Significance
Carbapenem resistance has become steadily more common in recent decades, and its prevalence in Enterobacteriaceae has been marked as an urgent antibiotic priority by both the World Health Organization and the US Centers for Disease Control and Prevention. In this paper, we use RNA sequencing to explore resistance development and engineer peptide nucleic acids (PNA) for bacterial growth inhibition. Our results mark the use of transcriptomics for the design of antibiotic PNA and demonstrate that this method of PNA design can be equally or more effective than standard genome-based design. Furthermore, we identify three genes that may point to antibiotic targets and two genes that offer clues about how Enterobacteriaceae acquire carbapenem resistance.
Keywords: carbapenem-resistant Escherichia coli, transcriptome, short RNA sequencing, genome sequence, antibiotic resistance
Abstract
In recent years, the prevalence of carbapenem-resistant Enterobacteriaceae (CRE) has risen substantially, and the study of CRE resistance mechanisms has become increasingly important for antibiotic development. Although much research has focused on genomic resistance factors, relatively few studies have examined CRE pathogens through changes in gene expression. In this study, we examined the gene expression profile of a CRE Escherichia coli clinical isolate that is sensitive to meropenem but resistant to ertapenem to explore transcriptomic contributions to resistance and to identify gene knockdown targets for carbapenem potentiation. We sequenced total and short RNA to analyze the gene expression response to ertapenem or meropenem treatment and found significant expression changes in genes related to motility, maltodextrin metabolism, the formate hydrogenlyase complex, and the general stress response. To validate these findings, we used our laboratory’s Facile Accelerated Specific Therapeutic (FAST) platform to create antisense peptide nucleic acids (PNAs), gene-specific molecules designed to inhibit protein translation. PNAs were designed to inhibit the pathways identified in our transcriptomic analysis, and each PNA was then tested in combination with each carbapenem to assess its effect on the antibiotics’ minimum inhibitory concentrations. We observed significant PNA–antibiotic interaction with five different PNAs across six combinations. Inhibition of the genes hycA, dsrB, and bolA potentiated carbapenem efficacy in CRE E. coli, whereas inhibition of the genes flhC and ygaC conferred added resistance. Our results identify resistance factors and demonstrate that transcriptomic analysis is a potent tool for designing antibiotic PNA.
In recent years, the emergence of carbapenem-resistant Enterobacteriaceae (CRE) has been marked by the World Health Organization as a critical priority for antibiotic development (1). Resistance to carbapenems, a subclass of the cell wall-targeting β-lactams that are often termed antibiotics of last resort, has become increasingly common in recent decades (2–4), resulting in rising threats of infection mortality (5, 6). In 2017, the Centers for Disease Control and Prevention estimated that, in the United States, 13,100 infections and 1,100 deaths per year were caused by CRE alone (7). Our study uses transcriptomics to better understand carbapenem resistance in a clinical isolate of multidrug-resistant (MDR) Escherichia coli (referred to from here as E. coli CUS2B), which is resistant to ertapenem but sensitive to doripenem and meropenem. Further, we use this transcriptomic analysis to design methods to combat the development of resistance. Each of these aims is facilitated by the application of our Facile Accelerated Specific Therapeutic (FAST) platform, a semiautomated pipeline for the design, synthesis, and testing of antisense peptide nucleic acids (PNAs), which allow for the sequence-specific knockdown of target genes. Here, PNAs allow us to validate our conclusions from our transcriptomic analysis and to restore carbapenem susceptibility to a CRE clinical isolate.
Many prior studies have sought to understand carbapenem resistance via the analysis of protein binding, mutation, and abundance. Structural modifications, particularly a trans-α-1-hydroxylethyl substituent at position 6, are believed to endow carbapenems with higher stability against β-lactamases and broader observed efficacy compared with other β-lactam drugs (8, 9). However, the increasing prevalence of carbapenemase enzymes in Enterobacteriaceae—such as oxacillinase-48, metallo-β-lactamases, and Klebsiella pneumoniae carbapenemases, which have been disseminated widely via plasmid conjugation (10)—provides mechanisms for the development of resistance, as these enzymes possess broad hydrolyzing activity against numerous β-lactams, including carbapenems (2, 4). Furthermore, multiple studies have found that pathogenic bacteria that do not possess carbapenemases may still acquire carbapenem resistance, and ertapenem resistance in particular, through the overproduction of extended-spectrum β-lactamases (ESBLs)—including both plasmid-encoded ESBLs and extended-spectrum chromosomal AmpC—when coupled with outer membrane porin deficiencies that discourage drug uptake (8, 11–13). Prior research has also shown that carbapenem resistance may be acquired through the expression of low-affinity or mutated penicillin-binding proteins (PBPs) (14, 15), the substrates for carbapenem action, or through the overproduction of efflux pumps (16, 17), which actively remove antibiotics from the bacterial cytoplasm.
Although fewer in number, other studies have sought to understand resistance by sequencing the transcriptome of carbapenem-resistant pathogens. Two studies on the gene expression profile of carbapenem-resistant Acinetobacter baumannii found significant up-regulation of transposable elements, recombinase, and other mutation-encouraging factors, in addition to the expected up-regulation of efflux pump and β-lactamase genes (18, 19). Four recent studies that examined transcriptomic profiles of CRE—Enterobacter cloacae, K. pneumoniae (two studies), and E. coli—identified less consistent responses, although down-regulation of porin genes and up-regulation of cell survival and β-lactamase genes were observed (20–23). Of this prior research, only one study (20) attempted to evaluate the transcriptomic response of a resistant Enterobacteriaceae strain under antibiotic challenge, the others examining only the constitutive gene expression levels of an untreated resistant pathogen.
Antisense technology offers an avenue for progress in such transcriptomic work, and likewise, transcriptomics offers targets for antisense antibiotics. As bacterial genome sequencing has become more accessible in recent years, nuclease-resistant antisense technologies—such as PNA and phosphorodiamidate morpholinos—have shown promise in targeting MDR pathogens, by binding to messenger RNA (mRNA) to inhibit translation of essential proteins for fatty acid synthesis (24) and cell division (25), as well as known resistance factors like β-lactamases (26, 27). However, a limitation of this approach, in which antisense molecules are designed against established gene targets, is the requirement that the genome of the pathogen in question be well characterized in terms of either gene essentiality or the presence of resistance genes. Measuring changes in gene expression presents an alternative semi-“black box” approach for finding such targets. Although transcriptomic analysis has recently been used to design antisense therapy against mammalian tumors (28), this approach has yet to be used for inhibiting bacterial growth or antibiotic resistance.
In this study, we demonstrate the utility of transcriptomics both as a tool to understand the role of transient gene expression in carbapenem resistance and as a strategy to discover gene targets to counter carbapenem resistance. To do this, we examined the short-term (<1-h) transcriptomic response, using both total and short RNA sequencing, of E. coli CUS2B to ertapenem and meropenem treatment. We identified genes that were potentially important to the phenotype and used our FAST platform to design and synthesize PNA molecules that bind to each gene’s corresponding mRNA and inhibit translation. Growth and cell viability assays of PNA–carbapenem combination treatments were used to validate each gene’s relevance to the resistance phenotype and to determine whether PNA designed using transcriptomics could make E. coli CUS2B susceptible to subminimum inhibitory concentration (sub-MIC) of carbapenem treatments.
Results
E. coli CUS2B: An MDR Enterobacteriaceae with Partial Carbapenem Resistance.
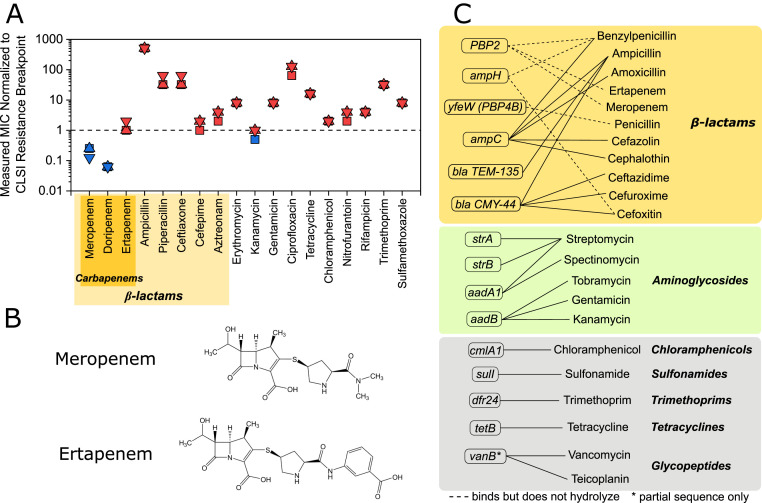
E. coli CUS2B was obtained from the University of Colorado Hospital Clinical Microbiology Laboratory’s organism bank. To validate the E. coli CUS2B resistance phenotype observed in the clinic, we measured the isolate’s MICs for a variety of antibiotics from different classes (Fig. 1A and SI Appendix, Fig. S1). We found that E. coli CUS2B was resistant to almost all antibiotics [based on break points defined by the Clinical and Laboratory Standards Institute (29)], including multiple penicillins and ertapenem. Two potent carbapenem antibiotics, meropenem and doripenem, were the only drugs to which the clinical isolate was susceptible. To investigate this partial carbapenem resistance, we focused on the E. coli CUS2B response to ertapenem and meropenem. The structures of meropenem and ertapenem differ in the pyrrolidinyl ring’s position 2 side chain (Fig. 1B). Meropenem’s substituent amide group is thought to be responsible for increased potency against gram-negative organisms in comparison with imipenem (30). At this position, ertapenem has a benzoate substituent group, which imbues the molecule with a net negative charge and increases its lipophilicity, resulting in increased plasma half-life but decreased affinity for membrane porins (31).
Fig. 1.
Resistance profile and resistance genes of E. coli CUS2B. (A) MIC of E. coli CUS2B for 18 antibiotics. The concentrations are shown normalized to the Clinical & Laboratory Standards Institute (CLSI) resistance break point for each of three replicates. Values greater than or equal to one indicate resistance, while values less than one indicate intermediate resistance or susceptibility. (B) Structure of meropenem and ertapenem. (C) Antibiotic resistance genes identified in CU2SB from whole-genome sequencing data. Solid lines: antibiotics to which the gene product confers resistance; dotted lines: antibiotics to which the gene product binds but has not been shown to hydrolyze.
Resistance Factor Identification via Whole-Genome Sequencing.
We performed whole-genome shotgun sequencing for two purposes: 1) to create a genome assembly that could be used for antisense PNA design and 2) to search for genomic contributions to the resistance phenotype. Using the Antibiotic Resistance Gene ANNOTation (ARG-ANNOT) database (32), we found that the strain encodes 15 genes related to antibiotic resistance (Fig. 1C), including 6 associated with β-lactams either as PBPs or β-lactamase enzymes. Four of these six—PBP2, PBP4B, ampH, and ampC—are encoded chromosomally and have high similarity with their homologs in E. coli reference strain MG1655 (SI Appendix, Table S1). PBP2 is known to bind both carbapenems tested and is the PBP with which meropenem and ertapenem demonstrate greatest activity (33–35). Neither PBP4B nor AmpH have been found to bind with carbapenems. The gene ampC encodes a β-lactamase enzyme that can contribute to an elevated carbapenem MIC when overexpressed by Enterbacteriaceae strains (16). The E. coli CUS2B copy of this gene has 10 altered amino acids from the reference sequence (SI Appendix, Table S1), and its −35 to −10 promoter region has four mismatched nucleotides from the same promoter region in MG1655, which could affect its expression level relative to the basal nonresistant levels in the reference strain. The remaining two genes, β-lactamases TEM-135 and CMY-44, are plasmid encoded, but neither have been associated with carbapenem resistance (36). We did not identify any gene encoding a carbapenemase or ESBL.
E. coli CUS2B also encodes the outer membrane porins OmpA, OmpC, and OmpF, the mutation or down-regulation of which may influence carbapenem efficacy (21, 37). These three proteins have 95, 90, and 90% nucleotide homology, respectively, with the corresponding genes in E. coli MG1655.
Profiling Gene Expression in Response to Ertapenem and Meropenem Treatment.
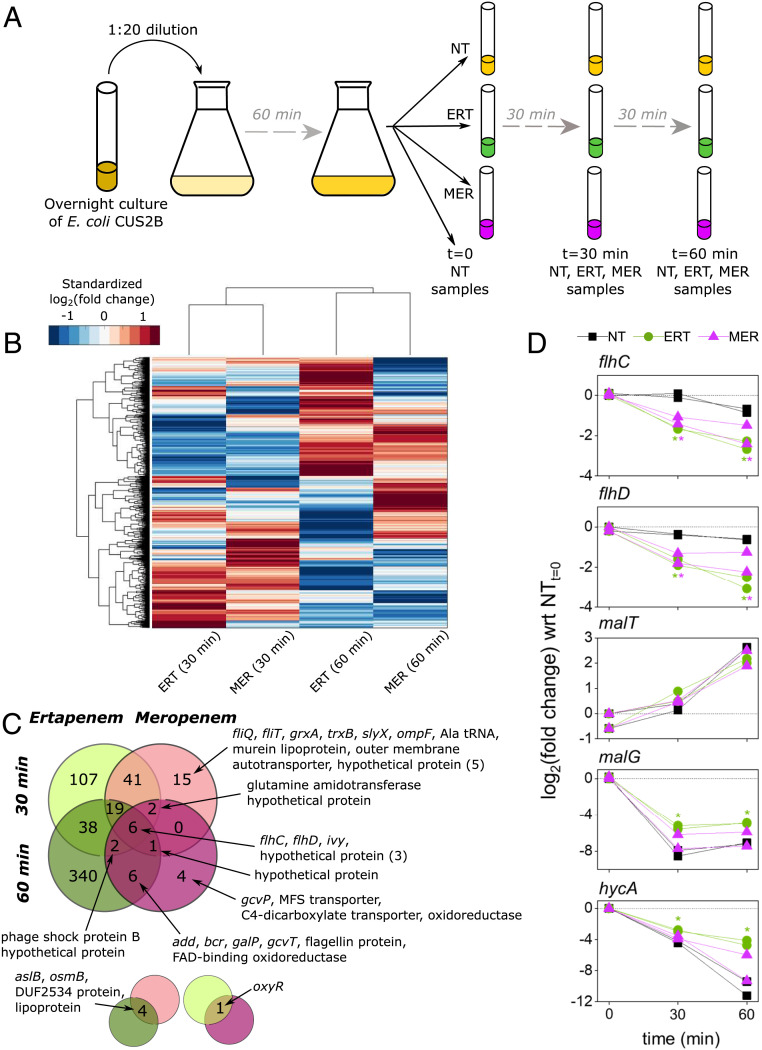
To explore possible transcriptomic contributions to the strain’s carbapenem resistance profile, we exposed exponentially growing E. coli CUS2B to ertapenem and meropenem and examined gene expression profiles after 30 and 60 min of treatment (Fig. 2A). This exposure time was selected based on observations from others that 30 to 60 min is favorable for locating gene expression changes specific to antibiotic exposure (38, 39). The short time frame also greatly reduces the likelihood that the gene expression signal will be confounded by the emergence of one or more mutant genotypes. E. coli CUS2B was diluted 1:20 from overnight cultures and grown for 1 h to exponential phase prior to treatment with 2 µg/mL of ertapenem or 1 µg/mL of meropenem. Each carbapenem concentration is half of that required to eradicate E. coli CUS2B under these culture conditions (conditions differ from Fig. 1A, MIC assay; SI Appendix, Fig. S2).
Fig. 2.
Total RNA sequencing uncovers ertapenem (ERT) response and identifies motility genes as resistance factors. (A) Overview of the sample collection protocol for RNA sequencing. Exponential-phase cells were grown with no antibiotic (NT), meropenem (MER), or ERT and collected after 0 (NT only), 30, and 60 min of growth in CAMHB. Two biological replicates were sampled for each condition. (B) Hierarchical clustering of gene expression values in ERT- and MER-treated E. coli CUS2B. Values are log2(fold change), with respect to the corresponding no treatment duplicates at the same time point. For visual clarity, the heat map has been standardized such that the mean is zero and the SD is one across each gene (each row). (C) The Venn diagram shows the degree to which significantly DE genes (expression levels compared with the no treatment condition at the given time point) overlapped across the four different conditions. Smaller pop outs are shown below to indicate overlaps on the main diagram diagonals. tRNA: transfer RNA; MFS: major facilitator superfamily; FAD: flavin adenine dinucleotide. (D) Time course of gene expression changes for the five transcripts that were explored further using PNA experiments. All conditions are normalized to the 0-min time point expression levels for the respective replicate, measured in duplicate just before antibiotic treatments were introduced. Asterisks indicate significant DE (q < 0.05) vs. the no treatment condition at the corresponding time point.
We identified differentially expressed (DE) genes by comparing the RNA-sequencing data from ertapenem- and meropenem-treated samples with an untreated control at the same time point (Dataset S1A). The DESeq R package (40) was used to evaluate significance and correct the significance statistic for multiple hypothesis testing (Materials and Methods). DE genes were identified using a q-value threshold of 0.05. General expression trends were evaluated using hierarchical clustering across genes and conditions (Fig. 2B and SI Appendix, Fig. S3). Conditions were found to cluster by time point rather than antibiotic, which suggests a generalized and transient antibiotic response. In our analysis, we detected 41 transcripts that were DE in both treatments after 30 min of exposure (Dataset S1H), 6 transcripts that were DE in both antibiotics after 60 min of exposure (Dataset S1I), and 6 transcripts that were DE in both treatments at 30 and 60 min (Fig. 2C).
Of these six genes with consistent differential expression, two, flhC and flhD, code for components of the transcriptional regulator FlhDC, which is responsible for regulating motility-associated functions such as swarming and flagellum biosynthesis (41). Both flhC and flhD genes were significantly down-regulated at 30 and 60 min (Fig. 2D). Perhaps relatedly, motility-associated gene ontology (GO) terms (GO:0040011: locomotion; GO:0071918: bacterial-type flagellum-dependent swarming motility) were significantly overrepresented within the 30-min overlapping set, accounting for 26 of the total 41 genes (SI Appendix, Table S2). All 26 were down-regulated. This effect was diminished by 60 min, with only flhC, flhD, and fliC [the gene encoding flagellin (42)] remaining significantly down-regulated.
The gene ivy, an inhibitor of bactericidal vertebrate lysozymes (43), was up-regulated in both treatments at 30 and 60 min. Both lysozymes and carbapenems disrupt peptidoglycan polymerization, although ivy is not known to interact with these antibiotics. Three other transcripts of unknown function were up-regulated in all conditions: BTW13_RS03610 (ymgD superfamily), BTW13_RS11940 (DUF1176 superfamily), and a transcript antisense to BTW13_RS17895 (putative lipoprotein, DUF1615 superfamily).
In the ertapenem response, we find many more DE genes than in the meropenem response, including 38 DE genes shared between the two time points (compared with none shared across both meropenem time points). Within this set, we observed significant overrepresentation of genes related to maltodextrin transport (mal operon, GO:0042956), under the control of the malT regulator, and the ferredoxin hydrogenase complex (hyc operon, GO:0009375), under the control of the hycA regulator (Fig. 2D) (44). All of these overrepresented genes were found to be up-regulated in ertapenem treatments, although the malT regulator itself was not found to be significantly DE between treatments (malG expression is shown in Fig. 2D as representative of the mal operon expression pattern). Only three genes were down-regulated in ertapenem at both 30 and 60 min: lptG, a member of the lipopolysaccharide transport system; phoH, an adenosine triphosphate (ATP)-binding protein; and cstA, a starvation-induced peptide transporter. Of the genes specific to the meropenem response, only the flagellar biosynthesis proteins fliQ and fliT are related, and the down-regulation of these genes did not continue to the 60-min time point.
We also searched for differential expression in outer membrane porin operon (omp) genes, previously linked to carbapenem resistance (21, 37), and resistance-related genes identified by ARG-ANNOT. Of the omp operon, only ompF was found to be significantly DE in any condition with respect to no treatment (down-regulated in meropenem, 30 min) (SI Appendix, Fig. S4), while ompA and ompC expression was not significantly different from the control in any condition. When expression levels of the ertapenem and meropenem experiments were directly compared at each time point, none of the three genes were found to be significantly DE. No resistance-related genes were DE in any condition (Dataset S1A and SI Appendix, Fig. S5).
Based on these observations, we chose three genes to target using PNA: hycA, malT, and flhC (Fig. 2D). The former two genes were chosen to probe the hyc and mal operons for their importance to resistance and their utility as antibiotic potentiation targets. The gene flhC was chosen to validate the consistent down-regulation of the FlhDC system and evaluate whether further knockout of the gene would confer greater carbapenem resistance.
Differential Expression of Short RNA Transcripts.
Next, we performed RNA sequencing in which shorter transcripts were enriched to search for resistance contributions and potential FAST PNA targets among short nucleic acids and small RNA (sRNA). sRNA have been previously shown to influence bacterial stress and antibiotic response (45, 46). We used an RNA isolation protocol designed to enrich for sRNA (Materials and Methods) prior to sequencing, and sequencing data were aligned to the E. coli UMN026 genome (the reference that maximized alignment homology) using the Rockhopper (47) pipeline, which allowed for identification of short RNA transcripts, as well as documented sRNAs.
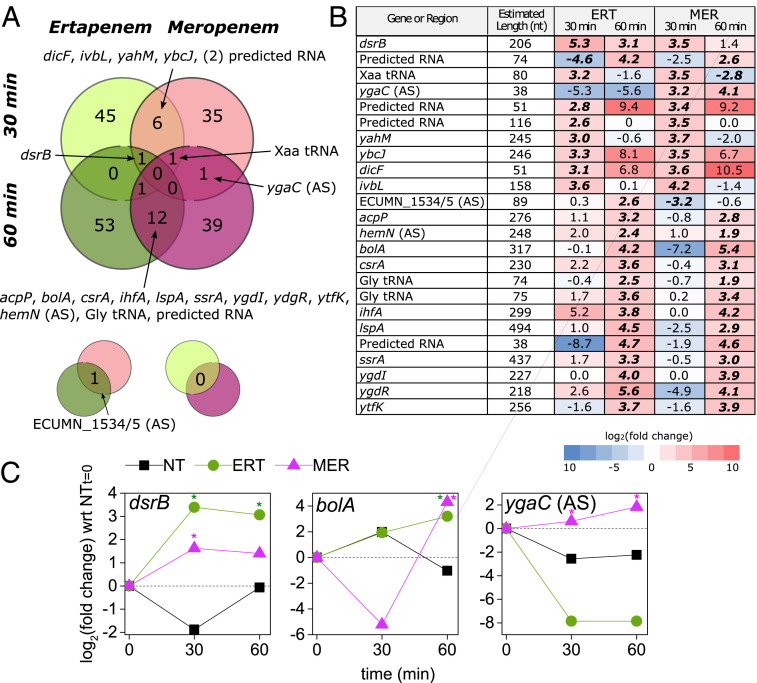
We observe more overlap of DE genes between single time points than between respective antibiotic treatments, suggesting a generalized and transient response similar to that of total RNA expression (Fig. 3A and Dataset S2A). We find 22 short RNA transcripts to be DE in at least two of the four conditions (Fig. 3B), including known regulatory sRNAs (DicF, SsrA), annotated short protein-coding genes (ilvB, acpP, bolA, csrA, ihfA, lspA), small putative protein-coding genes (dsrB, yahM, ybcJ, ygdI, ygdR, ytfK), small transcripts antisense (AS) to coding genes [ygaC (AS), hemN (AS), ECUMN_1534/5 (AS)], and predicted transcripts.
Fig. 3.
Short RNA sequencing identifies regulator genes and targets for PNA antibiotics. (A) Experimental setup was consistent between total RNA and short RNA sampling (Fig. 2A). Two biological replicates were sampled for each condition. The Venn diagram shows the degree to which significantly DE genes (expression levels compared with the no treatment [NT] condition at the given time point) overlapped across the four different conditions. Smaller pop outs are shown below to indicate overlaps on the main diagram diagonals. tRNA: transfer RNA. (B) Detail on the 22 RNAs that were DE in at least two conditions; log2(fold change) values here are with respect to the NT condition from the same time point. Bold italicized text indicates significant DE (q < 0.05) vs. the NT condition at the corresponding time point. (C) Time course of gene expression for three short RNA transcripts of interest. All conditions are normalized to the 0-min time point expression levels, measured in duplicate just before antibiotic treatments were introduced. ERT, ertapenem; MER, meropenem. *Significant DE (q < 0.05).
From these lists, we chose three genes to investigate with FAST PNA: bolA, dsrB, and ygaC. bolA was chosen based on its relation to PBPs, AmpC (48), and the cellular stress response (49), whereas the latter two were chosen to discriminate between the two carbapenem responses. The transient gene expression for bolA and dsrB is presented in Fig. 3C. We found bolA to be up-regulated in meropenem and ertapenem at 60 min, which may indicate that both antibiotics are being detected and able to activate bolA, but the subsequent response is only effective against ertapenem. dsrB was up-regulated in ertapenem at both time points but in meropenem, only at 30 min. Although the function of dsrB is unknown, it is controlled by σS, the general stress response and stationary-phase σ factor (50). Also shown in Fig. 3C is the transient gene expression for the RNA transcript antisense to ygaC (the gene itself was not found to be DE). This antisense transcript was up-regulated in meropenem at both time points but was not detected in ertapenem-treated populations. The function of ygaC is unknown, but it is controlled by the Fur transcriptional dual regulator (51).
PNA Antisense Inhibition of RNA-Sequencing Targets.
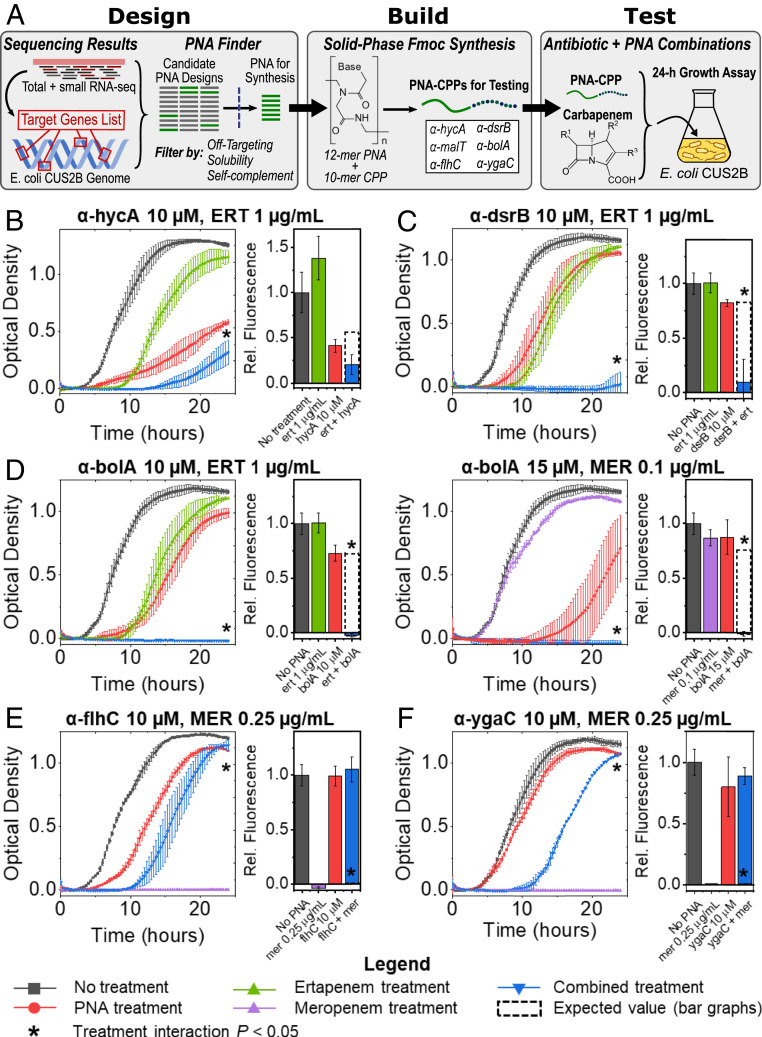
The FAST platform comprises Design, Build, and Test modules for the creation of antisense PNA (Fig. 4A). In this study, we began our design process using transcriptomic data to generate a list of target genes, which together with a whole-genome assembly and genome annotation, were used as inputs for the FAST tool PNA Finder. This tool was used to design multiple antisense PNA candidates for each gene target, with 12-mer sequences—a length that seeks to optimize both specificity (52, 53) and transmembrane transport (54)—that were complementary to mRNA nucleotide sequences surrounding the translation start codon. PNA Finder then filtered this set of candidates to minimize the number of predicted off targets within the E. coli CUS2B genome, to maximize solubility, and to avoid self-complementing sequences (Materials and Methods). For the FAST Build module, a single PNA for each gene target was selected and synthesized using fluorenylmethoxycarbonyl (Fmoc) chemistry, with a cell-penetrating peptide (CPP) composed of lysine and phenylalanine ((KFF)3K) attached at the N terminus to improve transport across the bacterial membrane. These PNAs were then tested in E. coli CUS2B cultures in combination with each carbapenem (three replicates for each condition) to determine whether the two treatments would interact as predicted. A two-way ANOVA test was used to assess interaction significance, and normalized S values (Materials and Methods) were used to compare the observed growth with the expected growth, as predicted by the Bliss Independence Model for drug combinations (55).
Fig. 4.
Application of the FAST platform to the gene targets identified in transcriptomic analysis. (A) Schematic of the FAST platform’s Design, Build, and Test modules, as they were applied to the conclusions generated by our gene expression experiments. CPP: cell-penetrating peptide. (B–F) Growth curves and end point cell viability assays for CRE E. coli PNA–antibiotic combinations, measured in triplicate. Curves are shown for experiments in which significant interaction was observed (two-way ANOVA, P < 0.05): (B) α-hycA (10 µM) combined with ertapenem (ERT; 1 µg/mL); (C) α-dsrB (10 µM) combined with ERT (1 µg/mL); (D) α-bolA (10 µM) combined with ERT (1 µg/mL) and α-bolA (15 µM) combined with meropenem (MER; 0.1 µg/mL); (E) α-flhC (10 µM) combined with MER (0.25 µg/mL); and (F) α-ygaC (10 µM) combined with MER (0.25 µg/mL). *Two-way ANOVA interaction (antibiotic and PNA conditions) P < 0.05, corrected for multiple hypothesis testing via the Benjamini–Hochberg procedure.
Based on the results of our transcriptomic analysis, we selected three genes identified by our total RNA-sequencing analysis (hycA, malT, and flhC) and three genes identified by our short RNA-sequencing analysis (bolA, dsrB, and ygaC) to be targeted by FAST PNA. Differential expression of flhC and bolA was observed in both carbapenems, while differential expression of hycA, dsrB, and the operon controlled by malT was prevalent in the ertapenem response. The transcript antisense to ygaC was up-regulated in both meropenem conditions, but the gene itself was not DE. While we suspect that the ygaC antisense transcript may regulate the gene ygaC, it has not been shown that PNA binding can interfere with antisense regulation. With this in mind, rather than attempt to inhibit transcription of the antisense transcript, we instead designed a PNA to bind to the ygaC sense transcript and inhibit protein translation to test the hypothesis that ygaC inhibition may confer greater meropenem resistance in a similar manner to the flhC-targeted PNA.
The genome assembly for the clinical isolate was used by FAST to design multiple PNA for each selected gene, which were then screened for high solubility, minimal self-complement, and zero off-target gene inhibition in E. coli CUS2B (selection criteria are detailed in Materials and Methods; SI Appendix, Table S4). PNAs were tested for binding specificity using an electrophoretic mobility shift assay (EMSA) gel shift assay and phenotypic knockdown specificity by assaying growth inhibition in corresponding Keio knockout strains (SI Appendix, Methods and Figs. S6 and S7). In these experiments, each transcriptome-derived PNA demonstrated two-mismatch discrimination in its binding, while α-ampC showed a slight reduction in binding to a two-mismatch oligonucleotide. No PNA significantly affected the strain in which its target was knocked out. Further, an anti-green fluorescent protein (GFP) PNA was designed and tested to confirm PNA direct inhibition of protein expression, and that inhibition of protein expression failed when two mismatches were introduced into the PNA sequence (SI Appendix, Fig. S8). Additionally, scrambled-sequence PNAs were designed and tested for each inhibitory PNA to control for base composition and any possible effects of the PNA or CPP independent of sequence. No effects were found with scrambled-sequence PNA alone or in any combination treatment between scrambled-sequence PNA and carbapenem antibiotic (SI Appendix, Figs. S9 and S10).
We also designed a PNA to inhibit the translation of the chromosomal β-lactamase AmpC (11, 12). This PNA was synthesized to assess the relative effectiveness of PNA targets selected using transcriptomic analysis, in comparison with those selected on a genomic basis. We have previously shown the ability to resensitize MDR bacteria by targeting β-lactamases (27). To assess PNA and antibiotic efficacy, we measured the optical density of bacterial cultures treated with each treatment for 24 h, as well as the end point cell viability via resazurin assay, which has been established both as a precise tool for measurement of drug efficacy and as an alternative to colony-forming units (SI Appendix, Fig. S15) (56, 57). In a combination of the PNA α-ampC (10 µM) with ertapenem (1 µg/mL), we observe significant synergistic interaction according to both metrics (Table 1 and SI Appendix, Fig. S11A). E. coli CUS2B cultures treated with a combination of 0.1 µg/mL meropenem with 10 µM α-ampC grew similarly to cultures treated with meropenem alone, as expected. Although the comparison of treatments in the cell viability assay did demonstrate significant interaction, the data do not seem to indicate a combinatorial effect, as the fluorescence level is virtually unchanged across the three treated conditions. Additionally, increasing the concentration of α-ampC to 15 µM could not resolve any effect, as the PNA alone was lethal at this concentration (SI Appendix, Fig. S11B).
Table 1.
Summary of combination treatment significant interactions
| PNA | 24-h growth | 24-h cell viability | ||
| Ertapenem | Meropenem | Ertapenem | Meropenem | |
| α-ampC 10 µM | 0.76 ± 0.11 | −0.06 ± 0.18 | 0.44 ± 0.22 | −0.2 ± 0.09 |
| α-hycA 10 µM | 0.23 ± 0.06 | — | 0.36 ± 0.20 | — |
| Α-dsrB 10 µM | 0.85 ± 0.06 | — | 0.73 ± 0.20 | — |
| Α-bolA 10 µM | 0.83 ± 0.02 | — | 0.75 ± 0.09 | — |
| Α-bolA 15 µM | — | 0.61 ± 0.13 | — | 0.77 ± 0.12 |
| Α-flhC 10 µM | — | −0.90 ± 0.02 | — | −1.11 ± 0.08 |
| Α-ygaC 10 µM | — | −0.93 ± 0.00 | — | −0.90 ± 0.04 |
Normalized S values (Materials and Methods) are calculated relative to drug combination predictions from the Bliss Independence Model. Plus/minus values represent SEs of the S values. Bold numbers indicate P < 0.05, after adjustment for multiple hypothesis testing.
To determine whether transcriptomic analysis could be used by FAST to produce similar antibiotic potentiation effects, E. coli CUS2B was treated with each of the six PNA at a concentration of 10 µM in combination with sub-MIC carbapenem treatments (1 µg/mL ertapenem, 0.1 µg/mL meropenem). We analyzed the cultures’ end point optical densities and cell viabilities (via resazurin assay) to assess interaction between the two treatments, based on a comparison with each individual PNA and carbapenem treatment. At these concentrations of PNA and antibiotic, we observed significant synergy between the PNAs α-hycA, α-dsrB, and α-bolA and ertapenem, with S values of 0.23, 0.85, and 0.83 for their end point optical densities, respectively (Fig. 4 B–D and Table 1). With each of these three PNAs, we performed additional interaction experiments at a PNA concentration of 15 µM with meropenem treatment to determine whether increased inhibition of these genes would demonstrate significant interaction. Of these combinations, only 15 µM α-bolA demonstrated significant synergistic interaction (S = 0.61) with meropenem (Fig. 4D, Table 1, and SI Appendix, Fig. S12). The PNA α-ygaC, α-malT, and α-flhC at concentrations of 10 µM did not exhibit significant interaction with ertapenem at 1 µg/mL or meropenem at 0.1 µg/mL (SI Appendix, Figs. S13 and S14A).
As noted above, we hypothesized that a combination of the PNA α-flhC or α-ygaC with carbapenem treatment would result in a recovery of growth and increased resistance. However, in the PNA–carbapenem combination treatments we did not observe growth recovery relative to the carbapenem-only treatment for the sub-MIC concentrations. We hypothesized that effects at such concentrations could be difficult to resolve, given that the growth curves of carbapenem-treated E. coli CUS2B reached end points similar to the untreated condition (SI Appendix, Figs. S11A and S14A). To examine this hypothesis, we treated the clinical isolate with α-flhC or α-ygaC at 10 µM in combination with ertapenem or meropenem at 2 and 0.25 µg/mL, respectively. With antibiotic alone, we observe no growth in either condition. However, we observed a recovery of growth when each PNA was added to the meropenem treatment, with the combination treatments showing significant antagonistic interaction (S = –0.9 for α-flhC and S = –0.93 for α-ygaC) (Fig. 4E and Table 1). Combination of either PNA with ertapenem showed no growth rescue (Table 1 and SI Appendix, Figs. S13A and S14B).
Discussion
In this study, we sought to combine transcriptomic analysis of carbapenem resistance with our FAST platform, both to better understand the partial carbapenem resistance profile of E. coli CUS2B and to engineer carbapenem potentiation in the clinical isolate. Our results identify genes that contribute to the strain’s resistance and introduce a strategy for the design of antisense molecules that does not rely on detailed genomic characterization of a pathogenic bacterial strain.
To begin our analysis of the clinical isolate, we first used whole-genome sequencing to search for any possible genomic resistance factors. We identified 15 genes related to antibiotic resistance, including 6 related to β-lactam antibiotics. None of these genes are dedicated carbapenemases, although the chromosomal E. coli AmpC β-lactamase is present. The basal expression of AmpC in wild-type E. coli is not enough to confer resistance to β-lactams, but mutation-induced overproduction of the chromosomal AmpC in Enterobacteriaceae has been shown to promote antibiotic resistance (11, 12, 21). Furthermore, when coupled with porin deficiencies, AmpC expression can increase enterobacterial resistance to ertapenem while retaining susceptibility to other carbapenems (12, 58, 59). Our genomic analysis identified 10 codon mutations in the E. coli CUS2B AmpC compared with E. coli MG1655, as well as four nucleotide mutations in the −35 to −10 promoter region. Furthermore, we identified numerous mutations of the E. coli CUS2B omp operon porin genes. It is possible that these mutations contribute to the ertapenem-resistant/meropenem-sensitive resistance profile that we observe in this clinical isolate. Using the FAST platform, we were able to design the PNA α-ampC to knock down the translation of the β-lactamase and determine whether it has a significant role in resistance. As expected, we observed a strong significant synergistic combination between α-ampC and ertapenem but not between α-ampC and meropenem. These observations agree with prior work from our laboratory that establish β-lactamases as viable potentiation targets and serve as a basis of comparison for our transcriptomics-based PNA.
Transcriptomic analysis of carbapenem-challenged E. coli CUS2B revealed a much greater gene expression change in response to ertapenem than to meropenem: 485 DE genes were unique to ertapenem treatment vs. 19 DE genes unique to meropenem. This may suggest the recognition of ertapenem and resultant response activation, in addition to innate basal resistance factors. In the DE analysis, we find no significant difference between the expression of outer membrane porin genes ompA, ompC, and ompF in the ertapenem experiments when compared with the meropenem experiments or the untreated conditions. Although ompF was found to be slightly down-regulated in meropenem after 30 min with respect to untreated conditions, the lack of differential expression in the ertapenem-treated conditions leads us to conclude that transcriptomic control of this gene is not a major resistance factor for E. coli CUS2B. As discussed above, the down-regulation and deletion of these genes have previously been linked to carbapenem resistance (21, 37), but if this mechanism is active in E. coli CUS2B, it does not appear to be regulated by transient gene expression. Similarly, none of the resistance-related genes identified in our genomic analysis were found to be DE in any carbapenem treatment condition, and we may conclude that transcriptomic control of these genes does not contribute to the E. coli CUS2B resistance phenotype. Notably, however, this does not rule out a constitutively higher expression of the AmpC β-lactamase nor a constitutively lower expression of porin genes as resistance factors.
To determine the transient gene expression changes that contribute to the carbapenem resistance phenotype, we analyzed differential expression overlaps between the each of the four samples (two carbapenems, two time points each). The most apparent trend was an overwhelming underexpression of motility-related genes after 30 min of exposure to both antibiotics, an effect that lessened after 60 min of exposure. This response is consistent with prior work that has noted a decreased expression of motility genes in response to generalized environmental stressors (60, 61). Two of these motility-related genes, flhC and flhD, were consistently DE in both treatments at both time points, along with the lysozyme inhibitor ivy, an RNA antisense to a putative lipoprotein, and two transcripts of unknown function.
We used antisense PNA to probe the resistance contributions of the flhDC operon. Interestingly, we observed significant antagonistic interaction between PNA inhibition of FlhC translation and meropenem treatment but not ertapenem treatment. The deletion of the FlhDC regulator has been shown in E. coli to eliminate motility and has been shown to cause down-regulation of a large number of genes that are primarily related to chemotaxis/motility and flagellar surface structures (60). It is striking that this resistance factor can be artificially induced to so effectively rescue the E. coli CUS2B strain from carbapenem challenge and that the effect is stronger than any response that the cell naturally activates, even over the course of 24 h. Furthermore, the dissonance between transcriptomic results (down-regulated in all conditions) and PNA inhibition results shows that FlhC is down-regulated as a general carbapenem response but does not universally improve survival in resistant strains. These observations point to avenues by which enterobacteria may acquire greater degrees of carbapenem resistance.
We next examined the ertapenem-specific response in an effort to use transcriptomics to engineer carbapenem potentiation. We identified a total of 38 transcripts that were uniquely DE in ertapenem-treated samples. In this set, we identified two operons—the mal operon (maltodextrin metabolism) and the hyc (formate hydrogenlyase complex) operon—that are consistently up-regulated. We designed and synthesized PNA inhibitors for malT, which while not DE, is an activator of the up-regulated mal operon, as well as the DE gene hycA, in order to evaluate and interfere with the clinical isolate’s response to ertapenem. We observed significant synergistic growth inhibition between α-hycA and ertapenem but not meropenem, while combinations of α-malT with the carbapenems did not produce significant treatment interaction.
The genes of the hyc operon code for formate hydrogenlyase complex, which mediates formate oxidation and has shown a potential connection to ATP synthesis (62). However, previous mutational analysis of hycA has provided evidence that the protein HycA works as a negative regulator of this system, as increased formate dehydrogenase activity was observed following its deletion (63, 64). Based on our observations, the up-regulation of the formate hydrogenlyase complex is likely important to the E. coli CUS2B’s ertapenem resistance but requires commensurate up-regulation of the HycA regulator. The absence of this regulator results in the observed synergistically toxic effect when α-hycA is combined with ertapenem, an effect not observed in the meropenem response because the system is not activated by the antibiotic. These results validate the role of formate metabolism in the carbapenem response and support the conclusion that transcriptomic data are valuable for engineering antibiotic potentiation in resistant enterobacteria.
In addition to applying FAST PNA to gene targets identified by total RNA sequencing, we also analyzed differential expression of short RNA transcripts in search of potential resistance factors. We identified 22 transcripts that were DE across one or more of the treatment conditions and time points and selected three genes to be targeted by FAST: dsrB, bolA, and ygaC. dsrB and bolA are translated into short proteins, but no annotation for the ygaC antisense complement has been identified, leading to the hypothesis that the transcript serves as a regulatory RNA for the gene.
Perhaps the most dramatic result of this study was the effectiveness of α-dsrB and α-bolA in restoring carbapenem susceptibility to E. coli CUS2B. At 10 µM, each PNA showed significant synergistic interaction with sub-MIC ertapenem, and α-bolA showed significant synergistic interaction with sub-MIC meropenem at a concentration of 15 µM. Surprisingly, each S value for these three combinations (0.85, 0.83, and 0.61) was comparable with the value of α-ampC (S = 0.76). Each of these results is consistent with the predictions of the RNA sequencing: dsrB was found to be up-regulated at both time points for ertapenem but only at 30 min for meropenem, and bolA was found to be up-regulated in both at 60 min. Although little is known about dsrB, it is reported to be regulated by the general stress response σ factor RpoS (50). Inhibition of DsrB translation abolished bacterial growth in combination with ertapenem (S = 0.85) (Fig. 4), which is consistent with previous research that has associated β-lactam antibiotics with induction of the RpoS general stress response regulon.
Importantly, our results confirm that activation of DsrB is specific to the transcriptome of a resistance phenotype and that its up-regulation is a necessary component of the CRE response in this clinical isolate. BolA is known to be induced under a variety of stress conditions (49) and is involved in biofilm induction (65) and protective morphological changes (48) for E. coli cells. Perhaps most important, however, is that BolA has been shown to control expression of the proteins PBP5 and PBP6 in E. coli, as well as the β-lactamase AmpC (48). Although neither E. coli PBP5 nor PBP6 have been shown to bind meropenem or ertapenem—these antibiotics are known to bind preferentially to PBP2 and PBP3 (14, 33)—our previous results indicate that AmpC contributes to the observed ertapenem resistance phenotype. The effect of α-bolA differs from that of α-ampC, however, in its effect on meropenem efficacy. This finding provides strong evidence that BolA is important to the general carbapenem response in E. coli and is not merely incidentally up-regulated in the meropenem response, as might be concluded from transcriptomic analysis alone. Additionally, the success of both the α-dsrB and α-bolA in PNA–carbapenem combination treatments—in three experiments fully abolishing growth (Fig. 4)—represents transcriptomic analysis that has been used to design PNA antibiotics and importantly, shows equal if not better efficacy than the genome-derived PNA α-ampC. Further development of this strategy will aid in the design of antisense inhibitors to target pathogenic species about which less information—essential genes, genome annotation, etc.—is readily available.
The final PNA for which we find interaction with carbapenem treatment is α-ygaC. Similar to α-flhC, this PNA showed the ability to rescue growth in E. coli CUS2B in cultures treated with previously lethal meropenem concentrations but did not replicate this effect in ertapenem-treated cultures. Although we observed overexpression of the unannotated RNA transcript antisense to ygaC, not the gene itself, the PNA demonstrated surprising effectiveness at inducing resistance. These results affirm that the ygaC antisense transcript is likely controlling the gene’s expression and suggest that ygaC down-regulation is associated with a more broadly resistant phenotype. The growth rescue in meropenem-treated conditions by both α-ygaC and α-flhC points to routes that resistant bacteria may take toward developing broad resistance. Furthermore, the success of α-ygaC, considering that it is a largely unstudied gene never before linked to antibiotic resistance, demonstrates the specific utility of transcriptomics in the design of PNA for antibiotic applications.
Resistance has been extensively studied at the genetic level, which has helped researchers to understand many genetic factors that can contribute to resistance. However, carbapenem resistance is infrequently studied at a transcriptomic level, and when it is, the research almost exclusively examines basal expression levels (18–23). In this work, we profiled the short-term transient transcriptomic responses of a partially carbapenem-resistant clinical isolate of E. coli and used our FAST platform to design PNA that uncovered the importance of multiple systems in the development of this phenotype, including the regulators flhC, hycA, and bolA. Furthermore, by inhibiting hycA, bolA, and dsrB with FAST PNA, we were able to potentiate sub-MIC carbapenem concentrations in cultures of the clinical isolate. There are no small molecules known to inhibit the action of any of these gene targets, which demonstrates the value of PNA inhibitors to fill in the gaps in modern antibiotic discovery. While much prior research into antibiotic PNA has picked specific gene targets based on well-established resistance factors or gene essentiality, our study uses transcriptomic analysis to identify targets for antibiotic PNA applications. The strategy shows a striking effectiveness in its induction of carbapenem susceptibility in the CRE E. coli clinical isolate: we find two such PNA–carbapenem combinations that are more effective than even the anti–β-lactamase PNA α-ampC. In future research, we will seek to develop these results into a strategy to consistently and systematically design PNAs that potentiate carbapenem efficacy in MDR pathogens and thus, maintain the efficacy of these antibiotics of last resort.
Materials and Methods
Strains and Culture Conditions.
E. coli CUS2B was provided by Nancy Madinger, University of Colorado Hospital Clinical Microbiology Laboratory’s organism bank (Aurora, CO). The isolate was obtained via rectal swab from a 29-y-old pregnant female patient. This strain was streaked on solid medium plates of cation-adjusted Mueller Hinton broth (CAMHB) with 100 µg/mL ampicillin and 15 g/L agar and grown at 37 °C for 16 h to produce individual colonies for biological replicates in growth experiments. For these growth experiments, unless otherwise mentioned, E. coli CUS2B was propagated in aerobic conditions at 37 °C in liquid cultures of CAMHB with shaking at 225 rpm.
For the knockout assays with PNA, six strains from the Keio collection (66)—knockouts for the genes flhC, hycA, dsrB, bolA, ygaC, and ampC—were streaked separately onto solid plates of Luria–Bertani (LB) broth with 50 µg/mL kanamycin and 15 g/L agar and grown for 16 h at 37 °C. For the fluorescence knockdown experiment, GFP (obtained from pAKgfp1; Addgene #14076) was cloned into E. coli DH5αZ1 (Expressys) under control of a pLac promoter, and this strain was streaked onto a solid plate of LB broth with 50 µg/mL kanamycin and 15 g/L agar and grown for 16 h at 37 °C.
MIC Assays.
Three colonies were picked from an E. coli CUS2B plate and used to inoculate three separate overnight cultures in 1 mL CAMHB each. After 16 h, the cultures were diluted to a 0.5 McFarland standard in a 96-well plate and were treated with each antibiotic (Fig. 1A) at a range of concentrations (serially diluted in twofold increments) that spanned the Clinical and Laboratory Standards Institute MIC resistance break point (29). Growth in the plate was monitored with a Tecan GENios (Tecan Group Ltd.) running Magellan software (v7.2) at an absorbance of 590 nm every 20 min for 16 h, with shaking between measurements. The MIC was identified as the lowest antibiotic concentration preventing growth.
Genome Sequencing.
Five colonies were picked from an E. coli CUS2B plate and resuspended in liquid culture. After 16 h, 1 mL of culture was used for genomic DNA isolation with the Wizard DNA Purification Kit (Promega). Approximately 2 µg of DNA was used to prepare a paired-end 250-bp sequencing library with the Nextera XT DNA library kit. The library was sequenced on an Illumina MiSeq, resulting in 407,910 reads (20× coverage). The de novo assembly is 5,325,941 bp in length with a guanine/cytosine content of 50.59%. The largest contig is 394,969 bp, and the N50 (length-weighted median) contig length is 100,215. The genome contains 5,360 protein-coding sequences, 114 RNA-coding sequences, 82 transfer RNAs, 11 non-coding RNAs, 260 pseudogenes, and two CRISPR arrays.
The FASTQ files were filtered for quality using Trimmomatic (v0.32) (67) in sliding window mode with a window size of 4 bases, a minimum average window quality of 15 (phred 33 quality score), and a read length of at least 36 bases. For resequencing, reads were aligned to various E. coli RefSeq reference genomes using Bowtie 2 (v2.2.3) (68). SAMTools (v0.1.19) (69) was used to remove PCR duplicates and create indexed, sorted binary alignment/map (BAM) files. Variants were called and filtered using the Genome Analysis Toolkit (v2.4–9) (70). To pass the filter, a single-nucleotide polymorphism had to meet the following criteria: Qualby Depth (QD) < 2.0, Fisher strand (FS)> 60, mapping quality < 40.0, ReadPosRankSum < −2.0, and MappingQualityRankSum < −12.5. Filter criteria for indels was QD < 2.0, FS > 60.0, and ReadPosRankSum < −2.0. A custom Python script was used to annotate variant call files using the corresponding general feature format (GFF) file from RefSeq. For de novo assembly, reads were assembled using SPAdes (v 3.5.0) (71), and annotation was performed with the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (v4.0) (72). Quality of the assembly was assessed using QUAST (73). The FASTA generated by SPAdes was used for multilocus sequence typing (74), identification of resistance genes with ARG-ANNOT (32), and locating plasmids with PlasmidFinder (75).
RNA Sequencing and Differential Expression Analysis.
Two colonies were picked from an E. coli CUS2B plate and resuspended in liquid culture. After 16 h of growth, the culture was diluted 1:20 into duplicate 15-mL cultures. These were grown for 1 h; then, 3 mL from each were preserved in 2 vol of RNAprotect. Each culture was divided into three equal parts. No antibiotic was added to one part, and antibiotics were added to the other two for final concentrations of 2 μg/mL of ertapenem or 1 μg/mL of meropenem, corresponding to 50% of the MIC under these growth conditions (note that these conditions are different from the procedures used to determine the MICs in Fig. 1). After 30 and 60 min of growth, 1.5 mL of culture was collected from each and stored in RNAprotect. Cultures were flash frozen in ethanol and dry ice and stored at −80 °C until the time of RNA extraction.
To extract RNA, samples were thawed and resuspended in 100 μL Tris ethylenediaminetetraacetic acid (EDTA) buffer with 0.4 mg/mL lysozyme and proteinase K. After incubation at room temperature for 5 min, 300 μL of lysis buffer with 20 μL/mL β-mercaptoethanol was added to each and vortexed to mix. Each lysis solution was split in half, with one-half being processed for total RNA isolation and the other for short RNA isolation. Total RNA was isolated using the GeneJet RNA Purification kit (Thermo Scientific) followed by deoxyribonuclease (DNase) treatment with the TURBO DNA-free kit (Ambion). Short RNA was isolated using the mirVana miRNA isolation kit (Thermo Scientific). Concentration and absorbance at 260 nm and 280 nm were measured on a Nanodrop 2000 (Thermo Scientific). A minimum of 130 ng of RNA per sample was submitted for sequencing library preparation. Quality was further assessed with a Bioanalyzer (Agilent). Libraries were prepared using the RNAtag-Seq protocol (76), wherein individual samples are bar coded and pooled prior to ribosomal RNA treatment and complementary DNA synthesis. Here, the total RNA samples were combined into one pool, and the short RNA-enriched samples were combined into a separate pool. The total RNA pool was fragmented via incubation with FastAP buffer at 94 °C. After another DNase treatment, bar-coded adapters were ligated with T4 DNA ligase; then, the samples were pooled and subjected to Ribo-Zero treatment. The prep for short RNA libraries was similar, without the fragmentation or ribosomal RNA treatment steps. Total RNA libraries were sequenced on a NextSeq 500 (Illumina) using a high-output cycle 75-cycle run. Short RNA libraries were sequenced on a NextSeq with a medium-output run, halted after 75 cycles.
FASTQ files were demultiplexed with the bar-code splitter function from the FastX toolbox (http://hannonlab.cshl.edu/fastx_toolkit; v0.0.13.2). The first seven base calls (containing the bar code) were trimmed using FastX; then, adapters were removed, and all reads were trimmed for quality using the sliding window mode in Trimmomatic (v0.32) (67). Quality of the resulting FASTQ files was validated with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). For total RNA data (including sense and antisense transcripts) and differential expression analysis, reads were aligned to the draft assembly of the CUS2B genome (available as RefSeq GCF_001910475.1) with Bowtie 2 (v2.2.3). An average of 16.5 ± 3.2 million reads was successfully mapped per sample. SAMTools (v0.1.19) was used to create BAM files. HTSeq (v0.6.1) (77) was used to build count tables for sense and antisense reads. DESeq 40 was used to determine DE genes, with a pooled dispersion metric and a parametric fit. Genes were considered significantly DE if the Benjamini–Hochberg-adjusted q value was less than 0.05. For short RNA data, trimmed FASTQ files were submitted to Rockhopper (47), which mapped to the E. coli UMN026 genome (90,152 ± 25,642 reads mapped per sample) and performed differential expression analysis. Short RNA transcripts were considered significantly DE if the q value was less than 0.05.
PNA Design.
PNA design was carried out using our laboratory’s PNA Finder toolbox. The toolbox is built using Python 2.7, the alignment program Bowtie 2 (68), the read alignment processing program SAMtools (69), and the feature analysis program BEDTools (78). The toolbox takes a user-provided list of gene identifications and cross-references the identifications against a genome annotation file to determine the feature coordinates for each identification. The toolbox then uses these coordinates to extract PNA target sequences of a user-specified length (12 bases in this study) and user-specified positions relative to the start codon from a genome assembly FASTA file. PNA Finder provides a list of PNA candidates (the reverse complements of the target sequences) and sequence warnings regarding solubility and self-complement. Based on design rules previously established by Gildea and Coull (79), PNAs were flagged as potentially insoluble and rejected if, within any stretch of 10 bases, they contained five purines in a row, four guanines in a row, or more than six total purines. PNAs were flagged as potentially self-complementing and rejected if they contained a stretch of more than seven bases that complemented another section of the same PNA, by forward or reverse alignment. Finally, PNA Finder screens the list of PNA candidates against a user-provided genome assembly (in this study, the genome for E. coli CUS2B) to search for off targets. Potential off targets for bacterial translation inhibition are defined as zero- or one-mismatch alignments within 20 bases of the translation start codon, a criterion that was developed in previous work (27). This analysis was used to select the candidates with the fewest potential off targets and create a final PNA list for synthesis. Scrambled-sequence versions of the selected PNA were designed by using a PNA Finder script to produce 100 different randomly shuffled sequences of the same base composition (respectively for each PNA). These were similarly screened for solubility, self-complements, and off targets.
PNA Synthesis.
PNAs were synthesized using an Apex 396 peptide synthesizer (AAPPTec, LLC) with solid-phase Fmoc chemistry at a 10-µmol scale on 4-methylbenzhydrylamine (MBHA) rink amide resin. Fmoc–PNA monomers were obtained from PolyOrg Inc., with A, C, and G monomers protected with Bhoc groups. PNAs were synthesized with the N-terminal cell-penetrating peptide (KFF)3K. Cell-penetrating peptide Fmoc monomers were obtained from AAPPTec, LLC, and lysine monomers were protected with Boc groups. PNA products were precipitated in diethyl ether and purified as trifluoroacetic acid salts via reverse-phase high-performance liquid chromatography using a C18 column. PNAs were stored at −20 °C dissolved in 5% vol/vol dimethyl sulfoxide in water.
PNA–Antibiotic Growth Curve Interaction Assays.
Three colonies were picked from an E. coli CUS2B plate and used to inoculate three separate overnight cultures in 1 mL CAMHB each. After 16 h, the culture was diluted 1:10,000 in a 384-well microplate using three biological replicates per condition. The total culture volume for each treatment was 50 µL. Growth in the plate was monitored with a Tecan GENios (Tecan Group Ltd.) plate reader running Magellan software (v 7.2) at an absorbance of 590 nm every 20 min for 24 h, with shaking between measurements.
Resazurin Cell Viability Interaction Assays.
Cell viability was measured at the end point of each 24-h PNA–antibiotic interaction growth curve. Assay conditions were optimized to improve sensitivity of fluorescence measurement and to avoid saturation of the resazurin dye during the incubation period. Cells were diluted 1:20 into a new plate and incubated with 22 μM resazurin (Sigma-Aldrich) for 2 h. Fluorescence was measured at an excitation wavelength of 485 nm and an emission wavelength of 610 nm using a Tecan GENios (Tecan Group Ltd.) plate reader running Magellan software (v 7.2).
Treatment Interaction Calculation.
Interaction effects for both 24-h end point optical density and cell viability (via resazurin) were evaluated for significance using a two-way ANOVA test and the Benjamini–Hochberg procedure to correct for multiple testing, and S values were calculated with respect for the expected growth inhibition as calculated by the Bliss Independence Model (55). The S value for a given time point was calculated as follows:
| [1] |
For a given time point (24 h were used in our analyses), the variable ODAB represents optical density with only carbapenem treatment, OD0 represents the optical density without treatment, ODPNA represents optical density with only antisense PNA treatment, and ODAB, PNA represents the optical density with a combination treatment of antibiotic (AB) and PNA. Plus/minus values for S values (Table 1) were calculated by propagating SE values for each term in Eq. 1.
Other Software and Resources Utilized.
The clustergram function from MATLAB’s Bioinformatics toolbox was used for building heat maps and dendrograms. A Euclidean distance metric, optimal leaf ordering, and average linkage function were used for clustering. Ecocyc (80) was used to gain gene names and descriptions and to define functional classes. NCBI’s BLAST (81) was used to predict gene function and to determine similarity of sequences in CUS2B to other bacterial strains. PANTHER (44) was used for statistical overrepresentation tests, with a Bonferroni correction applied to all reported P values for such tests.
Supplementary Material
Acknowledgments
This work was funded by University of Colorado Dean’s Graduate research grants (to T.R.A. and K.E.E.), the University of Colorado Graduate Assistance in Areas of National Need Fellowship (to T.R.A.), the W. M. Keck Foundation, Defense Advanced Research Projects Agency Young Faculty Award D17AP00024 (to A.C.), National Aeronautics and Space Administration Cooperative Agreement Notice–Translational Research Institute for Space Health Grant NNX16A069A (to A.C.), and Lab Venture Challenge Award (to A.C.). We thank the University of Colorado BioFrontiers Institute Next-Gen Sequencing Core Facility, which performed all Illumina sequencing and library construction.
Footnotes
Competing interest statement: T.R.A and A.C. have a patent on the FAST platform. A.C. is the founder of a start-up company Sachi Bioworks, Inc. based on this technology.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922187117/-/DCSupplemental.
Data Availability.
The whole-genome shotgun sequencing data have been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank (accession no. MSDR00000000.1) (82). The version described in this paper is version MSDR01000000. The RNA-sequencing data have been deposited in NCBI’s Sequence Read Archive (accession no. SRP101716) (83). PNA Finder scripts can be downloaded from GitHub at https://github.com/taunins/pna_finder.
References
- 1.Tacconelli E., Magrini N., Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization (2017). https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. Accessed 15 November 2019.
- 2.Gupta N., Limbago B. M., Patel J. B., Kallen A. J., Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 53, 60–67 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Johnson A. P., Woodford N., Global spread of antibiotic resistance: The example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 62, 499–513 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P., Naas T., Poirel L., Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) , Vital signs: Carbapenem-resistant Enterobacteriaceae. MMWR Morb. Mortal. Wkly. Rep. 62, 165–170 (2013). [PMC free article] [PubMed] [Google Scholar]
- 6.Tamma P. D., et al. , Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin. Infect. Dis. 64, 257–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC) , Biggest threats and data: 2019 AR threats report (2019). https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed 15 November 2019.
- 8.Zhanel G. G., et al. , Comparative review of the carbapenems. Drugs 67, 1027–1052 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Miyashita K., Massova I., Mobashery S., Quantification of the extent of attenuation of the rate of turnover chemistry of the TEM-1 beta-lactamase by the alpha-1R-hydroxyethyl group in substrates. Bioorg. Med. Chem. Lett. 6, 319–322 (1996). [Google Scholar]
- 10.Queenan A. M., Bush K., Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillon H., Tande D., Mammeri H., Emergence of ertapenem resistance in an Escherichia coli clinical isolate producing extended-spectrum beta-lactamase AmpC. Antimicrob. Agents Chemother. 55, 4443–4446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mammeri H., Nordmann P., Berkani A., Eb F., Contribution of extended-spectrum AmpC (ESAC) β-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol. Lett. 282, 238–240 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Lartigue M.-F., Poirel L., Poyart C., Réglier-Poupet H., Nordmann P., Ertapenem resistance of Escherichia coli. Emerg. Infect. Dis. 13, 315–317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhanel G. G., et al. , Ertapenem: Review of a new carbapenem. Expert Rev. Anti Infect. Ther. 3, 23–39 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Livermore D. M., Oakton K. J., Carter M. W., Warner M., Activity of ertapenem (MK-0826) versus Enterobacteriaceae with potent beta-lactamases. Antimicrob. Agents Chemother. 45, 2831–2837 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore D. M., Of Pseudomonas, porins, pumps and carbapenems. J. Antimicrob. Chemother. 47, 247–250 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Köhler T., Michea-Hamzehpour M., Epp S. F., Pechere J.-C., Carbapenem activities against Pseudomonas aeruginosa: Respective contributions of OprD and efflux systems. Antimicrob. Agents Chemother. 43, 424–427 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin H., et al. , Comparative transcriptomics of multidrug-resistant Acinetobacter baumannii in response to antibiotic treatments. Sci. Rep. 8, 3515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang K.-C., et al. , Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genomics 15, 815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low Y. M., et al. , Elucidating the survival and response of carbapenem resistant Klebsiella pneumoniae after exposure to imipenem at sub-lethal concentrations. Pathog. Glob. Health 112, 378–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majewski P., et al. , Altered outer membrane transcriptome balance with AmpC overexpression in carbapenem-resistant Enterobacter cloacae. Front. Microbiol. 7, 2054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong H.-K., et al. , Fine-tuning carbapenem resistance by reducing porin permeability of bacteria activated in the selection process of conjugation. Sci. Rep. 8, 15248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M., et al. , Network integrative genomic and transcriptomic analysis of carbapenem-resistant Klebsiella pneumoniae strains identifies genes for antibiotic resistance and virulence. mSystems 4, e00202-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otsuka T., et al. , Antimicrobial activity of antisense peptide-peptide nucleic acid conjugates against non-typeable Haemophilus influenzae in planktonic and biofilm forms. J. Antimicrob. Chemother. 72, 137–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh S., Loeffler A., Lloyd D. H., Nair S. P., Good L., Oxacillin sensitization of methicillin-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus pseudintermedius by antisense peptide nucleic acids in vitro. BMC Microbiol. 15, 262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sully E. K., et al. , Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo. J. Antimicrob. Chemother. 72, 782–790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Courtney C. M., Chatterjee A., Sequence-specific peptide nucleic acid-based antisense inhibitors of TEM-1 β-lactamase and mechanism of adaptive resistance. ACS Infect. Dis. 1, 253–263 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Annalora A. J., et al. , A k-mer based transcriptomics approach for antisense drug discovery targeting the Ewing’s family of tumors. Oncotarget 9, 30568–30586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute , Performance Standards for Antimicrobial Disk Susceptibility Tests (Clinical and Laboratory Standards Institute, Wayne, PA, 2015). [Google Scholar]
- 30.Craig W. A., The pharmacology of meropenem, a new carbapenem antibiotic. Clin. Infect. Dis. 24 (suppl. 2), S266–S275 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Hammond M. L., Ertapenem: A group 1 carbapenem with distinct antibacterial and pharmacological properties. J. Antimicrob. Chemother. 53 (suppl. 2), ii7–ii9 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Gupta S. K., et al. , ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., Bhachech N., Bush K., Biochemical comparison of imipenem, meropenem and biapenem: Permeability, binding to penicillin-binding proteins, and stability to hydrolysis by beta-lactamases. J. Antimicrob. Chemother. 35, 75–84 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Kohler J., et al. , In vitro activities of the potent, broad-spectrum carbapenem MK-0826 (L-749,345) against broad-spectrum beta-lactamase-and extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 43, 1170–1176 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odenholt I., Ertapenem: A new carbapenem. Expert Opin. Investig. Drugs 10, 1157–1166 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Alcock B. P., et al. , CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oteo J., et al. , Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int. J. Antimicrob. Agents 32, 534–537 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Bailey A. M., et al. , Exposure of Escherichia coli and Salmonella enterica serovar Typhimurium to triclosan induces a species-specific response, including drug detoxification. J. Antimicrob. Chemother. 64, 973–985 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Shaw K. J., et al. , Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5, 105–122 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soutourina O., et al. , Multiple control of flagellum biosynthesis in Escherichia coli: Role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181, 7500–7508 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohnishi K., Kutsukake K., Suzuki H., Iino T., Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221, 139–147 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Callewaert L., et al. , Purification of Ivy, a lysozyme inhibitor from Escherichia coli, and characterisation of its specificity for various lysozymes. Enzyme Microb. Technol. 37, 205–211 (2005). [Google Scholar]
- 44.Mi H., et al. , PANTHER version 7: Improved phylogenetic trees, orthologs and collaboration with the gene ontology consortium. Nucleic Acids Res. 38, D204–D210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoe C.-H., Raabe C. A., Rozhdestvensky T. S., Tang T.-H., Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 303, 217–229 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Dersch P., Khan M. A., Mühlen S., Görke B., Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8, 803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClure R., et al. , Computational analysis of bacterial RNA-seq data. Nucleic Acids Res. 41, e140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos J. M., Lobo M., Matos A. P. A., De Pedro M. A., Arraiano C. M., The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45, 1729–1740 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Santos J. M., Freire P., Vicente M., Arraiano C. M., The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 32, 789–798 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Sledjeski D. D., Gupta A., Gottesman S., The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15, 3993–4000 (1996). [PMC free article] [PubMed] [Google Scholar]
- 51.Vassinova N., Kozyrev D., A method for direct cloning of Fur-regulated genes: Identification of seven new fur-regulated loci in Escherichia coli. Microbiology (Reading) 146, 3171–3182 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Doyle D. F., Braasch D. A., Simmons C. G., Janowski B. A., Corey D. R., Inhibition of gene expression inside cells by peptide nucleic acids: Effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry 40, 53–64 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Good L., Nielsen P. E., Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 95, 2073–2076 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Good L., Awasthi S. K., Dryselius R., Larsson O., Nielsen P. E., Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19, 360–364 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Bliss C. I., The toxicity of poisons applied jointly. Ann. Appl. Biol. 26, 585–615 (1939). [Google Scholar]
- 56.Sarker S. D., Nahar L., Kumarasamy Y., Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 42, 321–324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rampersad S. N., Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors (Basel) 12, 12347–12360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livermore D. M., Sefton A. M., Scott G. M., Properties and potential of ertapenem. J. Antimicrob. Chemother. 52, 331–344 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Jacoby G. A., Mills D. M., Chow N., Role of beta-lactamases and porins in resistance to ertapenem and other beta-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 3203–3206 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao K., Liu M., Burgess R. R., Adaptation in bacterial flagellar and motility systems: From regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 35, 4441–4452 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erickson K. E., Otoupal P. B., Chatterjee A., Transcriptome-level signatures in gene expression and gene expression variability during bacterial adaptive evolution. mSphere 2, e00009-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDowall J. S., et al. , Bacterial formate hydrogenlyase complex. Proc. Natl. Acad. Sci. U.S.A. 111, E3948–E3956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauter M., Böhm R., Böck A., Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol. Microbiol. 6, 1523–1532 (1992). [DOI] [PubMed] [Google Scholar]
- 64.Leonhartsberger S., Korsa I., Böck A., The molecular biology of formate metabolism in enterobacteria. J. Mol. Microbiol. Biotechnol. 4, 269–276 (2002). [PubMed] [Google Scholar]
- 65.Dressaire C., Moreira R. N., Barahona S., Alves de Matos A. P., Arraiano C. M., BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio 6, e02352-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baba T., et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKenna A., et al. , The genome analysis toolkit: A mapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tatusova T., et al. , NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurevich A., Saveliev V., Vyahhi N., Tesler G., QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsen M. V., et al. , Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carattoli A., et al. , In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shishkin A. A., et al. , Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anders S., Pyl P. T., Huber W., HTSeqA python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gildea B. D., Coull J. M., “Methods for modulating the solubility of synthetic polymers.” US Patent 6770442 (2001).
- 80.Keseler I. M., et al. , EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39, D583–D590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson M., et al. , NCBI BLAST: A better web interface. Nucleic Acids Res. 36, W5–W9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Erickson K. E., Chatterjee A., Escherichia coli strain CUS2B, whole genome shotgun sequencing project. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MSDR00000000.1. Deposited 15 December 2016.
- 83.Erickson K. E., Chatterjee A., RNAseq: CRE E. coli strain CUS2B. NCBI Sequence Read Archive. https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP101716. Deposited 10 March 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome shotgun sequencing data have been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank (accession no. MSDR00000000.1) (82). The version described in this paper is version MSDR01000000. The RNA-sequencing data have been deposited in NCBI’s Sequence Read Archive (accession no. SRP101716) (83). PNA Finder scripts can be downloaded from GitHub at https://github.com/taunins/pna_finder.