Significance
Microglia, a type of macrophage, were discovered a little more than a century ago by Pío del Río-Hortega. Since that time, we have gained an immense amount of knowledge on their origin and multifaceted function with the aid of labeling techniques and animal models, among other tools. Only recently have macrophage cells been imaged in living humans. Here we characterize macrophage spatial distribution and temporal dynamics in live human eyes using a label-free adaptive optics imaging approach. This investigation lays a foundation to better understand the body’s immune response not only to ocular diseases like glaucoma, but also to a vast array of neurological diseases with ocular manifestations, including Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis.
Keywords: adaptive optics, optical coherence tomography, macrophage, microglia, glaucoma
Abstract
Microglia are resident central nervous system macrophages and the first responders to neural injury. Until recently, microglia have been studied only in animal models with exogenous or transgenic labeling. While these studies provided a wealth of information on the delicate balance between neuroprotection and neurotoxicity within which these cells operate, extrapolation to human immune function has remained an open question. Here we examine key characteristics of retinal macrophage cells in live human eyes, both healthy and diseased, with the unique capabilities of our adaptive optics–optical coherence tomography approach and owing to their propitious location above the inner limiting membrane (ILM), allowing direct visualization of cells. Our findings indicate that human ILM macrophage cells may be distributed distinctly, age differently, and have different dynamic characteristics than microglia in other animals. For example, we observed a macular pattern that was sparse centrally and peaked peripherally in healthy human eyes. Moreover, human ILM macrophage density decreased with age (∼2% of cells per year). Our results in glaucomatous eyes also indicate that ILM macrophage cells appear to play an early and regionally specific role of nerve fiber layer phagocytosis in areas of active disease. While we investigate ILM macrophage cells distinct from the larger sample of overall retinal microglia, the ability to visualize macrophage cells without fluorescent labeling in the live human eye represents an important advance for both ophthalmology and neuroscience, which may lead to novel disease biomarkers and new avenues of exploration in disease progression.
Glial cells are nonneuronal support cells in the central nervous system (CNS) (1, 2) that include macroglia (astrocytes, ependymal cells, oligodendrocytes, and radial cells) and microglia. These cells are derived during development from different sources: macroglia from the neuroectoderm and microglia from primitive yolk sac progenitors (3, 4). In the retina, glial cell types include Müller cells, a radial cell that spans the length of the entire retina (5, 6); astrocytes, found primarily in the nerve fiber layer (NFL) (7); and microglia, a type of macrophage that reside primarily in the inner plexiform layer (IPL) and outer plexiform layer (OPL), but also in a smaller proportion in the ganglion cell layer (GCL), in the NFL, and near the inner limiting membrane (ILM) (8). Vitreous macrophages, known as hyalocytes (9), exist in two distinct morphological (and functional) subgroups, depending on their location in the vitreous and anteriorly near the ciliary body (ovoidal cells with microvilli and short cytoplasmic protrusions) or posteriorly at the ILM (larger, elongated cells with thick processes) (10, 11). When hyalocytes are imaged together with retinal microglia, they have been described as cells without ramification (12). Thus, reports on vitreous macrophage characteristics vary, probably depending on the subgroup studied. Regardless of classification, there is strong evidence for their role in epiretinal membrane formation (13, 14).
While retinal microglia and ILM macrophages are typically defined according to the side of the ILM on which they reside (i.e., within or outside the CNS), they share similar features in terms of morphology, capability for migration, and process motility (15, 16). There is also strong evidence that labeled retinal microglia exist above the ILM in both mice and nonhuman primates (NHPs) (12, 17) and may be the same cells as ILM macrophages reported in the literature. In this paper, we use the terms “vitreous hyalocytes,” “ILM macrophage cells,” and “retinal microglia” to distinguish among free hyalocytes in the vitreous (and those on the epithelial surface of the ciliary body), macrophages that exist at the ILM surface, and microglia embedded within the retinal parenchyma. This nomenclature may require updating after further immunohistochemistry studies provide definitive classification of ILM macrophages as hyalocytes, retinal microglia, or a unique macrophage cell type. Furthermore, because ILM macrophage cells have such close morphological and functional correspondence to true retinal microglia embedded within the neuropil, we compare their characteristics with those of retinal microglia where appropriate throughout the paper.
In the retina, as in the rest of the CNS, microglia are the primary resident immune cells and play key roles in homeostasis, neuroprotection, and neuronal cell death (8, 18, 19). Microglia assume a variety of conformational shapes to fulfill these functions and are broadly active in two different phenotypes, depending on whether they serve a neuroprotective or a neurotoxic role (20). In the neuroprotective phenotype, microglia are ramified cells whose morphology takes on a dendritic appearance with processes that constantly probe the local environment, releasing anti-inflammatory and neurotrophic factors (21, 22). In contrast, in the neurotoxic phenotype, microglia retract their processes and take on an ameboid shape, migrating to the site of injury, infection, or disease where they release inflammatory factors and become highly phagocytotic. Activated, reprogrammed microglia that migrate to the retinal pigment epithelium also act in a cytoprotective fashion that indicates their complex, multifaceted nature (23). Microglia play roles in neuroplasticity (24), cell phagocytosis (25, 26), neuroinflammatory processes (27), antigen presentation (28), complement system activation (29), angiogenesis (30), and retinal homeostasis (31). Several recent studies have also indicated that retinal microglia have regional specialization (17, 23).
Microglia are involved in neurodegenerative disease processes (8, 19, 20) and, like other long-lived cells, change with senescence (18). In age-related diseases like Alzheimer’s disease, Parkinson’s disease, glaucoma, and age-related macular degeneration, the effects are interrelated (19, 20). Microglia are involved in the pathogenesis and progression of several retinal diseases (8, 20). In age-related macular degeneration, IPL and OPL microglia migrate to the subretinal space and participate in photoreceptor degeneration and drusen formation (8, 32, 33) or neovascularization (34), with subsequent inner retina replenishment from recruited monocytes (35). Elevation of proinflammatory cytokines in diabetic retinopathy is associated with microglia activation and aggregation in areas most affected during disease progression (36, 37). Microglia activation has a large role in pathogenesis of retinopathy of prematurity, in both hypoxia and hyperoxygenation (8, 38). In inherited retinal disorders such as retinitis pigmentosa, following rod photoreceptor apoptosis, microglia migrate from the inner to the outer retina, participate in rod phagocytosis, and may be implicated in subsequent cone photoreceptor cell death or phagoptosis (39). Microglia are also implicated in the pathogenesis of the family of axonopathies known as glaucoma, which result in retinal ganglion cell (RGC) death (3, 40). While glaucoma has historically been considered a disease caused primarily by the mechanical effects of elevated intraocular pressure or alternatively dysregulation of ocular blood flow (41), there are several lines of evidence indicating that neuroinflammation, including microgliosis, is closely involved in its pathogenesis (42, 43). Because microglia play such a pivotal role in disease progression, many studies have suggested that they are a prime target for neuroprotective strategies (1, 8, 20, 44).
Changes in microglia occur both with age-related neurological and retinal diseases like Alzheimer’s disease, Parkinson’s disease, glaucoma, and age-related macular degeneration, but also with natural aging processes (19, 20, 25). Studies in both mice and NHPs have shown that retinal microglia density increases with age (17, 34). Other microglial structural and dynamic changes with aging include reduced branching, shorter processes, and slower migration and motility, leading to an overall deficit of their surveillance function (45). This leads to a dysfunction in microglia injury response capabilities, also manifest by aggregation, rather than dispersion, at injury sites following activation (45). The disruption in the normal activation response of microglia may involve down-regulation of proinflammatory cytokines and other neuroprotective genes (46).
Until very recently, microglia required labeling for direct visualization, and so they have been studied predominantly in the mouse despite the essential roles they play in human disease progression and aging. Liu et al. (47) were the first to resolve macrophages at the ILM in the live human eye with the aid of adaptive optics–optical coherence tomography (AO-OCT). A more recent AO-OCT study has begun to explore their longer-term migration (48). Castanos et al. (16) imaged these same ILM macrophage cells in healthy controls and retinopathy patients using clinical OCT. In these early reports of human ILM macrophage imaging, extensive characterization of distribution, motility, and changes with aging and pathology were not examined.
Here we begin to address this gap using the powerful combination of AO-OCT imaging and our analysis approach. Adaptive optics provides the ocular aberration correction, high transverse resolution, and precise focusing necessary to resolve ILM macrophage soma, while OCT provides the optical depth sectioning needed to visualize cells and processes in their precise axial location above the ILM. Our analysis approach includes subcellular accuracy volumetric registration using the spatial contrast collectively provided by multiple retinal layers. Indeed, a fully registered RGC mosaic may have been a prerequisite to ILM macrophage visualization, owing to its proximity to the ILM and as a cellular target in the inner retina for precise registration. High-speed acquisition and optimal temporal sampling and averaging further enhance process contrast for motility quantification. Imaging live human ILM macrophage cells without fluorescent markers was also aided by their favorable position and optical properties relative to surrounding tissue. ILM macrophage cells are not embedded within the neural parenchyma as they are in all other CNS locations in which they reside.
The aim of this study was to exploit this new capability to begin to reexamine some of the characteristics of macrophage cells that have been gathered from decades of ex vivo and in vivo investigations. In particular, we sought to characterize the distribution of ILM macrophage cells across the macula, the effect of aging on distribution, the fast and slow dynamics of their motility, and their response to ocular disease. The ability to visualize and track these important immunocompetent cells in a live human eye represents an important advance in our ability to explore infection, disease, and aging processes.
Results and Discussion
ILM Macrophage Cells Resolved with AO-OCT.
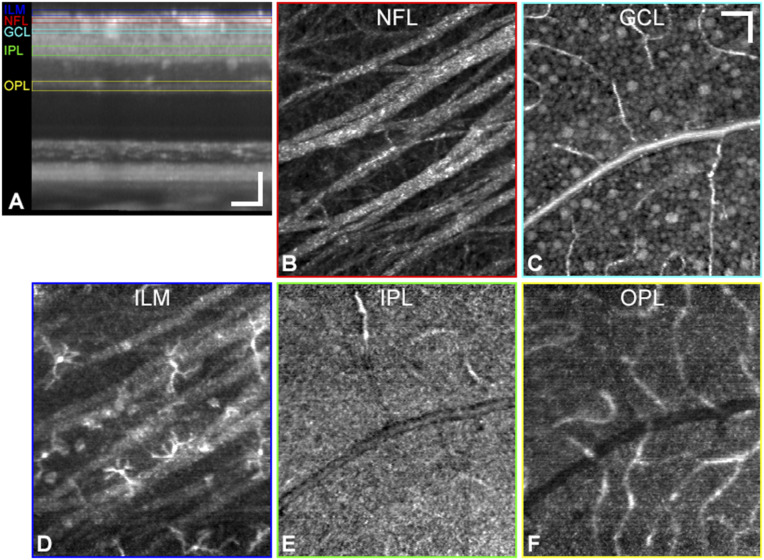
With capabilities provided by high-resolution AO-OCT axial sectioning and ocular aberration correction, as well as subcellular 3D registration and image averaging, retinal cells and structures were visualized in the living eye (Fig. 1), including nerve fiber bundles (Fig. 1B), RGCs (Fig. 1C), and retinal capillaries. Macrophage cells were resolved just above the ILM (Fig. 1D) but not in the plexiform layers (IPL and OPL) where they are known to exist in abundance (Fig. 1 E and F). Both the hyperreflective ILM macrophage soma as well as their thin processes are visible in the AO-OCT volumes. The exceptionally dense dendritic and synaptic composition of the plexiform layers give rise to fine OCT speckle patterns, which readily obscure lower spatial frequency signals from individual ILM macrophage cells, compared with the more spatially uniform refractive index (n) characteristic of the vitreous humor, which creates a dark OCT background. The refractive index differential between the vitreous (n = 1.34 to 1.36) (49, 50) and the various macrophage subcellular components (n = 1.36 to 1.6) (51, 52) creates strong overall macrophage contrast in this transparent media. Here the focus is set near the GCL, but even when the focus was set lower to optimize OPL contrast, we did not resolve microglia in this layer.
Fig. 1.
Macrophage cells are resolved above the ILM but not in the IPL and OPL. (A) Cross-sectional view of an averaged AO-OCT volume (n = 395 volumes) from a 35-y-old healthy control subject and en face projections (axial thickness) of the following inner retinal layers: NFL (8 µm) (B), GCL (7 µm) (C), ILM (10 µm) (D), IPL (15 µm) (E), and OPL (15 µm) (F). Layers are color-coded in corresponding cross-sectional regions of interest. The bottom row shows known retinal macrophage locations. (Scale bars: 50 µm.)
ILM Macrophage Distribution.
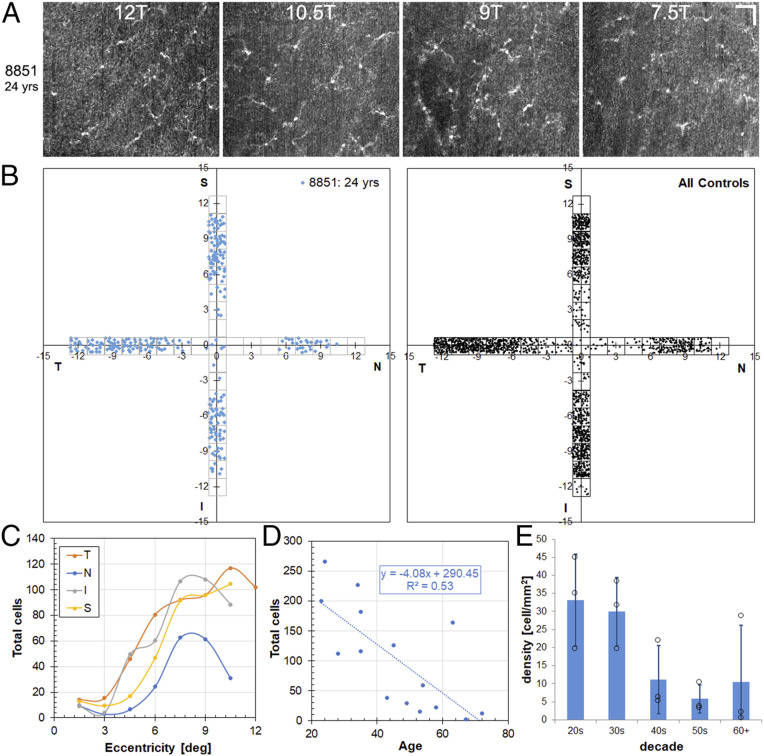
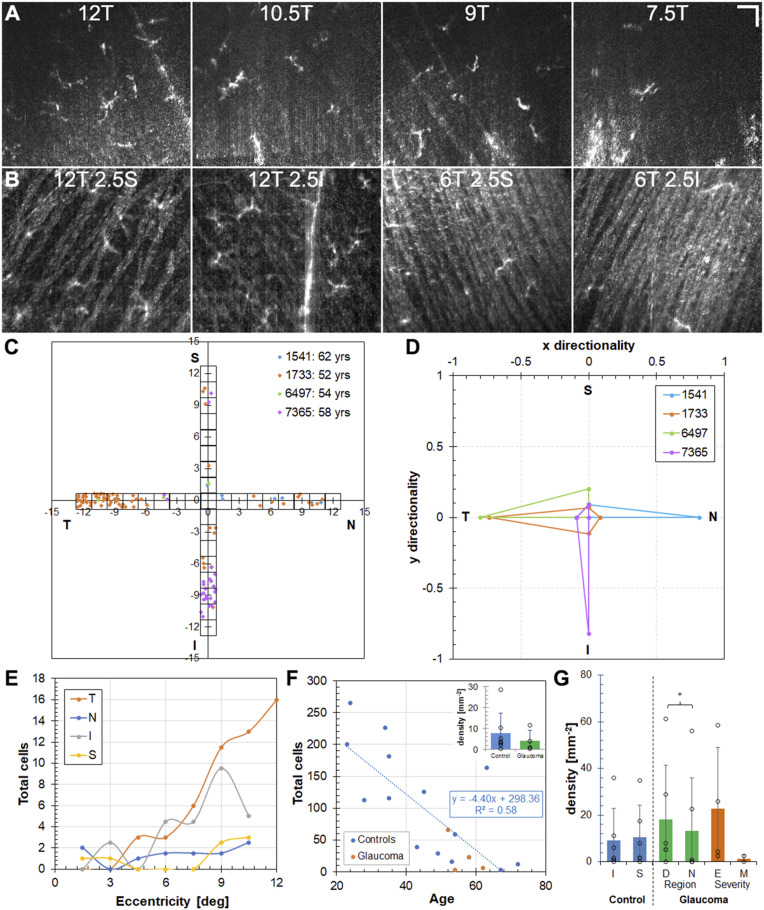
ILM macrophage distribution was quantified across the macula in 15 healthy control subjects and 4 subjects with glaucoma. Fig. 2 shows the distribution of results for the healthy control subjects. Example en face OCT axial projections (spanning 10 µm above the ILM) at four temporal locations for a 24-y-old control subject are shown in Fig. 2A. The distributions maps for that subject and for all healthy control subjects are shown in Fig. 2B. Example AO-OCT images for other subjects and distribution maps separated by decade are shown in SI Appendix, Fig. S3. Accounting for individual variation, ILM macrophage cells were absent or sparse in the central macula of the control subjects irrespective of age and increased monotonically to a peak at ∼8 to 11° in all four directions. Only 13% of ILM macrophage cells were counted from regions <5°, representing ∼30% of the total area surveyed. A previous study in NHPs found a similar lower density of microglia centrally, although their absence was confined to the foveal avascular zone (17), whereas our results indicate human ILM macrophage cells are lacking in a larger central region. The temporal and superior peaks (Fig. 2C) were at slightly higher eccentricities compared with nasal and inferior peaks, and the nasal region, bound by the optic nerve head, had a slightly lower amplitude peak compared with the other three quadrants. We observed a relatively symmetrical distribution of ILM macrophage cells in healthy control subjects (SI Appendix, Fig. S4). Irrespective of direction, ILM macrophage macular distribution in stasis is similar to NFL central macular thickness (i.e., sparse where the NFL is thin near the fovea and more concentrated in areas of thickened NFL further in the periphery), giving rise to the inference that their arrangement is designed to play a key role in axonal health, including phagocytosis of debris during normal aging and disease processes. However, a lower peak was observed in the nasal direction compared with the other directions, where the NFL is thickest. This suggests NFL thickness alone does not predict ILM macrophage density.
Fig. 2.
ILM macrophage cells are sparse in the central macula, and density decreases with age in healthy control subjects. (A) Example en face AO-OCT axial projections near the temporal peak in a 24-y-old healthy control subject. (Scale bar: 50 µm.) (B) ILM macular macrophage distribution maps (horizontal and vertical meridians) for the same subject and for all healthy control subjects. (C and D) Total ILM macrophage counts in the regions surveyed as a function of eccentricity for the four quadrants (C) and as a function of age (D) for the individual control subjects. (E) ILM macrophage density as a function of age (by decade). P = 0.037, ANOVA.
Human ILM macrophage density decreases monotonically with age at a rate of ∼2% per year from 20 to 70 (Fig. 2 D and E). The correlation with age improved even further (R2 = 0.53 to 0.72) when data from a single 63-y-old subject, considered an outlier because of exceptionally large counts compared with his age-matched counterparts, was removed. The loss of ILM macrophage cells with senescence runs counter to all evidence to date on retinal microglia in animal models, both NHPs and rodents (17, 19, 45), although without the ability to resolve microglia within the retinal layers, ours is a decidedly incomplete picture. It is not clear whether the loss of cells at the ILM is due to posterior migration (into the retina) or if the cells are merely consumed in their phagocytotic function. It is possible that there are regional variations that are not accurately captured in our distribution measurements. ILM macrophage loss with aging does indicate that cells are not replenished over time from the plexiform layers or hematopoietic sources (e.g., recruited monocytes). Thus, cautious interpretation of these results is warranted, as the findings represent examination of retinal macrophage cells in their sparsest location most likely without extrapolation to their counterparts in other retinal locations. Indeed, ILM macrophage cells may be specialized for NFL axonal homeostasis and phagocytosis.
ILM Macrophage Fast Dynamics.
Macrophage cells in the ramified state constantly probe their local environment, sprouting, extending, retracting, and sweeping their processes on timescales of a few minutes (53). To capture and quantify the motion of fine macrophage processes requires high 3D spatial resolution, fast acquisition speed and sampling, and accurate registration and averaging. Combining these capabilities, we were able to visualize and track the dynamic motion of ILM macrophage cells in our subjects. Movie S1 shows an example time-lapse video from a 33-y-old subject with macrophage endpoints marked. We observed multiple ramified macrophage cells rapidly extend, retract, and sweep their processes over their field of regard. While macrophage processes were in constant motion, most of the cell bodies remained relatively immobile over the short time durations of the time-lapse video (∼25 min).
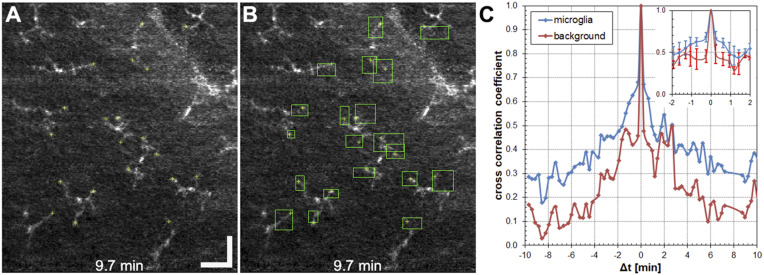
To quantify ILM macrophage process endpoint velocity and coverage area—the former a measure of motility and the latter a measure of active probing region size—we manually marked process endpoint positions (Fig. 3A) and, from the endpoint positions over the video time span, calculated their coverage area (Fig. 3B). The macrophage process motility measurements warranted further validation as follows. Accurate quantification of fast temporal dynamics (e.g., process velocity, vp) required a balance of sufficient averaging over a long enough duration to enhance process contrast without motion blur, while sampling over a short enough interval to accurately capture process movements.
Fig. 3.
Quantification of ILM macrophage process fast dynamics. (A and B) Example en face AO-OCT images of ILM macrophage cells in a 33-y-old subject showing manually marked process endpoint locations (A) and coverage area calculated from the endpoint motion over 20 min (B). (Scale bar: 50 µm.) (C) Average cross-correlation coefficient calculated on a 20-min time-lapse video for 12 ILM macrophage cells and three background locations for a healthy control subject. (Inset) Coefficients for Δt <±2.0 min. Error bars indicate standard deviation (SD).
We examined the temporal sampling and averaging conditions required to track process motility (SI Appendix). Regions in adjacent frames remain relatively highly correlated (coefficient > 0.5) when they contain stable, immobile structures (e.g., NFL or ILM background, macrophage cell soma), while motion (e.g., blood flow, macrophage process motility) will decorrelate a region on timescales commensurate with the motion. This can be further validated by direct visual observation of the stable cell soma locations in Movies S1 and S2. Correlation calculation over time-lapse AO-OCT videos in regions marked with ILM macrophage cells and background (Fig. 3C) indicate rapid decorrelation in background regions within 0.25 min (with residual correlation presumably from faint background structural features), while regions with macrophage cells decorrelate on slower timescales (∼1 to 2 min). Thus, fast macrophage dynamics were measured using time-lapse videos with timesteps (t) limited to <0.5 min.
The process velocity (vp) and coverage area (A) for 75 processes (36 ILM macrophage cells) from six healthy subjects are shown in Table 1. Macrophage process motion was sporadic over a wide range of instantaneous displacements, leading to high variability in the velocity measurements. The coverage area was similarly variable (e.g., Fig. 3B). The average velocity across all subjects was 15.9 µm/min, and the average process coverage area was 482 µm2 over ∼20 to 25 min. Repeat process velocity measurements in three control subjects over a period of months show good reproducibility (SI Appendix, Table S2). Movies S3 and S4 are time-lapse videos for two additional control subjects.
Table 1.
ILM macrophage process velocity (vp) and coverage area (A) for healthy controls measured from averaged videos with frames separated by timestep (t)
| Subject | Age | Cells | Processes | Density, mm−2 | vp(avg ± SD), µm/min | vp(max), µm/min | A(avg ± SD), µm2 | A(min), µm2 | A(max), µm2 | t(avg ± SD), min |
| 8851 | 24 | 6 | 9 | 84.0 | 18.4 ± 12.2 | 66.4 | 569 ± 287 | 318 | 1,166 | 0.26 ± 0.08 |
| 3228 | 28 | 4 | 10 | 63.0 | 17.9 ± 14.0 | 91.8 | 501 ± 352 | 43 | 1,271 | 0.22 ± 0.07 |
| 8009 | 33 | 10 | 20 | 78.7 | 16.0 ± 11.0 | 72.0 | 738 ± 357 | 138 | 1,478 | 0.30 ± 0.07 |
| 6581 | 35 | 5 | 12 | 63.0 | 13.5 ± 9.3 | 57.3 | 228 ± 167 | 82 | 668 | 0.32 ± 0.08 |
| 7822 | 35 | 8 | 18 | 94.5 | 14.1 ± 10.5 | 64.2 | 386 ± 328 | 83 | 1,408 | 0.29 ± 0.09 |
| 4136 | 50 | 3 | 6 | 31.5 | 15.5 ± 12.4 | 77.3 | 471 ± 125 | 303 | 668 | 0.32 ± 0.07 |
| Total | Σ = 36 | Σ = 75 | 15.9 ± 2.0 | 482 ± 172 | 0.28 ± 0.04 |
Previous animal studies have reported mixed results in terms of structural and motility changes with aging. In transgenic mice (GFP-labeled microglia), Damani et al. (45) observed decreased structural measures (dendritic field area, total branch points, and total dendritic length), as well as reduced dynamic behavior on stimulation (process branch sprouting, motility, and migration) for retinal microglia in older animals. Singaravelu et al. (17) observed unchanged microglia structure (total cell volume, dendritic length, branch points, and ellipticity) with aging in NHPs. We observed no qualitative differences in ramified-state macrophage structure with age (SI Appendix, Fig. S3), although the small number of subjects imaged and the inability to confidently resolve the entire dendritic tree for all subjects prevented quantitative confirmation. Nonetheless, we did observe an increased incidence of extracellular debris at this axial location near the ILM in older subjects (54). We also found evidence of activated macrophage cells without processes, particularly for older subjects (SI Appendix, Fig. S6).
Regarding macrophage dynamics, we did see a weak trend of slower velocities and smaller process coverage area with aging, confirmation of which will require further investigation. Overall, human ILM macrophage process velocity was approximately three times that measured previously for small rodents in an ex vivo preparation (53), a surprising result given the generally faster rate of some physiological parameters (e.g., heart rate) in small animals. It is not known how much the ex vivo preparation in the previous study (retinal explant whole-mount preparation) may have affected the process rate. It is also not clear whether our measured rate is similar for human microglia embedded within the retinal plexiform layers or if ILM macrophage cells are uniquely more motile than other CNS microglia.
ILM Macrophage Slow Dynamics.
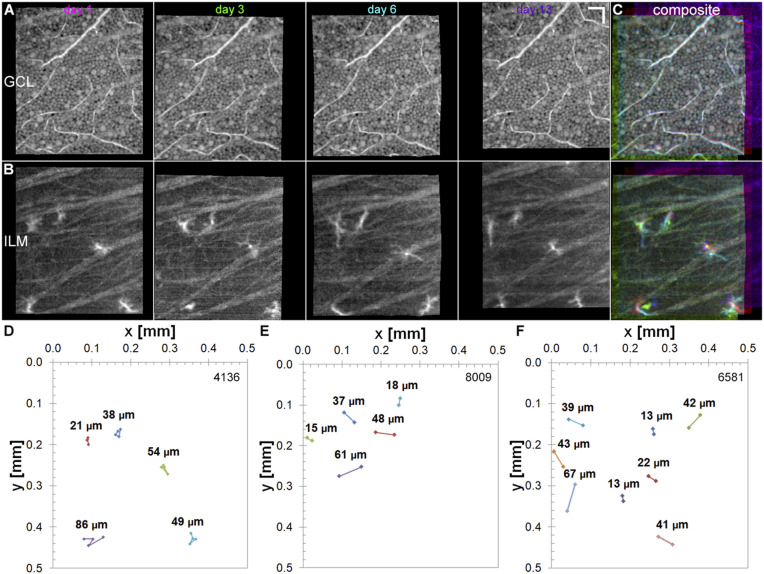
In addition to macrophage process motility on fast timescales to probe their local environment, ramified macrophage cells migrate more slowly across a retinal patch on timescales of days to months to probe a wider field and fulfill their immune system sensing function. We tracked ILM macrophage soma locations in three subjects over days to months to explore macrophage stability and the extent of migration (Fig. 4). To track macrophage migration to an independent, fixed location, collected volumes were first registered to the GCL (Fig. 4A), and then the locations of the ILM macrophage soma were manually marked from the registered volumes (Fig. 4 B and C). As expected, the RGC locations remained highly stable over 2 wk, further demonstrating the high registration accuracy for tracking ILM macrophage cells. The pathlength traveled for five to eight ILM macrophage somas tracked for the three subjects is shown in Fig. 4 D–F. In one or two cases, usually at the edge of the field, we observed ILM macrophage cells present in early timepoints that were missing at later timepoints, presumably from migration outside of the imaged field of view. While there were differences in migration among the three subjects, the ILM macrophage cells tracked over longer durations of days to months were relatively stationary, migrating from 13 to 86 µm. We also note that we tracked ramified (resting) ILM macrophage cells only in healthy subjects, and that reactive macrophage cells or macrophage cells in diseased eyes likely exhibit different migratory dynamics. Our results indicate that ILM macrophage cells are long-lived and have relatively small fields of regard during quiescent periods.
Fig. 4.
Slow ILM macrophage migration occurs from days to months. ILM macrophage migration was tracked for days to months in three healthy subjects by registering volumes to the GCL. In a 49-y-old subject, ILM macrophage cells were tracked at multiple timepoints over 2 wk. (A–C) Registered GCL (A) and ILM layers (B) and composite false color images (C) created from magenta (day 1), green (day 3), cyan (day 6), and purple (day 13). (Scale bar: 50 µm.) (D) Mean total pathlength (TPL) of macrophage soma migration (TPL: 49 ± 24 µm; range: 21 to 86 µm; n = 5 cells; 13 d). The mean pathlength (PL) using only start and end days was lower (PL: 20 ± 17 µm; range: 7 to 50 µm). (E) Macrophage migration in a 33-y-old healthy subject (PL: 36 ± 20 µm; range: 15 to 61 µm; n = 5 cells; 44 d). (F) Macrophage migration in a 35-y-old healthy subject (PL: 35 ± 17 µm; range: 13 to 67 µm; n = 8 cells; 112 d).
ILM Macrophage Cells in Glaucoma.
We observed some similarities as well as distinct differences in ILM macrophage characteristics in glaucoma subjects compared with control subjects that partially reveals their role in ocular disease. ILM macrophage cells for glaucoma subjects were acquired and counted in a manner similar to the healthy control subjects. Fig. 5 shows the distribution results for four glaucoma subjects. Example en face OCT images across the temporal peak density location (∼7.5 to 12°) and at 6T and 12T about the horizontal midline are shown in Fig. 5 A and B for a 52-y-old glaucoma subject with relatively high ILM macrophage density and a superior hemifield defect. While the counts were similar at 12T, this subject had five more ILM macrophage cells at 6T2.5S than at 6T2.5I. The corresponding distribution map for the four glaucoma subjects are shown in Fig. 5C. Like the control subjects, ILM macrophage density was lower in the central macula of glaucoma subjects to roughly an equivalent degree; 11% of ILM macrophage cells were counted from regions <5°, representing ∼30% of the total area surveyed. In contrast to the healthy control subjects, ILM macrophage density was highly asymmetric in the glaucoma subjects (Fig. 5D). The mean (± SD) symmetry for glaucoma subjects was 0.79 ± 0.04 (Student’s t test between control and glaucoma cohorts: P < 0.001), and all had symmetry >0.5 in any one direction. In terms of eccentricity for glaucoma subjects (Fig. 5E), the peak occurred in a similar range (9 to 11°) as in control subjects for temporal and inferior regions. In the nasal and superior regions, the ILM macrophage density for glaucoma subjects was low and flat, although the small number of glaucoma subjects included in this analysis may limit attribution of this finding more broadly to the disease. The ILM macrophage densities in glaucoma subjects were in line with those of their age-matched healthy counterparts (Fig. 5F and Inset), particularly in the similar slope and offset of fit when glaucoma subjects are included (compare Figs. 2E and 5F).
Fig. 5.
Macular ILM macrophage distribution in glaucomatous eyes is similar to that in controls but with regional differences. (A and B) Example AO-OCT images near the temporal distribution peak (A) and around the horizontal meridian (B) at 12T and 6T in a 52-y-old glaucoma subject. (Scale bar: 50 µm.) (C and D) ILM macrophage distribution maps (C) and symmetry plots (D) for four glaucoma subjects. (E and F) Total ILM macrophage counts in the regions surveyed as a function of eccentricity for the four quadrants (glaucoma subjects) (E) and age (all subjects) (F). (Inset) Average density across all distribution regions for four glaucoma subjects compared with seven age-matched control subjects. P = 0.27, Student’s t test. (G) Comparison of ILM macrophage density for 3°/6°/12° regions in six age-matched control and six glaucoma subjects. P = 0.25 for S-I, P < 0.01 for D-N, P = 0.23 for E-M, Student’s t test. S, superior; I, inferior; D, defect region; N, more normal region; E, early; M, moderate.
Further analysis of ILM macrophage density for the 3°/6°/12° regions (SI Appendix, Fig. S1) revealed interesting characteristics with respect to the hemifield defect in the glaucoma subjects (Fig. 5G). In six age-matched control subjects (mean age 61.1 ± 8.3 y), there was no difference between inferior and superior regions (P = 0.25), while in glaucoma subjects (58.1 ± 4.5 y old) we observed more ILM macrophage cells on the defect side compared with the more normal side (P < 0.01) of the hemifield defect. Despite a similar age range (early: 57.3 ± 5.4 y; moderate: 58.9 ± 4.5 y), disease severity was different (albeit not statistically significantly so) for the small number of subjects in each disease stage (P = 0.22).
These results indicate that ILM macrophage cells likely play a role in glaucoma disease progression. The distribution asymmetry (Fig. 5 C and D) and hemifield defect difference (Fig. 5G) may indicate early migration to active areas of disease. All glaucoma subjects had a higher ILM macrophage density on their defect side, except one subject with moderate disease that had no ILM macrophage cells in any region imaged. The early, active role is followed by losses, presumably as macrophage cells are consumed in their phagocytotic function during disease progression to more advanced stages. Although glaucoma disease stage classification is inexact, the results for glaucoma subjects separated by stage (Fig. 5G) also indicates early accumulation in response to the disease followed by losses with disease progression. To evaluate this hypothesis (i.e., peak macrophage cells in early to moderate disease, followed by losses in advanced disease), a sufficient number of subjects would need to be evaluated to control for normal age-related losses.
For two glaucoma subjects, there was sufficient density of ILM macrophage cells in the 12° temporal region to quantify process motility. Table 2 shows the ILM macrophage process velocity and coverage area for 32 processes (13 ILM macrophage cells) from the two glaucoma subjects. The average velocity for the two glaucoma subjects was 20.5 µm/min, and the average area was 283 µm2. For the small number of glaucoma subjects analyzed here, the process velocity was slightly higher than that of control subjects, while the average process coverage area was smaller (cf. Table 1), although the latter was probably due to the shorter tracking duration (8 to 12 min vs. 20 to 25 min). As with the control subjects, these measures had similarly high variability across the processes quantified. Movie S5 shows an average time-lapse videos for a subject with glaucoma.
Table 2.
ILM macrophage process velocity and coverage area for glaucoma subjects
| Subject | Age | Cells | Processes | Density, mm−2 | vp(avg ± SD), µm/min | vp(max), µm/min | A(avg ± SD), µm2 | A(min), µm2 | A(max), µm2 | t(avg ± SD), min |
| 1733 | 52 | 8 | 20 | 47.2 | 18.7 ± 14.2 | 86.2 | 139 ± 89 | 15 | 311 | 0.20 ± 0.04 |
| 4179 | 57 | 5 | 12 | 73.5 | 22.3 ± 13.7 | 70.3 | 427 ± 431 | 69 | 1,242 | 0.20 ± 0.04 |
| Total | Σ = 13 | Σ = 32 | 20.5 ± 2.5 | 283 ± 204 |
New Biomarkers to Study Ocular and Neurologic Diseases and Neuroprotection.
Microglia were discovered a little more than a century ago by the Spanish neuroscientist Pío del Río-Hortega (1, 55). Since that time, we have gained an immense amount of knowledge about their origin and multifaceted function with the aid of labeling techniques, animal models, and other tools (1, 2). The recent generation of microglia from human induced-pluripotent stem cells may provide a path to further investigate human microglial characteristics, physiology, and interactions in vitro (56, 57). Despite advances, however, these tools are limited in providing a platform by which microglia within their natural environment with complex interactions can be explored. Only recently have ILM macrophage cells in humans been resolved. Here we examine ILM macrophage distribution and dynamics in live human eyes without labeling using adaptive optics imaging. Macrophage structure (size, density, distribution, symmetry) and function (motility, migration) are potential cellular biomarkers that may provide earlier and finer scale metrics of disease initiation and progression and, with validation, may serve as future clinical endpoints for several retinal diseases, particularly glaucoma, which is closely related to inner retina function, but others related to aging (e.g., age-related macular degeneration) as well.
Microglia have received attention as potential targets for the treatment of neurologic and retinal diseases. Neuroprotective strategies that use microglia must consider the dual nature of their neuroprotective and neurotoxic roles, as well as target specific signaling pathways. A recent study has shown that removal and repopulation of microglia by pharmacologic inhibition of the colony-stimulating factor receptor (CSF1R) followed by drug withdrawal reverses neural deficits associated with aging (58). In the eye, minocycline has been used in a glaucoma rodent model to inhibit microglia activation and preserve the optic nerve (59). Triamcinolone and other corticosteroids are also known to inhibit microglial activity (60) and have been used to treat macular edema and retinal vein occlusions with intravitreal injection (61). In these examples, access to retinal microglia would allow direct observation of their response to novel pharmacologic treatments and other therapeutic strategies in humans, and possibly facilitate more rapid clinical translation.
It has become somewhat cliché to describe the retina as the “window to the brain”; however, in this case, optical access affords a unique opportunity to study these important immune cells within their natural environment but outside of the highly-scattering neural and retina parenchyma. With axial sectioning capability, AO-OCT allows 3D morphometric quantification of ILM macrophage soma and processes. This may help answer questions with respect to their role in retinal homeostasis, as contacts between their processes and other cells can be tracked in four dimensions. AO-OCT thus provides an important new capability for both neuroscience and ophthalmology.
Materials and Methods
Patients and AO Imaging.
A total of 22 subjects, including 16 healthy control subjects and 6 subjects with clinically diagnosed glaucoma were imaged on the FDA multimodal AO (mAO) imager (62). Glaucoma was diagnosed by an experienced glaucoma specialist, and severity was graded based on the Hodapp–Anderson–Parrish criteria (63). All glaucoma subjects were being treated for their disease. The healthy control subjects were selected across an age range of 23 to 72 y, with three subjects each in their 20s, 30s, 40s, 50s, and 60+. One additional young control subject was imaged for motility measurement only. The mean (±SD) age of the glaucoma subjects was 58.1 ± 4.5 y. A subset of age-matched control subjects with a mean age of 61.1 ± 8.3 y was used for comparison. All subjects provided consent according to a human subject protocol approved by the FDA Institutional Review Board in accordance with the principles of the Declaration of Helsinki. Subjects were coded with a four-digit number that is used throughout the text.
AO-OCT volumes of 1.5° × 1.5° were collected as described below for each experiment, registered in three dimensions with subcellular accuracy (47, 64), averaged, and flattened to the ILM. The AO-OCT system focus was pulled to the inner retina at approximately the level of the NFL-GCL during image acquisition. En face average intensity projection AO-OCT images of macrophage cells were extracted from the axial region within 10 µm of the ILM by manual segmentation. Short (∼7 s) adaptive optics scanning laser ophthalmoscopy (AOSLO) videos were also collected from each subject (SI Appendix, Fig. S2). Throughout the paper, retinal locations are denoted in degrees from the fovea in the temporal (T), nasal (N), superior (S), and inferior (I) directions.
ILM Macrophage Distribution.
For the ILM macrophage distribution experiment, AO-OCT volumes (n = 30) were collected at each eccentricity across the horizontal and vertical meridians with nonoverlapping spacing of 1.5° as shown in SI Appendix, Fig. S1A for 15 healthy control subjects and 4 subjects with glaucoma. AO-OCT volumes were registered and averaged, and ILM macrophage cells were counted manually in the average volumes by two graders using the criteria listed in SI Appendix with custom software developed in MATLAB that simultaneously provides 3D visualization of macrophage cells. Owing to uncorrected eye motion and other sources of field displacements, macrophage locations in the distribution maps represent approximations.
Fast Dynamics: ILM Macrophage Motility.
ILM macrophage fast dynamics were quantified by imaging subjects continuously for ∼20 to 25 min near their peak density temporal location (10.5T or 12T). AO-OCT volumes were registered and averaged across short durations (<0.5 min) to enhance ILM macrophage contrast without blurring their processes (SI Appendix, Fig. S5). En face AO-OCT images were extracted and combined to create a time-lapse video. Macrophage process endpoint positions were manually marked in the time-lapse videos with custom software for six healthy controls and two glaucoma subjects. In addition to process velocity calculated from their endpoint locations, coverage area was also calculated for each process from the minimum and maximum positions in both axes across the imaging session. The coverage area is a measure of the local retinal region probed by each macrophage process.
Slow Dynamics: ILM Macrophage Migration.
We quantified slow ILM macrophage cell migration in three healthy subjects. One 49-y-old subject was imaged at the same location (10.5T) four times over the course of 2 wk (days 1, 3, 6, and 13). Another two subjects (age 33 and 35 y) were imaged across months to quantify slow migration and other dynamics on even longer timescales. To track ILM macrophage migration to an independent, fixed location, collected volumes were first registered to the GCL by correcting the nonuniform pixel-level displacements (image warp), which were then applied to the ILM layer. Then the locations of the ILM macrophage soma were manually marked from the registered volumes with custom software. The path length of five to eight ILM macrophage somas from the three subjects were calculated across all image session dates (range: 2 to 112 d). For shorter time separations (up to 2 wk) with sparse density when migration was small, the same ILM macrophage cells were readily identified. For longer time separations (months), the relative pattern of distribution across the field of view, as well as their motility characteristics (from time-lapse videos), were used to identify the same ILM macrophage cells in the volumes.
Supplementary Material
Acknowledgments
We thank Donald Miller (Indiana University School of Optometry) for use of the OCT 3-D registration software, and Wai Wong (NIH National Eye Institute) and Furu Zhang (FDA) for helpful discussions. This study was supported by an FDA Critical Path Initiative (CPi) Grant and an NIH Career Development Grant (K23EY025014). O.S. has received financial and nonfinancial support from Heidelberg Engineering and a grant from Vasoptic Medical outside the scope of the submitted work. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the US Department of Health and Human Services.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.R.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010943117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Sierra A., Paolicelli R. C., Kettenmann H., Cien Años de Microglía: Milestones in a century of microglial research. Research Trends Neurosci. 42, 778–792 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Nimmerjahn A., Kirchhoff F., Helmchen F., Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Wang J. W., Chen S. D., Zhang X. L., Jonas J. B., Retinal microglia in glaucoma. J. Glaucoma 25, 459–465 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ginhoux F., Guilliams M., Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Distler C., Dreher Z., Glia cells of the monkey retina—II: Müller cells. Vision Res. 36, 2381–2394 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Distler C., Weigel H., Hoffmann K. P., Glia cells of the monkey retina, I: Astrocytes. J. Comp. Neurol. 333, 134–147 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Bringmann A., et al. , Müller cells in the healthy and diseased retina. Prog. Retin. Eye Res. 25, 397–424 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Rathnasamy G., Foulds W. S., Ling E. A., Kaur C., Retinal microglia—A key player in healthy and diseased retina. Prog. Neurobiol. 173, 18–40 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Balazs E. A., Toth L. Z., Eckl E. A., Mitchell A. P., Studies on the structure of the vitreous body, XII: Cytological and histochemical studies on the cortical tissue layer. Exp. Eye Res. 3, 57–71 (1964). [DOI] [PubMed] [Google Scholar]
- 10.Ogawa K., Scanning electron microscopic study of hyalocytes in the guinea pig eye. Arch. Histol. Cytol. 65, 263–268 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Sommer F., et al. , FACS as useful tool to study distinct hyalocyte populations. Exp. Eye Res. 88, 995–999 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Fischer F., Martin G., Agostini H. T., Activation of retinal microglia rather than microglial cell density correlates with retinal neovascularization in the mouse model of oxygen-induced retinopathy. J. Inflamm. 8, 120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarus H. S., et al. , Hyalocytes synthesize and secrete inhibitors of retinal pigment epithelial cell proliferation in vitro. Arch. Ophthalmol. 114, 731–736 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Kohno R. I., et al. , Possible contribution of hyalocytes to idiopathic epiretinal membrane formation and its contraction. Br. J. Ophthalmol. 93, 1020–1026 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto T., Ishibashi T., Hyalocytes: Essential cells of the vitreous cavity in vitreoretinal pathophysiology? Retina 31, 222–228 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Castanos M. V., et al. , Imaging of macrophage-like cells in living human retina using clinical OCT. Invest. Ophthalmol. Vis. Sci. 61, 48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singaravelu J., Zhao L., Fariss R. N., Nork T. M., Wong W. T., Microglia in the primate macula: Specializations in microglial distribution and morphology with retinal position and with aging. Brain Struct. Funct. 222, 2759–2771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hefendehl J. K., et al. , Homeostatic and injury-induced microglia behavior in the aging brain. Aging Cell 13, 60–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman S. M., Wong W. T., Microglia in the retina: Roles in development, maturity, and disease. Annu. Rev. Vis. Sci. 4, 45–77 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Ramirez A. I., et al. , The role of microglia in retinal neurodegeneration: Alzheimer’s disease, Parkinson, and glaucoma. Front. Aging Neurosci. 9, 214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silverman S. M., Ma W., Wang X., Zhao L., Wong W. T., C3- and CR3-dependent microglial clearance protects photoreceptors in retinitis pigmentosa. J. Exp. Med. 216, 1925–1943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todd L., et al. , Reactive microglia and IL1β/IL-1R1-signaling mediate neuroprotection in excitotoxin-damaged mouse retina. J. Neuroinflammation 16, 118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Koren E. G., et al. , Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity 50, 723–737.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens B., et al. , The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Li W., Phagocyte dysfunction, tissue aging and degeneration. Ageing Res. Rev. 12, 1005–1012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puñal V. M., et al. , Large-scale death of retinal astrocytes during normal development is non-apoptotic and implemented by microglia. PLoS Biol. 17, e3000492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivakumar V., Foulds W. S., Luu C. D., Ling E. A., Kaur C., Retinal ganglion cell death is induced by microglia derived pro-inflammatory cytokines in the hypoxic neonatal retina. J. Pathol. 224, 245–260 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Carson M. J., Thrash J. C., Walter B., The cellular response in neuroinflammation: The role of leukocytes, microglia and astrocytes in neuronal death and survival. Clin. Neurosci. Res. 6, 237–245 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo C., Chen M., Xu H., Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Mol. Vis. 17, 1588–1597 (2011). [PMC free article] [PubMed] [Google Scholar]
- 30.Checchin D., Sennlaub F., Levavasseur E., Leduc M., Chemtob S., Potential role of microglia in retinal blood vessel formation. Invest. Ophthalmol. Vis. Sci. 47, 3595–3602 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Langmann T., Microglia activation in retinal degeneration. J. Leukoc. Biol. 81, 1345–1351 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Ma W., Zhao L., Fontainhas A. M., Fariss R. N., Wong W. T., Microglia in the mouse retina alter the structure and function of retinal pigmented epithelial cells: A potential cellular interaction relevant to AMD. PLoS One 4, e7945 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sennlaub F., et al. , CCR2(+) monocytes infiltrate atrophic lesions in age-related macular disease and mediate photoreceptor degeneration in experimental subretinal inflammation in Cx3cr1-deficient mice. EMBO Mol. Med. 5, 1775–1793 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L., Heiduschka P., Alex A. F., Niekämper D., Eter N., Behaviour of CD11b-positive cells in an animal model of laser-induced choroidal neovascularization. Ophthalmologica 237, 29–41 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Ma W., et al. , Monocyte infiltration and proliferation reestablish myeloid cell homeostasis in the mouse retina following retinal pigment epithelial cell injury. Sci. Rep. 7, 8433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng H. Y., Green W. R., Tso M. O., Microglial activation in human diabetic retinopathy. Arch. Ophthalmol. 126, 227–232 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Vujosevic S., et al. , Hyperreflective intraretinal spots in diabetics without and with nonproliferative diabetic retinopathy: An in vivo study using spectral domain OCT. J. Diabetes Res. 2013, 491835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vessey K. A., Wilkinson-Berka J. L., Fletcher E. L., Characterization of retinal function and glial cell response in a mouse model of oxygen-induced retinopathy. J. Comp. Neurol. 519, 506–527 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Gupta N., Brown K. E., Milam A. H., Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp. Eye Res. 76, 463–471 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Liu S., et al. , Tracking retinal microgliosis in models of retinal ganglion cell damage. Invest. Ophthalmol. Vis. Sci. 53, 6254–6262 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Jones A., Kaplowitz K., Saeedi O., The role of autoregulation of blood flow in the development and progression of open-angle glaucoma. Expert Rev. Ophthalmol. 9, 487–501 (2014). [Google Scholar]
- 42.Wei X., Cho K. S., Thee E. F., Jager M. J., Chen D. F., Neuroinflammation and microglia in glaucoma: Time for a paradigm shift. J. Neurosci. Res. 97, 70–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosco A., et al. , Neurodegeneration severity can be predicted from early microglia alterations monitored in vivo in a mouse model of chronic glaucoma. Dis. Model. Mech. 8, 443–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlstetter M., et al. , Retinal microglia: Just bystander or target for therapy? Prog. Retin. Eye Res. 45, 30–57 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Damani M. R., et al. , Age-related alterations in the dynamic behavior of microglia. Aging Cell 10, 263–276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma W., et al. , Gene expression changes in aging retinal microglia: Relationship to microglial support functions and regulation of activation. Neurobiol. Aging 34, 2310–2321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z., Kurokawa K., Zhang F., Lee J. J., Miller D. T., Imaging and quantifying ganglion cells and other transparent neurons in the living human retina. Proc. Natl. Acad. Sci. U.S.A. 114, 12803–12808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurokawa K., Crowell J. A., Zhang F., Miller D. T., Suite of methods for assessing inner retinal temporal dynamics across spatial and temporal scales in the living human eye. Neurophoton 7, 015013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gross H., Blechinger F., Achtner B., “Human eye” in Handbook of Optical Systems, Vol. 4: Survey of Optical Instruments, Gross H., Ed. (Wiley-VCH, 2008), pp. 4–5. [Google Scholar]
- 50.Sardar D. K., Swanland G. Y., Yow R. M., Thomas R. J., Tsin A. T., Optical properties of ocular tissues in the near infrared region. Lasers Med. Sci. 22, 46–52 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Savage J. C., Picard K., González-Ibáñez F., Tremblay M. È., A brief history of microglial ultrastructure: Distinctive features, phenotypes, and functions discovered over the past 60 years by electron microscopy. Front. Immunol. 9, 803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu P. Y., et al. , Cell refractive index for cell biology and disease diagnosis: Past, present and future. Lab Chip 16, 634–644 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Lee J. E., Liang K. J., Fariss R. N., Wong W. T., Ex vivo dynamic imaging of retinal microglia using time-lapse confocal microscopy. Invest. Ophthalmol. Vis. Sci. 49, 4169–4176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammer D. X., Liu Z., Cava J. A., Carroll J., Saeedi O., On the axial location of Gunn’s dots. Am. J. Ophthalmol. Case Rep. 19, 100757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sierra A., et al. , The “Big-Bang” for modern glial biology: Translation and comments on Pío del Río-Hortega 1919 series of papers on microglia. Glia 64, 1801–1840 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Muffat J., et al. , Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 22, 1358–1367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasselmann J., Blurton-Jones M., Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 68, 721–739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elmore M. R. P., et al. , Replacement of microglia in the aged brain reverses cognitive, synaptic, and neuronal deficits in mice. Aging Cell 17, e12832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C., et al. , Neuroprotection of photoreceptors by minocycline in light-induced retinal degeneration. Invest. Ophthalmol. Vis. Sci. 45, 2753–2759 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Hong J., Kim B. K., Lim H., Lee S., Lee S. J., Identification and characterization of triamcinolone acetonide, a microglial-activation inhibitor. Immunopharmacol. Immunotoxicol. 34, 912–918 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Demir M., Oba E., Guven D., Acar Z., Cinar S., Results of intravitreal triamcinolone acetonide in patients with macular edema secondary to branch retinal vein occlusion. Int. J. Clin. Pharm. 36, 438–442 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Liu Z., Tam J., Saeedi O., Hammer D. X., Trans-retinal cellular imaging with multimodal adaptive optics. Biomed. Opt. Express 9, 4246–4262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hodapp E., Parrish R. K. II, Anderson D. R., Clinical Decisions in Glaucoma (Mosby, 1993), pp. 52–61. [Google Scholar]
- 64.Do N. H., “Parallel processing for adaptive optics optical coherence tomography (AO-OCT) image registration using GPU,” Master’s thesis, Indiana University–Purdue University, Indianapolis, Indiana (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.