Significance
Many phytopathogenic fungi need specialized structures, called appressoria, to penetrate the plant before infection starts. For these structures, the fungus must carefully control when and where its cells divide, and it has been reported in different fungal phytopathogens that appressorium formation is subordinated to the cell cycle control. However, the molecular details about how this control operates were unknown. We described in the corn smut fungus U. maydis that Biz1, a regulator for the formation of appressorium, is phosphorylated by the kinase responsible for cell cycle progression. Biz1 is part of a negative feedback loop responsible for the final decision to invade or not the plant tissue. Targeting this process will lead to the development of antipenetrant fungicides.
Keywords: cell cycle regulation, phytopathogenic fungi, appressorium, corn smut
Abstract
Plant pathogenic fungi often developed specialized infection structures to breach the outer surface of a host plant. These structures, called appressoria, lead the invasion of the plant by the fungal hyphae. Studies in different phytopathogenic fungi showed that appressorium formation seems to be subordinated to the cell cycle. This subordination ensures the loading in the invading hypha of the correct genetic information to proceed with plant infection. However, how the cell cycle transmits its condition to the genetic program controlling appressorium formation and promoting the plant’s invasion is unknown. Our results have uncovered how this process occurs for the appressorium of Ustilago maydis, the agent responsible for corn smut disease. Here, we described that the complex Clb2-cyclin-dependent kinase (Cdk)1, one of the master regulators of G2/M cell cycle progression in U. maydis, interacts and controls the subcellular localization of Biz1, a transcriptional factor required for the activation of the appressorium formation. Besides, Biz1 can arrest the cell cycle by down-regulation of the gene encoding a second b-cyclin Clb1 also required for the G2/M transition. These results revealed a negative feedback loop between appressorium formation and cell cycle progression in U. maydis, which serves as a “toggle switch” to control the fungal decision between infecting the plant or proliferating out of the plant.
Most antimicrobial treatments in plants aim at prevention rather than cure because once the infective agent, upon recognition of the host plant, penetrated the vegetal tissue, the possibilities to eradicate infection drastically decrease due to the low accessibility of the therapeutic agents within the plant (1). The period of greatest vulnerability to stop the infection is the window of time elapsing between the plant surface recognition by the pathogen and its penetration. In the case of microorganisms that infect the aerial parts, the plant’s cuticle represents a primary natural barrier in the defense against pathogens (2). However, many phytopathogenic fungi overcome this obstacle by producing specific infection structures termed appressoria (3). The morphology of appressoria is highly variable: These elements can be formed either by single-celled structures or by several cells forming structures known as infection cushions (4). In some cases, appressorium is a simple slight swelling of the germ tube apex that emerges from spores on the leaf surface; but it is a well-defined structure resembling a dome in other cases. Besides, the manner how appressorium guides plant penetration is also heterogeneous. For some fungi, the appressorium allows the localized secretion of enzymes that weaken the plant cuticle and cell wall, while in other cases, the fungus penetrates upon the physical rupture of the plant surface by using the turgor pressure produced inside the appressorium (5).
Appressoria could provide critical sites for therapeutic intervention. Learning about the molecular mechanisms required for the formation of these infectious structures is crucial in understanding how plant pathogens enter their host, and, therefore, it represents a predictable target for the design of antifungal substances. However, an important caveat for the search of common antifungals, is that all this diversity in form and function in appressoria most likely reflects distinct genetic programs in different fungi and, consequently, distinct cellular targets (6). Opportunely, all appressoria share, as a feature during their formation, the occurrence of more or less drastic morphological changes (7). The form that a fungus adopts, including the infectious structures, is dictated by its cell wall, which forms a kind of armor that must be produced while the fungus is growing and dividing. Because of this connection among morphogenesis, cell growth, and division (8), it is expected that appressorium formation should require, among other processes, the readjustment of the cell cycle to allow the formation of these structures.
Studies performed in, at least, three different model systems for fungal plant virulence (Magnaporthe oryzae, Colletotrichum orbiculare, and U. maydis) have provided evidence to support the notion that cell cycle regulation and appressorium morphogenesis are intricately linked (9–15). These connections seem to proceed in two directions: The genetic program resulting in appressorium formation controls the cell cycle, but at the same time, the cell cycle progression influences the formation and functionality of the appressorium.
How the induction of the virulence program impinges the cell cycle to allow the appressorium formation has already been addressed in these model systems. In M. oryzae (rice blast fungus) and C. orbiculare (cucumber anthracnose fungus), the appressorium uses a mechanical process driven by turgidity to breach the cuticle of the plant. The functionality of this kind of appressorium links to a genetic program that includes morphological changes producing a clearly defined structure with a thick, multilayered, and highly melanized cell wall as well as metabolic reprogramming that increases internal turgor pressure via glycerol accumulation (7). Elegant work demonstrated that, in these fungi, the G1 phase has to be enlarged to provide time to synthesize a septum at the neck of the developing appressorium that might be used for the accumulation of the massive amount of glycerol (11). In the corn smut fungus U. maydis, the appressoria are not dependent on turgor pressure to penetrate the plant tissue (16). In this case, the appressorium results in a localized area of secretion where plant cell wall-degrading enzymes, which help penetrate the cuticle (17), and specific effector proteins required for the precise signaling occurring during infection (18) are concentrated. Consequently, the appressorium morphology seems to be less complicated, relatively small swellings of the hyphal tip that point to the plant surface (19). In this case, the virulence program halted the mitosis entry (promoting the arrest at the G2 phase), avoiding in this way that the secretion machinery competes with the mitotic apparatus for cytoskeletal components (20).
Several pieces of evidence also indicated that the cell cycle controls the developmental program resulting in appressorium formation. In M. oryzae, transition through the S phase is mandatory for appressorium formation, and two distinct S phase checkpoints monitor the completion of DNA replication to allow the progression of appressorium differentiation (13). Besides, completion of the M phase in conidia is essential for developing functional appressoria that form the penetration peg (14, 15). On the contrary, in U. maydis, the progression through mitosis seems to halt the formation of the appressorium, which can only be produced during the G2 period (10). To summarize, the requirement for a specific cell cycle phase during appressorium formation has been noted in several fungi that produce very different kinds of appressoria. However, the intricacies for such control and the elements from the virulence program that are the actual targets of cell cycle regulators remain unclear.
Because of its simplicity, the appressorium from U. maydis seems to be well suited to address the elements linking cell cycle and appressorium formation (16). The appressorium differentiates from the infective filament, which consists of a single dikaryotic cell (resulting from the mating of two compatible yeastlike cells on the plant surface) that expands by apical growth (21). During this process, the cell composing the infective filament was arrested in its cell cycle at the G2 phase (22). How the cell cycle is regulated in U. maydis is well comprehended (23). The G2/M transition depends on two distinct CDK complexes Clb1-Cdk1 and Clb2-Cdk1 (24). Although the activity of both complexes is essential for G2/M transition, the central control is exerted by inhibitory phosphorylation of the kinase subunit Cdk1 when it is complexed to Clb2 (Clb1-Cdk1 is not sensitive to this inhibitory phosphorylation). This phosphorylation depends on the relative activity of the Wee1 kinase (which inhibits Cdk1) and the Cdc25 phosphatase (which activates Cdk1) (25, 26).
Not surprisingly, the mechanism required for the prolonged G2 cell cycle arrest in the infective filament involves the down-regulation of CDK activity via inhibitory phosphorylation of Cdk1 (27). This cell cycle arrest is sustained during the growth of the infective filament by the presence of the transcriptional regulator called the b factor, composed of two subunits (bW and bE, provided by each mating partner) (28). The transcriptional program led by the b factor activates the DNA damage response (20, 29) independently of the presence of DNA damage (30), resulting in the activation of the kinase Chk1, which, in turn, phosphorylates Cdc25 promoting its retention in the cytoplasm where is not functional (31). Simultaneously, the b factor represses the transcription of hsl1, which encodes a kinase that down-regulates Wee1 kinase. The combination of both processes increased the Cdk1 inhibitory phosphorylation and, consequently, the cell cycle arrest (9). Disabling the connection between the b-dependent program and the cell cycle arrest by the introduction of two independent mutations in these cell cycle regulators (chk1Δ- and hsl1tef1-producing constitutive expression of hsl1) resulted in the maintenance of a high CDK activity (and thereby an active cell cycle) and strikingly in the absence of appressorium formation (10). However, how this high CDK activity was interfering with the appressorium formation was unknown.
Previous results from our laboratory (SI Appendix, Fig. S1 and ref. 9) suggested that the inhibition of appressorium formation by an active cell cycle in U. maydis most likely occurred downstream the signaling pathway leading appressorium differentiation. This pathway involves two membrane proteins Sho1 and Msb2 (32) sensing plant-derived stimuli and transmitting the signal by a well-characterized mitogen-activated protein kinase cascade (33). A plethora of transcription factors seems to be genetically located downstream of this signaling cascade (34). Two of these transcription factors Biz1 and Hdp2 induced by the b-dependent program are required for appressorium formation (34, 35). Interestingly, a connection with cell cycle regulation was already described in one of these transcription factors: Biz1 binds and represses the promoter of the gene encoding the Clb1 cyclin, and its ectopic expression resulted in G2 cell cycle arrest. However, the actual role of this cell cycle connection was unclear because Biz1 was not required for the b-dependent G2 cell cycle arrest (35).
Here, we describe that Clb2-Cdk1, one of the master regulators of G2/M cell cycle progression and the b-dependent cell cycle arrest target, interacts and controls the Biz1 subcellular localization, inhibiting its activity. This finding uncovers how an active cell cycle disables the appressorium formation, and it opens the possibility to target the early steps during the infective process in terms of developing the next generation of antipenetrant fungicides.
Results
The Transcriptional Regulator Biz1 Is Involved in the Incompatibility between an Active Cell Cycle and Appressorium Formation in U. maydis.
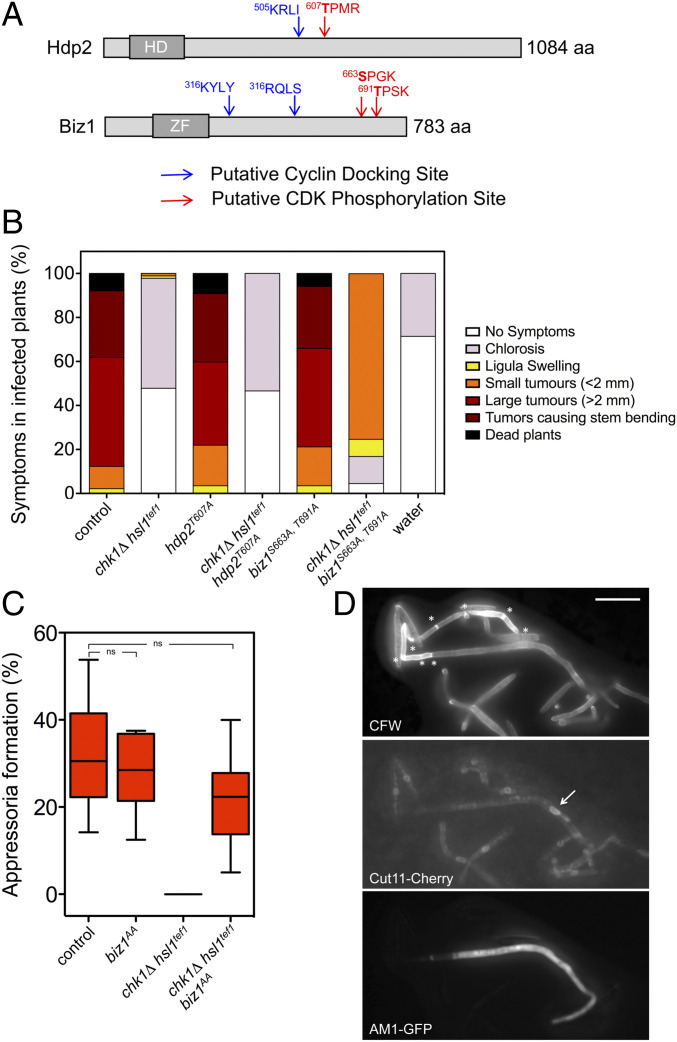
The transcriptional factors Biz1 and Hdp2 are promising candidates to be the targets of the observed interference by an active cell cycle in the appressorium formation in U. maydis. Both regulators are expressed in the infective filament by the b-dependent program (36). Genetic analyses located these factors downstream of the Mbs2-Sho1 cascade (34), and in both cases, loss-of-function mutations severely affected the induction of appressorium formation (34, 35). Moreover, from the analysis of its amino acid sequences, it can be predicted in both proteins, putative phosphorylation sites for CDK and cyclin docking sites (Fig. 1A). These cyclin-interacting motives promote efficient phosphorylation of CDK sites, and they do so in a cyclin-specific manner (37). Therefore, we considered the possibility of these transcriptional factors to be targeted and disabled by CDK phosphorylation, providing a manner to explain the inhibition of appressorium formation by an active cell cycle. To address this possibility, we have introduced Ser/Thr to Ala changes into the putative phosphorylation sites, and the resulting alleles (hdp2T607A and biz1S663A T691A [biz1AA]) were independently exchanged with the native ones in sexually compatible strains (a1 b1 and a2 b2 mating types) alone or combined with the two mutations disabling the b-dependent cell cycle arrest (chk1Δ and hsl1tef1). We have infected corn plants with compatible mixtures of the control and the respective mutant strains. We have observed that each mutant allele (either hdp2T607A or biz1AA) alone does not affect the ability of U. maydis to infect corn plants. However, we have found that the presence of the biz1AA allele recovered the capacity to infect plants by cells carrying the chk1Δ hsl1tef1 mutations (Fig. 1B). The suppression of the virulence defect in chk1Δ hsl1tef1 mutants required the two Ser/Thr to Ala substitutions in Biz1 (SI Appendix, Fig. S2). We have also constructed an allele carrying phosphomimetic substitutions (Ser/Thr to Asp, biz1DD), and it showed the same effect recovering the virulence in chk1Δ hsl1tef1 mutants as the biz1AA allele did (SI Appendix, Fig. S2).
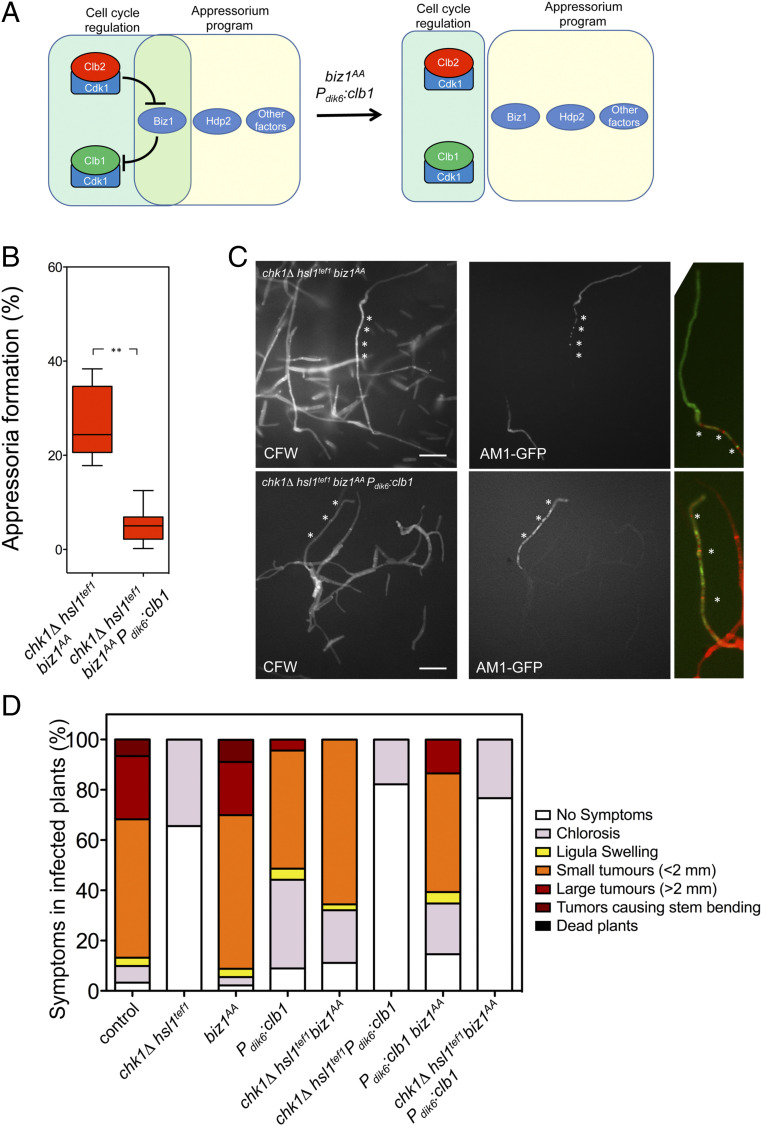
Fig. 1.
Biz1 is involved in the incompatibility between cell cycle and appressorium formation. (A) Scheme of the transcriptional regulators Hdp2 and Biz1 showing the putative CDK phosphorylation sites (red) as well as the putative cyclin docking sites (blue). The corresponding DNA-binding motives are also shown (HD, homeodomain; ZF, zinc finger). The putative CDK phosphorylation sites as well as the cyclin docking sites were predicted by the eukaryotic linear motif (ELM) algorithm (elm.eu.org/) using the default values. (B) Graph showing disease symptoms caused by crosses of wild type and the indicated mutant combinations. The symptoms were scored 14 d after infection. Three independent experiments were carried out, and the average values were expressed as percentages of the total number of infected plants (n: 30 plants in each experiment, raw data are provided in the SI Appendix). (C) Frequency of appressoria formation in vitro in strains carrying the appressorium-specific AM1-GFP reporter as well as the indicated mutations. Cells were sprayed on Parafilm M in the presence of 100 μM 16-hydroxyhexadecanoic acid. After 20 h, cells were stained with Calcofluor white (CFW) and analyzed for AM1 marker expression (AM1-GFP). The graph shows the result from three independent experiments, counting more than 50 filaments each (ns: not significative). Images of appressorium-forming filaments are provided in SI Appendix, Fig. S6. (D) Micrographs to show in vitro appressorium formation in the chk1Δ hsl1tef1 biz1AA mutant strain. Asterisks mark the presence of retraction septa. The arrow marks the nucleus from the appressorium, detected by the nuclear envelope membrane protein Cut11 fused to Cherry. (Scale bar, 20 μm.)
Following the virulence recovery, we have also observed that chk1Δ hsl1tef1 mutants carrying the biz1AA allele could produce appressoria on the plant (SI Appendix, Fig. S3) as well as penetrate the plant tissue (SI Appendix, Fig. S4). The observed symptoms in the triple mutant (chk1Δ hsl1tef1 biz1AA) infections were milder than those found in wild-type infections, although we believe that these defects could be attributable to the consequences of the lack of Chk1 kinase in the growth of the mutant fungus inside the plant (SI Appendix, Fig. S5) (29).
The above results suggested that the biz1AA allele’s presence bypassed the apparent requirement for a cell cycle arrest during plant infection by U. maydis. While chk1Δ hsl1tef1 mutant strains were unable to induce appressoria formation because of the absence of cell cycle arrest, we were curious about the appressoria formed in the triple mutant. Appressorium formation by U. maydis can be induced in vitro by incubating solopathogenic strains (carrying compatible alleles of mating-type genes, being, therefore, independent of the mating step, ref. 38) on hydrophobic surfaces in the presence of 16-hydroxyhexadecanoic acid (33). These strains also carried a fluorescent reporter (AM1-green fluorescent protein [GFP]) consisting of GFP under the control of a promoter induced during the appressorium formation, enabling a straightforward quantification of appressoria-forming filaments (33). We have found that the solopathogenic triple mutant strain (chk1Δ hsl1tef1 biz1AA) produces appressoria-forming filaments in vitro at a frequency not significantly lower than the wild-type control strain (Fig. 1C and SI Appendix, Fig. S6). Strikingly, analyzing the morphological appearance of the appressorium-forming filaments in the triple mutant chk1Δ hsl1tef1 biz1AA, we have observed that they showed a single mononucleated apical cell compartment carrying the characteristic retraction septa behind the cell body (Fig. 1D). All these features were compatible with a cell cycle arrested appressorium. These results suggested that the mutations in the putative CDK phosphorylation sites of Biz1 seemed to restore the cell cycle arrest rather than bypass the requirement of a cell cycle arrest during appressorium formation.
Biz1AA-Mediated Down-Regulation of Clb1 Cyclin Restores the Cell Cycle Arrest during the chk1Δ hsl1tef1 Mutant Filament Induction.
The formation of the U. maydis appressorium occurs in an infective filament already cell cycle arrested. If the infective filament is not cell cycle arrested (such as happens in the chk1Δ hsl1tef1 mutant strain), the appressorium is not induced. The above results showed that the biz1AA allele’s presence appears to restore the cell cycle arrest in appressorium-producing filaments from strains carrying mutations that disconnected the b-dependent program from cell cycle regulation and thereby unable to arrest the cell cycle during the induction of the infective filament. This observation prompted us to wonder whether the restoration of the cell cycle arrest occurred during the appressorium formation or before when the infective filament was induced. To address that question, we took advantage of the AB33 strain, a particular haploid strain expressing the b factor under the control of a nitrate-induced promoter, which upon growth in a minimal medium containing nitrate, resulted in the formation of the b-dependent G2-arrested filament (but appressorium formation is not induced) (39). We have introduced in this genetic background the biz1AA allele and the mutations disconnecting the b program from cell cycle arrest (chk1Δ hsl1tef1), and we have analyzed whether upon induction of the b factor the cell cycle was arrested or not (these strains also carried an NLS-GFP transgene as a nuclear marker). We have found (SI Appendix, Fig. S7) that the biz1AA allele’s presence restored the cell cycle arrest in the infective filaments carrying the chk1Δ hsl1tef1 mutations, independently of the formation of appressoria. This result suggested that, in the triple mutant strain (chk1Δ hsl1tef1 biz1AA), most likely, the appressorium formation was recovered because of the restoration of the cell cycle arrest in the infective filament.
The above results raised the question about how in infective filaments from chk1Δ hsl1tef1 biz1AA cells, the presence of Biz1AA could reestablish the cell cycle arrest. In infective filaments from wild-type cells, the b-induced cell cycle arrest resulted from the increase in the inhibitory phosphorylation of Cdk1 (20). However, in the infective filaments produced by cells carrying chk1Δ and hsl1tef1 mutations, the phosphatase Cdc25 cannot be repressed by Chk1, and the kinase Wee1 is down-regulated by the constitutive expression of hsl1. Consequently, in these mutant filaments, the cell cycle was not arrested at G2 because of the low level of inhibitory phosphorylation of Cdk1 (9). One possibility to explain the reestablishment of the cell cycle arrest by the presence of Biz1AA is that somehow, in the triple mutant strain, the Cdk1 inhibitory phosphorylation was restored to the levels from wild-type infective filaments. However, we have observed, using the AB33-derived strains, that this was not the case (SI Appendix, Fig. S8), indicating that the reason for the reinstatement of cell cycle arrest by the presence of Biz1AA was unrelated to the inhibitory phosphorylation of Cdk1.
Biz1 was initially isolated from a genetic screen devoted to looking for genes able to arrest the cell cycle at G2 when ectopically expressed. The Biz1-induced cell cycle arrest resulted from the transcriptional repression of clb1—one of the cyclins required for G2/M transition in U. maydis—because Biz1 binds to its promoter (35). The physiological reasons why Biz1 represses the expression of clb1 were unclear. Its deletion does not affect the b-induced cell cycle arrest, even though the expression of biz1 is induced in the infective filament under the b-dependent transcriptional program (35, 36). Because of that, it was proposed that the role of Biz1 during the b-induced cell cycle arrest was to be a redundant secondary cell cycle brake operating by decreasing the amount of Clb1-Cdk1 complex once the cell cycle was already arrested via inhibitory phosphorylation of the Cdk1 (22).
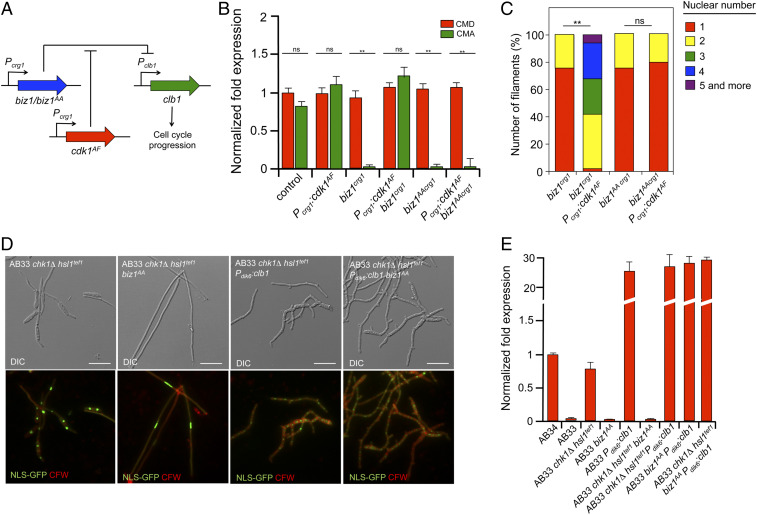
We considered that one way to understand the proposed secondary role of Biz1 in b-dependent cell cycle arrest as well as to explain the above-observed restoration of the cell cycle arrest by Biz1AA in the triple mutant was to hypothesize that the putative phosphorylation of Biz1 by CDK was affecting its ability to repress clb1. Following this explanation, in a wild-type filament, the down-regulation of CDK activity (by the inhibitory phosphorylation of Cdk1) impedes the phosphorylation of Biz1 (which remains active) and thereby the repression of clb1 expression by Biz1 assures the secondary brake. In the case of chk1Δ hsl1tef1 filaments because the CDK activity is high (resulting from the low Cdk1 inhibitory phosphorylation), Biz1 was phosphorylated and thereby inhibited, and as a consequence, clb1 expression was not repressed (maintaining the cell cycle active). However, in the triple chk1Δ hsl1tef1 biz1AA mutant, despite the high CDK activity, Biz1AA was not phosphorylated—and then not inhibited—and, therefore, it was able to repress clb1 and to reestablish the G2 cell cycle arrest. Supporting this hypothesis, we have observed that, upon ectopic expression of biz1 or biz1AA, the down-regulation of clb1 was differentially influenced by conditions mimicking low inhibitory phosphorylation of Cdk1. For that, we have constructed strains (using the haploid wild-type background FB1), which expressed biz1 or biz1AA alleles as well as cdk1AF (a cdk1 allele refractory to inhibitory phosphorylation, ref. 26) under the regulatable promoter crg1 promoter (induced by arabinose) (Fig. 2A). Coherently, with our hypothesis, we have found that the controlled expression of either biz1 or biz1AA alone was able to down-regulate the clb1 expression and, therefore, arrest the cell cycle in these cells. However, when expressed in the presence of cdk1AF, we have found that Biz1 was unable to repress clb1 while Biz1AA was still acting as a repressor of clb1 (Fig. 2B). Since the repression of clb1 mediated by Biz1 resulted in cell cycle arrest, we also have found a correlation between the cell cycle arrest and the ability or not to repress clb1 expression in the respective mutant backgrounds (Fig. 2C and SI Appendix, Fig. S9).
Fig. 2.
CDK-mediated phosphorylation of Biz1 inhibits its ability to down-regulated clb1 expression. (A) Scheme showing the experimental design to address in vivo whether the ability of Biz1 to repress the transcription of clb1 was sensitive to CDK activity. See the text for explanations. (B) Quantitative real-time-PCR of clb1 expression for the indicated strains. RNA was isolated after 6 h of induction of the crg1 promoter (arabinose complete medium [CMA]) or control conditions (glucose complete medium [CMD]). As an internal control, the expression of tub1 (encoding tubulin α) was used. Values are referred to the expression of clb1 in control strain (FB1) growing in CMD. Each column represents the mean value of three independent biological replicates (**P < 0.01, [ns] not significant). (C) Cultures of the indicated strains were incubated in CMA for 6 h and Biz1-induced filaments (cells larger than 30 μm, see SI Appendix, Fig. S9 for images of the cells) were counted and sorted in function of nuclear number. The graph shows the result from three independent experiments, counting more than 50 filaments each (**P < 0.01, ns). (D) Micrographs of the indicated AB33-derived strains grown in nitrate minimal medium to induce the b program for 8 h. All strains carried a transgene expressing a NLS-GFP fusion to detect nuclei. Cells were stained with CFW to detect the cell wall and septa. Filaments carrying a single nucleus were considered cell cycle arrested (see SI Appendix, Fig. S10 for quantification of arrested filaments). Note that filaments overexpressing clb1 showed winding morphology and a higher number of nuclei (most likely as a consequence of an accelerated cell cycle). (E) Quantitative real-time-PCR of clb1 expression for the indicated strains. RNA was isolated after 8 h of induction of the b factor in nitrate minimal medium. As an internal control, the expression of tub1 (encoding tubulin α) was used. Values are referred to the expression of clb1 in AB34 (a strain carrying noncompatible b subunits and, therefore, unable to activate the b program) growing in the same conditions. Each column represents the mean value of three independent biological replicates.
To summarize, we have found that Biz1AA restores the cell cycle arrest in filaments unable to activate the b-dependent Cdk1 inhibitory phosphorylation. Most likely, this restoration was a consequence of the ability of Biz1AA to repress the promoter of clb1. To support this assumption, we attempted to disrupt specifically the connection between Biz1 and clb1 regulations and to analyze how this disconnection would impact the b-induced cell cycle arrest. One direct manner of performing this was to mutate the binding sites of Biz1 in the regulatory region of clb1. However, the DNA sequence recognized by Biz1 is unknown, and besides, Biz1 seems to bind to several sites distributed in a broad region in the promoter of clb1 (35). Therefore, we took advantage of the previously reported Pdik6:clb1 transgene. In this construction, clb1 was under the dik6 promoter’s control, induced during the infective filament formation, and it was not down-regulated by Biz1 (35). We have introduced an ectopic copy of the Pdik6:clb1 transgene in AB33-derived strains carrying single and double combinations of hsl1tef1 chk1Δ and biz1AA mutations, and we have analyzed the presence or not of cell cycle arrested filaments (Fig. 2D and SI Appendix, Fig. S10) and the messenger RNA (mRNA) levels of clb1 (Fig. 2E). We have found that the filaments from hsl1tef1 chk1Δ biz1AA cells carrying the ectopic copy of the Pdik6:clb1 transgene showed a high level of clb1 mRNA and that they were not cell cycle arrested.
These results supported the idea that Biz1AA-mediated down-regulation of Clb1 cyclin caused the restablishment of the cell cycle arrest during the induction of mutant infective filaments that were impaired in the b-activated Cdk1 inhibitory phosphorylation.
The Clb2-Cdk1 Complex Interacts with Biz1.
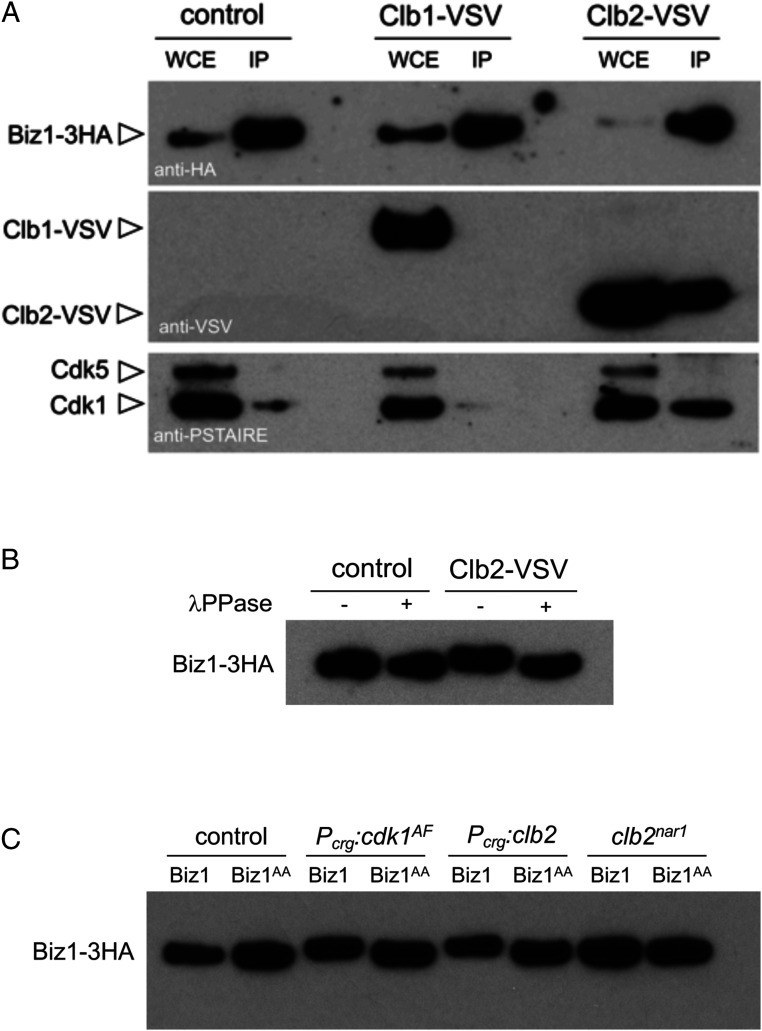
The results shown above can be included in a working hypothesis explaining the observed incompatibility between an active cell cycle and the appressorium formation. In an infective filament where the cell cycle was not arrested, the presence of an active CDK complex phosphorylates the transcription factor Biz1, promoting in some manner its inhibition. This inhibition disables Biz1 to sustain the cell cycle arrest and, most likely, to produce appressoria. To support this idea, we have tried to detect physical interaction between CDK and Biz1. We took advantage of the above-described FB1-derived strains expressing ectopically biz1 under the crg1 promoter’s control. The reason to use this genetic background was related to the fact that, in natural conditions, the expression of biz1 is dependent on the b factor (35, 36). Therefore, to induce the production of Biz1, we should express the genes encoding the b factor, which also resulted in the inhibition of CDK activity, impeding the study of the proposed CDK-Biz1 interactions. We have introduced in this genetic background vesicular stromatitis virus (VSV)-tagged versions of Clb1 and Clb2—the two U. maydis b cyclins—also under the control of the crg1 promoter (24) to bypass the observed down-regulation of Biz1 on clb1 expression. Also, the Biz1 protein was tagged with three copies of the hemagglutinin (HA) epitope (the tagging does not affect the functionality of the protein, ref. 35). Lysates from cells expressing Biz1-3HA and the respective b cyclins were submitted to immunoprecipitation with anti-HA antibodies. The corresponding immunoprecipitates were analyzed for the presence of Biz1 (using anti-HA antibodies), cyclins (using anti-VSV antibodies), and Cdk1 (using anti-PSTAIRE antibodies, which detects Cdk1 as well as Cdk5, ref. 24). We have found that Biz1 interacts specifically with the Clb2-Cdk1 complex: In the anti-HA immunoprecipitates, it was possible to recover both Cdk1 and Clb2 but not Clb1 (Fig. 3A). Note that in both control and Clb1-VSV expression conditions, it was also possible to observe a faint Cdk1 signal in the immunoprecipitates, most likely being a consequence of the Clb2 endogenous levels.
Fig. 3.
Biz1 interacts with Clb2-Cdk1. (A) Western blots to show interaction among Biz1, Clb1, and Cdk1. Soluble extracts from strains carrying Biz1-3HA and Clb1-VSV or Clb2-VSV under the control of a crg1 promoter were incubated with anti-HA antibodies coupled to magnetic beads to obtain immunoprecipitates. The immunoprecipitates were submitted to Western blot with anti-VSV (cyclins) and anti-HA (Biz1) antibodies in succession. The lower part of the membrane was excised and processed independently with anti-PSTAIRE to detect the Cdk1 protein. Cells were grown in inducing conditions for crg1 promoter (complete medium [CM] plus 1% arabinose, CMA) during 6 h. (B) Western blot to show the effect of treatment with λ protein phosphatase (λ PPase) of anti-HA immunoprecipitates from cultures expressing or not clb2-VSV. Immunoprecipitates were incubated at 30 °C for 20 min in the absence (−) or presence (+) of λ PPase. (C) Western blot (anti-HA) from immunoprecipitates obtained from extracts of cultures grown in inducing conditions (CMA, 6 h) for the cells carrying biz1-3HA or biz1AA-3HA alleles as well as the indicated constructions.
Interestingly, we have also noted a moderated but reproducible shift in the electrophoretic mobility of Biz1-3HA immunoprecipitated in Clb2-VSV expression conditions. We have treated the immunoprecipitates obtained from the control as well as from Clb2-VSV extracts with λ phosphatase, and we have found that the shift was corrected upon treatment with the phosphatase (Fig. 3B), strongly suggesting that Biz1-3HA was phosphorylated in cells in a manner dependent on Clb2. To gain further support in this observation, we analyzed immunoprecipitates obtained from cells with high Clb2-Cdk1 activity and carrying either a wild-type biz1 or a biz1AA allele (in both cases under the control of the crg1 promoter and tagged with HA). We have achieved this high Clb2-Cdk1 activity in two independent ways: either by the overexpression of clb2 or by the expression (also under the crg1 promoter) of a cdk1 allele refractory to inhibitory phosphorylation (cdk1AF, inhibitory phosphorylation only affects the Clb2-Cdk1, ref. 26). Besides, we have also included a control resulting in the absence of Clb2-Cdk1 activity by using a conditional allele of clb2 [clb2nar1, ref. 24]) grown in repressive conditions. We have found a correlation between the high Clb2-Cdk1 activity and the electrophoretic mobility shift in Biz1 and, more critical, the absence of mobility shift in any condition in the biz1AA allele. The observed electrophoretic mobility shift (which can also be observed in control conditions, although weakly) was also lost in cells carrying the clb2 conditional allele, regardless of the biz1 allele analyzed (Fig. 3C).
Taking together all these observations, we propose that Biz1 interacts specifically with the Clb2-Cdk1 complex. The electrophoretic mobility shift in Biz1 that can be suppressed by phosphatase treatment, its absence in the Biz1AA mutant, and its dependency on the presence of Clb2 function, lead us also to propose that the Clb2-Cdk1 complex phosphorylates Biz1. More direct proofs of these interactions, such as in vitro phosphorylation assays and phosphoproteomic analysis of in vivo infections by wild-type and mutant strains, would strengthen our conclusion, although technical reasons impede these approaches currently.
CDK-Mediated Phosphorylation Confines Biz1 into the Cytoplasm by the Promotion of Its Binding to 14–3-3 Proteins.
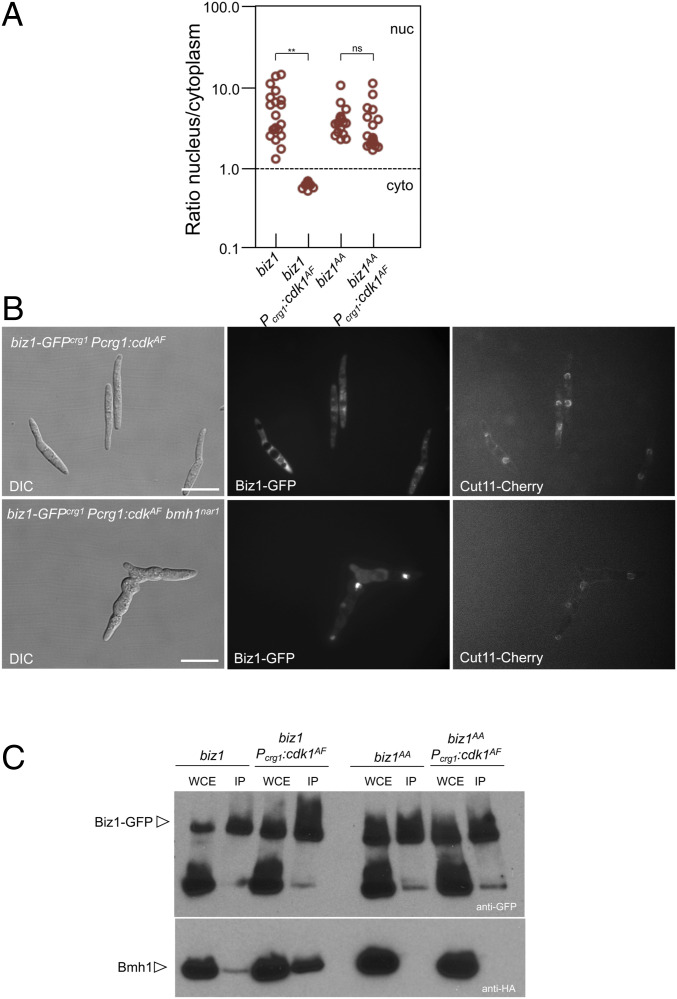
We were curious about the mechanism that inhibited the ability of Biz1 to repress the transcription of clb1 upon phosphorylation by Clb2-Cdk1. Disabling the Biz1-mediated repression of clb1 expression could be occurring in different not exclusive ways: inhibition of nuclear translocation of Biz1; inhibition of the ability of Biz1 to bind the clb1 promoter region; or inhibition of the repressive activity of Biz1. As a start, we decided to analyze whether the transport of Biz1 into the nucleus was affected by high Clb2-Cdk1 activity. For that, using the FB1 background, GFP fusions of biz1 and biz1AA alleles—under the control of the crg1 promoter—were performed, and its subcellular location was addressed in control strains and strains expressing the cdk1AF allele (i.e., high Clb2-Cdk1 activity). We have found (Fig. 4A and SI Appendix, Fig. S11) that high Clb2-Cdk1 activity excludes Biz1 from the nucleus, but that this effect was lost in cells carrying the allele refractory to CDK phosphorylation, biz1AA. We infer from these results that CDK inhibits Biz1 by keeping the protein in the cytoplasm.
Fig. 4.
CDK-mediated phosphorylation retains Biz1 at the cytoplasm. (A) Graph showing the nuclear versus cytoplasmic distribution of the GFP signal of the indicated strains. Micrographs used for quantification are shown in SI Appendix, Fig. S11. Cultures were incubated for 6 h in inducing conditions (CMA). The intensity of the nuclear and cytoplasmic GFP signals were determined by measuring pixel intensity in the nucleus and of an equivalent area in the cytoplasm, and the ratio was determined. A ratio higher than 1 means nuclear distribution, while a ratio lower than 1 indicates cytoplasmic distribution. Twenty cells were quantified for each experiment (two independent experiments, **P < 0.01, ns). (B) Bmh1 was required for CDK-dependent retention of Biz1 in the cytoplasm. Indicated strains were grown overnight in permissive conditions for the bmh1nar1 allele and restrictive for biz1-GFPcrg1 and Pcrg1:cdkAF (minimal medium amended with nitrate and glucose) and then transferred for 6 h to CMA medium, that represses nar1 expression and activates crg1 expression. Note that in the absence of the Bmh1 function, the Biz1-GFP signal can be colocated with the nucleus (marked by a Cut11-Cherry fusion). The Cut11-Cherry signal was very weak in the bmh1nar1 strain growing in restrictive conditions, and in some cells the nucleus seems not to be assembled. (C) Western blots to show interaction between Biz1 and Bmh1. Soluble extracts from strains carrying Bmh1-3HA and Biz1-GFP or Biz1AA-GFP tagged in their corresponding endogenous loci and carrying ectopic copies of cdk1AF under the control of a crg1 promoter were incubated with GFP-trap beads, and the immunoprecipitates submitted to Western blot with anti-HA (Bmh1) and anti-GFP (Biz1) antibodies in succession. Cells were grown in inducing conditions (CMA) for the crg1 promoter during 6 h.
One of the roles of the 14–3-3 protein in the cell is to sequester proteins in the cytoplasm upon recognizing phosphoserine or phosphothreonine residues in its targets (40). In U. maydis, there is a sole 14–3-3 protein, encoded by bmh1, which is essential for growth (31). We have considered that 14–3-3 proteins could be well suited to be one of the elements involved in the regulation of Biz1. To address this possibility, we have introduced in the strain expressing biz1-GFP and the cdk1AF alleles (both under the crg1 promoter), a bmh1nar1 conditional allele (which is induced when cells are growing in minimal nitrate medium and repressed in complete medium), and we analyzed the Biz1-GFP subcellular localization in restrictive conditions for the bmh1 conditional allele. In agreement with our hypothesis, we have found that when Bmh1 was down-regulated, Biz1 was not retained in the cytoplasm (Fig. 4B).
We also have attempted to analyze whether the CDK-dependent phosphorylation of Biz1 affected this protein’s ability to interact with Bmh1 in vivo. Using GFP-trap beads, we have isolated GFP fusions of Biz1 and Biz1AA versions from strains carrying a bmh1-3HA tagged allele and expressing or not an ectopic copy of cdk1AF· We have found that high Clb2-Cdk1 activity promoted the Biz1 and Bmh1 interaction and that this interaction was lost in the Biz1AA mutant (Fig. 4C). These results were coherent with the role of Bmh1 in the cytoplasmic retainment of Biz1.
To summarize, it seems that Biz1 phosphorylation by Clb2-Cdk1 precludes its activity as a transcriptional factor by promoting the interaction with Bmh1 and its cytoplasmic retention.
Disconnecting Biz1 and Cell Cycle Regulation Seems to Abrogate the Role of Biz1 in the Decision of Proliferation versus Appressorium Formation.
The above-described results help explain the role of Biz1 in the observed incompatibility between an active cell cycle and the appressorium formation. If, for some reason, during the induction of the infective filament, the primary cell cycle brake (composed of the Cdk1 inhibitory phosphorylation) is not operative, then the presence of an active Clb2-Cdk1 complex phosphorylates the transcription factor Biz1. This phosphorylation promotes the retention of Biz1 in the cytoplasm, precluding its role as a transcriptional regulator and affecting the filament’s ability to support the secondary cell cycle brake and to induce the appressorium as well.
Following this working model, we have reasoned that if we disentangled Biz1 from the cell cycle regulation, then, most likely, the observed incompatibility between active cell cycle and appressorium induction might be dismissed (Fig. 5A). To untie Biz1, we must disable both the upstream negative regulation of Biz1 by Clb2-Cdk1 (i.e., using the biz1AA allele) and the downstream repression of Biz1 on clb1 expression (i.e., using the Pdik6:clb1 transgene). Therefore, using a solopathogenic strain carrying the four mutations (hsl1tef1 chk1Δ biz1AA Pdik6:clb1) and the AM1-GFP reporter, we tried to detect in vitro the presence of appressoria. Encouragingly, we have found filaments showing GFP fluorescence, although in a much lower frequency than a control strain (hsl1tef1 chk1Δ biz1AA) (Fig. 5B). However, these fluorescent filaments were composed of several cell compartments separated by septa, which contrast to the control appressoria composed of a single apical cell compartment (Fig. 5C). We cannot strictly assure that these filaments were true appressoria, although the transcriptional program activating the AM1-GFP reported was operative in these filaments.
Fig. 5.
Disentangling Biz1 from cell cycle regulation. (A) Scheme showing the hypothesis to be tested: untieing Biz1 from cell cycle regulation by using the biz1AA and Pdik6:clb1 alleles should disconnect appressorium formation from the cell cycle. (B) Frequency of appressoria formation in vitro in strains carrying the indicated mutations. The graph shows the result from three independent experiments, counting more than 50 filaments each (**P < 0.01). (C) Micrographs showing appressoria formation in vitro of the indicated strains. Images show AM1 marker expression (AM1-GFP) and CFW stained samples as well as magnified merged images of the apical part from GFP positive filaments. Asterisks mark septa. (Scale bar: 20 μm.) (D) Graph showing disease symptoms caused by crosses of indicated solopathogenic strains. The symptoms were scored 14 d after infection. Three independent experiments were carried out, and the average values are expressed as percentages of the total number of infected plants (n: 30 plants in each experiment, Raw data from infections are provided in the SI Appendix).
We wondered whether these structures were functional to lead plant infection. Therefore, we have evaluated the virulence of the hsl1tef1 chk1Δ biz1AA Pdik6:clb1 mutant strain. As a control, we used SG200-derived strains carrying individual combinations (chk1Δ hsl1tef1; biz1AA; Pdik6:clb1) and double and triple combinations (Fig. 5D). We have observed that a strain in which Biz1 was disconnected from cell cycle regulation (biz1AA Pdik6:clb1) was still able to infect plants, although the symptoms caused seem to be weaker (we believe this is a side effect of the presence of the Pdik6:clb1 transgene as was already described, ref. 35, and it can be observed in the single SG200 Pdik6:clb1 mutant). However, when we disconnected Biz1 from cell cycle regulation in cells unable to arrest the cell cycle during the induction of the filament (chk1Δ hsl1tef1 biz1AA Pdik6:clb1 strain), we have found the unexpected result that this strain was avirulent.
We were also able to find GFP positive filaments on the plant (although very infrequently), showing a similar morphology as those found in vitro regarding the presence of several cell compartments showing GFP fluorescence (SI Appendix, Fig. S12A). However, we could not find any evidence supporting that the mutant strain could penetrate the plant tissue (SI Appendix, Fig. S12B). These data suggested that the structures (which we believe are some sort of appressoria) were not functional for plant invasion.
Considering these structures as aberrant appressoria, we would like to propose that disconnecting Biz1 from cell cycle regulation, abrogated its ability to work as a point of control to discriminate whether the cell cycle was arrested before allowing the induction of the formation of appressorium. We believe that, in the absence of this sort of checkpoint, the concurrence of an active cell cycle and the program’s activation leading to appressorium induction would result in an inactive appressorium (see Discussion for more details about possible factors affecting the functionality of the appressorium in this scenario).
Discussion
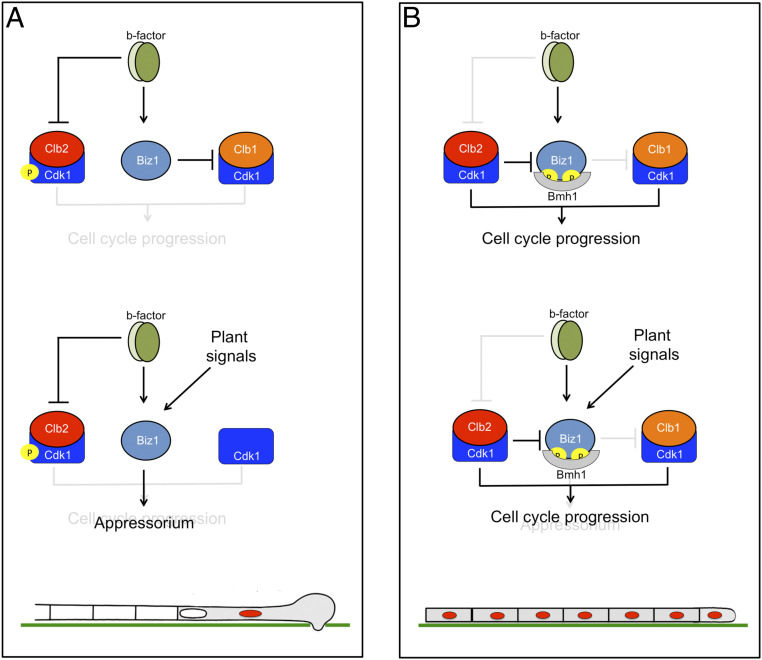
Appressorium formation seems to be subordinated to the cell cycle in those fungal phytopathogens where these connections have been studied (10, 41). There are cases in which particular cell cycle phases have to be completed before the appressorium formation, such as it has been described in M. oryzae where the completion of both S and M phases is mandatory for appressorium formation and functionality (41). However, in other cases, the cell cycle has to be arrested at some particular cell cycle phase to allow the appressorium formation. In U. maydis, the infective filament (where the appressorium will differentiate) must be arrested at the G2 phase. If the cell cycle was not arrested, then the appressorium was not formed. In this paper, we addressed the intricacies about how, in this fungus, the cell cycle status seems to determine the induction or not of the appressorium formation: The Clb2-Cdk1 complex (one of the CDK complexes involved in G2/M transition) phosphorylates and inhibits the activity of Biz1, a transcriptional factor required in appressorium induction. Therefore, to progress in the appressorium formation, the Clb2-Cdk1 complex has to be inhibited, resulting in a G2 cell cycle arrest.
The appressorium induction in U. maydis occurs in the b-dependent infective filament, which was cell cycle arrested. In a wild-type situation (Fig. 6A), during the formation of the infective filament, the transcriptional program promoted by the b factor results in the induction of biz1 expression (together with many other regulatory factors, ref. 36) as well as the inhibition of the Clb2-Cdk1 complex by the increase in the inhibitory phosphorylation of Cdk1. The second CDK complex required for G2/M transition (Clb1-Cdk1) is not sensitive to Cdk1 inhibitory phosphorylation, but without the concourse of Clb2-Cdk1, it was unable to induce mitosis. Because Clb2-Cdk1 is inhibited and Biz1 is not a target of Clb1-Cdk1, the Biz1 protein is not phosphorylated, and it travels to the nucleus where it down-regulates clb1, resulting in the inhibition of the second CDK complex required for G2/M transition. Once in the nucleus, eventually in response to inductive cues from the plant, Biz1 might act on other unknown targets to activate the appressorium formation (together with other factors, such as Hdp2, ref. 34). However, if for some reason, the Clb2-Cdk1 activity is not down-regulated during the formation of the infective filament (Fig. 6B), then the Clb2-Cdk1 complex phosphorylated Biz1, promoting its retainment in the cytoplasm (via its interaction with 14–3-3 proteins) and disabling the repression of the Clb1-Cdk1 complex (and thereby the cell cycle can progress) as the appressorium formation.
Fig. 6.
Model to explain the incompatibility between cell cycle progression and appressorium formation in U. maydis. A describes a wild-type situation, while B shows a scenario in which the ability of the b factor to arrest the cell cycle (via Cdk1 inhibitory phosphorylation) was not operative. See the text for explanations.
In our working model explaining the conflict between cell cycle progression and appressorium formation in U. maydis, the protein Biz1 plays a central role. This central role may induce assuming that Biz1 is some kind of specialized cell cycle regulator “evolutionarily designed” to link the cell cycle and appressorium formation. However, the role of Biz1 seems not to be circumscribed to be the link between cell cycle regulation and appressorium formation. In strains that properly arrest the cell cycle, the absence of Biz1 resulted in a defective formation of appressoria and defects in plant invasion, suggesting roles aside from cell cycle regulation (35). Moreover, Biz1 expression could be detected for more than 10 d postinfection, once the fungus was proliferating inside the plant, suggesting roles further than plant invasion (35). Indeed, since the symptoms observed in infections by strains carrying the biz1AA allele alone were similar to those observed in wild-type infections, we believe that the connections described in our paper between cell cycle and Biz1 phosphorylation may play a minor role, if any, at steps of the plant infection later than appressorium formation. In other words, we favor the hypothesis that along with the evolution, the cell cycle “trapped” one of the regulators involved in appressorium formation to transmit the information from the cell cycle to the appressorium formation program. Moreover, this connection between Biz1 and cell cycle goes further and established a two-direction way by the ability of Biz1 to repress the promoter of clb1. As a result, it created a negative feedback loop between appressorium formation and cell cycle progression in U. maydis. High CDK activity represses appressorium formation, but appressorium formation represses CDK activity. Mutual repression is a classic mechanism in developmental programs, and it has been suggested that it serves as a “toggle switch” to control decisions between two states (42). In this case, the two relevant states are to infect the plant or proliferate from the plant. There are several examples, mainly in metazoans, by which mutual repression between cell cycle regulators and transcription factors determine developmental transitions. One well-known example is the differentiation of a neuroblast from Drosophila melanogaster where the regulator Prospero represses the transcription of S-phase cyclin, blocking cell cycle progression (43) and, conversely, the S-phase CDK complex inhibits Prospero by changing its subcellular localization (44) in a similar way to that we have described here for Biz1.
Our results also proportionate clues to understanding how in fungal phytopathogens, the cell cycle informs the completeness or not of each specific phase to the genetic program resulting in appressorium formation. Cell cycle transitions are promoted by sequential activation of distinct CDK complexes. Each specific cell cycle phase is not activated until the corresponding CDK complex reaches a threshold activity. This increase in activity only occurs once the previous cell cycle phase is completed (45). Therefore, it makes sense that the CDK responsible for the decision to enter or not into a particular cell cycle phase (Clb2-Cdk1 in the case of U. maydis) was used directly to monitor the feasibility to produce or not appressorium. Applying this line of reasoning to other systems, we propose that in M. oryzae—where mitosis is mandatory for correct appressorium maturation (14)—critical regulators of the appressorium maturation step could be direct targets of CDK complexes involved in G2/M transition. However, in contrast to what we have observed in U. maydis appressorium, in M. oryzae, the corresponding CDK phosphorylation may have an activating role in appressorium formation.
A pertinent question is what are the reasons for the observed subordination of appressorium formation to cell cycle regulation. We envisioned several possibilities. In one case, this subordination seems reasonable considering that the appressorium’s primary role is to facilitate the penetration of the fungal hyphae inside its host to proliferate once it had invaded the plant tissue. Thereby it seems logical that the infection process assures the loading in the invading hypha of the correct genetic information and the importance of the subordination to the cell cycle. In M. oryzae, for instance, since a full division takes place in the appressorium cell before the penetration peg enters the plant, the completion of both S and M phases is mandatory for the process and for that, checkpoints control that these steps were completed correctly (41). The second class of reasons for the subordination of appressorium formation to cell cycle regulation could be related to mechanistic causes. For instance, the secretion of effectors by the appressorium was primordial for the invasion of the plant by U. maydis (18). Because of the competence for cytoskeletal components among the mitotic, the cell division apparatus, and the secretion machinery, the concurrent activation of the two processes (cell cycle progression and secretion) might result in defective secretion and, therefore, in a failed infection (10). The importance of a correct cytoskeleton for the appressorium functionality in U. maydis has been noted previously: The Aga1 kinase is required for the correct actin organization in U. maydis during appressorium formation (46). In the absence of Aga1, despite a correct activation of the transcriptional program leading to appressorium induction (assessed by the AM1-GFP reporter), the appressorium was not functional. Moreover, the consequences of the lack of Aga1 function during the appressorium formation could be mimicked by inhibiting the formation of actin filaments with Latrunculin A (46). Additional connections between secretion and specific cell cycle phases have also been provided by studies in other fungal systems, such as Saccharomyces cerevisiae. In this yeast, the secretion was affected upon the phosphorylation of the exocyst subunit Exo84 by the Clb2-Cdc28 complex (the ortholog of Clb2-Cdk1 from U. maydis) (47). We ignore whether this regulatory link exists in U. maydis, but it could be in line with the requirement of the down-regulation of CDK activity to enable a functional appressorium. The unappropriated cytoskeleton or the inhibition of secretion machinery as well as other nonenvisioned possibilities could help us explain the behavior of the chk1Δ hsl1tef1 biz1AA Pdik6:clb1 strain. In this strain, the b program could not arrest the filament in G2, and Biz1 was untied from cell cycle regulation. According to our expectations, in this strain, the program leading to appressorium induction seemed to be disconnected from cell cycle regulation, and it appears to produce some sort of appressoria. However, the fungus was avirulent, most likely because it was unable to penetrate the plant tissue, suggesting that the appressorium was not functional.
To summarize, the data provided in this paper lead us to propose that, in U. maydis, mechanistic reasons forced the incompatibility between cell cycle progression and appressorium functionality. Then, to avoid the concurrence of both processes, the evolution selected a surveillance point involving Biz1 to inhibit the induction of appressorium formation in an active cell cycle.
Materials and Methods
Strains and Growth Conditions.
U. maydis strains are derived from a FB1 background (48) and are listed in SI Appendix, Table S1. Cells were grown in YPD or CM (49). The controlled expression of genes under the crg1 and nar1 promoters was performed as described previously (39). To construct the different strains, transformation of U. maydis protoplasts with the desired constructions was performed as described previously (50). Integration of the corresponding construction into the corresponding loci was verified in each case by diagnostic PCR and subsequent Southern blot analysis or RT-PCR analysis of transcripts depending on the type of integrated mutant allele. U. maydis DNA isolation was performed as previously described (50).
Plasmid Construction.
Plasmid pGEM-T easy (Promega) and pJET1.2 (Thermo Fisher) were used for subcloning and sequencing of genomic fragments generated by PCR. Details of the construction for allelic replacement vectors for generating the different mutant alleles are given in the SI Appendix.
Mutant alleles of the following genes were already described: Pcrg1:cdk1AF (26); Pcrg1:clb1-VSV, Pcrg1:clb2-VSV, and clb2nar1 (24); cut11-cherry (51); bmh1-3HA and bmh1nar1 (31); Pcrg1:fuz7DD (52); Pcrg1:kpp6 P130A P131A (32).
RNA Analysis.
Total RNA was extracted with acidic phenol solution. After extraction, the RNA was cleaned using the High Pure RNA Isolation Kit (Roche Diagnostics GmbH). For RT-PCR, complementary DNA (cDNA) was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) employing 1 μg total RNA per sample. qRT-PCR was performed using the SsoAdvanced Universal SYBR Green Supermix (BioRad) in a CFX96 real-time PCR system (BioRad). Reaction conditions were as follows: 3 min 95 °C followed by 40 cycles of 10 s 95 °C/10 s 60 °C/30 s 72 °C.
Protein–Protein Interaction Analysis by Coimmunoprecipitation.
To perform immunoprecipitations, crude protein extracts were prepared. Briefly, cells were harvested by centrifugation at 4 °C and washed twice with ice-cold water. The cell pellet was resuspended in ice-cold hemoglobin (HB) buffer (25 mM Mops pH 7.2, 15 mM MgCl2, 15 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 1% Triton X-100, PhosSTOP [1 tablet per 10 mL], and Roche protease inhibitor mixture [1 tablet per 10 mL]), and cells were broken using a FastPrep FP120 cell disrupter. The lysate was recovered by punching a hole on the bottom of the tube, and the glass beads were further washed with ice-cold HB buffer, and the lysate was cleared by centrifugation (10 min/13,000 × g).
For coimmunoprecipitation analysis, ∼3.5 mg of total protein extracts (1 mL) were incubated with 1 μg of the monoclonal antibody for 2 h at 4 °C and then prewashed G protein coupled magnetic beads (50 μL, Dynabeads) were added and incubated for 30 min at 4 °C with agitation. For GFP trap, 50 μL GFP trap beads (Chromotek) were mixed with 3.5 mg of total protein extracts (1 mL) for 2 h at 4 °C with agitation. Immunoprecipitates were washed six times with 1 mL of HB buffer.
Western analysis was performed on total protein extracts or immunoprecipitates (50–100 μg), separated on TGX (4–20%) gels (BioRad) at constant 100v running conditions. Anti-HA and anti-VSV horseradish peroxidase (HRP) conjugated antibodies (Sigma-Aldrich, 1:10,000 dilution); anti-GFP (Living Colors, Clontech, 1:5,000 dilution) followed by anti-mouse-immunoglobulin (Ig)-HRP (Sigma-Aldrich, 1:10,000 dilution); anti-PSTAIRE (Santa Cruz Biotechnology, 1:5,000 dilution) followed by anti-rabbit-Ig-HRP (Sigma-Aldrich, 1:10,000 dilution) were used. All western analyses were visualized using enhanced chemiluminescence (Clarity ECL, BioRad). Western blots were repeated from, at least, three independent experiments in each case.
Plant Infections and Appressorium Formation Analysis.
Pathogenic development of wild-type and mutant strains was assayed by plant infections of the maize (Zea mays) variety Early Golden Bantam (Olds seeds) as described before (53). For appressoria in vitro formation we followed procedures previously described (33).
Microscopy.
Images were obtained using a Nikon Eclipse 90i fluorescence microscope with a Hamamatsu Orca-ER camera driven by Metamorph (Universal Imaging, Downingtown, PA). Images were further processed with ImageJ software.
Quantification and Statistical Analysis.
To determine the statistical significance of the differences, a two-tailed Student t test was used. P values were calculated with the GraphPad Prism 5.0 software.
Supplementary Material
Acknowledgments
We thank members of the J.P.-M. Laboratory for discussion and critical reading of the paper. S.C. was supported by a Marie Curie Initial Training Networks Grant (ARIADNE, PITN-GA-2009-237936). This work was supported by Grants from the Spanish Government (BIO2014-55398-R and BIO2017-88938-R).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006909117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Brauer V. S., et al. , Antifungal agents in agriculture: Friends and foes of public health. Biomolecules 9, 521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulze-Lefert P., Knocking on the heaven’s wall: Pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr. Opin. Plant Biol. 7, 377–383 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Deising H. B., Werner S., Wernitz M., The role of fungal appressoria in plant infection. Microbes Infect./Inst. Pasteur 2, 1631–1641 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Mendgen K., Deising H., Infection structures of fungal plant pathogens -a cytological and physiological evaluation. New Phytol. 124, 193–213 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Mendgen K., Hahn M., Deising H., Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu. Rev. Phytopathol. 34, 367–386 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Perez-Nadales E., et al. , Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 70, 42–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryder L. S., Talbot N. J., Regulation of appressorium development in pathogenic fungi. Curr. Opin. Plant Biol. 26, 8–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Martín J., Bardetti P., Castanheira S., de la Torre A., Tenorio-Gómez M., Virulence-specific cell cycle and morphogenesis connections in pathogenic fungi. Semin. Cell Dev. Biol. 57, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Castanheira S., Mielnichuk N., Pérez-Martín J., Programmed cell cycle arrest is required for infection of corn plants by the fungus Ustilago maydis. Development 141, 4817–4826 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Castanheira S., Pérez-Martín J., Appressorium formation in the corn smut fungus Ustilago maydis requires a G2 cell cycle arrest. Plant Signal. Behav. 10, e1001227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukada F., Kodama S., Nishiuchi T., Kajikawa N., Kubo Y., Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol. 222, 1909–1923 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Fukada F., Kubo Y., Colletotrichum orbiculare regulates cell cycle G1/S progression via a two-component GAP and a GTPase to establish plant infection. Plant Cell 27, 2530–2544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osés-Ruiz M., Sakulkoo W., Littlejohn G. R., Martin-Urdiroz M., Talbot N. J., Two independent S-phase checkpoints regulate appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Proc. Natl. Acad. Sci. U.S.A. 114, E237–E244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders D. G., Aves S. J., Talbot N. J., Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22, 497–507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders D. G., Dagdas Y. F., Talbot N. J., Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 2417–2428 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matei A., Doehlemann G., Cell biology of corn smut disease-Ustilago maydis as a model for biotrophic interactions. Curr. Opin. Microbiol. 34, 60–66 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Schirawski J., et al. , Endoplasmic reticulum glucosidase II is required for pathogenicity of Ustilago maydis. Plant Cell 17, 3532–3543 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanver D., et al. , Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15, 409–421 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Snetselaar K., Mims C. W., Infection of maize stigmas by Ustilago maydis: Light and electron microscopy. Phytopathology 83, 843–850 (1993). [Google Scholar]
- 20.Mielnichuk N., Sgarlata C., Pérez-Martín J., A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J. Cell Sci. 122, 4130–4140 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Vollmeister E., et al. , Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev. 36, 59–77 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Bardetti P., Castanheira S. M., Valerius O., Braus G. H., Pérez-Martín J., Cytoplasmic retention and degradation of a mitotic inducer enable plant infection by a pathogenic fungus. eLife 8, e48943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Martín J., et al. , Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol. Genet. Genomics 276, 211–229 (2006). [DOI] [PubMed] [Google Scholar]
- 24.García-Muse T., Steinberg G., Perez-Martín J., Characterization of B-type cyclins in the smut fungus Ustilago maydis: Roles in morphogenesis and pathogenicity. J. Cell Sci. 117, 487–506 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Sgarlata C., Pérez-Martín J., The cdc25 phosphatase is essential for the G2/M phase transition in the basidiomycete yeast Ustilago maydis. Mol. Microbiol. 58, 1482–1496 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Sgarlata C., Pérez-Martín J., Inhibitory phosphorylation of a mitotic cyclin-dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis. J. Cell Sci. 118, 3607–3622 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Perez-Martin J., “Cell cycle and morphogenesis connections during the formation of the infective filament in Ustilago maydis” in Morphogenesis and Pathogenicity in Fungi, Perez-Martin J., di Pietro A., Eds. (Springer-Verlag, Berlin, 2012), pp. 97–114. [Google Scholar]
- 28.Feldbrügge M., Kämper J., Steinberg G., Kahmann R., Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7, 666–672 (2004). [DOI] [PubMed] [Google Scholar]
- 29.de Sena-Tomás C., Fernández-Álvarez A., Holloman W. K., Pérez-Martín J., The DNA damage response signaling cascade regulates proliferation of the phytopathogenic fungus Ustilago maydis in planta. Plant Cell 23, 1654–1665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenorio-Gómez M., de Sena-Tomás C., Pérez-Martín J., MRN- and 9-1-1-independent activation of the ATR-chk1 pathway during the induction of the virulence program in the phytopathogen Ustilago maydis. PLoS One 10, e0137192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mielnichuk N., Pérez-Martín J., 14-3-3 regulates the G2/M transition in the basidiomycete Ustilago maydis. Fungal Genet. Biol. 45, 1206–1215 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Lanver D., Mendoza-Mendoza A., Brachmann A., Kahmann R., Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell 22, 2085–2101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendoza-Mendoza A., et al. , Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 71, 895–911 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Lanver D., et al. , Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog. 10, e1004272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flor-Parra I., Vranes M., Kämper J., Pérez-Martín J., Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell 18, 2369–2387 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heimel K., et al. , The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog. 6, e1001035 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Örd M., et al. , Multisite phosphorylation code of CDK. Nat. Struct. Mol. Biol. 26, 649–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolker M., Genin S., Lehmler C., Kahmann R., Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73, S320–S325 (1995). [Google Scholar]
- 39.Brachmann A., Weinzierl G., Kämper J., Kahmann R., Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42, 1047–1063 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Dougherty M. K., Morrison D. K., Unlocking the code of 14-3-3. J. Cell Sci. 117, 1875–1884 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Osés-Ruiz M., Talbot N. J., Cell cycle-dependent regulation of plant infection by the rice blast fungus Magnaporthe oryzae. Commun. Integr. Biol. 10, e1372067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasty J., McMillen D., Collins J. J., Engineered gene circuits. Nature 420, 224–230 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Choksi S. P., et al. , Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775–789 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Berger C., et al. , Cell cycle independent role of Cyclin E during neural cell fate specification in Drosophila is mediated by its regulation of Prospero function. Dev. Biol. 337, 415–424 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Lim S., Kaldis P., Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 140, 3079–3093 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Berndt P., Lanver D., Kahmann R., The AGC Ser/Thr kinase Aga1 is essential for appressorium formation and maintenance of the actin cytoskeleton in the smut fungus Ustilago maydis. Mol. Microbiol. 78, 1484–1499 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Luo G., Zhang J., Luca F. C., Guo W., Mitotic phosphorylation of Exo84 disrupts exocyst assembly and arrests cell growth. J. Cell Biol. 202, 97–111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banuett F., Herskowitz I., Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. U.S.A. 86, 5878–5882 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holliday R., “Ustilago maydis” in Handbook of Genetics, King R. C., Ed. (Plenum Press, New York, 1974), pp. 575–595. [Google Scholar]
- 50.Tsukuda T., Carleton S., Fotheringham S., Holloman W. K., Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8, 3703–3709 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez-Martín J., DNA-damage response in the basidiomycete fungus Ustilago maydis relies in a sole Chk1-like kinase. DNA Repair (Amst.) 8, 720–731 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Müller P., Weinzierl G., Brachmann A., Feldbrügge M., Kahmann R., Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2, 1187–1199 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kämper J., et al. , Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444, 97–101 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.