Abstract
Background
While circulating tumor cells may serve as minimally invasive cancer markers for bladder cancers, the relationship between primary bladder cancers and circulating tumor cells in terms of somatic mutations is largely unknown. Genome sequencing of bladder tumor and circulating tumor cells is highlighted to identify the somatic mutations of primary bladder cancer.
Methods
Bladder cancer tissue was collected by transurethral resection of the bladder and preserved by snap-freezing. Circulating tumor cells were Isolated from the blood obtained before treatment. We performed whole exome sequencing of 20 matched pairs of primary bladder cancers and circulating tumor cells to identify and compare somatic mutations of these two different genomic resources.
Results
We observed that mutation abundances of primary bladder cancers and circulating tumor cells were highly variable. The mutation abundance was not significantly correlated between matched pairs. Of note, the mutation concordance between two resources was only 3–24% across 20 pairs examined, suggesting that the circulating tumor cell genomes of bladder cancer patients might be genetically distinct from primary bladder cancers. A relative enrichment of mutations belonging to APOBEC-related signature and a depletion of C-to-G transversions were observed for primary- and circulating tumor cells specific mutations, respectively, suggesting that distinct mutation forces might have been operative in respective lesions during carcinogenesis.
Conclusions
The observed discrepancy of mutation abundance and low concordance level of mutations between genomes of primary bladder cancers and circulating tumor cells should be taken into account when evaluating clinical utility of circulating tumor cells for treatments and follow-up of bladder cancers.
Trial registration
Patients were selected and registered retrospectively, and medical records were evaluated.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-020-07684-6.
Keywords: Bladder cancer, Circulating tumor cells, Mutation
Background
Bladder cancer is a widespread and highly heterogeneous malignancy. Clinical outcome of this disease is poor because of its highly recurrent nature with frequent disease progression and treatment failure [1]. In clinical situation, patients with advanced bladder cancer are at high risk of disease recurrence or progression, with half of them having relapse after radical surgery. For example, 30–80% of bladder cancer patients with advanced disease below pT1 stages experienced disease recurrence and up to 45% of cases progressed to muscle invasion within 5 years [2–4]. The majority of relapses are distant metastasis and 10–15% of cases are already metastatic at diagnosis [5]. Consequently, bladder cancer is characterized with variable clinical outcomes requiring frequent follow-up and repeated treatments, making this disease obstinate and challengeable to overcome.
To overcome these hurdles, circulating tumor cells (CTCs) have been recently highlighted for “liquid biopsy” using the peripheral blood of patient as a resource of tumor cells. Ideally, CTCs can replace invasive tissue biopsies for serial monitoring of tumor characteristics and facilitate the revision of treatment or follow-up protocols [6].
High-throughput sequencing technologies have facilitated mutation screening of cancer genomes. Mutation profiles of individual cancer genomes have been traditionally used to prioritize cancer drivers (e.g., actionable alterations with available therapeutic options or predictive/prognostic markers). Moreover, an entire catalogue of mutations in given cancer genomes can be used as genetic markers to evaluate the extent of within-tumor heterogeneity between regional biopsies [7] or similarities between primary-vs.-metastatic lesions of given individuals [8]. In the case of CTC, it is challenging to estimate the extent as to how many mutations of primary tumors can be discovered by CTC which can be only evaluated by direct sequencing of primary tumor genomes and their matched CTC genomes. However, sequencing coverage to evaluate mutation-level concordance between primary tumors and CTC genomes has been largely limited due to the number of CTCs that is generally too few to obtain sufficient amount of DNA to perform exome-sequencing.
In this study, we isolated CTCs from 20 patients with urinary bladder cancers. For genomic analyses, we performed whole-exome sequencing of matched primary and CTC genomes. Somatic mutations were compared between primary tumor and CTC genomes to evaluate mutation-level concordance and identify the potential of CTCs as alternative cellular resources to identify somatic mutations of primary tumors.
Methods
Patient recruitment and collection of blood and tissue
A total of 20 patients were enrolled for this study from May 2016 to September 2017 under an Institutional Review Board–approved protocol of our institution, Seoul St. Mary’s Hospital, College of Medicine, the Catholic University of Korea and written informed consents were obtained (approval number: KC15TNSI0924). For each patient, bladder cancer tissue was collected by transurethral resection of the bladder (TURBT) and blood was collected before starting TURBT. A total of 15 cc of blood was divided into 5 ml for CTC enumeration and 10 ml for CTCs culture. All blood and tissue samples as well as medical data used were anonymous to ensure patient confidentiality. Primary tumor specimens were snap-frozen and histologically examined by pathologists after hematoxylin and eosin staining. Tissue blocks with tumor purity (> 70%) confirmed were used to extract genomic DNAs.
Isolation of CTCs and enrichment process
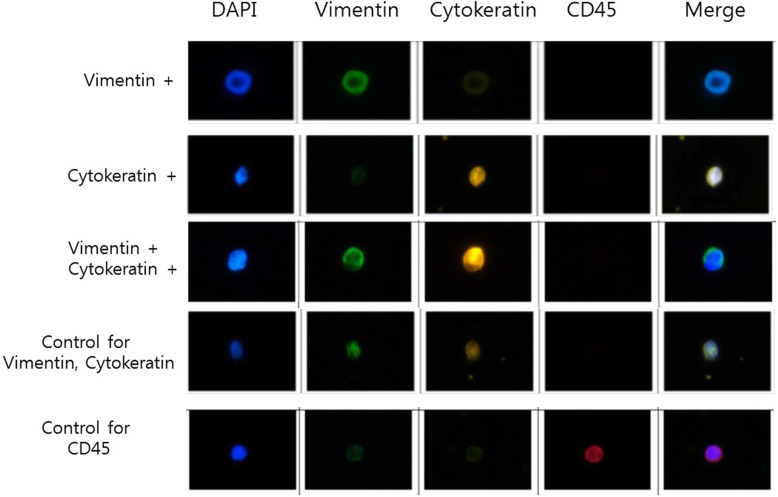
Blood samples were collected in acid citrate dextrose tubes from patients and processed within 4 h after sampling. All procedures of CTC isolation from blood were performed using Cell Isolation kit (#CIKW10; CytoGen, Inc., Seoul, Korea). Briefly, blood samples were incubated with an antibody complex against white blood cells (WBCs) and red blood cells at room temperature for 20 min and mixed with pre-activation buffer. WBC-depleted peripheral blood mononuclear cells (PBMC) were fractionated from whole blood by Ficoll density gradient centrifugation. The fraction of PBMC was diluted with a dilution buffer. Cell suspension was then filtered to isolate CTCs through a high-density microporous (HDM) chip. Enriched CTCs were fixed onto slides in 4% paraformaldehyde for 5 min at room temperature and kept at 4 °C until further processing. CTCs on slides were permeablized with 0.2% Triton X-100 in phosphate buffered saline (PBS) for 10 min at room temperature, blocked with 1% bovine serum albumin in PBS for 60 min, incubated with primary antibodies for 60 min, and then incubated with secondary antibody for 60 min. Primary antibodies used were rabbit anti-CD45 (Cell Signaling Technology), mouse anti-pan cytokeratin (Sigma), and mouse anti-vimentin Alexa Fluor® 488 conjugated (Cell Signaling Technology). Secondary antibodies used were goat anti-rabbit Alexa Fluor® 647 and goat anti-mouse Alexa Fluor® 546 (Thermo Fisher Scientific, Inc.). Slides were mounted using Fluoroshield with DAPI (ImmunoBioScience) and then detected using Nikon Eclipse Ti fluorescent microscope. Figure 1 shows representative fluorescent images of CTCs.
Fig. 1.
Representative immunostaining images showing circulating tumor cells (CTCs) with 4′,6-diamidino-2-phenylindole (DAPI), Vimentin, Cytokeratin, and CD45 positive
Primary short-term culture of CTCs
CTCs retrieved on the HDM chip were washed with PBS and cultured in 6-well Costar® Ultra-Low Attachment plates (Costar®; Corning Korea Company, Ltd., Seoul, Korea) containing mesenchymal stem cell growth medium (MSCGMTM, human Mesenchymal Stem Cell Growth BulletKitTM Medium and Supplements; Lonza Group, Basel, Switzerland) at 37 °C in an atmosphere containing 5% CO2. Following 16–18 days of culture, cells were collected for exome sequencing analysis.
Whole-exome sequencing
DNA extraction and library preparation were done as described previously [8]. To capture exonic DNA, Agilent SureSelect Human All Exome 50 Mb kit (Agilent, USA) was used. Paired-end 100 bp sequencing reads were generated on Illumina HiSeq 2500 platform (Illumina, USA) according to the manufacturer’s recommendation. Sequencing information is available in additional file 1.
Somatic mutations
Sequencing reads in FASTQ files were aligned with human reference genomes (UCSC hg19) using BWA (Burrows-Wheeler alignment) aligner [9]. Local realignment and score recalibration of initial alignment results were done using Genome Analysis ToolKit [10]. Overall management and processing of sequencing data were done using SamTools and Picard [11]. To identify somatic mutations, sequencing data of primary tumors and CTC were compared with data of matched normal blood for each patient. Point mutations and short insertions/deletions (indels) were identified using MuTect and Indelocator, respectively [12]. ANNOVAR software was used to curate somatic mutations regarding resulting changes in amino acids [13].
Results
Patients
A total of 20 patients with non-metastatic bladder cancer were enrolled in this study. Clinical characteristics of these 20 patients with counts and immunophenotypes of CTCs are shown in Table 1. Regarding pathological staging, there were 3 patients with Ta, 4 patients with T1, 4 patients with T2, 7 patients with T3, and 2 patients with T4. CTC counts at baseline were reported for all cohorts. There was a general tendency of increased number of CTC with tumor stage (pT1 to pT4). However, the immunophenotype of captured CTCs showed a wide distribution even for those with the same stage.
Table 1.
Clinical characteristics and immunophenotype of initial captured CTCs from bladder-cancer patients
| Immunophenotyping & Enumeration (5 ml) | |||||||
|---|---|---|---|---|---|---|---|
| Subject number | Pathologic T stage | LVI | Vein invasion | Vimentin (+), CK (−) | Vimentin (−), CK (+) | Vimentin (+), CK (+) | Total CTC |
| TCC14 | T4a | Pos | Neg | 1 | 17 | 0 | 18 |
| TCC16 | T1 | Neg | Neg | 1 | 7 | 1 | 9 |
| TCC17 | T3a | Pos | Neg | 1 | 5 | 2 | 8 |
| TCC18 | T3a | Neg | Neg | 0 | 17 | 1 | 18 |
| TCC20 | T2b | Pos | Pos | 6 | 1 | 21 | 28 |
| TCC21 | T3a | Pos | Pos | 0 | 0 | 15 | 15 |
| TCC27 | T3a | Neg | Neg | 0 | 11 | 0 | 11 |
| TCC28 | T2b | Pos | Neg | 0 | 12 | 5 | 17 |
| TCC31 | T2a | Pos | Pos | 9 | 1 | 4 | 14 |
| TCC32 | T3a | Pos | Pos | 3 | 11 | 18 | 32 |
| TCC37 | Ta | Neg | Neg | 1 | 1 | 0 | 2 |
| TCC38 | T3a | Neg | Neg | 1 | 12 | 17 | 30 |
| TCC39 | T1 | Neg | Neg | 0 | 4 | 4 | 8 |
| TCC40 | Ta | Neg | Neg | 2 | 1 | 4 | 7 |
| TCC41 | Ta | Neg | Neg | 0 | 1 | 2 | 3 |
| TCC43 | T1 | Neg | Neg | 1 | 2 | 7 | 10 |
| TCC44 | T1 | Neg | Neg | 1 | 2 | 5 | 8 |
| TCC45 | T3a | Pos | Pos | 0 | 19 | 8 | 27 |
| TCC46 | T2a | Neg | Neg | 4 | 2 | 4 | 10 |
| TCC47 | T2a | Neg | Neg | 2 | 1 | 7 | 10 |
LVI Lymphovascular invasion
Somatic mutations of primary bladder cancers and CTCs
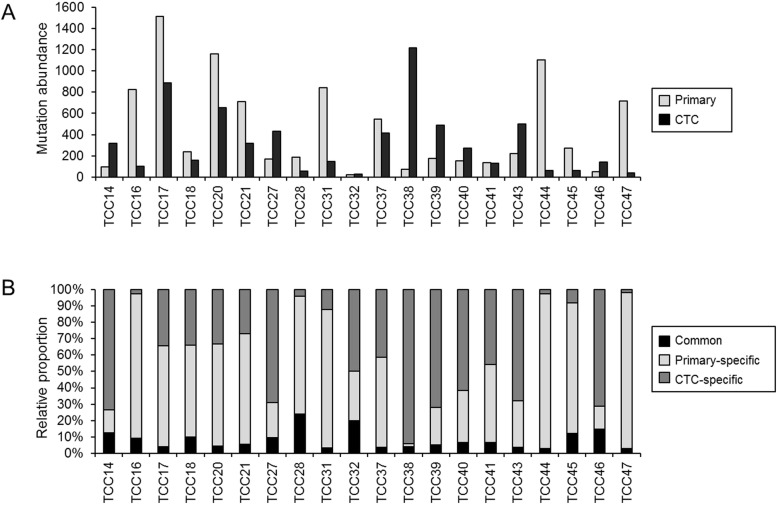
Somatic mutations were identified from primary tumor and CTC genomes by comparing them with matched normal genomes. For 20 pairs of primary and CTC genomes, we identified a total of 14,864 exonic mutations (Additional file 2). Mutation abundance was highly variable across tumors, i.e., 20–1515 exonic mutations (median of 231) for primary tumors and 28–1217 exonic mutations (median of 217) for CTC genomes (Fig. 2a). Mutation abundances of primary tumors and CTC genomes were not significantly correlated (r = 0.16, P = 0.499), suggesting that mutation abundance of primary tumors might not have been accurately inferred from CTC mutation profiles. Next, we classified mutations into common and primary−/CTC-specific mutations. Common mutations were those observed both in the primary and CTC genomes while primary and CTC-specific mutations were those observed only in the primary and CTC genomes, respectively. Relative proportions of common and primary−/CTC-specific mutations in 20 bladder cancers are shown in Fig. 2b. We identified 8 to 89 common mutations comprising 2.7 to 23.9% of mutations in a given case across 20 cases. The low level of mutation concordance and low correlation of mutation abundance between the primary and CTC genomes suggest that CTC might have diverged from primary tumors in early evolutionary stages and undergone distinct genome evolution.
Fig. 2.
Mutation abundance of primary tumors and CTC (circulating tumor cells) genomes. (a) Mutation abundance of primary tumors and their matched CTC genomes for 20 cases of UBC. (b) Relative proportion of common mutations (i.e., those commonly observed both for primary and CTC genomes) and region-specific mutations in given cases
Mutation features with respect to regional categories
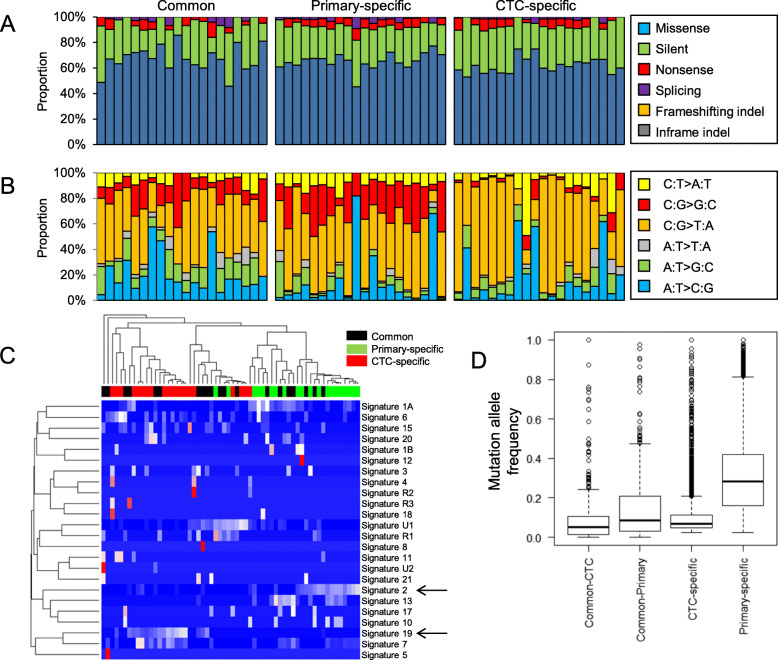
Common mutations as well as primary- and CTC-specific mutations were examined for functional consequence of somatic mutations on encoded peptides (e.g., missense and silent mutations) (Fig. 3a). We observed that ratios of missense mutations to silent mutations (NS/S ratios) were similar across regional categories (1.1–6.0, 1.2–4.2, and 1.1–3.7 for common, primary-specific, and CTC-specific mutations, respectively). The comparable level of NS/S ratios across different mutation categories and the comparable mutation abundance of primary tumors and CTC genomes indicate that mutations identified in CTC genomes are somatic mutations arising from tumors instead of germline variants [14]. However, mutation spectra analysis showed that CTC-specific mutations were distinct from common or primary-specific mutations (Fig. 3b). A depletion of C-to-G transversions and an overrepresentation of C-to-T transitions were observed specifically for CTC-specific mutations compared to common and primary-specific mutations. We further explored mutation signatures across the three mutation categories (Fig. 3c). Hierarchical clustering of signature levels revealed two clusters representing primary- and CTC-specific mutations were relatively enriched with mutation signatures 2 and 19, respectively (arrows in Fig. 3c). Mutation signature 2 representing the activity of endogenous APOBEC cytidine deaminase has been associated with UBC in TCGA analysis [15, 16]. Mutation signature 19 is characterized by C-to-G transversions, but distinct from C-to-G transversions occurring on CpG dinucleotide contexts (e.g., signature 1A and 1B representing spontaneous deamination of methylated CpG dinucleotides) [17]. This signature has been observed in pilocytic astrocytomas whose mutation etiology is largely known. Mutation spectra and signature analysis results suggest that mutational forces that have been operative in the generation of primary- and CTC-specific mutations are not identical, further highlighting that CTC genomes might have evolved separately from primary tumors. Clonality analysis results of mutations in terms of mutant allele frequencies are shown in Fig. 3d. Mutations in primary and CTC genomes were segregated into common and region-specific mutation. Of interests, region-specific mutations showed higher level of mutant allele frequencies compared to region-common mutations. These results suggest that common mutations observed in both primary tumors and CTCs might not represent clonal mutations and that the mutational subclonal architecture of two genomes might have been subjected to selection events.
Fig. 3.
Three mutation categories and mutation analysis. (a) Consequences of somatic mutations on encoded peptide sequences are categorized and relative proportions are shown across three regional mutation categories (common, primary-specific, and CTC-specific). (b) Similarly shown for six mutation spectra. (c) Mutation signature levels estimated across mutation categories are shown in a heatmap with a dendrogram. Two arrows indicate signatures of 2 and 13. (d). Mutation allele frequencies of mutations are shown as clonality measures
Cancer-related genes
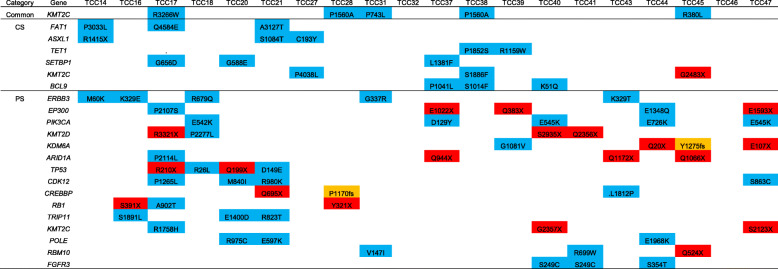
To identify functional driver mutations in each regional category, we first examined cancer-related genes (i.e., Cancer Census Genes [18]) that occurred in at least three cases in our cohort (Table 2). In region-common mutations, missense mutations of KMT2C encoding MLL3 were frequently observed (5 cases), suggesting that mutations on this epigenetic regulator might represent early genomic alterations in the development of bladder cancer. KMT2C gene has been frequently observed to be mutated in muscle invasive bladder cancer [19]. Consistently, KMT2C mutations were also frequently observed in CTC- and primary-specific mutations including one and two nonsense mutations, respectively. Frequent mutations on other epigenetic modifiers in CTC- and primary-specific mutations were not identical in that CTC-specific mutations were observed on ASXL1 and TET1 while primary-specific mutations were common in KMT2D, KDM6A, and ARID1A. Pathway-level convergent mutations targeting different context of epigenetic regulators highlight the importance of epigenetic dysregulation in the pathogenesis of bladder cancers. Of note, CTC-specific frequent mutations included those associated with hematologic malignancies such as ASXL1 [20] and SETBP1 [21] suggestive of environmental impact on the acquisition of mutation. Previously reported mutations that were frequent in muscle invasive bladder cancers such as TP53, PIK3CA, RB1, EP300, and FGFR3 were observed in primary-specific mutations.
Table 2.
Mutations on cancer-related genes
Discussion
In this study, we observed that the concordance rate of somatic mutations between CTC and corresponding primary tumor genomes was relatively low, suggesting that CTC genomes might have undergone distinct evolutionary pathway compared to primary tumors. Supporting this, CTC-specific mutations are distinct from common or primary-specific mutations in terms of sequence compositions (i.e., mutation signatures) and clonality measures such as allele frequencies, suggesting that mutational forces for CTC genomes might be distinct from those associated with primary tumors. In addition, cancer-related genes affected by CTC- and primary-specific mutations were also different from each other.
Research on clinical relevance of ‘liquid biopsy’ has been recently highlighted. CTC may be used as a substitute for tissue biopsy to evaluate drug responsiveness and predict an optimal therapy [22, 23]. CTC-driven genomic or transcriptomic findings may provide valuable information for predicting patients’ prognosis and further guide personalized treatment decision-making in bladder cancer. Until now, primary tumor specimens have served as major cellular sources to obtain genetic information. Some studies have compared genomic or transcriptomic profiles and reported a substantial level of heterogeneity between primary and matched metastatic lesions of given individual [24, 25]. Our study performed analyses of genomic profiles of cultured CTCs and corresponding primary tumor tissue. The current study presented some meaningful results. As demonstrated here, genomic profiling of primary tumors and CTCs is very different in other studies. Reports for colorectal cancer showed similar results. For example, Lyberopoulou et al. [26] have observed 52 patients with colorectal cancer and found discordance between primary tumor and CTCs for KRAS, BRAF, CD133, re3130, and Plastin3 rs6643869. This discordance was confirmed by Kondo et al. [27] and a recent meta-analysis including nine studies and 244 patients [28]. We expect that mutational heterogeneity of CTCs and corresponding primary bladder tumor may recapitulate the relationship between primary and metastatic lesions given that CTCs are major sources for distant metastases. To cope with metastasis and disease progression, genetic information from CTCs might be complementary for longitudinal management of bladder cancer.
To explain the discordance between CTCs and corresponding primary tumor, we need to pay attention to intra-tumor heterogeneity (ITH) and CTCs heterogeneity. Modeling genomic or mutational diversity between regional biopsies in a given individual using a tree structure of tumor growth has been proposed as a ‘trunk-branch model’ [29]. The trunk and branch/leaf in the tree represent founding ubiquitous driver mutations present in every tumor subclone and region and regionally heterogeneous mutations that are not present in every tumor cell or tumor region, respectively [29]. The number of ubiquitous and heterogeneous genetic events in the tumor can depend on the length of the trunk and size of the branch of the phylogenetic trees inferred from mutation profiles and their regional distribution. Thus, as disease progresses, in addition to mutations detected in primary tumors, CTC genomes can acquire mutations in a similar evolutionary process where minority subclones emerge from pre-existing clones. Tumor cells can shed from different and many other tumor sites. Hence, mutation profiles of CTC genomes may be substantially different from those of primary tumors as composite of genetically distinct tumor subject to ITH [30]. Thus, single site tumor biopsy may underestimate the clonal landscape of the overall tumor burden.
Among mutations commonly observed in both CTCs and corresponding primary tumors, KMT2C mutations were detected in five patients. All these five patients (patients TCC 17,28,31,38,45) were shown to be pathologically T2 or higher stage (muscle invasive bladder cancer). KMT2C gene has been frequently observed to be mutated in muscle invasive bladder cancers [19]. On the contrary, KMT2C mutations in CTCs were not observed in patients with T1 or Ta disease. In clinical practice for bladder cancer, T2 disease has a great clinical significance in decision-making for treatment. Although it requires further supporting evidence, the correlation of disease stages and certain mutations such as KMT2C is one of major applications in using CTCs for clinical practice.
To the best of our knowledge, this is the first study to conduct mutation-based comparison between CTCs and primary corresponding tumor mutations using whole-exome sequencing in bladder cancer. Although a number of studies have used high-throughput sequencing to characterize CTCs for bladder cancers [31], their results are only for CTC, not amenable for comparison between CTC and primary tumors. In addition, the present study suggests that the application of sequencing or CTCs could provide a potential clinical role to investigate biologic targets by peripheral blood sampling. Genetic characteristics of CTCs and corresponding primary tumor were shown to differ. Thus, identifying only genetic characteristics of primary tumors is not enough to cope with the disease’s progression. Clearly, CTCs are expected to play an important role. The need for genetic analysis using liquid biopsy such as CTCs can also play a role in handling bladder cancer in addition to convenience of sampling in liquid biopsy. Moreover, recent studies addressed the genomic alterations associated with distant metastases, e.g., ESR1 mutations and MYC, YAP1 amplifications enriched in metastatic breast cancer and lung cancer genomes, respectively [32, 33]. These studies also highlight the difference in terms of driver mutations. Given the CTCs represent the cells that have escaped the physical constrains of primary tissues, the comparison of somatic mutations and other genomic alterations between primary and CTC genomes would provide clues on the genetic drivers of distant metastases.
One limitation of this study is an incomplete establishment of CTC culture. Despite the importance of CTCs in prognosis of cancer patients, the clinical utility of CTCs was limited by its rareness in blood (one CTC in a billion normal blood cells) [34]. For this reason, expanding CTCs to large numbers is encouraged for performing CTC genotyping and phenotyping despite there are concerns that in vitro culture of CTCs may change characteristics of bona fide CTCs. It can be questionable whether cultured CTCs originating from primary cancer could retain their original identification. To verify if cultured CTCs could retain their original identification of primary bladder cancer, we have previously verified that cultured CTCs could retain their original identification of primary bladder cancer by applying FISH method although further studies are needed [35]. In addition, bladder cancer tissue specimens were obtained by TURBT in our study. Although tissue specimens were acquired with sufficient margins, which were removed before the tissue preparation, tissue and DNA damage may have occurred during TURBT. In order to minimize or avoid these concerns, samples were obtained with TURBT preferably thick and large as much as possible. Then, samples were frozen and stored after removing the boundary parts that are suspected of being damaged.
Conclusion
In this mutation-based comparison study, we observed a low concordance level of mutations between genomes of CTCs and primary corresponding bladder cancers, suggesting that CTC genomes might have undergone distinct evolutionary pathway compared to primary tumors. The current study presented some meaningful results to be taken into account when evaluating clinical utility of circulating tumor cells for treatments and follow-up of bladder cancers.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- CTCs
Circulating tumor cells
- TURBT
Transurethral resection of bladder tumor
- ITH
Intra-tumor heterogeneity
Authors’ contributions
TMK contributed writing, analysis and interpretation of the data; KJH, HWM edited the manuscript; JSY performed statistical analyses; YJS reviewed and revised the manuscript; JBC provided resources, review and editing; TMK, USH contributed conception, formal analysis, investigation and design of the study. All authors read and approved the submitted version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Award Number: NRF-2015R1C1A1 A01051802, 2018R1D1A1B07049542, 2019R1A5A2027588). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets analyzed during the current study would be available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was performed under Institutional Review Board–approved protocol of our institution, Seoul St. Mary’s Hospital, College of Medicine, the Catholic University of Korea (approval number: KC15TNSI0924) and written informed consents were obtained from all study participants, following the regulations outlined in the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ma G, Yang X, Liang Y, Wang L, Li D, Chen Y, et al. Precision medicine and bladder cancer heterogeneity. Bull Cancer. 2018;105(10):925–931. doi: 10.1016/j.bulcan.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou-Redorta J. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008;54(2):303–314. doi: 10.1016/j.eururo.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 3.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, et al. Predicting recurrence and progression in individual patients with stage ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49(3):466–477. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Chamie K, Litwin MS, Bassett JC, Daskivich TJ, Lai J, Hanley JM, et al. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer. 2013;119(17):3219–3227. doi: 10.1002/cncr.28147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg JE, Carroll PR, Small EJ. Update on chemotherapy for advanced bladder cancer. J Urol. 2005;174(1):14–20. doi: 10.1097/01.ju.0000162039.38023.5f. [DOI] [PubMed] [Google Scholar]
- 6.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TM, Jung SH, Baek IP, Lee SH, Choi YJ, Lee JY, et al. Regional biases in mutation screening due to intratumoural heterogeneity of prostate cancer. J Pathol. 2014;233(4):425–435. doi: 10.1002/path.4380. [DOI] [PubMed] [Google Scholar]
- 8.Kim TM, Jung SH, An CH, Lee SH, Baek IP, Kim MS, et al. Subclonal genomic architectures of primary and metastatic colorectal Cancer based on Intratumoral genetic heterogeneity. Clin Cancer Res. 2015;21(19):4461–4472. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171(5):1029–41.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alix-Panabières C, Mader S, Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J Mol Med (Berl) 2017;95(2):133–142. doi: 10.1007/s00109-016-1500-6. [DOI] [PubMed] [Google Scholar]
- 17.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15(9):585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder Cancer. Cell. 2018;174(4):1033. doi: 10.1016/j.cell.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, et al. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45(8):942–946. doi: 10.1038/ng.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 23.van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JM. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res. 2011;71(18):5955–5960. doi: 10.1158/0008-5472.CAN-11-1254. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Roca C, Raynaud CM, Penault-Llorca F, Mercier O, Commo F, Morat L, et al. Differential expression of biomarkers in primary non-small cell lung cancer and metastatic sites. J Thorac Oncol. 2009;4(10):1212–1220. doi: 10.1097/JTO.0b013e3181b44321. [DOI] [PubMed] [Google Scholar]
- 25.Sherwood J, Dearden S, Ratcliffe M, Walker J. Mutation status concordance between primary lesions and meastatic sites of advanced non-small-cell lung cancer and the impact of mutation testing methodologies: a literature review. J Exp Clin Cancer Res. 2015;34:92. [DOI] [PMC free article] [PubMed]
- 26.Lyberopoulou A, Aravantinos G, Efstathopoulos EP, Nikiteas N, Bouziotis P, Isaakidou A, et al. Mutational analysis of circulating tumor cells from colorectal cancer patients and correlation with primary tumor tissue. PLoS One. 2015;10(4):e0123902. doi: 10.1371/journal.pone.0123902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo Y, Hayashi K, Kawakami K, Miwa Y, Hayashi H, Yamamoto M. KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer. 2017;17(1):311. doi: 10.1186/s12885-017-3305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Meucci S, Sheng L, Keilholz U. Meta-analysis of the mutational status of circulation tumor cells and paired primary tumor tissues from colorectal cancer patients. Oncotarget. 2017;8(44):77928–77941. doi: 10.18632/oncotarget.18272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4(127):127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 30.Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alva A, Friedlander T, Clark M, Huebner T, Daignault S, Hussain M, et al. Circulating tumor cells as potential biomarkers in bladder Cancer. J Urol. 2015;194(3):790–798. doi: 10.1016/j.juro.2015.02.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertucci F, Ng CKY, Patsouris A, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 33.David JHS, Naema N, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet. 2020;52(4):371–377. doi: 10.1038/s41588-020-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Ramnath N, Nagrath S. Current status of CTCs as liquid biopsy in lung Cancer and future directions. Front Oncol. 2015;5:209. doi: 10.3389/fonc.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TJ, Moon HW, Kang S, Yang J, Hong SH, Lee JY, et al. Urovysion FISH could be effective and useful method to confirm the identity of cultured circulating tumor cells from bladder Cancer patients. J Cancer. 2019;10(14):3259–3266. doi: 10.7150/jca.30079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study would be available from the corresponding author on reasonable request.