Significance
NAP1 family proteins are evolutionarily conserved and important histone chaperones. However, because histone chaperones are very flexible, determination of their crystal structures, especially when complexed with histones, is very difficult. Here we obtained the co-crystal structure of Arabidopsis NAP1-Related Protein 1 (NRP1) and the H2A-H2B heterodimer at 3.0 Å, which led to the discovery of a previously unknown mode of interaction between the NAP1 family and H2A-H2B. Our results provide evidence of multiple interaction modes of a NAP1-family histone chaperone, thereby shedding light on how histone chaperones effectively shield H2A-H2B from nonspecific interaction with DNA.
Keywords: crystal structure, NAP1 family protein, H2A-H2B
Abstract
Nucleosome Assembly Protein 1 (NAP1) family proteins are evolutionarily conserved histone chaperones that play important roles in diverse biological processes. In this study, we determined the crystal structure of Arabidopsis NAP1-Related Protein 1 (NRP1) complexed with H2A-H2B and uncovered a previously unknown interaction mechanism in histone chaperoning. Both in vitro binding and in vivo plant rescue assays proved that interaction mediated by the N-terminal α-helix (αN) domain is essential for NRP1 function. In addition, the C-terminal acidic domain (CTAD) of NRP1 binds to H2A-H2B through a conserved mode similar to other histone chaperones. We further extended previous knowledge of the NAP1-conserved earmuff domain by mapping the amino acids of NRP1 involved in association with H2A-H2B. Finally, we showed that H2A-H2B interactions mediated by αN, earmuff, and CTAD domains are all required for the effective chaperone activity of NRP1. Collectively, our results reveal multiple interaction modes of a NAP1 family histone chaperone and shed light on how histone chaperones shield H2A-H2B from nonspecific interaction with DNA.
In eukaryotic cells, chromatin is the carrier of genetic and epigenetic information. The basic structural and functional unit of chromatin is the nucleosome, which consists of ∼146 bp of DNA wrapped around a histone octamer containing two H2A-H2B heterodimers and one (H3-H4)2 tetramer (1). The dynamic assembly and disassembly of nucleosomes regulates, both spatially and temporally, fundamental biological processes such as DNA replication, transcription, and repair (2). Histone chaperones, a large family of proteins with histone-binding activity, play a crucial role in nucleosome assembly/disassembly by escorting and evicting histones to and from DNA (3). In addition to the canonical histones H2A, H2B, H3, and H4, other diverse histone variant isoforms exist, including H2A.X, H2A.Z, H3.3, and CenH3. Based on their affinities to different histones, histone chaperones are classified as H2A/H2B type, H3/H4 type, and specific variant type chaperones (4, 5).
Nucleosome Assembly Protein 1 (NAP1), a H2A/H2B-type histone chaperone (3, 6–10), was first identified from HeLa cell extracts as a histone-binding protein capable of mediating nucleosome assembly in vitro (11). NAP1 was later found to be evolutionarily conserved in eukaryotes, including yeast, plants, and animals (12, 13). NAP1 family proteins have a wide range of roles in such processes as cell proliferation (14), chromosome segregation (15, 16), gene expression (17, 18), DNA replication (17), DNA repair (19), and somatic homologous recombination (20, 21). Unlike other families of histone chaperones that comprise one or a few isoforms per organism, the NAP1 family has multiple members in different organisms; for example, four NAP1s (NAP1L1 to NAP1L4) and one TAF-1β (also known as SET or INHAT) are present in humans (13), and four NAP1s (NAP1;1 to NAP1;4) and two NAP1-Related Proteins (NRP1 and NRP2) are known in Arabidopsis (5). NRP1/2 and TAF-1β proteins are closely related to yeast vacuolar protein sorting 75 (Vps75) (18) and together with Vps75 constitute a phylogenetic clade that is distinct from the canonical NAP1 clade (12).
Determination of the first crystal structure of yeast NAP1 (ScNAP1) revealed the formation of a homodimer via its long N-terminal α-helix (αN) (13). Crystal structures were subsequently obtained for TAF-1β (22), Vps75 (23), and NRP1 (18). Similar to ScNAP1, all these proteins display N-terminal homodimerization that assumes a headphone-like structure, and their earmuff domains form a central cleft for histone binding. Hydrogen–deuterium exchange experiments have suggested that one ScNAP1 dimer can accommodate two copies of H2A-H2B (7), while 6.7 Å resolution of the crystal structure of ScNAP1 complexed with H2A-H2B has indicated that the negatively charged surface of ScNAP1 dimer asymmetrically interacts with single H2A-H2B heterodimer, and that the oligomerization of ScNAP1-H2A-H2B likely plays an essential function in the deposition of H2A-H2B into nucleosomes (8). Recently, the crystal structure of Caenorhabditis elegans NAP1 (CeNAP1) in complex with H2A-H2B has revealed that the “acidic strip” within the concave surface of the earmuff domain is essential for H2A-H2B binding (24). Interestingly, in addition to the stoichiometric conflict, the key H2A-H2B interacting residues within ScNAP1 and CeNAP1 are not conserved in Arabidopsis NRP1, raising additional questions about the specificity of the binding between H2A-H2B and different NAP1 family proteins.
In this study, we determined the crystal structure of the NRP1-H2A-H2B complex, showing that H2A-H2B interacts with the αN domain of NRP1, and further confirmed the biological function of this unique interaction by both in vitro isothermal titration calorimetry (ITC) and cross-linking analyses and in vivo transgenic rescue experiments. Guided by the cross-linking results, we also mapped the H2A-H2B interacting interface within the earmuff domain of NRP1. Finally, we solved three crystal structures of C-terminal acidic domains (CTADs) of NRP1 and human NAP1 in complex with H2A-H2B, revealing a conserved histone binding mode among NAP1 family proteins. In combination with histone chaperone activity assay, our structural analyses suggest that a NAP1 family histone chaperone holds multiple modules of interaction with H2A-H2B, thereby flexibly shielding histones from nonspecific binding with DNA.
Results
Unique Mode of Interaction between NRP1 and H2A-H2B.
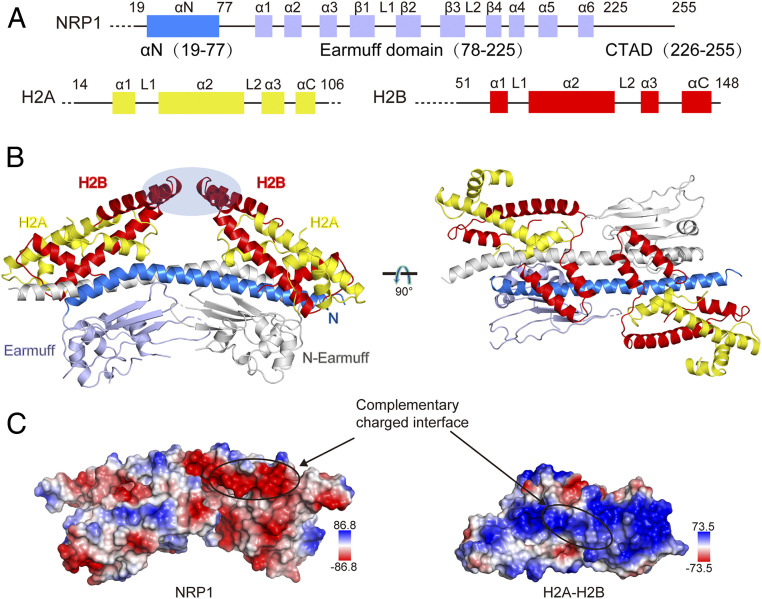
Arabidopsis NRP1 is organized into three domains: αN at the N terminus (amino acids 19 to 77), an earmuff domain in the middle (78 to 225), and a CTAD (225 to 255) at the C terminus (Fig. 1A). To investigate the molecular basis of how NRP1 interacts with histones, we purified NRP1 (residues 19 to 255) and solved the cocrystal structure of NRP1 in complex with the H2A-H2B histone global domain (H2A residues 14 to 106 and H2B residues 51 to 148) to a 3.0 Å resolution by the molecular replacement method (Table 1). In the structure (Fig. 1B), one NRP1 homodimer interacts with two H2A-H2B heterodimers, and interactions between each NRP1 monomer and one H2A-H2B heterodimer are similar. Interestingly, we found that in the crystal lattice, two H2A-H2B heterodimers form a tetramer through domain-swapping of H2B (SI Appendix, Fig. S1 A and B). The α3 and αC domains of H2B are swapped between two H2A-H2B heterodimers (SI Appendix, Fig. S1C), and the structure of the swapped H2A-H2B is identical to that of the apo H2A-H2B (SI Appendix, Fig. S1D). Because the domain swap of H2B occurs distal from its interface with NRP1 (Fig. 1B), it is unlikely to perturb how NRP1 and H2A-H2B interact.
Fig. 1.
Co-crystal structure of the NRP1–H2A-H2B complex. (A) Schematic view showing the domain architectures of Arabidopsis NRP1, H2A, and H2B. (B) Cartoon diagrams of the NRP1–H2A-H2B complex. The CTAD domain is invisible in the electron density map. For the NRP1 dimer, one NRP1 monomer is colored in gray and the other is in bright blue (αN domain) and light blue (earmuff domain). H2A and H2B are shown in yellow and red, respectively. The domain-swapped H2B regions that form crystal lattice contacts are indicated by an oval. (C) Surface representation of NRP1 and H2A-H2B. The interaction regions within NRP1 and H2A-H2B are marked by circles. Blue and red colors on the surface indicate positive and negative charges, respectively. The scales of the electrostatic potential are −86.8 (red) to +86.8 KT/e (blue) for NRP1 and −73.5 (red) to +73.5 KT/e (blue) for H2A-H2B.
Table 1.
Data collection and refinement statistics
| Parameter | AtH2A-H2B | AtNRP1-H2A-H2B | AtNRP1-CTAD- H2A-H2B | HsNAP1- CTAD1-H2A-H2B | HsNAP1- CTAD2-H2A-H2B |
| Data collection | |||||
| Space group | P31 | P212121 | P212121 | P212121 | P212121 |
| Cell parameter | |||||
| a, Å | 65.7 | 66.7 | 40.1 | 61.7 | 40.2 |
| b, Å | 65.7 | 128.3 | 62.2 | 62.2 | 62.7 |
| c, Å | 96.6 | 140.2 | 66.8 | 130.6 | 68.1 |
| α, ° | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| β, ° | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| γ, ° | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| Wavelength, Å | 0.9785 | 0.9793 | 0.9793 | 0.9793 | 0.9793 |
| Resolution, Å | 30.0–1.58 | 30.0–3.0 | 30.0–1.58 | 30.0–2.10 | 30.0–1.90 |
| Last shell, Å | 1.64–1.58 | 3.11–3.0 | 1.64–1.58 | 2.18–2.10 | 1.97–1.90 |
| Completeness, % | 99.9 (99.9) | 96.9 (85.0) | 99.7 (99.0) | 99.8 (98.5) | 96.8 (90.2) |
| Redundancy | 8.7 (7.1) | 7.6 (3.6) | 9.6 (7.0) | 9.4 (6.3) | 4.6 (3.1) |
| I/σ(I) | 42.2 (6.2) | 13.9 (2.3) | 28.5 (4.4) | 17.5 (2.4) | 13.2 (2.6) |
| Rmerge, % | 5.2 (32.1) | 9.3 (39.3) | 7.8 (38.2) | 13.3 (56.9) | 11.6 (42.4) |
| Refinement | |||||
| Resolution, Å | 30.0–1.58 | 30.0–3.0 | 30.0–1.58 | 30.0–2.10 | 30.0–1.90 |
| Rwork/Rfree, % | 15.5/18.6 | 22.3/25.8 | 16.2/20.0 | 18.6/22.6 | 16.8/21.6 |
| No. of atoms | |||||
| Protein | 2,722 | 5,454 | 1,408 | 2,736 | 1,376 |
| Ligand | 74 | 24 | 0 | 6 | 6 |
| Water | 392 | 24 | 141 | 157 | 159 |
| Rmsd | |||||
| Bond length, Å | 0.018 | 0.006 | 0.012 | 0.011 | 0.010 |
| Bond angle, ° | 1.295 | 1.540 | 1.666 | 1.676 | 1.587 |
| Ramachandran plot, % | |||||
| Most favored | 99.13 | 92.11 | 99.44 | 98.53 | 98.83 |
| Additional allowed | 0.87 | 7.89 | 0.56 | 1.47 | 1.17 |
| PDB ID code | 7BP2 | 7C7X | 7BP6 | 7BP4 | 7BP5 |
Values in parentheses are for the last resolution shell.
As observed in the complex structure (Fig. 1B), the positively charged surface composed of the first helices of H2A (α1) and H2B (α1) was attracted by the negatively charged surface of the NRP1 αN domain (Fig. 1C), thus establishing a 553-Å2 interacting interface (as calculated with PDBePISA) (25) per interacting partner. This binding mode between H2A-H2B and NRP1 mediated by the αN domain is completely different from those reported in previous studies, which showed that the earmuff domains of NAP1 family proteins engage in association with H2A-H2B (7, 8, 24).
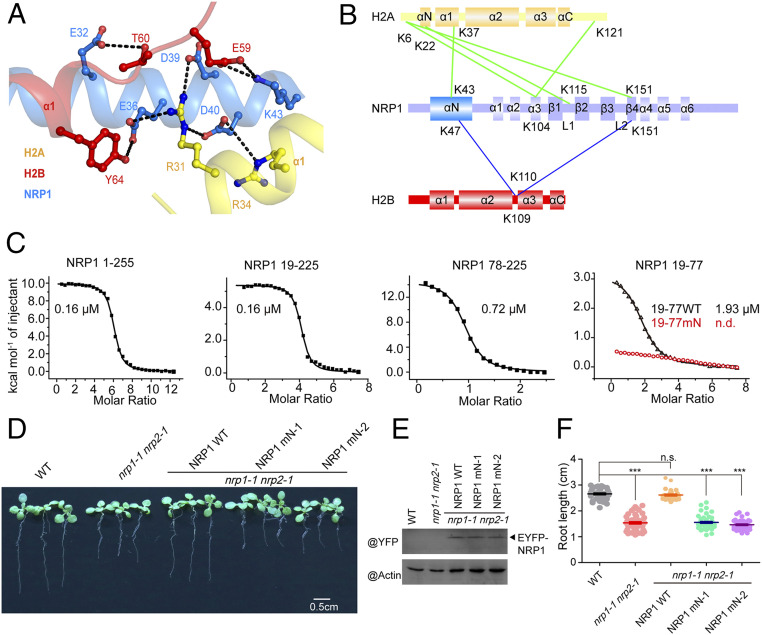
Based on the structure, we were able to identify the residues involved in the interaction between NRP1 αN and H2A-H2B (Fig. 2A). NRP1 αN interacts with α1 helices of both H2A and H2B. E36, D39, and D40 within NRP1 αN form hydrogen bonds (H-bonds) with R31 of H2A, and D40 of NRP1 αN also interacts with R34 of H2A. E32 of NRP1 αN forms one H-bond with H2B T60, and E36 of NRP1 recognizes H2B Y64 via an H-bond. In addition to these electronegative residues, the electropositive residue K43 of NRP1 αN forms a salt bridge with E59 of H2B. Indicated by the low rmsd value (0.91 Å over 261 pairs of Cα atoms), the overall structure of NRP1 in the complex with H2A-H2B is similar to the apo NRP1 structure (Protein Data Bank [PDB] ID code 5DAY) reported in our previous study (18). However, compared with the apo NRP1 structure, the N terminus of NRP1 αN (SI Appendix, Fig. S1E) in the complex turns ∼2.5° toward H2A-H2B, and the side chains of NRP1 αN involved in interactions with H2A-H2B also differ in the two structures (SI Appendix, Fig. S1F).
Fig. 2.
Binding to H2A-H2B mediated by the αN domain is important for the biological function of NRP1. (A) The detailed interactions observed in the NRP1–H2A-H2B complex. The hydrogen bonds are indicated by black dashed lines. (B) Cartoon schedule of the cross-linked sites between NRP1 and H2A-H2B. The linked sites of NRP1-H2A and NRP1-H2B are connected by green and blue lines, respectively. (C) ITC results showing the binding affinities between H2A-H2B (20 μM) and NRP1 1 to 255 (full-length NRP1; 800 μM), NRP1 19 to 225 (NRP1 αN plus earmuff domains; 600 μM), NRP1 78 to 225 (NRP1 earmuff domain; 300 μM), NRP1 19 to 77 (19-77 WT, wild-type NRP1 αN domain; 600 μM), and NRP1 19 to 77 E36A/D39A/D40A/K43A (19 to 77 mN; 600 μM). The integrated heats of each injection were fitted to a one-site binding model (solid line), and the dissociation constants (Kd) are shown. (D) The phenotypes of WT, nrp1-1 nrp2-1 double mutant, and transgenic Arabidopsis plants expressing EYFP fused NRP1 1 to 255 (NRP1 WT) or mutated NRP1 within αN (NRP1 mN: NRP1 1 to 255 E36A/D39A/D40A/K43A) in nrp1-1 nrp2-1 background, respectively (Scale bar: 0.5 cm). NRP1 mN-1 and NRP1 mN-2 are two independent Arabidopsis transgenic lines. (E) The expression levels of EYFP fused proteins in the plants of D were detected with anti-GFP antibody. (F) The statistics of the root length of the plants in D. ***P < 0.001; n.s., nonsignificant (P > 0.05).
To confirm the specific binding mode of NRP1 αN and H2A-H2B, we performed cross-linking experiments to detect the interface between NRP1 and H2A-H2B using discuccinimidyl suberate and bissulfosuccinimidyl suberate under physiological conditions (SI Appendix, Fig. S2). The cross-linking between NRP1 and H2A-H2B resulted in the formation of a shifted band with high molecular weight (∼250 kDa) (SI Appendix, Fig. S2A). To identify the cross-linked sites, we digested the cross-linked NRP1-H2A-H2B and analyzed the cross-linked peptides by mass spectrometry. Within the αN domain of NRP1, K43 was linked to K37 of H2A and K47 was linked to K109 of H2B (Fig. 2B and SI Appendix, Fig. S2B), which is consistent with our crystal structure analysis. Together, the crystal structure and cross-linking analyses support that NRP1 αN interacts with H2A-H2B in vitro.
To further determine the binding affinity between NRP1 αN and H2A-H2B, we performed isothermal titration calorimetry (ITC) experiments (Fig. 2C). We found that the full-length NRP1 (amino acids 1 to 255) and truncated NRP1 (amino acids 19 to 225) display the same binding affinity to H2A-H2B with an equilibrium dissociation constant (Kd) value of 0.16 μM. Compared with NRP1 19 to 225, the earmuff domain of NRP1 (amino acids 78 to 225) alone showed a decreased binding affinity to H2A-H2B with a Kd value of 0.72 μM, indicating that the αN domain of NRP1 (amino acids 19 to 77) contributes to H2A-H2B binding. Indeed, NRP1 19 to 77 alone binds to H2A-H2B with a Kd value of 1.93 μM, while the quadruple mutation NRP1 19 to 77 mN (NRP1 19 to 77 E36A/D39A/D40A/K43A) almost completely disrupted the interaction between αN and H2A-H2B, showing that these four amino acids are essential for the interaction between NRP1 αN and H2A-H2B. Size exclusion chromatography with multiangle light scattering and circular dichroism (CD) spectroscopy analyses showed that both NRP1 19 to 77 and NRP1 19 to 77 mN form dimers and present a proper folding in solution (SI Appendix, Fig. S3 A and B). The binding affinities of NRP1 19 to 77 (αN domain) and NRP1 78 to 225 (earmuff domain) to H2A-H2B are much lower than those of NRP1 19 to 225 containing both αN and earmuff domains (Fig. 2C), suggesting that a cooperation of different domains may exist in the interaction between NRP1 and H2A-H2B.
Plant rescue experiments were performed to analyze whether the interaction between NRP1 αN and H2A-H2B is important for the function of NRP1 in vivo. Consistent with our previous study (14), the short root phenotype of nrp1-1 nrp2-1 double mutant could be rescued by overexpressing enhanced yellow fluorescent protein (EYFP)-tagged full-length WT NRP1 1 to 255 (Fig. 2 D–F). In contrast, the EYFP-tagged NRP1 mN mutant (NRP1 1 to 255 E36A/D39A/D40A/K43A; SI Appendix, Table S1) failed to rescue the phenotype of the nrp1-1 nrp2-1 mutant, even though the expression levels of NRP1 and NRP1 mN proteins are similar in the transgenic plants (Fig. 2 D–F). Taken together, our results prove that the novel interaction mode between H2A-H2B and NRP1 mediated by αN domain is essential for the function of NRP1 in vitro and in vivo.
Interaction between the NRP1 Earmuff Domain and H2A-H2B.
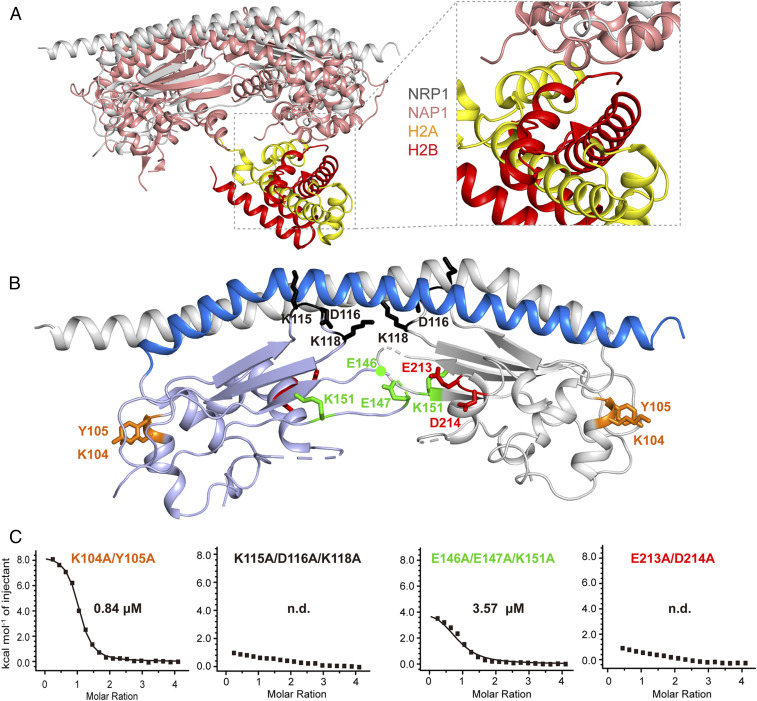
In addition to the αN domain, cross-linking analysis showed that the earmuff domain of NRP1 also binds to H2A-H2B (Fig. 2B), which was confirmed by ITC experiments (Fig. 2C) and consistent with previous studies showing that some NAP1 family proteins interact with H2A-H2B through their earmuff domains (7, 8, 24). However, structural superposition and sequence alignment revealed significant differences between the earmuff domains of NRP1 and other NAP1 proteins (Fig. 3A and SI Appendix, Fig. S4). For example, NRP1 lacks the corresponding histone binding surface observed in the CeNAP1-H2A-H2B complex (Fig. 3A). Previous studies showed that a single mutation in NAP1 proteins has a limited effect on their histone-binding activities (8, 22). Therefore, guided by the cross-linking analysis (Fig. 2B) and the crystal structure (Fig. 3B), we constructed several mutant proteins of NRP1 in which two or three residues close in space were simultaneously substituted by Ala. ITC analysis showed that K115/D116/K118 and E213/D214 sites are essential for binding of the NRP1 earmuff domain to H2A-H2B, whereas the E146/E147/K151 and K104/Y105 sites had a moderate effect and an unobvious effect on H2A-H2B binding, respectively (Fig. 3C). CD spectroscopy analysis supported a proper folding of these mutant proteins of NRP1 earmuff domain (SI Appendix, Fig. S3C). The K115/D116/K118 site locates at loop L1, which connects sheets β1 and β2, and the E213/D214 site exits at the terminus of helix α5, indicating that the key residues involved in interacting with H2A-H2B in the NRP1 earmuff domain are different from those in NAP1 proteins (Fig. 3C and SI Appendix, Fig. S4). Based on these observations, we constructed NRP1 mutant proteins within earmuff domain only (NRP1 19 to 225 mE, NRP1 19 to 225 K115A/D116A/K118A/E213A/D214A) and within both αN and earmuff domains (NRP1 19 to 225 mNE, NRP1 19 to 225 E36A/D39A/D40A/K43A/K115A/D116A/K118A/E213A/D142A). CD spectroscopy analysis supported a proper folding of the foregoing mutant proteins of NRP1 19 to 225 (SI Appendix, Fig. S3D). Compared with that of WT NRP1 19 to 225 (Kd = 0.16 μM; Fig. 2C), the H2A-H2B binding affinities of NRP1 19 to 225 mE (Kd = 1.66 μM) and NRP1 19 to 225 mNE (Kd = 11.86 μM) showed approximately 10-fold and 70-fold decreases, respectively (SI Appendix, Fig. S5), supporting that both αN domain and earmuff domain contribute to the interaction between NRP1 and H2A-H2B.
Fig. 3.
Mapping the key residues within NRP1 earmuff domain involved in H2A-H2B binding. (A) Structural superposition between Arabidopsis NRP1 and the CeNAP1–H2A-H2B complex. Ce, C. elegans. (B) The detailed orientations of K104, K115, and K151 and the residues close to them within the NRP1 earmuff domain (NRP1 78 to 225). (C) The residues close to K104, K115, and K151 were mutated in NRP1 78 to 225, resulting in NRP1 78 to 225 K104A/Y105A, NRP1 78 to 225 K115A/D116A/K118A, NRP1 78 to 225 E146A/E147A/K151A, and NRP1 78 to 225 E213A/D214A. The binding affinities between H2A-H2B (20 μM) and NRP1 78 to 225 mutant proteins (400 μM) were measured by ITC. Titrations were carried out using an initial injection volume of 0.5 μL (omitted from the analysis), followed by 20 injections of 2 μL each.
NRP1 CTAD Domain Interacts with H2A-H2B through a Conserved Histone-Binding Motif.
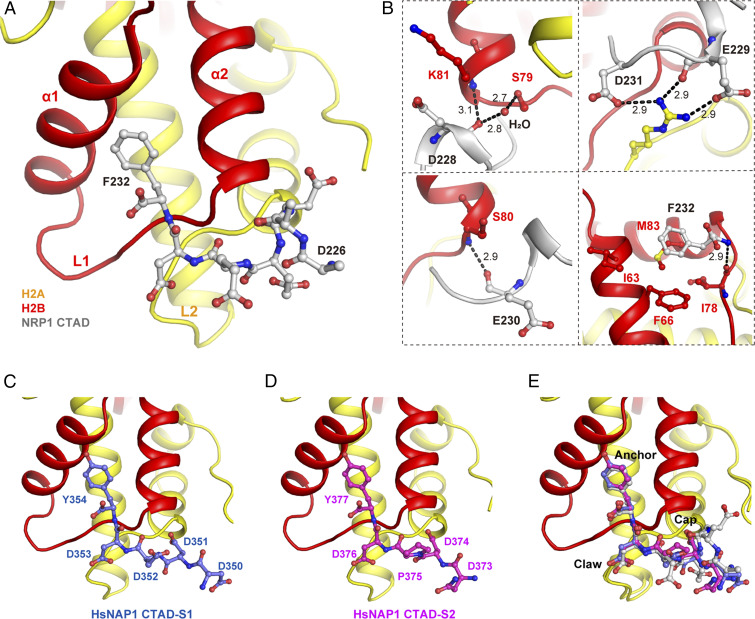
Although our previous study showed that NRP1 CTAD (amino acids 225 to 255) contributes to H2A-H2B binding and is important for the function of NRP1 in planta (18), the CTAD domain was disordered in the NRP1-H2A-H2B complex. Our ITC results showed that NRP1 19 to 225 lacking CTAD shares the same H2A-H2B–binding affinity as the full-length NRP1 1 to 255, but the stoichiometry of NRP1 to H2A-H2B decreases to 4, and that of NRP1 1 to 255 is 6 (Fig. 2B), indicating that the lack of CTAD affects the interaction between NRP1 and H2A-H2B. To reveal how NRP1 CTAD interacts with H2A-H2B, we solved the structure of H2A-H2B in complex with CTAD peptide D226-F232 (SI Appendix, Table S2). All seven residues (D226 to F232) of CTAD are well ordered in the cocrystal structure and engage H2A L2 and H2B L1 (Fig. 4A), and electronegative residues D228, E229, E230, and D231 within NRP1 CTAD are cocooned by the positively charged groove of H2A-H2B (SI Appendix, Fig. S6A). The contact interface between NRP1 CTAD and H2A-H2B is 703 Å2. As shown in Fig. 4B, D228 assembles like a cap and covers H2B at the N terminus of helix α2; the main chain O atom of D228 forms one direct H-bond with the main chain N atom of H2B K81 and one water-mediated H-bond with the side chain OG atom of H2B S79. E229 forms two H-bonds with H2A R79: one (2.9 Å) between the main chain O atom of E229 and the NH2 atom of H2A R79 and the other (2.9 Å) between the side chain OE2 atom of E229 and the NH1 atom of H2A R79. H2A R79 also interacts with the side chain OD1 atom of D231 by an H-bond (2.9 Å). In addition, E230 interacts with H2B, forming one H-bond (2.9 Å) between its main chain O atom and the N atom of H2B S80. Similar to E230, F232 also interacts with H2B, and its main chain N atom forms one H-bond (2.9 Å) with the main chain O atom of H2B I78. The side chain of F232 projects into the hydrophobic pocket, which is composed of H2B F66, I78, and M83, and interacts with H2B M83 by extensive hydrophobic interaction.
Fig. 4.
The conserved binding mode between H2A-H2B and CTAD domains of NAP1 family proteins. (A) The overall structure of the NRP1 CTAD (226 to 232) in complex with H2A-H2B. (B) H-bonds and hydrophobic interactions between key residues within NRP1-CTAD and H2A-H2B. The H-bonds are indicated by black dashed lines. (C and D) The structure of the HsNAP1 CTAD-1 (C) and HsNAP1 CTAD-2 (D) in complex with H2A-H2B. Hs. Homo sapiens. (E) Structural superposition of NRP1 CTAD with HsNAP1 CTAD-1 and HsNAP1 CTAD-2. The key resides are marked as Cap, Claw, and Anchor.
ITC experiments further confirmed that NRP1 CTAD binds to H2A-H2B with a Kd value of 1.85 μM, while the mutations on E229, D231, or F232 decrease this interaction (SI Appendix, Fig. S6B). The NRP1 CTAD-interacting residues within H2A-H2B are highly conserved in eukaryotes (SI Appendix, Fig. S6C) and display similar conformations in their crystal structures (SI Appendix, Fig. S6D). In addition, the NRP1 CTAD residues involved in H2A-H2B binding are also conserved in NAP1 family proteins and other histone-binding proteins, and here we named the conserved motif Cap-Claw-Anchor (cap residue: E, D, or S; claw residue: D or E; anchor residue: Y or F) (SI Appendix, Fig. S6E), which is in line with the previous study (26). Human NAP1 (HsNAP1) protein contains two CTAD domains, which we term CTAD-1 (E348 to Y354) and CTAD-2 (E371 to Y377), reported to play important roles in H2A-H2B binding in a previous study (10). We then solved the complex structures of both HsCTAD-1-H2A-H2B and HsCTAD-2-H2A-H2B at high resolution (Table 1 and SI Appendix, Table S2), showing that the overall conformations of the two complexes are nearly identical or highly similar to that of NRP1-CTAD-H2A-H2B (Fig. 4 C–E and SI Appendix, Fig. S6 F and G). In addition to NAP1 family proteins, structural analysis showed that many other histone chaperones, such as ANP32E, Spt16, and YL1, and histone remodeling factor SWR1 interact with histones through the Cap-Claw-Anchor motif (SI Appendix, Fig. S6 H–L). Taken together, our data indicate that NRP1 CTAD binds to H2A-H2B via a motif conserved in NAP1 family proteins.
αN, Earmuff, and CTAD Domains of NRP1 Are all Required for Chaperoning Histones H2A-H2B.
Within a nucleosome, DNA interacts with H2A-H2B at three different interfaces (SI Appendix, Fig. S7A): DNA interface 1 (IF1), IF2, and IF3, consisting of helix α1 of H2A and helix α1 of H2B, H2A L1 and H2B L2, and H2A L2 and H2B L1, respectively (1). Interestingly, structural analysis showed that the αN domain of NRP1 just binds to IF1 of H2A-H2B, and NRP1 CTAD interacts with IF3 (SI Appendix, Fig. S7B). To investigate whether the NRP1 earmuff domain interacts with the histone-DNA interfaces, we performed ITC experiments. The ITC data showed that only the mutation within IF2 of H2A-H2B decreased the interaction between the NRP1 earmuff domain and H2A-H2B, indicating that the NRP1 earmuff domain binds to IF2 but not to IF1 or IF3 (SI Appendix, Fig. S7C and Table S1). CD spectroscopy analysis supported a proper folding of H2A-H2B mutant proteins in solution (SI Appendix, Fig. S3E). Together, these results suggest that the αN, earmuff, and CTAD domains may all contribute to NRP1 function, since they bind to different DNA interfaces of H2A-H2B.
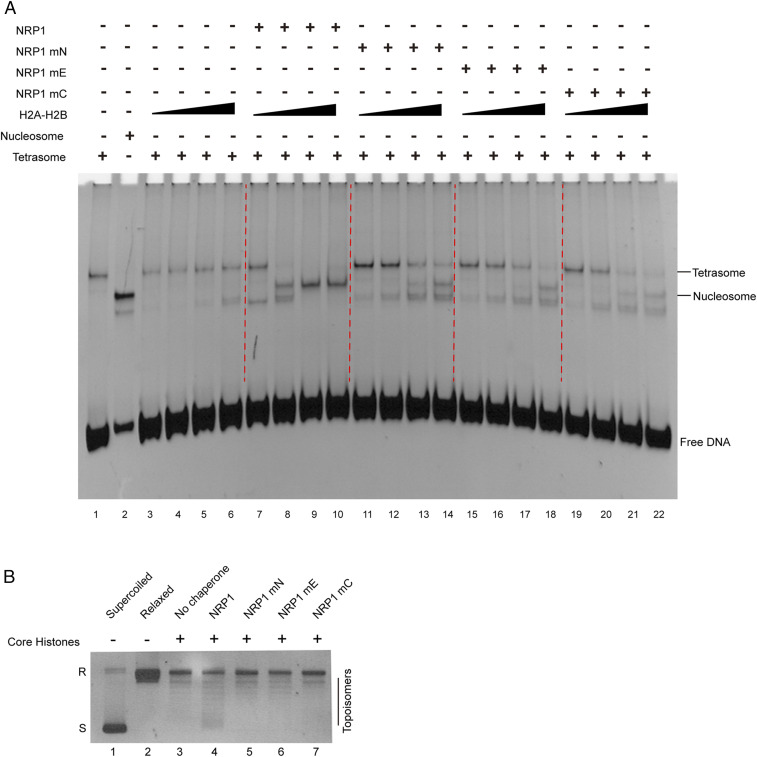
To investigate how the αN, earmuff, and CTAD domains contribute to NRP1 activity in chaperoning histones H2A-H2B, we performed electrophoretic mobility shift assay (EMSA) experiments to test NRP1 nucleosome assembly activity, using (H3-H4)2-DNA tetrasomes and H2A-H2B as the substrates. While increasing H2A-HB alone failed to promote nucleosome formation from tetrasomes (Fig. 5A, lanes 1 to 6), the presence of WT NRP1 in the reaction resulted in a decrease of tetrasomes and an accumulation of nucleosomes following the increase of H2A-H2B (Fig. 5A, lanes 7 to 10), indicating that NRP1 functions as an H2A-H2B chaperone. Compared with WT NRP1, the NRP1 mN, NRP1 mE, and NRP1 mC mutants (SI Appendix, Table S1) all showed decreased activity to promote H2A-H2B deposition with (H3-H4)2-DNA tetrasomes in nucleosome formation (Fig. 5A, lanes 11 to 22). CD spectroscopy analysis supported a proper folding of NRP1 mutant proteins in solution (SI Appendix, Fig. S3F). A plasmid supercoiling assay also consistently showed that NRP1, but not NRP1 mN, NRP1 mE, or NRP1 mC mutants, displayed histone chaperone activity (Fig. 5B). Based on these results, we conclude that the αN, earmuff, and CTAD domains are all required for the nucleosome assembly activity of NRP1.
Fig. 5.
αN, earmuff, and CTAD domains are all required for chaperoning histones H2A-H2B. (A) Nucleosome assembly activities of NRP1 and NRP1 mutants were detected using (H3-H4)2-DNA tetrasomes as the substrates. Lanes 1 and 2 are tetrasomes and nucleosomes, respectively. Tetrasomes were mixed with increasing H2A-H2B (0.06, 0.12, 0.18, and 0.24 μM; lanes 3 to 6), with WT NRP1 (1 μM) and increasing H2A-H2B (lanes 7 to 10), and with NRP1 mutant (1 μM) and increasing H2A-H2B (lanes 11 to 22). (B) Histone chaperone activities of NRP1 and NRP1 mutants were measured by plasmid supercoiling assay. Lanes 1 and 2 show untreated supercoiled plasmids and relaxed plasmids treated by topoisomerase I, respectively. Relaxed plasmids were mixed with core histones (lane 3), with both core histones and WT NRP1 (lane 4), and with both core histones and NRP1 mutants (lanes 5 to 7). Relaxed and supercoiled plasmids are indicated as R and S, respectively.
Our previous study found that NRP1 can also promote nucleosome disassembly in the transcriptional activation of target genes in planta (18). Thus, we performed further EMSA experiments to test how the αN, earmuff, and CTAD domains contribute to the histone-binding and nucleosome disassembly activities of NRP1 (SI Appendix, Fig. S8). Incubation with increasing amounts of NRP1 led to efficient release of H2A-H2B from the DNA-H2A-H2B aggregates (SI Appendix, Fig. S8A), indicating that NRP1 shields H2A-H2B and blocks their interaction with DNA. In comparison, NRP1 mN, NRP1 mE, and NRP1 mC mutants (SI Appendix, Table S1) all showed a reduced efficiency of H2A-H2B release (SI Appendix, Fig. S8A), indicating that the αN, earmuff, and CTAD domains all contribute to histone binding. To test the nucleosome disassembly activity of NRP1, we used a 229-bp fragment containing a core 146-bp Widom 601 DNA segment at the central and purified Arabidopsis core histones to reconstitute the recombinant nucleosomes. However, we could not detect an obvious histone disassembly activity when adding NRP1 into the nucleosomes (SI Appendix, Fig. S8B). A previous study showed that yeast NAP1 alone hardly removed histones from nucleosomes, and that the nucleosome disassembly capacity of yeast NAP1 was significantly increased in the presence of the chromatin remodeling complex (RSC) (27), because RSC and SWI2/SNF2 family helicases can mobilize nucleosome in an ATP-dependent manner (28, 29). A Decrease in DNA Methylation 1 (DDM1), a member of the Arabidopsis SWI2/SNF2 family, is reported to induce nucleosome reposition (30). Thus, we performed the nucleosome disassembly activity experiments in the presence of DDM1 and found that incubation with increasing amounts of NRP1 can evict H2A-H2B from the nucleosomes and generate increasing amounts of DNA-H2A-H2B complexes, while the properly folded mutants of NRP1 mN, NRP1 mE, and NRP1 mC cannot (SI Appendix, Fig. S8 B–D and Table S1), indicating that the αN, earmuff, and CTAD domains are all essential for the nucleosome disassembly activity of NRP1. Taken together, our results demonstrate that the αN, earmuff, and CTAD domains are all required for the effective H2A-H2B chaperone activity of NRP1.
Discussion
In this study, we identified three modes of interaction between NRP1 and H2A-H2B. The crystal structure of the NRP1-H2A-H2B complex reveals a previously unknown histone-binding mode mediated by the N-terminal α helix for NAP1 family histone chaperones. Cross-linking and ITC analyses confirmed that NRP1 earmuff domain also interacts with H2A-H2B as CeNAP1 does. The crystal structures of CTAD domains of NRP1 and human NAP1 in complexes with H2A-H2B show a conserved histone binding mode among H2A-H2B chaperones. In this way, the histone chaperone NRP1 is able to efficiently and flexibly bind to H2A-H2B and compete with nonspecific electrostatic histone–DNA interactions. Together, our results provide valuable insights into the molecular basis of how an NAP1 family member functions as histone H2A-H2B chaperone in higher eukaryotes.
Even though the overall structures of NRP1 and NAP1 are similar, they show significant differences in histone recognition. CeNAP1 and ScNAP1 mainly use the acidic strips within the earmuff domain (SI Appendix, Fig. S9 A and B) to interact with H2A-H2B. The surface of NRP1 αN also contains an acidic strip formed by consecutive negatively charged residues in space (SI Appendix, Fig. S9 C and D), and this acidic strip is important for H2A-H2B binding (Fig. 2A). However, such an acidic strip is not present in the αN domains of CeNAP1 and ScNAP1 (SI Appendix, Fig. S9 A and B). The αN and earmuff domains of NRP1 are closely connected (SI Appendix, Fig. S4), whereas the two domains in NAP1 are connected by a longer linker, which buries the H2A-H2B binding surface within NRP1 αN (SI Appendix, Fig. S9E). The earmuff domain of NRP1 is smaller than those of CeNAP1 and ScNAP1 and lacks the histone-binding sites observed in the concave surface of the CeNAP1-H2A-H2B complex (Fig. 3A and SI Appendix, Fig. S9 A and B). The key residues within the NRP1 earmuff domain involved in histone binding (Fig. 3 B and C) are distinct from those in the concave surface of NAP1 (8, 24). Consistent with their structural differences, NRP1 and NAP1 have different functions in Arabidopsis. For example, the nrp1-1 nrp2-1 double mutant, which lacks the redundant NRP1 and NRP2 genes, displays obvious defects in root and root hair development (14, 18), while the quadruple mutant lacking four NAP1 genes shows no obvious defects during plant growth and development under normal conditions (19, 20, 31, 32).
Different from the αN and earmuff domains, the CTAD domain of NRP1 binds to H2A-H2B in a conserved manner. CTAD domains of NAP1 family proteins are flexible and rich in acidic residues, and both NRP1 and human NAP1 bind to H2A-H2B via a conserved Cap-Claw-Anchor motif. In addition to NAP1 family proteins, we note that histone chaperones ANP32E, Spt16, YL1, and chromatin remodeling factor SWR1 all bind to H2A-H2B via the conserved Cap-Claw-Anchor motif (SI Appendix, Fig. S6 H–L), suggesting that the CTAD-binding mode is probably conserved in H2A-H2B binding proteins. Thus, the Cap-Claw-Anchor motif in the acidic region may act as a marker for H2A-H2B binding, which will help us to identify more H2A-H2B binding proteins.
Our EMSA experiments showed that NRP1 can efficiently remove H2A-H2B from DNA-H2A-H2B aggregates, but NRP1 alone is insufficient for evicting H2A-H2B from the nucleosome (Fig. 5). Previous work has demonstrated that the C-terminal docking segment of H2A and the conserved H2B repression region (HBR) (SI Appendix, Fig. S7B) are important for tying H2A-H2B to the nucleosome (33). We suspect that sequestering the H2A-docking segment and HBR may help remove histones from the nucleosome more efficiently; for example, the histone chaperone FACT contains SSRP1 and SPT16 subunits, which display extensive interactions with histones. Indeed, the structure of SPT16-H2A-H2B shows that the hMid domain of SPT16 occupies the H2A-docking surface of the nucleosome and competitively binds to the H2B HBR, promoting eviction of histones from the nucleosome (34). Moreover, the H2A.Z-specific chaperone ANP32E can directly remove H2A.Z-H2B dimer from the nucleosome by squeezing out the H2A.Z-docking segment from the H3-H4 surface (35). Compared with FACT and ANP32E, NRP1 does not directly interact with the H2A-docking segment or H2B HBR according to our cross-linking and structural analyses, which may explain the weak nucleosome disassembly activity of NRP1.
Several factors may be able to loosen the H2A-docking segment or detach the H2B HBR from the nucleosome during the DNA transaction process within chromatin. First, the DNA torsional strain processed by the polymerase can peel the histone–DNA interaction within nucleosomes (36–38). Second, the histone chaperones, such as FACT, compete with the H2A-docking domain and detach H2B HBR from the nucleosome, creating a space between histones and nucleosomal DNA (34). Third, ATP-dependent chromatin remodeling factors, such as RSC and SWI/SNF, can destroy the histone–DNA interaction within the nucleosome and generate DNA loops or bulges (29, 39–41). Such destruction may provide a space and promote the exposure of the NRP1-binding surface, triggering NRP1 invasion into the nucleosome. Like yeast NAP1, which efficiently peels histones from nucleosomes in the presence of RSC (27), our results suggest that adding ATP-dependent remodeling factor DDM1 will promote NRP1 to remove H2A-H2B from nucleosomes in vitro. Space creation between histones and DNA is probably important for the NRP1 activity of removing H2A-H2B from the nucleosome. Further research to analyze how NRP1 functions together with other histone chaperones or chromatin remodeling complexes will be interesting and important.
Methods
NRP1, Arabidopsis histones, and their mutant proteins were all expressed in Escherichia coli strain BL21 (DE3) and purified by affinity/cation exchange columns and size exclusion chromatography. Diffraction data of crystals were collected at the Shanghai Synchrotron Radiation Facility. Isothermal titration calorimetry (ITC) experiments were performed at 25 °C using a MicroCal iTC200 microcalorimeter. Electrophoretic mobility shift assay (EMSA) experiments were performed to test histone competition, nucleosome assembly, and disassembly activities. Further details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Wei Yang for a critical reading of the manuscript. We also thank the staff of beamlines BL17U1, BL18U1, and BL19U1 at the Shanghai Synchrotron Radiation Facility for their assistance during data collection. This work was supported by the National Natural Science Foundation of China (NSFC31930017 and 91519308) and the National Basic Research Program of China (2012CB910500). The research was conducted under the auspices of the International Associated Laboratory on Plant Epigenome Research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011089117/-/DCSupplemental.
Data Availability.
Structural factors and coordinates have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID codes 7BP2, 7C7X, 7BP6, 7BP4, and 7BP5 for apo AtH2A-H2B, ANRP1-H2A-H2B, AtNRP1-CTAD-H2A-H2B, HsNAP1-CTAD1-H2A-H2B, and HsNAP1-CTAD2-H2A-H2B, respectively). All study data are included in the main text and SI Appendix.
References
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Venkatesh S., Workman J. L., Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avvakumov N., Nourani A., Côté J., Histone chaperones: Modulators of chromatin marks. Mol. Cell 41, 502–514 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Zhou W., Zhu Y., Dong A., Shen W.-H., Histone H2A/H2B chaperones: From molecules to chromatin-based functions in plant growth and development. Plant J. 83, 78–95 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K., Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280, 1817–1825 (2005). [DOI] [PubMed] [Google Scholar]
- 7.D’Arcy S., et al. , Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol. Cell 51, 662–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar-Gurrieri C., et al. , Structural evidence for Nap1-dependent H2A-H2B deposition and nucleosome assembly. EMBO J. 35, 1465–1482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., et al. , Histone chaperone Nap1 is a major regulator of histone H2A-H2B dynamics at the inducible GAL locus. Mol. Cell. Biol. 36, 1287–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtomo H., et al. , C-terminal acidic domain of histone chaperone human NAP1 is an efficient binding assistant for histone H2A-H2B, but not H3-H4. Genes Cells 21, 252–263 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Ishimi Y., et al. , Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 142, 431–439 (1984). [DOI] [PubMed] [Google Scholar]
- 12.Dong A., et al. , Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta 216, 561–570 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Park Y. J., Luger K., The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. U.S.A. 103, 1248–1253 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., et al. , Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18, 2879–2892 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luebben W. R., Sharma N., Nyborg J. K., Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc. Natl. Acad. Sci. U.S.A. 107, 19254–19259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J. Y., et al. , NAP1L1 accelerates activation and decreases pausing to enhance nucleosome remodeling by CSB. Nucleic Acids Res. 45, 4696–4707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S., Frappier L., Nucleosome assembly proteins bind to Epstein-Barr virus nuclear antigen 1 and affect its functions in DNA replication and transcriptional activation. J. Virol. 83, 11704–11714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y., et al. , The histone chaperone NRP1 interacts with WEREWOLF to activate GLABRA2 in Arabidopsis root hair development. Plant Cell 29, 260–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., et al. , Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J. 59, 27–38 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Gao J., et al. , NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 24, 1437–1447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W., et al. , Distinct roles of the histone chaperones NAP1 and NRP and the chromatin-remodeling factor INO80 in somatic homologous recombination in Arabidopsis thaliana. Plant J. 88, 397–410 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Muto S., et al. , Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. U.S.A. 104, 4285–4290 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y., Meeth K., Jiang E., Luo C., Marmorstein R., Structure of Vps75 and implications for histone chaperone function. Proc. Natl. Acad. Sci. U.S.A. 105, 12206–12211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., et al. , Structural insights into ceNAP1 chaperoning activity toward ceH2A-H2B. Structure 27, 1798–1810.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Kemble D. J., McCullough L. L., Whitby F. G., Formosa T., Hill C. P., FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorch Y., Maier-Davis B., Kornberg R. D., Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. U.S.A. 103, 3090–3093 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masliah-Planchon J., Bièche I., Guinebretière J. M., Bourdeaut F., Delattre O., SWI/SNF chromatin remodeling and human malignancies. Annu. Rev. Pathol. 10, 145–171 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Ye Y., et al. , Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brzeski J., Jerzmanowski A., Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278, 823–828 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Liu Z. Q., Gao J., Dong A. W., Shen W. H., A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Mol. Plant 2, 688–699 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Wang P., Richardson C., Hawes C., Hussey P. J., Arabidopsis NAP1 regulates the formation of autophagosomes. Curr. Biol. 26, 2060–2069 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Nag R., Kyriss M., Smerdon J. W., Wyrick J. J., Smerdon M. J., A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast. Nucleic Acids Res. 38, 1450–1460 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsunaka Y., Fujiwara Y., Oyama T., Hirose S., Morikawa K., Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obri A., et al. , ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Teves S. S., Henikoff S., Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 21, 88–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheinin M. Y., Li M., Soltani M., Luger K., Wang M. D., Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat. Commun. 4, 2579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kujirai T., et al. , Structural basis of the nucleosome transition during RNA polymerase II passage. Science 362, 595–598 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Eustermann S., et al. , Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayala R., et al. , Structure and regulation of the human INO80-nucleosome complex. Nature 556, 391–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farnung L., Vos S. M., Wigge C., Cramer P., Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural factors and coordinates have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID codes 7BP2, 7C7X, 7BP6, 7BP4, and 7BP5 for apo AtH2A-H2B, ANRP1-H2A-H2B, AtNRP1-CTAD-H2A-H2B, HsNAP1-CTAD1-H2A-H2B, and HsNAP1-CTAD2-H2A-H2B, respectively). All study data are included in the main text and SI Appendix.