Abstract
Aspergillus fumigatus is a saprobic fungus that causes a range of pulmonary diseases, some of which are characterised by fungal persistence such as is observed in cystic fibrosis (CF) patients. Creation of genetic variation is critical for A. fumigatus to adapt to the lung environment, but biofilm formation, especially in CF patients, may preclude mutational supply in A. fumigatus due to its confinement to the hyphal morphotype. We tested our hypothesis that genetic variation is created through parasexual recombination in chronic biofilms by phenotypic and genetic analysis of A. fumigatus isolates cultured from different origins.
As diploids are the hallmark of parasex, we screened 799 A. fumigatus isolates obtained from patients with CF, chronic pulmonary lung disease and acute invasive aspergillosis, and from the environment for spore size. Benomyl sensitivity, nuclear content measurements through fluorescence-activated cell sorting and scanning electron microscopy were used to confirm the diploid state of large size spores. Whole genome sequencing was used to characterise diploid-associated genetic variation.
We identified 11 diploids in isolates recovered from six of 11 (55%) CF patients and from one of 24 (4%) chronic aspergillosis patients, but not in 368 isolates from patients with acute Aspergillus infection and the environment. Diploid formation was associated with accumulation of mutations and variable haploid offspring including a voriconazole-resistant isolate.
Parasexual recombination allows A. fumigatus to adapt and persist in CF patients, and plays a role in azole resistance development. Our findings are highly significant for understanding the genetics and biology of A. fumigatus in the human lung.
Short abstract
Aspergillus fumigatus can undergo parasexual recombination in the lungs of chronically colonised individuals. This is highly relevant, as the parasexual cycle can facilitate genetic variation and, consequently, in-host adaptation. https://bit.ly/3kAtrXd
Introduction
Aspergillus fumigatus is a ubiquitous saprobic fungus that causes a broad range of diseases in humans [1]. Aspergillus disease manifestations depend on host immunity and the presence of pre-existing lung diseases. The establishment of Aspergillus in the lung environment may lead to chronic and allergic fungal disease or invasive disease, including chronic pulmonary aspergillosis (CPA) [2, 3], subacute invasive aspergillosis (SAIA), allergic bronchopulmonary aspergillosis (ABPA), severe asthma with fungal sensitisation and Aspergillus bronchitis [3]. Treatment is aimed at reduction of symptoms and prevention of progression but may be challenging with a limited number of antifungal drug classes and the risk of accelerated disease progression with the use of immunocompromising agents [4].
For many of these Aspergillus diseases lung colonisation is a critical initial step. Chronic colonisation has been well documented in patients with cystic fibrosis (CF), where isogenic A. fumigatus isolates may be found in sputum from individual CF patients for many years [5]. However, the pathophysiology of fungal persistence in the lung environment is currently poorly understood. We recently postulated that Aspergillus may undergo an evolutionary trajectory during long-term colonisation that results in progeny that is better adapted to the lung environment and thus facilitates fungal persistence [6]. Understanding in-host fungal adaptation may help to design intervention strategies or identify new intervention targets in colonised patients aimed at preventing the development of Aspergillus disease and associated lung damage.
Evidence of fungal in-host adaptation can be observed by macroscopic abnormalities such as white colour (lack of sporulation) and small colony size (slow growth rate) in cultures of respiratory secretions, grown under laboratory conditions, taken from chronically colonised patients. Atypical colony morphology indicates that fungal adaptations to the lung environment come at the cost of growth and sporulation under laboratory conditions [6, 7]. Another marker of in-host adaptation is the development of antifungal drug resistance. Azole resistance develops in 5% to 13% of azole-treated CPA-patients [8] and complicates patient management as the azoles represent the only drug class that is active against Aspergillus species and can be administered orally. Azole resistance development is often associated with the presence of a pulmonary cavity [9]. The cavity allows A. fumigatus to undergo asexual sporulation, which is known to increase mutation supply [10] and the clonal progeny may thus harbour spontaneous mutations, some of which confer azole resistance which can be selected for through azole therapy [6].
In contrast, the epithelial mucus of CF patients is frequently colonised with A. fumigatus where the fungus is believed to produce mycelia and extracellular matrices [11]. Although this biofilm provides shielding from stressors like other microorganisms, antifungal drugs and host immune effectors [12], as a consequence the fungus cannot benefit from asexual sporulation to adapt to the lung environment. Nevertheless, antifungal drug resistance reported in azole-treated and azole-naïve CF patients indicates that A. fumigatus may employ other strategies that enable adaptation [13]. We hypothesise that during long-term hyphal colonisation A. fumigatus adapts through parasexual recombination, a process that involves fusion of compatible hyphae and nuclei leading to mitotic recombination [14, 15]. This process involves a temporary ploidy change from haploid to diploid, and diploidy is considered the hallmark of parasexual recombination [16].
To test our hypothesis, we screened a large collection of A. fumigatus isolates obtained from various patient groups for the presence of diploids as a marker of parasex and subsequently determined if new mutations had been generated in the progeny.
Methods
Strain selection and conidial size measurements
The laboratory information system, and subsequently the department's fungal culture collection, was searched for A. fumigatus isolates from three groups of patients and one environmental group as detailed in the supplementary material. No additional samples were taken for this study, and only stored A. fumigatus isolates from our fungal culture collection were used. All patients could object to the use of residual materials, and those were excluded from our study. Our institutional guidelines did not prescribe formal ethical approval for research with stored isolates. Initial screening of the A. fumigatus isolates was based on the size of the conidia as an indication for the presence of diploidy. The conidial size was measured with a Casy® TT cell counter (OLS OMNI Life Science, Bremen, Germany); for validation of this method see supplementary figure E1.
Confirmation of diploidy
Scanning electron microscopy (SEM) was performed for one selected haploid (V157-27) and one selected diploid (V133-16) isolate for confirmation of the size differences detected in the Casy® TT measurements. To exclude rare artefacts of large conidia such as bi-nucleation, a DAPI (4’,6-diamidino-2-phenylindole) staining was performed. The same haploid (V157-27) and diploid (V133-16) isolates from the SEM analysis were used for this. Benomyl susceptibility testing and fluorescence-activated cell sorting were used to confirm ploidy. Benomyl is a systemic benzimidazole fungicide, and the cytological effects in fungi are caused by binding to the tubulins, a major component of the microtubules, which are involved in segregating chromosomes during mitosis and meiosis [17]. Haploid colonies are generally more tolerant to benomyl in the medium than diploid colonies, and this difference in morphology can be used to distinguish the ploidy level in A. fumigatus isolates. As the nuclear content is doubled in diploid isolates compared to haploid isolates, staining the nuclear content of spores with propidium iodide and measurement by using fluorescence-activated cell sorting (FACS) technology can differentiate the ploidy level as already shown by de Lucas et al. in 1998 [18]. The same selection of isolates for benomyl susceptibility testing were also subjected to FACS analysis as previously described by Veselská et al. [19] with minor adaptations. Finally, as a proof of principle, a diploid isolate was haploidised resulting in reduced spore size, while this is not possible for a haploid isolate.
Genetic characterisation of patient isolates
To determine if subsequent A. fumigatus isolates recovered from individual patients represented single or variable genotypes, microsatellite genotyping was performed by using a panel of nine short tandem repeats as described previously [20]. To understand the genetic adaptation of the isolates that changed in ploidy over time whole genome sequencing analysis was performed. Genomic DNA was extracted from four A. fumigatus isolates from one CF patient, representing two haploid and two diploid isolates. Sequencing was performed on the BGISEQ-500 platform, with a minimum of 1.5 Gb clean data per sample, by the Beijng Genome Institute (BGI, Shenzhen, China). For mapping and variant calling see the supplementary material.
Results
Selection of A. fumigatus isolates
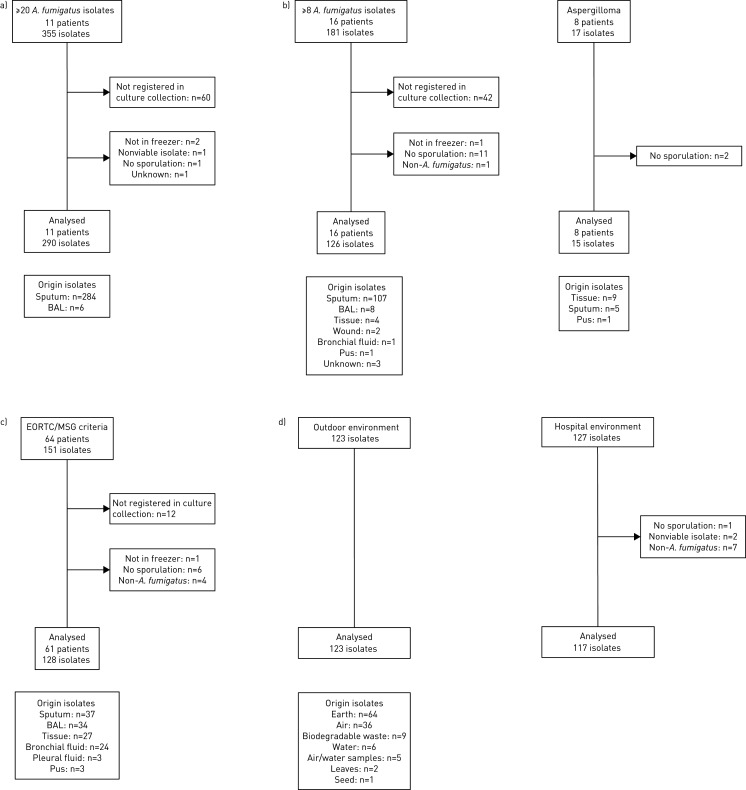
We selected 799 A. fumigatus isolates from four different groups: a) 290 isolates of 11 chronically colonised CF patients; b) 141 isolates of 24 chronically colonised patients with other respiratory diseases, including 15 CPA, 4 ABPA and 5 bronchiectasis; c) 128 isolates of 59 patients with acute invasive aspergillosis, including 23 with proven infection, 23 with probable and 15 with putative invasive aspergillosis [21]; and d) 240 environmental A. fumigatus isolates (figure 1). Inclusion criteria are described in more detail in the supplementary material.
FIGURE 1.
Aspergillus fumigatus isolates selection diagram. Isolates meeting the inclusion criteria are shown for all four groups. If isolates were excluded from the final selection, the reason for exclusion is indicated in the figure. The total number of isolates per group used for conidial size measurement is indicated per group as well as the origin of sample type. a) Cystic fibrosis (CF) patients; b) chronic lung disease; c) acute invasive aspergillosis; and d) environmental. BAL: bronchoalveolar lavage; EORTC/MSG: European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group.
Spore size analysis
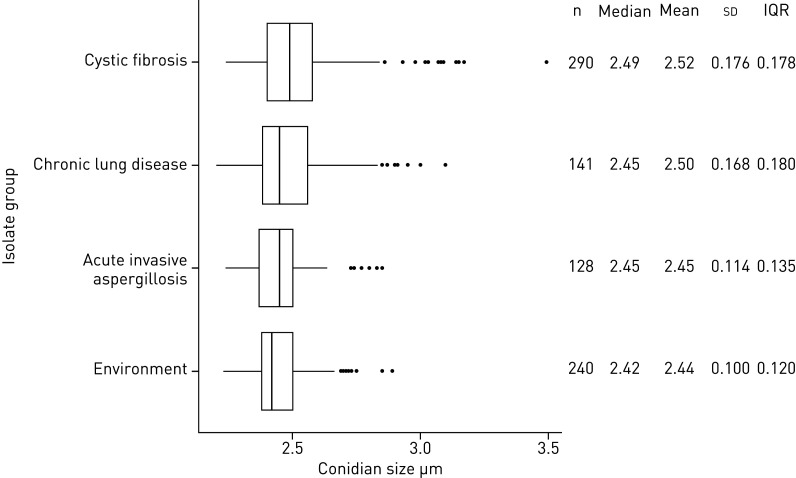
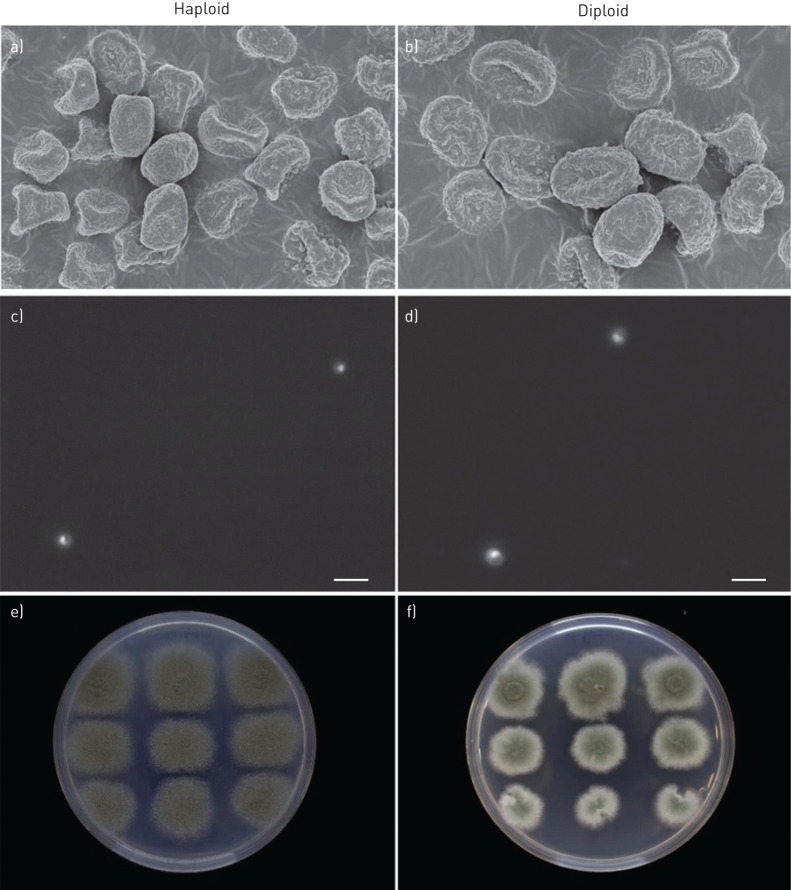
Initial conidial size screening resulted in measurements of 799 unique A. fumigatus isolates with conidial sizes ranging from 2.20 µm to 3.49 µm (figure 2). The mean conidial size of isolates recovered from chronically colonised CF patients was significantly larger than conidia of isolates from patients with invasive aspergillosis or from the environment (p<0.001), but not different from conidia of isolates of chronically colonised respiratory disease patients (p=0.13). To confirm the conidial size difference, scanning electron microscopy was performed on a haploid and its laboratory-derived diploid isolate (Laboratory of Genetics, Wageningen University & Research, the Netherlands, unpublished data). The conidial size of the haploid isolate was 2.41 µm measured by Casy® TT cell counter (triplicate) and 2.38 µm calculated by SEM (quintuplicate), while that of the diploid spores was 3.07 µm and 3.02 µm, respectively. SEM analysis of the haploid and diploid conidia thus confirmed the conidial size differences as measured by the Casy® TT cell counter (figure 3a and b).
FIGURE 2.
Boxplot analysis of 799 Aspergillus fumigatus isolates screened for conidial size. On the left boxplots are shown for each isolate group. On the right summary statistics from the boxplot analysis are depicted for each group (group size n, median, mean, standard deviation and interquartile range (IQR)). Due to the presence of diploids in the chronically colonised cystic fibrosis patients group and chronically colonised chronic lung disease patients group, the mean conidial size and the standard deviation is largest for these two groups.
FIGURE 3.
Conidial size confirmation by a and b) scanning electron microscopy (SEM), c and d) nuclear staining and e and f) benomyl susceptibility testing. The conidial size was assessed by using SEM and fluorescence microscopy, and both confirm a conidial size difference between the haploid and diploid isolate, while excluding the presence of bi-nucleated cells in the diploid isolate (d). The diploid isolate shows a reduced growth on benomyl and the so-called sectoring of the colony (f) (which are the haploidising conidia from an original diploid) that are both absent in the haploid isolate (e).
To further confirm the ploidy level of haploid versus diploid spores DAPI staining was performed, which specifically stains nuclear material. Both haploid and diploid conidia were uninucleate and haploid nuclei were smaller than diploid nuclei (figure 3c and d).
Confirmation of diploidy
Benomyl was used to distinguish between haploid and diploid Aspergillus strains [17, 22] and was performed on a selection of 107 of the 799 A. fumigatus isolates. Whereas the majority of samples could be clearly judged, in some isolates the ability to determine the ploidy level was compromised due to a natural variation in benomyl susceptibility. Therefore, nuclear content measurement through fluorescence-activated cell sorting (FACS) was used as an additional method to determine the ploidy level. To set the reference gating in the FACS analysis software, the haploid and diploid isolates used for SEM were used for comparison of the samples subjected to analysis during each sample run. If benomyl and FACS analyses were in agreement, ploidy level was considered conclusive, while if both analyses were not in agreement, the isolates were considered to be of inconclusive ploidy. The isolates were ranked according to conidial sizes; all 57 isolates with a conidial size of ≥2.70 µm were analysed and from other conidial size ranges (2.20 µm – 2.69 µm) 10 isolates per decimal group were analysed (n=50). The 10 isolates with the largest conidial sizes were confirmed to be diploid, of which nine were recovered from CF patients and one from a patient with chronic lung disease (table 1 and supplementary figure E2). These isolates were generally more sensitive to benomyl compared to haploid colonies (figure 3e) and grew irregularly with the typical sectoring of the colony (figure 3f). These sectors were haploid segregants from the original diploid colony. Haploid isolates are less sensitive to benomyl and do not show irregular growth and sectoring. The next group of 10 isolates was recovered from patients with chronic lung colonisation (five CF and five chronic lung disease): one diploid was identified, three were of inconclusive ploidy and six were haploids (table 1 and supplementary figure E2). From the 50 selected A. fumigatus isolates in the size ranges 2.20–2.69 µm only haploids were observed and never a diploid or inconclusive ploidy isolate.
TABLE 1.
Ploidy analysis of Aspergillus fumigatus isolates with a conidial size of ≥2.90 µm
| Isolate number | Group | Conidial size µm | Ploidy |
| V132-02 | CF | 3.49 | Diploid |
| V124-78 | CF | 3.17 | Diploid |
| V104-11 | CF | 3.15 | Diploid |
| V208-76 | CF | 3.14 | Diploid |
| V108-12 | CF | 3.14 | Diploid |
| V130-40 | CPA | 3.10 | Diploid |
| V194-19 | CF | 3.09 | Diploid |
| V109-60 | CF | 3.09 | Diploid |
| V209-70 | CF | 3.08 | Diploid |
| V133-16 | CF | 3.07 | Diploid |
| V138-11 | CF | 3.03 | Haploid |
| V183-71 | CF | 3.02 | Diploid |
| V113-61 | CF | 3.02 | Inconclusive ploidy |
| V214-14 | CPA | 3.00 | Inconclusive ploidy |
| V158-23 | CF | 2.98 | Inconclusive ploidy |
| V214-17 | CPA | 2.95 | Haploid |
| V172-54 | CF | 2.93 | Haploid |
| V224-65 | CPA | 2.91 | Haploid |
| V211-47 | CPA | 2.91 | Haploid |
| V130-14 | CPA | 2.90 | Haploid |
After conidial size measurements, ploidy level was confirmed by benomyl susceptibility and fluorescence-activated cell sorting (FACS) analysis. Ploidy levels were based on benomyl susceptibility and FACS ratio results. If the two markers were not in agreement, inconclusive ploidy was concluded. Diploid isolates were only detected in the cystic fibrosis (CF) and chronic pulmonary aspergillosis (CPA) groups. No isolates with a conidial size ≥2.90 µm were detected in the acute invasive aspergillosis or environmental group.
To further confirm their diploid state, two diploid isolates (isolate V183-71: 3.02 µm and V209-70: 3.08 µm) were haploidised using a benomyl-containing agar plate and sector growth was analysed for conidial size. Nine subcultured sectors showed six (V183-71) and three (V209-70) sectors with haploid fungus and three (V183-71) and six (V209-70) sectors that remained diploid. The conidial size was 2.30 µm for haploid segregants and 3.02 µm (V183-71) or 3.08 µm (V209-70) for diploid sectors (data not shown). The haploid and diploid conidial sizes did not show variable measurements (p<0.001) confirming the ability of a diploid isolate to haploidise to one specific haploid size.
Characteristics of patients with diploid isolates
Overall, 11 confirmed diploid isolates were identified in six of 11 CF patients (55%) and in one of 24 chronic lung disease patients (4%), while diploids were not found in any of the 368 isolates obtained from patients with acute invasive aspergillosis or from the environment.
To understand the implications of diploid formation we analysed the medical and microbiological records of the seven patients who harboured a diploid isolate. The time between the first cultured A. fumigatus isolate and diploid detection varied per patient (respectively 26, 464, 672, 928, 1241, 1299 and 1785 days). Diploid isolates were detected both in absence and presence of Pseudomonas isolates cultured in respiratory samples (data not shown). In two patients, diploid formation preceded the culture of an azole-resistant isolate; in a CF patient one isolate was highly resistant to voriconazole and isavuconazole (minimum inhibitory concentration (MIC) >16 mg·L−1), and in a patient with chronic lung disease one isolate was resistant to itraconazole (MIC >16 mg·L−1), posaconazole (MIC=1 mg·L−1) and isavuconazole (MIC=16 mg·L−1). The CF patient was homozygous for the Δf508 mutation and despite long-term A. fumigatus colonisation, Aspergillus diseases such as ABPA had not been diagnosed and the patient had no record of previous azole therapy. The second patient was known to have bronchiectasis and recurrent Pseudomonas aeruginosa pneumonia. Despite chronic A. fumigatus lung colonisation, repeated chest radiographs showed no evidence for CPA, in particular absence of nodular lesions and pulmonary cavities. The patient had not received antifungal therapy.
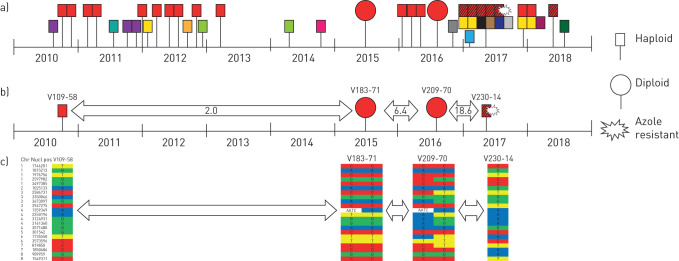
Sequencing of the cyp51A gene showed a wild-type sequence in both resistant isolates, indicating an unknown azole resistance mechanism. The medical records of the two patients indicated that neither had received any prior azole therapy. A. fumigatus isolates of the two patients were genotyped using microsatellite analysis to investigate the genetic relationships between the isolates within a patient. The five isolates of the chronic lung disease patient were isolated over a period of 7.5 years and were non-isogenic. Since it is assumed that only isogenic isolates can undergo a parasexual cycle, the isolates of this patient were not further investigated. Genotyping of 44 A. fumigatus isolates of the CF patient, cultured between 2010 and 2018, showed 15 different genotypes, including one genotype that had persisted over a period of 7.5 years. The persistent genotype was present in 23 isolates of which two were diploid, and one haploid isolate showed the azole-resistant phenotype (figure 4a). This haploid genotype differed at one of the nine loci used for microsatellite genotyping (supplementary table E1). But since microsatellites have a relatively high spontaneous mutation rate [23], it is likely that this new genotype originated from the persistent genotype, before, during or after the process of haploidisation. One of the six isogenic isolates of this variant genotype showed azole resistance (supplementary table E1) that could have originated through recombination from the preceding diploid isolate.
FIGURE 4.
Genotyping results of sequential Aspergillus fumigatus isolates of a cystic fibrosis patient. a) Microsatellite genotypes are each represented with a unique colour. The genotype marked in red with black stripes differs in one of the nine genetic markers from the red only. One isolate of the red with black stripes genotype developed voriconazole resistance due to an unknown mechanism. The four selected isolates of the red genotype that were analysed by using whole genome sequencing are shown. b) The arrows indicate the average number of single-nucleotide polymorphisms (SNPs) per year difference between the sequential isolates. c) The unique SNPs are indicated for each isolate with heterozygous accumulating SNPs in the second diploid isolate V209-70 segregating in the voriconazole-resistant isolate V230-14.
Relating diploidy to genetic variation and azole resistance
To investigate genetic changes in the successive isogenic isolates, whole genome sequencing (WGS) was performed on four isolates cultured from the CF patient, including the first haploid A. fumigatus isolate with the dominant genotype (strain ID V109-58), the two diploid isolates (V183-71 and V209-70) and the voriconazole-resistant haploid isolate (V230-14); for MIC values and microsatellite typing see supplementary table E1. General statistics on the WGS data of the four isolates can be found in the supplementary table E2. Isolates were sequenced with mean coverages ranging between 64.68× and 76.91× coverage. Although microsatellite typing showed one locus difference in the voriconazole-resistant isolate (V230-14) compared with previous isolates, WGS showed that they belonged to the same lineage.
We compared sequences between a first haploid (V109-58), two diploids (V183-71 and V209-70) and a second haploid strain (V230-14) (figure 4b and c). The two first isolates, haploid V109-58 and diploid V183-71, were cultured 5 years apart. We discovered 10 single-nucleotide polymorphisms (SNPs) and one indel that differed between these two samples, averaging 2.0 new SNPs per year. Four of these SNPs were found to be variable in either V109-58 or V183-71. Between the two diploid strains V183-71 and V209-70, 10 new SNPs were identified, while they have been cultured only 15 months apart, leading to an average of eight new SNPs per year. Interestingly, while all but two previously found variants were fixed in the haploid V230-14, only one new SNP was detected (figure 4b and c). This indicates that during the diploid phase, many more SNPs accumulated by mutation than during the haploid phase. Evolution therefore took place during the diploid phase by the generation of heterozygotes, while these variable sites were fixed in the haploid and resistant strain V230-14.
Discussion
Using phenotypic and genetic analyses we performed a comprehensive analysis to find evidence for parasexual recombination in A. fumigatus in patients with persistent pulmonary colonisation. The vast majority of filamentous fungi, including A. fumigatus [24, 25], possess a working machinery for parasexual recombination that has been shown many times in vitro [26–28]. Experimental evolution in A. nidulans showed that genetic variation generated by parasex accelerates adaptive evolution in isogenic diploid strains [29]. However, since the discovery of the parasexual cycle in filamentous fungi over 65 years ago, its occurrence and significance in nature has remained unclear, with heterokaryon incompatibility presenting a major barrier for parasex between non-isogenic clones [30]. Furthermore, the presumed unlikelihood of relevant isogenic variation and the low frequency of diploids would limit the relevance of parasex for isogenic clones [31]. In this study we demonstrate that the CF lung represents a niche environment in which A. fumigatus diploid frequency is high, and genetic variation is created through parasexual recombination. Given the potential of a broad range of filamentous fungi to undergo parasexual recombination, it is likely that specific environments exist in nature where fungi rely on parasexual recombination as an adaptive strategy. The characteristics of the environment that enable parasexual recombination in A. fumigatus may help to identify these in other filamentous fungi [32].
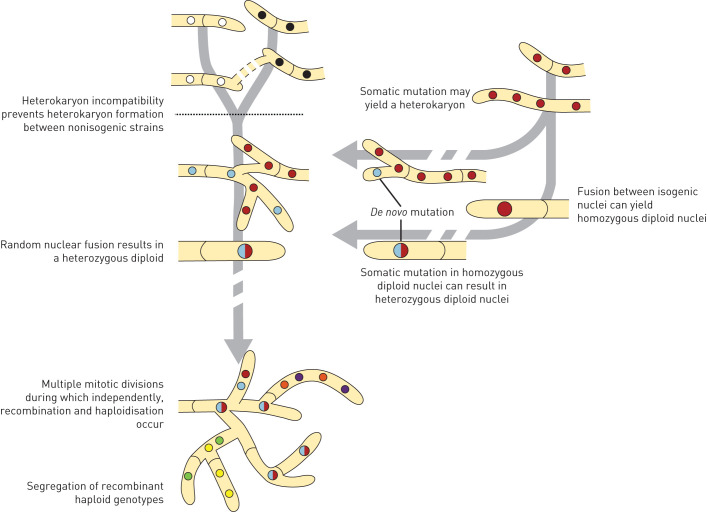
The observed high frequency of diploids in CF patients may be due to local stress factors that induce and/or select for diploidy in A. fumigatus crucial for parasexual recombination (figure 5). Indeed, nitrogen starvation, sublethal concentrations of the antifungal compound itraconazole or the inhibitory drug cerulenin were found to induce cell fusion and heterokaryon formation in isogenic A. fumigatus isolates in vitro [33]. In the absence of asexual and sexual sporulation as modes of generating genetic variation, the parasexual processes in long-lasting pulmonary mycelial biofilms can be highly relevant for gradual adaptation of A. fumigatus to the human host [34].
FIGURE 5.
The parasexual cycle in Aspergillus fumigatus. Parasexual recombination can be achieved for most fungi in laboratory experiments and is characterised by heterokaryon formation, heterozygous diploid formation, recombination and haploidisation. Heterokaryon incompatibility normally prevents parasexual recombination between non-isogenic strains in A. fumigatus (dashed black line), but in this study we show that during long-lasting fungal colonisation of the human lung, sufficient isogenic genetic variation is generated to make parasexual recombination of A. fumigatus effective. Somatic mutation may yield a heterokaryon or fusion of isogenic nuclei may yield a homozygous diploid which can both subsequently evolve into a heterozygous diploid by de novo mutations. A heterozygous diploid can continue to accumulate mutations or finally after haploidisation segregate into new recombinant genotypes.
Furthermore, we found evidence for a role of parasexual recombination in the development of azole resistance in A. fumigatus. Several studies have reported A. fumigatus azole resistance in CF patients, both in azole-naïve patients as well as those that have received antifungal therapy [13, 34, 35]. Previous azole therapy was shown to be associated with increased probability of recovery of azole-resistant isolates, and resistance rates of 20% have been reported in CF patients treated with itraconazole [35]. Analysis of resistance genotypes in CF patients indicated the presence of mutations that are associated with resistance selection in the environment, i.e. TR34/L98H and TR46/Y121F/T289A, as well as single resistance mutations such as M220 and G54, which are considered to be patient-derived, reflecting both environmental and in-host resistance selection. In-host resistance selection is furthermore supported by the recovery of isogenic A. fumigatus colonies with azole-resistant and azole-susceptible phenotypes from individual CF patients [34].
Although typing studies in CF patients indicate that a single Aspergillus genotype may colonise the lung for many years, our study indicates that isogenic isolates are able to create genetic variation through parasexual recombination. The in-host evolution from a population of genetically identical fungal cells (clone) to a population of genetically diverse cells has important implications. Among the progeny, cells may emerge that are better adapted to the lung environment. Having a survival advantage, these cells will eventually become dominant in the fungal population in the lung environment. It is likely that the obtained trait overcomes specific stress factors and that the cells will be better equipped to persist. Over time the continued selection of beneficial traits will decrease the ability of the host to eradicate the fungus. A genetically diverse Aspergillus population also increases the probability of antifungal drug-resistant cells developing. When a chronically colonised patient requires azole therapy, cells that harbour an azole resistance mutation will thrive through the selection pressure, potentially leading to lung colonisation with the resistant clone and treatment failure.
Chronic lung colonisation may thus serve as an environment that allows Aspergillus to obtain specific traits, which may subsequently be transmitted to the environment or to other patients. Recently aerosol transmission of A. fumigatus was documented in CF patients, which opens the possibility of patient-to-patient transmission, and identical A. fumigatus genotypes have been found in unrelated CF patients [36]. Although environmental exposure is considered the main origin for Aspergillus colonisation, patient-to-patient transmission might also take place and be facilitated by the adaptation traits that have been gained during lung colonisation. As biofilms may also be present in other clinical manifestations of Aspergillus disease including CPA, aspergilloma and sinus disease, further studies into the role of parasex in Aspergillus persistence are required [37].
The ability of A. fumigatus to engage in parasexual reproduction is highly significant for understanding the genetics and biology of A. fumigatus for adaptation and persistence in the human host, as well as for antifungal drug resistance development and management.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00020-2020.supplement (247.9KB, pdf)
Acknowledgements
The authors would like thank H. de Jong, R. Wagenaar and S. Blom for technical assistance, T. Franssen-Verheijen for performing the scanning electron microscopy analysis, and M. Maas for the design of figure 5 (all affiliated to Wageningen University and Research, Wageningen, The Netherlands).
Footnotes
This article has supplementary material available from openres.ersjournals.com
Conflict of interest: T. Engel has nothing to disclose.
Conflict of interest: P.E. Verweij reports grants from Gilead Sciences, MSD, Pfizer and F2G, and nonfinancial support from OLM and IMMY, outside the submitted work.
Conflict of interest: J. van den Heuvel has nothing to disclose.
Conflict of interest: D. Wangmo has nothing to disclose.
Conflict of interest: J. Zhang has nothing to disclose.
Conflict of interest: A.J.M. Debets has nothing to disclose.
Conflict of interest: E. Snelders has nothing to disclose.
References
- 1.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 1999; 12: 310–350. doi: 10.1128/CMR.12.2.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax 2015; 70: 270–277. doi: 10.1136/thoraxjnl-2014-206291 [DOI] [PubMed] [Google Scholar]
- 3.Gago S, Denning DW, Bowyer P. Pathophysiological aspects of Aspergillus colonization in disease. Med Mycol 2019; 57: Suppl. 2, S219–S227. doi: 10.1093/mmy/myy076 [DOI] [PubMed] [Google Scholar]
- 4.Denning DW, Cadranel J, Beigelman-Aubry C, et al. . Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur Respir J 2016; 47: 45–68. doi: 10.1183/13993003.00583-2015 [DOI] [PubMed] [Google Scholar]
- 5.de Valk HA, Klaassen CH, Yntema JB, et al. . Molecular typing and colonization patterns of Aspergillus fumigatus in patients with cystic fibrosis. J Cyst Fibros 2009; 8: 110–114. doi: 10.1016/j.jcf.2008.10.003 [DOI] [PubMed] [Google Scholar]
- 6.Verweij PE, Zhang J, Debets AJM, et al. . In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: a dilemma for clinical management. Lancet Infect Dis 2016; 16: e251–e260. doi: 10.1016/S1473-3099(16)30138-4 [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Snelders EE, Zwaan BJ, et al. . Relevance of heterokaryosis for adaptation and azole-resistance development in Aspergillus fumigatus. Proc Biol Sci 2019; 286: 20182886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bongomin F, Harris C, Hayes G, et al. . Twelve-month clinical outcomes of 206 patients with chronic pulmonary aspergillosis. PLoS One 2018; 13: e0193732. doi: 10.1371/journal.pone.0193732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camps SM, van der Linden JW, Li Y, et al. . Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother 2012; 56: 10–16. doi: 10.1128/AAC.05088-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Debets AJ, Verweij PE, et al. . Asexual sporulation facilitates adaptation: the emergence of azole resistance in Aspergillus fumigatus. Evolution 2015; 69: 2573–2586. doi: 10.1111/evo.12763 [DOI] [PubMed] [Google Scholar]
- 11.Bhargava V, Tomashefski JF Jr., Stern RC, et al. . The pathology of fungal infection and colonization in patients with cystic fibrosis. Hum Pathol 1989; 20: 977–986. doi: 10.1016/0046-8177(89)90269-4 [DOI] [PubMed] [Google Scholar]
- 12.Kaur S, Singh S. Biofilm formation by Aspergillus fumigatus. Med Mycol 2014; 52: 2–9. [DOI] [PubMed] [Google Scholar]
- 13.Stevens DA, Moss RB, Hernandez C, et al. . Effect of media modified to mimic cystic fibrosis sputum on the susceptibility of Aspergillus fumigatus, and the frequency of resistance at one center. Antimicrob Agents Chemother 2016; 60: 2180–2184. doi: 10.1128/AAC.02649-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pontecorvo G. The parasexual cycle in fungi. Annu Rev Microbiol 1956; 10: 393–400. doi: 10.1146/annurev.mi.10.100156.002141 [DOI] [PubMed] [Google Scholar]
- 15.Firon A, Beauvais A, Latge JP, et al. . Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 2002; 161: 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pontecorvo G, Roper JA, Forbes E. Genetic recombination without sexual reproduction in Aspergillus niger. J Gen Microbiol 1953; 8: 198–210. doi: 10.1099/00221287-8-1-198 [DOI] [PubMed] [Google Scholar]
- 17.Hastie AC. Benlate-induced instability of Aspergillus diploids. Nature 1970; 226: 771. doi: 10.1038/226771a0 [DOI] [PubMed] [Google Scholar]
- 18.De Lucas JR, Dominguez AI, Mendoza A, et al. . Use of flow-cytometry to distinguish between haploid and diploid strains of Aspergillus fumigatus. Fungal Genet Rep 1998; 45: 3. [Google Scholar]
- 19.Veselská T, Svoboda J, Růžičková Ž, et al. . Application of flow cytometry for genome size determination in Geosmithia fungi: a comparison of methods. Cytometry A 2014; 85: 854–861. doi: 10.1002/cyto.a.22500 [DOI] [PubMed] [Google Scholar]
- 20.de Valk HA, Meis JF, Curfs IM, et al. . Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J Clin Microbiol 2005; 43: 4112–4120. doi: 10.1128/JCM.43.8.4112-4120.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lestrade PP, Bentvelsen RG, Schauwvlieghe A, et al. . Voriconazole resistance and mortality in invasive aspergillosis: a multicenter retrospective cohort study. Clin Infect Dis 2019; 68: 1463–1471. doi: 10.1093/cid/ciy859 [DOI] [PubMed] [Google Scholar]
- 22.Upshall A, Giddings B, Mortimore ID. The use of benlate for distinguishing between haploid and diploid strains of Aspergillus nidulans and Aspergillus terreus. Microbiology 1977; 100: 413–418. [Google Scholar]
- 23.Li YC, Korol AB, Fahima T, et al. . Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 2002; 11: 2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x [DOI] [PubMed] [Google Scholar]
- 24.Pontecorvo G, Roper JA, Hemmons LM, et al. . The genetics of Aspergillus nidulans. Adv Genet 1953; 5: 141–238. doi: 10.1016/S0065-2660(08)60408-3 [DOI] [PubMed] [Google Scholar]
- 25.Stromnaes O, Garber ED. Heterocaryosis and the parasexual cycle in Aspergillus fumigatus. Genetics 1963; 48: 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debets AJM. Parasexuality in fungi: mechanisms and significance in wild populations In: Bridge PD, Couteaudier Y, Clarkson JM, eds. Molecular Variability of Fungal Pathogens. UK, CAB international Oxon, 1998; pp. 41–52. [Google Scholar]
- 27.Mehrabi R, Mirzadi Gohari A, Kema GHJ. Karyotype variability in plant-pathogenic fungi. Annu Rev Phytopathol 2017; 55: 483–503. doi: 10.1146/annurev-phyto-080615-095928 [DOI] [PubMed] [Google Scholar]
- 28.Nieuwenhuis BP, James TY. The frequency of sex in fungi. Philos Trans R Soc Lond B Biol Sci 2016; 371: 20150540. doi: 10.1098/rstb.2015.0540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoustra SE, Debets AJ, Slakhorst M, et al. . Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet 2007; 3: e68. doi: 10.1371/journal.pgen.0030068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aanen DK, Debets AJM, Glass NL, et al. . Biology and genetics of vegetative incompatibility in fungi In: Ebbole DJ, Borkovich K, eds. Cellular and Molecular Biology of Filamentous Fungi. Texas, ASM Press, 2010; pp. 274–288. [Google Scholar]
- 31.Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell 2003; 2: 1–8. doi: 10.1128/EC.2.1.1-8.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgel PR, Paugam A, Hubert D, et al. . Aspergillus fumigatus in the cystic fibrosis lung: pros and cons of azole therapy. Infect Drug Resist 2016; 9: 229–238. doi: 10.2147/IDR.S63621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macdonald D, Thomson DD, Johns A, et al. . Inducible cell fusion permits use of competitive fitness profiling in the human pathogenic fungus Aspergillus fumigatus. Antimicrob Agents Chemother 2019; 63: e01615–e01618. doi: 10.1128/AAC.00706-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortensen KL, Jensen RH, Johansen HK, et al. . Aspergillus species and other molds in respiratory samples from patients with cystic fibrosis: a laboratory-based study with focus on Aspergillus fumigatus azole resistance. J Clin Microbiol 2011; 49: 2243–2251. doi: 10.1128/JCM.00213-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgel PR, Baixench MT, Amsellem M, et al. . High prevalence of azole-resistant Aspergillus fumigatus in adults with cystic fibrosis exposed to itraconazole. Antimicrob Agents Chemother 2012; 56: 869–874. doi: 10.1128/AAC.05077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engel TGP, Erren E, Vanden Driessche KSJ, et al. . Aerosol transmission of Aspergillus fumigatus in cystic fibrosis patients in the Netherlands. Emerging Infect Dis 2019; 25: 797–799. doi: 10.3201/eid2504.181110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramage G, Rajendran R, Gutierrez-Correa M, et al. . Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol Lett 2011; 324: 89–97. doi: 10.1111/j.1574-6968.2011.02381.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00020-2020.supplement (247.9KB, pdf)