Significance
Fragile X syndrome (FXS), the most prevalent monogenic form of autism, is caused by the loss of FMRP, an RNA binding protein. In the absence of FMRP, translation is dysregulated, but restoration of translational homeostasis rescues the syndrome and thus could suggest new treatments for the disorder. Using ribosome profiling and RNA metabolic profiling, we show that, in the FMRP-deficient mouse brain, there are few specific translational disturbances. Instead, there is widespread imbalance of RNA levels. This imbalance is caused by destabilization of the FMRP targets and other mRNAs based on codon optimality. Rebalancing the transcriptome may therefore be a key to correcting syndrome-related pathophysiologies. This study establishes a role for FMRP in linking codon optimality to RNA stability.
Keywords: FMRP, codon optimality, RNA decay, ribosome profiling, CPEB1
Abstract
Fragile X syndrome (FXS) is caused by inactivation of the FMR1 gene and loss of encoded FMRP, an RNA binding protein that represses translation of some of its target transcripts. Here we use ribosome profiling and RNA sequencing to investigate the dysregulation of translation in the mouse brain cortex. We find that most changes in ribosome occupancy on hundreds of mRNAs are largely driven by dysregulation in transcript abundance. Many down-regulated mRNAs, which are mostly responsible for neuronal and synaptic functions, are highly enriched for FMRP binding targets. RNA metabolic labeling demonstrates that, in FMRP-deficient cortical neurons, mRNA down-regulation is caused by elevated degradation and is correlated with codon optimality. Moreover, FMRP preferentially binds mRNAs with optimal codons, suggesting that it stabilizes such transcripts through direct interactions via the translational machinery. Finally, we show that the paradigm of genetic rescue of FXS-like phenotypes in FMRP-deficient mice by deletion of the Cpeb1 gene is mediated by restoration of steady-state RNA levels and consequent rebalancing of translational homeostasis. Our data establish an essential role of FMRP in codon optimality-dependent mRNA stability as an important factor in FXS.
Fragile X syndrome (FXS) is the most common form of inherited intellectual disability caused by a single gene mutation (1). FXS is caused by a trinucleotide repeat expansion in the FMR1 locus, which results in transcriptional silencing and loss of its protein product, FMRP (2). In FMRP-deficient mice, protein synthesis in the brain is elevated by ∼15 to 20% (3, 4), indicating that FMRP represses translation. In the mouse brain, FMRP binds mostly to coding regions of ∼850 to 1,000 mRNAs (5–9) and cosediments with polyribosomes (5). FMRP has been proposed to repress translation by impeding ribosome translocation (5, 10–13). This hypothesis is based on the evidence that ribosomes associated with many of these mRNAs are resistant to puromycin treatment in vitro, which causes premature polypeptide release and thus is an indirect measure of ribosome translocation, and that ribosomes transit at faster rates in FMRP-knockout (KO) brain extracts compared to WT (5, 10). FMRP also regulates translation directly or indirectly at the level of initiation (14, 15), RNA splicing (13), editing (16, 17), nuclear export (18, 19), and m6A modifications (18–20). However, the relationship of these molecular impairments to the etiology of FXS, if any, is unknown.
Translation is also controlled by the supply and demand of available tRNAs (21). When particular codons represented in mRNA are not met by a sufficient supply of charged tRNA, ribosome transit slows or stalls, which in turn causes RNA destruction (22–27). Control of specific translation and RNA destruction by codon bias varies with tissue (24), time of development (23), and cell stress (28). Additionally, in yeast, transacting factors can influence codon bias-mediated RNA destruction (22), indicating a more complex regulation than simple codon–tRNA balance.
Here, we have used ribosome profiling and RNA sequencing (RNA-seq) to investigate translational dysregulation in the FMRP KO cortex and found that FMRP coordinates the link between RNA destruction and codon usage bias (codon optimality). We find that the apparent dysregulation of translational activity (i.e., ribosome occupancy) in FMRP KO cortex can be accounted for by commensurate changes in steady-state RNA levels. Down-regulated mRNAs in FMRP KO cortex are enriched for those that encode factors involved in neuronal and synaptic functions and are highly enriched for FMRP binding targets. These observations suggest that, in the cortex, FMRP directly or indirectly regulates RNA stability. Indeed, RNA metabolic profiling by 5-ethynyl uridine (5-EU) incorporation and whole-transcriptome sequencing reveals widespread RNA degradation in FMRP KO cortical neurons, while synthesis and processing rates remained substantially unchanged. Of the ∼700 mRNAs that degraded significantly faster in FMRP KO cortex compared to WT, those enriched for optimal codons were particularly affected. This widespread codon-dependent RNA destruction in FMRP-deficient neurons involves a massive reshuffling of the identities of stabilizing or destabilizing codons. Moreover, FMRP can distinguish between optimal and nonoptimal codon-containing mRNAs, probably through the associated translational machinery. Finally, we demonstrate that, in a genetic rescue paradigm of FXS where a double deficiency of FMRP and CPEB1 mitigates the disorder in mice, restoration of RNA levels drives the recovery of ribosome occupancy. These results indicate that a primary consequence of FMRP depletion from the brain is the uncoupling of codon bias from the RNA destruction machinery. This uncoupling may be a general mechanism that underlies FXS, and restoration of the RNA stability landscape could be a key to ameliorating the disorder.
Results
Steady-State RNA Level Changes Drive Translational Buffering in Fragile X Brain Cortex.
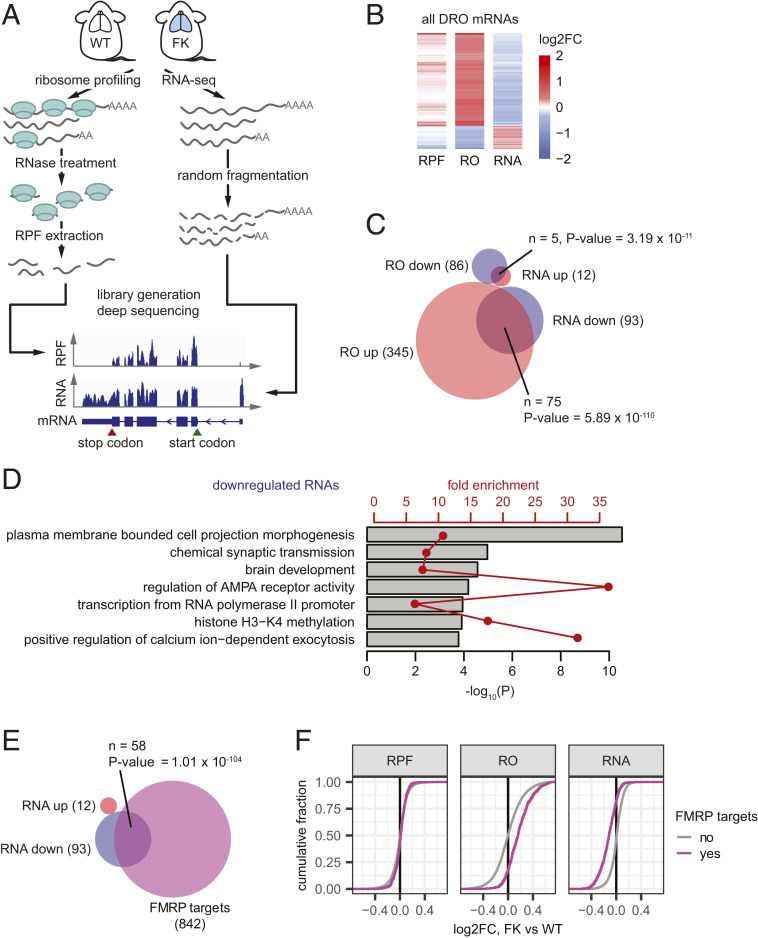
To identify mRNAs that are translationally dysregulated in FMRP-deficient mouse cortex, we performed ribosome profiling (29) and RNA-seq from WT and FMRP KO (FK) animals (Fig. 1A). Ribosome occupancy (RO), defined as ribosome protected fragments (RPFs) normalized to mRNA levels, is a measure of translational activity (29) and has served as a proxy for protein synthesis. Accumulating evidence suggests that one mechanism whereby FMRP inhibits translation is by stalling ribosome transit (5, 10, 12, 13), and indeed there is a moderately (10 to 15%) higher rate of protein synthesis in FMRP-deficient brain (3, 4, 30). Using Xtail (31), an algorithm that tests for differential ROs (DROs) between samples, we identified 431 mRNAs with DROs between FK and WT (false discovery rate [FDR] < 0.05; Fig. 1B, SI Appendix, Fig. S1A, and Dataset S1). Consistent with FMRP acting as a translation repressor, 80% of these mRNAs (345 of 431) have increased RO.
Fig. 1.
Steady-state RNA level changes drive translational buffering in Fragile X brain cortex. (A) Illustration of the experimental pipeline of ribosome profiling and RNA-seq for WT and FK mouse brain cortices. (B) Heat maps showing log2FC of RPF, RO, and RNA levels for mRNAs having statistical significant different ROs as calculated using Xtail (31) between FK and WT (two biological replicates for each genotype; FDR < 0.05). (C) Venn diagram showing the overlap of mRNAs with DRO and mRNAs that are DE. (D) Representative GO terms enriched for mRNAs down-regulated at the RNA level in the FK brain cortex. Gray bars and red points and lines show the −log10 P value and fold enrichments of each of these GO terms, respectively. Dataset S4 provides full lists of enriched GO terms. (E) Venn diagrams showing the overlap between the down-regulated mRNAs and FMRP binding targets (5). Numbers of RNAs in each group and in each overlap as well as P values of enrichment (hypergeometric test, upper tail) are indicated. (F) Empirical cumulative distribution function (ECDF) plots showing log2FC of RPF, RO, and RNA levels for FMRP binding targets (5) and other mRNAs.
To determine the underlying cause of DRO, we analyzed our RPF and RNA-seq data separately. DRO can result from translational dysregulation driven by differential RPF but with little change in RNA levels; conversely, translational buffering occurs when RPFs are unchanged but the DRO is driven by dysregulated RNA levels (32). The RPF changes in the DRO mRNAs are subtle; however, the changes in RNA levels strongly oppose the changes in RO (Fig. 1B). When taken as a group, these mRNAs with altered ROs have stronger changes at RNA levels than at RPF levels (SI Appendix, Fig. S1A).
We identified 12 and 21 mRNAs with strongest increase and decrease at RPF level (Dataset S2) and 12 and 93 mRNAs with strongest increase and decrease at steady-state RNA level (including Fmr1 mRNA; Dataset S3; see Materials and Methods). The 345 RNAs with increased RO in FK are significantly enriched for those with increased RPFs (n = 7; P = 4.17 × 10−11, hypergeometric test), but much more so for mRNAs with reduced steady-state levels (n = 75; P = 5.89 × 10−110, hypergeometric test). This is also the case for mRNAs with decreased RO (Fig. 1C). These data show that mRNA steady-state level changes drive the observed RO changes, likely via translational buffering in the FK brain cortex.
It is surprising that so few mRNAs have significantly altered RPFs; however, if FMRP stalls ribosomes as indicated by others (5, 10, 12, 13), then this may not be detected by steady-state ribosome profiling. To assess potential stalling changes in FK brain, we tested the variation of RPFs across transcripts measured by the coefficient of variation (CV) per transcript (12). With our very deep RPF libraries (∼19 million unique coding sequence mappable reads per sample), we did not detect any discernible changes in the CV per transcript in FK vs. WT brain (SI Appendix, Fig. S1B), indicating that steady-state ribosome profiling is unable to detect subtle changes in translation elongation. However, by blocking initiation and performing ribosome profiling during or after the ribosomes run off, Shah et al. (13) found that ∼50 mRNAs were associated with FMRP-stalled ribosomes.
Some of the top mRNAs down-regulated at the steady-state level include those that are involved in ion channel function [e.g., Pdzd2 (33) and Wnk2 (34)] and synapse development and communication [e.g., Bai2 (35) and Sipa1l3 (36)]. Gene Ontology (GO) analysis of down-regulated mRNAs shows an enrichment for functions related to neuronal dendrites, pre- and postsynapses, and channels and receptors (Fig. 1D and Dataset S4). The loss of these RNAs could contribute to the neurological defects related to FXS. The FMRP binding target RNAs in the brain are also highly enriched for these similar GO terms (5). Indeed, over two thirds of the down-regulated mRNAs (58 of 93) are FMRP targets (5) (enrichment P = 7.53 × 10−51, hypergeometric test; Fig. 1E), including the four example genes mentioned above. In contrast, there were no overlapping mRNAs among the 12 that were the most increased and the FMRP targets (enrichment P = 0.498, hypergeometric test). As a group, the FMRP target RNAs are significantly reduced [mean log2 fold change (FC) = −0.12; P < 2.2 × 10−16, one-tailed t test]. This observation indicates that the increased RO (mean log2FC = 0.17; P < 2.2 × 10−16, one-tailed t test) is caused by reduced RNA levels, although their changes at RPF level is very subtle (mean log2FC = 0.0093; P = 0.00292, one-tailed t test; Fig. 1F). These data show that, in the FMRP-deficient mouse brain cortex, translational buffering is driven by steady-state mRNA changes, particularly the down-regulation of the FMRP target mRNAs. Interestingly, the down-regulation of mRNAs, including FMRP binding targets, is observed across multiple FXS mouse and human models (SI Appendix, Fig. S1C), suggesting that reduction of these mRNAs could underlie FXS.
RNA Metabolic Profiling Reveals Disrupted RNA Stability in FMRP-Deficient Neurons.
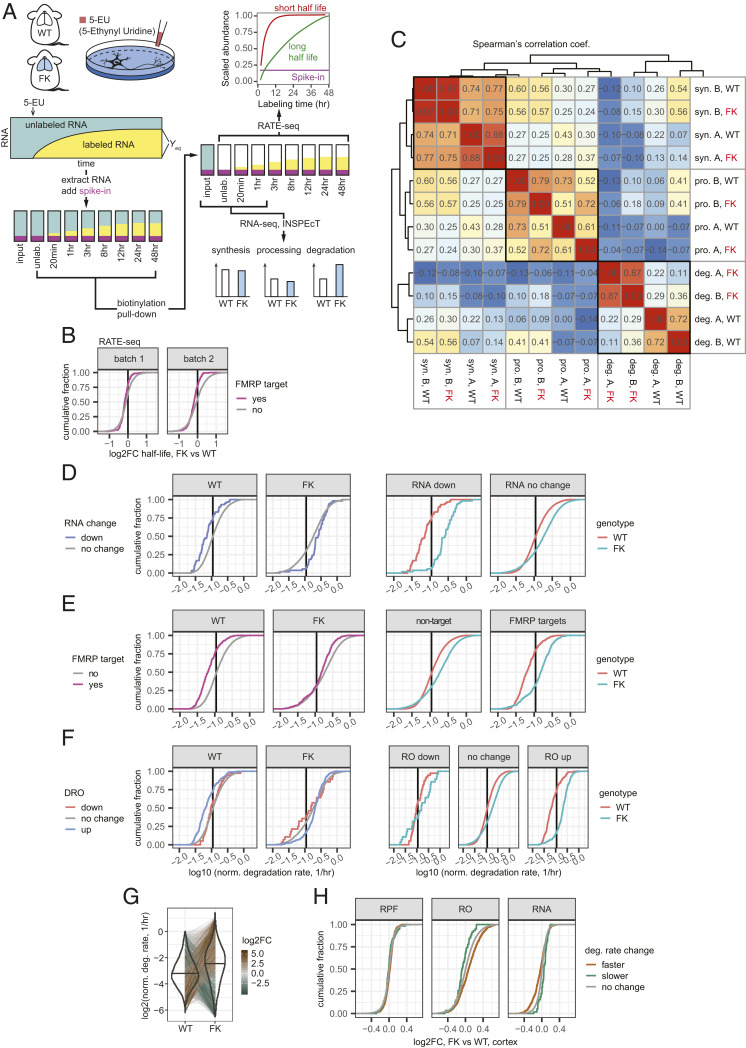
Because FMRP is mostly a cytoplasmic protein (11), we hypothesized that the down-regulation of its target (and other mRNAs) is due to a posttranscriptional mechanism, possibly destabilization upon loss of FMRP. To test this hypothesis, we have used RNA approach to equilibrium sequencing (RATE-seq) (37) to measure half-lives of the transcriptome in WT and FK neurons. This general method was developed over 40 y ago (38) and describes the fact that, upon incorporation of a nucleotide analog into nascent transcripts with increasing incubation periods, the fraction of transcripts that are labeled increases until a steady-state level has been reached. RNAs with faster degradation attain steady state at the earliest times.
To perform RATE-seq, we incubated WT and FK mouse cortical neurons [14 d in vitro (DIV)] with 5-EU for 0 min (for input and unlabeled control, or “unlab”), 20 min, and 1, 3, 8, 12, 24, and 48 h, after which the RNA was “clicked” to biotin and purified by streptavidin pull-down. The RNA was mixed with 5-EU–labeled Drosophila RNA and unlabeled yeast RNA as controls and sequenced together with total unenriched RNAs as input samples (Fig. 2A). The spike-in Drosophila and yeast RNAs for library generation were used as quality control measures (SI Appendix, Fig. S2 A–C). The sequencing reads mapped to the Drosophila transcriptome were then used as a normalization reference to confirm the approach to equilibrium curves of the labeled mouse transcripts (SI Appendix, Fig. S2D). After filtering (SI Appendix, Fig. S2E), we fit the normalized reads of each transcript over the labeling time course according to the approach-to-equilibrium equation and calculated the half-life of each transcript (Materials and Methods and SI Appendix, Fig. S3A).
Fig. 2.
Increased mRNA degradation rates in FK neurons correlate with the steady-state RNA depletion. (A) Illustration of the experimental pipeline of RNA metabolism profiling for WT and FK neurons. (B) ECDF plots of log2FC of half-lives in FK relative to WT neurons calculated using RATE-seq. (C) Heat map of Spearman’s correlation coefficients between synthesis (syn.), processing (pro.), and degradation (deg.) rates estimated from RNA-seq libraries generated from WT and FK neurons labeled for 20 min (A) or 1 h (B). Dendrogram shows the unsupervised hierarchical clustering using their Spearman’s correlation coefficients. (D–F) ECDF plots of normalized degradation rates for down-regulated mRNAs in FK cortex (D), FMRP binding targets (E), and mRNAs with DROs (F) and other mRNAs compared between the mRNA groups (Left) or between the genotypes (Right). The mean of the normalized degradation rates from libraries A and B of each mRNA was used. Black vertical lines denote the median degradation rates for all mRNAs. (G) Violin-and-line plot for the degradation rates of all mRNAs. The mean of the normalized degradation rates from libraries A and B of each mRNA was used. The black horizontal line in each violin denotes the median. Thin lines spanning WT and FK connect the values of the same mRNA in both genotypes. Brown-gray-green shades of the thin lines indicate the log2FC of the normalized degradation rates of each mRNA. (H) ECDF plots showing log2FC of RPF, RO, and RNA levels in FK cortex compared to WT for mRNAs with faster (brown) or slower (green) degradation rates.
We collected two independent biological replicates for each genotype (total of four sets of data). RNA-seq libraries were processed in two batches, each with one set of WT samples and one set of FK samples. We calculated half-lives for 3,194 mRNAs that are present in all four sets of libraries (Dataset S5), which include 163 FMRP CLIP targets. For each of the batches, more mRNAs in FK neurons show shorter half-lives compared to WT neurons [batch 1, 2,071 of 3,194 (∼65%) have decreased half-lives, mean log2FC = −0.10, P value for log2FC less than 0 < 2.2 × 10−16, one-tailed t test; batch 2, 1,774 of 3,194 (∼56%), mean = −0.037, P = 2.08 × 10−5]. This destabilization is even stronger for the FMRP targets. For batches 1 and 2, 130 of 163 (80%) and 127 of 163 (78%) FMRP targets show shortened half-lives (batch 1, mean log2FC for FMRP targets’ half-lives = −0.16, P value for FMRP target half-lives to reduce more than nontargets = 0.000665, one-tailed t test; batch 2, mean log2FC = −0.20, P = 1.71 × 10−12; Fig. 2B). This result confirms our hypothesis that neuronal mRNAs, especially FMRP targets, are less stable upon loss of FMRP.
One caveat of RATE-seq is that it is prone to batch effects (SI Appendix, Fig. S3B) (39). Consequently, we sequenced the input, unlabeled, 20-min labeled (A), and 1-h labeled (B) libraries to greater depth and used the inference of synthesis, processing and degradation rates from transcriptomic data (INSPEcT) program (40, 41) to estimate the RNA metabolic rates (synthesis, processing, and degradation) by comparing nascent and mature RNA concentrations in the 5-EU–labeled and input total RNA libraries. This strategy is fundamentally different from RATE-seq (Fig. 2A). We obtained metabolism rate information for 8,590 RNAs, which include 412 FMRP target mRNAs. The rates follow log-normal distributions with medians of 1.12 and 1.04 RPKM/h for synthesis, 6.84 and 6.60 h−1 for processing, and 0.13 and 0.14 h−1 for degradation for libraries A and B, respectively (SI Appendix, Fig. S4A). These values demonstrate the reproducibility of the assay.
We calculated Spearman’s correlation coefficients for all three metabolism rates for the two genotypes for both libraries (Fig. 2C). For synthesis, processing, and degradation, we observed decreasing correlation coefficients between WT and FK. For synthesis rates, WT and FK cluster together for the same labeling parameter (library A or B), indicating that there is little genotype difference. For libraries A and B, the correlation coefficients were 0.97 and 0.88 between WT and FK, again demonstrating that the synthesis rates between the two genotypes are similar. For processing rates, the two genotypes were also similar despite slightly lower Spearman’s correlation coefficients between WT and FK (0.79 and 0.61 for libraries A and B). Strikingly, the correlation coefficients for degradation rates between WT and FK were substantially lower (0.22 and 0.36 for libraries A and B), indicating that there is a major difference in RNA degradation between genotypes. The Spearman’s correlation coefficients between libraries A and B for each genotype (0.87 and 0.72, respectively) indicate high reproducibility. Therefore, the degradation rates for the four libraries are separated by genotype (Fig. 2C), demonstrating that RNA stability in FK neurons is disrupted.
We determined whether the following groups of mRNAs have altered degradation rates: 1) mRNAs that are down-regulated (Fig. 2D), 2) FMRP targets (Fig. 2E), and 3) mRNAs that have DROs (Fig. 2F). For convenience, we will refer to group 1 as down-regulated mRNAs and group 3 as RO-up/down mRNAs, respectively. Statistical tests were performed on log10-transformed degradation rates. In WT neurons, the down-regulated mRNAs degraded significantly slower than the other RNAs (P = 1.31 × 10−6, two-tailed t test). However, in FK neurons, the down-regulated mRNAs degraded significantly faster (P = 0.000684, two-tailed t test; Fig. 2 D, Left). Moreover, the mRNAs in FK neurons are globally destabilized relative to those in WT neurons (for the mRNAs not down-regulated in FK cortex, the difference of log10 degradation rate means in FK vs. WT = 0.17 h−1; P < 2.2 × 10−16, two-tailed t test); however, the down-regulated mRNAs are destabilized even more (difference of log10 degradation rate means in FK vs. WT = 0.53 h−1; P = 2.09 × 10−15, two-tailed t test; Fig. 2 D, Right). FMRP targets and RO-up mRNAs are also destabilized more than the global trend (Fig. 2 E and F). Interestingly, the RO-down mRNAs were resistant to mRNA destabilization upon loss of FMRP; their degradation rates are not significantly different in FK compared to WT neurons (P = 0.323, two-tailed t test; Fig. 2 F, Right). This resistance to transcriptome-wide destabilization could explain the observed steady-state up-regulation of these mRNAs (Fig. 1B).
We did not identify RNAs with significantly dysregulated synthesis or processing rates in FMRP-deficient neurons, but detected 748 RNAs with significantly altered degradation rates, of which 688 (92%) degraded faster in FK compared to WT (adjusted P value cutoff of 0.01; Fig. 2G and SI Appendix, Fig. S4B). The RNAs that degraded faster in FK neurons were significantly enriched for FMRP targets, as well as those that were down-regulated at the steady-state level in the cortex (SI Appendix, Fig. S4C). As groups, the RNAs that degraded faster or slower in FK neurons displayed decreased or increased steady-state levels (P = 5.49 × 10−14 and 1.47 × 10−4, two-tailed t test), as well as increased or decreased ROs in FK cortex, respectively (P < 2.2 × 10−16 and = 5.28 × 10−3), with no change at the RPF level (Fig. 2H). Based on these and the RATE-seq data, we conclude that there is widespread mRNA destabilization, including many of the FMRP targets, in the absence of FMRP; this in turn drives steady-state RNA level changes and translation buffering.
Loss of FMRP Uncouples the Link between Optimal Codons and mRNA Stability.
To identify features of mRNAs that are involved in this FMRP-dependent stability, we examined the correlation of brain cortex steady-state mRNA level changes or neuronal mRNA degradation rate changes with codon optimality [gene codon adaptation index (cAI) score; Materials and Methods and Datasets S6 and S7], coding sequence (CDS) guanine-cytosine (GC) content, and CDS 5′ and 3′ untranslated region (UTR) lengths and their minimal energy of folding (MEF; an indication of possible secondary structure; SI Appendix, Fig. S5A). The strongest and most consistent correlations with both cortical steady-state mRNA level changes and neuronal mRNA degradation rate changes were gene cAI scores and CDS GC content, which are correlated features themselves (42, 43). More specifically, the log2FC of cortical steady-state mRNA levels had a negative correlation with gene cAI scores (Spearman’s correlation coefficient = −0.22; P < 2.2 × 10−16), while the log2FC of the neuronal mRNA degradation rate had a positive correlation (Spearman’s correlation coefficient = 0.20; P < 2.2 × 10−16; SI Appendix, Fig. S5A). This means mRNAs which contain more optimal codons were the ones that had faster degradation rates and showed a consequent down-regulation in FK.
Codon optimality, a measure of the balance between the usage frequency of a given codon (demand) and supply of charged tRNAs encoding the complementary anticodon (21), is a major determinant of mRNA stability from yeast to vertebrates (22–27). Generally, mRNAs with more optimal codons (high gene cAI scores; presumably with faster decoding rates) are more stable than mRNAs using less optimal codons, connecting translation regulation to mRNA stability. Because FMRP regulates translation, codon optimality could be a mechanism that links FMRP-mediated translation to mRNA stability. Therefore, we investigated the role of codon optimality in FMRP-dependent mRNA stability.
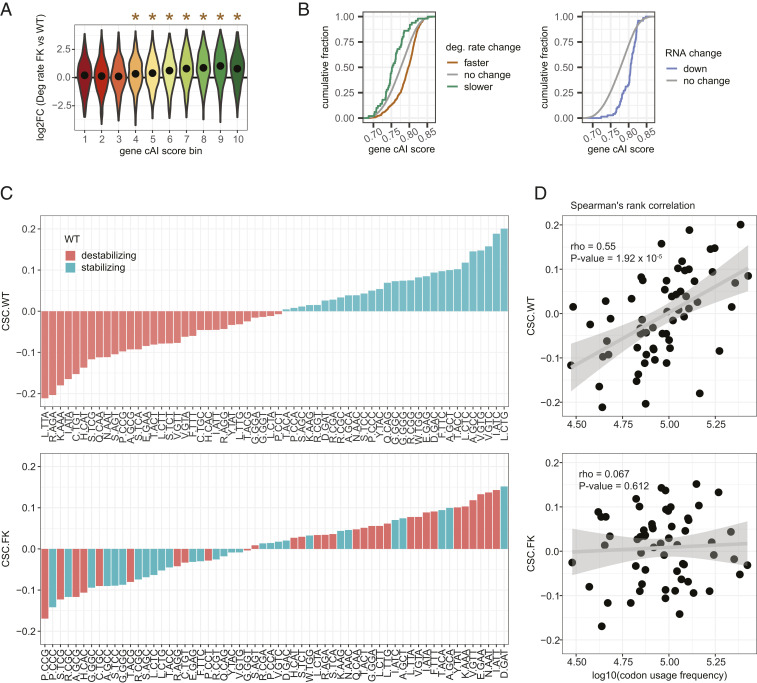
We grouped all detectable mRNAs into 10 equal-sized bins of increasing gene cAI score and examined which bins were particularly affected by mRNA degradation rates (Fig. 3A). mRNAs in bins 4 to 10 degraded significantly faster in FK neurons than in WT neurons (Holm-adjusted P < 0.01, one-tailed t test); the higher the cAI score bin, the faster the degradation rates become. However, mRNAs in bins 1 to 3 were barely changed, indicating that mRNAs containing nonoptimal codons are resistant to destabilization upon loss of FMRP.
Fig. 3.
Increased mRNA degradation rates in FK neurons are codon optimality-dependent. (A) All mRNAs were grouped into 10 equal bins based on their gene cAI scores. Bin 1 contains genes with gene cAI scores of the lowest quantile, and bin 10 contains genes of the highest quantile. Violin plots show log2FC of degradation rates in FK vs. WT neurons for mRNAs in each gene cAI score bin. The point in each violin denotes the median of the bin. Brown stars indicate the median of the bin greater than 0 (Holm-adjusted P < 0.01, one-tailed t test). No bin had a median less than 0. (B) ECDF plots of gene cAI scores for mRNAs with faster or slower degradation rates in FK neurons (Left) or reduced mRNA levels in FK cortex (Right). (C) Bar graphs for CSCs for each codon arranged from minimum to maximum in WT (Upper) and FK (Lower) neurons. The color of each bar indicates the codon as stabilizing (CSC > 0; green) or destabilizing (CSC < 0; orange) in WT neurons. The amino acid for each codon is indicated. (D) Scatter plots and linear regressions of the CSCs as a function of log10(codon usage frequency) of the top 10% of expressed genes in WT (Upper) and FK (Lower) neurons. Spearman’s rank correlation coefficients and P values of the correlations are indicated.
Compared to the general transcriptome (gene cAI scores = 0.78 ± 0.038, mean ± SD), mRNAs that degrade faster in FK neurons (gene cAI scores = 0.79 ± 0.033) and the mRNAs that are down-regulated in the cortex (gene cAI scores = 0.80 ± 0.023) have significantly more optimal codons (P < 2.2 × 10−16 and 1.12 × 10−14, two-tailed t test). However, the mRNAs that degrade slower in FK neurons (gene cAI scores = 0.76 ± 0.034) are significantly less optimal (P = 2.85 × 10−4, two-tailed t test; Fig. 3B). tRNA adaptation index (tAI), a metric that measures codon optimality by approximating tRNA abundance based on their gene copy number, leads to the same conclusions (SI Appendix, Fig. S5B). These results indicate that FMRP stabilizes mRNAs that have an optimal codon bias.
Because optimal codons may confer stability to mRNAs (22–27), we examined whether this link is uncoupled upon loss of FMRP. The codon-stability coefficient (CSC), which describes the link between mRNA stability and codon occurrence, has been calculated for each codon from yeast to human (23, 24, 26, 27). We determined whether this relationship is maintained in the absence of FMRP. The CSC values in WT neurons ranged from <−0.2 to >0.2 (Dataset S6), which is comparable to previously reported CSC values for human cell lines and mouse embryonic stem cells (SI Appendix, Fig. S5C). This differs from what has been described in Drosophila, where the neuronal CSC is attenuated relative to somatic cells (24). Of the 60 non-start or -stop codons, 29 had CSCs greater than 0 (stabilizing codons) and 31 had CSCs smaller than 0 (destabilizing codons; Fig. 3 C, Upper). Strikingly, 17 codons that are stabilizing in WT are destabilizing in FK neurons, and 21 codons changed in the opposite direction (Fig. 3 C, Lower, and Dataset S6). Optimal codons are prevalent in abundant mRNAs and are associated with positive CSCs (i.e., are stabilizing codons) (23, 24, 26, 27), as we have observed in WT neurons (Spearman’s r = 0.55; P = 1.92 × 10−5; Fig. 3 D, Upper). In FK neurons, however, this relationship breaks down. Codon usage frequencies in the top 10% of highly expressed mRNAs are nearly the same as in WT (Dataset S6), but the correlation between codon usage frequencies and CSCs is nearly random in FK (r = 0.067; P = 0.612; Fig. 3D). These results show that the link between codon-mediated RNA stabilization and codon usage bias is uncoupled in FMRP-deficient neurons.
FMRP May Stabilize Codon Optimality-Dependent mRNA Stability Directly and through Other Factors.
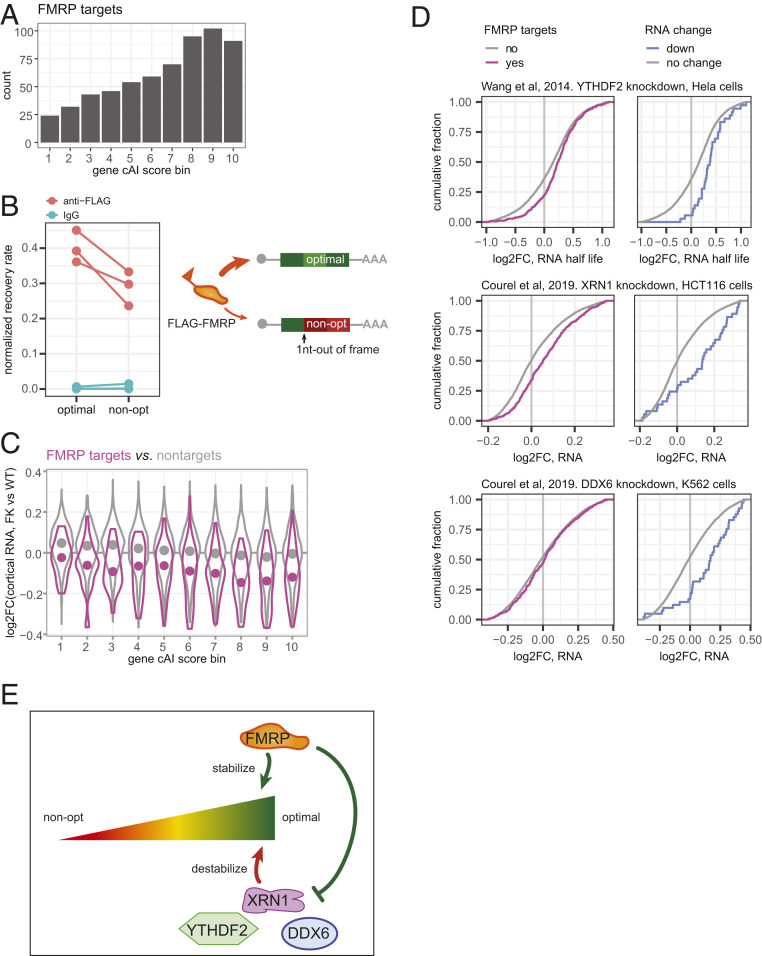
We considered possible mechanisms for how FMRP could regulate codon optimality-dependent mRNA stability. Because the down-regulated mRNAs are enriched for FMRP targets, FMRP may stabilize them through direct binding. We noticed a bias for high gene cAI scores of the FMRP targets (Fig. 4A) and hypothesized that FMRP may associate with mRNAs of higher codon optimality. To test this, we compared a pair of previously described reporter mRNAs that are enriched in optimal or nonoptimal codons (27). They differ by a single nucleotide insertion to induce a frame shift, and thus they have almost identical nucleotide compositions. HEK293T cells were transduced with these reporters together with a FLAG-tagged FMRP-expressing vector (44). Subsequent anti-FLAG immunoprecipitation and RT-qPCR showed that the reporter with optimal codons was more enriched compared to the reporter with nonoptimal codons (P = 0.0261, one-tailed paired t test; Fig. 4B). This result suggests that FMRP can indeed “identify” mRNAs with optimal codons as opposed to general nucleotide composition.
Fig. 4.
FMRP and other factors may regulate optimal codon-dependent mRNA stability. (A) Bar graph of count of FMRP target genes in each gene cAI score bin. (B) RNA immunoprecipitation (RIP) testing FMRP recruitment to reporter mRNAs that use optimal versus nonoptimal codons. The reporter that uses nonoptimal codons was generated using 1 nt insertion into the reporter that uses optimal codon (Left) (27). Recovery rate of the reporters and of Fmr1 RNA was calculated as the ratio of immunoprecipitated relative to input RNA. Normalized recovery rate was then calculated by normalizing the recovery rate of the reporter by the recovery rate of the Fmr1 mRNA, the latter as an internal control for the RIP experiment. Each point, a biological replicate, is an average of two or three technical replicates. Each line connects data from the two reporters in a paired experiment that was performed independently from other pairs (P = 0.0261, one-tailed paired t test). (C) Violin plots show log2FC of steady-state RNA levels in FK vs. WT cortex for all FMRP targets (purple) or nontargets (gray) in each gene cAI score bin. The point in each violin denotes the median of the bin. (D) ECDF plots showing log2FC of RNA levels for FMRP binding targets (Left) and mRNAs down-regulated in FK cortex (Right) after knock-down of YHDF2 (45), XRN1, and DDX6 (46) in human cell lines. Only genes with unique mouse orthologs were considered. (E) Model of how FMRP could stabilize mRNAs that use optimal codons directly and through other factors.
We next examined whether FMRP binding could impact RNA stability. As a group, the degradation rates of FMRP CLIP targets increased more than the nontarget RNAs (P = 1.36 × 10−9, one-tailed t test; SI Appendix, Fig. S6A). Because too few FMRP targets (30%) were detected in the degradation rate data set, we focused on the steady-state RNA changes and compared RNAs in each cAI score bin. Similar to the global trend discussed above (Fig. 3A), FMRP targets with more optimal codons were also more strongly down-regulated than those with nonoptimal codons (Spearman’s correlation coefficient of RNA log2FC and gene cAI score bin = −0.21; P = 8.27 × 10−8). However, FMRP targets in all but the lowest codon-optimality bin were significantly reduced in the absence of FMRP (Holm-adjusted P < 0.01, one-tailed t test; Fig. 4C, red). This is different from the codon optimality-matched nontargets (Fig. 4C, black). The nontarget mRNAs also showed an optimal codon-biased down-regulation (Spearman’s correlation coefficient of RNA log2FC and gene cAI score bin = −0.20; P < 2.2 × 10−16), but only RNAs in cAI bins 7, 8, and 9 are significantly down-regulated (Holm-adjusted P < 0.01, one-tailed t test). In all gene cAI score bins, the log2FC of the FMRP targets are significantly more negative than the nontarget RNAs (Holm-adjusted P < 0.01, one-tailed t test). These results suggest that the stability of the FMRP target RNAs is not only affected by their codon optimality, but also by the FMRP binding.
Not all mRNAs that are depleted in the FK cortex are FMRP CLIP targets, suggesting that FMRP could also stabilize mRNAs through other factors. For example, YTHDF2 was shown to antagonize FMRP’s stabilizing effect for some FMRP targets in mouse brain cortex (20), and knockdown of this protein in human cell lines (45) leads to longer half-lives of the FMRP targets [D = 0.14; P = 2.19 × 10−7, two-sided Kolmogorov–Smirnov (KS) test] and of mRNAs down-regulated in the FK cortex (D = 0.37; P = 3.56 × 10−5, two-sided KS test; Fig. 4D).
We examined RNA-seq datasets to assess other factors that might be involved in FMRP-mediated optimal codon-dependent mRNA stability. We selected factors that target mRNAs of higher GC content (46) or higher codon optimality (47), and therefore the RNA level changes upon depletion of these factors likely affects their stability (Fig. 4D and SI Appendix, Fig. S6B). Depletion of XRN1 (5′−3′ exonuclease) and DDX6 (DEAD-Box Helicase 6) in human cell lines (46) leads to up-regulation of the mRNAs down-regulated in FK cortex (D = 0.42, P = 4.05 × 10−8; D = 0.37, P = 2.24 × 10−6, two-sided KS test; Fig. 4D). On the other hand, depletion of FXR1 (Fragile X Mental Retardation Syndrome-Related Protein 1) (48) leads to down-regulation of the orthologs of FMRP targets (SI Appendix, Fig. S6B). These results suggest that XRN1 and DDX6 might antagonize FMRP while FXR1 might cooperate with it to stabilize mRNAs of high codon optimality (Fig. 4E).
CPEB1 Depletion Rescues FXS by Rebalancing RNA Homeostasis.
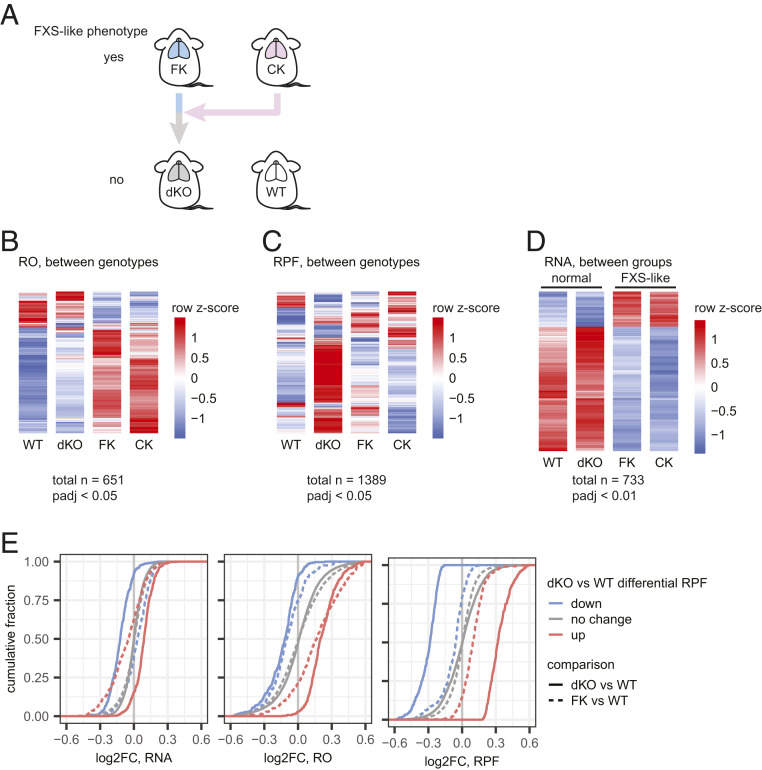
Nearly all pathophysiologies associated with FMRP deficiency are rescued in FMRP/CPEB1 double knockout (dKO) mice (10). To assess how this rescue could be achieved, we performed ribosome profiling and RNA-seq on CPEB1 KO (CK) and dKO mouse brain cortices in addition to the WT and FK samples as described above (Fig. 5A). We identified 651 genes with DROs among the four genotypes (FDR < 0.05; Fig. 5B and Dataset S1), including the 431 genes with DRO between FK and WT described above (Fig. 1B). Importantly, 425 of these DROs were rescued in the dKO cortex (i.e., not significantly different compared to WT). Unexpectedly, >50% of RNAs with DRO in CK (204 of 359) also had DROs in FK and were changed in the same direction (i.e., increased or decreased). These molecular data are consistent with previous observations such as dendritic spine number and metabotropic glutamate receptor-mediated long-term depression (mGLuR-LTD), which are nearly identically aberrant in the two single KO mice but rescued to normal in dKO animals (10). Because of the molecular similarities between WT and dKO, and between FK and CK, we henceforth refer to these two groups as “normal” and “FXS-like.”
Fig. 5.
Genetic rescue of FX by CPEB KO is rebalanced at RNA level. (A) Illustration of the genetic rescue paradigm described previously (10). (B and C) Heat maps showing mRNAs having differential RO (B) and RPF (C) between any two genotypes of the four genotypes noted above. (D) Heat maps showing mRNAs that are DE between the normal (WT and dKO) and FXS-like groups (FK and CK). Red and blue shades of the heat maps show high or low z-scores calculated for each mRNA (row) across all samples. Each genotype has two biological replicates. (E) ECDF plots for log2FC of RNA, RO, and RPF in FK (dotted lines) and dKO (solid lines) relative to WT for mRNAs with significantly up- (red) or down-regulated (blue) RPFs in dKO relative to WT.
To determine the underlying molecular driver of the DRO among the four genotypes, we analyzed our RPF and RNA-seq data separately. At an adjusted P value (Padj) <0.05, only 23 and 21 RPFs were significantly different between FK and WT and between CK and WT, respectively. Conversely, the dKO was the most different from WT, with 410 and 333 RPFs that were significantly higher or lower (Fig. 5C and Dataset S2). Therefore, this genetic rescue of FXS-like phenotypes in the dKO is not achieved by correcting translational activity as defined by the number of RPFs. In contrast, the RNA-seq heat map displayed a mirror image of the DRO heat map (SI Appendix, Fig. S7A). Compared to WT, the expression of 50 genes was dysregulated in FK (Padj < 0.05; 10 up-regulated, 40 down-regulated), along with 145 in CK (Padj < 0.05; 13 up-regulated, 132 down-regulated), but only 2 in dKO (Padj < 0.05; Cpeb1 and Fmr1; Dataset S3). The differentially expressed (DE) genes in FK and CK were largely identical. Among the 10 and 13 genes up-regulated in FK and CK, 7 overlap (P = 8.72 × 10−25, hypergeometric test, upper tail); among the 40 and 132 genes down-regulated, 35 overlap (P = 3.91 × 10−72, hypergeometric test, upper tail). Because the transcriptome profiles in FK and CK are as similar as the WT and dKO profiles, we performed an unsupervised hierarchical clustering to test for sample-to-sample similarities (SI Appendix, Fig. S7B). FK and CK formed one cluster while WT and dKO formed another, validating the FXS-like vs. normal grouping at the RNA level.
Having validated the grouping, we tested for DE RNAs in the FXS-like group (FK and CK) relative to the normal group (WT and dKO). The DE RNAs between the groups are changed the same direction (i.e., up or down) in the single KOs and are rescued in the dKO to WT levels. With greater statistical power than when comparing between single genotypes, we identified 733 DE RNAs in the FXS-like group (Padj < 0.01), 162 (22.1%) up-regulated and 571 (77.9%) down-regulated (Fig. 5D and Dataset S8). Almost all down-regulated RNAs in FK compared to WT are down-regulated in the FK-like group (87 of 93). Over 77% of the RNAs with up-regulated ROs in FK vs. WT (267 of 345) were significantly reduced in the FXS-like group (P = 0, hypergeometric test, upper tail). Similarly, 42% of the RNAs with down-regulated ROs in FK (36 of 86) were significantly increased steady-state levels (P = 3.28 × 10−51, hypergeometric test, upper tail). Therefore, we conclude that rescue of FXS in FMRP/CPEB1 dKO animals is achieved by rebalancing the transcriptome.
Many mRNAs show differential RPFs in dKO compared to WT. We investigated their RNA, RO, and RPF level changes in FK and dKO compared to WT (Fig. 5E). The RNA levels were only mildly changed (mean RNA log2FC for RPF-up mRNAs = −0.065, for RPF-down mRNAs = 0.019; Fig. 5 E, Left, dotted lines) in FK, but the slightly stronger change in RO (mean RO log2FC for RPF-up mRNAs = 0.199, for RPF-down mRNAs = −0.107; Fig. 5 E, Middle, dotted lines) suggests subtle translational dysregulation and consequent subtle changes in RPFs (mean RPF log2FC for RPF-up mRNAs = 0.105, for RPF-down mRNAs = −0.092; Fig. 5 E, Right, dotted lines). Upon deletion of CPEB1 in the dKO, the ROs remain about the same as in FK (mean RO log2FC for RPF-up mRNAs = 0.251, for RPF-down mRNAs = −0.153; Fig. 5 E, Middle, solid lines), suggesting the subtle translational dysregulation caused by loss of FMRP is not rescued. However, the RNA levels also changed, albeit mildly, in the same direction as their ROs (mean RNA log2FC for RPF-up mRNAs = 0.086, for RPF-down mRNAs = −0.127; Fig. 5 E, Left, solid lines). That is, in dKO, reduced RO is accompanied by reduced RNAs, and vice versa. These mild but additive changes are reflected in strong alterations in RPFs (mean RPF log2FC for RPF-up mRNAs = 0.363, for RPF-down mRNAs = −0.325; Fig. 5 E, Right, solid lines). These observations suggest there may be mild translational dysregulation in the FK brain, but it is not rescued by the deletion of CPEB1 in the dKO. The main change introduced by removing CPEB1 was indeed at the RNA level.
Discussion
It is widely assumed that excessive protein synthesis is not only a corollary of FXS, but a proximate cause of the disorder (49). Consequently, several studies have used high-resolution whole-transcriptome methods to identify mRNAs that are either bound by FMRP (5–9) or whose translation is dysregulated in FMRP-deficient mouse models (5, 12, 13, 50, 51). Although nearly 1,000 FMRP binding targets have been identified in the mouse brain, surprisingly few are dysregulated at the translational level, and, of those that are, the dysregulation is usually modest. Moreover, in even fewer cases has the dysregulation been linked to FXS pathophysiology (52). Therefore, the functional output of FMRP binding remains largely elusive.
Our study shows that, in the FK mouse brain cortex, steady-state mRNA levels are globally disrupted, which drives the dysregulated translational buffering (Figs. 1 and 5). FMRP targets are particularly down-regulated, which happens specifically in neurons (8, 50, 53). By metabolic labeling and RNA-seq in neurons, we show that the loss of FMRP results in reduced stability not only of its direct target substrates, but also of other mRNAs with an optimal codon bias (Figs. 2 and 3). This instability drives the changes in steady-state RNA levels and translational buffering in the cortex (Fig. 2). Moreover, our data demonstrate that RNA stability conferred by optimal codons is mediated by FMRP and possibly other transacting factors. The loss of FMRP leads to a massive reshuffling of the identities of stabilizing versus destabilizing codons (Fig. 3).
How does FMRP stabilize mRNAs that use optimal codons? One possibility is that FMRP can stabilize its targets by directly interacting with the translational machinery. We show that FMRP preferentially binds an mRNA reporter that contains optimal codons over a reporter that contains nonoptimal codons but has nearly the same nucleotide composition (Fig. 4B). Upon loss of FMRP, target mRNAs are destabilized more than nontargets matched for their codon optimality (Fig. 4C), suggesting FMRP binding could lead to enhanced stability in addition to that provided by optimal codons. In yeast, the Ccr4–Not complex directly binds the empty E site of the ribosome during ineffective decoding on a nonoptimal codon, thereby rendering the mRNA more susceptible to degradation (54). One can postulate that the E site on an optimal codon is more frequently occupied compared to a nonoptimal codon, and therefore is less likely to be recognized by the Ccr4–Not complex. Moreover, the N terminus of Drosophila FMRP was also shown to be able to bind directly to ribosomes (55). If these findings are conserved in mammalian systems, FMRP binding to ribosomes may further reduce the chance of Ccr4–Not binding.
We cannot rule out the possibility that FMRP may “sense” other features of mRNA, e.g., GC content of the coding sequence. In mammalian systems, mRNAs that contain many optimal codons inevitably have higher GC content in the coding sequence, which predicts a stronger secondary structure (43). Interestingly, mRNAs that have reduced steady-state RNA levels or have increased degradation rates tend to have long CDSs, which also predicts more CDS mRNA structures, as suggested by their low minimum free energy (MFE; SI Appendix, Fig. S3). In mammalian cells, strong secondary structure in the CDS stabilizes mRNA, possibly by preventing strand-specific endonucleolytic cleavage or ribosome collisions (43). FMRP binding may help stabilize such structures in the CDS, thereby reducing RNA degradation. Indeed, there is at least in vitro evidence that RNA secondary structure is recognized by FMRP (1). On the other hand, DDX6 was shown to mediate mRNA decay of GC-rich mRNAs, possibly by facilitating unwinding of the secondary structures throughout the transcripts and facilitating XRN1 progression (42). Therefore, it is possible that FMRP stabilizes the mRNAs by antagonizing DDX6 and XRN1. Indeed, the orthologs of the mRNAs down-regulated in the mouse FK brain are up-regulated upon loss of DDX6 and XRN1 in human cells (Fig. 4D), suggesting FMRP, DDX6, and XRN1 may share common targets. By interacting with DDX6, FMRP could regulate a much wider pool of mRNAs than those in the FMRP target list (5).
Our data show that nonoptimal codons can make mRNAs refractory to FMRP deficiency-induced destabilization (Fig. 3A and SI Appendix, Fig. S5B), while they are not essential. For instance, the mRNAs with reduced ROs in FK (not destabilized; Fig. 2F) are not particularly enriched for nonoptimal codons. However, they have short CDSs and 3′ and 5′ UTRs (SI Appendix, Fig. S8A) and are replete with 5′TOP mRNAs (56) (21 of 86; P = 7.5 × 10−34, hypergeometric test, upper tail). These features are in stark contrast to the RO-up mRNAs (destabilized in FK neurons; Fig. 2F), which have long UTRs and CDSs and are depleted of 5′TOP mRNAs (0 of 345; P = 0.80, hypergeometric test, upper tail). We speculate the RO-up and RO-down mRNAs are under the control of distinct posttranscriptional regulation programs.
In the dKO, rebalancing of RNA levels is accompanied by, and probably necessary for, mitigation of FXS-like phenotypes in mice by CPEB1 depletion (Fig. 5) (10). Among the mRNAs dysregulated between the FXS-like group (i.e., FK and CK) and the normal group (WT and FK/CK dKO), GO analysis shows that many up-regulated and rescued mRNAs have protein synthetic functions, including ribosome biogenesis, translation, and protein folding, while the down-regulated and rescued mRNAs have cell projection, synaptic transmission, as well as transcription and chromatin functions (SI Appendix, Fig. S8B and Datasets S9 and S10). Several important points come from this analysis. First, the up-regulated mRNAs are among the highest expressed in the brain (SI Appendix, Fig. S8C), and their protein products could promote general protein synthesis. This could contribute to the net increase in protein output in the FXS brain (3, 4, 30). Second, we find that FMRP regulates the levels of mRNAs that encode chromatin modifying factors, which is reminiscent of other observations showing that FMRP controls the synthesis of epigenetic regulators, albeit at the translational level (13, 52). Third, the brain- and neuron-related GO terms enriched for the down-regulated RNAs reflect the neural dysfunction that occurs in FXS. Indeed, the down-regulated RNAs are also significantly enriched for those related to autism, as compiled by the Simons Foundation Autism Research Initiative project (57) (P = 3.56 × 10−7, hypergeometric test, upper tail; SI Appendix, Fig. S8D). Therefore, restoring RNA homeostasis could be a key to FXS treatment.
The precise molecular mechanism(s) by which the lack of CPEB1 rescues FXS-like phenotypes in mice is likely to be complex. This is not surprising given that RNA-binding proteins can perform a variety of tasks that depend on the RNAs and/or proteins to which they are bound in specific tissues. For example, although FMRP inhibits protein synthesis, recent evidence indicates it also activates translation (9, 58) as well as transcription. In a similar vein, CPEB1 activates translation by elongating poly(A) tails (59) but also alters 3′ UTR length (60), which may affect mRNA translation, stability, or subcellular localization. Therefore, a simple balancing of protein synthesis by the two proteins as originally proposed by Udagawa et al. (10) is almost certainly too simplistic. This may also be the case with other mouse FXS rescue paradigms (61).
Materials and Methods
Animals.
WT, FK, CK, and dKO mice were as used previously (10). Specifically, FK (JAX stock no. 004624) and its WT controls (JAX stock no. 004828) were purchased from the Jackson Lab. CK were created in-lab (62). Mice were bred as previously described (10). All mice were maintained in a temperature- (25 °C), humidity- (50 to 60%), and light-controlled (12 h light/dark cycle) pathogen-free environment. Animal protocols (no. 1158) were approved for use by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Ribosome Profiling and RNA-Seq in Cortex.
Two mice per genotype were used for ribosome profiling and RNA-seq. The brain was rapidly removed from postnatal day (P) 28 to P35 mice, rinsed in ice-cold dissection buffer (1× HBSS + 10 mM Hepes-KOH), and rapidly dissected in dissection buffer ice/liquid mixture to collect cerebral cortex as described previously (63). Both cortex hemispheres were homogenized in 900 µL of homogenization buffer (63). Five hundred microliters of the resulting ∼700 µL supernatant (cytoplasmic lysate) was used for ribosome profiling using RNase A and T1 digestion (64), and the rest was used for RNA-seq. Ribosome profiling and RNA-seq libraries were prepared following published protocols (65) and sequenced with Illumina NextSeq. More details are provided in SI Appendix, Supplementary Text.
Spike-In RNA for RNA Metabolism Profiling.
Drosophila melanogaster (fly) Schneider 2 cells were grown in 12 mL Schneider’s insect medium (Sigma-Aldrich, cat. no. S0146) containing 10% (vol/vol) fetal bovine serum (Sigma-Aldrich, cat. no. F2442) at 28 °C until confluent. Cells were incubated with 200 µM 5-EU for 24 h and were washed, pelleted, and snap-frozen in liquid nitrogen. RNA was extracted using TRIzol.
Saccharomyces cerevisiae (yeast) cells were grown in 10 mL YEP medium containing 3% glucose at 30 °C until OD600 reached 0.5. Cells were then pelleted, and RNA was extracted using hot acidic phenol (66).
RNA Metabolic Profiling with Cortical Neuron Cultures.
DIV14 mouse cortical neurons were used for 5-EU labeling experiments. One million cells in each 60-mm dish were incubated with 200 µM 5-EU (Click-iT Nascent RNA Capture Kit; Invitrogen, cat. no. C10365) for 0 min (input and unlabeled), 20 min, and 1, 3, 8, 12, 24, and 48 h. Two dishes were used for unlabeled and 20-min-labeled samples, respectively. Neuron RNA was extracted using TRIzol (Invitrogen, cat. no. 15596018). The 5-EU–labeled RNA was enriched, and the RNA-seq library was prepared by adapting the Forrest et al. protocol (67). For each WT and FK, two independent batches of neurons were prepared, and each batch resulted in one of each input, unlabeled and 20-min– to 48-h–labeled libraries (Dataset S11). SI Appendix, Supplementary Text, includes more details.
Differential Translation and RNA Expression Analysis.
Brain cortex ribosome profiling and RNA-seq reads were processed as previously described (51). Batch-corrected counts were used to identify differentially translated/expressed genes with DESeq2 (68) (RPF and RNA) or Xtail (31) (RO). To identify mRNAs with significant DROs, an FDR < 0.05 was used as cutoff. To identify mRNAs with strongest changes at RPF and RNA levels, a Padj < 0.1 was used as cutoff. More details are provided in SI Appendix, Supplementary Text.
Ribosome Coverage Variability Analysis.
P-site offsets were calculated by examining the cumulative distribution of 27- to 34-nt reads aligning at start codons using the package Plastid (69). After applying the respective offset to reads of each size, only in-frame reads were used to determine the P-site codon. The first 15 and the last 5 codons of each transcript were removed from the reference annotation. Scripts are available at https://github.com/elisadonnard/riboEPAsite. For each gene, the highest-expressed isoform in WT samples was used. Mean codon coverage per transcript was calculated for each sample (number of RPFs per codon), as was the coefficient of variation (CV) in codon coverage (SD in the number of RPFs per codon divided by mean), similar to Das Sharma et al. (12). To compare WT and FMRP KO samples, the log2(CV in codon RPFs) per transcript was plotted versus log2(mean codon RPF coverage). A linear model fit per sample was calculated using geom_smooth from the ggplot2 package [method = “gam”, formula = y ∼ s(x, bs = “cs”)].
GO Analysis.
GO enrichment analysis was performed using Cytoscape with the ClueGo (70) plug-in (v2.3.3), with genes that are expressed in the mouse cortex as the reference gene set. Specifically, biological-function GO terms of levels 6 to 13 were tested for enrichment at Padj value <0.05 (right-sided hypergeometric test). Enriched GO terms that are similar were then fused to a group based on their Kappa score, which quantifies percentage of common genes between terms. The leading group terms, which are the terms with highest significance in each group, are presented in Fig. 1D and SI Appendix, Fig. S8B. All enriched terms are in Datasets S4, S9, and S10.
RNA Metabolism Profiling Analysis.
Reads generated from the RNA metabolism profiling libraries were processed as for cortical RNA-seq libraries described above, except that a mouse–fly–yeast merged genome (mm10 + dm6 + sacCer3) was used as the reference genome for reads mapping by hisat2 (71). The mapping statistics here were used for quality control and filtering purposes (SI Appendix, Fig. S2 A–E). The RATE-seq analysis was then performed using tools in the Determination of Rates Using Intron Dynamics pipeline (39). For INSPEcT analysis, uniquely mapped reads that are depleted of rRNA, tRNA sequences, and PCR duplicates were again mapped to mm10 genome with hisat2. Intron and exon read quantification and RNA metabolism rate (synthesis, processing, and degradation) estimation was performed using INSPEcT (40) (v1.10.0), with the degDuringPulse parameter set to TRUE. SI Appendix, Supplementary Text, provides more details.
tRNA and Codon Adaptation Index (tAI and cAI).
The codon adaptation index was calculated for a given sample as described by Sharp and Li (72). Briefly, for each sample, a set of the top 10% expressed genes was defined using batch-corrected TPM; the relative synonymous codon usage was then calculated, dividing the observed frequency of each codon by the frequency expected assuming all synonymous codons for a given amino acid are used equally; the cAI is then calculated by comparing the frequency of each codon to the frequency of the most abundant (or optimal) codon for a given amino acid. All codes used to perform this analysis are available on GitHub (https://github.com/elisadonnard/CodonOPT). This pipeline also includes codon tAI and calculates gene tAI scores. Codon cAI and gene cAI scores were calculated for both brain cortex and neurons using their respective transcriptomes (Datasets S6 and S7). Because the results from cortex and neurons are largely identical, only WT neuron codon cAI and gene tAI and cAI scores are presented.
CSC Analysis.
CSCs were calculated as previously described (23, 24, 26, 27). SI Appendix, Supplementary Text, provides more details.
MFE Analysis.
The sequences of 5′UTR, CDS, and 3′UTR were first extracted from the highest-expressing isoform of each gene using Biostrings (v2.54.0) (73). MFE of each sequence was then calculated using the RNALfold module in ViennaRNA (v2.4.14) (74) with the default parameters.
RNA Immunoprecipitation.
Cultured HEK 293T cells (ATCC) were transfected with Lipofectamine 3000 (Invitrogen) based on manufacturer instructions in a six-well plate. At 48 h post transfection, cytoplasmic fraction was collected by lysing the cells with polysome buffer, as was used for ribosome profiling (see above), plus 1% Nonidet P-40. Ten percent of the cleared lysate was put aside as input sample, and the rest of the lysate was diluted 1:10 with NT2 buffer (75) and used for immunoprecipitation using antibody-precoated Protein G Dynabeads (Invitrogen) for 40 min at 4 °C by end-to-end rotation. The antibodies used were monoclonal ANTI-FLAG M2 antibody (Sigma-Aldrich) or normal mouse IgG (Santa Cruz). After washing three times with NT2 buffer, RNA was extracted using the TRIzol Reagent (Invitrogen) from the beads and from the input lysate. cDNA was made with a QuantiTect Reverse Transcription Kit (Qiagen). qPCR was performed to quantify the reporters (27) with mCherry sequences and Fmr1 gene, with primers mCherry-F gtacgggtcaaaagcttacgtt, mCherry-R ccttctggaaatgaaagtttaagg, Fmr1-F cgcggtcctggatatacttc, and Fmr1-R tggagctaatgaccaatcactg. More details are provided in SI Appendix, Supplementary Text.
Code Availability.
Codes used to perform ribosome coverage variability analysis and cAI analysis are available on GitHub (https://github.com/elisadonnard/riboEPAsite and https://github.com/elisadonnard/CodonOPT). Other customized R scripts for data analysis are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Emmiliano Ricci for sharing his experience and protocol for ribosome profiling, Mariya Ivshina for advice and help with the mouse breeding, Lindsay Romo for sharing her protocol for 5-EU labeling of neuron culture, and Jeff Coller (Case Western Reserve University) for Click-iT EU tagging–RNA-seq protocol. We also thank Ariel Bazzini (Stowers Institute for Medical Research) for his very helpful discussion and input. Nathan Gioacchini and Yongjin Lee helped in preparing yeast and fly spike-in RNA samples. Victor Ambros gave great advice on how to improve the manuscript. This work was supported by the NIH (U54HD82013 and GM46770), the Simons Foundation, and the Charles H. Hood Foundation (J.D.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009161117/-/DCSupplemental.
Data Availability.
The data supporting the findings of this study have been deposited in the publicly accessible GEO repository with the accession codes GSE140565 and GSE140642.
References
- 1.Santoro M. R., Bray S. M., Warren S. T., Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 7, 219–245 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Pieretti M., et al. , Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66, 817–822 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Dölen G., et al. , Correction of fragile X syndrome in mice. Neuron 56, 955–962 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya A., et al. , Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron 76, 325–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell J. C., et al. , FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurin T., et al. , HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 46, 6344–6355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driesche S. J. V., et al. , FMRP binding to a ranked subset of long genes is revealed by coupled CLIP and TRAP in specific neuronal cell types. bioRxiv (September 9, 2019), 10.1101/762500. [DOI]
- 8.Sawicka K., et al. , FMRP has a cell-type-specific role in CA1 pyramidal neurons to regulate autism-related transcripts and circadian memory. eLife 8, e46919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabet R., et al. , Fragile X mental retardation protein (FMRP) controls diacylglycerol kinase activity in neurons. Proc. Natl. Acad. Sci. U.S.A. 113, E3619–E3628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udagawa T., et al. , Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat. Med. 19, 1473–1477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y., et al. , Fragile X mental retardation protein: Nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 17, 1539–1547 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das Sharma S., et al. , Widespread alterations in translation elongation in the brain of juvenile Fmr1 knockout mice. Cell Rep. 26, 3313–3322.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah S., et al. , FMRP control of ribosome translocation promotes chromatin modifications and alternative splicing of neuronal genes linked to autism. Cell Rep. 30, 4459–4472.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napoli I., et al. , The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134, 1042–1054 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Gonatopoulos-Pournatzis T., et al. , Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Mol. Cell 77, 1176–1192.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Tran S. S., et al. , Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 22, 25–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamay-Ramot A., et al. , Fmrp interacts with adar and regulates RNA editing, synaptic density and locomotor activity in zebrafish. PLoS Genet. 11, e1005702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu P. J., et al. , The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edens B. M., et al. , FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F., et al. , Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pechmann S., Frydman J., Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20, 237–243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radhakrishnan A., et al. , The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122–132.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bazzini A. A., et al. , Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 35, 2087–2103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burow D. A., et al. , Attenuated codon optimality contributes to neural-specific mRNA decay in Drosophila. Cell Rep. 24, 1704–1712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishima Y., Tomari Y., Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol. Cell 61, 874–885 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Presnyak V., et al. , Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Q., et al. , Translation affects mRNA stability in a codon-dependent manner in human cells. eLife 8, e45396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saikia M., et al. , Codon optimality controls differential mRNA translation during amino acid starvation. RNA 22, 1719–1727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., Weissman J. S., The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross C., et al. , Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Rep. 11, 727–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Z., Zou Q., Liu Y., Yang X., Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 7, 11194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oertlin C., et al. , Generally applicable transcriptome-wide analysis of translation using anota2seq. Nucleic Acids Res. 47, e70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao D., et al. , A multi PDZ-domain protein Pdzd2 contributes to functional expression of sensory neuron-specific sodium channel Na(V)1.8. Mol. Cell. Neurosci. 42, 219–225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinehart J., et al. , WNK2 kinase is a novel regulator of essential neuronal cation-chloride cotransporters. J. Biol. Chem. 286, 30171–30180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duman J. G., Tu Y.-K., Tolias K. F., Emerging roles of BAI adhesion-GPCRs in synapse development and plasticity. Neural Plast. 2016, 8301737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolnik A., et al. , Sipa1l3/SPAR3 is targeted to postsynaptic specializations and interacts with the Fezzin ProSAPiP1/Lzts3. J. Neurochem. 136, 28–35 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Neymotin B., Athanasiadou R., Gresham D., Determination of in vivo RNA kinetics using RATE-seq. RNA 20, 1645–1652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg J. R., High stability of messenger RNA in growing cultured cells. Nature 240, 102–104 (1972). [DOI] [PubMed] [Google Scholar]
- 39.Lugowski A., Nicholson B., Rissland O. S., DRUID: A pipeline for transcriptome-wide measurements of mRNA stability. RNA 24, 623–632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Pretis S., et al. , INSPEcT: A computational tool to infer mRNA synthesis, processing and degradation dynamics from RNA- and 4sU-seq time course experiments. Bioinformatics 31, 2829–2835 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Mukherjee N., et al. , Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 24, 86–96 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Courel M., et al. , GC content shapes mRNA storage and decay in human cells. eLife 8, e49708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauger D. M., et al. , mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. U.S.A. 116, 24075–24083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ascano M., Jr, et al. , FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., et al. , N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courel M., et al. , GC content shapes mRNA storage and decay in human cells. eLife 8, e49708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hia F., et al. , Codon bias confers stability to human mRNAs. EMBO Rep. 20, e48220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan Y., et al. , FXR1 regulates transcription and is required for growth of human cancer cells with TP53/FXR2 homozygous deletion. eLife 6, e26129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelleher R. J. 3rd, Bear M. F., The autistic neuron: Troubled translation? Cell 135, 401–406 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Thomson S. R., et al. , Cell-Type-specific translation profiling reveals a novel strategy for treating fragile X syndrome. Neuron 95, 550–563.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B., et al. , Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E11397–E11405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korb E., et al. , Excess translation of epigenetic regulators contributes to fragile X syndrome and is alleviated by Brd4 inhibition. Cell 170, 1209–1223.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donnard E., Shu H., Garber M., Single cell transcriptomics reveals dysregulated cellular and molecular networks in a fragile X syndrome model. bioRxiv (February 13, 2020), 10.1101/2020.02.12.946780. [DOI] [PMC free article] [PubMed]
- 54.Buschauer R., et al. , The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368, eaay6912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen E., Sharma M. R., Shi X., Agrawal R. K., Joseph S., Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, 407–417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thoreen C. C., et al. , A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simons Foundation Autism Research Initiative , SFARI Gene. https://gene.sfari.org/database/animal-models/genetic-animal-models/. Accessed 15 November 2017.
- 58.Greenblatt E. J., Spradling A. C., Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 361, 709–712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Udagawa T., et al. , Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell 47, 253–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bava F.-A., et al. , CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495, 121–125 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Richter J. D., Bassell G. J., Klann E., Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 16, 595–605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tay J., Richter J. D., Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell 1, 201–213 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Stefani G., Fraser C. E., Darnell J. C., Darnell R. B., Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cenik C., et al. , Integrative analysis of RNA, translation and protein levels reveals distinct regulatory variation across humans. Genome Res. 25, 1610–1621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heyer E. E., Ozadam H., Ricci E. P., Cenik C., Moore M. J., An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res. 43, e2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collart M. A., Oliviero S., Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 23, 13.12.1–13.12.5 (1993). [DOI] [PubMed] [Google Scholar]
- 67.Forrest M. E., et al. , Codon and amino acid content are associated with mRNA stability in mammalian cells. PLoS One 15, e0228730 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunn J. G., Weissman J. S., Plastid: Nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics 17, 958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bindea G., et al. , ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharp P.M., Li W.H., The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pagès H., Aboyoun P., Gentleman R., DebRoy S., Biostrings: efficient manipulation of biological strings, version 3.10 (Bioconductor, 2020).
- 74.Lorenz R., et al. , ViennaRNA package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keene J. D., Komisarow J. M., Friedersdorf M. B., RIP-chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302–307 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study have been deposited in the publicly accessible GEO repository with the accession codes GSE140565 and GSE140642.