aneuploid human cancer cells to inhibition of the spindle assembly checkpoint. BioRXiv: 10.1101/2020.06.18.159038 (19 June 2020). [DOI]
- 43.Yang W., et al. , Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-David U., et al. , Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560, 325–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seashore-Ludlow B., et al. , Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 5, 1210–1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall M. D., et al. , Say no to DMSO: Dimethylsulfoxide inactivates cisplatin, carboplatin, and other platinum complexes. Cancer Res. 74, 3913–3922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundquist W. I., Ahmed K. J., Hollis L. S., Lippard S. J., Solvolysis reactions of cis- and trans-diamminedichloroplatinum(II) in dimethyl sulfoxide. Structural characterization and DNA binding of trans-bis(ammine)chloro(DMSO)platinum(1+). Inorg. Chem. 26, 1524–1528 (1987). [Google Scholar]
- 48.Santaguida S., Amon A., Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Davoli T., et al. , Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isola J. J., Helin H. J., Helle M. J., Kallioniemi O. P., Evaluation of cell proliferation in breast carcinoma. Comparison of Ki-67 immunohistochemical study, DNA flow cytometric analysis, and mitotic count. Cancer 65, 1180–1184 (1990). [DOI] [PubMed] [Google Scholar]
- 51.Kawai T., et al. , Proliferating cell nuclear antigen and Ki-67 in lung carcinoma. Correlation with DNA flow cytometric analysis. Cancer 74, 2468–2475 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Taylor A. M., et al. , Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zasadil L. M., et al. , Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 6, 229ra43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dall’Acqua A., et al. , CDK6 protects epithelial ovarian cancer from platinum-induced death via FOXO3 regulation. EMBO Mol. Med. 9, 1415–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eljack N. D., et al. , Mechanisms of cell uptake and toxicity of the anticancer drug cisplatin. Metallomics 6, 2126–2133 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Kuo M. T., Chen H. H. W., Song I.-S., Savaraj N., Ishikawa T., The roles of copper transporters in cisplatin resistance. Cancer Metastasis Rev. 26, 71–83 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Salgueiro L., et al. , Acquisition of chromosome instability is a mechanism to evade oncogene addiction. EMBO Mol. Med. 12, e10941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beach R. R., et al. , Aneuploidy causes non-genetic individuality. Cell 169, 229–242.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaffer S. M., et al. , Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arora S., Kothandapani A., Tillison K., Kalman-Maltese V., Patrick S. M., Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair (Amst.) 9, 745–753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salvador-Barbero B., et al. , CDK4/6 inhibitors impair recovery from cytotoxic chemotherapy in pancreatic adenocarcinoma. Cancer Cell 37, 340–353.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Tsherniak A., et al. , Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghandi M., et al. , Next-generation characterization of the cancer cell line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.