Significance
Dysregulation of the evolutionarily conserved Hippo pathway has been implicated in multiple diseases including cancer. Here we identified the RNA-binding protein (RBP) Rox8 as a regulator of Hippo signaling-mediated tumorigenesis. Rox8 not only directly binds to the 3′ UTR of yki mRNA, but also interacts with miR-8 to recruit miRNA-loaded RISC to degrade yki mRNA and therefore impedes Yki-induced tissue growth. We further revealed that TIAR, the human ortholog of Rox8, has retained a conserved regulatory function in yki/YAP mRNA stability. Our work uncovers a collaborative action of RBP and miRNA in regulating Hippo signaling.
Keywords: Hippo pathway, Rox8, TIAR, Yki, YAP
Abstract
The Hippo pathway is an evolutionarily conserved regulator of organ growth and tumorigenesis. In Drosophila, oncogenic RasV12 cooperates with loss-of-cell polarity to promote Hippo pathway-dependent tumor growth. To identify additional factors that modulate this signaling, we performed a genetic screen utilizing the Drosophila RasV12/lgl−/− in vivo tumor model and identified Rox8, a RNA-binding protein (RBP), as a positive regulator of the Hippo pathway. We found that Rox8 overexpression suppresses whereas Rox8 depletion potentiates Hippo-dependent tissue overgrowth, accompanied by altered Yki protein level and target gene expression. Mechanistically, Rox8 directly binds to a target site located in the yki 3′ UTR, recruits and stabilizes the targeting of miR-8–loaded RISC, which accelerates the decay of yki messenger RNA (mRNA). Moreover, TIAR, the human ortholog of Rox8, is able to promote the degradation of yki mRNA when introduced into Drosophila and destabilizes YAP mRNA in human cells. Thus, our study provides in vivo evidence that the Hippo pathway is posttranscriptionally regulated by the collaborative action of RBP and microRNA (miRNA), which may provide an approach for modulating Hippo pathway-mediated tumorigenesis.
The Hippo signaling pathway, initially discovered in Drosophila, has emerged as an evolutionarily conserved mechanism that controls tissue growth and organ size (1–4). Deregulation of this pathway has been implicated in multiple types of human cancers (5–10). The core components of this pathway comprise a kinase cascade consisting of the kinase Drosophila Hippo (Hpo)/mammalian MST1/2 (11–15) that phosphorylates the downstream kinase Warts (Wts)/LATS1/2 (16, 17), which subsequently results in the phosphorylation and inactivation of the oncoprotein Yorkie (Yki)/YAP/TAZ (18, 19). When the Hippo-signaling activity is compromised, unphosphorylated Yki/YAP/TAZ translocates into the nucleus and acts as a coactivator for the transcription factor Scalloped (Sd)/TEAD1-4 to up-regulate the expression of well-described target genes involved in cell proliferation and cell death (20–22). Over the past two decades, more than 20 conserved components or regulators of this pathway have been identified, yet the mechanism by which yki/YAP/TAZ activity is regulated at the messenger RNA (mRNA) level remains poorly understood.
To identify modulators of the Hippo pathway, we performed a systematic genetic screen utilizing the Hippo pathway-dependent RasV12/lgl−/− Drosophila tumor model (23) and identified the RNA-binding protein (RBP) Rox8 as a regulator of yki. RBPs play pivotal roles in posttranscriptional regulation, while dysregulated RBPs have been associated with various cancers (24–26). RBPs regulate their target genes through a wide array of mechanisms including mRNA stability, translation, and alternative splicing, underscoring the utmost importance of RBP-RNA regulatory interaction in cancers (26). We have further characterized Rox8 and its human ortholog TIAR as a crucial regulator of mRNA stability of Drosophila yki and human YAP, respectively. First, overexpression of Rox8 attenuates tumor hyperplasia caused by RasV12/lgl−/− oncogenic cooperation, while loss of Rox8 acts cooperatively with RasV12 to trigger tumor overgrowth. Second, Rox8 overexpression potently diminishes whereas Rox8 depletion dramatically increases the expression of Hippo pathway-responsive target genes. In addition, Rox8 genetically functions in parallel with Yki and biochemically binds to yki mRNA via its 3′ UTR, accelerating mRNA decay that results in decreased Yki protein level. Moreover, Rox8 executes this function by recruiting and stabilizing the targeting of miR-8–loaded RISC onto yki mRNA. Significantly, introduction of human TIAR into Drosophila executes a similar regulatory function on yki mRNA, Hippo pathway target genes, and tissue growth. Furthermore, TIAR destabilizes YAP mRNA via its 3′ UTR in human cells and inhibits YAP-induced cell proliferation and colony formation. Thus, our study reveals that Rox8/TIAR is an evolutionarily conserved regulator of the Hippo pathway from Drosophila to human.
Results
Rox8 Inhibits Cell Proliferation and Tissue Growth in Drosophila.
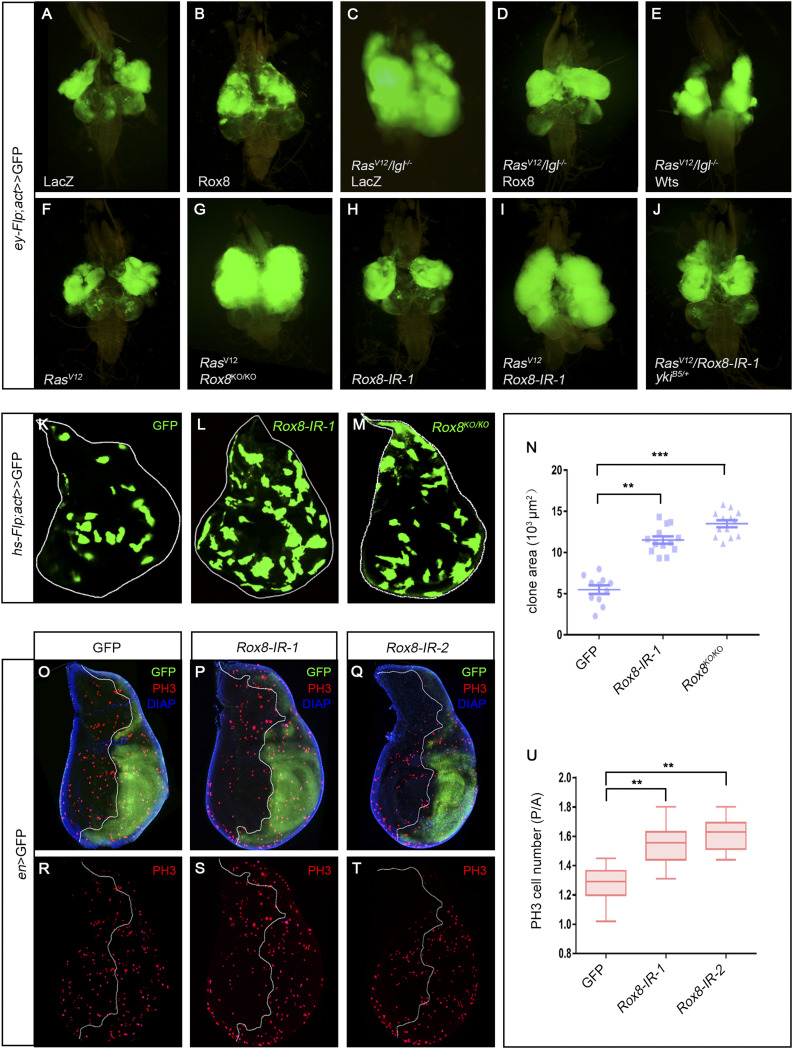
The oncogenic cooperation between RasV12 and loss-of-cell polarity genes (scrib, lgl, dlg) in larval eye-antennal clones has been well-established as a Drosophila tumor model that includes the massive tumor-like overgrowth and invasion into the ventral nerve cord (27). While impaired Hippo pathway is required for the overgrowth (23), JNK signaling is necessary for the invasion (9). To identify additional factors critical for tumor progression in vivo, we previously utilized this model to perform a genetic screen and characterized multiple regulators of JNK-mediated cell invasion (27–31). From the same screen, we found that RasV12/lgl−/−-induced tumor overgrowth was significantly impeded by Rox8EY02351, an UAS-bearing EP element inserted in the Rox8 5′ UTR (SI Appendix, Fig. S1A) that is able to drive Rox8 overexpression in RasV12/lgl−/− clones (Fig. 1 C and D), whereas the growth of control clones was not inhibited by Rox8 overexpression (Fig. 1 A and B). As a positive control, activation of the Hippo pathway by ectopic Wts expression was sufficient to block RasV12/lgl−/−-induced tumor growth (Fig. 1E). On the other hand, Rox8 knockdown clones synergized with RasV12 to promote tumorigenesis, which was confirmed by a Rox8 null mutant generated by the CRISPR/Cas9 technique (Fig. 1 F–I and SI Appendix, Figs. S1 A, B, and D and S2A), indicating a tumor suppressor function for Rox8. Consistent with its tumor suppressor function, Rox8 null mutant clones in the eye imaginal discs grow much larger than the wild-type controls (SI Appendix, Fig. S3). To verify the tumor suppressor function of Rox8 in other cellular contexts, we generated Rox8 knockdown or mutant clones in the wing imaginal discs and found that Rox8 depletion leads to increased clonal size (Fig. 1 K–N). To determine whether Rox8 plays a vital role in cell proliferation, we performed immunohistochemistry staining against PH3, a common mark for mitosis. We found that knockdown of Rox8 in the posterior compartment of wing discs promotes cell proliferation, as indicated by ectopic PH3 incorporation (Fig. 1 O–U and SI Appendix, Fig. S2B). In addition, knockdown of Rox8 in the midgut intestinal stem cells leads to increased clonal size and PH3 incorporation, coupled with enlarged gut width (SI Appendix, Fig. S4 M–P). The increased PH3 staining and gut width were confirmed in Rox8 null mutants (SI Appendix, Fig. S4 Q–T), which were viable and fertile with no other discernible phenotype. To examine the gain-of-function effect of Rox8 in vivo, we used nub-Gal4 to overexpress Rox8 (nub > Rox8EY02351) and found that the wing size significantly decreased (SI Appendix, Fig. S4 A–C). Rox8s−572, a GS line inserted in the first intron (SI Appendix, Fig. S1A), appears relatively stronger than Rox8EY02351as measured by RT-qPCR (SI Appendix, Fig. S1C), probably due to its proximity to the translation initiation site. Intriguingly, ptc > Rox8s−572 resulted in a reduced area between L3 and L4 in the wings and diminished scutellum (SI Appendix, Fig. S4 D–I), while GMR > Rox8s−572 produced a small-eye phenotype (SI Appendix, Fig. S4 J–L). Collectively, our findings suggest that Rox8 curbs cell proliferation and tissue growth in development.
Fig. 1.
Rox8 synergizes with RasV12 to promote tumorigenesis. (A–J) Fluorescence micrographs of GFP-labeled clones in eye-antennal discs dissected from larvae 7 d after egg laying are shown. Compared with the control (A), RasV12/lgl−/−-induced massive tumor overgrowth (C), which was dramatically suppressed by overexpression of Rox8 (D) or Wts (E). Meanwhile, ectopic Rox8 by itself caused no discernible phenotype (B). RasV12 cooperated with Rox8 mutant (G) or RNAi (I) to trigger tumor-like overgrowth, whereas expression of RasV12 (F) or Rox8.RNAi (H) alone was not sufficient to produce such a phenotype. RasV12/Rox8-IR-induced overgrowth was suppressed in the heterozygous yki mutant (J). (K–M) and (O–T) Fluorescence micrographs of third instar larval wing discs. Compared with the controls (K), Rox8 knockdown (L) or mutant (M) clones displayed augmented clonal size. (N) Quantification of clonal size is shown in K–M. Compared with the control (O and R), Rox8 knockdown in the posterior compartment of the wing disk led to increased PH3 staining (P, Q, S, and T). (U) Quantification of PH3-positive cell numbers in O–T. ***P < 0.001, **P < 0.01.
Rox8 Negatively Regulates Hippo Target Gene Expression.
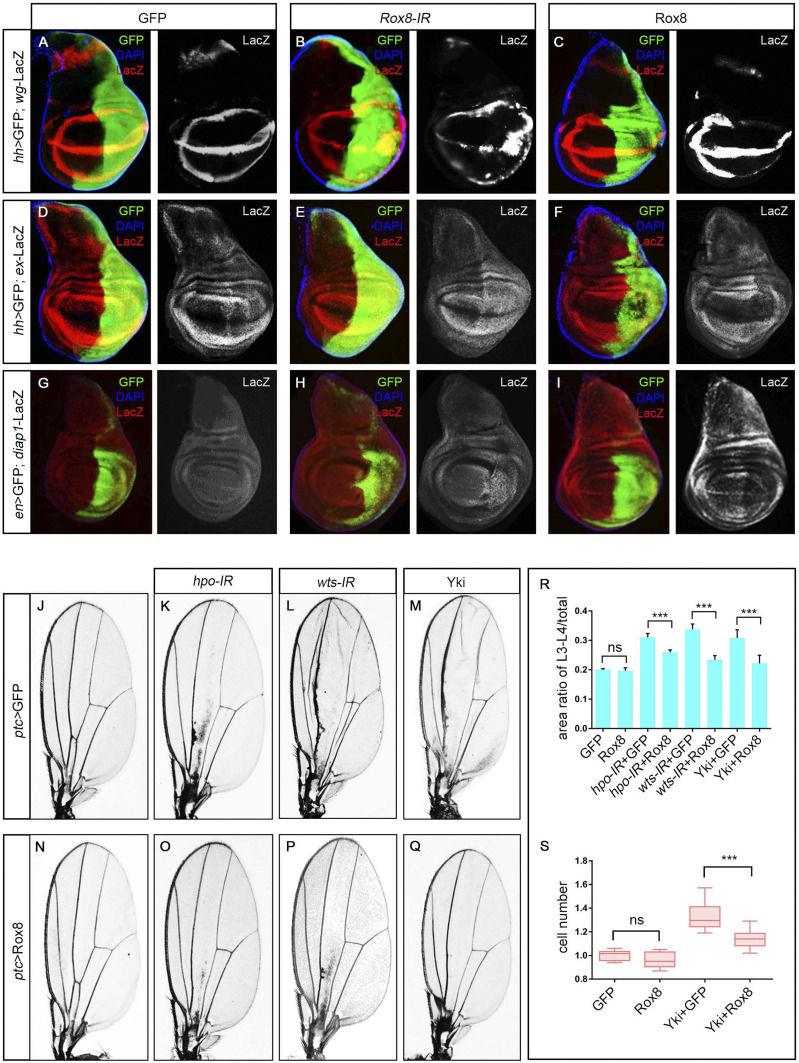
As the Hippo pathway plays an indispensable role in RasV12/loss-of-cell-polarity–triggered tumorigenesis (23), and overexpression of Rox8 mimics that of Wts to inhibit RasV12/lg−/−-induced tumor growth (Fig. 1 D and E), we decided to test the genetic interaction between Rox8 and the Hippo pathway. Intriguingly, RasV12/Rox8-IR-induced overgrowth was strongly suppressed in heterozygous yki background (Fig. 1J), suggesting that Rox8 might execute its tumor suppressor function via the Hippo pathway. To verify this hypothesis, we checked the expression of three well-characterized Hippo pathway target genes: wingless (wg), expanded (ex), and diap1 (21, 32, 33). We found that knockdown of Rox8 notably enhanced, whereas overexpression of Rox8 significantly diminished, the transcription of these target genes in third instar larval wing discs (Fig. 2 A–I). Consistently, Diap1 and ex-LacZ expression were up-regulated in Rox8 mutant clones in the wing imaginal discs (SI Appendix, Fig. S5). In addition, depletion of Rox8 in midgut intestinal stem cells resulted in up-regulated bantam expression, another well-described target gene of the Hippo pathway (SI Appendix, Fig. S6). Taken together, these data suggest that Rox8 positively modulates the Hippo pathway.
Fig. 2.
Rox8 regulates Hippo pathway in development. (A–I) Fluorescence micrographs of third instar larval wing discs are shown. Compared with the control (A, D, and G), expression of wg-LacZ (A–C), ex-lacZ (D–F), or diap1-LacZ (G–I) was enhanced by Rox8 depletion (B, E, and H) but impeded by Rox8 overexpression (C, F, and I) in the posterior compartment. (J–M) and (N–Q) Light micrographs of Drosophila adult wings are shown. In comparison with the control (J), depletion of hpo (K) or wts (L) or overexpression of Yki (M) driven by ptc-Gal4 increased the area size between L3 and L4, which were significantly suppressed by Rox8 expression (O–Q). (R) Quantification of the area ratio of L3 to L4/total in adult wings shown in J–Q. (S) Quantification of cell number in the area between L3 and L4 shown in J, M, N, and Q. ***P < 0.001. ns, no significant difference. Magnification for J–Q: 4×.
To corroborate the genetic interaction between Rox8 and the Hippo pathway in vivo, we examined the adult eye and observed a synergistic effect on eye-size enlargement between Rox8 depletion and heterozygous wts mutation (SI Appendix, Fig. S7 A–E). Moreover, nub > Hpo-induced wing pouch reduction was significantly suppressed upon Rox8 knockdown (SI Appendix, Fig. S7 F–J). Collectively, these data support the conclusion that Rox8 positively regulates the Hippo pathway in development.
Rox8 Regulates the Hippo Pathway in Parallel with Yki.
To unravel the mechanism by which Rox8 regulates the Hippo pathway, we performed genetic epistasis analysis between Rox8 and the Hippo pathway core components. Knockdown of hpo or wts, or overexpression of Yki along the anterior/posterior (A/P) compartment boundary by ptc-Gal4, dramatically enlarged the area between L3 and L4 in the adult wings (Fig. 2 J–M), which was significantly attenuated by mild expression of Rox8 (Rox8EY02351, Fig. 2 N–R). Consistent with previous studies that Hippo pathway regulates tissue growth by affecting cell number (18, 34, 35), we found that cell number in the area between L3 and L4 was indeed increased by ptc > Yki, which was significantly suppressed upon Rox8 coexpression (Fig. 2S). In agreement with the adult phenotype, ptc > Yki-induced tissue overgrowth in the larval wing discs was potently suppressed by Rox8 (SI Appendix, Fig. S8 I–L). Furthermore, mild expression of Rox8 (Rox8EY02351) impeded the adult-eye hyperproliferation phenotype induced by depletion of hpo or wts or overexpression of Yki (SI Appendix, Fig. S8 A–H and M). Collectively, these data suggest that Rox8 may act genetically downstream of, or in parallel with, yki. However, RasV12/Rox8-IR–triggered tumor overgrowth and Rox8-IR–induced ex-LacZ expression were robustly attenuated by the heterozygous yki mutation (Fig. 1J and SI Appendix, Fig. S9), suggesting that Rox8 may act upstream of, or in parallel with, yki. Collectively, our genetic epistasis data support the notion that Rox8 regulates the Hippo pathway in parallel with yki.
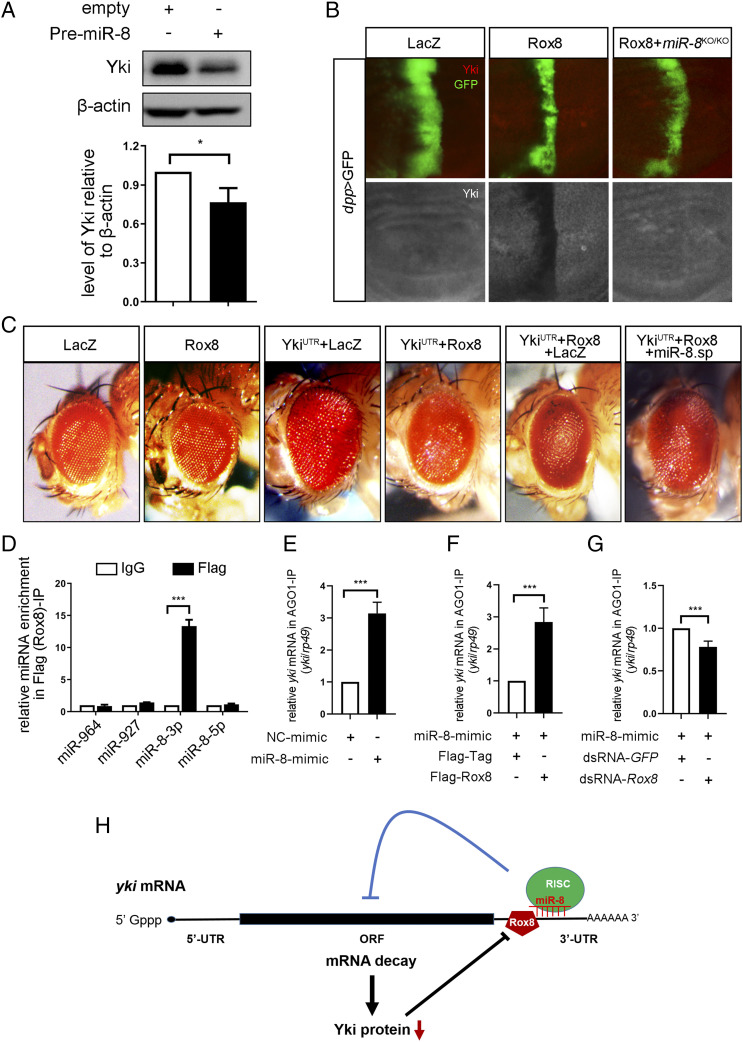
Rox8 Impedes yki mRNA and Protein Expression.
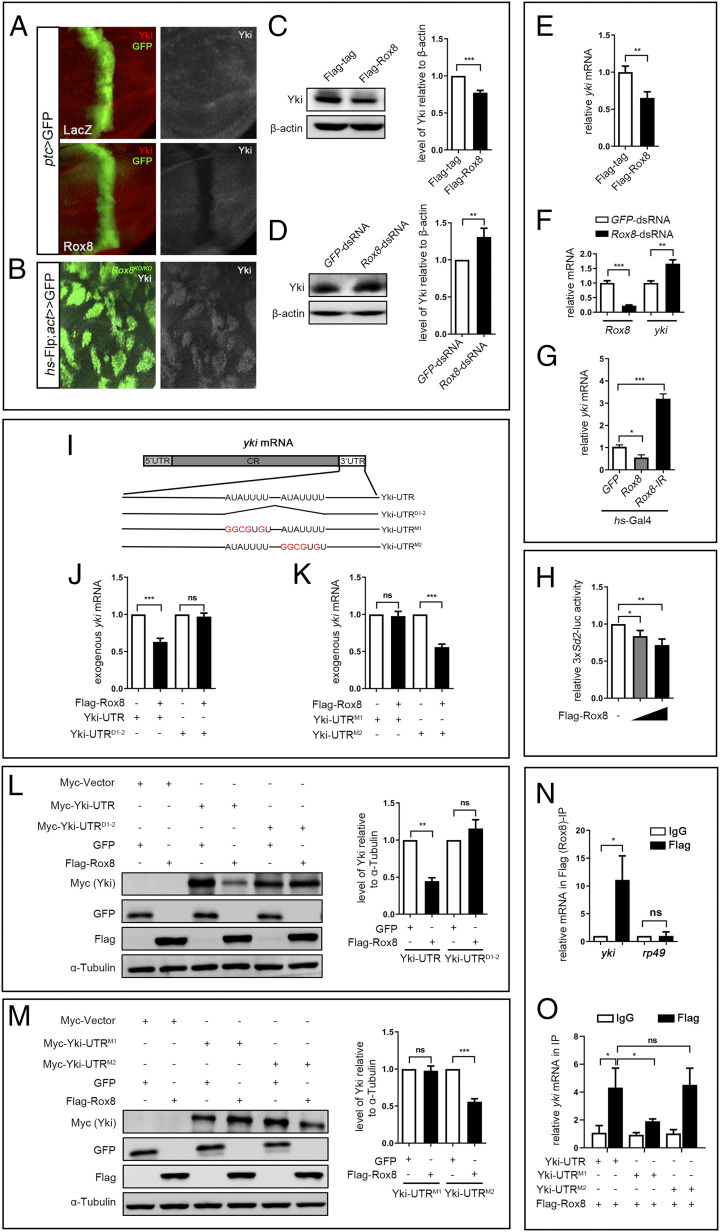
Given that Rox8 encodes an RNA-binding protein that regulates mRNA splicing, stability, or translation (9, 26), it is plausible that Rox8 modulates the Hippo pathway via yki mRNA. To test this possibility, we first checked whether Rox8 regulates Yki protein expression in vivo and in vitro. In third instar larval wing discs, Yki protein level, as judged by immunohistochemistry staining with antibody against Yki, was dramatically reduced upon Rox8 overexpression along the A/P compartment boundary driven by ptc-Gal4 (Fig. 3A), but significantly increased in Rox8 mutant clones (Fig. 3B). Consistently, in S2 cells, Rox8 expression significantly diminished whereas Rox8 knockdown increased endogenous Yki protein level (Fig. 3 C and D). Next, we examined if deregulated Rox8 influences yki mRNA level utilizing the RT-qPCR assay. We found that yki mRNA decreased upon Rox8 overexpression (Fig. 3E), but was elevated by Rox8 knockdown in S2 cells (Fig. 3F). We further confirmed these results by RT-qPCR that yki mRNA was indeed negatively regulated by Rox8 in the wing discs (Fig. 3G). On the other hand, Rox8 expression did not affect sd mRNA level (SI Appendix, Fig. S10), suggesting a specific regulation of yki mRNA by Rox8. In agreement with the above data, ectopic Rox8 suppressed the luciferase expression of 3×Sd2-luc, a reporter of Yki-Sd transcriptional activity in S2 cells (20), in a dosage-dependent manner (Fig. 3H). Taken together, these data indicate that Rox8 negatively regulates yki expression both in vitro and in vivo.
Fig. 3.
Rox8 accelerates yki mRNA decay. (A and B) Fluorescence micrographs of wing discs are shown. Yki protein level in the wing disk was attenuated by Rox8 overexpression (A), but was up-regulated in Rox8 mutant clones (B). Yki protein level in S2 cells was decreased by Rox8 overexpression (C), but was increased by Rox8 knockdown (D). (E) yki mRNA level was significantly down-regulated by Rox8 overexpression in S2 cells. (F) The level of yki mRNA was up-regulated by knockdown of Rox8 in S2 cells. (G) RT-qPCR data showing yki mRNA level in larval wing discs was reduced or elevated by Rox8 overexpression or depletion driven by hs-Gal4. (H) The expression of 3× Sd2-luc, a reporter for Yki/Sd activity, was inhibited by Rox8 in a dosage-dependent manner. (I) Schematic view of yki mRNA with the 5′ UTR, coding region (CR), and 3′ UTR, which contains two potential Rox8-binding motifs (AUAUUUU). Both motifs are deleted in UTRD1-2, while the first or second motif was mutated in UTRM1 or UTRM2, respectively. (J–M) Yki-UTR, Yki-UTRD1-2, Yki-UTRM1, or Yki-UTRM2 was subcloned into pUAST-Myc-tag vector. To detect exogenous yki mRNA, qPCR primers were designed to span the Myc-tag and Yki coding region. Overexpressing Rox8 resulted in reduction of exogenous Yki-UTR mRNA but not Yki-UTRD1-2 (J). Rox8 overexpression attenuated exogenous Yki-UTRM2 mRNA but not Yki-UTRM1 (K). (L and M) Immunoblot analysis of Myc-tagged-Yki-UTR, UTRD1-2, UTRM1, or UTRM2 expression upon coexpression of Flag-Rox8 in S2 cells. (Left) Immunoblot staining. (Right) Quantification data. As shown in L, expression of Myc-Yki-UTR but not of Myc-Yki-UTRD1-2 was dramatically inhibited by Rox8 overexpression. (M) Expression of Myc-Yki-UTRM2 but not Myc-Yki-UTRM1 was attenuated by overexpression of Rox8. (N and O) RIP assay was performed to detect the physical interaction between Rox8 and yki mRNA. Rox8 specifically bound to endogenous yki mRNA but not to rp49 mRNA that served as a negative control (N). The binding of Rox8 to yki 3′ UTR was blocked by M1 mutation, but not by M2 in S2 cells (O). ***P < 0.001, **P < 0.01, *P < 0.05. ns, no significant difference.
Rox8 Promotes yki mRNA Decay via Its Binding Site in yki 3′ UTR.
RBPs often promote mRNA decay through direct binding to their target sequences, usually located within the 3′ UTR (26). Intriguingly, we noted two potential Rox8-binding sites (AUAUUUU) in the 3′ UTR of yki mRNA (http://cisbp-rna.ccbr.utoronto.ca/index.php) (Fig. 3I), raising the possibility that Rox8 might physically interact with yki mRNA via these two sites. To verify whether these two sites are responsible for Rox8-induced yki mRNA decay, we constructed Myc-tagged Yki-UTR (with intact 3′ UTR), Yki-UTRD1-2 (both sites are deleted from the 3′ UTR), Yki-UTRM1 (site 1 is mutated), and Yki-UTRM2 (site 2 is mutated) (Fig. 3I). Cotransfection of Rox8 in S2 cells decreased the expression of Yki-UTR at both mRNA and protein levels, but had no effect on that of Yki-UTRD1-2 (Fig. 3 J and L), suggesting that one or both sites are necessary for Rox8’s activity on yki mRNA. Furthermore, mutation of the first site (M1), but not the second one (M2), fully blocked Rox8-triggered yki mRNA and protein reduction (Fig. 3 K and M), suggesting that the first site is essential for Rox8 to negatively regulate the yki mRNA level.
To examine the physical interaction between Rox8 and yki mRNA, we performed a RNA immunoprecipitation (RIP) assay. We found that endogenous yki mRNA, but not rp49 mRNA, was significantly enriched by Flag-Rox8 (Fig. 3N). Consistently, ectopic Yki-UTR mRNA was also enriched by Flag-Rox8 (Fig. 3O), and this enrichment was significantly abrogated by M1, but not by M2 (Fig. 3O), suggesting that Rox8 physically interacts with yki mRNA, most probably by direct binding to the first AUAUUUU motif located in the 3′ UTR of yki mRNA.
Rox8 Requires Its Binding Site to Suppress yki-Induced Overgrowth.
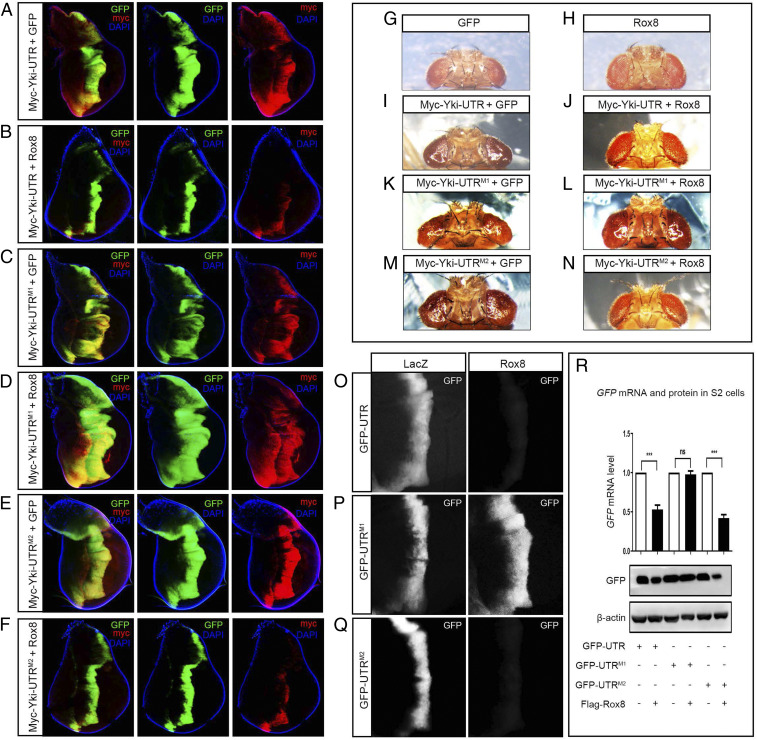
To corroborate the role of the Rox8-binding site in vivo, we generated transgenic flies expressing Myc-tagged Yki-UTR, Yki-UTRM1, or Yki-UTRM2. In agreement with the in vitro results, Rox8 overexpression in the wing discs driven by ptc-Gal4 dramatically inhibited the hyperplastic overgrowth induced by Myc-Yki-UTR or Myc-Yki-UTRM2, but not that by Myc-Yki-UTRM1 (Fig. 4 A–F). In line with the growth phenotype, ectopic Rox8 was able to reduce Myc-Yki protein produced by Myc-Yki-UTR or Myc-Yki-UTRM2, but not that by Myc-Yki-UTRM1 (Fig. 4 A–F). Furthermore, ectopic Rox8 suppressed adult eye overgrowth induced by Myc-Yki-UTR or Myc-Yki-UTRM2, but not by Myc-Yki-UTRM1 (Fig. 4 G–N), confirming that the first Rox8-binding site in yki 3′ UTR is necessary for Rox8 to impede Yki expression and Yki-induced tissue growth in vivo.
Fig. 4.
The yki 3′ UTR is necessary and sufficient for Rox8-mediated mRNA decay in vivo. (A–F) Fluorescence micrographs of wing discs are shown. Rox8 overexpression dramatically suppressed the overgrowth induced by Myc-Yki-UTR (A and B) or Myc-Yki-UTRM2 (E and F), but not that by Myc-Yki-UTRM1 (C and D). Consistently, Myc-Yki expression from Myc-Yki-UTR or Myc-Yki-UTRM2, but not from Myc-Yki-UTRM1, was impeded by Rox8 (A–F). (G–N) Light micrographs of Drosophila adult eyes are shown. Compared with the GMR > GFP control (G), GMR > Rox8 displayed no obvious phenotype (H). However, overexpression of Rox8 significantly impeded tumor-like overgrowth triggered by ectopic expression of Myc-Yki-UTR (I and J) or Myc-Yki-UTRM2 (M and N), but not of Myc-Yki-UTRM1 (K and L). (O–Q) Fluorescent images of third instar larval wing discs. Expressing Rox8 strikingly attenuated the expression of GFP from GFP-UTR (O) or GFP-UTRM2 (Q), but not that from GFP-UTRM1 (P). (R) RT-qPCR and immunoblot assays were performed in cultured S2 cells. Rox8 overexpression decreased the expression of GFP-UTR and GFP-UTRM2 at both the mRNA and protein level, but had no significant effect on that of GFP-UTRM1. ***P < 0.001. ns, no significant difference. Magnification for I–N: 6.3×.
Yki 3′ UTR Is Sufficient for Rox8-Mediated mRNA Degradation.
The above results demonstrate that the 3′ UTR is necessary for yki mRNA to be regulated by Rox8. However, RBPs could regulate mRNA level by multiple means, including transcription, splicing, and stability. To investigate whether Rox8 regulates yki mRNA by affecting its stability, and whether yki 3′ UTR is sufficient for this activity of Rox8, we made UAS− transgenes in which the 3′ UTR, UTRM1, or UTRM2 was placed after the GFP-coding region, respectively. In this setting, the transcription of GFP is solely controlled by the Gal4/UAS binary system, and no splicing event is involved in the production of GFP mRNA; hence, any effect of Rox8 on GFP mRNA should be achieved via influencing mRNA stability. Intriguingly, we found that both GFP mRNA and protein expression of GFP-UTR or GFP-UTRM2, but not that of GFP-UTRM1, was significantly suppressed by Rox8 in cultured S2 cells (Fig. 4R). These results were confirmed in vivo, where GFP and Rox8 were coexpressed along the A/P compartment boundary by ptc-Gal4 (Fig. 4 O–Q) or in the dorsal compartment by ap-Gal4 in the wing imaginal discs (SI Appendix, Fig. S11). Thus, yki 3′ UTR, which carries a Rox8-binding motif, is both necessary and sufficient for Rox8-mediated mRNA degradation.
Rox8 Promotes yki mRNA Degradation via miR-8.
To reveal the molecular mechanism by which Rox8 facilitates yki mRNA decay, we hypothesized that other factors affecting the stability of yki mRNA might be involved. The microRNA miR-8 has been previously reported to regulate yki mRNA stability (36, 37). Consistently, PremiR-8 overexpression has resulted in reduced yki mRNA and protein levels (SI Appendix, Fig. S12A and Fig. 5A), which was confirmed by miR-8 mimics (SI Appendix, Fig. S12C). Consistently, overexpression of miR-8 in the wing discs driven by dpp-Gal4 inhibited the expression of ban-LacZ (SI Appendix, Fig. S12B), an in vivo reporter of Yki activity. To test whether miR-8 is involved in Rox8-mediated yki degradation, we expressed Rox8 in miR-8 null mutant and found that Rox8-mediated Yki reduction was dramatically inhibited by loss of miR-8 (Fig. 5B). Consistently, the inhibitory effect of Rox8 on Yki-induced enlarged eye size was potently cancelled by miR-8 depletion (Fig. 5C). These data suggest that miR-8 is required for Rox8-mediated yki mRNA degradation. Consistently, RIP analysis showed that Rox8 specifically interacted with miR-8–3p, but not with other miRNAs (Fig. 5D) predicated to target yki 3′ UTR via the miRanda target prediction algorithm. Furthermore, Rox8 overexpression enhanced, whereas Rox8 depletion suppressed, the binding between yki mRNA and the miR-8/RISC complex (Fig. 5 E–G). Together, these results suggest that Rox8 promotes yki mRNA decay by recruiting and/or stabilizing the targeting of miR-8–loaded RISC to yki mRNA (Fig. 5H).
Fig. 5.
Rox8 promotes yki mRNA decay via miR-8. (A) Overexpression of PremiR-8 in S2 cultured cells resulted in reduced Yki protein level. (B) Fluorescent images of third instar larval wing discs. Ectopic Rox8-mediated Yki reduction was dramatically inhibited in miR-8 null mutants. (C) Light micrographs of Drosophila adult eyes are shown. Rox8 overexpression suppressed the enlarged eye size caused by YkiUTR, which was cancelled by depletion of miR-8. Magnification for C: 8×. (D) miR-8–3p was specifically enriched in the Flag (Rox8)-IP reaction. (E) yki mRNA is a target of miR-8. (F) Overexpression of Rox8 enhanced the binding ability of miR-8 to yki mRNA. (G) Depletion of Rox8 decreased miR-8 binding to yki mRNA. (H) Proposed model of Rox8-mediated yki mRNA degradation by recruiting or stabilizing miR-8–loaded RISC into yki mRNA 3′ UTR. ***P < 0.001, *P < 0.05.
Rox8 Function in the Hippo Pathway Is Retained by Its Human Ortholog TIAR.
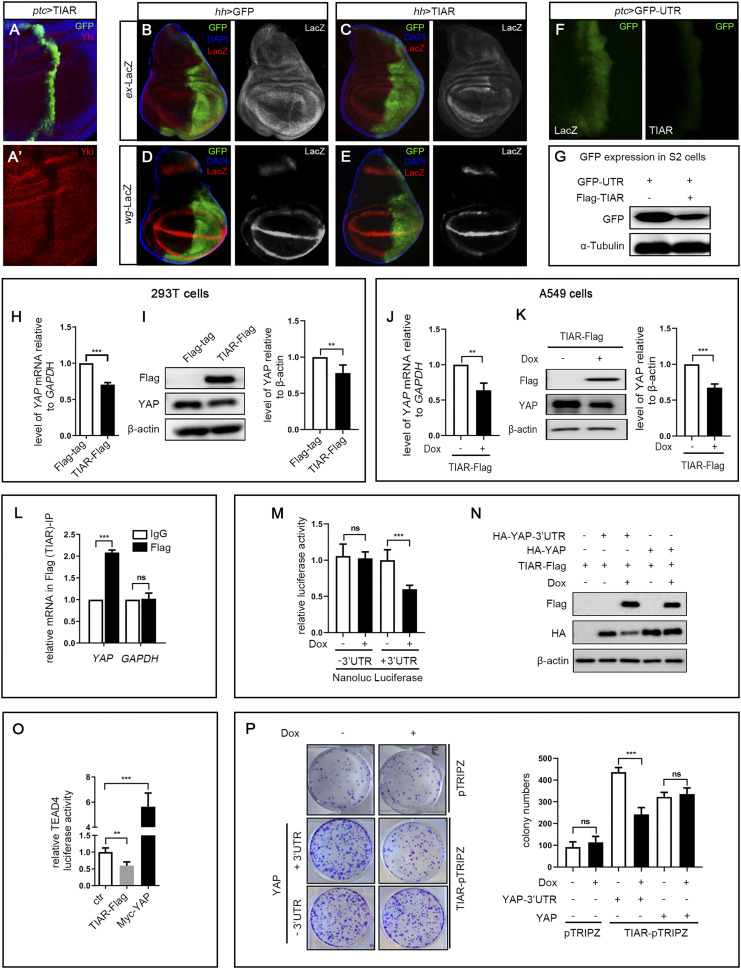
As the Hippo pathway is evolutionarily conserved from fly to human, we next asked whether TIAR, the human ortholog of Rox8, plays a similar role in the Hippo pathway. To this end, we created UAS-TIAR transgenic flies and checked whether ectopic TIAR could regulate the Hippo pathway in Drosophila. We found that TIAR expression driven by ptc-Gal4 in the wing discs potently reduced Yki protein (Fig. 6A). Consistently, TIAR expression driven by hh-Gal4 in the posterior compartment of wing discs significantly reduced the expression of Hippo target genes, as judged by the staining of ex-LacZ and wg-LacZ (Fig. 6 B–E). Additionally, we found that the ptc > Yki-UTR–induced wing phenotype was suppressed by a mild expression of TIAR from a weak UAS-TIAR transgene, which by itself exhibited no obvious phenotype (SI Appendix, Fig. S13 A–E). Interestingly, the fact that TIAR could also suppress the expression of GFP-UTR both in vivo (Fig. 6F) and in vitro (Fig. 6G) suggests that it is able to bind the Rox8 target site in 3′ UTR and promote the degradation of mRNA. Together, these data demonstrate that TIAR could functionally substitute for Rox8 to disrupt yki mRNA stability in Drosophila.
Fig. 6.
Human TIAR has retained Rox8’s activity on the Hippo pathway in Drosophila and human cells. (A–F) Fluorescent images of third instar larval wing discs. (A) ptc > TIAR resulted in down-regulation of Yki protein. Compared with the control (B and D), ectopic TIAR driven by hh-Gal4 led to reduced expression of ex-lacZ (C) and wg-LacZ (E). (F) ptc > TIAR dramatically suppressed the expression of GFP-UTR. (G) In cultured S2 cells, TIAR overexpression decreased GFP-UTR expression. Overexpression of TIAR transiently transfected into 293T cells inhibited YAP mRNA (H) and protein (I) expression. (Right) Quantification data of immunoblot analysis. (J–K) Overexpression of TIAR decreased YAP mRNA (J) and protein (K) level in A549 cells. A549-TIAR-Flag was untreated (−Dox) or treated (+Dox) for 2 d, followed by RT-qPCR and immunoblot assay. (Right) Quantification of immunoblot analysis. (L) Compared with IgG, YAP mRNA was significantly enriched in Flag (TIAR) immunoprecipitates, whereas GAPDH mRNA remained unchanged. (M) Nanoluc luciferase reporter assay. pMir-Nanoluc vector (–3′ UTR) or pMir-Nanoluc with YAP 3′ UTR (+3′ UTR) was cotransfected with pGL3-control plasmid into TIAR-Flag-pTRIPZ cells. Cells were cultured in the absence (−Dox) or presence (+Dox) of Dox for 2 d, followed by luciferase assays. The ratio of relative luciferase activity for +Dox/–Dox is shown as fold changes. (N) TIAR suppressed YAP stability in A549 cells through 3′UTR region. Plasmid expressing HA-YAP with or without 3′ UTR was cotransfected with TIAR-Flag plasmid into A549 cells, followed by no treatment or treatment with Dox for 2 d. Western blot was used to examine the levels of YAP-HA in cells. (O) TIAR overexpression inhibited the TEAD4-luc activity, and Myc-YAP as a positive control up-regulated the luciferase activity. (P) TIAR suppressed YAP-induced increased colony formation. Dox-inducible pTRIPZ vector or TIAR-pTRIPZ was cotransfected with plasmid expressing YAP with or without 3′ UTR. The transfected cells were selected in puromycin. About 1,000 cells were plated into each well of a six-well plate, followed by no treatment or treatment with Dox. Nine days later, the colonies were fixed and stained with crystal violet. (Right) Quantification data of colony formation assays. ***P < 0.001, **P < 0.01. ns, no significant difference.
TIAR Regulates YAP Expression in Human Cells.
To examine whether TIAR regulates YAP mRNA in human cells, we transiently transfected TIAR into HEK293T cells and found that excessive TIAR led to decreased YAP expression at both the mRNA and protein levels (Fig. 6 H and I). As YAP encodes a bona fide oncogene implicated in a wide spectrum of human cancers (6), we investigated the interaction between TIAR and YAP in cancer cells. Lentiviral infection of lung cancer A549 cells with TIAR resulted in reduced YAP mRNA and protein (Fig. 6 J and K), accompanied by decreased cell proliferation (SI Appendix, Fig. S13F). Moreover, RIP analysis showed that TIAR specifically bound to YAP mRNA, but not to GAPDH mRNA, served as a negative control (Fig. 6L). Consistent with our finding that Rox8 promotes yki mRNA decay via its 3′ UTR in Drosophila, YAP 3′ UTR is also necessary for YAP mRNA degradation by TIAR, as measured by dual luciferase assay in A549 cells (Fig. 6M). Furthermore, we carried out an immunoblot assay in cultured A549 cells and found that TIAR overexpression attenuated the expression of YAP from complementary DNA (cDNA) with the 3′ UTR (HA-YAP-3′ UTR), but not from cDNA without it (HA-YAP) (Fig. 6N). Finally, we checked whether TIAR could attenuate YAP activity in A549 cells and found that TIAR overexpression significantly suppressed the expression of TEAD4-luciferase (Fig. 6O), a mammalian Hippo pathway reporter (38, 39), and inhibited YAP-UTR (with 3′ UTR) but not YAP (without 3′ UTR)-induced cell proliferation, as measured by colony formation assay (Fig. 6P). Hence, we conclude that Rox8/TIAR modulate the Hippo pathway through the regulation of yki/YAP mRNA stability in a conserved manner from Drosophila to human cells.
Discussion
The Hippo pathway was initially identified in Drosophila and was subsequently proved to be highly conserved in mammals. Numerous studies have demonstrated that dysfunction of the Hippo pathway is closely associated with various cancers (5, 40–42). For the past two decades, multiple types of regulators of this pathway have been successfully identified, most of which are posttranscriptional-modification enzymes including kinases (43–48), E3 ubiquitin ligases (33, 49–51), methyltransferases (52), and deubiquitinating enzymes (53, 54). In the current study, we performed a genetic screen in Drosophila using a RasV12/lgl−/− trigged, Hippo pathway-dependent tumor overgrowth model and identified the RNA-binding protein Rox8 as a crucial regulator of the Hippo pathway. Mechanistically, Rox8 binds to the 3′ UTR of yki mRNA and promotes its decay, which ultimately results in decreased Yki protein and reduced expression of target genes. Intriguingly, this activity of Rox8 has been retained by its human ortholog TIAR, which attenuates YAP activity by promoting mRNA degradation in a similar manner. RBPs have been reported to regulate transcription, mRNA procession, and translation, yet the role of RBPs in the Hippo pathway has remained largely elusive. A recent study found that Drosophila RBP Hrb27C regulates Yki phosphorylation indirectly via an unknown mechanism (55). In addition, two RBPs, Dnd1 and FUS, were shown to stabilize LATS mRNA in cultured hepatocellular carcinoma cells (56, 57). Yet, our findings provide in vivo and physiological evidence that an RBP directly regulates the Hippo pathway and that this regulatory mechanism has been evolutionarily conserved from fly to human.
Recent studies suggest that RBPs can either cooperate or compete with microRNAs to regulate target gene expression (58–60). In Drosophila, removal of a potential miR-8 seed sequence in yki mRNA 3′ UTR leads to yki mRNA accumulation (36). Therefore, it would be intriguing to determine whether Rox8 acts in concert with miR-8 to modulate yki mRNA stability. In this study, we found that miR-8 is required for Rox8 to modulate yki expression and that Rox8 mechanistically promotes the decay of yki mRNA through facilitating the targeting of miR-8–loaded RISC into yki mRNA. Since our previous study showed that Yki negatively regulates Rox8 expression via bantam (9), it appears that there exists an exquisite regulatory circuit between Yki and Rox8 (Fig. 5H), which might fine-tune tissue homeostasis in development and ensure that Yki activity is turned off when Hippo signaling is active.
While the current study suggests that Rox8 acts as a tumor suppressor, Rox8 null mutants are viable and fertile with no discernible phenotype, implying that Rox8 is dispensable for normal development, which has been reported for other tumor suppressors, such as P53. However, loss of Rox8 synergizes with RasV12 to induce yki-dependent tumorigenesis. Consistently, it is convincingly accepted that oncogenic cooperation between RasV12 and loss of tumor suppressor genes triggers tumor growth and progression (23, 61–63). In addition, a previous study has demonstrated that the Hippo pathway interacts with Ras signaling to synergistically promote hyperproliferation and tumor development (64). These results indicate that Rox8 mutants are more prone to tumor formation and that the regulatory mechanism of Rox8-Yki may be more important under stressful conditions or in diseases. Given that mutated or deregulated RAS family genes (HRAS, NRAS, and KRAS) are frequently associated with various cancers (65, 66), our data suggest that TIAR may be a potential therapeutic target for not only the Hippo pathway but also for RAS-relevant cancer treatment.
Materials and Methods
Drosophila Stocks and Genetics.
Flies were raised on standard Drosophila media, and crosses were performed at 25 °C unless otherwise indicated. For experiments involving tub-Gal80ts, larvae were raised at 18 °C to restrict Gal4 activity for 7 d and shifted to 29 °C for 2 d. The following fly stocks have been described previously (9, 27): w1118; GMR-Gal4, ptc-Gal4, ap-Gal4, dpp-Gal4, UAS-RasV12, lgl4, UAS-GFP, and UAS-Dcr. Strains obtained from the Bloomington Drosophila Stock Center are UAS-LacZ (#3956), Rox8EY02351 (#15865), UAS-Wts (#30099), UAS-Hpo (#27105), UAS-mir-8 (#41176), mir-8Δ2 (#58932), and mir8-sp (#61374). Strains received from the Vienna Drosophila RNAi Center are UAS-Rox8-RNAi (#41439, referred as UAS-Rox8-IR-1), UAS-wts-RNAi (#106174), and UAS-hpo-RNAi (#104169). UAS-Rox8 (GS17980) and UAS-Rox8 (s-572) were acquired from the Kyoto Stock Center. UAS-Rox8-RNAi (#5422R-1, referred as UAS-Rox8-IR-2) was obtained from the National Institute of Genetics. Rox8KO mutation was generated by a germline-specific Cas9/single-guide RNA system. Fluorescently labeled invasive tumors were produced by the following strains: y w, ey-Flp; tub-Gal80, FRT40A; act>y+>Gal4, UAS-GFP (40A tester), lgl4 FRT40A UAS-RasV12, and ey-Flp, act > y+>Gal4, UAS-GFP. UAS-yki, ex-lacZ, ban-LacZ, and Diap1-lacZ were gifts from Lei Zhang, Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China. wts mutant was a gift from Shian Wu, Nankai University, Tianjin, China.
Immunostaining of Discs.
Immunostaining of discs was performed as previously described. In brief, third-instar larvae were dissected in phosphate-buffered saline (PBS) and fixed in freshly made 4% formaldehyde in PBS at room temperature for 20 min and then washed three times with PBST (PBS plus 0.1% Triton X-100). Larvae were incubated overnight with primary antibodies in PBST at 4 °C, then washed with PBST three times, and incubated with the corresponding fluorophore-conjugated secondary antibody for 2 h at room temperature. After being washed three times in PBST, discs were dissected and mounted in 40% glycerol. Images were captured with an Olympus stereo microscope SZX16. Antibodies used in this study were as follows: mouse anti–β-Gal (1:500) (Developmental Studies Hybridoma Bank); rabbit anti-PH3 (1:400) (Cell Signaling Technology); rabbit anti-Yki (1:1,000, a gift from Lei Zhang; and mouse anti-Myc) (1:500, Santa Cruz). Secondary antibodies used in this study were purchased from Life Technologies and were diluted at 1:500.
Supplementary Material
Acknowledgments
We thank Bloomington Drosophila Stock Center; Vienna Drosophila RNAi Center; the Kyoto Stock Center; Fly Stocks of National Institute of Genetics; the Core Facility of Drosophila Resource and Technology at the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences; Developmental Studies Hybridoma Bank; Lei Zhang for fly stocks and antibodies; Rejie Jiao, Lei Zhang, and Fan Zhang for plasmids; and Xinyao Li for technical assistance. This work is supported by the National Natural Science Foundation of China (Grants 31771595 and 31970536) and the Shanghai Committee of Science and Technology (Grants 09DZ2260100, 18430711600, and 18140900400) (to L.X.); the China Postdoctoral Science Foundation (Grant 2000229071) (to X.G.); the Canadian Institute of Health Research (Grants MOP119325 and MOP148629); the Canadian Cancer Society (X.Y.); and the National Natural Science Foundation of China (Grant 81671716) (to Z.L.). X.M. is partially supported by “Team for Growth Control and Size Innovative Research” (Grant 201804016).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013449117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information. Some study data are available upon request.
References
- 1.Zhang L., Yue T., Jiang J., Hippo signaling pathway and organ size control. Fly (Austin) 3, 68–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halder G., Johnson R. L., Hippo signaling: Growth control and beyond. Development 138, 9–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan D., Hippo signaling in organ size control. Genes Dev. 21, 886–897 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Yin M., Zhang L., Hippo signaling: A hub of growth control, tumor suppression and pluripotency maintenance. J. Genet. Genomics 38, 471–481 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Yu F. X., Zhao B., Guan K. L., Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B., Li L., Lei Q., Guan K. L., The hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 24, 862–874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan D., The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma X., et al. , Hippo signaling promotes JNK-dependent cell migration. Proc. Natl. Acad. Sci. U.S.A. 114, 1934–1939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B., Lei Q. Y., Guan K. L., The hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20, 638–646 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S., Huang J., Dong J., Pan D., Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G., Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Harvey K. F., Pfleger C. M., Hariharan I. K., The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Pantalacci S., Tapon N., Léopold P., The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Jia J., Zhang W., Wang B., Trinko R., Jiang J., The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J., The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Xu T., Wang W., Zhang S., Stewart R. A., Yu W., Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Huang J., Wu S., Barrera J., Matthews K., Pan D., The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Zhao B., et al. , Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., et al. , The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S., Liu Y., Zheng Y., Dong J., Pan D., The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Zhao B., et al. , TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atkins M., et al. , An ectopic network of transcription factors regulated by hippo signaling drives growth and invasion of a malignant tumor model. Curr. Biol. 26, 2101–2113 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Kechavarzi B., Janga S. C., Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 15, R14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Liu Q., Shyr Y., Dysregulated transcription across diverse cancer types reveals the importance of RNA-binding protein in carcinogenesis. BMC Genomics 16 (suppl. 7), S5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira B., Billaud M., Almeida R., RNA-binding proteins in cancer: Old players and new actors. Trends Cancer 3, 506–528 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Zhang S., et al. , Wingless modulates activator protein-1-mediated tumor invasion. Oncogene 38, 3871–3885 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Ma X., et al. , Myc suppresses tumor invasion and cell migration by inhibiting JNK signaling. Oncogene 36, 3159–3167 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Sun Y., et al. , MKK3 modulates JNK-dependent cell migration and invasion. Cell Death Dis. 10, 149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma X., et al. , Rho1-Wnd signaling regulates loss-of-cell polarity-induced cell invasion in Drosophila. Oncogene 35, 846–855 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Ma X., et al. , dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila. Dev. Biol. 380, 211–221 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Cho E., et al. , Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38, 1142–1150 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Ma X., Guo X., Richardson H. E., Xu T., Xue L., POSH regulates Hippo signaling through ubiquitin-mediated expanded degradation. Proc. Natl. Acad. Sci. U.S.A. 115, 2150–2155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H., Jiang D., Chi F., Zhao B., The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein Cell 3, 291–304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy B. V., Irvine K. D., Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development 138, 5201–5212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umegawachi T., et al. , Control of tissue size and development by a regulatory element in the yorkie 3'UTR. Am. J. Cancer Res. 7, 673–687 (2017). [PMC free article] [PubMed] [Google Scholar]
- 37.Sander M., Eichenlaub T., Herranz H., Oncogenic cooperation between Yorkie and the conserved microRNA miR-8 in the wing disc of Drosophila. Development 145, dev153817 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Jiao S., et al. , A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Zhang W., et al. , VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 24, 331–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Y., Analysis of the role of the Hippo pathway in cancer. J. Transl. Med. 17, 116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon S., Yeon Park S., Woo Park H., Regulation of the Hippo pathway in cancer biology. Cell. Mol. Life Sci. 75, 2303–2319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvey K. F., Zhang X., Thomas D. M., The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Chung H. L., Augustine G. J., Choi K. W., Drosophila Schip1 links expanded and tao-1 to regulate hippo signaling. Dev. Cell 36, 511–524 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Boggiano J. C., Vanderzalm P. J., Fehon R. G., Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21, 888–895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon C. L., Zhang X., Lin J. I., Manning S. A., Harvey K. F., Homeodomain-interacting protein kinase regulates Hippo pathway-dependent tissue growth. Curr. Biol. 22, 1587–1594 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Chen J., Verheyen E. M., Homeodomain-interacting protein kinase regulates Yorkie activity to promote tissue growth. Curr. Biol. 22, 1582–1586 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Huang H. L., et al. , Par-1 regulates tissue growth by influencing hippo phosphorylation status and hippo-salvador association. PLoS Biol. 11, e1001620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho Y. S., et al. , Regulation of Yki/Yap subcellular localization and Hpo signaling by a nuclear kinase PRP4K. Nat. Commun. 9, 1657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu L., et al. , The Drosophila F-box protein Slimb controls dSmurf protein turnover to regulate the Hippo pathway. Biochem. Biophys. Res. Commun. 482, 317–322 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Ribeiro P., Holder M., Frith D., Snijders A. P., Tapon N., Crumbs promotes expanded recognition and degradation by the SCF(Slimb/β-TrCP) ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 111, E1980–E1989 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma B., et al. , Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 17, 95–103 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Fang L., et al. , SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell 34, 103–118.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Sun X., et al. , Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat. Commun. 10, 411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toloczko A., et al. , Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the hippo pathway. Cancer Res. 77, 4921–4933 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mach J., et al. , Modulation of the Hippo pathway and organ growth by RNA processing proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 10684–10689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W., Gong F., Zhang T., Chi B., Wang J., RNA-binding protein Dnd1 inhibits epithelial-mesenchymal transition and cancer stem cell-related traits on hepatocellular carcinoma cells. Biotechnol. Lett. 39, 1359–1367 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Bao L., et al. , A FUS-LATS1/2 Axis inhibits hepatocellular carcinoma progression via activating hippo pathway. Cell. Physiol. Biochem. 50, 437–451 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Ciafrè S. A., Galardi S., microRNAs and RNA-binding proteins: A complex network of interactions and reciprocal regulations in cancer. RNA Biol. 10, 935–942 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang P., Singh M., Coller H. A., Computational assessment of the cooperativity between RNA binding proteins and microRNAs in transcript decay. PLOS Comput. Biol. 9, e1003075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kedde M., Agami R., Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 7, 899–903 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Ma X., et al. , PP6 disruption synergizes with oncogenic Ras to promote JNK-dependent tumor growth and invasion. Cell Rep. 19, 2657–2664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu M., Pastor-Pareja J., Xu T., Interaction between RasV12 and scribble clones induces tumour growth and invasion. Nature 463, 545–548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagliarini R. A., Xu T., A genetic screen in Drosophila for metastatic behavior. Science 302, 1227–1231 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Pascual J., et al. , Hippo reprograms the transcriptional response to Ras signaling. Dev. Cell 42, 667–680.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Ryan M. B., Der C. J., Wang-Gillam A., Cox A. D., Targeting RAS-mutant cancers: Is ERK the key? Trends Cancer 1, 183–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogelstein B., et al. , Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information. Some study data are available upon request.