Abstract
BACKGROUND:
Biomarkers are needed for noninvasive early detection of gastric cancer (GC). We investigated salivary extracellular RNA (exRNA) biomarkers as potential clinical evaluation tools for GC.
METHODS:
Unstimulated whole saliva samples were prospectively collected from 294 individuals (163 GC and 131 non-GC patients) who underwent endoscopic evaluation at the Samsung Medical Center in Korea. Salivary transcriptomes of 63 GC and 31 non-GC patients were profiled, and mRNA biomarker candidates were verified with reverse transcription quantitative real-time PCR (RT-qPCR). In parallel, microRNA (miRNA) biomarkers were profiled and verified with saliva samples from 10 GC and 10 non-GC patients. Candidate biomarkers were validated with RT-qPCR in an independent cohort of 100/100 saliva samples from GC and non-GC patients. Validated individual markers were configured into a best performance panel.
RESULTS:
We identified 30 mRNA and 15 miRNA candidates whose expression pattern associated with the presence of GC. Among them, 12 mRNA and 6 miRNA candidates were verified with the discovery cohort by RT-qPCR and further validated with the independent cohort (n = 200). The configured biomarker panel consisted of 3 mRNAs (SPINK7, PPL, and SEMA4B) and 2 miRNAs (MIR140-5p and MIR301a), which were all significantly down-regulated in the GC group, and yielded an area under the ROC curve (AUC) of 0.81 (95% CI, 0.72–0.89). When combined with demographic factors, the AUC of the biomarker panel reached 0.87 (95% CI, 0.80–0.93).
CONCLUSIONS:
We have discovered and validated a panel of salivary exRNA biomarkers with credible clinical performance for the detection of GC. Our study demonstrates the potential utility of salivary exRNA biomarkers in screening and risk assessment for GC.
Gastric cancer (GC)8 is the fourth most common cancer diagnosed globally and the third leading cause of cancer-related deaths (1). It is the leading cancer type diagnosed in East Asian countries (2). In Korea, genetics (3), diets containing salted and preserved foods (4), smoking (5), and a high prevalence of Helicobacter pylori infections (6) play a role in a large percentage of the population with GC. In Korea, most early-stage GCs are identified in asymptomatic individuals (74.2%–78.1%) compared with symptomatic individuals (25.9%–35.7%) (7). Once the disease progresses and results in serious symptoms and complications, the prognosis is poor and the survival rate decreases from approximately 65% (when found at an early stage) to <20% (8). In 1999, owing to the high prevalence of GC, the National Cancer Screening Program in Korea implemented an ongoing early detection program that recommends everyone over the age of 40 years undergo an upper endoscopy every other year (9). However, an upper endoscopy for GC detection is costly, time-consuming, and invasive. Since the screening program began, <30% of the targeted population participated (10). Thus, there is a need for predictive biomarkers that can be used as a credible screening tool for early detection of GC. These biomarkers are highly desirable to improve the outcome of the disease and reduce unnecessary endoscopies.
Several studies have looked for potential biomarkers in serum as a noninvasive method for screening GC. The serum pepsinogens I and II at low concentrations and their ratio have been considered indicators for preneoplastic gastric lesions, but results have varied depending on the location of cancer and the cutoff values used in different studies (11). The most frequently used gastric tumor markers, such as carcinoembryonic antigen, CA19-9, CA-50, and CA72-4, have reported area under the ROC curve (AUC) values of 0.54 to 0.73; thus, they are not sensitive or specific enough to screen for GC patients (12, 13). In addition to proteins, various types of RNA are emerging biomarkers for GC. A few studies have performed systematic microRNA (miRNA) profiling of blood samples from patients with GC. The expression patterns of MIR2219, MIR744, and MIR376c in serum showed value as biomarkers to distinguish GC patients from healthy individuals (14). Plasma miRNA biomarkers for GC were found by reverse transcription quantitative real-time PCR (RT-PCR), and the AUCs were 0.65 to 0.75 for MIR185, MIR20a, MIR210, MIR25, and MIR92b (15). In another study, MIR181a-1 and KAT2B mRNA were identified as a combined predictor with AUC >0.95 (16). These studies suggested RNAs as potential biomarkers in the diagnosis and prognosis of GC; however, no study with definitive validation of these biomarkers in a large enough cohort has been conducted. Thus, the value of these biomarkers needs to be further confirmed in human GC patients.
Salivary extracellular RNA (exRNA) biomarkers including miRNAs have been developed for detecting various local and systemic diseases such as oral cancer (17, 18), Sjögren syndrome (19), pancreatic cancer (20), breast cancer (21), and lung cancer (22). In this study, we developed salivary exRNA biomarkers for GC detection in a Korean high-risk population based on prospective specimen collection (before the tumor diagnosis) and retrospective blinded evaluation (PRoBE) guidelines (23).
Materials and Methods
SAMPLE COLLECTION AND STUDY DESIGN
This study was performed at the University of California, Los Angeles (US) and Samsung Medical Center (South Korea) with approval from the institutional review boards from both institutions (UCLA IRB 06–07-018–11, SMC IRB 2008–01-028 –016). The study design followed the principles of the PRoBE design (23). All study participants were recruited from the Samsung Medical Center, and 294 saliva samples (163 GC and 131 non-GC) were prospectively collected before endoscopic examination. The detailed patient enrollment and cell-free saliva collection procedure can be found in the Methods section of the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol64/issue10. We have performed sample randomization based on the demographic information of all patients. The age, ethnicity, and alcohol consumption of healthy individuals used in this study were balanced in the patient group, as shown in Table 1. Two demographic factors, smoking and the presence of H. pylori, could not be balanced between the groups, as these are known risk factors for GC (5, 24).
Table 1.
Demographic information on subjects in the discovery phase.
| Demographic variable | Characteristic | Transcriptomic biomarker discovery phasea | miRNA biomarker discovery phaseb | ||||
|---|---|---|---|---|---|---|---|
| GC (n = 63) | Non-GC (n = 31) | P value | GC (n = 10) | Non-GC (n = 10) | P value | ||
| Age, years | Mean ± SD | 56.2 ± 11.1 | 54.8 ± 10.4 | 0.56 | 58.4 ± 7.6 | 52.4 ± 7.9 | 0.10 |
| Gender, n (%) | Male | 43 (68.3) | 13 (41.9) | 0.02 | 6 (60.0) | 4 (40.0) | 0.66 |
| Female | 20 (31.7) | 18 (59.1) | 4 (40.0) | 6 (60.0) | |||
| Ethnicity, n (%) | Asian | 63 (100) | 31 (100) | 10 (100) | 10 (100) | ||
| Smoking, n (%) | 27 (42.8) | 5 (16.1) | 0.01 | 3 (30.0) | 0 (0.0) | 0.21 | |
| Drinking, n (%) | 28 (44.4) | 9 (29.0) | 0.15 | 4 (40.0) | 5 (50.0) | 1.00 | |
| H. pylori, n (%) | 37 (58.7) | 8 (25.8) | 0.003 | 5 (50.0) | 4 (40.0) | 1.00 | |
Transcriptomic profiling (n = 94).
miRNA profiling (n = 20).
The biomarker development study consisted of 2 parts: discovery and validation. The first part was the discovery and verification phase of biomarkers using 2 different platforms: transcriptomic and miRNA (Fig. 1). The salivary transcriptomes of 63 GC samples and 31 non-GC controls (Table 1) were profiled using Affymetrix HG U133 + 2.0 microarrays. The identified exRNA candidates were verified by RT-qPCR using all 94 of the original samples. In the discovery phase for the miRNA biomarkers, 10 early-stage GC samples and 10 non-GC controls were selected (Table 1). The salivary miRNAs of these samples (n = 20) were profiled using the TaqMan miRNA array (Applied Biosystems). The miRNA candidates were verified using TaqMan miRNA assay (Thermo Scientific). The second part of the study was to validate these verified exRNA biomarker candidates with exRNA samples extracted from an independent cohort of 100 GC and 100 non-GC saliva samples (Table 2). The cohort was not balanced for demographics on sex and smoking history but more accurately reflected the diagnostic setting where our proposed final model could be implemented. Table 1 in the online Data Supplement shows the histological classification of GC individuals in the validation cohort.
Fig. 1.
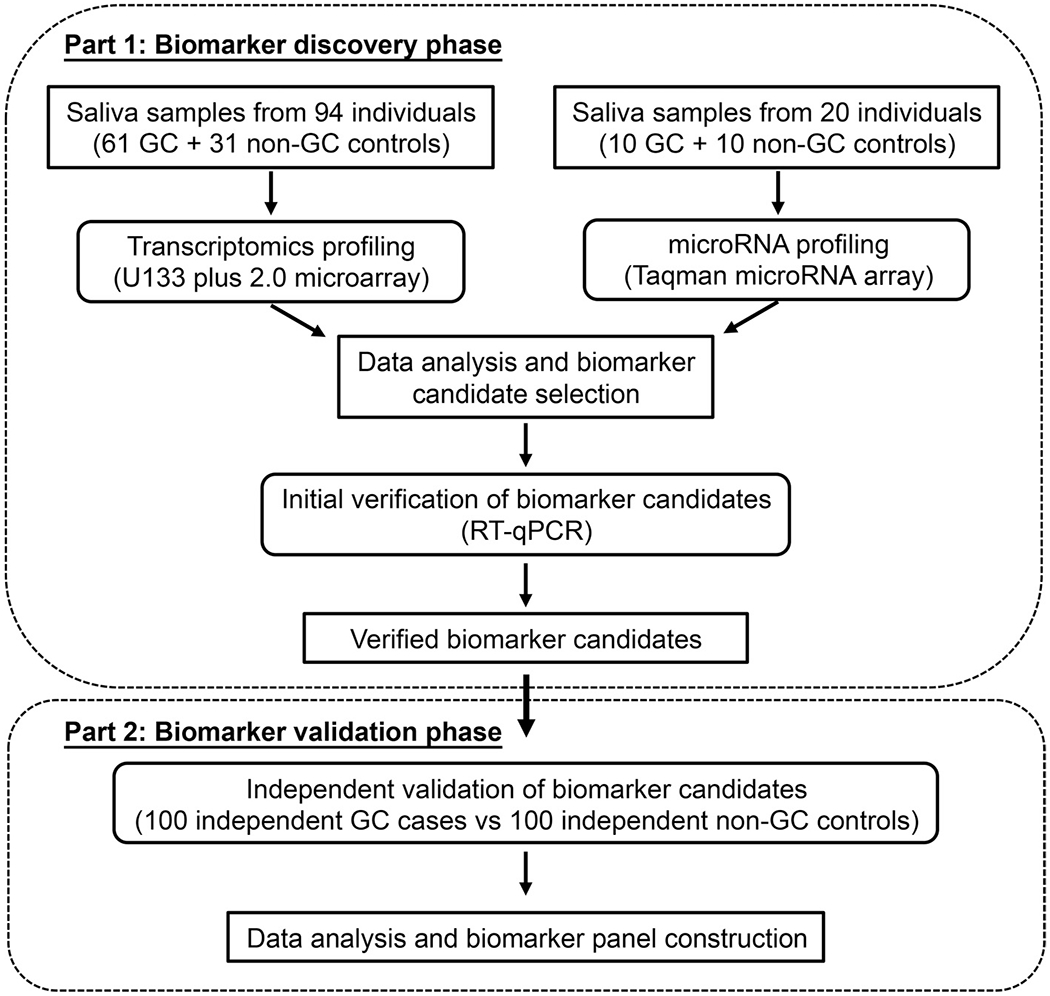
Schematic diagram of the study design for the 2 phases of salivary exRNA biomarker development for GC.
Table 2.
Demographic information on subjects in the validation phase (n = 200).
| Demographic variable | Characteristic | Validation phase | ||
|---|---|---|---|---|
| GC (n = 100) | Non-GC (n = 100) | P value | ||
| Age, years | Mean ± SD | 51.9 ± 9.2 | 55.7 ± 8.1 | 0.003 |
| Gender, n (%) | Male | 79 (79.0) | 47 (47.0) | <0.001 |
| Female | 21 (21.0) | 53 (53.0) | ||
| Ethnicity, n (%) | Asian | 100 (100) | 100 (100) | |
| Smoking, n (%) | 66 (44.0) | 33 (33.0) | <0.001 | |
| Drinking, n (%) | 55 (55.0) | NA | ||
| H. pylori, n (%) | NAa | NA | ||
NA, not available.
SALIVARY TRANSCRIPTOMIC PROFILING AND DATA ANALYSIS
Total RNA was isolated from 300 μL of saliva supernatant using the miRNeasy micro kit (QIAGEN). The method to avoid RNase contamination can be found in the Methods section of the online Data Supplement. The extracted RNA was treated with DNase I (Ambion) to remove contaminating DNA. The quality of salivary mRNA was evaluated by detecting expression levels of a saliva internal reference gene (GAPDH) using RT-qPCR (25). Isolated salivary mRNA (approximately 10 ng) was amplified using the RiboAmp RNA Amplification kit (Molecular Devices) and biotin-labeled using GeneChip Expression 3′-Amplification Reagents for in vitro transcription labeling (Affymetrix). Biotin-labeled complementary RNA (approximately 20 μg) was subsequently fragmented and sent to the University of California, Los Angeles microarray core facility for chip hybridization and scanning. The Affymetrix Human Genome U133 Plus 2.0 Array, which represents >47 000 transcripts and variants, was used for the salivary transcriptomic profiling. The microarray data have been uploaded to the GEO database (access no. GSE64951) based on the Minimum Information About a Microarray Experiment guidelines (26)
mRNA BIOMARKER VERIFICATION USING RT-qPCR
The selected candidate mRNA biomarkers (12) generated by microarray profiling were verified by nested RT-qPCR (RT-PCR followed by a separate SYBR green qPCR) on the same set of samples used for the microarray analysis (n = 94). The qPCR primers were designed using Primer3 software and synthesized by Sigma-Genosys after performing a Primer-BLAST search (27). The primer sequences were designed to avoid any known single-nucleotide polymorphism region in the target gene. All the amplicons were intron spanning. The RT-qPCR assay followed the Minimum Information for Publication of Quantitative Real-Time PCR Experiment guidelines and was performed in duplicate with each biomarker candidate (28). The detailed protocol can be found in the Methods section of the online Data Supplement. The specificity of the PCR product for each gene was confirmed with melting curve analysis and 3% agarose gel analysis. We calculated ∆Cq by subtracting the Cq value of the housekeeping gene (GAPDH/ACTB) from the raw Cq value of each biomarker candidate. The gene accession numbers and primer sequences used for transcriptomic biomarker verification and validation are shown in Table 2 of the online Data Supplement.
SALIVARY miRNA PROFILING
Total RNA was extracted from 300 μL of saliva supernatant using the mirVana PARIS extraction kit (Ambion). On-column DNase treatment (Qiagen) was used to remove contaminating DNA during RNA extraction. Total RNA (3 ng) was converted to complementary DNA using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems). Two different sets of stem-loop RT primers (human pool A and human pool B) were used (Megaplex RT primers, Applied Biosystems). After reverse transcription, the RT product was preamplified using TaqMan PreAmp Master Mix (Applied Biosystems) and Megaplex PreAmp primers (Applied Biosystems). The preamp product was not diluted before miRNA quantification. The TaqMan Human miRNA array set version 3.0 (Applied Biosystems) and TaqMan Universal PCR Master Mix, no AmpErase uracil N-glycosylase were used for miRNA quantification. All reactions were performed on a 7900HT Fast Real-Time PCR System containing a special cardholder (Applied Biosystems). Data were analyzed using RQ Manager software version 1.2 and DataAssist software version 3.0 (Applied Biosystems). Similarly, the ∆Cq value was computed using RNA polymerase III transcribed U6 small nuclear RNA as the reference gene.
VERIFICATION AND VALIDATION OF SALIVARY miRNA BIOMARKERS
The biomarker candidates generated by the TaqMan miRNA array profiling were verified by TaqMan miRNA assays (Applied Biosystems) on the same set of samples used for the miRNA array analysis (n = 20). TaqMan miRNA assays containing specific miRNA genes were ordered from Applied Biosystems. The protocol was like that recommended by the manufacturer for creating custom RT and preamplification pools using TaqMan miRNA assays. There was no dilution of sample before the real-time PCR reaction. The qPCR reactions for each candidate miRNA were performed in duplicate on a Roche LightCycler 480 II (Roche). The average threshold cycle (Cq) was examined, and U6 small nuclear RNA was used as the reference gene for normalizing the data. The TaqMan miRNA assay was also used to assay miRNAs in the validation cohort.
STATISTICAL ANALYSIS
Initial analyses summarized demographic characteristics within each cohort. Next, χ2 tests and t-tests were used to compare demographic characteristics between cancer and noncancer participants within cohorts.
Microarray analysis: The CEL files from all data sets were imported into the statistical software R 3.0.2 (29) using Bioconductor 2.2 (30). The data preprocessing was performed using the Probe Logarithmic Intensity Error Estimation expression measures after background correction and quantile normalization for each microarray expression data set. Probe set-level quantile normalization was performed across all samples. Finally, for every probe set, the Wilcoxon rank-sum test was used to compare gene expression between GC patients and non-GC controls.
The candidate salivary mRNA and miRNA biomarkers met the following criteria: (a) P value from Wilcoxon test <0.05 and (b) fold change >1.2. The top 30 ranking mRNA and 15 ranking miRNAs with the smallest P values were selected for verification. In the RT-qPCR verification step, ∆Cq values for mRNA and miRNA were compared between groups using the Wilcoxon rank-sum test. Twelve mRNA and 6 miRNA markers with P < 0.05 were included in the panel for evaluation in the validation step.
VALIDATION AND MODEL BUILDING
For each of the 12 mRNA and 6 miRNA candidates chosen from the discovery set, the Wilcoxon rank-sum test was used to compare markers between groups in the validation set of patients (100 GC vs 100 non-GC individuals). First, the Wilcoxon rank-sum test was used to compare ∆Cq transformed values for each marker between the GC and non-GC groups. Next, we created a multiple logistic model to identify the best combination of markers that could discriminate GC from non-GC samples.
We used the LASSO variable selection technique/estimation to construct the logistic regression model (31). The tuning parameter (λ) was chosen via 10-fold cross-validation with the GLMnet package in R. The diagnostic ability of the model was assessed using the AUC computed based on the predicted probabilities from the model. Statistical analyses were carried out using R 3.0.2 and SPSS V22 (IBM Corp). Values (in tables) are reported as mean (SD), and P values <0.05 were considered statistically significant.
Results
DISCOVERY OF SALIVARY TRANSCRIPTOMIC CANDIDATE BIOMARKERS
Gene expression profiles of saliva samples from GC patients (n = 63) and non-GC controls (n = 31) were examined in the discovery phase using Affymetrix Human Genome U133 Plus 2.0 Array (Fig. 1). To ensure accuracy of the microarray profiling, the quantity and quality of RNA in each saliva sample were assessed. On average, 117.51 ± 70.67 ng (n = 94) of total RNA was obtained from 300 μL of saliva supernatant. There was no significant difference in the total RNA isolated between the GC patients and non-GC controls (P = 0.39; n = 94). The RT-qPCR results of a saliva internal reference gene GAPDH in all saliva samples showed no significant difference in expression levels between GC patients and non-GC controls (P = 0.71; n = 94). A consistent amplification magnitude was obtained after 2 rounds of amplification, yielding an average of 58.58 ± 14.76 μg of biotinylated complementary RNA. There was no significant difference in the yield of complementary RNA between GC patients and non-GC controls (P = 0.23; n = 94).
Expression microarray results revealed 38 extracellular mRNAs significantly up-regulated and 2601 extracellular mRNAs significantly down-regulated in the saliva of GC patients when compared with the saliva of non-GC controls (n = 94; P < 0.05; fold change > 1.2). A heat map built from the microarray analysis of the top 150 genes resulted in an unadjusted P value cutoff of 0.002, revealing a potentially different saliva profile between the 2 groups (see Fig. 1 in the online Data Supplement).
VERIFICATION OF mRNA CANDIDATE BIOMARKERS FOR GC DETECTION
The candidate mRNA markers from microarray profiling were verified before validation. The top 30 ranking mRNA candidates (25 down-regulated and 5 up-regulated) with the smallest P values were selected for verification. Then, RT-qPCR was performed to verify the results on the discovery sample set (n = 94) and confirmed the differential RNA expression level of 12 of the 30 exRNAs, which were consistent with the microarray data and showed significant differences (P < 0.05) between GC patients and non-GC controls. Using GAPDH gene, the Cq from the GC patients was 24.85 ± 1.53, whereas the Cq from the non-GC controls was 25.03 ± 2.32, showing no significant difference (P = 0.65). As shown in Table 3 of the online Data Supplement, 11 down-regulated exRNAs (S100A10, ANXA1, CSTB, KRT6A, ERO1A, PPL, SPINK7, RANBP9, KRT4, CD24, and SEMA4B) and 1 up-regulated exRNA (EIF3G) were verified (P < 0.05; n = 94). The expression patterns of the verified biomarkers were consistent with the microarray profiling and exhibited AUC values of 0.63 to 0.74.
SALIVARY miRNA EXPRESSION PROFILES, CANDIDATE DISCOVERY, AND VERIFICATION
The miRNA expression profiles of 10 early-stage GC patients (stage 1a or 1b) and 10 non-GC controls were used for miRNA discovery. Both TaqMan Human miRNA A and B array version 3.0 cards were used to profile each sample (40 cards total). Before data normalization, only miRNAs with Cq values <35 in at least 80% of samples (16 of 20 patients) were included to ensure accuracy of miRNA verification by RT-qPCR. The numbers of detectable miRNAs between the saliva of GC patients and non-GC controls were similar (218 ± 3). Using the aforementioned criteria, 15 miRNA candidates (12 down-regulated and 3 up-regulated) were selected for verification by RT-qPCR using the discovery phase sample set. For the U6 small nuclear RNA reference gene, the Cq values acquired were 18.79 ± 1.37 from the GC patients and 18.67 ± 1.90 (P = 0.87) from the non-GC controls. RT-qPCR confirmed that the relative expression levels of 6 miRNAs were consistent with the TaqMan miRNA array data and showed significant differences (P < 0.05) between GC patients and non-GC controls. As shown in Table 4 of the online Data Supplement, 6 down-regulated miRNAs (MIR140-5p, MIR374a, MIR454, MIR15b, MIR28-5p, and MIR301a) were verified. These miRNAs exhibited AUC values of 0.79 to 0.88.
VALIDATION OF mRNA AND miRNA CANDIDATE BIOMARKERS
From the top 30 ranking mRNA and 15 ranking miRNAs candidates, the 12 verified mRNA candidates and 6 verified miRNA candidates were chosen for further validation using an independent cohort (100 GC patients and 100 matched non-GC controls). As shown in Table 3, 9 of the 12 mRNAs were validated, including ANXA1, CD24, CSTB, ERO1A, KRT4, KRT6A, PPL, S100A10, and SPINK7 (P < 0.05), yielding AUC values of 0.59 to 0.64 (Table 3). Four of 6 miRNA candidates were validated, including MIR140-5p, MIR374a, MIR454, and MIR15b. All 4 miRNA biomarkers showed significant differences between GC patients and non-GC controls (P < 0.05; n = 200), yielding AUC values of 0.63 to 0.70 (Table 3).
Table 3.
Quantitative RT-qPCR results and ROC-plot AUC values of the 12 mRNA and 6 miRNA biomarker candidates in saliva.
| Gene | GC (n = 100) vs non-GC control (n = 100) ∆Cq | |||
|---|---|---|---|---|
| GC, mean (SD) | Non-GC, mean (SD) | P valuea | AUC (95% CI) | |
| ANXA1 | −2.58 (2.07) | −3.36 (1.63) | 0.008 | 0.61 (0.53, 0.69) |
| CD24 | 1.20 (1.90) | 0.32 (1.66) | 0.001 | 0.63 (0.56, 0.71) |
| CSTB | −2.83 (2.15) | −3.74 (1.79) | 0.004 | 0.62 (0.54, 0.70) |
| EIF3G | 6.98 (3.08) | 7.08 (3.21) | 0.945 | 0.50 (0.42, 0.58) |
| ERO1A | 4.53 (2.07) | 3.70 (1.96) | 0.002 | 0.63 (0.55, 0.71) |
| KRT4 | −2.28 (2.35) | −3.02 (2.00) | 0.035 | 0.59 (0.51, 0.67) |
| KRT6A | −0.34 (2.34) | −1.21 (2.15) | 0.001 | 0.63 (0.56, 0.71) |
| PPL | 1.08 (2.23) | 0.34 (2.20) | 0.007 | 0.61 (0.53, 0.69) |
| RANBP9 | 4.26 (3.11) | 3.56 (2.77) | 0.157 | 0.56 (0.48, 0.64) |
| S100A10 | 2.21 (2.02) | 1.55 (2.04) | 0.006 | 0.61 (0.54, 0.69) |
| SEMA4B | 11.47 (3.98) | 10.57 (4.14) | 0.149 | 0.56 (0.48, 0.64) |
| SPINK7 | 2.37 (2.72) | 1.18 (1.98) | 0.001 | 0.64 (0.56, 0.72) |
| MIR140-5p | 1.54 (3.68) | −1.08 (3.27) | <0.001 | 0.70 (0.63, 0.78) |
| MIR374a | 6.95 (5.69) | 4.26 (4.59) | <0.001 | 0.65 (0.57, 0.73) |
| MIR454 | 4.61 (3.40) | 3.14 (3.40) | 0.003 | 0.63 (0.55, 0.70) |
| MIR15b | 2.92 (3.52) | 1.00 (3.42) | <0.001 | 0.65 (0.57, 0.72) |
| MIR28-5p | 5.15 (4.17) | 3.59 (3.94) | 0.024 | 0.59 (0.51, 0.67) |
| MIR301a | 8.46 (4.17) | 6.95 (3.82) | 0.01 | 0.61 (0.53, 0.69) |
All 15 biomarker candidates with P < 0.05 have q values (FDR-adjusted P values) of also <0.05.
PREDICTION MODEL CONSTRUCTION USING VALIDATED SALIVARY exRNA BIOMARKERS
From the 12 mRNA and 6 miRNA candidates, 3 mRNA biomarkers (SPINK7, PPL, and SEMA4B) and 2 miRNA markers (MIR140-5p and MIR301a) selected by the LASSO procedure yielded an AUC value of 0.81 (95% CI, 0.72–0.89) (Fig. 2, black dashed ROC curve). The point on the ROC curve that maximizes sensitivity and specificity results in a test being 75% sensitive and 83% specific. Setting the sensitivity at 80% or 90% yields specificity estimates of 54% and 40%, respectively. To assess the prognostic ability of our markers, we constructed a demographic characteristic-only model from our GC database repository. The model applied to our validation data set resulted in an AUC of 0.69 (95% CI, 0.59–0.79) (Fig. 2, dark gray solid ROC curve) with coefficients summarized in Table 5 of the online Data Supplement (smoking, sex, age). The combination of assessments of the exRNA panel and the demographic variables (smoking, sex, and age) provided an AUC of 0.87 (95% CI, 0.80–0.93) (Fig. 2, gray long dashed ROC curve; see also Table 6 in the online Data Supplement). The calibration tests of 3 presented models using the Hosmer–Lemeshow test is shown in Table 7 of the online Data Supplement. All 3 models have P > 0.05, indicating no significant lack of calibration. The comparison of the AUCs of these 3 models was performed with DeLong’s test (see Table 8 of the online Data Supplement). Only the markers plus demographics model (AUC = 0.87) vs demographics-only model (AUC = 0.69) showed a significant difference in the ∆ AUC. The point on the ROC curve of this model with maximum sensitivity and specificity is sensitivity = 82% and specificity = 77%. The positive predictive value was 82% and the negative predictive value was 77%. If we set a threshold with high sensitivity (90%), the respective specificity, positive predictive value, and negative predictive value are 65%, 76%, and 84%, respectively.
Fig. 2. Clinical utility of 3 salivary mRNA biomarkers (SPINK7, PPL, and SEMA4B) and 2 miRNA biomarkers (MIR140-5p and MIR301a) combinations.
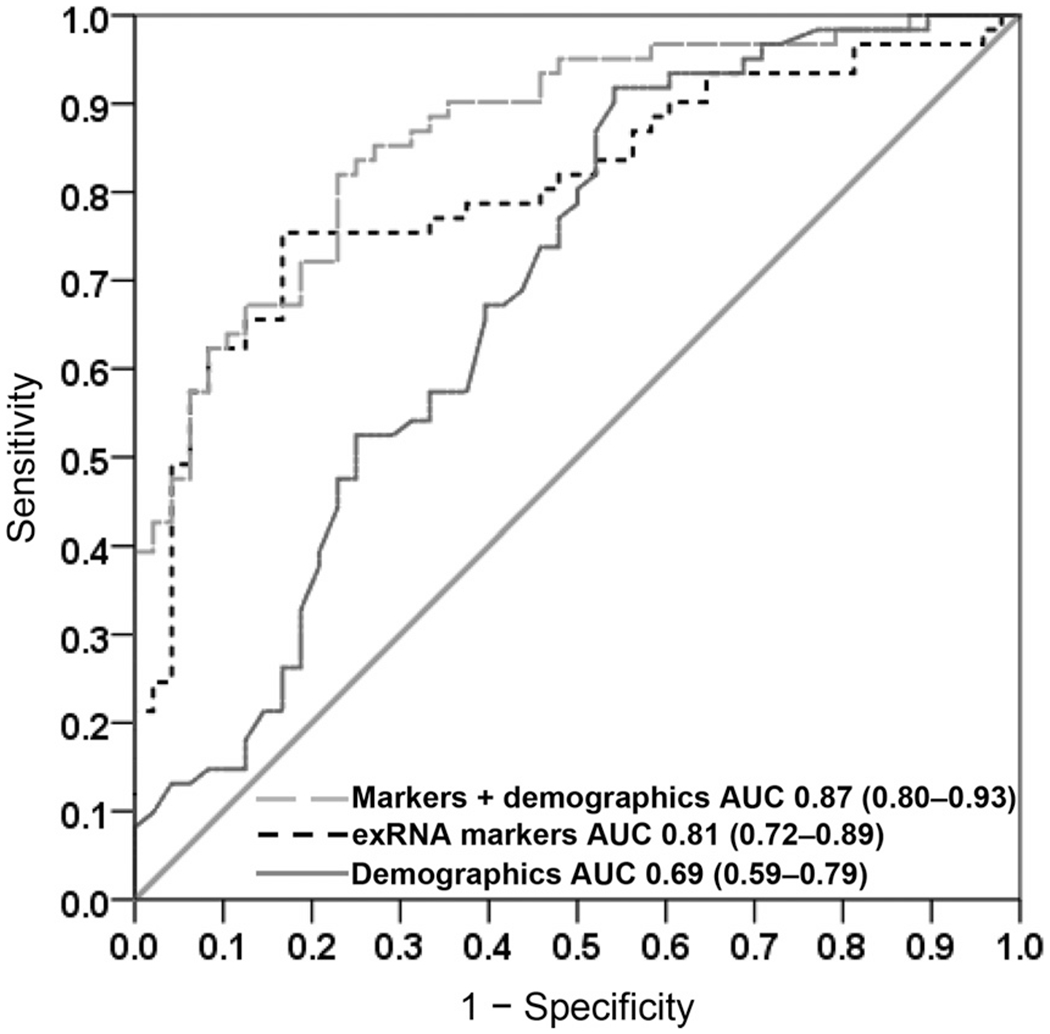
AUC values of ROC curves computed with 5 biomarkers (black dashed curve), demographic characteristics (dark gray solid curve), and combination of biomarkers plus demographic information (gray long dash curve) are 0.81, 0.69, and 0.87, respectively.
Discussion
This biomarker development study identified salivary biomarkers that can be definitively validated for GC detection. Nine of the 30 (30%) top-ranking salivary mRNA candidates and 4 of the 15 (27%) top-ranking miRNA salivary biomarkers were discovered based on a prospective clinical design, compliant with the PRoBE guidelines for biomarker development, and were validated. The discovered salivary mRNA and miRNA biomarkers were first individually validated in a cohort of 100 GC participants and 100 non-GC controls to yield the best performance panel of 3 mRNAs (SPINK7, PPL, and SEMA4B) and 2 miRNAs (MIR140-5p and MIR3014) with an AUC value of 0.81. Combined with demographic variables, the performance of the panel reached an AUC of 0.87 (95% CI, 0.80–0.93). We also analyzed whether validated biomarker candidates could distinguish different stages of GC. As shown in Table 9 of the online Data Supplement, only expression of PPL showed significant differences between early and late stages of GC.
One of the limitations of this study is that all participants included in this study were from a Korean cohort; the performance of this panel in other populations, especially in Western countries, needs to be further determined. Another limitation is that our model coefficients were constructed and validated on the same cohort. Thus, a further validation with an independent cohort from a multisite study would be required before clinical usage. H. pylori is known as 1 of the most potent risk factors for GC, although we could not include it in our final model because the screening population data were not available for much of the validation cohort. We do not think the absence of H. pylori index would be a confounding factor for the performance of our markers, but it may be something to further explore in a follow-up study.
Intriguingly, the most significant discriminative marker SPINK7, which is also named ECRG2 (esophagus cancer-related gene 2), was found dramatically down-regulated in primary esophageal squamous carcinoma (32). It is a tumor-suppressor gene that inhibits invasion of cancer cells through the urokinase-type plasmin activator receptor/β1 integrin pathway (33). Periplakin (PPL) has been reported to be down-regulated in esophageal squamous carcinoma and urothelial carcinoma (34). It also can act as a tumor suppressor in colon cancer progression (35). SEMA4B can work as a tumor suppressor to inhibit the invasion of non–small cell lung cancer through the PI3K/AKT pathway (36). Recently, MIR140 was found significantly decreased in breast cancer and non–small cell lung cancer tissues and cell lines. It also functions as a tumor suppressor in these cancers (37). The discovery of these down-regulated tumor-suppressor genes in saliva from GC patients may reflect their tumor-suppressor functions in GC tissues or just the indirect reactions of the human body to GC. This also needs to be determined in future study.
It is notable that the AUCs of carcinoembryonic antigen and CA19-9, which are currently regarded as the most valuable serum protein markers for the diagnosis of early-stage GC, were 0.73 and 0.68, respectively (12, 13). The performance of these validated salivary exRNA biomarkers is better than that of many existing and clinically used biomarkers. Thus, the merit of this study is not necessarily the development of validated biomarkers with outstanding performance; rather, it is the development of salivary biomarkers that are validated with discriminatory performance. This is of value to the emerging field of salivary diagnostics, as all efforts hinge on the biomarkers achieving regulatory approval by surviving definitive clinical validation trials. This study assures that salivary biomarkers, when properly developed, can be definitively validated for translational and clinical utilities.
An important rationale for this study was to determine the translational validity of salivary biomarkers for systemic disease detection. To date, no salivary biomarkers have been developed de novo and then definitively validated in a specific clinical context (23). Our study is the first to profile exRNA in saliva samples from individuals with a systemic disease and advance toward definitive validation. The validated salivary biomarkers, which are discriminatory for GC, can be used for screening GC patients and reduce unnecessary endoscopies. Our findings enhance the prospect for salivary diagnostics in the detection of systemic diseases.
Supplementary Material
Acknowledgments
J. Yoshizawa, provision of study material or patients; K. Kim, provision of study material or patients; T.R. Grogan, statistical analysis; D. Elashoff, statistical analysis; D.M. Akin, provision of study material or patients; X. Yan, provision of study material or patients; S.-M. Kim, administrative support, provision of study material or patients; J.-M. Bae, provision of study material or patients; T.-S. Sohn, provision of study material or patients; J.-H. Lee, provision of study material or patients; S. Kim, administrative support, provision of study material or patients; D.T.W. Wong, financial support.
Research Funding: F. Li, Hunan Provincial Administration of Traditional Chinese Medicine (201818); Y. Kim, University of California Los Angeles RSRF Grant; D.T.W. Wong, the National Institutes of Health (grant numbers: UH3-TR000923, T32-DE007296), University of California Los Angeles RSRF Grant.
Role of Sponsor: The funding organizations played a direct role in the design of study, preparation of manuscript, and final approval of manuscript. The funding organizations played no role in the choice of enrolled patients or review and interpretation of data.
Footnotes
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: D.T.W. Wong, UCLA, RNAmeTRIX.
Consultant or Advisory Role: D.T.W. Wong, RNAmeTRIX
Stock Ownership: D.T.W. Wong, RNAmeTRIX The University of California also holds equity in RNAmeTRIX.
Honoraria: D.T.W. Wong, EZLife Bio.
Expert Testimony: None declared.
Patents: D.T.W. Wong, UCLA Case 2011-347. Intellectual property that D.T.W. Wong invented and was patented by the University of California has been licensed to RNAmeTRIX.
Nonstandard abbreviations: GC, gastric cancer; ROC, receiver operating characteristic; AUC, area under the ROC curve; miRNA, microRNA; RT-qPCR, reverse transcription quantitative real-time PCR; exRNA, extracellular RNA; PRoBE, prospective-specimen collection and retrospective blinded evaluation.
Human Genes: MIR221, microRNA 221; MIR744, microRNA 744; MIR376c, microRNA 376c; MIR185, microRNA 185; MIR20a, microRNA 20a; MIR210, microRNA 210; MIR25, microRNA 25; MIR92b, microRNA 92b; MIR181a-1, microRNA 181a-1; KAT2B, lysine acetyltransferase 2B; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ACTB, actin beta; S100A10, S100 calcium binding protein A10; ANXA1, annexin A1; CSTB, cystatin B; KRT6A, keratin 6A; ERO1A, endoplasmic reticulum oxidoreductase 1 alpha; PPL, periplakin; SPINK7, serine peptidase inhibitor, Kazal type 7; RANBP9, RAN binding protein 9; KRT4, keratin 4; CD24, CD24 molecule; SEMA4B, semaphorin 4B; EIF3G, eukaryotic translation initiation factor 3 subunit G; MIR140-5p, microRNA 140-5p; MIR374a, microRNA 374a; MIR454, microRNA 454; MIR15b, microRNA 15b; MIR28-5p, microRNA 28-5p; MIR301a, microRNA 301a; ECRG2, esophagus cancer-related gene 2.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, Won YJ, Kong HJ, Oh CM, Lee DH, Lee JS. Cancerstatistics in Korea: incidence, mortality, survival, and prevalence in 2011. Cancer Res Treat 2014;46:109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristescu R, Lee J, Nebozhyn M,Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 4.Shikata K, Kiyohara Y, Kubo M, Yonemoto K, Ninomiya T, Shirota T, et al. A prospective study of dietary salt intake and gastric cancer incidence in a defined Japanese population: the Hisayama study. Int J Cancer 2006;119:196–201. [DOI] [PubMed] [Google Scholar]
- 5.Nomura AM, Wilkens LR, Henderson BE, Epplein M, Kolonel LN. The association of cigarette smoking with gastric cancer: the multiethnic cohort study. Cancer Causes Control 2012;23:51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin A, Kim J, Park S. Gastric cancer epidemiology in Korea. J Gastric Cancer 2011;11:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong SH, Park DJ, Lee HJ, Jung HC, Lee KU, Choe KJ, Yang HK. Clinicopathologic features of asymptomatic gastric adenocarcinoma patients in Korea. Jpn J Clin Oncol 2004;34:1–7. [DOI] [PubMed] [Google Scholar]
- 8.Herrero R, Parsonnet J, Greenberg ER. Prevention of gastriccancer. JAMA 2014;312:1197–8. [DOI] [PubMed] [Google Scholar]
- 9.Park CH. Proposal of a screening program for gastric cancer in Korea. J Korean Med 2002;45:964–71. [Google Scholar]
- 10.Lee KS, Oh DK, Han MA, Lee HY, Jun JK, Choi KS, Park EC. Gastric cancer screening in Korea: report on the national cancer screening program in 2008. Cancer Res Treat 2011;43:83–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol 2008;9:279–87. [DOI] [PubMed] [Google Scholar]
- 12.Bagaria B, Sood S, Sharma R, Lalwani S. Comparative study of CEA and CA19–9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol Med 2013;10:148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pectasides D, Mylonakis A, Kostopoulou M, Papadopoulou M, Triantafillis D, Varthalitis J, et al. CEA, CA 19-9, and CA-50 in monitoring gastric carcinoma. Am J Clin Oncol 1997;20:348–53. [DOI] [PubMed] [Google Scholar]
- 14.Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, et al. Identification of serum microRNAs as novel noninvasive biomarkers for early detection of gastric cancer. PloS one 2012;7:e33608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer 2014;110:2291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Zhu J, Yu F, Tian Y, Ma S, Liu X. Combination of miRNA and RNA functions as potential biomarkers for gastric cancer. Tumour Biol 2015;36:9909–18. [DOI] [PubMed] [Google Scholar]
- 17.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clinical Cancer Research 2009;15:5473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elashoff D, Zhou H, Reiss J, Wang J,Xiao H, Henson B, et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev 2012;21:664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Wang J, Meijer J, leong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum 2007;56:3588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010;138:949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L,Xiao H, Karlan S, Zhou H, Gross J, Elashoff D, et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the noninvasive detection of breast cancer. PloS One 2010;5:e15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao H, Zhang L, Zhou H, Lee JM, Garon EB, Wong DTW. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol Cell Proteomics 2012;11:M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst 2008;100:1432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 2010;10:403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res 2004;83:199–203. [DOI] [PubMed] [Google Scholar]
- 26.Edgar R, Barrett T. NCBI GEO standards and services for microarray data. Nat Biotechnol 2006;24:1471–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.NCBI/Primer-BLAST. Finding primers specific to your PCR template. https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (Accessed July 2017).
- 28.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009;55:611–22. [DOI] [PubMed] [Google Scholar]
- 29.R Core Team. R: a language and environment for statistical computing. https://www.R-project.org (Accessed July 2017).
- 30.Bioconductor core team. Bioconductor: an open source software for bioinformatics. https://www.bioconductor.org/about/ (Accessed July 2017).
- 31.Tibshirani R Regression shrinkage and selection via the lasso. J Royal Statist Soc B 1996;58:267–88. [Google Scholar]
- 32.Cui Y, Bi M, Su T, Liu H, Lu SH. Molecular cloning and characterization of a novel esophageal cancer related gene. Int J Oncol 2010;37:1521–8. [DOI] [PubMed] [Google Scholar]
- 33.Cheng X, Shen Z, Yin L, Lu SH, Cui Y. ECRG2 regulates cell migration/invasion through urokinase-type plasmin activator receptor (UPAR)/beta1 integrin pathway. J Biol Chem 2009;284:30897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimori T, Tomonaga T, Matsushita K, Oh-Ishi M, Kodera Y, Maeda T, et al. Proteomic analysis of primary esophageal squamous cell carcinoma reveals down-regulation of a cell adhesion protein, periplakin. Proteomics 2006;6:1011–8. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Zhang G, Wang Y, Elgehama A, Sun Y, Li L, et al. Loss of periplakin expression is associated with the tumorigenesis of colorectal carcinoma. Biomed Pharmacother 2017;87:366–74. [DOI] [PubMed] [Google Scholar]
- 36.Jian H, Zhao Y, Liu B, Lu S. SEMA4b inhibits growth of non-small cell lung cancer in vitro and in vivo. Cell Signal 2015;27:1208–13. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Eades G, Yao Y, Li Q, Zhou Q. Estrogen receptor alpha signaling regulates breast tumorinitiating cells by down-regulating miR-140 which targets the transcription factor SOX2. J Biol Chem 2012;287:41514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


