Abstract
Background
Six Sarcocystis species are known to use cattle (Bos taurus) as the intermediate host, two of which, S. hominis and S. heydorni, are zoonotic. There is a need for a method that will enable rapid identification of the Sarcocystis species in cattle.
Methods
The diaphragm muscles of 102 cattle from Lithuania were examined for the presence of Sarcocystis spp., using two different methods for species identification. Individual sarcocysts were isolated from squash preparations of the diaphragm muscle under the light microscope, followed by genetic characterisation of excised cysts using sequence analysis of the 18S rRNA (18S rRNA) and cytochrome c oxidase subunit I (cox1) genes. The same cattle muscle samples were digested and species-specific PCR analyses targeting cox1 were developed to identify the Sarcocystis isolates to the species level.
Results
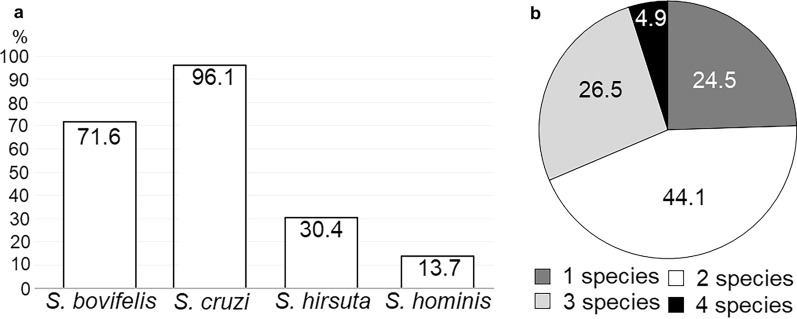
Under the light microscope, sarcocysts were detected in 87.3% of animals, and Sarcocystis infection was verified in all digested samples. Three species, namely S. cruzi (n = 20), S. bovifelis (n = 23) and S. hirsuta (n = 6), were identified by DNA sequence analysis of isolated sarcocysts. Based on sequence analysis of cox1, the level of genetic variability depended on Sarcocystis species and geographical location. Four Sarcocystis species, S. cruzi (96.1%), S. bovifelis (71.6%), S. hirsuta (30.4%) and S. hominis (13.7%), were confirmed in the digested samples. In individual samples, the most common finding was two species of Sarcocystis (44.1%), followed by three species (26.5%), a single species (24.5%) and four species (4.9%).
Conclusions
Although examination of tissue preparations under the light microscrope did not detect any sarcocysts belonging to S. hominis, this species was identified in the digested samples subjected to a cox1-specific PCR analysis. These results demonstrate the need for effective molecular diagnosis techniques to detect Sarcocystis spp., which may be present at a lower prevalence and not detectable among the limited number of sarcocysts identified individually under the light microscope. 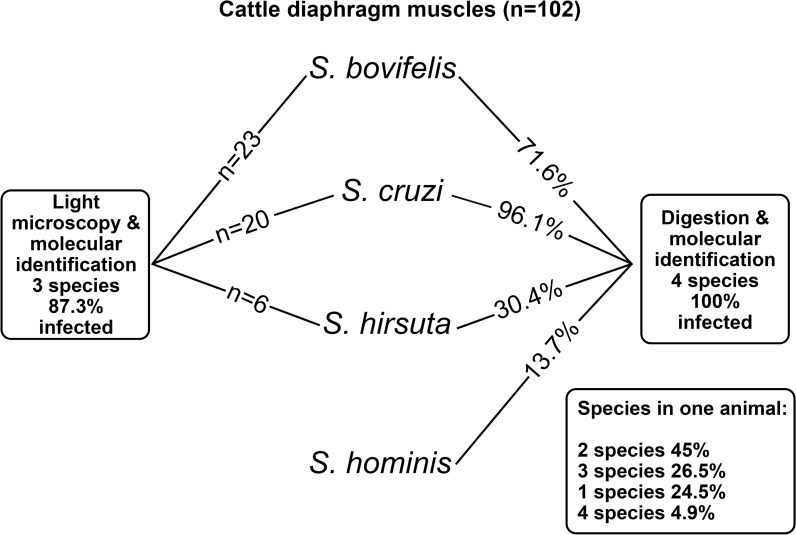
Keywords: Cattle, Sarcocystis hominis, Trypsin digestion, Molecular identification, cox1, 18S rRNA gene
Background
Protozoan parasites of the genus Sarcocystis (Apicomplexa: Sarcocystidae) infect mammals, birds and reptiles. The genus is characterised by an obligatory two-host life-cycle and endogenous sporulation. These parasites form sarcocysts mainly in the muscle tissues of the intermediate hosts and develop oocysts and sporocysts in the small intestine of the definitive hosts [1].
Three Sarcocystis species, S. suihominis, S. hominis and S. heydorni, are known to infect humans as definitive hosts [2–4]. The latter two species can be found in the muscles of cattle (Bos taurus), while humans may become infected by consuming raw/undercooked beef containing mature sarcocysts of S. hominis and S. heydorni. There has been much debate about the validity and classification of several Sarcocystis species from cattle, as well as their specificity to the intermediate host [5–8]. Current consensus is that that cattle may harbour up to six Sarcocystis species, namely S. bovifelis, S. bovini, S. cruzi, S. heydorni, S. hirsuta and S. hominis [4, 8–11]. As morphological analysis is not commonly sufficient to identify Sarcocystis species in cattle [12–14], the need for molecular methods is increasing. Sarcocystis spp. from cattle have been molecularly characterised at the genes for 18S rRNA (18S rRNA), 28S rRNA (28S rRNA) and cytochrome c oxidase subunit I (cox1) and at nuclear rDNA internal transcribed spacer 1 (ITS1) [8, 10, 11, 15]. Databases contain many 18S rRNA gene sequences of the genus Sarcocystis used for species identification [16–18]. However, studies have revealed that cox1 is the preferable target to differentiate taxonomically related Sarcocystis spp. whose intermediate hosts are such ruminants as cattle, sheep, goats, deer and others [19].
In Lithuania, the prevalence of Sarcocystis infection ranges from 44.9 to 98.1%, and infection has been recorded in six different muscle types of cattle [20]. However, the parasite species in the area under investigation have not yet been identified. In the study reported here, we isolated individual sarcocysts from squash preparations of muscle and then characterised the sarcocysts based on sequence analysis of 18S rRNA and cox1. We also report out development of a diagnostic technique to identify Sarcocystis species in cattle based on tissue digestion and PCR analysis using species-specific primers targeting cox1.
Material and methods
Samples and microscopic examination of sarcocysts
In 2017–2018, the diaphragm muscles of 102 cattle were collected from farmers and meat processing plants from all over Lithuania. The prevalence and density of Sarcocystis infection were evaluated in methylene blue-stained muscle specimens by counting sarcocysts per gram of sample according to Kirillova et al. [21]. The morphological analysis of sarcocysts was performed in freshly squashed preparations as described by Prakas et al. [22]. Sarcocysts were isolated from muscle fibres under a Nikon ECLIPSE 80i light microscope (Nikon Corp., Tokyo, Japan), at 40× or 100× magnification, and morphologically differentiated according to size and shape of sarcocysts, as well as the structure of the sarcocyst wall. Overall, 49 sarcocysts excised from the muscle tissues were separately preserved in 70% ethanol for molecular analysis.
Molecular analysis of sarcocysts
Genomic DNA was extracted from individual sarcocysts using the QIAamp® DNA Micro Kit (Qiagen, Hilden, Germany) tissue protocol according to the manufacturer’s recommendations. Sarcocystis species were identified by sequence analysis of 18S rRNA and cox1. The SarCF/SarDR primer pair was applied for 18S rDNA sequence amplification [23], while the SF1/SR9 primer pair was used to amplify cox1 sequences [19, 24]. The PCR conditions and product evaluation were as described previously [25]. The samples were sequenced directly with the 3500 Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA, USA) using the same forward and reverse primers as for the PCR. The obtained sequences were compared with those of various Sarcocystis spp. by Nucleotide BLAST (megablast option). A total of 213 cox1 and 126 18S rRNA sequences were selected and aligned using the MUSCLE algorithm [26] included in the MEGA7 software [27]. TOPALi v2.5 software [28] was used to select a nucleotide substitution model with the best fit to the aligned sequence datasets and to construct phylogenetic trees under Bayesian inference. The intraspecific genetic variation indices, including the number of haplotypes (h), the number of segregating sites (S), haplotype diversity (Hd) and nucleotide diversity (π), were calculated for whole dataset and separate samples using DnaSP v6 [29]. Genetic differentiation for Sarcocystis spp. sample pairs was assessed with ΦST (i.e. the proportion of genetic diversity due to differences among populations) based on the Tamura-Nei distance [30] with Arlequin v. 3.5.2.2 [31]. The statistical significance of ΦST was tested by 10,000 permutations and at the 0.05 confidence level.
Trypsin digestion
Digestion of cattle diaphragm muscles was performed according to the optimised protocols of Verma et al. [32] and Chiesa et al. [33]. Following trypsin digestion of 20-g samples each in 50 ml of digestion solution (trypsin 1:250, 2.5 g/l phosphate buffered saline [PBS]) for 16 h at 37 °C in an incubator with stirring, the suspension was centrifuged for 5 min at 7000 rpm, following which the pellet was resuspended in 5 ml PBS and centrifuged for 3 min at 5000 rpm. The resulting pellet was resuspended in 5 ml PBS, and 300 µl of the suspension was used for DNA extraction.
Molecular analysis of digested samples
Genomic DNA was extracted from the digested samples using the GeneJet Genomic DNA Purification Kit (Thermo Fisher Scientific Baltics, Vilnius, Lithuania) according to the manufacturer’s protocol. The resulting DNA samples were kept frozen at − 20 °C until used as templates for PCR amplification of cox1.
Primers targeting cox1 were designed with the help of Primer3Plus [34]. The primers designed for S. cruzi, S. bovifelis, S. bovini, S. heydorni, S. hirsuta and S. hominis are listed in Table 1. Each PCR was performed in a final volume of 25 μl containing 0.5 μM of each primer, dNTP mix (0.2 mM of each), 10× DreamTaq buffer with 20 mM MgCl2, 1.25 U DreamTaq polymerase (Thermo Fisher Scientific Baltics), 1 μg genomic DNA and nuclease-free water. The cycling conditions began with one cycle at 95 °C for 5 min, followed by 40 cycles at 94 °C for 45 s, 56 °C for 45 s and 72 °C for 45 s, and ending with one cycle at 72 °C for 10 min. PCR products were visualised by 1% agarose gel electrophoresis. The selected PCR products were purified using the GeneJET PCR Purification Kit (Thermo Fisher Scientific Baltics) according to the manufacturer’s recommendations. Sequencing of the purified PCR fragments was performed, and the obtained sequences were compared by BLAST analysis. Differences in the prevalence of the identified Sarcocystis species were evaluated using Chi-squared test.
Table 1.
Oligonucleotide primers used to amplify cox1 of Sarcocystis spp. isolated from cattle
| Primer name | Orientation | Primer sequence (5′→3′) | Species | Amplified product (bp) |
|---|---|---|---|---|
| GaBfEF | Forward | ATCAACTTCCTAGGTACAGCGGTATT | S. bovifelis | 523 |
| GaBfER | Reverse | CCACATCATTGGTGCTTAGTCTAGTA | ||
| GaBnEF | Forward | ATGGAGATGTGGTATTCTGTACGTCT | S. bovini | 485 |
| GaBnER | Reverse | ACAGTGCAACATCATTGGTATGTATC | ||
| GaCrEF | Forward | GCTATGTATCTACTTACGGCAGGTATC | S. cruzi | 531 |
| GaCrER | Reverse | GAATATAATGGCCCAGGTAAATAATG | ||
| GsSheyF | Forward | GGTATCCGGTATGAAGCATACAAC | S. heydorni | 443 |
| GsSheyR | Reverse | GATCCGCTGTCAGTGTACGATATT | ||
| GaHiEF | Forward | GTTGTGCGGTATGAATTATCAACCT | S. hirsuta | 513 |
| GaHiER | Reverse | GGTAAGAACTGGAATGGTTAATATCAG | ||
| GaHoEF | Forward | TCTCTGGTTTTGGTAACTACTTCGT | S. hominis | 551 |
| GaHoER | Reverse | CAGACACTGGGATATAATACCGAAC |
Results
Morphological examination of sarcocysts
Examination of methylene blue-stained squash preparations of diaphragm muscles from cattle reared in Lithuania revealed the presence of sarcocysts in 87.3% (89/102) of the samples collected. The number of sarcocysts per gram of muscle ranged from one to 79 (mean 10.5/g muscle, median 5.0/g muscle). Three types of sarcocysts were detected under the light microscope. Type I sarcocysts were thread-shaped, about 800 × 75 μm in size (n = 58) and had thin 3-µm-long hair-like protrusions; these sarcocysts were characterised as being S. cruzi-like. Type II sarcocysts were spindle- or fusiform-shaped, about 1600 × 130 μm in size (n = 40) and had 5- to 6-µm-long finger-like protrusions; these cysts were characterised as being S. bovifelis- and S. bovini-like. Type III sarcocysts were thread-shaped, about 2600 × 500 µm in size (n = 2) and had about 7-µm-long finger-like protrusions; these cysts were considered as being S. hirsuta-like.
The molecular analysis revealed that cysts belonging to types I–III were sarcocysts of S. cruzi (n = 20), S. bovifelis (n = 23) and S. hirsuta (n = 6), respectively. In some cases, sarcocysts morphologically assigned to S. hirsuta were identified as S. bovifelis or vice versa. In two cases, fragments of sarcocysts appeared to be smooth; however, based on molecular methods these cysts were assigned to S. cruzi.
Molecular results on isolated sarcocysts
The obtained 18S rRNA sequences of S. bovifelis (865 bp), S. cruzi (867 bp) and S. hirsuta (874 bp) were deposited in GenBank under accession numbers MT792432–MT792480. The analysed 18S rRNA fragments correspond to position 924–1774 of the S. neurona (U07812) sequence. Comparison of the 18S rRNA sequences of S. bovifelis and S. cruzi did not reveal intraspecific genetic variability. In contrast, one transition (C/T) at nucleotide position 588 was detected between the obtained isolates of S. hirsuta. Twenty sequences of S. cruzi showed 99.8–100% identity with other sequences of this species available in GenBank (AB682779–AB682780, AF017120, JX679467–JX679468, KC209738, KT901167, LC171827–LC171830). Among the Sarcocystis spp. isolated from cattle, the 18S rRNA sequences of S. cruzi demonstrated the greatest similarity (99.1–99.2%) with S. heydorni (KX057996–KX057997). The BLAST analysis revealed that six 18S rRNA gene sequences of S. hirsuta obtained in the present study shared 99.3–100% identity with those of S. hirsuta (AF017122, JX855283, KC209741, KT901156–KT901166, LC171839, MT706003–MT706004) and showed < 98% similarity with those of other Sarcocystis spp. from cattle. Based on 18S rRNA sequence analysis, Lithuanian isolates of S. bovifelis displayed 99.8–100% identity with S. bovifelis (KT901117–KT901138, KC209742–KC209744) and 99.2–99.5% similarity with S. bovini (KT901139–KT901155). It should be noted that these two species could be separated based on three fixed nucleotide positions: at nucleotide positions 441, 457 and 587 of the region studied, S. bovifelis had A, T and A, and S. bovini—G, A and T, respectively.
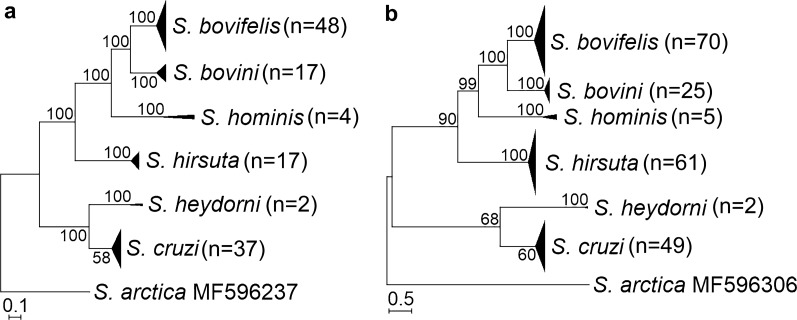
Phylogenetic analysis showed that the 18S rRNA fragment used in this study was suitable for the discrimination of Sarcocystis spp. forming cysts in the muscles of cattle (Fig. 1a). It should be noted that four species (S. bovifelis, S. bovini, S. hominis and S. hirsuta) characterised on the basis of sarcocysts with finger-like protrusions [8] were grouped into one cluster. The same topology was shown by the phylogenetic tree generated by sequence analysis of cox1 (Fig. 1b).
Fig. 1.
The phylogenetic placement of Sarcocystis spp. from cattle based on sequence analysis of the 18S rRNA (18S rRNA; a) and cytochrome c oxidase subunit I (cox1; b) genes. Trees were scaled according to the branch length and rooted on S. arctica
The genetic comparison of cox1 sequences of S. bovifelis, S. cruzi and S. hirsuta obtained in the present study are presented in Table 2. The highest sequence differences were found within S. cruzi. The 18S rRNA region used in this study was characterised by minimal intraspecific genetic variability among the analysed Sarcocystis species. Therefore, S. bovifelis, S. cruzi and S. hirsuta from different geographical areas were compared only for cox1. The highest overall intraspecific genetic variability was detected in S. cruzi, while a similar level of overall genetic variation was calculated in S. bovifelis and S. hirsuta (Table 3). Differences in the level of genetic variation were observed between samples of the same species. Relatively low variation was established for S. bovifelis from Lithuania and S. hirsuta from New Zealand, and relatively high variation was noticed for S. cruzi from China. The overall ΦST values showing genetic differentiation between populations were high for S. hirsuta (ΦST = 0.401, P < 0.001) and S. cruzi (ΦST = 0.255, P < 0.001) and medium for S. bovifelis (ΦST = 0.141, P < 0.01).
Table 2.
Molecular examination of Sarcocystis species from cattle based on cox1 sequence analysis
| Sarcocystis species | Sample type | Cox1 GenBank accession no. (length in bp) | Differences between obtained sequences (%) | Sequence identity (%) | |
|---|---|---|---|---|---|
| Comparison of isolates of the same species | Comparison of isolates with other closely related species | ||||
| S. bovifelis | Sarcocyst | MT796903–MT796925 (1038) | 0–1.0 | 98.4–100% to S. bovifelis (KT900961–KT900998, KC209690–KC209696, MK962347–MK962348) | 93.0–93.9% to S. bovini (KT900999–KT901022, LC171858) |
| Digested | MT796952–MT796954 (471) | 0 | 97.9–100% to S. bovifelis | 92.4–93.0% to S. bovini | |
| S. cruzi | Sarcocyst | MT796926–MT796945 (1038) | 0.2–1.2 | 96.1–99.9% to S. cruzi (KC209597–KC209600, KT901078–KT901095, LC171859–LC171862, MG787071–MG787076) | 92.6–93.9 to S. levinei (KU247874–KU247885, MH255771–MH255781) |
| Digested | MT796955–MT796957 (478) | 0–0.4 | 97.7–99.8% to S. cruzi | < 95% similarity with other Sarcocystis spp. | |
| S. hirsuta | Sarcocyst | MT796946–MT796951 (1038) | 0–0.7 | 99.2–100% to S. hirsuta (KC209634, KT901023–KT901077, LC171863) | 93.4–93.7 to S. buffalonis (KU247868–KU247873, MG792800–MG792802) |
| Digested | MT796958–MT796960 (461) | 0–0.2 | 99.4–100% to S. hirsuta | 95.6–96.3% to S. buffalonis | |
| S. hominis | Digested | MT796961–MT796964 (501) | 0 | 98.2–98.6% to S. hominis (MH021119, MK497840–MK497843) | < 90% similarity with other Sarcocystis spp. |
Table 3.
Measurements of intraspecific genetic variability of three Sarcocystis species from cattle based on cox1 sequence analysis
| Sarcocystis species | Country of origin | GenBank | n/h | S | Hd ± SD | π ± SD |
|---|---|---|---|---|---|---|
| S. bovifelis | Argentina | KC209690–KC209696, KT900961–KT900993 | 40/17 | 25 | 0.937 ± 0.018 | 0.00525 ± 0.00029 |
| Germany | MK962348, KT900994–KT900998 | 6/5 | 17 | 0.933 ± 0.122 | 0.00635 ± 0.00281 | |
| Lithuania | MT796903–MT796925 | 23/12 | 19 | 0.779 ± 0.091 | 0.00275 ± 0.00064 | |
| Overall | 69/32 | 45 | 0.919 ± 0.025 | 0.00491 ± 0.00043 | ||
| S. cruzi | Argentina | KC209597–KC209600, KT901078–KT901095 | 22/13 | 23 | 0.931 ± 0.036 | 0.00687 ± 0.00093 |
| China | MG787071–MG787076 | 6/6 | 50 | 1.000 ± 0.096 | 0.02667 ± 0.00390 | |
| Lithuania | MT796926–MT796945 | 20/20 | 33 | 1.000 ± 0.016 | 0.00718 ± 0.00054 | |
| Overall | 48/38 | 76 | 0.984 ± 0.010 | 0.01124 ± 0.00170 | ||
| S. hirsuta | Brazil | KT901042–KT901056 | 15/5 | 5 | 0.810 ± 0.059 | 0.00256 ± 0.00028 |
| Germany | KT901057–KT901077 | 21/6 | 10 | 0.795 ± 0.051 | 0.00399 ± 0.00037 | |
| Lithuania | MT796946–MT796951 | 6/5 | 9 | 0.933 ± 0.122 | 0.00308 ± 0.00084 | |
| New Zeeland | KT901023–KT901041 | 19/2 | 5 | 0.199 ± 0.112 | 0.00096 ± 0.00054 | |
| Overall | 61/15 | 19 | 0.863 ± 0.028 | 0.00387 ± 0.00029 |
n/h, Number of isolates/number of haplotypes; S, number of segregating sites; Hd, haplotype diversity; π, nucleotide diversity; SD standard deviation
Molecular results of digested samples
Cox1 fragments amplified using the GaBfEF/GaBfER, GaCrEF/GaCrER, GaHiEF/GaHiER and GaHoEF/GaHoER primer pairs were examined by DNA electrophoresis. To ascertain whether the used primers were species-specific, three PCR products amplified with each primer pair were purified and sequenced. Primer-annealing sequences were not included in the BLAST analysis. The sequence identity values are given in Table 2. DNA of S. bovifelis, S. cruzi and S. hirsuta extracted from individual sarcocysts were further used as positive or negative controls in the analysis of the digested samples. The control DNA sample of S. hominis was isolated from the muscular tissue of cattle in Italy [11] and subjected to 18S rRNA amplification and sequencing using the SarAF/SarBR and SarCF/SarDR primer pairs [23]. The obtained 1796-bp-long 18S rRNA sequence (MT792481) showed 99.4–99.7% similarity with S. hominis (JX679470–JX679471, KF954731) and up to 97.3% similarity with other Sarcocystis spp.
Sarcocystis spp. were detected in all 102 beef diaphragm samples examined using the newly developed molecular identification technique based on muscle digestion and species-specific PCR analysis targeting cox1. The Lithuanian cattle analysed in this study were most frequently infected with S. cruzi, which was identified in 98 samples (96.1%) (Fig. 2a). The prevalence of S. cruzi in the analysed cattle was statistically significantly higher than that of S. bovifelis (71.6%; χ2 = 22.59, P < 0.0001), the presence of which was confirmed in 73 samples. The third most common species was S. hirsuta (30.4%), detected in 31 samples. The prevalence of S. hirsuta was statistically significantly lower than of S. bovifelis (χ2 = 34.60, P < 0.0001) and higher than that of S. hominis (13.7%; χ2 = 8.24, P < 0.01), which was detected in 14 samples. Although the molecular technique did not identify S. hominis among the individual sarcocysts excised from the diaphragm samples, it was identified in the digested samples. These results show that individual cattle may be infected up to four species of Sarcocystis at any one time (Fig. 2b). The most common finding was the presence of two species of Sarcocystis (45 samples, 44.1%); three species were detected in 27 samples (26.5%), a single species was identified in 25 samples (24.5%) and four species were identified in five samples (4.9%). In cases of a single species being detected, S. cruzi was identified in 21 samples and S. bovifelis was confirmed in four samples. When two species of Sarcocystis were observed in the samples, co-infection with S. bovifelis/S. cruzi was the most common finding (86.7%), whereas S. cruzi/S. hirsuta (8.9%) and S. cruzi/S. hominis (4.4%) co-infections were much rarer. Three different species were identified in 27 samples, with S. bovifelis/S. cruzi/S. hirsuta (74.1%) being the most common combination, while mixed infections with S. bovifelis/S. cruzi/S. hominis (18.5%) and S. cruzi/S. hirsuta/S. hominis (7.4%) were rarely detected. It is noteworthy that S. hominis was established in different districts of Lithuania: Pasvalys (two cases), Tauragė (two cases), Kupiškis (two cases), Anykščiai (two cases), Jonava (one case), Tauragė (one case), Šakiai (one case), Šilalė (one case), Skuodas (one case) and Kėdainiai (one case).
Fig. 2.
Identification of Sarcocystis spp. in digested muscle samples of cattle from Lithuania. a Prevalence of Sarcocystis species, b Distribution of the number of species in samples
Discussion
In the present study sarcocysts isolated from the diaphragm muscles of cattle were attributed to three morphological types, with molecular methods identifying the types as S. cruzi, S. bovifelis and S. hirsuta. Of the six Sarcocystis species known to use cattle as an intermediate host, two species, S. cruzi and S. heydorni, can be relatively easily differentiated by morphological characteristics [4]. Sarcocysts of S. cruzi are distinguished by hair-like protrusions, while sarcocysts belonging to S. heydorni have an apparently smooth cyst wall; sarcocysts of the other four species (S. bovifelis, S. bovini, S. hirsuta and S. hominis) are characterised by finger-like protrusions [4, 8, 9, 35]. During the course of this study, morphological examination of fragments of two sarocysts released from muscle fibres appeared to be smooth and to be similar to S. heydorni; however, based on molecular methods, these were identified as S. cruzi. In these cases, the protrusions of S. cruzi were likely torn off during manipulation of the sarcocysts with preparation needles. Moreover, sarococyts belonging to S. bovifelis were sometimes misidentified as S. hirsuta and vice versa. Sarcocysts of S. hominis were not found during the light microscopy study, but this species was confirmed in 13.7% of digested samples using molecular methods. These results suggest that the isolation of individual sarcocysts is ineffective when the prevalence of infection of the species is low and, therefore, that the digestion protocol may be preferable in epidemiological studies.
The 18S rRNA fragment used in the present study showed sufficient variability for the discrimination of Sarcocystis spp. found in cattle (Fig. 1a). Studies involving population genetic research on Sarcocystis species from cattle are scarce. One study demonstrated a low genetic variability among various isolates of S. cruzi from different geographical regions [36]. In the present study, analysis of cox1 sequences implied that the level of genetic variability depended on the Sarcocystis species and geographical area. Also, a relatively high genetic differentiation between the samples of compared species was revealed.
This study established that the GaBfEF/GaBfER, GaCrEF/GaCrER, GaHiEF/GaHiER and GaHoEF/GaHoER primer pairs were specific to S. bovifelis, S. cruzi, S. hirsuta and S. hominis, respectively. In all cases, no PCR product was generated using the negative DNA controls. In summary, the developed molecular technique based on the digestion of the muscle samples and species-specific PCR was found to be suitable for the detection of S. bovifelis, S. cruzi, S. hirsuta and S. hominis from beef. Methylene-blue staining of squash preparations of the muscle samples detected sarcocysts in 87.3% of preparations; in comparison, the molecular methods confirmed the presence of Sarcocystis spp. in all 102 of the digested specimens examined. A very high prevalence of Sarcocystis infection, exceeding 90%, has also been reported in cattle from Argentina, Belgium, the Czech Republic, Egypt, Germany, Iran, Italy, Japan, Mexico, New Zealand, Sudan, Thailand and Turkey (reviewed by Dubey et al. [1]). The findings of the present study are consistent with the data obtained by other researchers demonstrating a different infection prevalence depending on the method used. For example, Moré et al. [37] reported a higher level of prevalence when molecular methods were used (67.7% PCR, 69.6% real-time PCR) in comparison to microscopy analysis (40.0%). Daptardar et al. [38] calculated 44% infection prevalence using intact cyst isolation, 58% infection using pepsin-HCl digestion and 68% infection prevalence using PCR analysis. Hence, current Sarcocystis infection prevalence data should be considered as being critically dependent on the method employed to detect prevalence. The high prevalence of Sarcocystis spp. detected in cattle in Lithuania could be explained by husbandry practices, as carcasses of slaughtered cattle in small farms are fed to dogs and cats, especially in rural areas. Also, a raw diet, which has become popular among dog breeders in recent years, where pets are fed fresh meat, is one possible pathway for the spread of sarcocysts [39].
Our examination of the diaphragm samples of 102 cattle bred in Lithuania identified S. cruzi in 83.7% of samples, S. bovifelis in 71.6%, S. hirsuta in 30.4%, and S. hominis in 13.7% (Fig 2a). The prevalence and abundance of Sarcocystis species may depend on the type of the muscle being analysed. For example, S. heydorni tissue tropism was noted in cattle from China [35], with the highest prevalence (9.7%) recorded in skeletal muscles, followed by 3.4% prevalence in the oesophagus, 2.5% in the diaphragm and 0.1% in the tongue muscles; however, S. heydorni was absent in the heart muscle. This zoonotic species has also been identified in cattle from the Netherlands (1%) and Turkey [4, 10]. It should be noted that due to the different morphological, serological or molecular methods used in studies, it is difficult to compare the distribution of Sarcocystis species identified in cattle. Nevertheless, several trends in the prevalence of Sarcocystis species can be indicated. Most studies carried out in cattle reported the highest prevalence of S. cruzi employing canids as definitive hosts [10, 14, 15, 18, 37, 40–44]. By contrast, a relatively low prevalence of S. hirsuta using felids as definitive hosts was often recorded [14, 15, 37, 41, 45]. Research performed in cattle from the Netherlands and Italy showed S. hirsuta infection rates of < 2% [10, 40]. By contrast, using PCR-restriction fragment length polymorphism, S. hirsuta infection in Iran was recorded as reaching 58.9% [46], whereas microscopy analysis noted a prevalence of S. hirsuta as high as 94.0% in Brazil [47]. Sarcocystis hirsuta has been identified in different geographic regions [8, 10, 14, 15, 40, 44, 45]. Felids are also known to be definitive hosts of S. bovifelis and S. bovini [8, 48]; however, there is a lack of knowledge of the geographical distribution of these two Sarcocystis species. It should be pointed out that S. bovini has not been identified in Europe yet; S. bovifelis and S. bovini have been confirmed in Argentina, and only S. bovini has been found in New Zealand [8] and in beef from New Zealand imported to Japan [15]. In addition, based on comparison of 18S rRNA sequences S. bovini has also been found to be common in China [8]. Differences in the distribution of Sarcocystis species in cattle across geographical regions are possibly related to the abundance of definitive hosts.
We detected from one single Sarcocystis species up to four Sarcocystis species simultaneously in individual samples, with the highest prevalence noted for two species concurrently in one animal (Fig 2b). Two species forming sarcocysts in cattle, namely S. bovifelis and S. bovini, were (re)described only in 2016 [8]. Therefore, there is a lack of reliable data on the number of Sarcocystis species in individual bovine samples. However, mixed infections of different Sarcocystis species in cattle is a common occurrence [10, 37, 44]. Further studies are needed to reveal the rate of combinations of Sarcocystis species in individual samples from different countries.
The high level (> 40%) of S. hominis infection reported in cattle from countries such as Italy and Iran [33, 46] is most likely overestimated due to the confusion in identifying S. hirsuta and S. bovifelis/S. bovini. The majority of the 18S rRNA sequences formerly presented in GenBank as S. hominis actually belonged to S. bovini, S. bovifelis or S. sinensis infecting the water buffalo [8]. There is a lack of detailed S. hominis molecular data [49] and the first cox1 sequence of this species became available in the GenBank database only in 2018. The prevalence of S. hominis infection in different countries based on molecular methods is poorly studied. A 0.2% prevalence of S. hominis was detected in China [50], 0.5% in Argentina [42], 3.5% in Iran [44], 6.2 % in Germany [37] and 12.5% in the Netherlands [10]. The molecular technique established in this study is an important step in the development of zoonotic S. hominis diagnosis in both cattle and humans.
Conclusion
Based on molecular methods, the presence of S. cruzi (96.1%), S. bovifelis (71.6%), S. hirsuta (30.4%), and S. hominis (13.7%) was confirmed in Lithuanian cattle meat for the first time. A rapid, accurate and relatively cheap technique was developed for the identification of Sarcocystis species in cattle meat. Further research on S. hominis epidemiology in various regions is needed.
Acknowledgements
This work was supported by the Open Access to research infrastructure of the Nature Research Centre under Lithuanian open access network initiative.
Authors’ contributions
PP, FC, ES, SP and DB conceived and designed the laboratory tests. ŽSŽ, VJ, AB and ERL performed the experiments. PP, VJ, FC, ES, SP and DB contributed reagents/materials/analysis tools. PP, ŽSŽ, FC, DB, EJN, ERL, ES and DB drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work received financial support from the Research Council of Lithuania (Grant number S-MIP-17-45).
Availability of data and materials
The sequences generated in the present study are available in GenBank database with accession numbers MT792432–MT792481 (18S rRNA) and MT796903–MT796964 (cox1).
Ethics approval and consent to participate
For this type of formal study consent is not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Petras Prakas, Email: prakaspetras@gmail.com.
Živilė Strazdaitė-Žielienė, Email: genetike@gmail.com.
Vytautas Januškevičius, Email: vytautasjanusk@gmail.com.
Francesco Chiesa, Email: francesco.chiesa@unito.it.
Agnė Baranauskaitė, Email: agne.terese@gmail.com.
Eglė Rudaitytė-Lukošienė, Email: egle.rudaityte@gmail.com.
Elena Servienė, Email: serviene@gmail.com.
Saulius Petkevičius, Email: saulius.petkevicius@lsmuni.lt.
Dalius Butkauskas, Email: dalius.butkauskas@gamtc.lt.
References
- 1.Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R. Sarcocystosis of animals and humans. 2. Boca Raton: CRC Press; 2016. [Google Scholar]
- 2.Heydorn AO. Beiträge zum Lebenszyklus der Sarkosporidien. IX. Entwicklungszyklus von Sarcocystis suihominis n. spec. Berl Münch Tierärztl Wochenschr. 1977;90:218–224. [PubMed] [Google Scholar]
- 3.Heydorn AO, Gestrich R, Janitschke K. Beiträge zum Lebenszyklus der Sarkosporidien. VIII. Sporozysten von Sarcocystis bovihominis in den Fäzes von Rhesusaffen (Macaca rhesus) und Pavianen (Papio cynocephalus) Berl Münch Tierärztl Wochenschr. 1976;89:116–120. [PubMed] [Google Scholar]
- 4.Dubey JP, van Wilpe E, Calero-Bernal R, Verma SK, Fayer R. Sarcocystis heydorni, n. sp. (Apicomplexa: Sarcocystidae) with cattle (Bos taurus) and human (Homo sapiens) cycle. Parasitol Res. 2015;114:4143–4147. doi: 10.1007/s00436-015-4645-2. [DOI] [PubMed] [Google Scholar]
- 5.Dubey JP, Fayer R, Rosenthal BM, Calero-Bernal R, Uggla A. Identity of Sarcocystis species of the water buffalo (Bubalus bubalis) and cattle (Bos taurus) and the suppression of Sarcocystis sinensis as a nomen nudum. Vet Parasitol. 2014;205:1–6. doi: 10.1016/j.vetpar.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 6.Dubey JP, Moré G, van Wilpe E, Calero-Bernal R, Verma SK, Schares G. Sarcocystis rommeli, n. sp. (Apicomplexa: Sarcocystidae) from cattle (Bos taurus) and its differentiation from Sarcocystis hominis. J Eukaryot Microbiol. 2016;63:62–68. doi: 10.1111/jeu.12248. [DOI] [PubMed] [Google Scholar]
- 7.Zuo YX, Yang ZQ. The validity of Sarcocystis sinensis. Dongwuxue Yanjiu. 2015;36:109–111. doi: 10.13918/j.issn.2095-8137.2015.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gjerde B. Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis) Parasitol Res. 2016;115:1473–1492. doi: 10.1007/s00436-015-4881-5. [DOI] [PubMed] [Google Scholar]
- 9.Gjerde B. The resurrection of a species: Sarcocystis bovifelis Heydorn et al., 1975 is distinct from the current Sarcocystis hirsuta in cattle and morphologically indistinguishable from Sarcocystis sinensis in water buffaloes. Parasitol Res. 2016;2016(115):1–21. doi: 10.1007/s00436-015-4785-4. [DOI] [PubMed] [Google Scholar]
- 10.Hoeve-Bakker BJA, van der Giessen JWB, Franssen FFJ. Molecular identification targeting cox1 and 18S genes confirms the high prevalence of Sarcocystis spp. in cattle in the Netherlands. Int J Parasitol. 2019;49:859–866. doi: 10.1016/j.ijpara.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Rubiola S, Chiesa F, Zanet S, Civera T. Molecular identification of Sarcocystis spp in cattle: partial sequencing of cytochrome c oxidase subunit 1 (COI) Ital J Food Saf. 2019;7:7725. doi: 10.4081/ijfs.2018.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubey JP, Calero-Bernal R, Verma SK, Mowery JD. Pathology, immunohistochemistry, and ultrastructural findings associated with neurological sarcocystosis in cattle. Vet Parasitol. 2016;223:147–152. doi: 10.1016/j.vetpar.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Choi TI, Hong EJ, Ryu SY, Sim C, Chae JS, Kim HC, et al. Detection and identification of Sarcocystis cruzi (Protozoa: Apicomplexa) by molecular and ultrastructural studies in naturally infected Korean cattle (Bos taurus coreanae) from Daejeon Korea. Korean J Parasitol. 2018;56:121–127. doi: 10.3347/kjp.2018.56.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue R, Yan W, Qian W, Wang T, Zhang M, Wei Z, et al. Prevalence and molecular characterization of Sarcocystis infections of retail beef products from central China. Acta Trop. 2019;190:339–343. doi: 10.1016/j.actatropica.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Murata R, Suzuki J, Hyuga A, Shinkai T, Sadamasu K. Molecular identification and characterization of Sarcocystis spp. in horsemeat and beef marketed in Japan. Parasite. 2018;25:27. doi: 10.1051/parasite/2018026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakas P, Rudaitytė E, Butkauskas D, Kutkienė L. Sarcocystis entzerothi n. sp. from the European roe deer (Capreolus capreolus) Parasitol Res. 2017;116:271–279. doi: 10.1007/s00436-016-5288-7. [DOI] [PubMed] [Google Scholar]
- 17.Rudaitytė-Lukošienė E, Prakas P, Butkauskas D, Kutkienė L, Vepštaitė-Monstavičė I, Servienė E. Morphological and molecular identification of Sarcocystis spp. from the sika deer (Cervus nippon), including two new species Sarcocystis frondea and Sarcocystis nipponi. Parasitol Res. 2018;117:1305–1315. doi: 10.1007/s00436-018-5816-8. [DOI] [PubMed] [Google Scholar]
- 18.Imre K, Dărăbuş G, Tîrziu E, Morariu S, Imre M, Plutzer J, et al. Sarcocystis spp. in Romanian slaughtered cattle: molecular characterization and epidemiological significance of the findings. Biomed Res Int. 2019;4123:154. doi: 10.1155/2019/4123154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol. 2013;43:579–591. doi: 10.1016/j.ijpara.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Januškevičius V, Januškevičienė G, Prakas P, Butkauskas D, Petkevičius S. Prevalence and intensity of Sarcocystis spp. infection in animals slaughtered for food in Lithuania. Vet Med Czech. 2019;64:149–157. doi: 10.17221/151/2017-VETMED. [DOI] [Google Scholar]
- 21.Kirillova V, Prakas P, Calero-Bernal R, Gavarāne I, Fernández-García JL, Martínez-González M, et al. Identification and genetic characterization of Sarcocystis arctica and Sarcocystis lutrae in red foxes (Vulpes vulpes) from Baltic States and Spain. Parasite Vectors. 2018;11:173. doi: 10.1186/s13071-018-2694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prakas P, Butkauskas D, Rudaitytė E, Kutkienė L, Sruoga A, Pūraitė I. Morphological and molecular characterization of Sarcocystis taeniata and Sarcocystis pilosa n. sp. from the sika deer (Cervus nippon) in Lithuania. Parasitol Res. 2016;115:3021–3032. doi: 10.1007/s00436-016-5057-7. [DOI] [PubMed] [Google Scholar]
- 23.Kutkienė L, Prakas P, Sruoga A, Butkauskas D. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis) Parasitol Res. 2010;107:879–888. doi: 10.1007/s00436-010-1945-4. [DOI] [PubMed] [Google Scholar]
- 24.Gjerde B. Sarcocystis species in red deer revisited: with a redescription of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology. 2014;141:441–452. doi: 10.1017/S0031182013001819. [DOI] [PubMed] [Google Scholar]
- 25.Prakas P, Kirillova V, Calero-Bernal R, Kirjušina M, Rudaitytė-Lukošienė E, Habela MÁ, et al. Sarcocystis species identification in the moose (Alces alces) from the Baltic States. Parasitol Res. 2019;118:1601–1608. doi: 10.1007/s00436-019-06291-0. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne I, Wright F, Rowe G, Marshall D, Husmeier D, McGuire G. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20:1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- 29.Rozas J, Ferrer-Mata A, Sánchez-Del Barrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA Sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 31.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 32.Verma SK, Lindsay DS, Grigg ME, Dubey JP. Isolation, culture and cryopreservation of Sarcocystis species. Curr Protoc Microbiol. 2017;45:11–127. doi: 10.1002/cpmc.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiesa F, Muratore E, Dalmasso A, Civera T. A new molecular approach to assess the occurrence of Sarcocystis spp. in cattle and products thereof: preliminary data. Ital J Food Saf. 2013;2:148–151. [Google Scholar]
- 34.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu JJ, Wen T, Chen XW, Liu TT, Esch GW, Huang S. Prevalance, morphology, and molecular characterization of Sarcocystis heydorni sarcocysts from cattle (Bos taurus) in China. J Parasitol. 2016;102:545–548. doi: 10.1645/16-49. [DOI] [PubMed] [Google Scholar]
- 36.Rosenthal BM, Dunams DB, Pritt B. Restricted genetic diversity in the ubiquitous cattle parasite Sarcocystis cruzi. Infect Genet Evol. 2008;8:588–592. doi: 10.1016/j.meegid.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Moré G, Pantchev A, Skuballa J, Langenmayer MC, Maksimov P, Conraths FJ, et al. Sarcocystis sinensis is the most prevalent thick-walled Sarcocystis species in beef on sale for consumers in Germany. Parasitol Res. 2014;113:2223–2230. doi: 10.1007/s00436-014-3877-x. [DOI] [PubMed] [Google Scholar]
- 38.Daptardar M, Singh BB, Aulakh RS, Gill JP. Prevalence and first molecular identification of Sarcocystis species in cattle and water buffaloes in India. Acta Parasitol. 2016;61:523–528. doi: 10.1515/ap-2016-0069. [DOI] [PubMed] [Google Scholar]
- 39.Januškevičius V. Protozoa of the genus Sarcocystis in cattle: identification, distribution and impact on the host organism. PhD thesis. Kaunas: Lithuanian University of Health Sciences; 2020.
- 40.Domenis L, Peletto S, Sacchi L, Clementi E, Genchi M, Felisari L, et al. Detection of a morphogenetically novel Sarcocystis hominis-like in the context of a prevalence study in semi-intensively bred cattle in Italy. Parasitol Res. 2011;109:1677–1687. doi: 10.1007/s00436-011-2441-1. [DOI] [PubMed] [Google Scholar]
- 41.Moré G, Abrahamovich P, Jurado S, Bacigalupe D, Marin JC, Rambeaud M, et al. Prevalence of Sarcocystis spp. in Argentinean cattle. Vet Parasitol. 2011;177:162–165. doi: 10.1016/j.vetpar.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 42.Moré G, Schares S, Maksimov A, Conraths FJ, Venturini MC, Schares G. Development of a multiplex real time PCR to differentiate Sarcocystis spp. affecting cattle. Vet Parasitol. 2013;197:85–94. doi: 10.1016/j.vetpar.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 43.Hornok S, Mester A, Takács N, Baska F, Majoros G, Fok É, et al. Sarcocystis-infection of cattle in Hungary. Parasites Vectors. 2015;8:69. doi: 10.1186/s13071-015-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hooshyar H, Abbaszadeh Z, Sharafati-Chaleshtori R, Arbabi M. Molecular identification of Sarcocystis species in raw hamburgers using PCR-RFLP method in Kashan, central Iran. J Parasit Dis. 2017;41:1001–1005. doi: 10.1007/s12639-017-0925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vangeel L, Houf K, Geldhof P, De Preter K, Vercruysse J, Ducatelle R, et al. Different Sarcocystis spp. are present in bovine eosinophilic myositis. Vet Parasitol. 2013;197:543–548. doi: 10.1016/j.vetpar.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Hajimohammadi B, Dehghani A, Moghadam Ahmadi M, Eslami G, Oryan A, Khamesipour A. Prevalence and species identification of Sarcocystis in raw hamburgers distributed in Yazd, Iran using PCR-RFLP. J Food Qual Hazards Control. 2014;1:15–20. [Google Scholar]
- 47.Pena HF, Ogassawara S, Sinhorini IL. Occurrence of cattle Sarcocystis species in raw kibbe from Arabian food establishments in the city of São Paulo, Brazil, and experimental transmission to humans. J Parasitol. 2001;87:1459–1465. doi: 10.1645/0022-3395(2001)087[1459:OOCSSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 48.Hu JJ, Huang S, Wen T, Esch GW, Liang Y, Li HL. Morphology, molecular characteristics, and demonstration of a definitive host for Sarcocystis rommeli from cattle (Bos taurus) in China. J Parasitol. 2017;103:471–476. doi: 10.1645/16-187. [DOI] [PubMed] [Google Scholar]
- 49.Rubiola S, Civera T, Ferroglio E, Zanet S, Zaccaria T, Brossa S, et al. Molecular differentiation of cattle Sarcocystis spp. by multiplex PCR targeting 18S and COI genes following identification of Sarcocystis hominis in human stool samples. Food Waterborne Parasitol. 2020;18:e00074. doi: 10.1016/j.fawpar.2020.e00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Dong H, Su R, Wang Y, Wang R, Jiang Y, et al. High prevalence of Sarcocystis spp. infections in cattle (Bos taurus) from central China. Parasitol Int. 2018;67:800–804. doi: 10.1016/j.parint.2018.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences generated in the present study are available in GenBank database with accession numbers MT792432–MT792481 (18S rRNA) and MT796903–MT796964 (cox1).