Abstract
Polo-like kinases (Plks) are the key regulators of cell cycle progression, the members of which share a kinase domain and a polo-box domain (PBD) that serves as a protein-binding module. While Plk1 is a promising target for antitumor therapy, Plk2 is regarded as a tumor suppressor even though the two Plks commonly recognize the S-pS/T-P motif through their PBD. Herein, we report the crystal structure of the PBD of Plk2 at 2.7 Å. Despite the overall structural similarity with that of Plk1 reflecting their high sequence homology, the crystal structure also contains its own features including the highly ordered loop connecting two subdomains and the absence of 310-helices in the N-terminal region unlike the PBD of Plk1. Based on the three-dimensional structure, we furthermore could model its interaction with two types of phosphopeptides, one of which was previously screened as the optimal peptide for the PBD of Plk2.
Keywords: polo-box domain, Polo-like kinase 2, tumor suppressor, structure, cell cycle
INTRODUCTION
Polo-like kinases (Plks) are serine/threonine kinases that regulate a variety of aspects of the cell cycle process, including M-phase entry, centrosome biogenesis, mitosis and cytokinesis, as well as response to genotoxic stress and neuron differentiation.1,2 To date, five Plk proteins (Plk1-Plk5) have been identified in mammals, which share an N-terminal kinase domain and a noncatalytic C-terminal polo-box domain (PBD) except the PBD-only protein human Plk5.2,3 The kinase domain of Plks is highly homologous to the catalytic domain of other serine/threonine kinases. On the other hand, the PBD, which is critical for the function of Plks by mediating protein-protein interaction and subcellular localization,4 is present uniquely in the Plk protein family.5,6 The PBD of Plk1, Plk2, and Plk3 recognizes the S-pS/T-P motif in common, in which phosphorylation is critical for the binding interaction.7,8 The cryptic polo-box domain (CPB) of Plk4 plays a similar role, but it shares limited sequence homology with the other PBDs and requires neither the motif nor phosphorylation to bind a target protein.9
In association with their role in controlling cell cycle, the correlation of Plks and cancer has been under intense investigation. Intriguingly, despite the high sequence homology between Plk1 and Plk2 (nearly 50% in the kinase domain and >30% in the PBD), the two proteins function oppositely in the tumor development. Plk1, the most extensively characterized member, is the master regulator of mitosis and also a promising cancer target that is generally overexpressed in human cancers and implicated in aggressiveness and bad prognosis.10–12 Plk2, also known as serum-inducible kinase or Snk, was first identified as an immediate-early gene with a potential role in early mitogenic signaling.13 Later studies have suggested that Plk2 functions in centriole duplication14,15 and controls cell proliferation.16,17 Considerable evidence indicates that Plk2 is a tumor suppressor; it is significantly downregulated in various B-cell neoplasms and hepatocellular carcinoma cells,18,19 and it is targeted by microRNA-126, which promotes acute myeloid leukemia cell survival.20 Recent reports have shown that Plk2 is also involved in nerve growth factor-driven neuronal cell differentiation21 and ADAM17-mediated inflammatory signaling.22
With the intention of overcoming the cross-reactivity usually accompanied with the inhibition of canonical kinase domain, a number of studies have focused on the PBD of Plk1 as an attractive target for the development of an antitumor drug candidate.23–28 However, the PBDs of Plk1 and Plk2 share remarkable similarity in their sequence and in motif they recognize, implying the possibility of another cross-reactivity between them. For this reason, accurate information of the PBD of Plk2 would be of importance and necessity, including its binding targets during cell cycle and its three-dimensional structure. Herein, we report the 2.7 Å resolution structure of the PBD of Plk2, which exhibits considerable similarity with that of Plk1 as their high sequence identity but also some structural features discriminating the two proteins. This structure allows us to model the complex structure bound to the phosphopeptide, which contains the optimal binding motif for the PBD of Plk2 found in the previous high-throughput screening study.
MATERIALS AND METHODS
Preparation, crystallization, and structure determination of the PBD of Plk2
The DNA fragment coding for the PBD of human Plk2 (residues 468–685) was amplified by polymerase chain reaction and cloned into the pPROEX HTa plasmid (Invitrogen). The protein was produced in the Escherichia coli BL21(DE3) RIL strain (Novagen) at 18°C and purified using a Ni-NTA column (QIAGEN) first. After removal of the N-terminal (His)6-tag by TEV protease treatment, the protein was further purified using a HiPrep 26/60 Sephacryl S-100 HR gel filtration column (GE Healthcare), equilibrated with a buffer solution containing 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 10% (v/v) glycerol, and 2 mM dithiothreitol. Crystals were obtained by the sitting-drop vapor diffusion method at 18°C by mixing and equilibrating 0.4 μL samples of the protein solution (20 mg/mL) and a precipitant solution containing 1.26 M sodium phosphate and 0.14 M potassium phosphate. Before data collection, the crystals were immersed briefly in a cryoprotectant solution, which was the reservoir solution plus 20% glycerol. Diffraction data were collected on the beamline 7A at the Pohang Accelerator Laboratory, Korea, and processed using the program HKL 2000.29 The structure was determined by the molecular replacement method with the program Phaser30 using the structure of the PBD of Plk131 as a search model. The programs Coot32 and PHENIX33 were used for the model building and refinement, respectively. Crystallographic data statistics are summarized in Table I. The coordinates of the structure together with the structure factors have been deposited in the Protein Data Bank with the accession code of 4XB0.
Table I.
Data Collection and Structure Refinement Statistics
| Data collection | Plk2-PBD |
|---|---|
| Space group | /23 |
| Unit cell dimensions | |
| a, b, c (Å) | 152.29, 152.29, 152.29 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 0.9795 |
| Resolution (Å) | 50.0–2.7 (2.75–2.70)b |
| Rsyma | 7.2 (35.7) |
| I/σ (I) | 48.1 (5.1) |
| Completeness (%) | 99.6 (99.4) |
| Redundancy | 22.1 (7.9) |
| Refinement | |
| Resolution (Å) | 50.0–2.7 |
| No. of reflections | 16,221 |
| Rworkc/Rfree | 22.5/26.3 |
| No. atoms | |
| Protein | 3467 |
| Water and ions | 29 |
| RMS deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.048 |
| Ramachandran plot (%) | |
| Most favored region | 96.0 |
| Additionally allowed region | 4.0 |
| Generously allowed region | |
| Average B-values (Å2) | |
| Protein | 49.1 |
| Water and ions | 48.3 |
, where Iobs is the observed intensity of individual reflection and Iavg is average over symmetry equivalents.
The numbers in parentheses are statistics from the highest resolution shell.
, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 10% of the data.
Size exclusion chromatography-multiangle light scattering (SEC-MALS)
The PBD of human Plk1 (Residues 371–603) and the CPB of human Plk4 (Residues 581–808) were prepared and purified as reported earlier.9,34 For the SEC-MALS experiments, samples were diluted to a concentration of 1.5 mg/mL in a buffer solution containing 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 5% (v/v) glycerol, and 2 mM dithiothreitol. SEC-MALS was performed using a BioSep-SEC-S2000 column (phenomenex), DAWN HELEOS-II (Wyat Technology Corporation), Optilab T-rEX (Wyat Technology Corporation), and ASTRA version 6.1 (Wyat Technology Corporation) coupled with high-performance liquid chromatography (Shimadzu).
RESULTS AND DISCUSSION
Structure of the Plk2-PBD
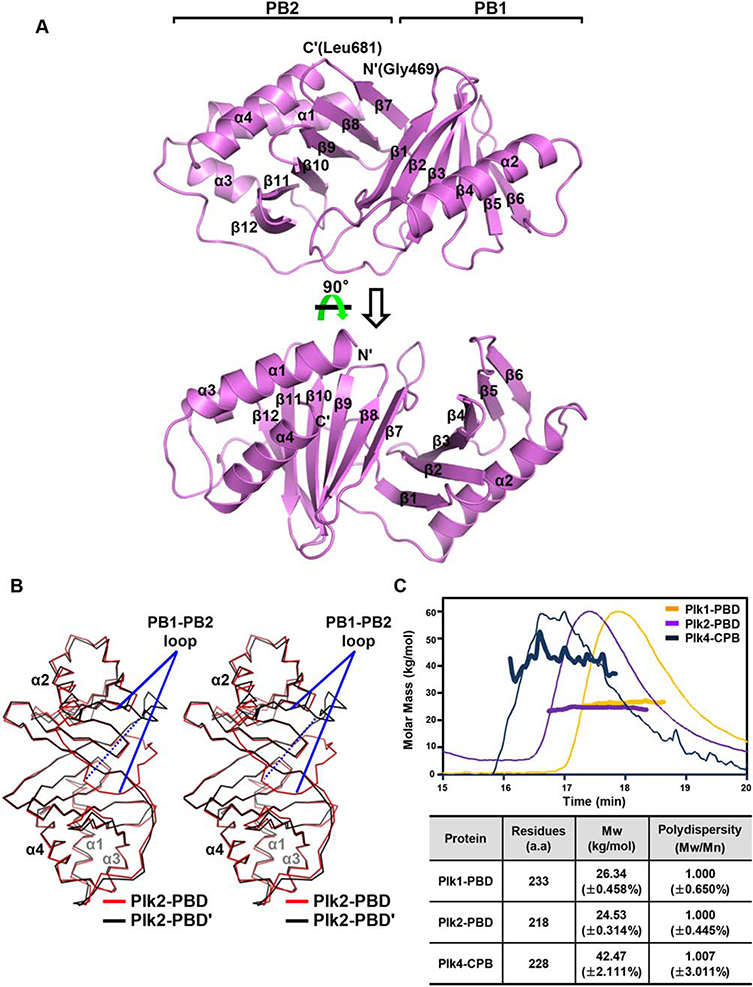
The structure of the PBD of Plk2, referred to as Plk2-PBD, was determined to 2.7 Å resolution (Table I). The asymmetric unit of the crystal contains two molecules of Plk2-PBD (Plk2-PBD and Plk2-PBD′, respectively; Supporting Information Fig. S1). The Plk2-PBD monomer is composed of 4 α-helices and 12 β-strands that are arranged to form two polo-box subdomains, PB1 (Residues 505–582) and PB2 (Residues 603–681), and an N-terminal extension (including α1 lying on PB2 and α1-β1 loop; Residues 469–504). Each subdomain adopts a canonical PB fold: a six-stranded antiparallel β-sheet (composed of β1–β6 in PB1 and β7–β12 in PB2) with an α-helix (α2 in PB1 and α4 in PB2) packed against one side of the sheet [Fig. 1(A)]. The N-terminal extension region, also known as “Polo-cap,” folds around PB2 and thus appears to support PB1-PB2 tethering, as the region does in Plk1-PBD.8 The two Plk2-PBD monomers in the asymmetric unit overlap each other fairly well when superposed, with the RMSD value of 1.00 Å over 197 aligned residues out of total 213 residues [Fig. 1(B)]. One noticeable discrepancy is exhibited with a loop segment linking α2 of PB1 and β7 of PB2 (Residues 583–602), hereafter referred to as the PB1-PB2 loop, which shows a significant translocation within a distance of 19.5 Å [Fig. 1(B)]. This is presumably due to the differences of the two loops in the intramolecular interaction with the rest of the protein and in the intermolecular crystal packing contacts with a symmetry-related Plk2-PBD molecule (Supporting Information Fig. S2).
Figure 1.
Crystal structure of Plk2-PBD. (A) Two views of the monomeric structure. Plk2-PBD is presented as ribbon drawings with the labels of secondary structures according to the order of their appearance in the primary sequence. Also labeled at the top are two subdomains of the molecule. (B) Structural superposition of two subunits in the asymmetric unit shown in Cα traces. For clarity, α-helices of the two monomers are labeled only. PB1-PB2 loops showing dramatically different location are indicated. The dashed line highlights the distance of the Cα atoms of two Gly586 residues where the loops are furthest apart. (C) SEC-MALS analysis of three PBDs. (Top) Molar masses (in kg/mol) are plotted against the elution time (in min) from the size exclusion column. (Bottom) Plk4-CPB, but not Plk1-PBD or Plk2-PBD, forms a dimer. Mw, weight-average molar mass; Mn, number-average molar mass.
At a glance, the two Plk2-PBDs seem to interact with each other and form a homodimeric structure (Supporting Information Fig. S1). This dimerization is yet questionable because the PBD of Plk1 (referred to as Plk1-PBD), sharing 33% sequence identity and 54% similarity with Plk2-PBD based on the structure-based sequence alignment (Supporting Information Fig. S3), has been reported as a monomer in a number of previous structural studies.4,8,34–36 To investigate this issue, SEC-MALS was performed, which is a powerful tool for determining the molecular weight of macromolecules in solution. The SEC-MALS data confirmed that Plk2-PBD and Plk1-PBD exist as a monomer in solution, in contrast with the CPB of Plk4 (referred to as Plk4-CPB) that forms a dimer as previously determined [Fig. 1(C)].9 Therefore, the observed homodimerization of Plk2-PBD in the asymmetric unit should be a crystallographic artifact rather than a functional form of the protein.
Structural comparison with Plk1-PBD and Plk4-CPB
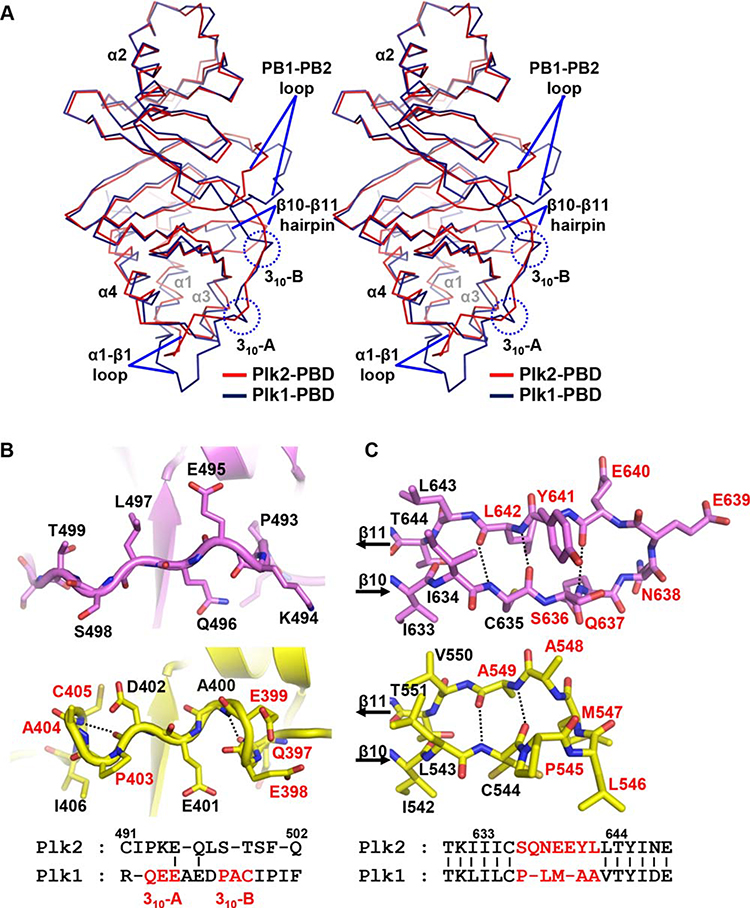
Reflecting a high sequence homology, a Dali search with Plk2-PBD in the Protein Data Bank identified the Plk1-PBD structures preferentially, with the Z-scores of more than 23.5. Structural alignment confirmed that Plk2-PBD and Plk1-PBD superpose each other well overall, with the RMSD value of 1.51 Å over 198 aligned residues [Fig. 2(A)]. But in detail, we were able to find several structural discrepancies between the two PBDs. First, α1-β1 loop (Residues 486–502) in the N-terminal extension of Plk2-PBD does not match well with that of Plk1-PBD both in sequence and in structure [Fig. 2(A,B)]. Especially, while Plk1-PBD contains two 310-helices (Residues 397–399 and 403–405, respectively) in this region, the corresponding region of Plk2-PBD does not build such turns [Fig. 2(B)]. Second, the PB1-PB2 loop is usually disordered in part or entirely in the structures of Plk1-PBD.34–36 However, in the crystal structure we identified, both the loops from Plk2-PBD and Plk2-PBD′ are ordered well (Supporting Information Fig. S2). Third, two more residues and one additional hydrogen bond are inserted in the β10-hairpin-β11 region of Plk2-PBD compared with the corresponding region of Plk1-PBD [Fig. 2(C)].
Figure 2.
Structural comparison with Plk1-PBD. (A) Superposition of Plk2-PBD (red) onto Plk1-PBD (deepblue; PDB code 3HIH) both shown in Cα trace representation. Labeled are α-helices of the two monomers. Regions showing prominent structural discrepancies are indicated: α1-β1 loop (including two 310-helices only present in Plk1-PBD), the PB1-PB2 loop and β10-β11 hairpin. (B) Two 310-helices present in Plk1-PBD (yellow) are not conserved in Plk2-PBD (violet). The Plk1 residues constituting the 310-helices are labeled in red in both ribbon drawings (top) and sequence alignment (bottom). Dashed lines highlight i → I + 3 hydrogen bonding patterns. (C) β10-β11 hairpin loop. Hydrogen bonds between the β10 and β11 strands of Plk2-PBD (violet) and Plk1-PBD (yellow) are represented as dotted lines. Residues that are not conserved between the two proteins are labeled in red (seven in Plk2 and five in Plk1), and the others are in black, in both stick presentation (top) and sequence alignment (bottom).
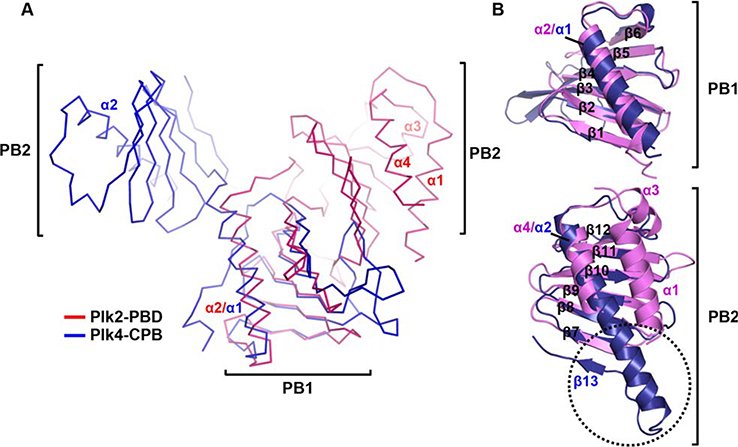
In the Dali search we performed, the structures of the CPB of Plk4 and ZYG-1, the Plk4 homologue from Caenorhabditis elegans, followed Plk1-PBD, with the Z-scores of 8.6–10.4. Therefore, we structurally aligned Plk2-PBD with a single molecule from the Plk4-CPB dimer. Despite quite different arrangement of the subdomains between Plk2-PBD and Plk4-CPB [Fig. 3(A)], each PB1 and PB2 of Plk2-PBD could be matched to the corresponding subdomain of Plk4-CPB with the RMSD values of 2.12 Å over 71 aligned residues and 1.30 Å over 70 aligned residues [Fig. 3(B)]. Distinguishingly, Plk4-CPB contains an “additional region” in PB2 composed of the second half of α2 and following β13, which is not shown in Plk2-PBD [Fig. 3(B)]. Since this part plays a critical role in forming the homodimeric interface of Plk4-CPB,9 the absence of such a region in Plk2-PBD and also in Plk1-PBD accounts for their monomeric state in solution [see Fig. 1(C)]. The absence of the N-terminal extension (containing α1 of Plk2-PBD) and the replacement of the PB1-PB2 loop with a two-residue linker (S700-P701 of Plk4) are also notable differences of Plk4-CPB compared with Plk2-PBD.
Figure 3.
Structural comparison with Plk4-CPB. (A) Plk2-PBD (violet) and Plk4-CPB (blue; PDB code 4N7Z), both shown in Cα trace representation with labels for α-helices of the two domains, are aligned by superposing their PB1 subdomains. (B) Subdomain comparison. Dotted circle indicated the second half of α2 and following β13 of Plk4-CPB that is critical for its homodimerization but absent in Plk2-PBD.
Modeling of phosphopeptide binding to Plk2-PBD
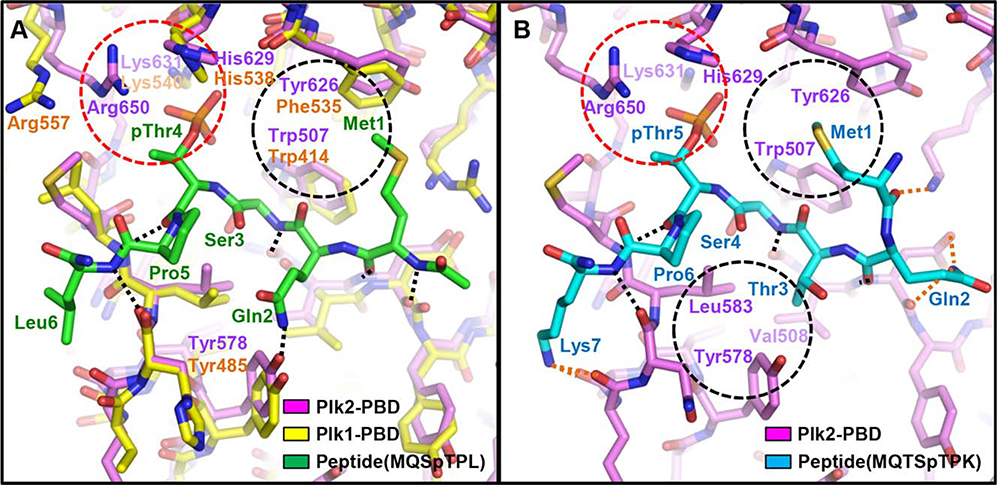
To structurally elucidate the binding of Plk2-PBD to the S-pS/T-P motif, we first tried to crystallize Plk2-PBD in a complex with the motif-containing phosphopeptide, but we were unsuccessful to obtain the complex crystal. Previous studies indicated that peptide binding does not induce any dramatic conformational change in the PBD domains.4,9,34 Therefore, we instead modeled a phosphopeptide-bound Plk2-PBD structure by superposing Plk2-PBD onto Plk1-PBD in a complex with a peptide MQSpTPL. Upon structural alignment, we found that the MQSpTPL motif, the core of the Plk1-PBD-optimal peptide sequence,7,28 is also able to associate well with Plk2-PBD [Fig. 4(A)]. The phosphothreonine residue in the consensus motif, which was proved to be inevitable for the peptide to binding Plk2,28 mediates ionic interactions with positively charged Plk2 residues (His629, Lys631, and Arg650). Together with a number of hydrogen bonds, hydrophobic interaction between Met1 of the peptide and the bulky side chains of Trp507 and Tyr626 of Plk2 also contributes the complex formation in this model. Intriguingly, through a high-throughput screening study, Reindl et al.28 previously reported that alteration of the motif from MQSpTPL (Plk1-PBD-optimal) to MQTSpTPK (Plk2-PBD-optimal) increases the Plk2-PBD binding affinity nearly 30-fold (from KD of 1580 to KD of 56 nM). We thus next modeled the complex structure of Plk2-PBD bound to this Plk2-PBD-optimized peptide [Fig. 4(B)]. In this complex model, the reinforcement of the interaction between the modified peptide and Plk2-PBD could be accounted for by three major points. First, insertion of a threonine residue between glutamine and serine induces a consequent shift of Met1, pushing its side-chain into the hydrophobic pocket more deeply and thus strengthening hydrophobic interactions with the bulky Plk2 residues Trp507 and Tyr626. Second, the insertion of threonine possibly provides the opportunity for hydrophobic interactions between its Cγ atom and Val508, Tyr578 and Leu583 of Plk2. Third, movement of Gln2 by the threonine insertion and lysine substitution of C-terminal leucine result in the additional formation of intermolecular electrostatic interactions. Further biochemical and structural studies, together with solid information of Plk2-PBD-binding proteins in cells which are largely unknown to date, would be needed to determine the precise discrepancy between the PBDs in assigning the binding specificity.
Figure 4.
Phosphopeptide-binding models of Plk2-PBD. Plk2-PBD (violet) in a complex with a peptide MQSpTPL (green; A) or MQTSpTPK (cyan; B) are modeled based on the structural superposition on Plk1-PBD (yellow) bound to the MQSpTPL peptide (PDB code 3P34). Labeled are the whole peptide residues and the Plk1 or Plk2 residues involving the intermolecular interactions. Dotted lines represent intermolecular hydrogen bonds. Among them, bonds only shown in (B) are in orange. Dashed red circle highlights ionic interactions mediated by phosphothreonine, and black circles exhibited intermolecular hydrophobic interactions.
Next, we compared the phosphopeptide-bound Plk2-PBD complex model with the Plk4-CPB structure in a complex with a Cep192 fragment. In both the structures, residues from both PB1 and PB2 are involved in intermolecular interactions (Supporting Information Fig. S4). However, the details are quite different from each other, especially due to the aforementioned distinct subdomain arrangement of Plk2-PBD and Plk4-CPB [see Fig. 3(A)]. The 29 residues-shown Cep192 fragment is quite longer than the 7-mer phosphopeptide modeled to bind Plk2. Furthermore, the Cep192 fragment has an aspartate-rich motif that associates with the lysine/arginine-enriched crater of Plk4-CPB and an α-helix region that mediates tight hydrophobic interactions with α1 of Plk4-CPB [Supporting Information Fig. S4(B)], but the Plk2-PBD-bound phosphopeptide does not contain any of them [Supporting Information Fig. S4(A)]. Therefore, such structural differences between the Plk2-PBD and Plk4-CPB complexes should prevent the two Plk proteins from sharing their binding partners and discriminate their role in cell cycle and in other cellular processes that they involve.
In summary, we presented the crystal structure of Plk2-PBD and delineated its structural features discriminating it from other PBDs, especially Plk1-PBD. Structure-based modeling of phosphopeptide-bound complexes provides an account for the drastic improvement in the binding affinity for Plk2-PBD found in the previous peptide screening study. We believe that this structural information will eventually contribute to the development of specific Plk-selective peptides or molecules, which are under intense investigation.
During the revision of this article, an article describing a similar structure was reported.37
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Kyoung-Seok Ryu and Jung-Hye Ha (Korea Basic Science Institute, Korea) for helping the SEC-MALS experiments.
Grant sponsor: National Research Foundation of Korea; Grant number: 2011-0030027; Grant sponsor: Korea Research Institute of Bioscience and Biotechnology Research Initiative Program.
Abbreviations:
- CPB
cryptic polo-box domain
- Plk
Polo-like kinase
- PBD
polo-box domain
- RMSD
root mean square deviation
- SEC-MALS
size exclusion chromatography-multiangle light scattering
Footnotes
This study made use of the beamline 7A at the Pohang Accelerator Laboratory in Korea.
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 2009; 10:265–275. [DOI] [PubMed] [Google Scholar]
- 2.de Carcer G, Manning G, Malumbres M. From plk1 to plk5: functional evolution of polo-like kinases. Cell Cycle 2011; 10:2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Carcer G, Escobar B, Higuero AM, Garcia L, Anson A, Perez G,Mollejo M, Manning G, Melendez B, Abad-Rodriguez J, Malumbres M. Plk5, a polo box domain-only protein with specific roles in neuron differentiation and glioblastoma suppression. Mol Cell Biol 2011; 31:1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JE, Soung NK, Johmura Y, Kang YH, Liao C, Lee KH, Park CH, Nicklaus MC, Lee KS. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci 2010; 67:1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol 2004; 5:429–440. [DOI] [PubMed] [Google Scholar]
- 6.Lowery DM, Lim D, Yaffe MB. Structure and function of Polo-like kinases. Oncogene 2005; 24:248–259. [DOI] [PubMed] [Google Scholar]
- 7.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing plk1 to mitotic substrates. Science 2003; 299:1228–1231. [DOI] [PubMed] [Google Scholar]
- 8.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of plks by the Polo-box domain. Cell 2003; 115:83–95. [DOI] [PubMed] [Google Scholar]
- 9.Park SY, Park JE, Kim TS, Kim JH, Kwak MJ, Ku B, Tian L, Murugan RN, Ahn M, Komiya S, Hojo H, Kim NH, Kim BY, Bang JK, Erikson RL, Lee KW, Kim SJ, Oh BH, Yang W, Lee KS. Molecular basis for unidirectional scaffold switching of human plk4 in centriole biogenesis. Nat Struct Mol Biol 2014; 21:696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strebhardt K Multifaceted polo-like kinases: drug targets and anti-targets for cancer therapy. Nat Rev Drug Discov 2010; 9:643–660. [DOI] [PubMed] [Google Scholar]
- 11.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer 2006; 6:321–330. [DOI] [PubMed] [Google Scholar]
- 12.Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence? Cancer Res 2013; 73:6848–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmons DL, Neel BG, Stevens R, Evett G, Erikson RL. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol Cell Biol 1992; 12:4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warnke S, Kemmler S, Hames RS, Tsai HL, Hoffmann-Rohrer U, Fry AM, Hoffmann I. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr Biol 2004; 14:1200–1207. [DOI] [PubMed] [Google Scholar]
- 15.Cizmecioglu O, Warnke S, Arnold M, Duensing S, Hoffmann I. Plk2 regulated centriole duplication is dependent on its localization to the centrioles and a functional polo-box domain. Cell Cycle 2008; 7:3548–3555. [DOI] [PubMed] [Google Scholar]
- 16.Matthew EM, Yen TJ, Dicker DT, Dorsey JF, Yang W, Navaraj A, El-Deiry WS. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle 2007; 6:2571–2578. [DOI] [PubMed] [Google Scholar]
- 17.Ma S, Charron J, Erikson RL. Role of plk2 (snk) in mouse development and cell proliferation. Mol Cell Biol 2003; 23:6936–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syed N, Smith P, Sullivan A, Spender LC, Dyer M, Karran L, O’Nions J, Allday M, Hoffmann I, Crawford D, Griffin B, Farrell PJ, Crook T. Transcriptional silencing of Polo-like kinase 2 (SNK/plk2) is a frequent event in B-cell malignancies. Blood 2006; 107:250–256. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrino R, Calvisi DF, Ladu S, Ehemann V, Staniscia T, Evert M, Dombrowski F, Schirmacher P, Longerich T. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology 2010; 51:857–868. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA 2008; 105:15535–15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draghetti C, Salvat C, Zanoguera F, Curchod ML, Vignaud C, Peixoto H, Di Cara A, Fischer D, Dhanabal M, Andreas G, Abderrahim H, Rommel C, Camps M. Functional whole-genome analysis identifies Polo-like kinase 2 and poliovirus receptor as essential for neuronal differentiation upstream of the negative regulator alphaB-crystallin. J Biol Chem 2009; 284:32053–32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz J, Schmidt S, Will O, Koudelka T, Kohler K, Boss M, Rabe B, Tholey A, Scheller J, Schmidt-Arras D, Schwake M, Rose-John S, Chalaris A. Polo-like kinase 2, a novel adam17 signaling component, regulates tumor necrosis factor alpha ectodomain shedding. J Biol Chem 2014; 289:3080–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasrao G, Park JE, Kim S, Ahn M, Cheong C, Nam KY, Gunasekaran P, Hwang E, Kim NH, Shin SY, Lee KS, Ryu E, Bang JK. Design and synthesis of a cell-permeable, drug-like small molecule inhibitor targeting the polo-box domain of polo-like kinase 1. PLoS One 2014; 9:e107432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reindl W, Yuan J, Kramer A, Strebhardt K, Berg T. Inhibition of polo-like kinase 1 by blocking polo-box domain-dependent protein-protein interactions. Chem Biol 2008; 15:459–466. [DOI] [PubMed] [Google Scholar]
- 25.Yin Z, Song Y, Rehse PH. Thymoquinone blocks pSer/pThr recognition by plk1 Polo-box domain as a phosphate mimic. ACS Chem Biol 2013; 8:303–308. [DOI] [PubMed] [Google Scholar]
- 26.Murugan RN, Park JE, Lim D, Ahn M, Cheong C, Kwon T, Nam KY, Choi SH, Kim BY, Yoon DY, Yaffe MB, Yu DY, Lee KS, Bang JK. Development of cyclic peptomer inhibitors targeting the polo-box domain of polo-like kinase 1. Bioorg Med Chem 2013; 21: 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Kramer A, Eckerdt F, Kaufmann M, Strebhardt K. Efficient internalization of the polo-box of polo-like kinase 1 fused to an antennapedia peptide results in inhibition of cancer cell proliferation. Cancer Res 2002; 62:4186–4190. [PubMed] [Google Scholar]
- 28.Reindl W, Graber M, Strebhardt K, Berg T. Development of high-throughput assays based on fluorescence polarization for inhibitors of the polo-box domains of polo-like kinases 2 and 3. Anal Biochem 2009; 395:189–194. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sledz P, Stubbs CJ, Lang S, Yang YQ, McKenzie GJ, Venkitaraman AR, Hyvonen M, Abell C. From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1. Angew Chem Int Ed Engl 2011; 50:4003–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun SM, Moulaei T, Lim D, Bang JK, Park JE, Shenoy SR, Liu F, Kang YH, Liao C, Soung NK, Lee S, Yoon DY, Lim Y, Lee DH, Otaka A, Appella E, McMahon JB, Nicklaus MC, Burke TR Jr, Yaffe MB, Wlodawer A, Lee KS. Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat Struct Mol Biol 2009; 16:876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J 2003; 22:5757–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murugan RN, Ahn M, Lee WC, Kim HY, Song JH, Cheong C, Hwang E, Seo JH, Shin SY, Choi SH, Park JE, Bang JK. Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1. PLoS One 2013; 8: e80043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan HM, Wang T, Quan JM. Crystal structure of the polo-box domain of polo-like kinase 2. Biochem Biophys Res Commun 2015; 456:780–784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.