Highlights
-
•
CHO cells are more efficient than 293 cells for expressing trimeric SARS-CoV-2 spike protein.
-
•
A trimerization domain and elimination of the S1/S2 furin site are essential to obtain spike trimers.
-
•
Unprecedented trimeric spike expression levels of 150 mg/L are obtained in CHO cells 7 days post-transfection.
Keywords: SARS-CoV-2, Trimeric spike, HEK293, CHO, Transient gene expression, Polyethylenimine
Abstract
Recombinant forms of the spike protein of SARS-CoV-2 and related viruses have proven difficult to produce with good yields in mammalian cells. Given the panoply of potential COVID-19 diagnostic tools and therapeutic candidates that require purified spike protein and its importance for ongoing SARS-CoV-2 research, we have explored new approaches for spike production and purification. Three transient gene expression methods based on PEI-mediated transfection of CHO or HEK293 cells in suspension culture in chemically-defined media were compared for rapid production of full-length SARS-CoV-2 spike ectodomain. A high-cell-density protocol using DXB11-derived CHOBRI/55E1 cells gave substantially better yields than the other methods. Different forms of the spike ectodomain were expressed, including the wild-type SARS-CoV-2 sequence and a mutated form (to favor expression of the full-length spike ectodomain stabilized in pre-fusion conformation), with and without fusion to putative trimerization domains. An efficient two-step affinity purification method was also developed. Ultimately, we have been able to produce highly homogenous preparations of full-length spike, both monomeric and trimeric, with yields of 100−150 mg/L in the harvested medium. The speed and productivity of this method support further development of CHO-based approaches for recombinant spike protein manufacturing.
1. Introduction
The availability of recombinant forms of the SARS-CoV-2 spike protein is becoming increasingly important for development of novel strategies to combat the current COVID-19 pandemic. In the human immune response to the SARS coronavirus, the spike protein is a key antigen and is often the target of neutralizing antibodies found in convalescent patients (Qiu et al., 2005). Recombinant spike protein has a wide range of potential applications, including in the development of novel therapeutics (e.g. neutralizing antibodies or other spike-targeting biologics), clinical diagnostic tools (e.g. to evaluate post-infection immunity) or subunit vaccines (different forms of recombinant SARS spike protein were effective as vaccines in animal models) (Liu et al., 2020; Wang et al., 2020).
The SARS-CoV-2 spike protein is closely related to that of SARS-CoV-1, the virus responsible for the 2003 SARS outbreak. Both proteins are large, multi-domain glycoproteins with transmembrane domains that traverse the viral envelope and are proteolytically processed into S1 and S2 subunits. Notably, while the SARS-CoV-1 spike is only cleaved during infection of target cells, the SARS-CoV-2 spike contains a conserved furin recognition site at the S1/S2 junction, such that significant cleavage occurs during biosynthesis in host cells; this difference may impact the route of entry of the two virus types into host cells (Xia et al., 2020). The SARS-CoV-1 spike protein was shown to assemble into homo-trimeric complexes that are found on mature viral particles (Gui et al., 2017). When expressed in the absence of its transmembrane and C-terminal domains, the spike ectodomain (ECD) is reported to be mostly monomeric; fusion of the spike ECD C-terminus to the trimerization domain of bacteriophage T4 fibritin (T4-Fib or foldon) favors its trimerization and increases its capacity to elicit neutralizing antibodies (Li et al., 2013).
In the literature on SARS-CoV-1 and other related coronaviruses, there are reports of various approaches for producing recombinant spike proteins. Individual domains of the spike, including the receptor-binding and hemagglutinin-esterase domains, have been produced in CHO, HEK293, Vero and insect cells (Li et al., 2013; Du et al., 2010; Huang et al., 2015). It is also possible to express the full-length spike, including transmembrane and C-terminal domains, which can be purified following membrane solubilisation of expressing cells (Kam et al., 2007; Coleman et al., 2014). Finally, expression of the full-length soluble forms of the coronavirus spike ECDs has also been reported in HEK293 and insect cells (Li et al., 2013; Tortorici et al., 2019; Chun et al., 2019; Kirchdoerfer et al., 2018; Wrapp et al., 2020; Walls et al., 2020). Importantly, however, in cases where this data is reported, yields were extremely low, ranging from 0.5 to 1.5 mg per litre of culture media (for expression of constructs containing the full-length ECD) (Kam et al., 2007; Wrapp et al., 2020). More recently, for the SARS-CoV-2 spike, several production and purification parameters were optimized to obtain yields up to 5 mg/L from transiently transfected HEK293 cells (Esposito et al., 2020). In addition, the tendency of the SARS-CoV-2 trimeric spike to disassemble during storage at 4 °C has motivated the development of disulfide-stabilized thermostable variants, but at the cost of a 3-fold drop in volumetric yield (∼400 μg/L) in Expi293 cells (Xiong et al., 2020). Higher titers have been achieved using a highly engineered, stabilized trimeric spike ECD variant, reaching up to 33 mg/L by transient gene expression (TGE) in CHO (Hsieh et al., 2020). Another recent paper used optimized processes and expression vectors to obtain yields of trimeric spike ECD of up to 10 and 6 mg/L by TGE in 293 and CHO, respectively (Johari et al., 2020). Using methionine sulfoximine (MSX) selection to establish a stable CHO pool, the same authors reported a further increase in titers of up to 53 mg/L post-IMAC purification. While significant improvements have recently been achieved, these productivities are well below desirable levels for mass production, in particular for development and manufacturing for potential diagnostic or vaccine applications at commercial-scale.
Recently, we have attempted to produce the full-length SARS-CoV-2 spike ECD using three rapid protein production platforms established previously in our group, based on polyethylenimine (PEI)-mediated plasmid transfection of CHO or HEK293 cells. Yields of soluble ECD from the two CHO-based methods were substantially higher than other reported methods, with one method yielding approximately 100−150 mg/L (depending on the form of the spike expressed) at 6–7 days post-transfection. Both the wild-type spike and a stabilized form with mutated furin cleavage site (Wrapp et al., 2020) were produced and purified; both forms were expressed with and without fusion to various putative trimerization domains. The CHO-secreted wild-type protein is a mix of unprocessed precursor and cleaved forms, likely including free S1, S2 and S1/S2 complexes. The stabilized protein (with furin site mutated) with or without trimerization domain fusion is secreted in a full-length, unprocessed form. Notably, for the wild-type (non-stabilized with furin site present) spike ECD constructs, we have observed dissociation of S1 from the monomeric and trimeric S1/S2 complexes during purification and storage, indicating that the furin site-deleted mutant is a better choice for ongoing production of the full-length ECD. The yield and purity we have obtained support continued method development to further improve titers and downstream processes for eventual mass production in CHO cells.
2. Materials & methods
2.1. Expression vectors
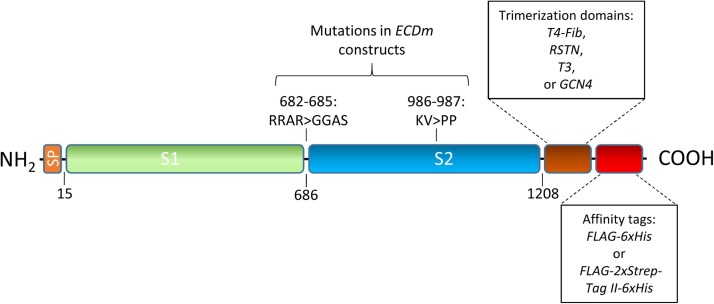
SARS-CoV-2 spike protein sequences were derived from Genbank accession number MN908947. Protein-coding DNA sequences were codon-optimized for CHO cells, synthesized and cloned into the pTT5™ vector (Shi et al., 2005) by Genscript. The specific regions of the spike ECD contained in each construct and the sequences and locations of the stabilizing mutations in ECDm are indicated in Fig. 1 . Trimerization domains were fused to the C-termini of the spike protein coding sequences followed by FLAG and 6xHis tags. Two constructs (pTT5-ECD-T4Fib and pTT5-ECDm-T4Fib) contain additional dual StrepTag-II tags between the FLAG and 6xHis tags. Amino acid sequence of the various constructs are shown in Supplementary Table 1.
Fig. 1.
Organization of SARS-CoV-2 spike protein constructs. Full-length ectodomain (ECD) constructs consist of the native SARS-CoV-2 spike N-terminal signal peptide (SP) and remaining sequence up to amino acid 1208, including full S1 and S2 domains (native transmembrane and C-terminal domains were removed). Mutations present in the prefusion-stabilized, furin site-mutated ECD (ECDm) are shown above. For purification, affinity tags (FLAG/His or FLAG-Twin-Strep-tag-His) were introducedat the C-terminus. For those constructs containing trimerization domains, these are located between the ECD and affinity tag sequences. Amino acid sequence of the various constructs is shown in Supplementary Table 1.
2.2. Expression platforms
Transfection and post-transfection culture of CHO-3E7 and 293-6E cells in FreeStyle F17 media were performed as described previously (Stuible et al., 2018a; L’Abbé et al., 2018).
The method for transient expression in CHOBRI/55E1 cells (Poulain et al., 2017) is based on a published method for high-density transfection of CHO-3E7 cells (Stuible et al., 2018a), with several modifications. Cells were maintained in shake flasks in a humidified, 5% CO2, 37 °C incubator, rotating at 120 rpm in a chemically-defined proprietary media formulation. Cells were seeded in the same media at 1 × 106 cells/mL 2 days prior to transfection to achieve a cell density of ∼8 × 106/mL at the time of transfection. Before transfection, cells were diluted with 25 % fresh media and dimethylacetamide was added to 0.083 % (v/v). Transfections were performed by adding PEI-DNA polyplexes (10 % final culture volume) to diluted cells (90 % final culture volume). To prepare polyplexes, PEI-Max (Polysciences) and plasmid DNA were diluted separately to 200 μg/mL and 28 μg/mL, respectively, in a volume of growth media equal to 5% of the final culture volume. Plasmid DNA consisted of 85 % of the different pTT5™-spike constructs, 10 % pTT™-Bcl-XL (anti-apoptotic effector) and 5% pTT™-GFP. The diluted PEI-Max was added to the diluted DNA and incubated for 7 min at room temperature before adding to cells. At 24 h post-transfection, cultures were shifted to 32 °C and supplemented with Anti-Clumping Supplement (1:500 dilution) and Feed 4 (2.5 % v/v), both from Irvine Scientific. Additional Feed 4 (5%) was added at 5 days post-transfection. Glucose concentrations were monitored every 2–3 days to maintain >10 mM. Cell supernatants were harvested at 6–7 days post-transfection (except for time-course experiments shown in Fig. 3 and Supplementary Fig. 1).
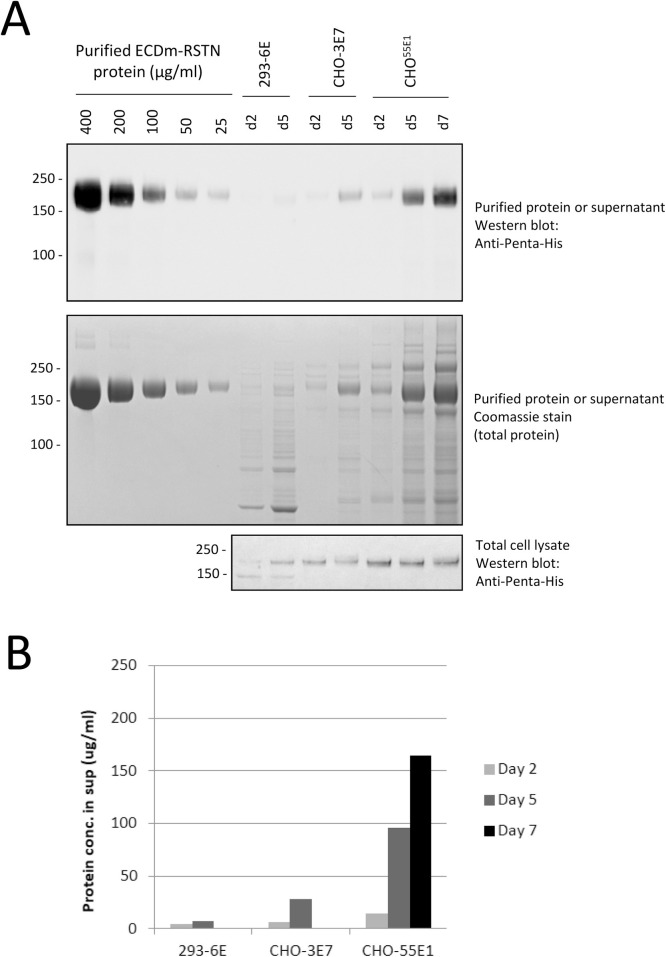
Fig. 3.
Evaluation of recombinant ECDm-RSTN concentrations in culture supernatants from transfected CHO-3E7, CHOBRI/55E1 and 293-6E cells. Cells were transfected with the ECDm expression plasmid. Samples were taken at 2, 5 or 7 days post-transfection for preparation of cleared culture supernatants and cell pellets (for lysis and total protein extraction). (A) Cleared supernatants (10 μL) and total protein extracts (equal amounts of total protein for all samples) were analyzed by western blotting along with known amounts of IMAC/FLAG-purified ECDm-RSTN protein. Densitometry analysis of the purified protein bands was used to prepare a standard curve and estimate ECD protein concentrations in the supernatants (B).
SDS-PAGE was performed using NuPAGE 4–12 % Bis-Tris gels (Invitrogen). Protein samples were heat-denatured at 70 °C for 10 min under reducing conditions prior to separation by SDS-PAGE. Total protein staining (Coomassie Blue) and western blotting were performed using standard methods. Anti-Penta-His Alexa Fluor 488 conjugate was from Qiagen.
2.3. Purification
All protein constructs were initially purified by IMAC on Nickel Sepharose Excel resin (GE Healthcare). 0.2 μm-filtered supernatants were applied to columns by gravity flow. The column volume used for a given volume of supernatant was variable for different productions; for maximum recovery of ECD constructs from cell supernatant, a packed resin volume of 1 mL per 0.5 mg of protein is optimal. Columns were washed with 50 mM sodium phosphate, pH 7.0, containing 300 mM NaCl and 25 mM imidazole and eluted with the same buffer containing 300 mM imidazole.
In order to achieve near homogeneity, a second step purification was required. For most constructs, ANTI-FLAG M2 affinity gel (Sigma Aldrich) was used in batch mode. IMAC-purified proteins were incubated with the gel for ∼ 2 h with gentle overhead mixing. One ml of gel was used for every 0.5 mg of IMAC-purified protein. Resin was collected by filtration in an empty column (Qiagen) and washed 3 times with 10 x CV of calcium- and magnesium-free Dulbecco’s PBS (DPBS). Bound proteins were eluted four times by competition with 100 μg/mL FLAG peptide in 2 CV of DPBS (Sigma Aldrich). Purified proteins were formulated in DPBS by buffer exchange through desalting columns (GE Healthcare), 0.2 μm filtered (Millipore) and stored at −80 °C at protein concentration not exceeding 2 mg/mL.
For constructs containing the T4-Fib trimerization domain, a StrepTactin column was used for the second purification step instead of anti-FLAG. IMAC eluate (in IMAC elution buffer) was loaded on a 5-ml StrepTrap HP column (Cytiva) at a flow rate of 5 mL/minute. The column was washed with 5 column volumes of DPBS and bound protein was eluted with DPBS containing 2.5 mM desthiobiotin. Buffer exchange into DPBS was performed as described above.
Concentrations of purified proteins in DPBS were determined by spectrophotometry (A280) using extinction coefficients calculated based on their amino acid composition.
2.4. UPLC-SEC/MALS
UPLC-SEC-MALS analyses were conducted on a 4.6 × 150 mm BEH200 SEC column with 1.7 μm particles (Waters, Milford, MA) connected to an Acquity H-Class Bio UPLC system (Waters) with a miniDAWN™ MALS detector and Optilab® T-rEX™ refractometer (Wyatt Technology, Santa Barbara, CA, USA). The column temperature was 30 °C and the mobile phase was DPBS (HyClone SH30028.01) with 0.02 % Tween 20 added. The flow rate was 0.4 mL/min. Weighted average molecular mass (M MALS) was calculated in ASTRA 6.1 software (Wyatt) using a protein concentration determined from the refractive index signal with a dn/dc value of 0.185.
3. Results and discussion
For protein expression, we used plasmid vectors containing CHO codon-optimized sequences encoding the full ECD of the SARS-CoV-2 spike protein. C-terminal affinity tags were added for purification. Two versions of the protein were expressed: one encoding an unmodified ECD protein sequence (ECD) and a second with two modifications, to block furin-mediated S1/S2 cleavage and to stabilize the pre-fusion conformation (ECDm), as recently described (Wrapp et al., 2020). To attempt to mimic the native trimeric structure of the spike protein, as found on the virus surface, trimerization domains of T4-Fibritin (T4-Fib) (Li et al., 2013), human resistin (RSTN) (Patel et al., 2004), or GCN4 (Harbury et al., 1993) were fused to the C-terminus of the ECD in certain constructs. A fourth, proprietary trimerization sequence (T3) was also tested.
We rapidly assessed the potential of three expression platforms for production of SARS-CoV-2 spike: two methods, based on EBNA1-expressing CHO (CHO-3E7) (Stuible et al., 2018a) and HEK293 (293-6E) (L’Abbé et al., 2018) have served as core platforms for recombinant protein production in our group for several years, and generally perform very well for a wide range of recombinant antibodies and other proteins. For both methods, cells are cultured in chemically-defined F17 media and transfected at low cell density using PEI. A third method uses a CHO-DXB11-derived clone (CHOBRI/55E1) that expresses machinery for cumate-inducible protein expression (Poulain et al., 2017); CHOBRI/55E1 cells are used by our group for stable pool and cell line development for biologics manufacturing. Recently, we have developed a method for high-cell-density PEI-mediated transfection of these cells which performs well with both constitutive and cumate-inducible promoters (unpublished data). For the CHO-3E7 and CHOBRI/55E1 methods, cultures are shifted to 32 °C from 37 °C at 24 h post-transfection, while for 293-6E cells, the temperature is maintained at 37 °C.
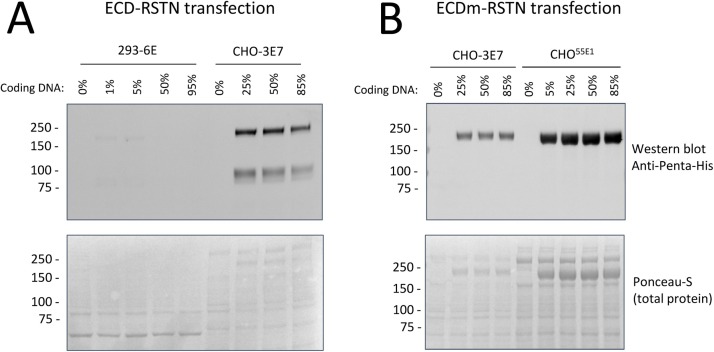
We began by transfecting CHO-3E7 and 293-6E cells with various amounts of plasmid DNA encoding the spike ECD fused to the resistin trimerization domain (ECD-RSTN). Expression levels were compared at 5 days post-transfection; for other proteins, we frequently observe that while keeping the total amount of transfected DNA constant, reducing the amount of coding vs. non-coding (sheared salmon sperm) DNA can improve yields (L’Abbé et al., 2018). As shown in Fig. 2 A, western blotting of bulk supernatants with an anti-penta-His antibody shows two clear bands in the CHO-3E7 supernatants transfected with 25–85 % coding DNA, but little signal in the 293-6E supernatants transfected with 1–95 % coding DNA. Two bands are detected in the anti-penta-His blots, at ∼80 kDa and ∼180 kDa, which likely correspond to the S2 subunit (after cleavage at the S1/S2 site) and the unprocessed precursor, respectively. These two proteins can be observed as faint bands by Ponceau S staining of membranes prior to western blotting. Based on amino acid sequence, the predicted molecular weight of the unprocessed spike is 143 kDa. The higher observed molecular weight is very likely due to glycosylation: the SARS-CoV-2 spike protein contains 22 confirmed N-linked glycosylation sites (Watanabe et al., 2020). In addition, for the SARS-CoV-1 spike, enzymatic removal of glycans decreases apparent molecular weight by SDS-PAGE by 30−40 kDa (Song et al., 2004).
Fig. 2.
Evaluation of mammalian cell expression platforms for expression of SARS-CoV-2 ectodomain. HEK293, CHO-3E7, or CHOBRI/55E1 cells were transfected with the indicated percentages of ECD-coding plasmid DNA. (A) Comparison of ECD-RSTN production in HEK293 and CHO-3E7 cells. Samples of culture supernatant were taken at 5 days post-transfection (dpt). (B) Comparison of ECDm-RSTN production in CHO-3E7 and CHOBRI/55E1 cells. Samples of culture supernatant were taken at At 5 dpt (CHO-3E7) or 6 dpt (CHOBRI/55E1). For all samples, 10 μL of culture supernatant was analyzed by western blotting. Ponceau S staining of membranes before western blotting is shown in lower panels.
Similar expression tests were performed for the stabilized spike protein (ECDm-RSTN) with the CHO-3E7 and CHOBRI/55E1 platforms, using a range of coding DNA ratios. As shown in Fig. 2B, an anti-penta-His western blot using unpurified supernatants gave a single band at ∼180 kDa, which corresponds to the size of the unprocessed precursor protein observed with expression of the non-stabilized spike (Fig. 2A). The CHOBRI/55E1 method gives substantially higher yields, based on band intensity on the anti-penta-His blot and Ponceau S stained membrane. For both cell lines, there was little difference in expression between 25 and 85 % of coding DNA.
To estimate the concentration of spike protein in the culture supernatants, we performed western blots with unpurified supernatants from HEK293, CHO-3E7 and CHOBRI/55E1 cells transfected with ECDm-RSTN along with a series of dilutions of highly purified ECDm protein (Fig. 3). Samples were taken up to 7 days post-transfection for CHOBRI/55E1 cells and up to 5 days post-transfection for CHO-3E7 and HEK293 cells (CHO-3E7 and HEK293 cell viability dropped to <70 % on Day 5). As shown in Fig. 3B, densitometry analysis of this blot allows us to estimate that yields in the transfected CHOBRI/55E1 supernatants reach >150 μg/mL at 7 days post-transfection. As expected, concentrations in CHO-3E7 supernatants were lower (∼25 μg/mL at Day 5) while they were nearly undetectable in 293-6E supernatants. A similar comparison was performed for the ECDm protein without trimerization domain in CHOBRI/55E1 and 293-6E cells (Supplementary Fig. 1); in this case CHOBRI/55E1 yields were ∼100 μg/mL and 293-6E yields were ∼20 μg/mL.
The differences in yields from the three cell lines were reproducible and were not due to differences transfection efficiency: for all transfections, we either co-transfected 5% of a GFP expression vector or included a GFP-only transfected external control, and we monitored GFP expression at 24−48 h post-transfection (data not shown). Also, we did not observe any obvious negative effect of spike protein expression on cell growth or viability.
The differences in the amounts of spike protein secreted by the three cell lines are not reflected by intracellular spike protein levels. As shown in Fig. 3A and Supplementary Fig. 1C, recombinant spike protein is easily detectable in total protein extracts from all three cell lines by western blotting. Intracellular protein levels are slightly lower in CHO-3E7 and 293-6E cells compared to CHOBRI/55E1 in Fig. 3A, but certainly, these differences do not reflect the substantial differences in concentrations of secreted protein. These results confirm that all three cell lines were transfected and appear to be able to produce intracellular spike protein at similar levels; however, an explanation for the low level of protein secretion, especially in 293-6E cells, is still elusive.
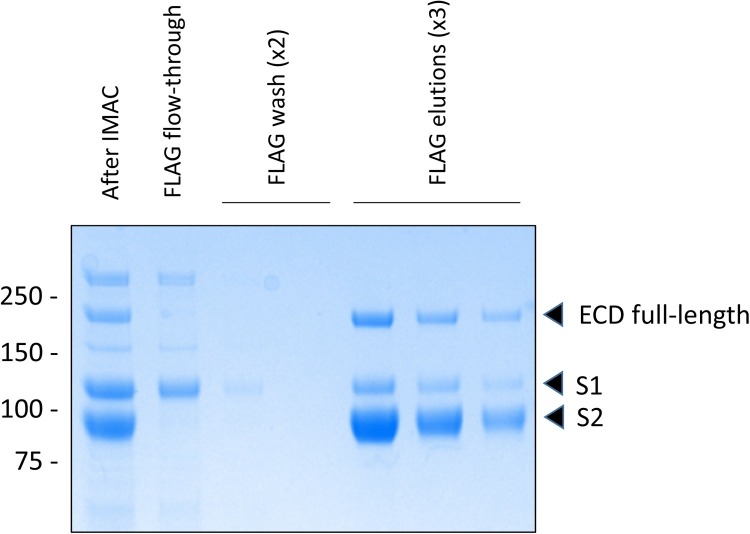
Based on these results, we decided to pursue the CHOBRI/55E1 TGE platform for ECD expression, and we proceeded to develop a purification method for supernatants from these cells. Bulk supernatants were first passed on an IMAC (nickel-Sepharose excel) column; as shown in Fig. 4 for the ECD-T3 construct, the spike protein is prominent in the eluate from this column; as expected, three forms of the protein are observed: full-length S1/S2 as well as the processed S1 and S2 subunits. Several other non-specific proteins are also present. Following a second-step purification of this eluate on an anti-FLAG column, these contaminating proteins were effectively removed. Importantly, we observed a significant amount of S1 subunit in the flow-through and washes from the anti-FLAG column for the ECD-T3 construct as well as all other constructs with intact protease cleavage sites. We believe that this represents slow dissociation of S1 from S2 (the FLAG/His tag is fused to S2); S1 does not immediately dissociate from S2 after S1/S2 processing, but it is evident over time with multiple steps of purification/storage. Certainly, this result supports the use of the mutated form of the spike ECD (ECDm), lacking the S1/S2 cleavage site, for production of full-length recombinant spike protein.
Fig. 4.
Loss of S1 subunit during second-step FLAG purification of ECD-T3. Proteins eluted from IMAC column (After IMAC) were purified with anti-FLAG beads. Flow-through, washes and elutions from FLAG column were analyzed by SDS-PAGE and Coomassie Blue staining.
Our purification method has evolved with our growing experience purifying different spike ECD constructs. In particular, in our initial IMAC purification runs, a large proportion (∼50 %) of the protein was found in the column flow-through. The standard binding capacity for the Ni-Sepharose excel IMAC column is >10 mg/mL, but after considerable effort troubleshooting this step, we have concluded that the capacity is much lower for the spike ECD proteins (∼0.5 mg/mL). We do not believe that this low binding capacity is due to interfering substances or low pH, because supernatant buffer exchange by tangential flow filtration prior to purification did not improve binding (data not shown); rather, we suspect the size and/or shape of the spike constructs may prevent diffusion into the pores of the Ni-Sepharose excel resin. With an appropriate amount of supernatant loaded on the IMAC column, we are now able to capture the expected amount of ECD protein based on its abundance in the bulk supernatant.
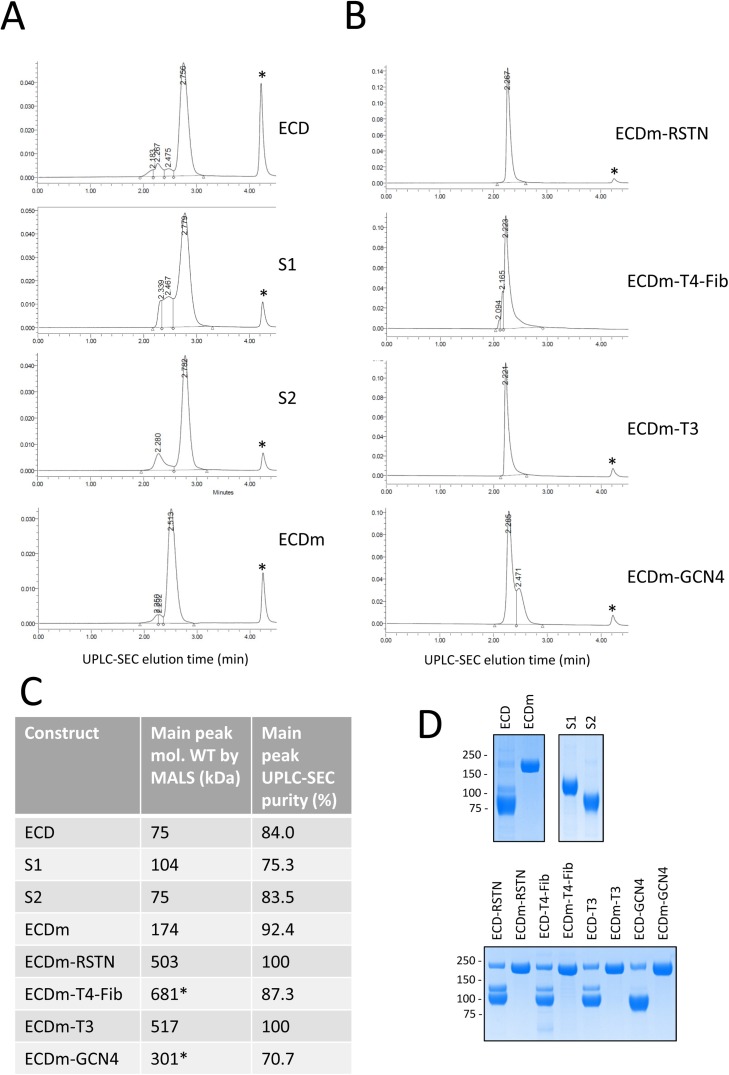
Several different SARS-CoV-2 spike ECD constructs were purified in this manner, including full-length, S1, and S2 forms of the wild-type spike (ECD, S1 and S2), the stabilized full-length spike (ECDm) and ECD and ECDm fused to the different trimerization domains. The constructs with the T4-Fib trimerization domain contain an additional Twin-Strep-Tag® between the FLAG and 6xHis sequences and were purified by IMAC followed by a Streptrap™ HP column. As shown in Fig. 5 , the purified proteins were analyzed by UPLC-SEC (Fig. 4A and B) with a MALS detector for molecular weight determination (Fig. 5C), as well as by SDS-PAGE/Coomassie staining (Fig. 5D). As expected, for all of the non-mutated ECD constructs (with and without trimerization domains), the purified protein consists of the S2 subunit with lower levels of S1 and uncleaved full-length protein. By SDS-PAGE, the sizes of S1 and S2 that result from processing of the full-length ECD match well the sizes of S1 and S2 when expressed individually. By UPLC-SEC, purified ECD, S1 and S2 show single predominant peaks of UV absorbance, with lower-abundance, fast-eluting peaks which are likely multimers or aggregates (or uncleaved full-length protein, in the case of ECD). Notably, the MALS molecular weight estimate for the principal peak in the ECD preparation is 75 kDa, which matches the MALS estimate for S2 expressed by itself, supporting the assertion that after purification, the ECD preparation is composed mostly of S2. The mutated ECD (ECDm) is estimated to have a molecular weight of 174 kDa, which is approximately equal to that of the sum of S1 and S2 expressed alone.
Fig. 5.
Characterization of purified spike ectodomain constructs by UPLC-SEC/MALS and SDS-PAGE. (A and B) Proteins purified by IMAC and anti-FLAG (or IMAC and StrepTrap for T4-Fib constructs) were separated by UPLC on a BEH200 SEC column. Protein elution was monitored by measuring UV absorbance of eluate (vertical axis). The position of the FLAG peptide elution peak is marked with an asterisk. (C) The molecular weight of the major elution peak for each sample was estimated by MALS. Values marked with asterisks are likely inaccurate due to peak shouldering. (D) 2 μg of each purified protein was analyzed by SDS-PAGE/Coomassie Blue staining.
Purified constructs containing trimerization domains were also analyzed. By SDS-PAGE (Fig. 5D, lower panel), they are quite similar, although the amount of S1 lost during purification of the ECD constructs is variable. The mutated (ECDm) trimerization domain fusions all appear to be highly pure by SDS-PAGE. Notably, for the UPLC-SEC analysis, due to varying degrees of loss of S1 from the trimeric ECD constructs, the UPLC-SEC profiles and MALS data are very difficult to interpret (data not shown). For the mutated constructs, the RSTN and T3 fusions give highly uniform purified preparations by UPLC-SEC that are estimated by MALS to have molecular weights of 503 and 517 kDa, respectively. This is consistent with the expected molecular weight of a trimer of ECDm (174 kDa). For the T4-Fib and GCN4 fusions, the principal peaks by UPLC have similar retention times as the other trimerization domains; however, the presence of shoulders on the main peak indicates that it could be composed of more than one species, and therefore, the MALS molecular weight estimates for the trimeric protein are likely inaccurate. Notably, T4-Fib is the most commonly used trimerization domain used in the literature for coronavirus spike proteins. Together, these results suggest that RSTN and T3 fusions could be better for preparing uniform preparations of SARS-CoV-2 spike ECD with trimeric stoichiometry. Very recently, an ELISA based assay using ECDm-RSTN was developed for sensitive detection of anti-spike antibodies in COVID-19 patients (Isho et al., 2020), as has already been reported for the T4-Fib fusion (Amanat et al., 2020a). Additional analyses will be required to evaluate how well the different trimerization domain fusions mimic the conformation of the transmembrane spike protein in its native state.
In summary, we have developed a method for transient production in CHO cells of full-length SARS-CoV-2 spike ECD, yielding 100−150 μg/mL within 7 days of plasmid transfection. This is significantly higher than yields reported previously for production of recombinant coronavirus spike proteins. It is important to note that our estimates of spike ECD yields are based on concentrations in harvested supernatants evaluated by western blotting/densitometry while other have reported yields of purified protein (eg. Johari et al., 2020), so our values are not directly comparable. Our results demonstrate that full-length recombinant spike protein is not inherently difficult to produce at high levels in mammalian cells, but that productivity is greatly dependent on host cell line selection: consistent with literature reports, we obtained low yields from transfected HEK293 cells. Between the two CHO host cell lines that were tested, CHOBRI/55E1 performed better than CHO-3E7; we cannot conclude whether this is due to differences in protocols (transfection cell density, media and feed are different), cell background (CHOBRI/55E1 and CHO-3E7 are derived from different parental cells) or other factors.
The mutated construct (ECDm) is likely the best starting point for ongoing development of recombinant protein for therapeutic or diagnostic applications: it can be readily expressed in CHO cells and can be purified to near homogeneity, as assessed by SEC and SDS-PAGE. To mimic the conformation of the spike protein on the virus surface, the RSTN and T3 fusions were effective at promoting uniform, trimeric stoichiometry. It is notable that the stabilized trimeric spike was recently shown as the most potent spike construct for generating neutralizing antibodies in mice (Amanat et al., 2020b).
The challenge of manufacturing sufficient amounts of spike protein is likely an important reason why alternative methods, such as mRNA-, adenovirus-, and AAV-based approaches to induce spike protein expression in vaccinated individuals, have gained prominence with COVID-19 vaccine developers. However, production of vaccine antigen in the form of recombinant spike protein in CHO cells has an intrinsic advantage over these newer technologies: the extensive, decades-long safety record of CHO-derived biologics. The improved yields we have demonstrated in the current study support the feasibility of recombinant, subunit-based vaccines for COVID-19.
Based on past experience with other recombinant proteins, we expected that TGE yields could be further improved in stably transfected CHO pools or clones (Poulain et al., 2017); indeed, more recently, we have generated CHO pools that can reach yields >800 mg/L for ECDm-RSTN (manuscript in preparation). Nonetheless, the speed at which we were able to generate substantial amounts of spike protein by CHO TGE highlights how this approach could complement other methods, in particular for rapid response to health emergencies. As detailed previously (Stuible et al., 2018b), we believe that development of alternatives to clone-based CHO production methods, including stable pools and TGE, should be prioritized as potential novel methods for rapid, large-scale manufacturing of biologics.
Credit authors statement
Experiments were designed by YD, MS, and CG. Experiments were performed by CG, SLD, SP, DL, GSL, JS and MS. YD supervised the work. MS wrote the manuscript, YD, CG and JS revised the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We thank the Quality Attributes Characterization team at the National Research Council of Canada for performing UPLC-SEC/MALS analyses. This is NRC publication #53486.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jbiotec.2020.12.005.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Amanat F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020:1–4. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F. Introduction of two prolines and removal of the polybasic cleavage site leads to optimal efficacy of a recombinant spike based SARS-CoV-2 vaccine in the mouse model. Biorxiv. 2020 doi: 10.1128/mBio.02648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J. Effect of fc fusion on folding and immunogenicity of middle east respiratory syndrome coronavirus spike protein. J. Microbiol. Biotechnol. 2019;29(5):813–819. doi: 10.4014/jmb.1903.03043. [DOI] [PubMed] [Google Scholar]
- Coleman C.M. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32(26):3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L. A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral Immunol. 2010;23(2):211–219. doi: 10.1089/vim.2009.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr. Purif. 2020 doi: 10.1016/j.pep.2020.105686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P.B. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hsieh C.-L. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. bioRxiv. 2020 doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89(14):7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5(52) doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johari Y.B. Production of trimeric SARS‐CoV‐2 spike protein by CHO cells for serological COVID‐19 testing. Biotechnol. Bioeng. 2020 doi: 10.1002/bit.27615. [DOI] [PubMed] [Google Scholar]
- Kam Y.W. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcγRII-dependent entry into B cells in vitro. Vaccine. 2007;25(4):729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Abbé D. Recombinant Protein Expression in Mammalian Cells. Springer; 2018. Transient gene expression in suspension HEK293-EBNA1 cells; pp. 1–16. [DOI] [PubMed] [Google Scholar]
- Li J. Immunogenicity and protection efficacy of monomeric and trimeric recombinant SARS coronavirus spike protein subunit vaccine candidates. Viral Immunol. 2013;26(2):126–132. doi: 10.1089/vim.2012.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. ACS Publications; 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.D. Disulfide-dependent multimeric assembly of resistin family hormones. Science. 2004;304(5674):1154–1158. doi: 10.1126/science.1093466. [DOI] [PubMed] [Google Scholar]
- Poulain A. Rapid protein production from stable CHO cell pools using plasmid vector and the cumate gene-switch. J. Biotechnol. 2017;255:16–27. doi: 10.1016/j.jbiotec.2017.06.009. [DOI] [PubMed] [Google Scholar]
- Qiu M. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7(5–6):882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C. Purification and characterization of a recombinant G-protein-coupled receptor, Saccharomyces cerevisiae Ste2p, transiently expressed in HEK293 EBNA1 cells. Biochemistry. 2005;44(48):15705–15714. doi: 10.1021/bi051292p. [DOI] [PubMed] [Google Scholar]
- Song H.C. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2004;78(19):10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuible M. Optimization of a high-cell-density polyethylenimine transfection method for rapid protein production in CHO-EBNA1 cells. J. Biotechnol. 2018;281:39–47. doi: 10.1016/j.jbiotec.2018.06.307. [DOI] [PubMed] [Google Scholar]
- Stuible M. Beyond preclinical research: production of CHO-derived biotherapeutics for toxicology and early-phase trials by transient gene expression or stable pools. Curr. Opin. Chem. Eng. 2018;22:145–151. [Google Scholar]
- Tortorici M.A. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26(6):481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00298. p. 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020 doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020:1–13. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X. A thermostable, closed SARS-CoV-2 spike protein trimer. Nat. Struct. Mol. Biol. 2020;27(10):934–941. doi: 10.1038/s41594-020-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.